Abstract
In view of risk coupled with synthetic polymer waste, there is an imperative need to explore biodegradable polymer. On account of that, six PHAs producing bacteria were isolated from mangrove forest and affilated to the genera Bacillus & Pseudomonas from morpho-physiological characterizations. Among which the potent PHAs producer was identified as Bacillus megaterium OUAT 016 by 16S rDNA sequencing and in-silico analysis. This research addressed a comparative account on PHAs production by submerged and solid-state fermentation pertaining to different downstream processing. Here, we established higher PHAs production by solid-state fermentation through sonication and mono-solvent extraction. Using modified MSM media under optimized conditions, 49.5% & 57.7% of PHAs were produced in submerged and 34.1% & 62.0% in solid-state fermentation process. Extracted PHAs was identified as a valuable polymer PHB-co-PHV and its crystallinity & thermostability nature was validated by FTIR, 1H NMR and XRD. The melting (Tm) and thermal degradation temperature (Td) of PHB-co-PHV was 166 °C and 273 °C as depicted from DTA. Moreover, FE-SEM and SPM surface imaging indicated biodegradable nature, while FACS assay confirmed cytocompatibility of PHB-co-PHV.
Keywords: PHAs, Bacillus megaterium, PHB-co-PHV, DTA, AFM
1. Introduction
Petrochemical based polymer is a matter of great concern due to its detrimental consequence on the environment, which demands a need to develop alternate biodegradable polymer. The eco-friendly polymer is nothing but polyhydroxyalkanoates (PHAs) synthesized by a wide array of Gram-positive and negative bacteria (Mohapatra et al., 2015b) and accumulated as carbon & energy storage inclusion in the cytoplasm. The PHAs granule is synthesized under certain stress conditions such as nitrogen, phosphorus, trace elements and oxygen (Mohapatra et al., 2016a). In particular, more than 150 different monomers of PHAs have been reported (Tan et al., 2014) from bacteria that are water-insoluble, UV resistant, non-toxic, impermeable to oxygen, biodegradable and biocompatible (Mayeli et al., 2015). These polymers have multifarious applications such as domestic plastic, skin tissue & other implants, drug coating, 3D printing in various photographic materials, nutritional dietary supplements, drugs and minute chemical substances (Maity et al., 2017).
Till date, the commercial PHAs production is exclusively dependent upon Gram-negative bacteria. However, production of endotoxin based PHAs by these bacteria holds back its successful commercialization and diverse biomedical applications (Sultanpuram et al., 2010). As a matter of fact, the genus Bacillus is widely used by academia due to their faster generation time, genomic stability, utilization of economical substrate that provides carbon resources for the production of endotoxin-free PHAs for different biomedical applications. Thus, for successful commercialization of endotoxin-free PHAs production, more efforts are indispensable to make the bioprocess technology, such as, submerged and solid-state fermentation process economically viable in contrast to both upstream and downstream processing. In addition, 50% costs of PHAs production crucially depends on upstream processing and the rest on downstream processing (Gomaa, 2014, Kunasundari and Sudesh, 2011). Nevertheless, inadequate research reports are available in the public domain for recovery of endotoxin-free PHAs produced under submerged and solid-state fermentation process by Bacillus species. In this research, we analyzed the quality and quantity of endotoxin free PHAs produced by Bacillus megaterium under submerged and solid-state fermentation process with special reference to downstream processing.
2. Materials and methods
2.1. Cultivation and screening of PHAs producers
Sediment samples were collected aseptically from the Bhitarkanika mangrove forest (19N & 22N and between 85E & 87E) of Odisha, India and processed in the laboratory for cultivation of aerobic heterotrophic bacteria using standard bacteriological techniques. Cultivated bacterial isolates were then induced for synthesis & accumulation of PHAs granules using minimal salt medium (2% glucose) and incubated at 37 °C for 24–72 h for screening of PHAs producer.
Subsequently, intracellular PHAs granule accumulation by bacteria were detected by Sudan black B followed by Nile blue staining under bright-field microscopic (1000X, Leica DM5000B) and UV-trans illuminator imaging (Maity et al., 2017). Further, PHAs accumulation and morphological (cell shape and size) transformation of bacteria during growth at different time interval was observed through field emission scanning electron microscopic (FE-SEM) imaging (Quanta 200 FEG) (Bhagowati et al., 2015). For conformation, transmission electron microscopic (TEM) (JEOL JEM 1200, JEOL Ltd., Tokyo, Japan) imaging was also conducted for detection of intracellular PHAs granule in the bacterial cytosol (Mohapatra et al., 2016a).
2.2. Morpho-physiological characterization
The morpho-physiological characterization of selected PHAs producers were studied based on standard bacteriological tests prescribed in Bergey’s manual of determinative bacteriology. The selected bacterial isolates were subjected to morphological, biochemical, enzymatic, sugar utilization and antibiotic sensitivity tests for generic level identification (Holt et al., 1994).
2.3. Evolutionary analysis of the bacterial isolate
PCR amplified 16S rDNA product was sent to Eurofin Genomics Pvt. Ltd., Bangalore, India and sequenced with 16SF (5′ AGAGTTTGATCCTGGCTCAG 3′) and 16SR (3′ AAGGAGGTGATCCACCGCA 5′) primer using ABI 3730xl genetic analyzer (Mohapatra et al., 2016a). Consensus sequence was analyzed by bio-edit software (v7.0.5.3) and submitted to NCBI (http://blast.ncbi.nlm.nih.gov/blast.cgi) to get accession number. Alignment of desired Bacillus sp. and its closely related homologous 16S rDNA sequences were carried out by using ClustalW (v1.6) for evolutionary analysis. The phylogenetic tree was constructed by neighbour-joining method (Saitou and Nei, 1987) using MEGA v7.0 (Molecular Evolutionary Genetics Analysis) software (Tamura et al., 2013). The tree topologies were evaluated by bootstrap analysis (1000 replications). Further, secondary structure analysis of rRNA was conducted by M-fold web server (http://www.bioinfo.rpi.edu/application/mfold) (Mathews and Turner, 2002) to get more stable structure of RNA with lowest free energy (Mohapatra et al., 2016a).
2.4. Optimization of growth parameters for higher biomass and PHAs yield
Bacterial cell biomass yield is regulated by several growth parameters. Further, PHAs production is parallal to bacterial cell biomass yield. Thus, growth parameters like cultivation medium, pH, temperature, carbon & nitrogen source & their concentration and inoculum size for PHAs producing bacteria were optimized by one factor at a time (OFAT) approach (Maity et al., 2017). As the primary growth factor, culture medium such as growth and modified minimal salt medium were prepared, 1.7x108 cells/ml of starter culture (0.5 McFarland standards) was inoculated and incubated at 37 °C for 24 h. Then, optimum bacterial cell biomass yield was estimated by measuring OD at 600 nm using UV–Vis spectrophotometer (λ35-Perkin-Elmer). Culture medium depicting higher bacterial cell biomass yield was kept constant and then other parameters including pH (5–9), temperature (25–55 °C), carbon source (maltose, lactose, sucrose, glucose, fructose), glucose concentration (1–5%), nitrogen sources (NH4Cl, KNO3, urea, (NH4)2SO4), (NH4)2SO4 (1–3 g/l) and inoculum age (5–30 h) were varied one by one sequencially. Then, the data obtained from the optimization studies were again validated by statistical analysis (p < 0.5).
2.5. PHAs production and extraction
PHAs production was carried out by submerged and solid-state fermentation process under optimized condition. Submerged fermentation was accomplished through one stage batch cultivation method (Singh et al., 2013). In brief, 1L of modified minimal salt medium (KH2PO4 0.5 g/l, K2HPO4 0.5 g/l, glutamic acid 1.5 g/l, malic acid 2.7 g/l, yeast-extract 4 g/l, glucose 30 g/l, NaCl 10 g/l, (NH4)2SO4 2.5 g/l) at pH 8.0 was inoculated with 20 h of inoculum age (1.5x108cells/ml) and incubated at 35 °C/ 72 h. Correspondingly, solid-state fermentation process was conducted by lawn culture method (Sharma and Bajaj, 2016). In the solid-state fermentation process, nutritionally inert material agar agar was used as a carrier that imparts solid support for attachment of bacteria. Briefly, 1L of modified minimal salt agar medium with pH 8.0 was taken in 50 different plates, 20 h of inoculum age (0.1 ml/ plate & 1.5x108 cells/ml) was spread on the plate and incubated at 35 °C/ 72 h. Cell biomass was then collected and PHAs was recovered by following different downstream processing like sonication-mono-solvent and sodium hypochlorite digestion-multi-solvent extraction as described earlier (Singh et al., 2009, Arikawa et al., 2017).
2.6. Structural and thermal characterization of PHAs
2.6.1. FTIR
FTIR spectroscopic analysis was conducted to determine the functional group of PHAs. Extracted PHAs film (2 mg) was placed on a diamond-based plate of ATR (attenuated total reflectance). Then, IR spectra were recorded using single beam spectrometer (Perkin- Elmer RX I) within the scanning range 4000–400 cm−1 (Mohapatra et al., 2014).
2.6.2. NMR
1H NMR was carried out to elucidate the proton configuration and it’s sequence distribution in the polymer chain of PHAs. Briefly, extracted PHAs film (10 mg/ml) was dissolved with CdCl3 (1 ml) and subjected to NMR (JEOL JNM-LA). NMR spectra were recorded at 500 MHz with 5 ms pulse width, 32,000 data points and 32 accumulations were collected (Wang et al., 2016).
2.6.3. XRD
Structural integrity (degrees of crystallinity) of extracted PHAs was analyzed by X-ray diffractometer (PANalytical). PHAs sample was used in a capillary (d; 2 mm) tube, data were recorded in 2θ range (2–30°), scan rate (2° /min) with nickel filtered Cu-K-α (λ = 0.1542 nm) beam and operated at 30 kV (Wang et al., 2016).
2.7. Thermal characterization of PHAs
Thermostability of PHAs was determined by differential thermo analysis (DTA) for its processing & application. Extracted PHAs was subjected to DSC Q20 and TGA Q50 analyzer for thermal analysis. Moisture free purified PHAs (10 mg) was taken with a sequential program of heating (heating rate 10 °C/min and temperature range 0 to 600 °C) in the presence of N2 flow of 200 ml/min. Then, melting temperature (Tm), degradation temperature (Td), glass transition temperature (Tg) and enthalpy of fusion (ΔHm) of PHAs was recorded. Crystallinity (Xc) of PHAs was calculated by;
(ΔHf = heat of fusion of PHAs and ΔHf° = 146 J/g, the heat of fusion of standard PHB, the most common homopolymer of PHAs) (Wang et al., 2016).
2.8. Morphological analysis of PHAs film
Surface morphological properties of PHAs film was observed under field emission scanning electron microscopic (FE-SEM) and atomic force microscopic (AFM) (Shrivastav et al., 2014) imaging. In brief, extracted PHAs films were fixed on aluminum stumps, followed by gold coating through Polaron SC7620 sputter coater for FE-SEM (QUANTA 200 FEG, Netherlands). Correspondingly, aluminum stumps fixed PHAs film was processed for AFM imaging and surface morphology was studied using surface probe & laser beam.
2.9. Cytotoxicity assay of PHAs
Cytocompatibility of extracted PHAs was evaluated by cytotoxicity assay along with standard PHB (Sigma Aldrich, USA). In brief, 10% (w/v) each of extracted PHAs and standard PHB solutions with concentration from 100 to 500 μg/ml were prepared with chloroform. Extracted polymer (PHB-co-PHV) film was cast in 24 well tissue culture plates and then seeded with adherent mouse fibroblast 3 T3 cell line (106 cells/well). Cell culture was maintained in the Dulbecco’s Modified Eagle Medium (DMEM) for 24 h [10% FBS, 2 mM glutamine and 1X antibiotic-antimycotic solution (Invitrogen, USA)]. Cells were collected with Trypsin-EDTA and survivability was estimated by propidium iodide (0.2 µg/106 cells) staining. Data acquisition was conducted by fluorescent activated cell sorting FACS caliber (BD) using cell quest software.
3. Result and discussion
3.1. Screening and morpho-physiological characterization of PHAs producing bacteria
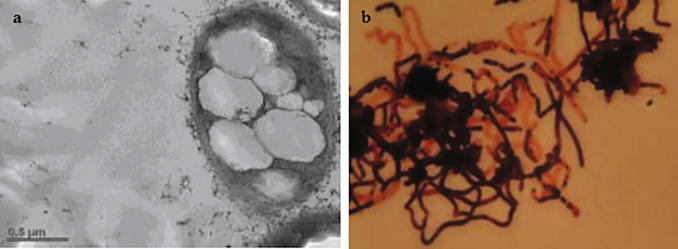
Although earlier studies reported that a wide array of bacterial species produces biopolymer (PHAs), the sheer diversity of the microbial world attracts for exploration of high PHAs yielding bacteria from different ecological niches. In light of the above, 06 PHAs producing bacteria were isolated from the rich, lush green, vibrant ecosystem, Bhitarkanika mangrove forest of Odisha, India. These bacterial isolates were capable of accumulating PHAs granule as carbonosome which was primarily confirmed by Sudan black staining (Fig. 1a). Among all, the bacterial isolate (SP8) was selected for PHAs production based on high intensity of light emitted under nile red staining (Fig. S1, Supplementary file). The bacterial isolate was affiliated to genus Bacillus sp. (SP8) based on morpho-physiological, biochemical, sugar utilization and antibiotics characterization (Fig. S2, Table S1 & Table S2, Supplementary file). Rod shape and smooth cell surface of the screened bacteria was confirmed by FE-SEM. Notably, rod shape of the bacteria transformed to marginally oval due to increase in number and size of PHAs granules accumulated (Fig. 2) in the cytosol.
Fig. 1.
PHAs accumulated B. megaterium OUAT 016 cells (a) under TEM and (b) BFM.
Fig. 2.
Morphological modification (a, b & c) of B. megaterium OUAT 016 cell during PHAs accumulation from 24 to 72 h.
Similar shape changes in Lysinibacillus sp. 3HHX (Mohapatra et al., 2016b), Bacillus sp. CS-605 & Bacillus cereus AC-1 (Bhagowati et al., 2015) and Cupriavidus necator (Mravec et al., 2016) were observed during accumulation of PHAs granule. Moreover, the earlier report also suggested that long rod shapes of Bacillus megaterium facilitated the accumulation of a maximum number of PHAs granule as compared to other Bacillus species (Wu et al., 2016). In contrast to our observation, similar PHAs producing Bacillus species were also found in numerous environmental niches such as waste-water, sludge and marine ecosystems (Mohapatra et al., 2015b); (Bhagowati et al., 2015); (Pandian et al., 2010). This fact can be correlated with high organic carbon and less nitrogen, phosphorus, potassium content of the Bhitarkanika mangrove forest sediment is due to either direct incorporation of organic matter generated from anthropogenic activity or decomposition of plant materials (Thatoi et al., 2012), which creates a selective pressure for formation of PHAs granules (Mohapatra et al., 2015b). Moreover, Bacillus is the predominant genera of soil, genetically stable, generally regarded as safe and capable of growing in presence of cheap raw materials than other bacterial isolates (Khiyami et al., 2011). Here, we explored for the first time PHAs producing Bacillus sp. from the Bhitarkanika mangrove forest.
3.2. Evolutionary analysis of the bacterial isolate
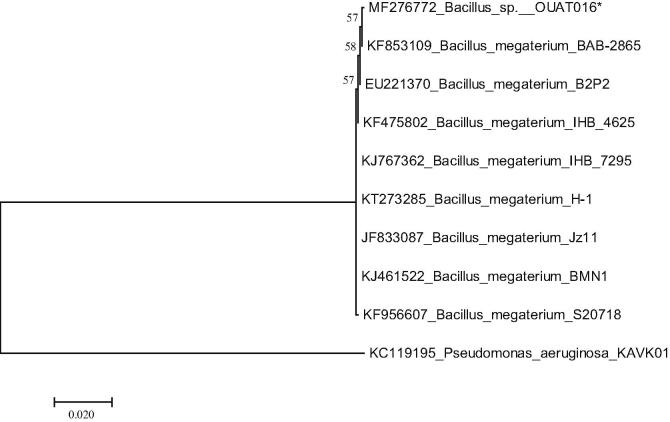
The predicted neighbour joining (NJ) based phylogenetic tree (Fig. 3) with the sum of branch lenghth 0.02 depicted an evolutionary relationship among isolated bacterial strain Bacillus sp. OUAT 016 with other eight closely related 16S rDNA sequences belonging Bacillus species retrived from NCBI database. The sequences are clussered into one major group and diverged distinctly from the outgroup (Pseudomonas aeruginosa KAVK01). There are two sub-groups are present within the major group, where all nine bacterial strains are clustered together. In sub-group I bacterial strain such as Bacillus sp. OUAT 016 (MF276772), Bacillus megaterium BAB 2865 (KF853109), Bacillus megaterium B2P2 (EU221370) and Bacillus megaterium IHB 4625 (KF475802) whereas, in sub-group II Bacillus megaterium IHB 7295 (KJ767362), Bacillus megaterium H 1 (KT273285), Bacillus megaterium Jz11 (JF833087), Bacillus megaterium BMN1 (KJ461522), Bacillus megaterium S20718 (KF956607) are clustered together. Moreover, our desired bacterial strain Bacillus sp. OUAT 016 (MF276772) and Bacillus megaterium BAB 2865 (KF853109) strains are strongly related to each other within the sub-group I. On the other hand, Bacillus megaterium B2P2 (EU221370) and Bacillus megaterium IHB 4625 (KF475802) strains are related to each other under sub-group I. However, within the sub-group II, all the bacterial strains seem to be related to each other without any divergence. The PHAs producing bacterial strain was identified as Bacillus megaterium OUAT 016 by in silico analysis with Gen-bank accession numbers MF276772. Strong evolutionary relationship of Bacillus megaterium OUAT 016 with other closely related strains of Bacillus megaterium was also confirmed from secondary rRNA structure in terms of the highly conserved loop and stem. The bacterium is energetically (free energy −583 kcal/mol) stable (Fig. S3 & Table S3, Supplementary file) as confirmed from secondary rRNA structure analysis.
Fig. 3.
Phylogram of B. megaterium OUAT 016 with its homologous sequences of BLAST search analysis.
3.3. PHAs production
Growth parameters play a key role in biomass as well as PHAs production by bacteria in-vitro. Optimization study revealed higher cell biomass production in modified minimal salt medium at pH 8.0, temperature 35 °C, glucose 3% as carbon source, (NH4)2SO4 (2.5 g/l) as nitrogen source and 20 h inoculum age were optimum at P < 0.05 significance level (Fig. S4a–g, Supplementary file). Under submerged fermentation, Bacillus megaterium OUAT016 produced 1.98 g/l and 2.31 g/l of PHAs from 4.0 g of dry cell weight (DCW) in 72 h by sodium hypochlorite digestion multi-solvent and sonication mono-solvent extraction method. Correspondingly, in solid-state fermentation 2.05 g/l and 3.72 g/l of PHAs were produced from 6.0 g of DCW in 72 h. Comparatively, higher PHAs production (62%) was obtained in solid-state fermentation than submerged fermentation (57.7%). Interestingly, sonication mono-solvent was proved to be an efficient method for PHAs recovery in both cases.
Several reports are available in support of PHAs production such as 2.5 g/l by Bacillus sp. S1 2013b (Mohapatra et al., 2014), 1.62 g/l by Bacillus subtilis KP172548 (Mohapatra et al., 2017b), 4.1 g/l by Bacillus thuringiensis KJ206079 (Desouky et al., 2014) 1.60 g/l by Bacillus megaterium (Israni and Shivakumar, 2013), 3.0 g/l by Bacillus cereus SPV (Valappil et al., 2007) and 6.07 g/l by Bacillus sp. (Musa et al., 2016) under submerged fermentation through sodium hypochlorite digestion multi-solvent extraction. So far our knowledge is concerned, inadequate attention has been provided towards the recovery of PHAs using mono-solvent extraction method. An earlier report (Arikawa et al., 2017) suggested that C. necator KNK-005 produced 15.6 g/l of PHAs from 18.6 g/l of biomass DCW under submerged fermentation through sonication mono-solvent extraction method. Besides submerged fermentation, solid-state fermentation can be used as an alternative strategy for PHAs production (Sindhu, 2015). As a matter of fact, Bacillus sphaericus NII0838 (Ramadas et al., 2009) and Bacillus megaterium MSBN04 (Sathiyanarayanan et al., 2013) produced 0.169 g/l and 8.637 g/l of PHAs under solid-state fermentation through sodium hypochlorite digestion multi-solvent extraction. Notably, research findings are not available on the recovery of PHAs by sonication mono-solvent extraction under solid-state fermentation using Bacillus species. Here, we report for the first time higher PHAs recovery through sonication mono-solvent extraction, which prevent degradation of PHAs and reduction in molecular weight as compared to sodium hypochlorite digestion multi-solvent extraction method (Rawte and Mavinkurve, 2002).
3.4. Structural and thermal analysis of PHAs
3.4.1. FTIR analysis
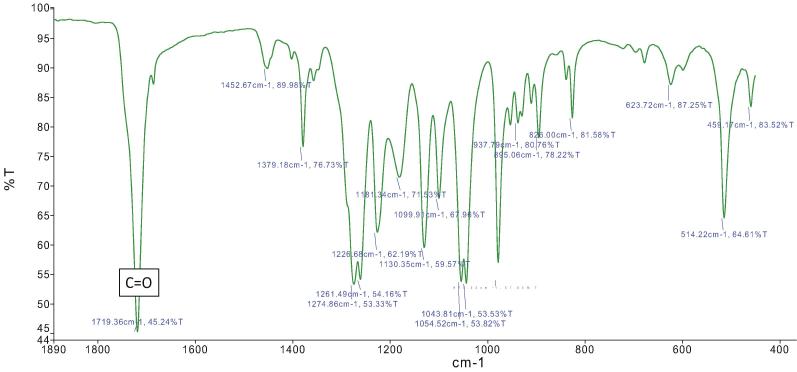
FTIR spectra showed four intense absorption bands at 1181, 1379, 1452 and 1261 cm−1 corresponding to R-CO-O-C2H5 stretch, —CH3, —CH2, and —CH groups respectively (Fig. 4). However, the high intense absorption band obtained at 1719.36 cm−1 assigned to the C O stretch mode of crystalline parts in PHB-co-PHV (Fig. 4). These major annotated vibrational peaks were also comparable with the peaks of PHB-co-PHV as reported earlier (Wang et al., 2016, Masood et al., 2012). Thus, IR analysis provided a correct insight for the chemical structure of PHB-co-PHV by reflecting the monomeric units that are predominantly present in the PHAs.
Fig. 4.
FTIR spectra at 1719.36 cm−1 depicting C O functional group of PHAs.
3.4.2. 1H NMR analysis
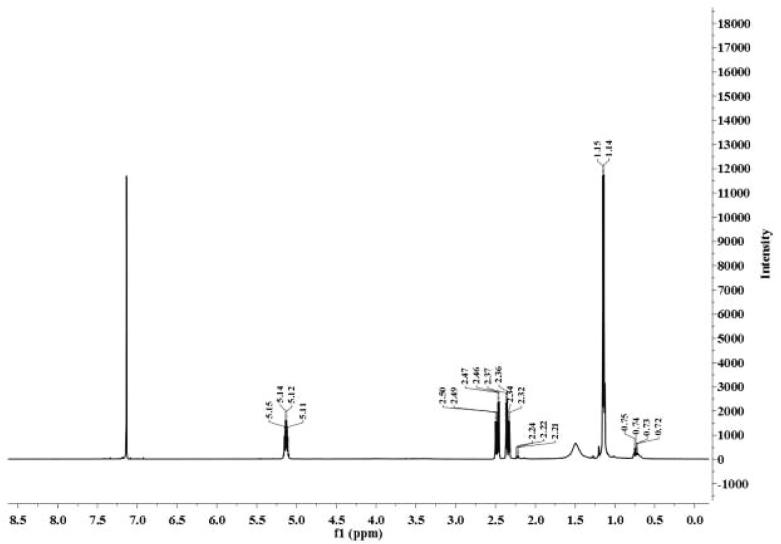
1H NMR spectroscopic (Fig. 5) patterns of various chemical shifting values ranging from 0.72 to 0.75 ppm correspond to the methyl terminal groups of 3HV monomer unit. Apart from that doublet at 1.14, 1.15 ppm corresponds to —CH3, small multiplet peaks at 2.21–2.24 ppm and quadrate of doublet peaks at 2.32–2.50 ppm indicates —CH2 group of HV & HB present in the PHAs polymer. Another chemical shift at 5.11–5.15 ppm detonates presence of —CH group in the extracted polymer. The 1H NMR spectrum of the purified polymer was consistent with previous findings of other studies. More specifically, the presence of valerate monomer in the polymer was confirmed by the existence of —CH3 at 0.9 & 0.92 ppm; a methylene at 1.5 & 1.63 ppm; —CH2 at 2.38–2.43 ppm & 2.55 ppm and —CH at 5.18–5.20 & 5.1 ppm (Wang et al., 2016, Masood et al., 2012, Wei et al., 2014). The 1H NMR spectrum depict presence of HV unit in the PHAs. These signals clearly describe the presence of PHB-co-PHV in the extracted polymer.
Fig. 5.
1H NMR chemical shift pattern established the extracted polymer is a co-polymer PHB-co-PHV.
3.4.3. Thermal characterization of PHAs
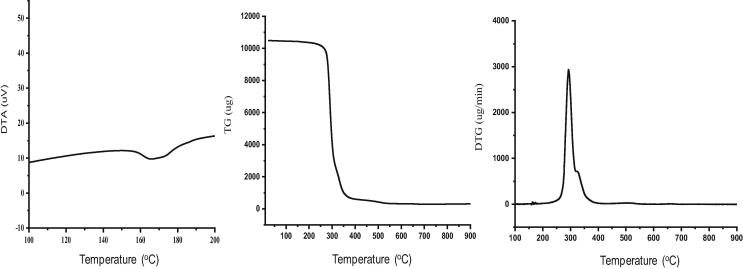
DTA analysis revealed that the Tm of extracted PHB-co-PHV was 166 °C (62 mj/mg ΔH; endothermic reaction). However, the glass transition temperature (Tg) was not detected. The PHB-co-PHV degradation was observed between 100 and 400 °C and weight loss (%) was gradually increased with temperature. Remarkably, 0.4, 2.4, 61.2, and 93.0% weight loss of PHB-co-PHV was detected at 100, 248, 300 and 371 °C respectively (Fig. 6). Optimum thermal degradation (2.92 mg/min) was detected at 273 °C (Td) as revealed from DTG curve (Fig. 6). Crystallinity (Xc) of extracted PHB-co-PHV was found to be 42.46%. Our results coincide with the thermal properties such as Tm (155 to 169 °C), Td (284 °C) and Xc (46.2%) of PHB-co-PHV (Brunel et al., 2014). The Tm, Xc of extracted PHB-co-PHV is lower than the PHB-co-PHV reported earlier which indicated the polymer containing more amount of HB than HV. Hence this result depicting the amorphous part of this polymer will decompose faster however the crystalline part of this polymer can provide durability. The thermal and crystallinity characteristics confer the quality as well as the purity of the polymer.
Fig. 6.
DTA elucidating melting (Tm-166 °C) and thermal degradation (Td-273 °C) temperature of PHB-co-PHV.
3.4.4. X-ray diffraction analysis
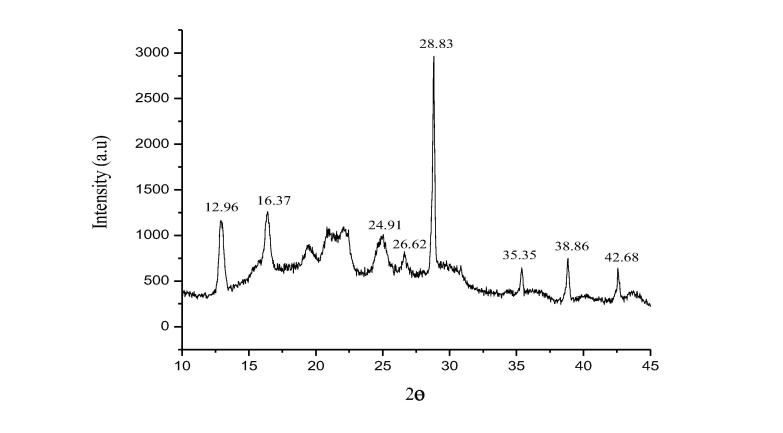
The degree of crystallinity of PHB-co-PHV was estimated from X-ray diffractogram. The 2θo values obtained at 12.96°, 16.37°, 24.91°, 26.62°, 28.83°, 35.35°, 38.86° and 42.68° (Fig. 7) are corresponding to PHB-co-PHV. Among all, five distinct peaks were observed at 12.96°, 16.37°, 35.35°, 38.86°, 42.68° and the high intense peak at 28.83° indicating crystalline nature of PHB-co-PHV. Our result also corroborates with the result of few other studies (Wei et al., 2014, Brunel et al., 2014) where the obtained absorbance points were similar.
Fig. 7.
X-ray diffractogram indicating crystalline nature of PHB-co-PHV.
3.4.5. Morphological analysis of PHAs film
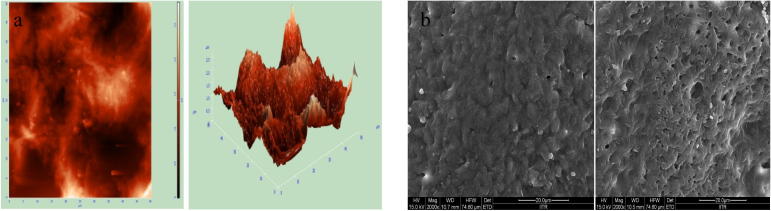
PHB-co-PHV film exterior was uneven and fairly regular as confirmed by FE-SEM (Fig. 8b) and AFM (Fig. 8a) imaging, which indicating its bio-degradable nature. Rough exterior of PHB-co-PHV film supports tenderness of microbial cells and subsequently formation of pores (micro size) on the surface (Shah, 2012, Padermshoke et al., 2004). Further, crystalline nature (Spyros et al., 1997) of the PHAs film also boosts tenderness of microbes, fronting to degradation of polymer to CO2 & H2O in oxic and CO2 & CH4 in anoxic condition (Ym and Savitha, 2011).
Fig. 8.
Surface morphology of PHB-co-PHV film under (a) AFM and (b) FE-SEM.
3.4.6. Cytotoxicity assay
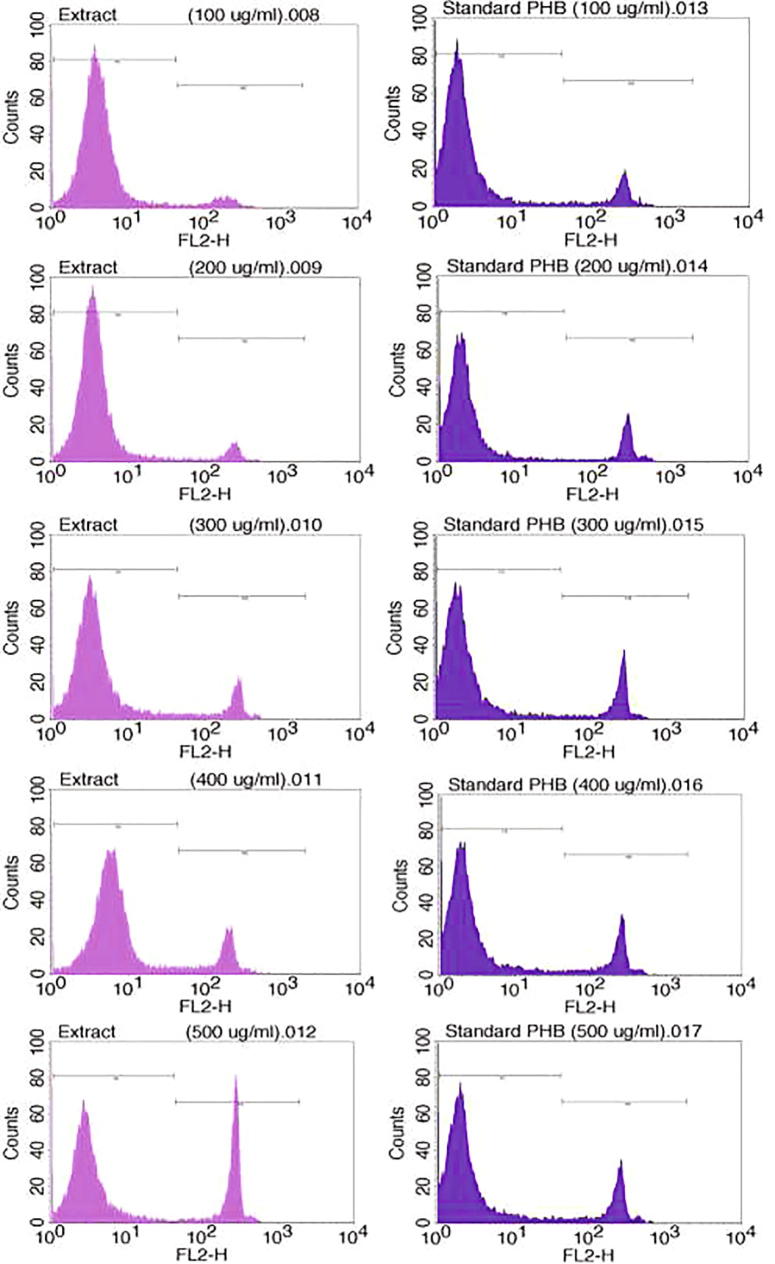
FACS assay data implied remarkable cell survivability between 91.02 and 65.58% and the death rate 6.74–31.31% in 100–500 µg/ml of extracted PHB-co-PHV. However, cell survivability and death rate ranged from 81.86 to 73.40% and 11.27–18.50% in standard PHB (Fig. 9 & Table S4 in supplementary file). In general, higher cell survivability was perceived in extracted PHB-co-PHV than standing up to 400 µg/ml, which confirmed noncytotoxic nature of extracted PHB-co-PHV on adherent mouse fibroblast 3T3 cells. Moreover, cell viability above 50% is clear evident of noncytotoxic nature of PHAs (Mohapatra et al., 2016b). In corroboration to our result, noncytotoxic effect of P(3HB-co-3HDD-co-3HTD) and PHB recovered from Lysinibacillus sp. and Bacillus subtilis were also reported earlier (Mohapatra et al., 2016b, Mohapatra et al., 2017b). Lower bioactivity and acidity than glycolic & lactic make PHB noncytotoxic to different cells (Chuah et al., 2013). Elevated cytocompatibility indicates diverse applications of PHB-co-PHV, which could be exploited for various biomedical implications in future.
Fig. 9.
Comparative cytotoxicity assay of extracted PHB-co-PHV and standard PHB.
4. Conclusion
In this research, reorientation of PHAs (PHB-co-PHV) production from submerged to solid state fermentation was validated. Production of PHB-co-PHV through solid state fermentation increases purity, productivity and minimizes use of energy. Additionally, agar generated from the bioprocess technology can be reused for further applications, which balances overall economy of its production. This research established a novel noncytotoxic biopolymer PHB-co-PHV production (3.72 g/l) from Bacillus megaterium OUAT 016 through solid state fermentation. Further, presence of different propertions of HV monomers and rough surface of the extracted PHB-co-PHV supports microbial tenderness and greatly enhance the biodegradibilty performance. The low melting temperature and crystallinity of PHB-co-PHV depicted its toughness, impact resistance and flexibility over other polymers. As PHB-co-PHV is extremely less or null toxic to diversified cells, thereby indicating its aptness for several biomedical applications including as a drug delivery carrier. Hence, further research is highly indispensable prior to biomedical applications of the PHB-co-PHV.
Acknowledgement
This research did not receive grant from any funding agencies. The authors are thankful to Dr. S.K. Dash, Dr. S. Acharya, Dr. G.S. Acharya (OIC, CIF) and Dr. S. K. Pradhan, HoD, Department of Bioinformatics, OUAT, Bhubaneswar, Odisha for providing laboratory facilities during the period of research work. The authors have no conflict of interest to declare.
Footnotes
Peer review under responsibility of King Saud University.
Supplementary data to this article can be found online at https://doi.org/10.1016/j.sjbs.2020.02.001.
Appendix A. Supplementary material
The following are the Supplementary data to this article:
References
- Arikawa H., Sato S., Fujiki T., Matsumoto K. Simple and rapid method for isolation and quantitation of polyhydroxyalkanoate by SDS-sonication treatment. J. Biosci. Bioeng. 2017;124:250–254. doi: 10.1016/j.jbiosc.2017.03.003. [DOI] [PubMed] [Google Scholar]
- Bhagowati P., Pradhan S., Dash H.R., Das S. Production, optimization and characterization of polyhydroxybutyrate, a biodegradable plastic by Bacillus spp. Biosci. Biotechnol. Biochem. 2015;79:1454–1463. doi: 10.1080/09168451.2015.1034651. [DOI] [PubMed] [Google Scholar]
- Brunel D.G., Pachekoski W.M., Carla-Dalmolin J.A.M.A. Natural additives for poly (hydroxybutyrate - CO - hydroxyvalerate) - PHBV: effect on mechanical properties and biodegradation. Mater. Res. 2014;17:1145–1156. doi: 10.1590/1516-1439.235613. [DOI] [Google Scholar]
- Chuah J., Yamada M., Taguchi S., Sudesh K., Doi Y., Numata K. Biosynthesis and characterization of polyhydroxyalkanoate containing 5-hydroxyvalerate units: effects of 5HV units on biodegradability, cytotoxicity, mechanical and thermal properties. Polym. Degrad. Stab. 2013;98:331–338. [Google Scholar]
- Desouky S.E., El-Shiekh H.H., Elabd M.A., Shehab A.M. Screening, optimization and extraction of polyhydroxyalkanoates (PHAs) from Bacillus thuringienesis. J. Adv. Biol. Biotechonol. 2014;1(1):40–54. [Google Scholar]
- Gomaa E.Z. Production of polyhydroxyalkanoates (PHAs) by Bacillus subtilis and Escherichia coli grown on cane molasses fortified with ethanol. Braz. Arch. Biol. Technol. 2014;57(1):145–154. [Google Scholar]
- Holt J.G., Krieg N.R., Sneath P.H.A., Stanley J.T., Willium S.T. Williamsons and Wilkins; Balitomore: 1994. Bergey’s Manual of Determinative Bacteriology. [Google Scholar]
- Thatoi H., Behera B.C., Dangar T.K., RanjanMishra R. Microbial biodiversity in mangrove soils of bhitarkanika. Odisha. India. Int. J. Environ. Biol. 2012;2:50–58. [Google Scholar]
- Khiyami M.A., Al-fadual S.M., Bahklia A.H. Polyhydroxyalkanoates production via Bacillus plastic composite support (PCS) biofilm and date palm syrup. J. Med. Plants. Res. 2011;5(14):3312–3320. [Google Scholar]
- Kunasundari B., Sudesh K. Isolation and recovery of microbial polyhydroxyalkanoates. Express Polym. Lett. 2011;5:620–634. doi: 10.3144/expresspolymlett.2011.60. [DOI] [Google Scholar]
- Maity S., Das S., Samantaray D.P. Effect of vitamin on accumulation of PHB by Zobellella species under submerged fermentation process. Int. J. Curr. Microbiol. Appl. Sci. 2017;6:1310–1316. [Google Scholar]
- Masood F., Hasan F., Ahmed S., Hameed P.C.A. Biosynthesis and characterization of poly- (3-hydroxybutyrate-co- 3-hydroxyvalerate) from Bacillus cereus S10. J. Polym. Environ. 2012;20:865–871. doi: 10.1007/s10924-012-0457-y. [DOI] [Google Scholar]
- Mathews D.H., Turner D.H. Dynalign: An algorithm for finding the secondary structure common to two RNA sequences. J. Mol. Biol. 2002;317:191–203. doi: 10.1006/jmbi.2001.5351. [DOI] [PubMed] [Google Scholar]
- Mayeli N., Motamedi H., Heidarizadeh F. Production of polyhydroxybutyrate by Bacillus axaraqunsis BIPC01 using petrochemical wastewater as carbon source. Braz. Arch. Biol. Technol. 2015;58:643–650. doi: 10.1590/S1516-8913201500048. [DOI] [Google Scholar]
- Mohapatra, S., Mohanta, P.R., Sarkar, B., Daware, A., Kumar, C., Samantaray, D.P., 2015. Production of polyhydroxyalkanoates (PHAs) by Bacillus strain isolated from waste water and its biochemical characterization. Proc. Natl. Acad. Sci. USA. http://dx.doi.org/10.1007/s40011-015-0626-6.
- Mohapatra S., Rath S.N., Pradhan S.K., Samantaray D.P., Rath C.C. Secondary structural models (16S rRNA) of polyhydroxyalkanoates producing Bacillus species isolated from different rhizospheric soil: phylogenetics and chemical analysis. Int. J. Bioautomation. 2016;20:329–338. [Google Scholar]
- Mohapatra S., Samantaray D.P., Samantaray S.M. Phylogenetic heterogeneity of the rhizospheric soil bacterial isolates producing PHAs revealed by comparative analysis of 16s-rRNA. Int. J. Curr. Microbiol. App. Sci. 2014;3:680–690. [Google Scholar]
- Mohapatra S., Samantaray D.P., Samantaray S.M., Mishra B.B., Das S., Majumdar S., Pradhan S.K., Rath S.N., Rath C.C., Akthar J., Achary K.G. Structural and thermal characterization of PHAs produced by Lysinibacillus sp. through submerged fermentation process. Int. J. Biol. Macromol. 2016;93:1161–1167. doi: 10.1016/j.ijbiomac.2016.09.077. [DOI] [PubMed] [Google Scholar]
- Mohapatra S., Sarkar B., Samantaray D.P., Daware A., Maity S., Pattnaik S., Bhattacharjee S. Bioconversion of fish solid waste into PHB using Bacillus subtilis based submerged fermentation process. Environ. Technol. 2017;1–8 doi: 10.1080/09593330.2017.1291759. [DOI] [PubMed] [Google Scholar]
- Mravec F., Obruca S., Krzyzanek V., Sedlacek P., Hrubanova K., Samek O., Kucera D., Benesova P., Nebesarova J. Accumulation of PHA granules in Cupriavidus necator as seen by confocal fluorescence microscopy. FEMS Microbiol. Lett. 2016;363(10):1–18. doi: 10.1093/femsle/fnw094. [DOI] [PubMed] [Google Scholar]
- Israni N., Shivakumar S. Combinatorial screening of hydrolytic enzyme/s and PHA producing Bacillus spp. for cost effective production of PHAs. Int. J. Pharma Bio. Sci. 2013;4:934–945. [Google Scholar]
- Padermshoke A., Sato H., Katsumoto Y., Ekgasit S., Noda I., Ozaki Y. Thermally induced phase transition of poly(3-hydroxybutyrate-co-3-hydroxyhexanoate) investigated by two-dimensional infrared correlation spectroscopy. Vib. Spectrosc. 2004;36:241–249. doi: 10.1016/j.vibspec.2003.11.016. [DOI] [PubMed] [Google Scholar]
- Musa H., Bolanle B.B., Kasim F.H., Arbain D. Screening and production of polyhydroxybutyrate (PHB) by bacterial strains isolated from rhizosphere soil of groundnut plants. Sains Malaysiana. 2016;45(10):1469–1476. [Google Scholar]
- Ramadas N.V., Singh S.K., Soccol C.R., Pandey A. Polyhydroxybutyrate production using agro-industrial residue as substrate by Bacillus sphaericus NCIM 5149. Braz. arch. biol. technol. 2009;52(1):17–23. doi: 10.1590/S1516-89132009000100003. [DOI] [Google Scholar]
- Pandian S.R.K., Deepak V., Kalishwaralal K., Rameshkumar N., Jeyaraj M., Gurunathan S. Optimization and fed-batch production of PHB utilizing dairy waste and sea water as nutrient sources by Bacillus megaterium SRKP-3. Bioresour. Technol. 2010;101:705–711. doi: 10.1016/j.biortech.2009.08.040. [DOI] [PubMed] [Google Scholar]
- Rawte T., Mavinkurve S. A rapid hypochlorite method for extraction of polyhydroxyalkanoates from bacterial cells. Ind. J. Exp. Biol. 2002;40:924–929. [PubMed] [Google Scholar]
- Saitou N., Nei M. The neighbor-joining method- a new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 1987;4:406–425. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- Sathiyanarayanan G., Kiran G.S., Selvin J., Saibaba G. Optimization of polyhydroxybutyrate production by marine Bacillus megaterium MSBN04 under solid state culture. Int. J. Biol. Macromol. 2013;60:253–261. doi: 10.1016/j.ijbiomac.2013.05.031. [DOI] [PubMed] [Google Scholar]
- Sharma P., Bajaj B.K. Economical production of poly-3-hydroxybutyrate by Bacillus cereus under submerged and solid state fermentation. J. Mater. Environ. Sci. 2016;7:1219–1228. [Google Scholar]
- Shah K.R. FTIR analysis of polyhydroxyalkanoates by novel Bacillus sp. AS 3–2 from soil of Kadi region, North Gujarat. Ind. J. Biochem. Technol. 2012;3:380–383. [Google Scholar]
- Shrivastav A., Biotech A., Kumar S., Algallio M., Private B. Biodegradability studies of polyhydroxyalkanoate (PHA) film produced by a marine bacteria using Jatropha biodiesel. World J. Microbiol. Biotechnol. 2014;27:1531–1541. doi: 10.1007/s11274-010-0605-2. [DOI] [Google Scholar]
- Sindhu R. Solid-state fermentation for the production of poly(hydroxyalkanoates) Chem. Biochem. Eng. Q. 2015;29:173–181. doi: 10.15255/CABEQ.2014.2256. [DOI] [Google Scholar]
- Singh M., Kumar P., Patel S.K.S., Kalia V.C. Production of polyhydroxyalkanoates co-polymer by Bacillus thuringiensis. Int. J. Microbiol. 2013;53:77–83. doi: 10.1007/s12088-012-0294-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh M., Patel S.K.S., Kalia V.C. Bacillus subtilis as potential producer for polyhydroxyalkanoates. Microb. Cell Fact. 2009;8:38–46. doi: 10.1186/1475-2859-8-38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spyros A., Kimmich R., Briese B.H., Jendrossek D. 1H NMR imaging study of enzymatic degradation in poly(3-hydroxybutyrate) and poly(3-hydroxybutyrate- co -3-hydroxyvalerate): evidence for preferential degradation of the amorphous phase by PHB depolymerase-B from Pseudomonas lemoignei. Macromolecules. 1997;30:8218–8225. doi: 10.1021/ma971193m. [DOI] [Google Scholar]
- Sultanpuram M.T., Reddy V., Mahmood S.K. Production and characterization of PHB from two novel strains of Bacillus spp. isolated from soil and activated sludge. J. Ind. Microbiol. Biotechnol. 2010;37:271–278. doi: 10.1007/s10295-009-0670-4. [DOI] [PubMed] [Google Scholar]
- Tamura K., Stecher G., Peterson D., Filipski A., Kumar S. MEGA 6: molecular evolutionary genetics analysis version 6.0. Mol. Biol. Evol. 2013;30:2725–2729. doi: 10.1093/molbev/mst197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan G.Y., Chen C.L., Li L., Ge L., Wang L., Razaad I., Li Y., Zhao L., Mo Y., Wang J.-Y. Start a research on biopolymer polyhydroxyalkanoate (PHA): A review. Polymers (Basel). 2014;6:706–754. doi: 10.3390/polym6030706. [DOI] [Google Scholar]
- Valappil S.P., Peiris D., Langley G.J., Herniman J.M., Boccaccini A.R., Bucke C., Roy I. Polyhydroxyalkanoate (PHA) biosynthesis from structurally unrelated carbon sources by a newly characterized Bacillus spp. J. Biotechnol. 2007;127:475–487. doi: 10.1016/j.jbiotec.2006.07.015. [DOI] [PubMed] [Google Scholar]
- Wang C., Zheng Y., Sun Y., Fan J., Qin Q., Zhao Z. Polymer chemistry mechanical properties and hemocompatibility. R. Soc. Chem. 2016;7:6120–6132. doi: 10.1039/c6py01131d. [DOI] [Google Scholar]
- Wei L., Guho N.M., Coats E.R., McDonald A.G. Characterization of poly(3-hydroxybutyrate- co -3-hydroxyvalerate) biosynthesized by mixed microbial consortia fed fermented dairy manure. J. Appl. Polym. Sci. 2014;131(11):1–12. doi: 10.1002/app.40333. [DOI] [Google Scholar]
- Wu H., Fan Z., Jiang X., Chen J., Chen G. Enhanced production of polyhydroxybutyrate by multiple dividing E. coli. Microb. Cell Fact. 2016;15:1–13. doi: 10.1186/s12934-016-0531-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ym V., Savitha R. Microbial & biochemical technology overview on polyhydroxyalkanoates: A promising biopol. J. Microbial. Biochem. Technol. 2011;3(5):99–105. doi: 10.4172/1948-5948.1000059. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.