Graphical abstract
Keywords: Computational Modeling, Glycosaminoglycans, Sulfotransferase, Enzyme Specificity, Heparin Biosynthesis
Abstract
Pharmaceutical heparin’s activity arises from a key high affinity and high selectivity antithrombin binding motif, which forms the basis for its use as an anticoagulant. The current problems with the supply of pig heparin raises the emphasis of understanding heparin biosynthesis so as to control and advance recombinantly expressed agent that could bypass the need for animals. Unfortunately, much remains to be understood about the generation of the antithrombin-binding motif by the key enzyme involved in its biosynthesis, 3-O-sulfotransferase–1 (3OST-1). In this work, we present a novel computational approach to understand recognition of oligosaccharide sequences by 3OST-1. Application of combinatorial virtual library screening (CVLS) algorithm on hundreds of tetrasaccharide and hexasaccharide sequences shows that 3OST-1 belongs to the growing number of proteins that recognize glycosaminoglycans with very high selectivity. It prefers very well defined pentasaccharide sequences carrying distinct groups in each of the five residues to generate the antithrombin binding motif. CVLS also identifies key residues including His271, Arg72, Arg197 and Lys173, which interact with 6-sulfate, 5-COO¯, 2-/6-sulfates and 2-sulfate at the −2, −1, +2, and +1 positions of the precursor pentasaccharide, respectively. Additionally, uncharged residues, especially Gln163 and Asn167, were also identified as playing important roles in recognition. Overall, the success of CVLS in predicting 3OST-1 recognition characteristics that help engineer selectivity lead to the expectation that recombinant enzymes could be designed to help resolve the current problems in the supply of anticoagulant heparin.
1. Introduction
Glycosaminoglycans (GAGs) are linear polysaccharide chains composed of repeating disaccharide units, which can be used to define the superfamily into heparin/heparan sulfate (Hp/HS), chondroitin sulfate (CS), dermatan sulfate (DS), keratan sulfate (KS) and hyaluronan [1]. GAGs interact with a large number of diverse proteins, such as proteases and protease inhibitors, growth factors and growth factor receptors, chemokines and chemokine receptors, and extracellular matrix proteins [2], [3], [4]. These interactions position GAGs as key players in the regulation of many processes, such as coagulation, angiogenesis, cell proliferation, viral invasion, etc [2], [3], [4].
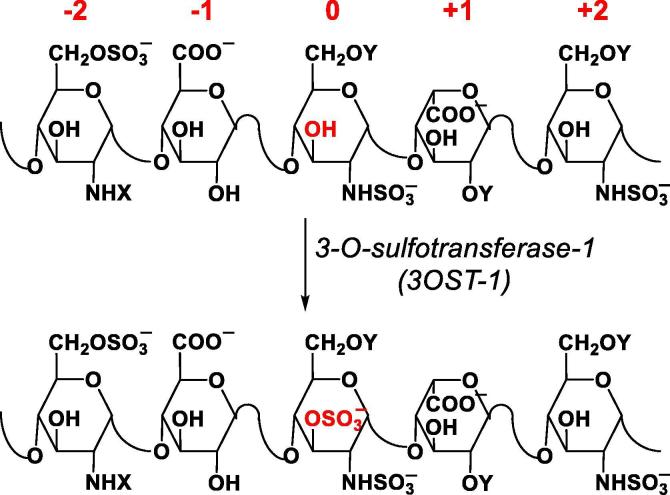
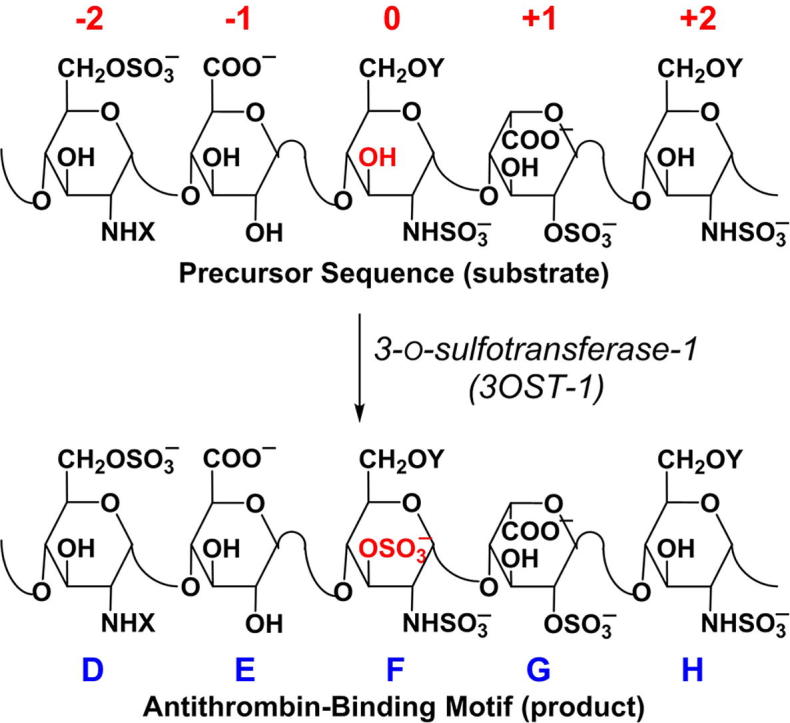
Heparin is a lifesaving drug that has been in use as a blood thinner for most of the past century [5], [6]. Its anticoagulant activity arises from the presence of a key five residue sequence, which binds to antithrombin (AT) with high affinity and high selectivity resulting in a large increase in the rate of inactivation of coagulation proteases, especially thrombin and factor Xa [6]. This five residue sequence is called the antithrombin binding pentasaccharide and contains three glucosamine residues and two uronic acids carrying several key structural components, including the 6-O- sulfate on the non-reducing end glucosamine, the glucuronic acid (GlcA) residue, the N- and 3-O-sulfates on central glucosamine, the iduronic acid (IdoA) residue and the N-sulfate (GlcNS) on the glucosamine at the reducing end (Fig. 1).
Fig. 1.
The antithrombin-binding motif (ABM) in heparin/heparan sulfate (Hp/HS) is introduced by 3-O-sulfotransferase isoform 1 (3OST-1; also called HS3ST-1) when it acts on the precursor sequence to convert the 3-OH group of central GlcN residue (red) to the 3-O-sulfate group (OSO3−). In this work, the precursor sequence residues are labeled as −2, −1, 0, +1, +2 residues (red labels), where the site of 3-O-sulfation by 3OST-1, identified as the 0th residue, is the centerpiece of biosynthetic modification. X can be SO3− or COCH3 and Y can be H or SO3−. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
The biosynthetic machinery of Hp/HS generates various types of 3-O-sulfated sequences with the aid of seven major 3-O-sulfotransferase enzymes (3OST-1, 2, 3a, 3b, 4, 5, and 6) [7]. Of these, 3OST-1 is known to be fairly selective for generating the five-residue antithrombin-binding motif (ABM, Fig. 1). 3OST isoforms 2, 3a, 3b, and 6 are known to generate glycoprotein D-binding motifs (gDBM), whereas 3OST-5 generates both ABM and gDBM. The two groups of 3OSTs, i.e., the ABM and gDBM generating enzymes, display three-dimensional similarity of the overall catalytic site but present important differences in the placement of several residues [7].
Much work has been undertaken in understanding the substrate and binding specificity of 3OST-1, given its importance in the generation of pharmaceutical Hp [8], [9], [10]. A number of 3OST-1 residues, including Arg67, Arg72, Lys123, Asn163, Asn167, His168, Lys171, Lys173, Arg197, Thr256, His271, Ser273, Lys274 and Trp283, have been implicated as important for recognition of the oligosaccharide substrate. From the perspective of the oligosaccharide sequences recognized by 3OST-1, a seminal study shows strong preference of GlcA residue at position −1 [10]. Generally, it has been assumed that 3OST-1 prefers GlcNAc6S residue at the −2 position and GlcNS6S at the +2 position based on crystal structure studies [8]. However, rigorous studies using a fairly large library of oligosaccharides sequences of varying lengths have not been undertaken. Considering the dearth in the availability of such a diverse library of sequences, comprehensive studies are needed to rigorously test the assumption of selectivity and disprove the possibility of elements of promiscuity, if any [7].
Theoretically, nature can generate 15,552 pentasaccharide sequences from 12 possible GlcN and 3 uronic acid (UA) residues. Of these, 16 possible sequences are the ‘motifs’ that can bind antithrombin with high affinity and high selectivity. This approximates to a maximal theoretical proportion of one ABM for every 1000 possible pentasaccharides assuming that the 3-O-sulfation reaction was fully random. However, natural preparations of pharmaceutical Hp show a much higher proportion of the ABM. It is estimated that one in three commercial unfractionated Hp chains carry this sequence, which approximates to 1 in 30 pentasaccharide sequences2 considering that the average chain of Hp could be thought of as 10 pentasaccharide units long.
This raises interesting questions. How does a template-less process generate higher level of ABM? Is it driven by one or more enzymes? If so, what are the key elements of the recognition process that engineers this selectivity? How are less optimal sequences filtered out? What is the optimal length of the precursor sequence that confers selectivity in generation of the ABM? Is it the pentasaccharide precursor? Understanding recognition of precursor sequences by biosynthetic enzymes could lead to rational modification of biosynthetic pathway for cellular expression of Hp/HS. The current problems revolving around the supply of Hp from pigs, which is highly susceptible to infection and disease in these animals, could be averted if bacterial Hp could be expressed to have defined levels of ABM. This would bypass the need for pigs as factories for world’s supply of pharmaceutical Hp.
In this work, we present a novel computational approach to understand recognition of oligosaccharide sequences by isoform 1 of 3-O-sulfotransferase (3OST-1; now renamed as HS3ST-1), the key enzyme that generates the ABM in Hp/HS. We utilize combinatorial virtual library screening (CVLS) algorithm, developed in our lab in the mid-2000s [11], [12] and demonstrated to identify GAG sequences that bind to target proteins with high affinity and high selectivity [13]. We apply this technology to understand substrate specificity of 3OST-1, which offers key insights into how nature engineers higher level of expression of ABM, despite a massive possibility of indiscriminate generation of sequences. We find that 3OST-1 is an enzyme that recognizes the precursor sequence and its few variants with high selectivity (Fig. 1). Based on this work, we infer that a recombinant biosynthetic process that generates this five residue precursor sequence in larger proportion is likely to yield recombinant heparins with higher proportions of ABMs.
2. Experimental methods
2.1. Protein modeling
The crystal structure coordinates of 3OST-1 were extracted from the 3OST-1–HS heptasaccharide complex (PDB ID: 3UAN) [8]. The 3OST-1 protein from the co-complex was extracted; hydrogens added; side chain amides checked for any steric clashes; protonation states of acidic and basic groups assigned; and the protein minimized with fixed heavy-atom coordinates using the Tripos force field for a maximum of 10,000 iterations subject to a termination gradient of 0.05 kcal/(mol·Å), a fixed dielectric constant of 80, and a non-bonded cutoff radius of 8 Å in SYBYLX 2.1 (Tripos Associates, St. Louis, MO). 3′-phosphoadenosine-5′-phosphosulfate (PAPS) is an obligatory co-factor in the 3-O-sulfation reaction and hence 3OST-1 was prepared in both PAPS bound and unbound forms. Oligosaccharide docking and screening experiments (see below) showed no variation in results with either forms (not shown). Hence, the PAPS free form of 3OST-1 was used for all experiments described in this work.
2.2. Native saccharide modeling
The coordinates for HS oligosaccharide from 3OST-1–heptasaccharide co-complex (PDB ID: 3UAN) were extracted using SYBYLX 2.1, then hydrogen atoms added, atom types fixed, ring conformations evaluated, inter-glycosidic torsion angles calculated, charges assigned and then the oligosaccharide was energy minimized using Gasteiger–Hückel charges for a maximum of 100,000 iterations. Note that the summation of Gasteiger–Hückel partial charges for atoms in a charged moiety, e.g., the sulfate group, equals the naked charge of the moiety. This optimized structure was used for re-docking of the protein–GAG complex using GOLD V. 5.2 [14].
2.3. Virtual library generation
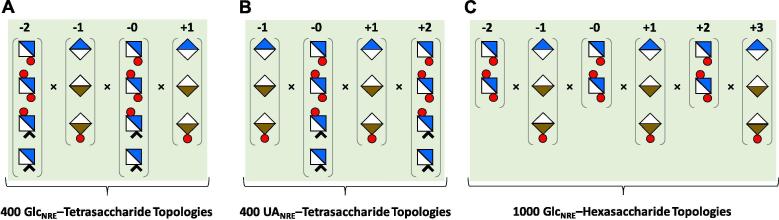
Two libraries of tetrasaccharide and one library of hexasaccharide topologies were constructed using naturally occurring saccharide residues including GlcNS, GlcNS6S, GlcNAc, GlcNAc6S, IdoA, IdoA2S and GlcA. For tetrasaccharide sequences, the non-reducing end (NRE) residue was either a GlcN or a UA (Fig. 2). Since IdoA exists in multiple conformations, of which the chair (1C4) and skew-boat forms (2SO) dominate at room temperature in aqueous solution [15], IdoA and IdoA2S were modeled in silico in both forms [13], [15] As observed in aqueous solution, both GlcA and GlcN residues were modelled in 4C1 chair forms. Thus, a total of 400 tetrasaccharide topologies were built for GlcNRE- (Fig. 2A) and UANRE- (Fig. 2B) libraries each from the possible 4 GlcN and 3 UA residues. The topologies were built in an automated manner using the in-house scripts that operate in SYBYLX 2.1. In theory, the two UANRE and GlcNNRE tetrasaccharide libraries could consist of 2592 unique topologies; however, residues not possible in precursor tetrasaccharides because of the known specificity of prior biosynthetic enzymes, e.g., 2OST (also known as HS2ST), were discarded.
Fig. 2.
Libraries of tetrasaccharide and hexasaccharide sequences were generated from naturally occurring saccharide residues including GlcNS ( ), GlcNS6S (
), GlcNS6S ( ), GlcNAc (
), GlcNAc ( ), GlcNAc6S (
), GlcNAc6S ( ), IdoA (
), IdoA ( ), IdoA2S (
), IdoA2S ( ) and GlcA (
) and GlcA ( ). IdoA and IdoA2S were modeled in 1C4 and 2SO forms, which increase the number of topologies to be studied. Labels −2, −1, 0, +1, and +2 refer to position of the residue from the site of 3-O-sulfation by 3OST-1, which is numbered 0. A) shows the generation of a tetrasaccharide library with a GlcN residue at the non-reducing end (position −2); B) show the tetrasaccharide library with a UA residue at the non-reducing end (position −1); and C) show the hexasaccharide library with a GlcN residue at the non-reducing end (position −2).
). IdoA and IdoA2S were modeled in 1C4 and 2SO forms, which increase the number of topologies to be studied. Labels −2, −1, 0, +1, and +2 refer to position of the residue from the site of 3-O-sulfation by 3OST-1, which is numbered 0. A) shows the generation of a tetrasaccharide library with a GlcN residue at the non-reducing end (position −2); B) show the tetrasaccharide library with a UA residue at the non-reducing end (position −1); and C) show the hexasaccharide library with a GlcN residue at the non-reducing end (position −2).
The hexasaccharide sequences were built with a GlcN residue at the NRE (Fig. 2C) because these would encompass the entire length of the pentasaccharide irrespective of the NRE residue. Thus, the library of hexasaccharide sequences consisted of 1000 topologies. The procedure for generation of GAG libraries can be found in detail from our earlier work [13], [16], [17]. The sequences were energy minimized using Gasteiger–Hückel charges for a maximum of 10,000 iterations. Each sequence from the library was analyzed using automated scripts for the parameters corresponding to the three filters of the computational algorithm.
2.4. Genetic algorithm-based docking and scoring protocol
The molecular docking of the library of Hp/HS sequences onto 3OST-1 was performed using GOLD V. 5.2. software [14]. For GAGs, the inter-glycosidic bonds were constrained and the rest of the molecule was treated completely flexible during the docking step. Based on our earlier GAG studies [11], [13], [17], the parameters were optimized, the binding site was defined as 10 Å around the crystal structure substrate binding site for tetrasaccharide, and 12 Å for hexasaccharide binding site to make sure enough space for sampling. Each GAG structure was docked using 100 genetic algorithm runs, each consisting of 100,000 iterations. The early termination option was turned on in the docking step if the top three solutions displayed an RMSD of 2.5 Å or lower. Each experiment was carried out in triplicate and two best poses were analyzed from each run. This yields six solutions for each sequence from three different experiments, which were used for further analysis.
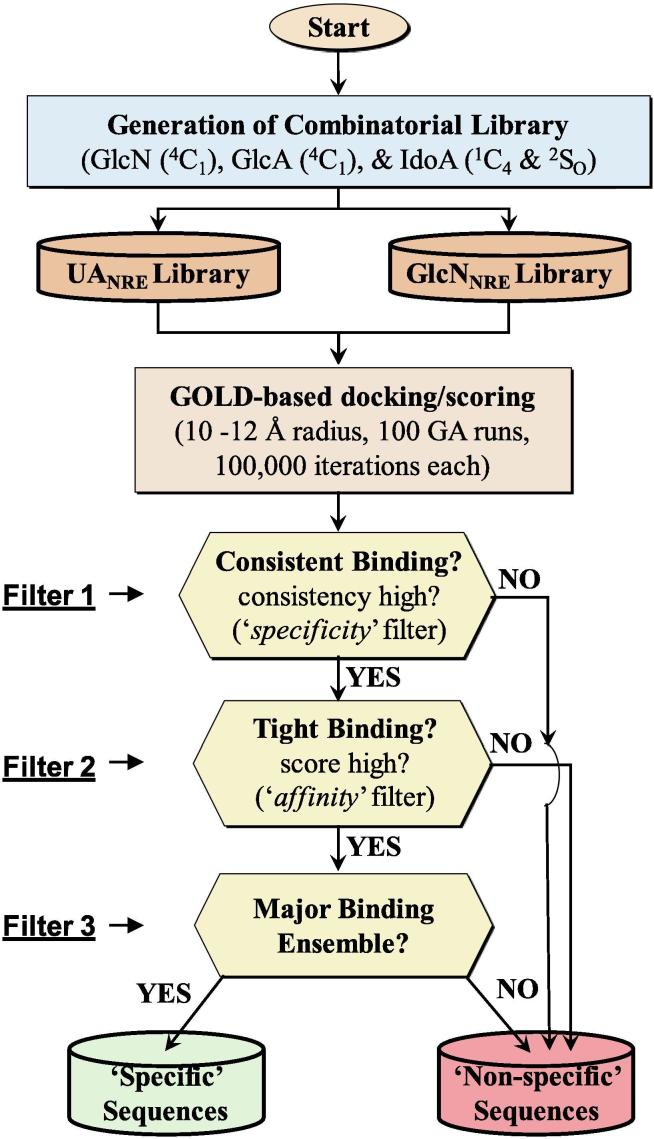
The identification of high affinity/high specificity sequences binding to 3OST-1 utilized our CVLS algorithm (Fig. 3) [13]. The CVLS utilizes more than one filter to select sequences that bind with high interaction score (the ‘affinity’ filter) and high consistency of binding (the ‘specificity/selectivity’ filter). For this work, we used GOLDScore as the ‘affinity’ filter and root mean square deviation (RMSD) between several docking runs as the ‘specificity/selectivity’ filter (Fig. 3). Briefly, multiple genetic algorithm (GA)-docking runs were performed and the RMSD between best six solutions for each sequence was calculated. If RMSD was found to be less than 2.5 Å, the sequences are deemed to be binding with high consistency, and therefore selectivity. Among the selective sequences, those with higher GOLDScore are deemed as ‘high affinity and high selectivity/specificity’ sequences. For this work, we introduced a third filter to deduce the ‘major binding ensemble’, which is the largest cohort of sequences that display identical binding pose among the most selective sequences (Fig. 3). Alternatively, we analyzed all the predicted binding poses following the application of the first two filters into bins of similar poses and identified the largest cohort as the major binding ensemble. This advanced CVLS algorithm was optimized and implemented to distinguish substrate recognition that yields productive binding from the vast number of non-productive interactions.
Fig. 3.
Combinatorial virtual library screening (CVLS) algorithm used to study the 3OST-1 binding to Hp/HS sequences. Our CVLS protocol evaluated tetrasaccharide and hexasaccharide topologies (Fig. 2) using a triple-filter strategy that relied on the consistency of binding (the ‘specificity/selectivity’ filter), the GOLDScore (the ‘affinity’ filter) and the size of ‘binding ensemble’ to assess selectivity of substrate recognition by 3OST-1 of the Hp/HS biosynthetic pathway.
3. Results and discussion
3.1. Rationale for selection of 3OST-1 as the enzyme for selectivity/specificity study
Experiments conducted over the past three decades have delineated the molecular mechanism of Hp/HS biosynthesis, which could be thought off primarily in terms of N-deacetylation/sulfation, C5-epimerization, 2-O-sulfation, 6-O-sulfation and 3-O-sulfation [18], [19], [20], [21]. Compilation of a large body of information have led to the conclusion that N-sulfation of GlcN is required for C5-epimerization; O-sulfation (but not N-sulfation) inhibits C5-epimerization; 2-O sulfation of IdoA residues occurs before 6-O-sulfation; and 3-O-sulfation of GlcN is dependent upon adjacent UA residues [7], [22]. Interestingly, nearly all biosynthetic enzymes, except for C5-epimerase and 2OST, exist in nature in multiple isoforms, which are likely to influence distribution of sequences being biosynthesized. For example, there are seven isoforms of 3OSTs, including 3OST-1, −2, −3a, −3b, −4, −5, and −6, which catalyze the conversion of a 3-OH group of an appropriate GlcN residue to its 3-O-sulfated form in vertebrates [7]. Considering that 3-O-sulfation is a rare modification, it is interesting that nature has engineered a large number of isoforms. Alternatively, it is highly probable that each of these isoforms leads to structural motifs that are important for selective biological functions, as exemplified by 3OST-1 generating the ABM for anticoagulant function. Thus, we reasoned that among all the biosynthetic enzymes, computational analysis of 3OST-1 recognition of precursor sequences should be undertaken first to not only understand substrate specificity but avail of the opportunity of pinpointing directions for rational design of better recombinant heparins.
3.2. Rationale for using CVLS as a tool to understand substrate specificity
For computational studies, we utilized the CVLS algorithm [11], [12], [13], [16], [17], [23], which we hypothesized would be particularly suitable for understanding substrate specificity of enzymes. The genetic algorithm-based CVLS strategy attempts to emphasize ‘consistency of binding’ of each sequence, via the RMSD parameter, to offer a comprehensive view of how a library of unique sequences would recognize the ligand binding site of a protein [13]. Because highly negatively charged sequences, such as the precursor oligosaccharides, tend to rely more on electrostatics, a majority bind in a promiscuous manner [24]. Such promiscuity would not allow the 0th residue, i.e., the GlcN unit to be 3-O-sulfated by 3OST-1, to bind in an optimal orientation resulting in non-productive binding. Only those precursor sequences that bind consistently, i.e., display low RMSD; with high enough affinity, i.e., display good interaction score, and bind an optimal orientation in the active site, i.e., display optimal ensemble, would be potentially 3-O-sulfated (see Fig. 3). We hypothesized that these three filters would help parse precursor sequences that serve as optimal substrates of 3OST-1 from the majority that bind in a non-productive manner.
3.3. Validation of docking protocol for 3OST-1
To assess whether CVLS could be implemented for studying 3OST-1 recognition of its potential substrates, we first performed re-docking of the Hp/HS sequence present in the crystal structure 3OST-1–heptasaccharide co-complex. For this, the coordinates of 3OST-1 were extracted from the PDB (ID: 3UAN) [8], the oligosaccharide was prepared for molecular docking in terms of ring puckering, torsional angles, atomic charges, etc. [13]. GOLD-based docking, wherein the inter-glycosidic bonds were constrained within ±30° of the average and the substituents accorded full flexibility, as described in our earlier works [13], led to several very similar poses as the highest scoring geometries. The all-atom RMSD between the docked poses and the native crystallographic heptasaccharide was found to be 0.83 Å (Fig. 4). An RMSD of less than 2.5 Å is typically considered as indicative of equivalence [25], which implies that the observed RMSD between docked poses and the crystal structure conveys high predictability of the molecular modeling protocol. The results indicated that the GOLD-based docking protocol and parameters could be used for CVLS studies.
Fig. 4.
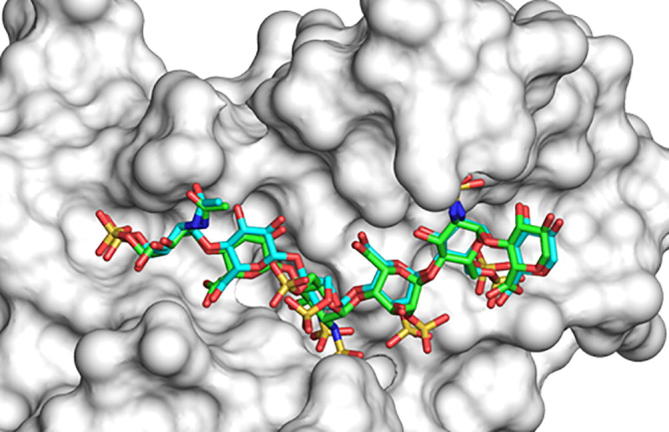
Comparison of GOLD-based modeled native heptasaccharide (cyan) docked onto 3OST-1 with the geometry of the same sequence found in the co-crystal structure of 3OST-1 (green). An RMSD of 0.83 Å was calculated between the docked poses and the crystal structure pose. Note: The crystal structure displays only the hexasaccharide part of the heptasaccharide. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
3.4. CVLS study of libraries of precursor tetrasaccharides
We designed two libraries of precursor tetrasaccharides based on the known substrate specificity of 2OST, the enzyme that has only one known isoform and contributes to biosynthesis 2-O-sulfated sequences present in Hp/HS [18], [26], [27]. This enzyme preferentially introduces a 2-O-sulfate group onto IdoA residues to yield IdoA2S. In contrast, a GlcA residue is very ineffectively converted to GlcA2S residue [19]. This preference of 2OST for IdoA, but not GlcA, results in GlcA2S occurring rarely in HS chains. However, in the presence of excessive sulfate donor (PAPS), 2OST can generate GlcA2S, as exemplified by published works [7], [28]. We also reasoned that GlcN that is unsubstituted at the 2-position is extremely rare and unlikely to contribute to generation of the ABM. Hence, we designed two tetrasaccharide libraries that are devoid of both GlcA2S and GlcNH2 residues. The two in silico libraries were generated by having either GlcN or UA at the NRE with GlcA/IdoA/IdoA2S and GlcNS/GlcNS6S/GlcNAc/GlcNAc6S residues (see Fig. 3).
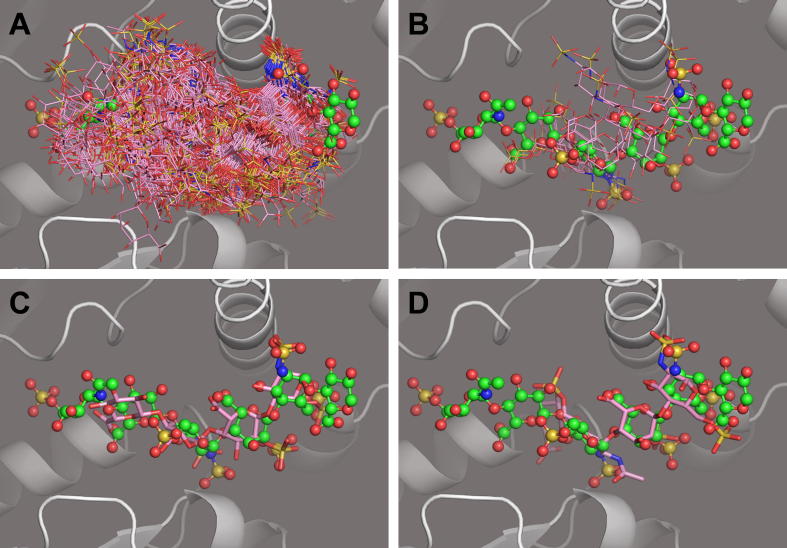
Application of CVLS to each of the 400 unique sequences from the tetrasaccharide libraries gave very interesting results. The vast majority of sequences from both libraries failed to satisfy either of the first two filters and interacted in essentially random orientation (Fig. 5A). Only five sequences passed both ‘selectivity’ filter 1 and ‘affinity’ filter 2 (Fig. 5B, Table 1); however majority of these bound in an orientation that did not allow for productive binding, as indicated by the oligosaccharide present in the crystal structure. In contrast, sequence GlcA-GlcNS6S-IdoA2S-GlcNS6S passed all three filters with a binding pose comparable to the crystal structure (Fig. 5C, Table 1).
Fig. 5.
CVLS analysis of library of tetrasaccharides binding to 3OST-1 (grey ribbons). A) Overlay of the best predicted poses of best 100 tetrasaccharide sequences (colored sticks). B) The five sequences that satisfied the first two filters of the CVLS approach but did not satisfy filter 3, the major binding ensemble. For comparison, the hexasaccharide sequence (green ball and stick) of the crystal structure is shown. C) The predicted binding pose of the only sequence [GlcA–GlcNS6S–IdoA2S–GlcNS6S; pink sticks] that satisfied all three filters and its comparison with the oligosaccharide sequence [GlcNAc6S–GlcA–GlcNS6S–IdoA2S–GlcNS6S–GlcA; green ball and sticks] of the crystal structure. D) The predicted binding pose of the tetrasaccharide carrying GlcNAc6S residue at the ‘0’th position (pink sticks) is similar to that of the hexasaccharide in the crystal structure, but this sequence did not pass filter1 of CVLS. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
Table 1.
CVLS analysis of most promising tetrasaccharide sequences binding into the active site of 3OST-1.
| Sequence | GOLD Score (IdoA form1) | CVLS Results |
|||
|---|---|---|---|---|---|
| Filter 1 (Selectivity) | Filter 2 (Affinity) | Filter 3 (Binding ensemble) | |||
| 1 | IdoA-GlcNS6S-GlcA-GlcNS6S | 102.6 (1C4) | |||
| 2 | IdoA2S-GlcNS6S-GlcA-GlcNS6S | 94.4 (1C4) & 106.8 (2SO) | |||
| 3 | IdoA2S-GlcNS6S-IdoA-GlcNS6S | 83.9 (1C4 and 2SO) & 98.5 (2SO and 2SO) | |||
| 4 | IdoA2S-GlcNS6S-IdoA2S-GlcNS6S | 87.1 (2SO and 1C4) | |||
| 5 | GlcA-GlcNS6S-IdoA2S-GlcNS6S | 102.6 (1C4) & 103.0 (2SO) | |||
| 6 | GlcA-GlcNAc6S-IdoA2S-GlcNS6S | 71.0 (1C4) & 75.4 (2SO) | |||
*Forms (1C4 or 2SO) are listed for IdoA at the NRE;  – passed filter;
– passed filter;  – failed to pass filter.
– failed to pass filter.
Interestingly, sequence GlcA-GlcNAc6S-IdoA2S-GlcNS6S, which has an acetyl group instead of the sulfate group in the 0th residue, did not satisfy the ‘affinity’ filter (Fig. 5D, Table 1) although it was generally predicted to bind in an orientation that matched the crystal structure. A closer inspection of the four sequences that do not satisfy filter 3 shows that variation in the −1 and +1 positions, especially the former (IdoA residue), induces non-productive binding. In combination, 3OST-1 selectivity appears to arise primarily from GlcA and GlcNS6S residues in the −1 and 0 positions of the tetrasaccharide. In other words, selectivity characteristics for the generation of the highly specific ABM are being induced at the tetrasaccharide level.
3.5. CVLS Study of the library of precursor hexasaccharides
Of the two precursor libraries possible to study, i.e., GlcNNRE- or UANRE- hexasaccharides, we studied only one because either would accommodate the five residue ABM. As for the case of tetrasaccharide libraries, we did not include GlcA2S and GlcNH2 residues in the library. Thus, the hexasaccharide library prepared with GlcA/IdoA/IdoA2S and GlcNS/GlcNS6S/GlcNAc/GlcNAc6S residues contained 1000 distinct sequences (Fig. 3).
CVLS analysis offered some very interesting insight into 3OST-1 recognition. First, sequences devoid of sulfation at the 6- and 2-positions, i.e., having GlcNS–GlcA/IdoA–GlcNS–GlcA/IdoA–GlcNS–GlcA/IdoA backbone, did not pass the ‘specificity’ filter. Second, sequences devoid of sulfation at the 6-position, but carrying the 2-sulfate, displayed selective binding depending upon the location of the sulfate group. For example, GlcNS–GlcA–GlcNS–IdoA2S–GlcNS–GlcA/IdoA2S and GlcNS–IdoA2S–GlcNS–IdoA2S–GlcNS–IdoA2S satisfied the ‘specificity/selectivity’ filter. Of these, only the former bound in an orientation similar to the crystal structure and satisfied filter 3 (not shown). Yet, the in silico ‘affinity’ (GOLDScore) of this sequence was not high (∼70 units), which implied reduced preference for 3OST-1. In comparison, the high specificity tetrasaccharide sequences displayed GOLDScore of >100 units. Third, we analyzed sequences containing one or more 6-O-sulfate groups, but devoid of the 2-sulfate group. Herein, only two sequences passed the specificity (RMSD < 2.5 Å), affinity (∼90 GOLDScore) and binding ensemble filters including GlcNS6S–GlcA–GlcNS6S–IdoA–GlcNS6S–UA, where UA is either GlcA or IdoA (Fig. 6). Finally, we analyzed 6-O- and 2-O- sulfated sequences to find that only four sequences satisfied all three filters including GlcNX6S–GlcA–GlcNS6S–IdoA2S–GlcNX6S–UAZ, where X = S or Ac and UAZ = GlcA or IdoA (Fig. 6, Table 2). Interestingly, the GOLDScores of these sequences were higher than any of the other hexasaccharide sequence group (∼100 units or higher; Table 2).
Fig. 6.
CVLS study of precursor hexasaccharides binding to 3OST-1. A) Overlay of predicted poses for GlcNS6S-GlcA-GlcNS6S-IdoA2S-GlcNS6S-IdoA2S, GlcNS6S-GlcA-GlcNS6S-IdoA2S-GlcNS6S-GlcA, and GlcNS6S-GlcA-GlcNS6S-IdoA-GlcNS6S-IdoA, which passed all three filters. B) Overlay of predicted poses for GlcNS6S-GlcA-GlcNS6S-IdoA-GlcNS6S-GlcA, GlcNAc6S-GlcA-GlcNS6S-IdoA2S-GlcNAc6S-IdoA2S, and GlcNAc6S-GlcA-GlcNS6S-IdoA2S-GlcNS6S-GlcA, each of which passed the three filters. Sequences are shown as sticks (pink) and the native crystal structure oligosaccharide is shown as ball and stick (green). C) and D) show hydrogen bond interactions of a representative sequence (pink ball and stick) from panels A) and B), respectively. The representative sequences are shown as ball and stick (pink color by atom); hydrogen bond interacting residues are green sticks, and hydrophobic non-bonded interactions are white sticks. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
Table 2.
HS hexasaccharide sequences identified by CVLS as 3OST-1 specific substrates.
| S. No | Sequences that passed CVLS | GOLD Score | RMSD (in Å) |
|---|---|---|---|
| 1 | GlcNS6S-GlcA-GlcNS6S-IdoA2S(2SO)-GlcNS6S-IdoA2S(1C4) | 106.7 | 1.61 |
| 2 | GlcNS6S-GlcA-GlcNS6S-IdoA2S(2SO)-GlcNS6S-GlcA | 111.8 | 1.52 |
| 3 | GlcNS6S-GlcA-GlcNS6S-IdoA(2SO)-GlcNS6S-IdoA(1C4) | 100.1 | 1.38 |
| 4 | GlcNS6S-GlcA-GlcNS6S-IdoA(2SO)-GlcNS6S-GlcA | 102.0 | 1.37 |
| 5 | GlcNAc6S-GlcA-GlcNS6S-IdoA2S(2SO)-GlcNAc6S-IdoA2S(1C4) | 102.7 | 1.56 |
| 6 | GlcNAc6S-GlcA-GlcNS6S-IdoA2S(2SO)-GlcNS6S-GlcA | 104.8 | 1.17 |
The above results provide some very interesting insight into recognition by 3OST-1 in terms of generation of the ABM. The constant feature of the sequences that pass all the three filters, irrespective of the GOLDScore, is the GlcA-GlcNS6S-IdoA2S microstructure at the −1, 0 and +1 positions, respectively. Of these, the −1 and 0 positions are identical to those deduced from the tetrasaccharide libraries. Another key point is that both GlcNS6S and GlcNAc6S are reasonably well accommodated at the −2 and +2 positions. In terms of the biosynthetic mechanism, 3OST-1 prefers epimerized form of UA at only one of the two positions. Second, 6-O-sulfation appears to be more important in terms of enhancing selectivity and binding affinity of recognition than 2-O-sulfation. In fact, 2-O-sulfation of the IdoA residue is not a strict requirement for 3OST-1 recognition.
3.6. Minimal pharmacophore analysis for generation of antithrombin-binding motif
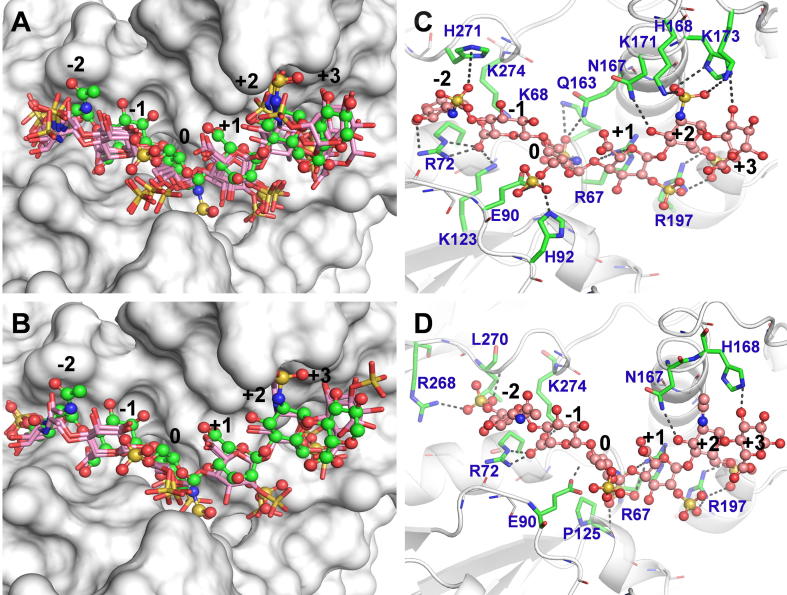
To deduce key interactions between 3OST-1 and the preferred precursor sequences, we performed detailed analysis of each of the predicted co-complexes (Fig. 6). Major interactions that were repeatedly observed included a) the 6-O-sulfate at the −2 position forming a hydrogen bond with His271, b) the carbonyl group at the −1 position forming a Coulombic bond with Arg72, c) the 3-hydroxyl group at the 0 position forming a hydrogen bond with Glu90, d) the 2-hydroxyl or 2-O-sulfate group at the +1 position and the 6-O-sulfate group at the + 2 position forming hydrogen bonds with Arg197, and e) the 2-O-sulfate group at the +2 position forming hydrogen bond with Lys173.
It is important to note that Glu90 is the catalytic site residue, which implies that the predicted co-complexes show 3-OH group of GlcN residue at the 0 position to be well aligned for modification by the 3OST-1. Thus, poses different from this prediction would be non-productive. Also, the enhanced flexibility of the residue at the +3 position, which can either be GlcA or IdoA, results in inconsistent binding interactions. Thus, the conformation of the residue located at +3 position only marginally influenced binding. Finally, several other polar but uncharged residues, e.g., Gln163, Asn167, etc. appear to be involved in precursor sequence recognition. Although it is difficult to ascribe a quantitative role for these polar residues at this time, the literature reports that many GAG–protein systems rely on recognition of polar residues to enhance selectivity [29].
In combination, the pharmacophore for selective recognition of 3OST-1 lies within the pentasaccharide motif. The origin of selectivity appears to be in the −1, 0 and +1 positions, which when present in either a tetrasaccharide or a hexasaccharide generates optimal affinity and orientation for modification at the 3-OH group. Additional pharmacophore elements are likely to enhance affinity of binding in water, thereby enabling biosynthesis at low concentration levels.
4. Conclusions and significance
This work shows for the first time that it is possible to identify elements of selectivity/specificity in recognition of GAG sequences by biosynthetic enzymes, e.g., 3OST-1, using virtual screening analysis. This is highly valuable because generating a large library of homogeneous GAG sequences is difficult [30], [31], which places a major barrier to understanding detailed substrate specificity features of important enzymes such as 3OSTs.
The CVLS algorithm developed in this work consisted of three sequential filters, which advance the application of this technology to elucidating GAG sequences that bind proteins with high selectivity. Following in silico construction of libraries of tetrasaccharide and hexasaccharide sequences carrying differences in type and position of functional groups (sulfate or acetyl), epimerization state (IdoA or GlcA) and length of chain, analysis of consistency of binding (low RMSD), high affinity (GOLDScore) and optimal geometry of binding (the binding ensemble) afforded identification of structural components that engineer selective recognition. Together, the three filters present a mechanism to understand efficiency of catalysis, i.e., kCAT/KM. Whereas RMSD represents a surrogate for the intrinsic catalysis rate kCAT, GOLDScore attempts to reflect KM. Either of these two terms, i.e., RMSD and GOLDScore, would not carry much significance, if the preferred pose of binding was non-productive. Alternatively, only those poses that can lead to product formation (i.e., 3-OH → 3-OSO3− at the 0 position) can lead to meaningful results on selective recognition by 3OST-1.
Both the precursor tetrasaccharide and hexasaccharide sequences led to the conclusion that the vast majority of sequences are not recognized well by 3OST-1. Although not rigorously demonstrated earlier, this result was anticipated because 3OSTs are known to generate rare sequences, which could arise primarily from good selectivity of substrate recognition. Yet, only a very few sequences of the several possible precursor substrates are recognized by 3OST-1 suggests that this enzyme demonstrates a much higher level of selectivity. This also implies that other likely sequences not sulfated by 3OST-1 are possible substrates for other 3OSTs, thus confirming a divergent recognition and 3-O-sulfate motif generation phenomenon.
The encoding of the rare ABM in heparin is ascertained by 3OST-1, which is the final enzyme of the biosynthetic pathway. Our results suggest that the structural elements that govern tight antithrombin binding [6], [32] are essentially identical to those necessary for 3OST-1, except for the presence of 3-sulfate group at the 0th position. Thus, the CVLS studies predict an extremely interesting one-to-one correspondence between the pharmacophores of antithrombin and 3OST-1. In turn, each occurrence of sequences shown in Table 2 in a precursor chain is highly likely to be converted into ABM by 3OST-1. This can have major consequences in terms of biosynthetic expression of ABM-enriched heparin preparation.
GAGs have been presumed to recognize proteins with rather poor selectivity [1], [2], [3], [4]. However, the CVLS technology is proving this to be a false assumption. In addition to antithrombin [13], our CVLS work has shown exquisite recognition of heparin cofactor II by a unique hexasaccharide sequence [23]. Likewise, we have recently shown that GAGs modulate cancer stem cells in highly chain length and structure specific manner [33]. This work shows that 3OST-1 is part of the growing number of proteins that recognize GAGs with a higher level of selectivity.
Although interesting in terms of recognition, the results raise the key question on why the ABM is present at high levels in pig heparin. This is especially intriguing because typically tight selectivity, as for 3OST-1, is expected to reduce the proportion of ABM in polymeric chains. We hypothesize that nature bioengineers the precursor pentasaccharide sequences at elevated levels. This could be brought about by the specificity of prior biosynthetic enzymes, e.g., epimerase, 2OST and 6OSTs. In other words, nature tends to encode higher levels of the 3OST-1 precursor sequences in a preferred manner, rather than utilize 3OST-1 to encode higher levels of ABM. Another possibility is the processive action of HS modification enzymes, which are known to generate non-random distribution of different sequences [34]. Both these hypotheses could be put to test by exhaustive sequencing of Hp/HS. Considering the advances being made today in mass spectrometry-based sequencing of GAGs, the level of ABM and the preferred 3OST-1 substrate sequence(s) should be possible to measure quantitatively in pharmaceutical heparin and/or cell-surface HS, which could help identify the dominant mechanism at work to explain higher levels of ABM in pharmaceutical Hp.
Our CVLS work identifies key residues for further studies including His271, Arg72, Arg197 and Lys173, which interact with 6-sulfate, 5-COO¯, 2-/6-sulfates and 2-sulfate at the −2, −1, +1, and +2 positions, respectively. Several polar residues, especially Gln163 and Asn167, were also identified as playing a key role in recognition. Based on crystal structure studies, these residues were known to contribute to precursor binding [8]. Further, at least one site directed mutant (Asn167Ala) has been shown to be defective in catalytic reactivity. Yet, to correlate in silico predictions on consistency and affinity of binding with catalytic efficiency (kCAT and KM, respectively), it would be important to express and study site directed mutants. More importantly, it may be possible to develop better 3OST-1 mutants with enhanced catalytic efficiency for generation of the ABM. We predict that the in vitro recombinant heparin technology, which is currently in developmental stages [35], [36], could benefit greatly by opting for such particularly efficient 3OST-1 mutant(s).
In summary, our work shows that GAG recognition of 3OST-1 is highly selective; there is a one-to-one correspondence between selectivity elements of antithrombin and 3OST-1; and an understanding on why high levels of ABM exist in heparin could be found at the level of other HS biosynthetic or metabolizing enzymes, rather than through 3OST-1. Our work on CVLS study of 3OST-1 should be of particular use to other HS biosynthetic enzymes, e.g., 2OST and 6OST. It is possible that such an analysis could help alter selectivity features so that recombinant heparins could be produced with higher proportion of ABMs.
CRediT authorship contribution statement
Nehru Viji Sankaranarayanan: Methodology, Software, Validation, Investigation, Data curation, Writing - original draft, Writing - review & editing, Visualization, Formal analysis. Yiling Bi: Validation, Investigation, Writing - original draft, Writing - review & editing, Visualization, Formal analysis. Balagurunathan Kuberan: Conceptualization, Writing - review & editing, Supervision. Umesh R. Desai: Conceptualization, Software, Resources, Writing - review & editing, Supervision, Project administration, Funding acquisition.
Acknowledgments
Acknowledgements
We thank the availability of research resources from National Center for Research Resources (S10 RR027411) to VCU. This work was supported in part by grants from the NIH including HL107152 (URD and KB), HL090586 (URD) and CA241951 (URD).
Conflict of interest statement
Authors declare they have no conflicts of interest with the work described herein.
Footnotes
Dedicate this article to the memory of Professor Robert D. Rosenberg.
Commercial unfractionated heparin is known to have an average molecular weight in the range of 12,000 – 18,000 Da and the pentasaccharide is about 1,800 Da, approximately 10 pentasaccharide units are likely to be present in an average chain. Because one in three heparin chains carry the high-affinity pentasaccharide sequence, effectively one such sequence is present every 30 pentasaccharide units.
Supplementary data to this article can be found online at https://doi.org/10.1016/j.csbj.2020.03.008.
Contributor Information
Balagurunathan Kuberan, Email: kuby.balagurunathan@utah.edu.
Umesh R. Desai, Email: urdesai@vcu.edu.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
References
- 1.Gandhi N.S., Mancera R.L. The structure of glycosaminoglycans and their interactions with proteins. Chem Biol Drug Des. 2008;72:455–482. doi: 10.1111/j.1747-0285.2008.00741.x. [DOI] [PubMed] [Google Scholar]
- 2.Capila I., Linhardt R.J. Heparin-protein interactions. Angew Chem Int Ed Engl. 2002;41:391–412. doi: 10.1002/1521-3773(20020201)41:3<390::aid-anie390>3.0.co;2-b. [DOI] [PubMed] [Google Scholar]
- 3.Xu D., Esko J.D. Demystifying heparan sulfate-protein interactions. Annu Rev Biochem. 2014;83:129–157. doi: 10.1146/annurev-biochem-060713-035314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pomin V.H., Mulloy B. Current structural biology of the heparin interactome. Curr Opin Struct Biol. 2015;34:17–25. doi: 10.1016/j.sbi.2015.05.007. [DOI] [PubMed] [Google Scholar]
- 5.Oduah E.I., Linhardt R.J., Sharfstein S.T. Heparin: past, present, and future. Pharmaceuticals (Basel) 2016;9:38. doi: 10.3390/ph9030038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Desai Umesh R. New antithrombin-based anticoagulants. Med Res Rev. 2004;24(2):151–181. doi: 10.1002/med.10058. [DOI] [PubMed] [Google Scholar]
- 7.Thacker B.E., Xu D., Lawrence R., Esko J.D. Heparan sulfate 3-O-sulfation: a rare modification in search of a function. Matrix Biol. 2014;35:60–72. doi: 10.1016/j.matbio.2013.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Moon A.F., Xu Y., Woody S.M., Krahn J.M., Linhardt R.J., Liu J., Pedersen L.C. Dissecting the substrate recognition of 3-O-sulfotransferase for the biosynthesis of anticoagulant heparin. Proc Natl Acad Sci. 2012;109(14):5265–5270. doi: 10.1073/pnas.1117923109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Edavettal S.C., Lee K.A., Negishi M., Linhardt R.J., Liu J., Pedersen L.C. Crystal structure and mutational analysis of heparan sulfate 3-O-sulfotransferase isoform 1. J Biol Chem. 2004;279:25789–25797. doi: 10.1074/jbc.M401089200. [DOI] [PubMed] [Google Scholar]
- 10.Zhang L., Lawrence R., Schwartz J.J., Bai X., Wei G., Esko J.D. The effect of precursor structures on the action of glucosaminyl 3-O-sulfotransferase-1 and the biosynthesis of anticoagulant heparan sulfate. J Biol Chem. 2001;276:28806–28813. doi: 10.1074/jbc.M100204200. [DOI] [PubMed] [Google Scholar]
- 11.Raghuraman A., Mosier P.D., Desai U.R. Finding a needle in a haystack: development of a combinatorial virtual screening approach for identifying high specificity heparin/heparan sulfate sequence(s) J Med Chem. 2006;49:3553–3562. doi: 10.1021/jm060092o. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Raghuraman A., Mosier P.D., Desai U.R. Understanding dermatan sulfate-heparin cofactor II interaction through virtual library screening. ACS Med Chem lett. 2010;1:281–285. doi: 10.1021/ml100048y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sankaranarayanan N.V., Desai U.R. Toward a robust computational screening strategy for identifying glycosaminoglycan sequences that display high specificity for target proteins. Glycobiology. 2014;24:1323–1333. doi: 10.1093/glycob/cwu077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jones G., Willett P., Glen R.C., Leach A.R., Taylor R. Development and validation of a genetic algorithm for flexible docking. J Mol Biol. 1997;267:727–748. doi: 10.1006/jmbi.1996.0897. [DOI] [PubMed] [Google Scholar]
- 15.Munoz-Garcia J.C., Lopez-Prados J., Angulo J., Diaz-Contreras I., Reichardt N., de Paz J.L. Effect of the substituents of the neighboring ring in the conformational equilibrium of iduronate in heparin-like trisaccharides. Chemistry (Weinheim an der Bergstrasse Germany) 2012;18:16319–16331. doi: 10.1002/chem.201202770. [DOI] [PubMed] [Google Scholar]
- 16.Sankaranarayanan N.V., Sarkar A., Desai U.R., Mosier P.D. Designing “high-affinity, high-specificity” glycosaminoglycan sequences through computerized modeling. Methods Mol Biol (Clifton, NJ) 2015;1229:289–314. doi: 10.1007/978-1-4939-1714-3_24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sankaranarayanan N.V., Nagarajan B., Desai U.R. So you think computational approaches to understanding glycosaminoglycan-protein interactions are too dry and too rigid? Think again! Curr Opin Struct Biol. 2018;50:91–100. doi: 10.1016/j.sbi.2017.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.(a) Petitou M., Casu B., Lindahl U. 1976–1983, a critical period in the history of heparin: the discovery of the antithrombin binding site. Biochemie. 2003;85:83–89. doi: 10.1016/s0300-9084(03)00078-6. [DOI] [PubMed] [Google Scholar]; (b) Lawrence R., Kuberan B., Lech M., Beeler D.L. Rosenberg RD Mapping critical biological motifs and biosynthetic pathways of heparan sulfate. Glycobiology. 2004;14:467–479. doi: 10.1093/glycob/cwh057. [DOI] [PubMed] [Google Scholar]
- 19.Kuberan B., Lech M.Z., Beeler D.L., Wu Z.L., Rosenberg R.D. Enzymatic synthesis of antithrombin III-binding heparan sulfate pentasaccharide. Nat Biotechnol. 2003;21:1343–1346. doi: 10.1038/nbt885. [DOI] [PubMed] [Google Scholar]
- 20.Carlsson P., Kjellén L. Heparin biosynthesis. Handb Exp Pharmacol. 2012;207:23–41. doi: 10.1007/978-3-642-23056-1_2. [DOI] [PubMed] [Google Scholar]
- 21.Li J.P., Kusche-Gullberg M. Heparan sulfate: Biosynthesis, structure, and function. Int Rev Cell Mol Biol. 2016;325:215–273. doi: 10.1016/bs.ircmb.2016.02.009. [DOI] [PubMed] [Google Scholar]
- 22.Nguyen T.K., Arungundram S., Tran V.M., Raman K., Al-Mafraji K., Venot A. A synthetic heparan sulfate oligosaccharide library reveals the novel enzymatic action of D-glucosaminyl 3-O-sulfotransferase-3a. Mol Biosyst. 2012;8:609–614. doi: 10.1039/c1mb05221g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sankaranarayanan N.V., Strebel T.R., Boothello R.S., Sheerin K., Raghuraman A., Sallas F. A hexasaccharide containing rare 2-O-sulfate-glucuronic acid residues selectively activates heparin cofactor II. Angew Chem Int Ed Engl. 2017;56:2312–2317. doi: 10.1002/anie.201609541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mosier P.D., Krishnasamy C., Kellogg G.E., Desai U.R. On the specificity of heparin/heparan sulfate binding to proteins. Anion-binding sites on antithrombin and thrombin are fundamentally different. PLoS One. 2012;7 doi: 10.1371/journal.pone.0048632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Guedes I.A., de Magalhaes C.S., Dardenne L.E. Receptor-ligand molecular docking. Biophys Rev. 2014;6:75–87. doi: 10.1007/s12551-013-0130-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rong J., Habuchi H., Kimata K., Lindahl U., Kusche-Gullberg M. Substrate specificity of the heparan sulfate hexuronic acid 2-O-sulfotransferase. Biochemistry. 2001;40:5548–5555. doi: 10.1021/bi002926p. [DOI] [PubMed] [Google Scholar]
- 27.Merry C.L., Wilson V.A. Role of heparan sulfate-2-O-sulfotransferase in the mouse. Biochim Biophys Acta. 2002;1573:319–327. doi: 10.1016/s0304-4165(02)00399-9. [DOI] [PubMed] [Google Scholar]
- 28.Boothello R.S., Sarkar A., Tran V.M., Nguyen T.K., Sankaranarayanan N.V., Mehta A.Y. Chemoenzymatically prepared heparan sulfate containing rare 2-O-sulfonated glucuronic acid residues. ACS Chem Biol. 2015;10:1485–1494. doi: 10.1021/acschembio.5b00071. [DOI] [PubMed] [Google Scholar]
- 29.Sarkar A., Desai U.R. A simple method for discovering druggable, specific glycosaminoglycan-protein systems. Elucidation of key principles from heparin/heparan sulfate-binding proteins. PLoS One. 2015;10 doi: 10.1371/journal.pone.0141127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhang X., Lin L., Huang H., Linhardt R.J. Chemoenzymatic synthesis of glycosaminoglycans. Acc Chem Res. 2019 doi: 10.1021/acs.accounts.9b00420. [DOI] [PubMed] [Google Scholar]
- 31.Lu W., Zong C., Chopra P., Pepi L.E., Xu Y., Amster I.J. Controlled chemoenzymatic synthesis of heparan sulfate oligosaccharides. Angew Chem Int Ed Engl. 2018;57:5340–5344. doi: 10.1002/anie.201800387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jin L., Abrahams J.P., Skinner R., Petitou M., Pike R.N., Carrell R.W. The anticoagulant activation of antithrombin by heparin. Proc Natl Acad Sci USA. 1997;94:14683–14688. doi: 10.1073/pnas.94.26.14683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Patel N.J., Sharon C., Baranwal S., Boothello R.S., Desai U.R., Patel B.B. Heparan sulfate hexasaccharide selectively inhibits cancer stem cells self-renewal by activating p38 MAP kinase. Oncotarget. 2016;7:84608–84622. doi: 10.18632/oncotarget.12358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Préchoux A., Halimi C., Simorre J.P., Lortat-Jacob H., Laguri C. C5-epimerase and 2-O-sulfotransferase associate in vitro to generate contiguous epimerized and 2-O-sulfated heparan sulfate domains. ACS Chem Biol. 2015;10:1064–1071. doi: 10.1021/cb501037a. [DOI] [PubMed] [Google Scholar]
- 35.Jin P., Zhang L., Yuan P., Kang Z., Du G., Chen J. Efficient biosynthesis of polysaccharides chondroitin and heparosan by metabolically engineered Bacillus subtilis. Carbohydr Polym. 2016;140:424–432. doi: 10.1016/j.carbpol.2015.12.065. [DOI] [PubMed] [Google Scholar]
- 36.Leroux M., Priem B. Chaperone-assisted expression of KfiC glucuronyltransferase from Escherichia coli K5 leads to heparosan production in Escherichia coli BL21 in absence of the stabilisator KfiB. Appl Microbiol Biotechnol. 2016;100:10355–10361. doi: 10.1007/s00253-016-7745-8. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.