Abstract
Infertility is a major health problem across the world. One of the main reasons for male infertility are defects in sperm. Semen analysis is the most common test utilized to evaluate male fertility and since it suffers from multiple drawbacks, reproduction scientists have tried to find new molecular markers for detecting sperm defects. MicroRNAs (miRNAs) are small molecules in cells which take part in regulating gene expression. Various studies have confirmed miRNAs to have a role in defining multiple sperm characteristics, including sperm count, motility, and morphology. In this paper, we have systematically reviewed the role of miRNAs in infertile men with sperm defects including azoospermia, oligospermia, asthenozoospermia, and teratozoospermia. Also, we have assembled various bioinformatics tools to come up with a pipeline for predicting novel miRNAs which could possibly participate in sperm count, motility, and morphology. Also, related KEGG and GO terms for predicted miRNAs have been included in order to highlight their role in sperm function. Our study emphasizes the potential role of miRNAs in male infertility and provides a general overview for future studies aiming to find robust molecular markers for this condition.
Electronic supplementary material
The online version of this article (10.1007/s10815-019-01674-9) contains supplementary material, which is available to authorized users.
Keywords: MicroRNA, Azoospermia, Oligospermia, Asthenozoospermia, Teratozoospermia, Bioinformatics
Introduction
Infertility, defined as the inability to conceive after one year of unprotected intercourse, is a common reproductive health problem which affects almost 15% of couples globally [1]. Male infertility is responsible for 50% of infertility cases, which can be due to acquired or congenital abnormalities. A considerable proportion of male infertility cases are defined as “unexplained male infertility” (UMI). Prevalence of UMI ranges from 6 to 37% and includes infertile men with reduced semen quality without a history of fertility-associated problems and normal physical/hormonal examination results [2]. Currently, diagnosis of UMI is restricted to semen analysis which includes quantitative (azoospermia, oligospermia) and qualitative (asthenozoospermia, teratozoospermia) abnormalities [3]. Semen analysis is the backbone of male infertility evaluation which suffers from multiple drawbacks: (1) normal semen results do not guarantee conception, (2) there is a significant overlap between semen parameters of fertile and infertile men, (3) semen analysis does not provide information about the intracellular functions of sperm [4]. Considering the disadvantages of semen analysis mentioned above, it is of utmost importance to look for new tests and biomarkers in order to diagnose and characterize male infertility.
Spermatogenesis, a complex procedure, contains three main steps: self-renewal and proliferation of spermatogonia, meiotic division of spermatocytes, and postmeiotic differentiation of spermatids into mature spermatozoa [5]. These intricate processes are regulated by the different types of regulatory mechanisms including piwi-interacting RNAs, long noncoding RNAs and microRNAs (miRNAs) [6].
MiRNAs are important regulatory molecules in cells participating in significant cellular processes including embryonic development, cell differentiation, and apoptosis [7, 8]. MiRNAs are none-coding RNAs which consist of 18–24 nucleotides with a single-stranded structure and are generated endogenously in cells [9]. Biosynthesis of miRNAs has been studied widely in a different type of cells. In brief, miRNAs are produced by RNA polymerase II in the nucleus with a hairpin structure called pri-miRNA. This structure is further processed by a member of the RNpAase III enzymes called Drosha resulting in the pre-miRNA. The pre-miRNA is exported to the cytoplasm by exportin-5 and is later processed by Dicer, another member of the RNAase III enzymes, leading to the generation of the mature miRNA. The mature miRNA is loaded to RNA-induced silencing complex (RISC) which represses the expression of target genes by degrading its mRNA or repressing translation [10]. There is a complex interplay between genes and miRNAs, one miRNA can regulate multiple RNAs or one gene can be regulated by multiple miRNAs [11].
Regarding the important role of miRNAs in the cell, dysregulation of miRNAs has been observed in various types of diseases, including infertility [12]. Ostermeier et al. (2004) identified miRNAs in the spermatozoa for the first time [13]. Until now, multiple studies have been conducted to find miRNAs with different expression patterns in infertile males as a new promising biomarker and also elucidate its function in spermatogenesis and fertility; still, many questions remain about the function of miRNAs in sperm cells [14]. In the current study, we have reviewed published research about the role of miRNA in four main sperm abnormality categories including azoospermia, oligospermia, asthenozoospermia, and teratozoospermia. Also, we have used bioinformatics tools to predict novel miRNAs in each category based on reported genes. Results of this study can guide future studies about the role of miRNAs in male infertility.
Methods
Search strategy
The current scoping review was performed according to the PRISMA statement [15]. The following electronic databases were searched in order to identify published articles up to 25 Oct 2019: PubMed, Embase, and Scopus. The search on the google scholar database was conducted to find gray literature. The search strategy for Embase database is shown in the Supplementary Table 1.
Study selection and assessment of studies
Following the aforementioned database search, all resulting papers were loaded into EndNote version X and duplicate studies were removed. Titles and abstracts of the studies were assessed and the full text of the remaining articles were evaluated with the inclusion criteria. The inclusion criteria of this review were studies that analyzed the expression level of miRNAs in reproductive system tissues and fluids (testicular tissue, sperm cells, and seminal plasma and its exosomes) in patients with sperm characteristics defects including azoospermia, asthenozoospermia, oligospermia, and teratospermia.
Data extraction
The required data were extracted using a self-constructed data extraction table. Author and publication year, origin, age and gender of the patients, sperm characteristics of patients, and identified miRNAs were extracted from the studies. The extracted data included identified miRNAs with different expression levels in infertile men with sperm defects.
Bioinformatics tools
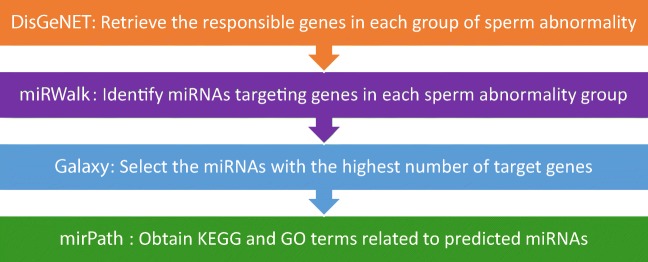
In this study, an important and useful database, DisGeNET (http://www.disgenet.org/), a database of gene-disease relations, has been used to retrieve the responsible genes in each group of sperm abnormality [16]. Reported genes in each group were used for predicting miRNAs that can potentially have an effect on the phenotype. The miRWalk database (http://www.ma.uni-heidelberg.de/apps/zmf/mirwalk/), an open-source database containing both predicted and validated miRNA, was employed to identify miRNAs targeting genes in each sperm abnormality group. Also, other databases in miRWalk including miRDB, PITA, MicroT4, miRMap, RNA22, miRanda, miRNAMap, RNAhybrid, miRBridge, PICTAR2, and Targetscan were used [17]. Galaxy (http://usegalaxy.org) was used to find and select the miRNAs with the highest number of interactions [18].
Galaxy is used to calculate the number of genes targeted by each predicted miRNA. Then miRNAs with the highest number of target genes were chosen and given to mirPath v.3 to obtain the Kyoto Encyclopedia of Genes and Genomes (KEGG) and gene ontology (GO) terms related to the predicted miRNAs (P value < 0.05) [19]. mirPath v.3 can only process 100 gene/miRNAs at a time, so based on this restriction, we chose the top five miRNAs with the highest number of target genes in each category for analysis (miRNAs with the same number of the target gene were included simultaneously). So, in azoospermia (7 miRNAs and its target genes), OS (9 miRNAs and its target genes), AZS (67 miRNAs and its target genes), and TS (100 miRNAs and its target genes) were included in miRPath v.3, respectively.
Consequently, in order to visualize miRNAs and their target genes, Cytoscape (https://cytoscape.org/) was used to visualize the interactions of miRNAs with the highest number of target genes [20]. The pipeline employed in this study has been depicted in Fig. 1.
Fig. 1.
Pipeline employed in this study
Results
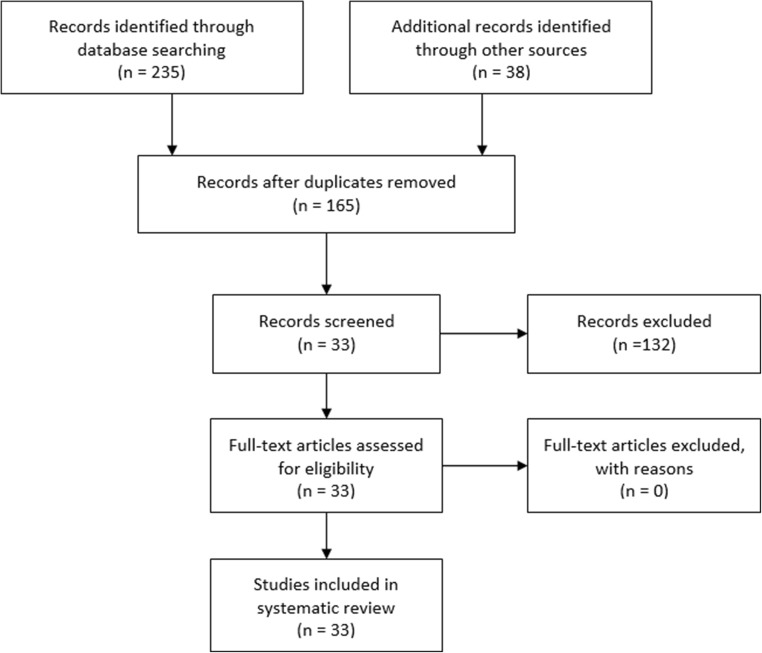
A total of 165 studies were identified by searching PubMed, Emabase, and proquest databases. After removing duplicated articles, 95 studies remained. Titles and abstracts of the studies were evaluated resulting in 42 studies. The full text of the articles based on our inclusion criteria were evaluated and 33 studies remained for our systematic review.
Between included studies, 18 studies were Chinese [21–38], 6 studies were German [39–44], 3 were Iranian [45–47], 3 were Spanish [48–50], and one study was Egyptian [51] and Dutch [52]. Different samples from urogenital tract and gonads were used in studies including testicular tissue [21, 24–28, 31, 39, 40, 45], sperm cell [22, 23, 32, 33, 38, 42, 43, 46, 47, 49–53], semen exosomes [44, 48], seminal plasma [30, 34–37, 41], and samples from different spermatogenesis steps [29]. The detail of the included studies is summarized in Table 1. Also, the flow chart of the study is depicted in Fig. 2.
Table 1.
Details of included studies. NOA = non-obstructive azoospermia, obstructive azoospermia = OA, RT-PCR = real-time PCR, not mentioned = NM, upregulation = ↑, downregulation = ↓
| Author year | Origin | Age | miRNA measurement technique | Sample type | Studied patients | Identified miRNAs/conclusion | REF |
|---|---|---|---|---|---|---|---|
| Lian et al. 2009 | China | 22–30 | Microarray assay validation = RT-PCR | Testicular tissue |
NOA = 3 Control = 2 |
Microarray = 19 ↑–154↓ Confirmed with RT-PCR= hsa-hsa-miR-302a, hsa-miR-491-3p, hsa-miR-520d-3p, hsa-miR-383 |
[21] |
| Wu et al. 2013 | China |
Case = 28.20 ± 4.02 Controls = 29.25 ± 3.03 |
TaqMan low density array validation = qRT–PCR | Sperm cell |
NOA = 100 Control = 100 |
Hsa-miR-141↑, hsa-miR-429↑, hsa-miR-7-1-3p↑ | [22] |
| Khosravi Mashizy et al. 2019 | Iran | NM | RT-PCR | Testicular tissue |
NOA = 40 Control = 40 |
let7↑ | [45] |
| Abu-Halima et al. 2014 | Germany | 25.7 ± 3.6 | Microarray assay validation = RT-PCR | Testicular tissue (compared with difference in histological pattern) | AZ = 56 | hsa-mir-34b*, hsa-mir-34b, hsa-mir-34c-5p, hsa-mir-449a, and hsa-mir-449b | [39] |
| Abu-Halima et al. 2014 | Germany | NM | RT-PCR | Testicular tissue | NOA = 40Control = 16 |
(hsa-miR-429) ↑ (hsa-miR-34b*, hsa-miR-34b, hsa-miR-34c-5p, hsa-miR-122) ↓ |
[40] |
| Zhou et al. 2019 | China |
Case = 26.40 ± 3.98 Control = 28.20 ± 3.56 |
RT-PCR | Sperm cell |
AZ = 30 Control = 30 |
Hsa-miR-525-3p ↓ | [23] |
| Tang et al. 2016 | China | 18–41 | RT-PCR | Testicular tissue |
NOA = 19 Control (OA) = 6 |
Hsa-miR-210 ↑ | [24] |
| Zhang et al. 2019 | China |
Validation group: NOA = 31.000 ± 4.621 Control = 32.531 ± 5.858 |
Microarray assay validation = RT-PCR | Testicular tissue |
Validation group: NOA = 32 Control = 32 |
Hsa- miR-10b-3p ↑ Hsa- miR-34b-5p ↓ |
[25] |
| Fang et al. 2018 | China | 22–45 | Microarray assay validation = RT-PCR | Testicular tissue |
SSR = 5 USR = 5 (All were from NOA patients) |
miRNAs related to sperm function in both groups were identified. | [26] |
| Tang et al. 2018 | China |
post-cryptorchidopexy NOA = 23, 26, 28 post-cryptorchidopexy OA = 24, 25, 36 |
Next-generation sequencing (NGS) Validation = RT-PCR |
Testicular tissue |
Post-cryptorchidopexy NOA = 3 post-cryptorchidopexy OA = 3 |
152 miRNAs ↑ 297 miRNAs ↓ |
[27] |
| Barceló et al. 2018 | Spain | 21–45 |
Real-time PCR profiling Validation = RT-qPCR |
Semen exosomes |
AZ (spermatogenic failure) = 14 OA = 13 Control = 9 |
hsa-miR-539-5p and hsa-miR-941 | [48] |
| Song et al. 2016 | China | NM | RT-PCR | Testicular tissue |
AZ = 16 Control = 8 |
Hsa-miR-188-3p | [28] |
| Yao et al. 2017 | China | 25–40 |
RNA deep sequencing Validation = RT-qPCR |
Human spermatogonia, pachytene spermatocytes, round spermatids |
OA = 20 NOA = 60 |
378 microRNAs were differentially expressed in human round spermatids between OA patients and NOA patients | [29] |
| Mostafa et al. 2016 | Egypt | 23–39 | RT-PCR | Sperm cell |
Normozoospermic = 52 Fertile normozoospermic Men with Vx = 43 Infertile OAT men without Vx = 62 Infertile OAT men with Vx = 63 |
hsa-miRNA-122, hsa-miRNA-181a, and hsa-miRNA-34c5 were positively correlated with sperm concentration, sperm motility, sperm morphology |
[51] |
| Radtke et al. 2019 | Germany |
Case: 29.5 (11.5) Control: 36.5 (8.25) Median (IQR) |
RT-PCR | Seminal plasma |
Patient = 38 Control = 30 |
hsa-miR-371-a-3p is correlated with sperm concentration | [41] |
| Wu et al. 2012 | China | 25.42–33.41 | RT-PCR | Seminal plasma |
NOA = 96 OS = 96 Control = 96 |
hsa-miR-19b let-7a ↑ | [30] |
| Boellaard et al. 2019 | Netherlands | NM | RT-PCR | Sperm cell | Different samples from urogenital tract | hsa-miR-371a-3p related to sperm concentration | [52] |
| Abu-Halima et al. 2012 | Germany |
N = 32.78 ± 5.63 AZS = 34.67 ± 7.87 OA = 31.33 ± 3.94 |
Microarray assay Validation = RT-PCR |
Sperm cell |
N = 9 AZS = 9 OA = 9 |
AS = hsa-miR-34b, hsa-miR-122, hsa-miR-1973 OA = hsa-miR-34b, hsa-miR-34b*, hsa-miR-15b, hsa-miR-34c-5p, hsa-miR-122, hsa-miR-449a, hsa-miR-1973, hsa-miR-16, and hsa-miR-19a |
[42] |
| Abhari et al. 2014 | Iran | 27.5 ± 4.8 | RT-PCR | Sperm cell |
OS = 43 Control = 43 |
hsa-miR-21, hsa-miR-22 ↑ | [46] |
| Li et al. 2016 | China |
OS = 23–37 OA = 25–28 |
Microarray assay Validation = RT-PCR |
Testicular tissue |
OS = 3 OA = 3 |
33 DE-hsa-miRNAs | [31] |
| Abu-Halima et al. 2019 | Germany | 20–35 |
RT-PCR Validation = RT-PCR |
Sperm cell |
OAS = 33 Control = 33 |
hsa-miR-23a/b-3p ↑ | [43] |
| Tian et al. 2017 | China | 20–45 | RT-PCR | Sperm cell |
AZS = 36 OS = 36 AZ = 36 OSAS = 36 |
hsa-miR-10a, hsa-miR-10b, hsa-miR-135a, hsa-miR-135b, hsa-miR-888, hsa-miR-891a ↓ | [32] |
| Abu-Halima et al. 2016 | Germany | 24–39 |
RT-PCR Validation = RT-PCR |
Semen exosomes |
OAS = 12 Control = 12 |
hsa-miR-765 and hsa-miR-1275 ↑ hsa-miR-15a ↓ |
[44] |
| Zhou et al. 2015 | China |
N = 29.00 ± 4.804 AZS = 30.13 ± 5.841 |
RT-PCR | Sperm cell | ASZ = 48Control = 42 | hsa-miR-27b ↓ | [33] |
| Zhou et al. 2015 | China | NM | RT-PCR | Seminal plasma |
AZS = 50 Control = 50 |
hsa-miR-101-3p, let-7b-5p ↓ sp-miR-151a-5p ↑ |
[34] |
| Wang et al. 2011 | China |
AZ = 29.07 ± 4.67 AZS = 29.41 ± 4.35 OS = 28.82 ± 5.31 Control = 28.63 ± 4.40 |
RT-PCR | Seminal plasma |
AZ = 73 AZS = 79 OS = 34 Control = 68 |
AZ = hsa-miR-34c-5p, hsa-miR-122, hsa-miR-146b-5p, hsa-miR-181a, hsa-miR-374b, hsa-miR-509–5p, and hsa-miR-513a-5p ↓ AZS = hsa-miR-34c-5p, hsa-miR-122, hsa-miR-146b-5p, hsa-miR-181a, hsa-miR-374b, hsa-miR-509–5p, and hsa-miR-513a-5p ↑ |
[35] |
| Qing et al. 2017 | China |
AZS = 33.73 ± 6.63 N = 33.00 ± 7.22 |
RT-PCR | Seminal plasma |
AZS = 22 N = 22 |
hsa-miR-891b, hsa-miR-892b, hsa-miR-892a, hsa-miR-888, hsa-miR-890 | [36] |
| Zhou et al. 2018 | China | NM |
TaqMan low-density array Validation = RT-PCR |
Seminal plasma |
AZS = 50 N = 50 |
let-7b-5p ↓ | [37] |
| Heidary et al. 2018 | Iran |
AZS = 36.7 ± 6.87 Control = 35.7 ± 5.05 |
Illumina high-throughput sequencing Validation = RT-PCR |
Sperm cell |
AS = 39 Control = 35 |
Hsa-miR-888-3p ↑ | [47] |
| Salas-Huetos et al. 2014 | Spain | NM | TaqMan array | Sperm cell | 10 males with normal seminogram | None of the miRNAs correlated with sperm motility, morphology or concentration | [53] |
| Zhou et al. 2017 | China |
AZS = 33.30 ± 1.46 Control = 29.60 ± 1.05 |
RT-PCR | Sperm cell |
AZS = 20 Control = 20 |
hsa-miR-27a ↑ | [38] |
| Salas-Huetos et al. 2015 | Spain | 19–50 | RT-PCR | Sperm cell |
AZS = 10 TS = 10 OS = 10 |
AZS = 32 DE-miRNAs TS = 19 DE-miRNAs OS = 18 DE-miRNAs |
[49] |
| Corral-Vazquez et al. 2019 | Spain | NM | RT-PCR | Sperm cell |
AZS = 10 TS = 10 OS = 10 |
AZS = hsa-miR-942-5p/hsa-miR-1208 TS = hsa-miR-296-5p/hsa-miR-328-3p OS = hsa-miR-139-5p/hsa-miR-1260a |
[50] |
Fig. 2.
Study flowchart
Azoospermia
Azoospermia (AZ) is defined as the complete absence of sperm in ejaculated semen; it affects 1% of the male population in the world and is responsible for 10–15% of infertility cases. AZ is clinically divided into two categories, obstructive azoospermia (OA) or non-obstructive azoospermia (NOA) [54, 55]. Genetic factors are responsible for 21–29% of infertility cases in men, but still, 12–41% of AZ cases have unknown etiology which most probably relate to unknown genetic factors [56]. Studies have indicated that different types of genetic disorders are responsible for AZ including chromosomal aberrations, monogenic disorders, multifactorial genetic diseases, epigenetics defects, and genetic abnormities at the primordial germ cell [57].
Multiple studies have evaluated the role of miRNAs in azoospermic infertile patients. In 2009, for the first time, expression of miRNAs in a testicular sample of NOA patients compared to fertile control samples evaluated by microarray technology, identified 19 upregulated and 154 downregulated miRNAs [21]. Hsa-miR-141, hsa-miR-429, and hsa-miR-7-1-3p expression levels were significantly increased in 100 NOA sperm samples compared to controls [22]. The expression of let7 and hsa-miR-30 in azoospermia patients showed higher expression of former while later showed no significant changes [45]. Abu-Halima et al. (2014) evaluated the expression level of miRNAs in four different groups of testicular tissue samples of azoospermic patients with histopathological defects. The results indicated different levels of expression in each group which participate in main cellular functions including apoptosis, cell proliferation, and differentiation [39]. Four downregulated miRNAs (hsa-miR-34b*, hsa-miR-34b, hsa-miR-34c-5p, hsa-miR-122) and one upregulated miRNA (hsa-miR-429) was identified in testicular tissue samples of NOA patients [40]. Based on luciferase experiments miR-525-3p targets 3′-untranslated region of SEMG1 gene and lower expression of this miRNA result in higher expression of SEMG1 gene in azoospermia patients [23] Zhuang et al. 2015, on NOA and normozoospermic OA patients, identified differential expression of 93 miRNAs and 4172 mRNAs compared to normal samples. Functional classification of the miRNA/mRNA pairs using bioinformatics tools indicated that the genes with different expression patterns play a role in spermatogenesis, cell meiosis, cell cycle, and development of secondary male sexual charac hsa-miR-210 has been shown to be upregulated in NOA patients which target insulin-like growth factor II (IGF2) [24]. Using microarray technology, Zhang et al. identified 129 miRNAs with different expression levels in testicular tissue samples of NOA patients which were involved in spermatogenesis, cell cycle, and mitotic prometaphase [25]. Microdissection testicular sperm extraction (micro-TESE) is used for NOA patients in order to retrieve sperm cells to be used in intracytoplasmic sperm injection (ICSI). Fang et al. 2018 evaluated the level of miRNAs in testicular tissue samples including successfully retrieved sperm (SRS) and unsuccessfully retrieved sperm (URS) as a new biomarker before micro-TESE. They identified different expression levels of miRNAs between SRS and URS groups which participate in important cellular processes related to spermatogenesis [26]. MiRNAs play a vital role in cryptorchidism in addition to the reproductive disorders mentioned above [27]. Attempts to find miRNAs inside small extracellular vesicles as a non-invasive diagnostic tool revealed different expression patterns of multiple miRNAs with two miRNAs, hsa-miR-539-5p and hsa-miR-941, promising to be useful markers for predicting the presence of spermatozoa in azoospermic patients before testicular biopsy [48]. Further studies in order to reveal the role of miRNAs in sperm cells of azoospermic patients identified downregulation of hsa-miR-188-3p resulting in upregulation of MLH1 and induction of apoptosis in spermatozoa [28]. The expression level of miRNAs in human spermatogonia, pachytene spermatocytes, and round spermatids between NOA and OA patients showed differences between mentioned groups and concluded that miRNA mimics and miRNA inhibitors could be used in order to down- and upregulate miRNAs respectively in order to restore normal spermatogenesis in AZ patients [29].
Oligospermia
Oligospermia (OS) is defined as sperm cell count lower than 15 million/ml in male ejaculate [58]. OS is usually associated with defects in sperm motility and morphology [59]. This categorization based on sperm density has limited diagnostic value because no threshold value has been defined, for instance, 5% of the fertile male are OS or even men with 1–5 million sperm count can have natural conception [60]. Deletions in the Y chromosome and karyotype anomalies have been reported in oligospermic patients, but still, further research is required in order to reach a better understanding of molecular elements playing role in OS [61].
Expression of hsa-miR-122, hsa-miR-181a, and hsa-miR-34c5 is positively correlated with sperm concentration, motility, and morphology [51]. Also, the expression of hsa-miR-371a-3p in seminal plasma is correlated with sperm concentration [41]. hsa-miR-19b and let-7a are reported to have a higher expression level in NOA patients, but no significant difference was observed in OS patients [30]. The analysis on expression level of miR-371a-3p on different urogenital tract tissues indicated that only testis parenchyma and semen samples had detectable expression levels of this miRNAs and moreover, its expression is correlated with sperm concentration [52]. Study of miRNA expression levels using microarrays show upregulation of 50 miRNAs and downregulation of 27 miRNAs in asthenozoospermic males, and also upregulation of 42 miRNAs and downregulation of 44 miRNAs in oligoasthenozoospermic men. Further bioinformatic analysis also indicated that these miRNAs regulate important genes which play a role in spermatogenesis, apoptosis, sperm chromatin compaction, and sperm motility [42]. Estrogen plays an important role in sperm capacitation and fertilization potential and since increased expression levels of hsa-mir-21 and hsa-mir-22 have been associated with lower expression of estrogen receptor beta in oligospermic patients, miRNAs have been indicated to have a regulatory role in male infertility [46]. Another study on the expression levels of miRNAs and mRNAs on testicular tissue samples of severe oligospermia and obstructive azoospermia indicated differential expression of mRNAs and miRNAs. One such miRNA-mRNA pair, namely hsa-miR-34c-3p and PLCXD3 (phosphatidylinositol-specific phospholipase C, X domain containing 3), is of special interest since hsa-miR-34c-3p expression in mouse and human cell lines reduced PLCXD3 levels which is known to regulate cytosolic calcium levels important for male fertility [31]. A study on oligoasthenospermic patients indicated that the expression of miR-23a/b-3p is negatively correlated with sperm motility, morphology, and sperm count and some of its identified direct targets including PFKFB4, HMMR, SPATA6, and TEX15 have fundamental role in sperm function [43]. Another Chinese study has evaluated the expression level of six miRNAs (hsa-miR-10a, hsa-miR-10b, hsa-miR-135a, hsa-miR-135b, hsa-miR-888, and hsa-miR-891a), in different infertile male groups including asthenozoospermia, OS, azoospermia, OS, and asthenozoospermia (OSAS) and reported lower expression levels of the abovementioned miRNAs in all infertile groups compared to fertile men [32]. Moreover, studies about semen or testicular tissue samples and extracellular microvesicle miRNA levels in the seminal plasma of patients with oligoasthenozoospermia have been published reporting 7 upregulated and 29 downregulated miRNAs in oligoasthenozoospermic cases and suggested that these miRNAs participate in spermatogenesis or its related processes [44].
Asthenozoospermia
Males harboring sperm motility defects such as reduced progressive motility or lack of motility are considered to have asthenozoospermia (AZS) [58]. AZS is responsible for almost 20% of male infertility cases and in 60% of the cases AZS is accompanied by OS (oligoasthenozoospermia) or teratozoospermia (oligoasthenoteratozoospermia, asthenoteratozoospermia) [62]. Multiple genetic factors are involved in AZS etiology including chromosomal aberrations, mutations in genes involved in sperm motility, mitochondrial DNA mutations, epigenetic aberration, and miRNAs [63].
Various studies have attempted to show the role of miRNAs in sperm motility defects. CRISP2 (cysteine-rich secretory protein 2) regulates calcium influx through ryanodine receptors and is located in sperm acrosome and tail. Studies on AZS semen samples have shown lower expression of CRISP2 and higher expression of hsa-miR-27b and have indicated the regulatory role of this gene-miRNA pair in sperm motility [33]. Studies on seminal miRNAs in AZS patients showed higher expression of hsa-miR-151a-5p and lower expression of hsa-miR-101-3p and let-7b-5p compared to control samples. Transfecting GC-2 cells with hsa-miR-151a-5p mimics resulted in decreased levels of adenosine triphosphate (ATP) and cytochrome b (Cytb) protein and mRNA [34]. Interestingly, study of Wang et al. identified 7 miRNAs (hsa-miR-34c-5p, hsa-miR-122, hsa-miR-146b-5p, hsa-miR-181a, hsa-miR-374b, hsa-miR-509–5p, and hsa-miR-513a-5p) which was upregulated in AZS patients but downregulated in AZ patients [35]. Expression of five miRNAs (hsa-miR-891b, hsa-miR-892b, hsa-miR-892a, hsa-miR-888, and hsa-miR-890) was dysregulated in ASZ patients which were correlated to sperm motility. Bioinformatics analysis identified these miRNAs to be important in regulating sperm motility-related pathways [36]. Zhoua et al. (2018) showed lower expression of let-7b-5p in sever ASZ compared to control. They also indicated that let-7b-5p acts in glycolysis metabolism by targeting aurora kinase B (AURKB) [37]. High-throughput sequencing on the sperm samples of idiopathic AZS patients revealed 18 miRNAs with altered expression levels which via validation with real-time PCR showed hsa-miR-888-3p to be highly upregulated compared to controls [47].
Teratozoospermia
Teratozoospermia (TS) is defined as having more than 85% morphologically abnormal sperm cell in ejaculate [64]. TS is divided into two subtypes, monomorphic and polymorphic. In monomorphic TS, all sperm cells harbor the same morphological abnormality while in polymorphic type, there are different kinds of morphological abnormalities [65]. Monomorphic type of TS is divided into two main categories: macrozoospermia (sperm cells with large misshaped heads) and globozoospermia (sperm cells with an oval-shaped head) [66]. Studies have demonstrated that various genetic factors play a role in morphologically abnormal sperm cells including aneuploidy, sperm DNA fragmentation, and mutations [67]. One of these genetic factors, as mentioned above, are miRNAs involved in morphogenesis.
Salas-Huetos et al. (2014) studied 736 miRNAs in human spermatozoa but reported no correlation between any of these miRNAs and sperm concentration, motility, and morphology [53]. Study on asthenoteratozoospermic semen samples indicated that the upregulation of hsa-miR-27a results in lower levels of cysteine-rich secretory protein 2 (CRISP2), an important protein for sperm motility [38]. Two studies by using TaqMan quantitative polymerase chain reaction (PCR) identified multiple miRNAs in different sperm characteristics defects, including teratozoospermia. Salas-Huetos et al. identified 32 miRNAs in AZS, 19 miRNAs in TS, and 18 miRNAs in OS group with altered expression which ontological analysis showed these miRNAs may have role in proper sperm cell function [49]. Corral-Vazquez et al. reported that hsa-miR-942-5p/hsa-miR-1208 in asthenozoospermia, hsa-miR-296-5p/hsa-miR-328-3p in teratozoospermia, and hsa-miR-139-5p/hsa-miR-1260a in oligozoospermia had altered expression compared to control samples [50]
In silico prediction of miRNAs in AZ, OS, AS, and TS
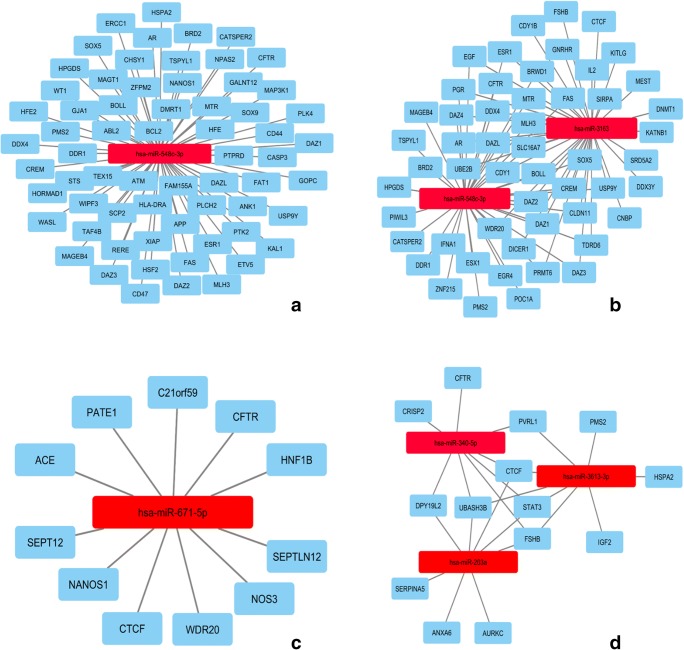
The DisGeNET database reported 173 genes associated with AZ (Supplementary Table 2). Based on these genes, we reached seven predicted miRNAs with the highest number of targeting genes (Supplementary Table 3). Between these miRNAs, hsa-miR-548c-3p had the greatest number of target genes (65 genes). The only reported involvement of this miRNA in human diseases has been in human alcoholic cardiomyopathy [68]. Interactions between this miRNA and its targeted genes are shown in Fig. 3 a.
Fig. 3.
Predicted miRNAs A: AZ, B: OS, C: AZS, D: TS
The DisGeNET database reports 108 genes associated with OS (Supplementary Table 2) and after further bioinformatics analysis, 9 miRNAs with the highest number of targeted genes were selected for further analysis (Supplementary Table 4). Between miRNAs with the highest number of targets, hsa-miR-3163 and hsa-miR-548c-3p had the greatest number of target genes. hsa-miR-548c-3p is shared with the AZS group. hsa-miR-3163 has been reported to be involved in non-small-cell lung cancer cell growth by suppressing the translation of Skp2 to inhibit NSCLC cell growth [69]. The interaction network of the abovementioned miRNAs and their target genes is shown in Fig. 3 b.
The DisGeNET database reports 34 genes associated with AZS (Supplementary Table 2) and further bioinformatic analysis revealed 67 miRNAs with the highest number of targets (Supplementary Table 5). hsa-miR-671-5p had the most target genes among predicted miRNAs in AZS (11 genes). This miRNA has been reported to participate in various cancers including hepatocellular carcinoma, pediatric chordomas, soft tissue sarcomas, glioblastoma multiforme, breast cancer, and epithelioid sarcoma [70–75]. The interaction network of hsa-miR-671-5p and its target genes is shown in Fig. 3 c.
The DisGeNET database reported 18 genes associated with TS (Supplementary Table 2) and 100 miRNAs were chosen between miRNAs with the highest number of targets for further analysis (Supplementary Table 6). Three miRNAs including hsa-miR-203a, hsa-miR-340-5p, and hsa-miR-3613-3p had the most target genes. Previous studies have reported these miRNAs to be important in various types of diseases including rheumatic carditis [76], nasopharyngeal carcinoma [77], influenza A [78], Parkinson’s disease [79], and hepatocellular carcinoma [80]. MiRNA-gene interaction network of the abovementioned miRNAs is shown in Fig. 3 d.
Pathways and GO terms
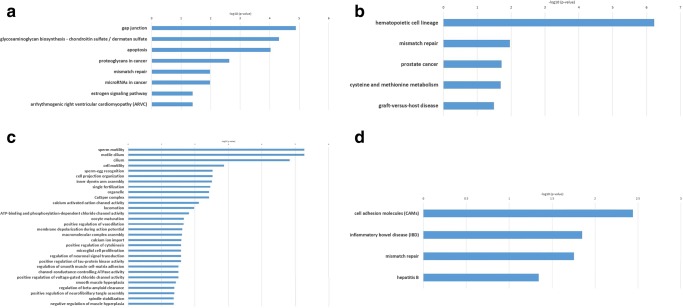
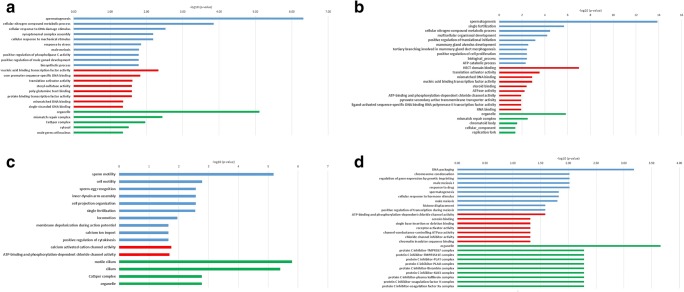
KEGG enrichment analysis of genes and predicted miRNAs involved in AZ results in eight pathways including gap junction, glycosaminoglycan biosynthesis, chondroitin sulfate/dermatan sulfate, apoptosis, proteoglycans in cancer, mismatch repair, miRNAs in cancer, estrogen signaling pathway, and arrhythmogenic right ventricular cardiomyopathy (ARVC) (Fig. 4 a). The most significantly enriched terms on BP, CC, and MF are listed in Fig. 5 a. Most BP terms are involved in sperm cell generation including spermatogenesis, synaptonemal complex assembly, male meiosis, and positive regulation of male gonad development.
Fig. 4.
Kegg pathways A: AZ, B: OS, C: AZS, D: TS
Fig. 5.
GO terms A: AZ, B: OS, C: AZS, D: TS. (blue: biological process, red: molecular function, green: cellular component)
In OS group, KEGG enrichment analysis resulted in five pathways including hematopoietic cell lineage, mismatch repair, prostate cancer, cysteine, and methionine metabolism and graft-versus-host disease (Fig. 4 b). Also, GO enrichment analysis results including BP, MF, and CC terms are depicted in Fig. 5 b.
In AZS group, the first 30 KEGG enrichment results are summarized in Fig. 4 c. The first four pathways are related to sperm motility based on p value score (p < 0.05). Also, multiple pathways are related to calcium and chloride ion transportation which have both been shown to have a fundamental role in sperm motility and male fertility [81, 82]. Similar to KEGG enrichment results, most of the GO enrichment terms are related to sperm motility which indicates predicted miRNAs and their target genes may have important roles in sperm mobility (Fig. 5 c).
In the TS group, KEGG enrichment analysis resulted in four pathways including cell adhesion molecules (CAMs), inflammatory bowel disease (IBD), mismatch repair, and hepatitis B (Fig. 4 d). GO enrichment analysis results are summarized in Fig. 5 d which mostly emphasize spermatogenesis.
Conclusion
Despite numerous efforts to find responsible underlying factors for male infertility, a great proportion of these patients are still reported as unexplained infertility. MiRNAs have been reported as novel biomarkers that can clarify ambiguities about male infertility [31]. Spermatogenesis is regulated by almost 1500 to 2000 genes and genetic alteration of these genes and also their regulatory molecules including miRNAs are emerging research topics for infertility [83, 84]. Understating of responsible molecular changes in infertility is in the utmost importance to find new therapeutic ways and avoid time-consuming and painful treatments [67]. In the current study, we have systematically reviewed the potential role of miRNAs in infertile male with azoospermia, asthenozoospermia, teratozoospermia, and oligospermia. Using different samples, different patient characteristics and inclusion criteria result in high heterogeneity between the included studies, so we were unable to conduct meta-analysis in order to find exact correlation between miRNAs and sperm characteristics. We have also used bioinformatics tools to predict novel miRNAs in each group and their role in biological processes and pathways.
Between the identified pathways, mismatch repair was in common between all groups. Sperm cells undergo multiple stages of mitosis and miosis cycles and DNA repair is essential for maintaining the genomic integrity of the sperm. Defects in components of this system have been reported both in infertile males and in animal models [85]. Predicted miRNAs in this study are involved in regulating this important system, and thus further studies on these miRNAs and their target genes may help clarify new aspects in male infertility.
A number of previously validated miRNAs in male infertility, discussed in previous sections of this review, are among the miRNAs predicted by our pipeline. These include, hsa-mir-141 [22], hsa-miR-34b [40], hsa-miR-34c-5p [40], hsa-miR-122 [40], hsa-miR-429 [40], hsa-miR-942 [48] among the AZ group, Let-7a [30], hsa-miR-21 [46], hsa-miR-22 [46], hsa-miR-34c-3p [31], hsa-miR-135a [32], hsa-miR-135b [32], hsa-miR-888 [32] among OS group and let-7b-5p [34], hsa-miR-891b [36], hsa-miR-892b [36], hsa-miR-892a [36], hsa-miR-888 [36], hsa-miR-890 [66] among the AZS group. This helps show the potential of using our pipeline for future experimental studies. Using bioinformatics tools, novel miRNAs have been predicted in different infertile groups of patients including AZ (hsa-miR-548c-3p), OS (hsa-miR-3163, hsa-miR-548c-3p), AZS (hsa-miR-671-5p), and teratozoospermia (hsa-miR-203a, hsa-miR-340-5p, hsa-miR-3613-3p). Predicted miRNAs of this study are involved in major pathways and GO terms related to sperm function. Regarding the high rate of false-positive results in target prediction algorithms, still more studies are required to validate identified miRNAs [86]. Also, these miRNAs can be further studied experimentally to be used as novel possible biomarkers for infertile male.
Electronic supplementary material
(DOCX 27 kb)
Acknowledgments
This article is the result of a student dissertation approved by Tabriz University of Medical Sciences code 59077. We also want to thank the Stem Cell Research Center of Tabriz University of Medical Sciences for their collaboration.
Compliance with ethical standards
Conflict of interest
The authors declare that they have no conflict of interest.
Footnotes
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.World Health Organization %J WHO G. Report of the meeting on the prevention of infertility at the primary health care level. 1983:12–6.
- 2.Hamada A, Esteves SC, Agarwal A. Unexplained male infertility: potential causes and management. Hum Androl. 2011;1(1):2–16. doi: 10.1097/01.Xha.0000397686.82729.09. [DOI] [Google Scholar]
- 3.Hargreave TB. Genetic basis of male fertility. Br Med Bull. 2000;56(3):650–671. doi: 10.1258/0007142001903454. [DOI] [PubMed] [Google Scholar]
- 4.Hamada A, Esteves SC, Nizza M, Agarwal A. Unexplained male infertility: diagnosis and management. Int Braz J Urol. 2012;38(5):576–594. doi: 10.1590/S1677-55382012000500002. [DOI] [PubMed] [Google Scholar]
- 5.Neto FT, Bach PV, Najari BB, Li PS, Goldstein M. Spermatogenesis in humans and its affecting factors. Semin Cell Dev Biol. 2016;59:10–26. doi: 10.1016/j.semcdb.2016.04.009. [DOI] [PubMed] [Google Scholar]
- 6.Mukherjee A, Koli S, Reddy KV. Regulatory non-coding transcripts in spermatogenesis: shedding light on 'dark matter'. Andrology. 2014;2(3):360–369. doi: 10.1111/j.2047-2927.2014.00183.x. [DOI] [PubMed] [Google Scholar]
- 7.Le Bot N. miRNAs and cell-cycle control in ESCs. Nat Cell Biol. 2012;14:658. doi: 10.1038/ncb2544. [DOI] [Google Scholar]
- 8.Cheng AM, Byrom MW, Shelton J, Ford LP. Antisense inhibition of human miRNAs and indications for an involvement of miRNA in cell growth and apoptosis. Nucleic Acids Res. 2005;33(4):1290–1297. doi: 10.1093/nar/gki200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tahmasbpour E, Balasubramanian D, Agarwal A. A multi-faceted approach to understanding male infertility: gene mutations, molecular defects and assisted reproductive techniques (ART) J Assist Reprod Genet. 2014;31(9):1115–1137. doi: 10.1007/s10815-014-0280-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Harchegani AB, Shafaghatian H, Tahmasbpour E, Shahriary A. Regulatory functions of microRNAs in male reproductive health: a new approach to understanding male infertility. Reprod Sci. 2018;1933719118765972. 10.1177/1933719118765972. [DOI] [PubMed]
- 11.Chen X, Li X, Guo J, Zhang P, Zeng W. The roles of microRNAs in regulation of mammalian spermatogenesis. J Anim Sci Biotechnol. 2017;8:35. doi: 10.1186/s40104-017-0166-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tesfaye D, Salilew-Wondim D, Gebremedhn S, Sohel MM, Pandey HO, Hoelker M, et al. Potential role of microRNAs in mammalian female fertility. Reprod Fertil Dev. 2016;29(1):8–23. doi: 10.1071/rd16266. [DOI] [PubMed] [Google Scholar]
- 13.Ostermeier GC, Goodrich RJ, Moldenhauer JS, Diamond MP, Krawetz SA. A suite of novel human spermatozoal RNAs. J Androl. 2005;26(1):70–74. [PubMed] [Google Scholar]
- 14.Khazaie Y, Nasr Esfahani MH. MicroRNA and male infertility: a potential for diagnosis. Int J Fertil Steril. 2014;8(2):113–118. [PMC free article] [PubMed] [Google Scholar]
- 15.Moher D, Liberati A, Tetzlaff J, Altman DG. Preferred reporting items for systematic reviews and meta-analyses: the PRISMA statement. PLoS Med. 2009;6(7):e1000097. doi: 10.1371/journal.pmed.1000097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Piñero J, Bravo À, Queralt-Rosinach N, Gutiérrez-Sacristán A, Deu-Pons J, Centeno E, García-García J, Sanz F, Furlong LI. DisGeNET: a comprehensive platform integrating information on human disease-associated genes and variants. Nucleic Acids Res. 2016;45(D1):D833–D8D9. doi: 10.1093/nar/gkw943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sticht C, De La Torre C, Parveen A, Gretz N. miRWalk: an online resource for prediction of microRNA binding sites. PLOS ONE. 2018;13(10):e0206239. doi: 10.1371/journal.pone.0206239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Afgan E, Baker D, Batut B, van den Beek M, Bouvier D, Cech M, Chilton J, Clements D, Coraor N, Grüning BA, Guerler A, Hillman-Jackson J, Hiltemann S, Jalili V, Rasche H, Soranzo N, Goecks J, Taylor J, Nekrutenko A, Blankenberg D. The galaxy platform for accessible, reproducible and collaborative biomedical analyses: 2018 update. Nucleic Acids Res. 2018;46(W1):W537–Ww44. doi: 10.1093/nar/gky379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Vlachos IS, Zagganas K, Paraskevopoulou MD, Georgakilas G, Karagkouni D, Vergoulis T, Dalamagas T, Hatzigeorgiou AG. DIANA-miRPath v3.0: deciphering microRNA function with experimental support. Nucleic Acids Res. 2015;43(W1):W460–W4W6. doi: 10.1093/nar/gkv403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D, Amin N, Schwikowski B, Ideker T. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13(11):2498–2504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lian J, Zhang X, Tian H, Liang N, Wang Y, Liang C, Li X, Sun F. Altered microRNA expression in patients with non-obstructive azoospermia. Reprod Biol Endocrinol. 2009;7(1):13. doi: 10.1186/1477-7827-7-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wu W, Qin Y, Li Z, Dong J, Dai J, Lu C, Guo X, Zhao Y, Zhu Y, Zhang W, Hang B, Sha J, Shen H, Xia Y, Hu Z, Wang X. Genome-wide microRNA expression profiling in idiopathic non-obstructive azoospermia: significant up-regulation of miR-141, miR-429 and miR-7-1-3p. Hum Reprod. 2013;28(7):1827–1836. doi: 10.1093/humrep/det099. [DOI] [PubMed] [Google Scholar]
- 23.Zhou QZ, Guo XB, Zhang WS, Zhou JH, Yang C, Bian J, Chen MK, Guo WB, Wang P, Qi T, Wang CY, Yang JK, Liu CD. Expressions of miR-525-3p and its target gene SEMG1 in the spermatozoa of patients with asthenozoospermia. Andrology. 2019;7(2):220–227. doi: 10.1111/andr.12573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tang D, Huang Y, Liu W, Zhang X. Up-regulation of microRNA-210 is associated with spermatogenesis by targeting IGF2 in male infertility. Med Sci Monit. 2016;22:2905–2910. doi: 10.12659/msm.897340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhang HT, Zhang Z, Hong K, Tang WH, Liu DF, Mao JM, et al. Altered microRNA profiles of testicular biopsies from patients with nonobstructive azoospermia. Asian J Androl. 2019. 10.4103/aja.aja_35_19. [DOI] [PMC free article] [PubMed]
- 26.Fang N, Cao C, Wen Y, Wang X, Yuan S, Huang X. MicroRNA profile comparison of testicular tissues derived from successful and unsuccessful microdissection testicular sperm extraction retrieval in non-obstructive azoospermia patients. Reprod Fertil Dev. 2018. 10.1071/rd17423. [DOI] [PubMed]
- 27.Tang D, Huang Z, He X, Wu H, Peng D, Zhang L, Zhang X. Altered miRNA profile in testis of post-cryptorchidopexy patients with non-obstructive azoospermia. Reprod Biol Endocrinol. 2018;16(1):78. doi: 10.1186/s12958-018-0393-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Song WY, Meng H, Wang XG, Jin HX, Yao GD, Shi SL, et al. Reduced microRNA-188-3p expression contributes to apoptosis of spermatogenic cells in patients with azoospermia. Cell Prolif. 2017;50(1). 10.1111/cpr.12297. [DOI] [PMC free article] [PubMed]
- 29.Yao C, Yuan Q, Niu M, Fu H, Zhou F, Zhang W, Wang H, Wen L, Wu L, Li Z, He Z. Distinct expression profiles and novel targets of microRNAs in human spermatogonia, Pachytene spermatocytes, and round spermatids between OA patients and NOA patients. Mol Ther Nucleic Acids. 2017;9:182–194. doi: 10.1016/j.omtn.2017.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wu W, Hu Z, Qin Y, Dong J, Dai J, Lu C, Zhang W, Shen H, Xia Y, Wang X. Seminal plasma microRNAs: potential biomarkers for spermatogenesis status. Mol Hum Reprod. 2012;18(10):489–497. doi: 10.1093/molehr/gas022. [DOI] [PubMed] [Google Scholar]
- 31.Li Z, Zheng Z, Ruan J, Li Z, Zhuang X, Tzeng C-M. Integrated analysis miRNA and mRNA profiling in patients with severe oligozoospermia reveals miR-34c-3p downregulates PLCXD3 expression. Oncotarget. 2016;7(33):52781–52796. doi: 10.18632/oncotarget.10947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tian H, Lv M, Li Z, Peng D, Tan Y, Wang H, Li Q, Li F, Liang W. Semen-specific miRNAs: suitable for the distinction of infertile semen in the body fluid identification? Forensic Sci Int Genet. 2018;33:161–167. doi: 10.1016/j.fsigen.2017.12.010. [DOI] [PubMed] [Google Scholar]
- 33.Zhou JH, Zhou QZ, Lyu XM, Zhu T, Chen ZJ, Chen MK, Xia H, Wang CY, Qi T, Li X, Liu CD. The expression of cysteine-rich secretory protein 2 (CRISP2) and its specific regulator miR-27b in the spermatozoa of patients with asthenozoospermia. Biol Reprod. 2015;92(1):28. doi: 10.1095/biolreprod.114.124487. [DOI] [PubMed] [Google Scholar]
- 34.Zhou R, Wang R, Qin Y, Ji J, Xu M, Wu W, Chen M, Wu D, Song L, Shen H, Sha J, Miao D, Hu Z, Xia Y, Lu C, Wang X. Mitochondria-related miR-151a-5p reduces cellular ATP production by targeting CYTB in asthenozoospermia. Sci Rep. 2015;5:17743. doi: 10.1038/srep17743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wang C, Yang C, Chen X, Yao B, Yang C, Zhu C, Li L, Wang J, Li X, Shao Y, Liu Y, Ji J, Zhang J, Zen K, Zhang CY, Zhang C. Altered profile of seminal plasma microRNAs in the molecular diagnosis of male infertility. Clin Chem. 2011;57(12):1722–1731. doi: 10.1373/clinchem.2011.169714. [DOI] [PubMed] [Google Scholar]
- 36.Qing X, Shi J, Dong T, Wu C, Hu L, Li H. Dysregulation of an X-linked primate-specific epididymal microRNA cluster in unexplained asthenozoospermia. Oncotarget. 2017;8(34):56839–56849. doi: 10.18632/oncotarget.18076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhou R, Zhang Y, Du G, Han L, Zheng S, Liang J, et al. Down-regulated let-7b-5p represses glycolysis metabolism by targeting AURKB in asthenozoospermia. Gene. 2018;663:83–87. doi: 10.1016/j.gene.2018.04.022. [DOI] [PubMed] [Google Scholar]
- 38.Zhou JH, Zhou QZ, Yang JK, Lyu XM, Bian J, Guo WB, Chen ZJ, Xia M, Xia H, Qi T, Li X, Liu CD. MicroRNA-27a-mediated repression of cysteine-rich secretory protein 2 translation in asthenoteratozoospermic patients. Asian J Androl. 2017;19(5):591–595. doi: 10.4103/1008-682x.185001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Abu-Halima M, Backes C, Leidinger P, Keller A, Lubbad AM, Hammadeh M, et al. MicroRNA expression profiles in human testicular tissues of infertile men with different histopathologic patterns. Fertil Steril. 2014;101(1):78–86.e2. doi: 10.1016/j.fertnstert.2013.09.009. [DOI] [PubMed] [Google Scholar]
- 40.Abu-Halima M, Hammadeh M, Backes C, Fischer U, Leidinger P, Lubbad AM, et al. Panel of five microRNAs as potential biomarkers for the diagnosis and assessment of male infertility. Fertil Steril. 2014;102(4):989–97.e1. doi: 10.1016/j.fertnstert.2014.07.001. [DOI] [PubMed] [Google Scholar]
- 41.Radtke A, Dieckmann KP, Grobelny F, Salzbrunn A, Oing C, Schulze W, Belge G. Expression of miRNA-371a-3p in seminal plasma and ejaculate is associated with sperm concentration. Andrology. 2019;7(4):469–474. doi: 10.1111/andr.12664. [DOI] [PubMed] [Google Scholar]
- 42.Abu-Halima M, Hammadeh M, Schmitt J, Leidinger P, Keller A, Meese E, et al. Altered microRNA expression profiles of human spermatozoa in patients with different spermatogenic impairments. Fertil Steril. 2013;99(5):1249–55.e16. doi: 10.1016/j.fertnstert.2012.11.054. [DOI] [PubMed] [Google Scholar]
- 43.Abu-Halima M, Ayesh BM, Hart M, Alles J, Fischer U, Hammadeh M, et al. Differential expression of miR-23a/b-3p and its target genes in male patients with subfertility. Fertil Steril. 2019;112(2):323–35.e2. doi: 10.1016/j.fertnstert.2019.03.025. [DOI] [PubMed] [Google Scholar]
- 44.Abu-Halima M, Ludwig N, Hart M, Leidinger P, Backes C, Keller A, et al. Altered micro-ribonucleic acid expression profiles of extracellular microvesicles in the seminal plasma of patients with oligoasthenozoospermia. Fertil Steril. 2016;106(5):1061–9.e3. doi: 10.1016/j.fertnstert.2016.06.030. [DOI] [PubMed] [Google Scholar]
- 45.Mashizy SKSM, Mokhtari M, Khatamsaz S. The relationship between expression of Mir30 and Let7 genes in infertile males and non-obstructive azoospermia. Biosci EJ. 2019;13(1).
- 46.Abhari A, Zarghami N, Farzadi L, Nouri M, Shahnazi V. Altered of microRNA expression level in oligospermic patients. Iran J Reprod Med. 2014;12(10):681–686. [PMC free article] [PubMed] [Google Scholar]
- 47.Heidary Z, Zaki-Dizaji M, Saliminejad K, Khorram Khorshid HR. MicroRNA profiling in spermatozoa of men with unexplained asthenozoospermia. Andrologia. 2019;51(6):e13284. doi: 10.1111/and.13284. [DOI] [PubMed] [Google Scholar]
- 48.Barcelo M, Mata A, Bassas L, Larriba S. Exosomal microRNAs in seminal plasma are markers of the origin of azoospermia and can predict the presence of sperm in testicular tissue. Hum Reprod. 2018;33(6):1087–1098. doi: 10.1093/humrep/dey072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Salas-Huetos A, Blanco J, Vidal F, Godo A, Grossmann M, Pons MC, et al. Spermatozoa from patients with seminal alterations exhibit a differential micro-ribonucleic acid profile. Fertil Steril. 2015;104(3):591–601. doi: 10.1016/j.fertnstert.2015.06.015. [DOI] [PubMed] [Google Scholar]
- 50.Corral-Vazquez C, Salas-Huetos A, Blanco J, Vidal F, Sarrate Z, Anton E. Sperm microRNA pairs: new perspectives in the search for male fertility biomarkers. Fertil Steril. 2019. 10.1016/j.fertnstert.2019.07.006. [DOI] [PubMed]
- 51.Mostafa T, Rashed LA, Nabil NI, Osman I, Mostafa R, Farag M. Seminal miRNA relationship with apoptotic markers and oxidative stress in infertile men with varicocele. Biomed Res Int. 2016;2016:4302754. doi: 10.1155/2016/4302754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Boellaard WPA, Gillis AJM, van Leenders G, Stoop H, van Agthoven T, Dorssers LCJ, Dinkelman-Smit M, Boormans JL, Looijenga LHJ. Cellular origin of microRNA-371a-3p in healthy males based on systematic urogenital tract tissue evaluation. Andrology. 2019;7(4):463–468. doi: 10.1111/andr.12595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Salas-Huetos A, Blanco J, Vidal F, Mercader JM, Garrido N, Anton E. New insights into the expression profile and function of micro-ribonucleic acid in human spermatozoa. Fertil Steril. 2014;102(1):213–22.e4. doi: 10.1016/j.fertnstert.2014.03.040. [DOI] [PubMed] [Google Scholar]
- 54.Anderson JE, Farr SL, Jamieson DJ, Warner L, Macaluso M. Infertility services reported by men in the United States: national survey data. Fertil Steril. 2009;91(6):2466–2470. doi: 10.1016/j.fertnstert.2008.03.022. [DOI] [PubMed] [Google Scholar]
- 55.Huang IS, Huang WJ, Lin AT. Distinguishing non-obstructive azoospermia from obstructive azoospermia in Taiwanese patients by hormone profile and testis size. J Chin Med Assoc. 2018;81(6):531–535. doi: 10.1016/j.jcma.2017.09.009. [DOI] [PubMed] [Google Scholar]
- 56.Hernandez Uribe L, Hernandez Marin I, Cervera-Aguilar R, Ayala AR. Frequency and etiology of azoospermia in the study of infertile couples. Ginecol Obstet Mex. 2001;69:322–326. [PubMed] [Google Scholar]
- 57.Hamada AJ, Esteves SC, Agarwal A. A comprehensive review of genetics and genetic testing in azoospermia. Clinics (Sao Paulo) 2013;68(Suppl 1):39–60. doi: 10.6061/clinics/2013(Sup01)06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.World Health Organization . World Health Organization laboratory manual for the evaluation and processing of human semen. 5. Cambridge: Cambridge University Press; 2010. [Google Scholar]
- 59.Liu DY, Baker HW. Defective sperm-zona pellucida interaction: a major cause of failure of fertilization in clinical in-vitro fertilization. Hum Reprod. 2000;15(3):702–708. doi: 10.1093/humrep/15.3.702. [DOI] [PubMed] [Google Scholar]
- 60.McLachlan RI. Approach to the patient with oligozoospermia. J Clin Endocrinol Metab. 2013;98(3):873–880. doi: 10.1210/jc.2012-3650. [DOI] [PubMed] [Google Scholar]
- 61.McLachlan RI, O'Bryan MK. Clinical review#: state of the art for genetic testing of infertile men. J Clin Endocrinol Metab. 2010;95(3):1013–1024. doi: 10.1210/jc.2009-1925. [DOI] [PubMed] [Google Scholar]
- 62.Asero P, Calogero AE, Condorelli RA, Mongioi L, Vicari E, Lanzafame F, et al. Relevance of genetic investigation in male infertility. J Endocrinol Investig. 2014;37(5):415–427. doi: 10.1007/s40618-014-0053-1. [DOI] [PubMed] [Google Scholar]
- 63.Heidary Z, Saliminejad K, Zaki-Dizaji M, Khorram Khorshid HR. Genetic aspects of idiopathic asthenozoospermia as a cause of male infertility. Hum Fertil (Camb). 2018:1–10. 10.1080/14647273.2018.1504325. [DOI] [PubMed]
- 64.Perrin A, Huong Nguyen M, Douet-Guilbert N, Morel F, De Braekeleer M. Motile sperm organelle morphology examination: where do we stand 12 years later? 2014. [Google Scholar]
- 65.Perrin A, Morel F, Moy L, Colleu D, Amice V, De Braekeleer M. Study of aneuploidy in large-headed, multiple-tailed spermatozoa: case report and review of the literature. Fertil Steril. 2008;90(4):1201.e13–1201.e17. doi: 10.1016/j.fertnstert.2007.09.013. [DOI] [PubMed] [Google Scholar]
- 66.Perrin A, Coat C, Nguyen MH, Talagas M, Morel F, Amice J, de Braekeleer M. Molecular cytogenetic and genetic aspects of globozoospermia: a review. Andrologia. 2013;45(1):1–9. doi: 10.1111/j.1439-0272.2012.01308.x. [DOI] [PubMed] [Google Scholar]
- 67.De Braekeleer M, Nguyen MH, Morel F, Perrin A. Genetic aspects of monomorphic teratozoospermia: a review. J Assist Reprod Genet. 2015;32(4):615–623. doi: 10.1007/s10815-015-0433-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Jing L, Jin C, Lu Y, Huo P, Zhou L, Wang Y, Tian Y. Investigation of microRNA expression profiles associated with human alcoholic cardiomyopathy. Cardiology. 2015;130(4):223–233. doi: 10.1159/000370028. [DOI] [PubMed] [Google Scholar]
- 69.Su L, Han D, Wu J, Huo X. Skp2 regulates non-small cell lung cancer cell growth by Meg3 and miR-3163. Tumour Biol. 2016;37(3):3925–3931. doi: 10.1007/s13277-015-4151-2. [DOI] [PubMed] [Google Scholar]
- 70.Dou C, Zhou Z, Xu Q, Liu Z, Zeng Y, Wang Y, Li Q, Wang L, Yang W, Liu Q, Tu K. Hypoxia-induced TUFT1 promotes the growth and metastasis of hepatocellular carcinoma by activating the Ca(2+)/PI3K/AKT pathway. Oncogene. 2019;38(8):1239–1255. doi: 10.1038/s41388-018-0505-8. [DOI] [PubMed] [Google Scholar]
- 71.Malgulwar PB, Pathak P, Singh M, Kale SS, Suri V, Sarkar C, Sharma MC. Downregulation of SMARCB1/INI1 expression in pediatric chordomas correlates with upregulation of miR-671-5p and miR-193a-5p expressions. Brain Tumor Pathol. 2017;34(4):155–159. doi: 10.1007/s10014-017-0295-7. [DOI] [PubMed] [Google Scholar]
- 72.Sapi Z, Papp G, Szendroi M, Papai Z, Plotar V, Krausz T, et al. Epigenetic regulation of SMARCB1 by miR-206, -381 and -671-5p is evident in a variety of SMARCB1 immunonegative soft tissue sarcomas, while miR-765 appears specific for epithelioid sarcoma. A miRNA study of 223 soft tissue sarcomas. Genes Chromosome Cancer. 2016;55(10):786–802. doi: 10.1002/gcc.22379. [DOI] [PubMed] [Google Scholar]
- 73.Barbagallo D, Condorelli A, Ragusa M, Salito L, Sammito M, Banelli B, et al. Dysregulated miR-671–5p / CDR1-AS / CDR1 / VSNL1 axis is involved in glioblastoma multiforme. Oncotarget. 2016;7(4):4746–4759. doi: 10.18632/oncotarget.6621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Tan X, Fu Y, Chen L, Lee W, Lai Y, Rezaei K, et al. miR-671-5p inhibits epithelial-to-mesenchymal transition by downregulating FOXM1 expression in breast cancer. Oncotarget. 2016;7(1):293–307. doi: 10.18632/oncotarget.6344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Papp G, Krausz T, Stricker TP, Szendroi M, Sapi Z. SMARCB1 expression in epithelioid sarcoma is regulated by miR-206, miR-381, and miR-671-5p on both mRNA and protein levels. Genes Chromosome Cancer. 2014;53(2):168–176. doi: 10.1002/gcc.22128. [DOI] [PubMed] [Google Scholar]
- 76.Gumus G, Giray D, Bobusoglu O, Tamer L, Karpuz D, Hallioglu O. MicroRNA values in children with rheumatic carditis: a preliminary study. Rheumatol Int. 2018;38(7):1199–1205. doi: 10.1007/s00296-018-4069-2. [DOI] [PubMed] [Google Scholar]
- 77.Li S, Hang L, Ma Y, Wu C. Distinctive microRNA expression in early stage nasopharyngeal carcinoma patients. J Cell Mol Med. 2016;20(12):2259–2268. doi: 10.1111/jcmm.12906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Zhao L, Zhang X, Wu Z, Huang K, Sun X, Chen H, Jin M. The Downregulation of MicroRNA hsa-miR-340-5p in IAV-infected A549 cells suppresses viral replication by targeting RIG-I and OAS2. Mol Ther Nucleic Acids. 2019;14:509–519. doi: 10.1016/j.omtn.2018.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Patil KS, Basak I, Dalen I, Hoedt E, Lange J, Lunde KA, et al. Combinatory microRNA serum signatures as classifiers of Parkinson’s disease. Parkinsonism Relat Disord. 2019. 10.1016/j.parkreldis.2019.04.010. [DOI] [PubMed]
- 80.Zhang D, Liu E, Kang J, Yang X, Liu H. MiR-3613-3p affects cell proliferation and cell cycle in hepatocellular carcinoma. Oncotarget. 2017;8(54):93014–93028. doi: 10.18632/oncotarget.21745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Ren D, Navarro B, Perez G, Jackson AC, Hsu S, Shi Q, Tilly JL, Clapham DE. A sperm ion channel required for sperm motility and male fertility. Nature. 2001;413(6856):603–609. doi: 10.1038/35098027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Liu S-W, Li Y, Zou L-L, Guan Y-T, Peng S, Zheng L-X, Deng SM, Zhu LY, Wang LW, Chen LX. Chloride channels are involved in sperm motility and are downregulated in spermatozoa from patients with asthenozoospermia. Asian J Androl. 2017;19(4):418–424. doi: 10.4103/1008-682X.181816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Matzuk MM, Lamb DJ. The biology of infertility: research advances and clinical challenges. Nat Med. 2008;14(11):1197–1213. doi: 10.1038/nm.f.1895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Papaioannou MD, Nef S. microRNAs in the testis: building up male fertility. J Androl. 2010;31(1):26–33. doi: 10.2164/jandrol.109.008128. [DOI] [PubMed] [Google Scholar]
- 85.Mukherjee S, Ridgeway AD, Lamb DJ. DNA mismatch repair and infertility. Curr Opin Urol. 2010;20(6):525–532. doi: 10.1097/MOU.0b013e32833f1c21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Pinzón N, Li B, Martinez L, Sergeeva A, Presumey J, Apparailly F, Seitz H. microRNA target prediction programs predict many false positives. Genome Res. 2017;27(2):234–245. doi: 10.1101/gr.205146.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(DOCX 27 kb)