Abstract
Abuse of alcohol and tobacco could exacerbate HIV pathogenesis by transferring materials through exosomes (small nanovesicles). Exosomes present a stable and accessible source of information concerning the health and/or disease status of patients, which can provide diagnostic and prognostic biomarkers for myriad conditions. Therefore, we aimed to study the specific exosomal proteins, which are altered in both HIV-infected subjects and alcohol/tobacco users. Exosomes were isolated from plasma of the following subjects: a) HIV-negative subjects (healthy), b) HIV-positive subjects (HIV), c) HIV-negative alcohol drinkers (drinkers), d) HIV-negative tobacco smokers (smokers), e) HIV-positive drinkers (HIV+drinkers), and f) HIV-positive smokers (HIV+smokers). Quantitative proteomic profiling was then performed from these exosomes. Sixteen proteins were significantly altered in the HIV group, ten in drinkers, four in HIV+drinkers, and fifteen in smokers compared to healthy subjects. Only one protein, fibulin-1 (FBLN1), was significantly altered in HIV+smokers. Interestingly, hemopexin was not significantly altered in drinkers or HIV patients but was significantly altered in HIV+drinkers. Further, our study is the first to show properdin expression in plasma exosomes, which was decreased in HIV+smokers and HIV+drinkers compared to HIV patients. The present findings suggest that hemopexin and properdin show potential as markers for physiological effects that may arise in HIV-infected individuals who abuse alcohol and tobacco.
Keywords: HIV, plasma exosome, alcohol, tobacco, proteomics
Graphical Abstract
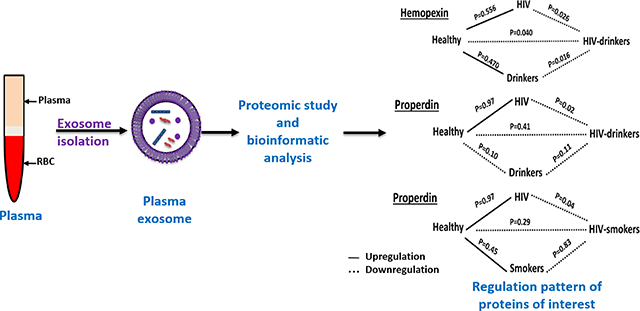
This study presents a proteomic analysis of plasma-derived exosomes from HIV-infected alcohol drinkers and smokers. Among the proteins altered due to drug-abuse, hemopexin and properdin were of highest significance. These proteins can be potential biomarkers for co-morbid conditions associated with drug abuse in HIV-patients.
Introduction
Despite vast improvements in diagnosis, monitoring, and therapy in recent years, human immunodeficiency virus (HIV) remains a significant public health concern. According to the CDC, more than one million Americans are currently HIV-positive, with approximately 40,000 new cases being diagnosed annually as of 2016 (HIV Surveillance Report 2016). As such, the HIV-positive population represents a significant patient pool with specific and unique healthcare needs, owing both to the presence of antiretroviral therapy (ART) and to comorbidities that may have deleterious interactions with the virus, or with ART.
Substance abuse, particularly of alcohol and tobacco, is a common secondary health issue for HIV patients. HIV-positive individuals are nearly twice as likely to regularly consume alcohol as the healthy population (Galvan et al. 2002). Alcohol abuse is well established to worsen outcomes in HIV therapy by reducing ART adherence (Hicks et al. 2007) and contributing to ART-induced hepatotoxicity (Barve et al. 2010). The metabolism of alcohol can also cause oxidative stress, damaging cells and promoting HIV replication (Kumar et al. 2012; Ande et al. 2015b). These factors combine to make alcohol abuse a significant contributor to HIV pathogenesis and transmission, and to ART failure (Pandrea et al. 2010).
Approximately 50–70% of HIV patients smoke tobacco, compared to 19% in the general population (Cioe 2013). Tobacco smoking also greatly impacts health and treatment outcomes in the HIV-positive population. HIV patients who smoke are at greater risk of developing non-AIDS-associated cancers, as well as cardiovascular disease (Calvo et al. 2015). Even among populations with free and available access to healthcare and ART, HIV patients who smoke are at significantly greater risk of tobacco-associated death than uninfected individuals, and have lower life expectancies than HIV-positive nonsmokers (Helleberg et al. 2013). Metabolism of tobacco smoke constituents produces oxidative stress and increases viral load, contributing to HIV pathogenesis (Ande et al. 2015a).
Because of the overlap between HIV and substance abuse-associated morbidities and the greater risk of enhanced viral pathogenesis and/or secondary conditions in those patients, superior patient surveillance is required. Therefore, there is a need for an accessible and reliable biomarker that can serve as an early indicator of ART failure, known alcohol and tobacco-associated conditions, or morbidities arising from the confluence of multiple harmful agents.
Exosomes are small (30–100 nm) cell-derived vesicles that transport proteins, mRNA, microRNA, and small molecules between cells. They are produced by most cell types, and can transport their molecular cargo between diverse cell types by diffusion through bodily fluids such as blood, saliva, and spinal fluid (Armstrong and Wildman 2018). These qualities make them key mediators of intercellular communication both in healthy and disease conditions(2014). Though it remains to be verified, some reports suggest that the chronic consumption of drugs of abuse can alter exosomal protein content, often with pro-inflammatory or toxic downstream effects. Tobacco and alcohol have also both been shown to increase net secretion of exosomes from multiple cell types, a phenomenon likely mediated by oxidative stress (Wang et al. 2009; Momen-Heravi et al. 2015; Atienzar-Aroca et al. 2016; Chen et al. 2018). HIV pathogenesis itself is partly mediated by exosomes as well. The retrovirus utilizes exosomal packaging machinery to facilitate its own reproduction, releasing mature viruses through interaction with the exosomal pathway (Gould et al. 2003; Madison and Okeoma 2015), as well as exosomes containing viral proteins such as Tat and Nef and viral RNA elements(Lenassi et al. 2010; Narayanan et al. 2013; Rahimian and He 2016).
Due to their relative stability, availability in biofluids, and stimulus-inducible cargo changes, exosomes are ideal candidates for potential biomarkers of these disease states (Revenfeld et al. 2014). Cho et al. recently published a review concerning the potential for use of exosomes as biomarkers of alcohol and drug-induced liver injury (Cho et al. 2018). It is highly likely that alcohol, tobacco, and HIV modulate exosomal cargo, i.e. exosomes in the plasma of HIV-positive substance users contain unique factors, which can serve as biomarkers of HIV-1-substance use interaction. Therefore, our aim is to identify these proteins in people with HIV-positive who use alcohol or tobacco because we are particularly interested in potential candidate molecules that are altered in patients with comorbid conditions but not patients under only one condition (alcohol or tobacco abuse). Thus, this study sought to investigate such potential by proteomic profiling of plasma exosomes from HIV-infected patients who regularly smoke tobacco or drink alcohol.
Results
Characterization of exosomes
In this study we implemented a double isolation method to improve the quality of exosomes for proteomic analysis. The size of the exosomes after double isolation method was in the range of 30–120nm (Figure 1a). Though the total protein yield was reduced by approximately half with the double isolation method (Figure 1b) relative to single isolation, the expression of the exosomal marker protein CD63 did not change significantly (Figure 1c). The purpose of implementing the double isolation method was to remove abundantly secreted free proteins such as albumin from the exosome samples, allowing for greater purity and superior results during proteomic analysis. The double isolation method was successful as the albumin expression was reduced by approximately by 4-fold when compared to the single isolation method (Figure 1c). However, the acetylcholine esterase (AchE) activity was also reduced, by approximately 6-fold compared to the single isolation method (Figure 1d). We have also measured the total exosomal protein concentration in different groups and compared it with a healthy group (Figure 1e). Total exosomal protein concentration varied significantly between healthy and HIV+ groups. We then verified the presence of the exosomal marker protein CD63 by western blotting from each group (Figure 1f) by loading equal amount of protein. Despite loading equal amount of protein, we observed that CD63 expression is relatively high in subjects who were either HIV-positive or HIV+smokers, possibly due to enrichment of CD63 in their exosomes indicating that exosomal marker, CD63, expression varies in different disease conditions. Expression of CD63 and CD81 levels were reported to vary between exosomes derived from different type of cell lines (Andreu and Yáñez-Mó 2014). There is no generally accepted exosomal house-keeping protein reported in the literature yet. Hence, until ‘THE’ exosome marker is identified, it will be necessary to combine several different methods and targets to verify the presence of exosomes. We have verified the exosomes by using CD63, CD81, and Alix markers previously in our lab (Haque et al. 2017). We have also identified and validated human plasma-derived exosomes by transmission electron microscopy (TEM) (Figure 1g).
Figure 1. Characterization of exosomes after single and double isolation.
a. The average size and size distribution of plasma exosomes after double isolation method was measured using zetasizer.
b. Comparison of total protein levels of exosomes after single and double isolation. Total protein levels were quantified using BCA protein assay kit.
c. Protein expressions of exosomal marker protein CD63 and plasma protein albumin.
Protein expressions of single- and double-isolated exosomes were quantified by western blot. Protein relative expressions of double-isolated exosomes were reported as fold changes of the single-isolated exosomes.
d. Acetylcholine esterase activity of exosomes after single and double isolation.
e. Comparison of total exosomal protein levels in different study groups. Bars indicate mean±SEM values.
f. Detection of exosomal marker protein, CD63, in different subjects from each study group by western blotting. C-control, S-smoker, H-HIV, HS-HIV+smoker, D-drinkers, HD-HIV+drinkers. The number on each blot represents the different subject in that particular group.
g. Identification and validation of human plasma-derived exosomes by transmission electron microscopy (TEM).
HIV infection and drugs of abuse alter the expression of plasma exosomal proteins and HIV-related exosomal proteins are associated with HIV Gag and Tat encoded proteins
A total of 343 proteins were identified and quantified in exosomes of all the studied groups (Supplementary Table S1 online). A heat map demonstrates significant quantitative changes in exosomal protein cargo derived from HIV and groups containing substance-abuse subjects (Figure 2)
Figure 2. Heatmap showing log (2) fold change of significant proteins in exosomes isolated from the plasma of present study groups.
a. Differences in exosomal proteins between control (healthy; n=5), and HIV (n=5), cigarette smokers (n=5), alcohol drinkers (n=5), HIV+drinkers (n=3), HIV+smokers (n=4).
b. Differences in exosomal proteins between drug abusers and HIV+drug abusers
c. Differences in exosomal proteins between HIV and HIV+drug abusers.
The heatmap includes proteins that had significant fold change in at least one of the study groups compared with control groups. Log (2) fold change of proteins were presented in the heatmap with downregulation depicted in blue and upregulation in red. Each small square in the heat map represents a protein, and the color represents the fold change of the protein; the greater the fold change, the darker the color. Each line represents the expression of each protein in different groups. Each column represents the expression of all the differentially expressed proteins in each group. The details of the significant proteins of figure 2 are provided in supplementary file 2.
We compared the relative expression of proteins between healthy subjects and the different study groups. A total of sixteen proteins were significantly altered in the HIV group, ten in drinkers, four in HIV+drinkers, and fifteen in smokers (Figure 3a–d; Supplementary Table S2a–d online), compared to healthy subjects. Only one protein, fibulin-1 (FBLN1), was altered significantly in HIV+smokers compared to healthy subjects. Compared with drinkers, twelve proteins were significantly down-regulated and two were significantly up-regulated in HIV+drinkers (Figure 3e; Supplementary Table S2e online). Five proteins were significantly upregulated and two proteins were down-regulated in HIV+smokers compared to smokers (Figure 3f; Supplementary Table S2f online). The proteins that were significantly altered between the HIV group and HIV-positive drug abusers are presented in Figure 3g–h; Supplementary Table S29-h online. The altered exosomal proteins from each subject group were validated against an exosome database, ExoCarta (Keerthikumar et al. 2016). A list of proteins that have been reported previously in exosomes and those we found in our study, is presented in Tables 1 & 2. The association of HIV-associated exosomal proteins with HIV-1 Gag and Tat encoded proteins were investigated using the most recent HIV-Human Protein Interaction Database (Ako-Adjei et al. 2015) (Tables 1 & 2).
Figure 3. Bar diagram showing significantly up/down-regulated proteins based on fold change in exosomes isolated from the present study groups.
a-d. Proteins significantly altered in HIV, drug abusers (alcohol and smokers), and HIV+drug abusers respectively compared to healthy subjects.
e&f. Proteins significantly altered in HIV+drug abusers compared to drug abusers
g&h. Proteins significantly altered in HIV+drug abusers compared to HIV.
Table 1.
Associations and variations between human plasma exosomal proteins reported in this study and HIV viral proteins, alcohol, and tobacco. Previous reports of exosomal packaging were checked using ExoCarta(Keerthikumar et al. 2016). Protein associations with HIV were determined using the HIV-1, Human Protein Interaction Database (Ako-Adjei et al. 2015).
| Accession | Gene symbol | Description | aReported previously in exosomes | Reported previously in plasma exosomes | Associated with HIV (Protein / replication interaction s) | Related to alcohol | Related to smoking | C vs H | C vs S | C vs D | C vs H+D | C vs H+S |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| P01023 | A2M | Alpha-2-macroglobulin | Yes | No | Gp41, Tat, retropepsin | Higher in smokers (Suriyaprom et al. 2007),patien ts with COPD (Arellano-Orden et al. 2017) | - | - | ↓ | - | - | |
| P01031 | C5 | Complement C5 | Yes | Yes | Gp120, Vif | Contributes to ethanol-induced fatty liver in mice (Pritchard et al. 2007) | Higher in smokers (Wyatt et al. 1981) | - | - | ↓ | - | - |
| P02748 | C9 | Complement component C9 | Yes | Yes | Gp120 | Protective against alcohol-induced rat liver damage (Bykov et al. 2004) | Higher in smokers (Wyatt et al. 1981; Bridges et al. 1986) | - | - | ↓ | - | - |
| P02751 | FN1 | Fibronectin | Yes | No | Gp120, gp160, precursor, gp41, Nef, Tat, retropepsin ), gp41 | Correlated with the number of years of smoking (Santoro Belangero et al. 2018) | - | - | ↓ | - | - | |
| P04278–1 | SHBG | Sex hormonebinding globulin | Yes | Yes | Associated with alcohol consumption (Jensen et al. 2014; Hirko et al. 2014) | Not associated (Wang et al. 2013) | - | - | ↓ | - | - | |
| P07357 | C8A | Complement component C8 alpha chain | Yes | Yes | Biomarker candidates for hepatocellular carcinoma (Awan et al. 2015) | - | - | ↓ | - | - | ||
| P08603–1 | CFH | Complement factor H | Yes | Yes | Gp120,gp41 | - | - | ↓ | - | - | ||
| P23142 | FBLN1 | Fibulin-1 | Yes | Yes | ↓ | ↓ | ↓ | - | ↓ | |||
| Q9BXR6 | CFHR5 | Complement factor H-related protein 5 | Yes | Replication interaction | - | - | ↓ | - | - | |||
| A0A075B6K4 | IGLV3-10 | Immunoglobuli n lambda variable 3–10 | No | Yes | ↑ | - | - | - | - | |||
| A0A087WSY6 | IGKV3 D-15 | Immunoglobuli n kappa variable 3D-15 | Yes | Yes | ↑ | - | - | - | - | |||
| A0A0A0MR Z8 | IGKV3 D-11 variable 3D-11 | Immunoglobuli n kappa | No | Yes | ↑ | - | - | - | - | |||
| P00736 | C1R | Complement C1r subcomponent | Yes | No | ↓ | - | - | - | - | |||
| P01700 | IGLV1-47 | Immunoglobuli n lambda variable 1–47 | No | Yes | ↑ | - | - | - | - | |||
| P01768 | IGHV3-30 | Immunoglobuli n heavy variable 3–30 | No | Yes | ↑ | - | - | - | - | |||
| P01824 | IGHV4-39 | Immunoglobuli n heavy variable 4–39 | No | Yes | ↑ | - | - | - | - | |||
| P01834 | IGKC | Immunoglobuli n kappa constant | Yes | Yes | ↑ | - | - | - | - | |||
| P04196 | HRG | Histidine-rich glycoprotein | Yes | Yes | ↓ | - | - | - | - | |||
| P05090 | APOD | Apolipoprotein D | Yes | No | ↓ | ↓ | - | - | - | |||
| P08519 | LPA | Apolipoprotein(a) | Yes | No | ↓ | ↓ | - | - | - | |||
| P0DOX5 | IGG1 | Immunoglobuli n gamma-1 heavy chain | No | Yes | ↑ | - | - | - | - | |||
| P0DOY2 | IGLC2 | Immunoglobuli n lambda constant 2 | Yes | ↑ | - | - | - | - | ||||
| P19827–1 | ITIH1 | Inter-alphatrypsin inhibitor heavy chain H1 | Yes | Yes | ↓ | - | - | - | - | |||
| Q13790 | APOF | Apolipoprotein F | No | Yes | ↓ | ↓ | - | - | - | |||
| Q9Y6R7 | FCGBP | IgGFc-binding protein | Yes | No | ↓ | - | - | - | - | |||
| P02652 | AP0A2 | Apolipoprotein A-II | Yes | No | - | - | - | ↓ | - | |||
| Q9H0U4 | RAB1B | Ras-related protein Rab-1B | Yes | Yes | Gp41, replication interaction | Decreased in ethanol-treated pituitaries (Ren et al. 2005) | - | - | - | ↑ | - | |
| P01024 | C3 | Complement C3 | Yes | No | Gp120, gp160, precursor, Nef, Tat | Induced by ethanol (Kalinin et al. 2018) | - | - | - | ↓ | - | |
| A0A0C4DH3 4 | IGHV4-28 | Immunoglobuli n heavy variable 4–28 | No | Yes | - | ↓ | - | - | - | |||
| O00187–1 | MASP2 | Mannan-binding lectin serine protease 2 | Yes | No | - | ↓ | - | - | - | |||
| P00734 | F2 | Prothrombin | Yes | Gp120, gp160, precursor, replication interaction | - | ↓ | - | - | ||||
| P01714 | IGLV3-19 | Immunoglobuli n lambda variable 3–19 | No | Yes | - | ↓ | - | - | - | |||
| P01743 | IGHV1-46 | Immunoglobuli n heavy variable 1–46 | No | Yes | - | ↓ | - | - | - | |||
| P01861 | IGHG4 | Immunoglobuli n heavy constant gamma 4 | Yes | Yes | - | ↑ | - | - | - | |||
| P05452 | CLEC3B | Tetranectin | Yes | Yes | - | ↑ | - | - | - | |||
| P0DP01 | IGHV1-8 | Immunoglobuli n heavy variable 1–8 | No | Yes | - | ↓ | - | - | - | |||
| P11597–1 | CETP | Cholesteryl ester transfer protein | Yes | Yes | - | ↓ | - | - | - | |||
| P21333 | FLNA | Filamin-A | Yes | Yes | Gp120), Gap-Pol, Nef, Pr55, Tat, Vpr, retropepsin | - | ↑ | - | - | - | ||
| Q13103 | SPP2 | Secreted phosphoprotein 24 | Yes | Yes | - | ↓ | - | - | - | |||
| P02790 | HPX | Hemopexin | Yes | Yes | Biomarker candidates of alcohol abu se (Lai et al. 2009) | - | - | - | ↓ | - |
Reported previously in exosomes derived from various sources, could be cell lines/plasma/saliva etc. C-Control, H-HIV, D-drinkers, S-smokers, H+D = HIV+drinkers, H+S = HIV+smokers; ‘-’ No change, ↓ -down regulation, ↑ -up regulation.
Table 2.
Associations and variations between human plasma exosomal proteins reported in this study from HIV-infected individuals and/or substance abusers. Exosome localization was checked against ExoCarta. Protein associations with HIV were identified from the HIV, Human Protein Interaction Database (Ako-Adjei et al. 2015).
| Description | aReported previously in exosomes | Reported previously in plasma exosomes | Associated with HIV (Protein interactions/ replication interactions) | Associated with alcohol | Associated with smoking | H vs H+D | H vs H+S |
|---|---|---|---|---|---|---|---|
| Immunoglobulin kappa variable 1–16 | No | Yes | ↓ | - | |||
| Ceruloplasmin | Yes | Yes | Tat | Serum levels higher in smokers (Milnerowic z and Slowinska 1997; Tungtrongchitr et al. 2002) | ↓ | - | |
| Collagen alpha-l(VI) chain | Yes | Yes | ↓ | ↓ | |||
| Immunoglobulin heavy variable 1 −69 | No | Yes | ↓ | - | |||
| Properdin | No | Yes | Gp120, gp41 | - | ↓ | ||
| Keratin, type I cytoskeletal 9 | Yes | Yes | ↓ | - | |||
| Immunoglobulin gamma-1 heavy chain | No | Yes | ↓ | - | |||
| Complement factor I | Yes | Yes | ↓ | - | |||
| Immunoglobulin kappa constant | Yes | Yes | ↓ | - | |||
| Vimentin | Yes | Yes | Gp120, Gag-Pol, Nef, Pr55, Vif, Vpr, matrix, retropepsin | ↑ | - | ||
| Alpha-1B-glycoprotein | Yes | Yes | ↓ | ↓ | |||
| Gelsolin | Yes | Yes | Replication interactions, gp120, Tat, Vpr | Loss of gelsolin expression in non-small cell lung cancers of heavy smokers (Dosaka-Akita et al. 1998) | ↑ | - | |
| Immunoglobulin kappa variable 2D-40 | No | Yes | ↓ | ↓ | |||
| Immunoglobulin kappa variable 1–5 | Yes | Yes | ↓ | - | |||
| Immunoglobulin kappa variable 3D-11 | No | Yes | ↓ | - | |||
| Histone H3.3 | Yes | Yes | Replication interactions, Tat | ↑ | - | ||
| Complement C3 | Yes | No | Gp120, gp160, precursor, Nef, Tat | Alcohol increases C3 (Kalinin et al. 2018) | ↓ | ↓ | |
| C4b-binding protein alpha chain | Yes | No | ↑ | - | |||
| Hemopexin | Yes | Yes | Biomarker candidate of alcohol abuse (Lai et al. 2009) | ↓ | - | ||
| Isoform 2 of Fructose-bisphosphate aldolase A | Yes | Yes | Vpr, retropepsin | ↑ | - | ||
| Complement factor H | Yes | Yes | Gp120, gp41 | ↓ | - | ||
| Immunoglobulin heavy variable 1–46 | No | Yes | ↓ | - | |||
| Immunoglobulin heavy variable 1–8 | No | Yes | ↓ | - | |||
| Complement C5 | Yes | Yes | Gp120, Vif | Ethanol-induced fatty liver in mice (Pritchard et al. 2007) | Higher in smokers (Wyatt et al. 1981) | ↓ | - |
| Isoform 2 of Insulinlike growth factorbinding protein complex acid labile subunit | Yes | Yes | ↓ | - | |||
| Isoform 2 of Src substrate cortactin | Yes | Yes | Pr55 | ↑ | ↑ | ||
| Immunoglobulin lambda variable 4–60 | No | Yes | ↓ | - | |||
| Histone H2A type 1 -B/E | Yes | Yes | Tat | ↑ | - | ||
| Ras-related protein Rab-1B | Yes | Yes | Gp41, replication interactions | Ethanol-induced alterations in Rab proteins (Ren et al. 2005) | ↑ | - | |
| Immunoglobulin heavy constant gamma 3 | Yes | Yes | ↓ | - | |||
| Complement factor H-related protein 1 | No | Yes | - | ↑ | |||
| Heat shock protein beta-1 | Yes | Yes | Vif, Vpr | - | ↑ | ||
| Protein S100-A8 | Yes | Yes | - | ↑ | |||
| Cystatin-A | Yes | Yes | - | ↑ | |||
| Apolipoprotein(a) | Yes | No | - | ↑ | |||
| Inter-alpha-trypsin inhibitor heavy chain H4 | Yes | Yes | - | ↓ | |||
| Immunoglobulin lambda variable 1–51 | Yes | - | ↓ | ||||
| Immunoglobulin heavy variable 3–72 | No | Yes | - | ↓ | |||
| HLA class I histocompatibility antigen, B-51 alpha chain | Yes | Yes | Replication interactions | - | ↑ | ||
| Protein AMBP | Yes | Yes | - | ↓ | |||
| Desmocollin-1 | Yes | Yes | - | ↑ | |||
| 14–3-3 protein sigma | Yes | Yes | - | ↑ | |||
Reported previously in exosomes derived from various sources-could be cell lines/plasma/saliva etc. H = HIV, H+D = HIV+drinkers, H+S = HIV+smokers ‘-’No change, ↓ -down regulation,↑ -up regulation.
Alterations in expression of exosomal proteins associated with HIV infection and drug abuse, impact various biological processes
To gain a broader and more accurate understanding of the potential relevance of the significantly altered proteins in biology, we applied Gene Ontology (GO) analysis using Panther software (Thomas et al. 2003). When considering biological processes (BP) in the HIV group, the greatest number of proteins were involved in immune system processes, followed by those involved in general biological regulation (Figure 4a). Among the drinkers, the largest percentage of proteins was associated with cellular processes, followed by metabolic processes (Figure 4b). In the HIV+drinker group, the highest percentage of proteins were involved in metabolism (Figure 4c). In smokers, the altered proteins were associated with the immune system, metabolism, and response to stimulus processes (Figure 4d). A comparison of HIV+smokers with healthy subjects showed that FBLN1 was the only protein whose level was altered. FBLN1 may play a role in cell adhesion and migration along protein fibers within the extracellular matrix.
Figure 4. Pie chart of gene ontology analysis of significantly altered exosomal proteins in different study groups based on biological processes.
To gain broader and more accurate understanding of the significantly altered proteins and their putative role in biology, we applied Gene Ontology (GO) analysis. Pie charts were created in excel by importing analyzed data from Panther. Biological processes are represented by color and percentage of each biological process are depicted in their respective sections of the pie chart.
a. HIV, b.Drinkers, c.HIV+drinkers and d. Smokers compared to healthy subjects.
e&f. HIV-positive drug users compared to HIV-negative drug users
g&h. HIV-positive drug users compared HIV-positive non-drug users
A comparison of HIV+drinkers with HIV-negative drinkers showed that most of the proteins were involved in the immune system and metabolic and cellular processes (Figure 4e). Majority of the significantly altered proteins in HIV+smokers group were enriched in cellular process compared to uninfected smokers (Figure 4f). Compared to the HIV non-abuser group, the majority of enriched proteins in both HIV+drinkers and HIV+smokers were involved in cellular, immunoregulatory, and metabolic processes (Figure 4g & h).
Next, we performed GO-term enrichment analysis using DAVID software (Huang et al. 2009) to determine the relative abundance of specific GO-terms for the significant proteins found in the present study. In the HIV group, immune response, antigen binding, and enzyme inhibitory components were enriched in the biological processes (BP) and molecular function (MF) categories (Figure 5a). In the group of drinkers, ten proteins which were significantly altered, were considered for GO-term enrichment analysis. In the BP category, inflammatory and immune response proteins were enriched, whereas in the MF category, enzyme regulatory activity was enriched (Figure 5b). In smokers, lipid transport was enriched in the BP category, while lipid transport and enzyme activity were enriched in MF category (Figure 5c). In HIV+drinkers, proteins associated with inflammatory and immune response processes were again enriched in BP category, and those associated with enzyme inhibitory activity were enriched in the MF category (Figure 5d).
Figure 5. Histogram showing the significantly enriched GO terms according to DAVID analysis.
DAVID tool was used to identify groups of genes sharing common biology or, alternatively, to identify groups of biological terms sharing common genes.
a-d. HIV, Drinkers, Smokers, and HIV+drinkers respectively compared to healthy subjects.
The proteins which distinguished HIV+drinkers from HIV-negative drinkers, were found to be enriched in regulation of immune response proteins in the BP category and in lipid-binding proteins in the MF category (Supplementary Table S3a online). A comparison of HIV+smokers with HIV-positive non-smokers suggests receptor-mediated endocytosis and vesicle-mediated transport proteins were enriched in BP and protein binding in MF categories (Supplementary Table S3b online). Further, a comparison of HIV+drinkers and non-drinkers indicated that complement activation proteins were enriched in the BP category and serine endopeptidase activity in the MF category (Supplementary Table S3c online). Finally, a comparison of HIV+smokers and non-smokers demonstrated that proteins responsible for regulation of endopeptidase activity were enriched in the BP category, and enzyme inhibitory activity enriched in MF (Supplementary Table S3d online).
Analysis of proteins in all groups showed enrichment for proteins associated with the extracellular compartment in the CC category (Figure 5a–d & Supplementary Table S3a–d online). This observation is not surprising given the origins of exosomes from the plasma membrane via the endosomal pathway.
Exosomal proteins altered in HIV-positive drug abusers but not in subjects with either condition alone
One of our areas of interest is proteins, which can potentially serve as markers for co-morbid conditions associated with drug abuse in HIV patients. Therefore, we looked for proteins which were not significantly altered in the HIV group or the two-drug abuse groups but were altered in HIV-positive drug abusers (Figure 6a). Hemopexin (HPx), apolipoprotein A-II (APOA2), ras-related protein Rab-1 (RAB1B), and complement C3 (C3) were significantly altered in HIV+drinkers but not in subjects with either condition alone. HPx and RAB1B proteins are newly found in plasma exosomes (Table 1). FBLN1was the only significantly altered protein in HIV+smokers compared to healthy individuals, but it was also significantly changed in patients with either of the two conditions (Figure 6b). Another protein of note, properdin, showed significant change in HIV+drinkers and HIV+smokers when compared with the HIV group (Table 2).
Figure 6. Regulation pattern of a. hemopexin, b. fibulin-1, c. Alpha-2-macroglobulin, d.Properdin and validation of those proteins by western blotting in different study groups.
‘−’ represent up-regulation and ‘....’ represent down regulation. C-control, H-HIV, D-drinkers, HD-HIV+drinkers, S-Smokers, HS-HIV+Smokers. p<0.05 is considered significant.
Validation of hemopexin and fibulin-1 proteins by western blotting
Of the significant proteins in HIV+drinkers, HPx was considered based on the literature (Lai et al. 2009) to be a potential candidate marker to diagnose co-morbid conditions in HIV+drinkers. Therefore, we validated HPx in study groups by western blotting (Figure 6a). Due to the overlap in its down-regulation between the HIV+smoker and non-smoker groups, we also validated FBLN1 in our clinical samples (Figure 6b). The results showed a slight decrease in the level of HPx in the HIV and HIV+drinker groups, while the level of FBLN1 was slightly decreased in HIV and HIV+smokers groups, compared to their respective controls. Overall, the results from proteomic analysis, at least in part, was consistent with the results obtained from the Western blot.
Discussion
HIV infection and drugs of abuse contribute to profound changes in cellular protein expression. Exosomes are emerging as important biological markers because they are involved in inter-cellular communication and often reflect physiological and/or pathological changes occurring in their donor cells. Our aim was to identify potential candidate protein biomarkers in plasma exosomes that may serve as indicators of drug-abuse related diseases that perhaps enhance the progression of HIV to AIDS. Therefore, the present study investigated the protein content of exosomes derived from HIV-infected patients and drug abusers. To accomplish this aim, we sought to enrich our plasma samples by removing the contaminating abundant, unbound, circulating soluble proteins utilizing a double isolation method. Characterization of exosomes after single and double isolation methods demonstrated that the double isolation enriched the samples for exosomal proteins by reducing soluble protein contaminants. This was evidenced by the decrease in total protein yield and expression of the highly abundant soluble plasma protein albumin without any significant decrease in the presence of the exosomal marker protein CD63 (Figure 1b, c). However, another exosomal marker, AchE activity, was reduced by the double isolation method. The exosomal membrane is enriched in the GPI-anchored proteins including AchE (Gutiérrez-Vazquez et al. 2013) which is located on the extracellular surface of the cell membrane (Taylor et al. 2009). The reduction in activity due to the double isolation method could be attributed to inactivation of enzyme activity as it is a surface protein and bound to membrane lipids. We observed that total exosomal protein concentration in the HIV and HIV+smoker groups was higher than the other groups (Figure 1e). Detection of exosomal marker protein, CD63, was also high in this group, indicating an increased release of exosomes in such conditions (Figure 1f).
In the present study, we found that the expression of multiple proteins in exosomes is altered under various stress conditions (Figure 2 & 3). We have verified the finding of altered exosomal proteins in HIV and drug abuse subjects in the exosome database, ExoCarta (Keerthikumar et al. 2016). Most of the proteins we detected, have been previously reported in exosomes from different cell lines. Our study is the first to show their presence in plasma exosomes (Table 1 and 2). The finding that plasma exosomal proteins may potentially serve as markers associated with drug abuse in HIV patients is novel. Further rigorous studies are necessary, however, to replicate our findings. If validated, it may be possible to develop a practical tool for risk/treatment stratification, early diagnosis, prognosis, or assessment of disease severity.
Further, we explored the general trends in functional changes of exosomal proteins identified in the present study subjects via GO analysis using DAVID (Huang et al. 2009). In the HIV group, most of the altered proteins in the “biological process” category is involved in immune system processes (Figure 5a). The analysis of the “biological process” category indicated that, in the group of drinkers, the majority of the significantly altered proteins in exosomes were associated with the acute inflammatory response (Figure 5b) and their levels were reduced compared to levels in healthy subjects. Among these, alpha-2-macroglobulin (A2M) is an interesting candidate for future investigation due to its ability to inhibit inflammatory cytokines, leading to disruption of inflammatory cascades. A2M can also inhibit proteases and transport cytokines. Its dysfunction is implicated in Alzheimer’s disease due to its ability to mediate the clearance and degradation of beta amyloid, a hallmark of the disease(ref). Further, Varma et al. have reported that patients with increased susceptibility to Alzheimer’s disease had high serum A2M levels (Varma et al. 2017). This increase could be a compensatory mechanism to facilitate the degradation of beta amyloid. The decreased A2M levels in the plasma exosomes of drinkers (Figure 6c) indicate that these subjects may be at greater risk of developing neurodegenerative diseases. Therefore, further research needs to be conducted to study the role of A2M in neurocognitive dysfunction induced by drinking alcohol in patients with HIV.
In smokers, the biological process that is most enriched was lipid transport (Figure 5c). In particular, CETP protein levels are decreased compared to the control group, a factor previously reported in association heart disease in smokers (Ritsch et al. 2010). In HIV+drinkers, proteins involved with regulation of immune effectors were enriched in the biological processes category (Figure 5d), and two of the proteins associated with it, apolipoprotein A-II (APOA2) and complement C3 (C3), have also been found in exosomes derived from the urine of HIV-positive subjects (Anyanwu et al. 2018) and plasma of HIV-infected and HIV/HCV co-infected individuals (Shetty et al. 2011). C3 was found to have direct interaction with HIV (Ako-Adjei et al. 2015) (Table 1). Another protein, hemopexin (HPX), is not altered in drinkers or HIV patients, but was significantly downregulated when these conditions overlapped. Further, HPX is altered in reports concerning HIV-positive methamphetamine abusers (Pottiez et al. 2012) and alcohol abusers (Lai et al. 2009). HPX binds to free heme in circulation and this complex is delivered to hepatocytes for degradation. The HPX is recirculated via the bloodstream after the HPX-heme complex is degraded in the liver. It may also possess an anti-inflammatory role via the negative regulation of IL-6 and TNF-α secretion by activated macrophages (Liang et al. 2009; Vinchi et al. 2016). HPX has a protective role in mouse models of sepsis and is decreased in plasma in association with the increased mortality in sepsis (Larsen et al. 2010). Since HPX serves as an extracellular antioxidant, its decreased level in the exosomes of HIV+drinkers are consistent with its role in protection against alcohol- induced oxidative stress. HPX is also an important antioxidant in the brain (Ma et al. 2016). Decreased levels of exosomal HPX in HIV-positive drinkers could exacerbate or lead to neuroaids in subjects. Similarly, in drinkers proteins that regulate immune responses were enriched in the BP category. Two of the proteins associated with this category are APOA2 and HPX (Supplementary Table S2a online).
In HIV+smokers, receptor-mediated endocytosis is enriched in the BP category compared to smoking alone, in which CD9, CETP, and vitronectin (VTN) proteins were significantly altered (Supplementary Table S3b online). Cigarette smoking down-regulates CD9 and CD81 expression in macrophages and ablation of CD9 function suppresses cell motility and protease production of macrophages (Takeda et al. 2008). Moreover, knockout of CD9 in mice results in macrophage accumulation, increased activity of proteases in the lung, and progressive development of pulmonary emphysema (Takeda et al. 2008). Further, CD9 has been reported to play an important role in membrane fusion induced by HIV, as knocking down CD9 expression results in increased syncytia formation and viral entry (Gordon-Alonso et al. 2006). Downregulation of exosomal CD9 level in HIV+smokers may serve as a biomarker for progression of pulmonary diseases such as emphysema and lung cancers in patients with HIV (Diaz et al. 2000; Ranjit and Kumar 2018). Plasma proteomic profiling of smokers by Bortner et al. revealed that VTN is down-regulated compared to non-smokers (Bortner et al. 2011). We observed this trend in our study, although it did not reach statistical significance. Interestingly, VTN levels are increased in HIV+smokers relative to uninfected smokers. At this stage, the role of VTN in tobacco-induced HIV replication is not understood. Out of all the quantified proteins, only one protein, FBLN1, was altered significantly in HIV+smokers compared to healthy subjects. Due to the overlap in its down-regulation between the HIV+smoker and non-smoker groups, we also validated FBLN1 in our clinical samples (Figure 6b). However, a prospective study suggested that the FBLN1 levels were significantly high in HIV-positive South Africans who were also drug abusers (46% of them were tobacco users and 26% were alcohol users) compared to HIV-negative subjects with the history of drug abuse (40% tobacco users, 29% alcohol users). Further, the percentage change in FBLN1 levels were associated with change in TG/HDL-c ratio, which may contribute to probable vascular changes in HIV-positive subjects (Pretorius 2014). Differences in study design, inclusion/exclusion criteria and sample size could have contributed to differences in regulation pattern of FBLN1 between our study and the reported prospective study.
Compared to HIV-infected non-drug abusers, serine-type endopeptidase activity (Supplementary Table S3c online) and enzyme inhibitor activity (Supplementary Table S3d online), were enriched in HIV+drinkers and smokers, respectively. Properdin (CFP) is a component of the human complement system that interacts directly with the HIV viral proteins gp120 and gp41 (Susal et al. 1994, 1996). Our study is the first to identify CFP in plasma exosomes; we found that it was decreased in HIV+smokers and HIV+drinkers compared to HIV-positive controls. Decreased levels of CFP in those groups may indicate that alcohol drinking and tobacco smoking increase the risk of secondary infections in patients with HIV.
Our study has limitations which require consideration. Firstly, we have a relatively low donor sample size which may have allowed potential proteins of interest to escape detection. Also, our subject pool consists only of West Africans from Cameroon. It is possible, due to genetic differences in expression of drug- metabolizing enzymes, other populations may have different exosomal proteomic profiles under otherwise similar recruitment conditions. With these limitations, our study provides new information in terms of packaging of these proteins in exosomes, which are altered by HIV and/or drugs of abuse.
Conclusion
In this study, we performed proteomic analysis of exosomes collected from plasma of subjects infected with HIV, who regularly consumed alcohol or tobacco, or both. We discovered several exosomal proteins that were affected by the combination of HIV and drugs of abuse, which may be of clinical significance. Altered levels of hemopexin, alpha-2-macroglobulin, and properdin in the exosomes of the HIV+drinker and HIV+smoker groups compared to either condition alone suggest that these proteins may be of interest as potential markers. This study is an important first step towards finding exosomal protein biomarkers in HIV-positive patients who abuse drugs.
Future Directions
Proteomic profiling of exosomes derived from HIV patients and drug abusers who are receiving ART could provide valuable information to enable the elucidation of the role of drugs of abuse in HIV pathogenesis and its manifestations (e.g. HAND). Proteomic profiling of exosomes collected from other accessible bio-fluids, such as urine, saliva, or semen, may also yield novel biomarkers not found in the blood. Both avenues of investigation would contribute to our understanding of the role of exosomes in HIV comorbidities, leading to better treatment outcomes.
Methods
Study population
Among previously recruited subjects in Cameroon, Africa (Ande et al. 2015a), we studied 27 subjects from which we had sufficient plasma for exosome isolation. We assigned them to six different cohorts: a) Five healthy HIV-negative control subjects who reported that they were non-smokers/drinkers (healthy), b) Five HIV-positive subjects who reported that they were non-smokers/drinkers (HIV), c) Five HIV-negative alcohol drinkers (drinkers), d) Five HIV-negative tobacco smokers (smokers), e) Four HIV-positive drinkers (HIV+drinkers), f) Three HIV-positive smokers (HIV+smokers). Participants were recruited in Cameroon, Africa, upon approval of both the Institutional Review Board (IRB) from the University of Missouri-Kansas City and the IRB/Institutional Ethic Committee (IEC) from Provincial Regional Hospital, Ministry of Public Health, Bamenda, Cameroon (Ande et al. 2015a). Mild-to-moderate smokers with a smoking history of less than 20 pack years (a pack year: smoking at least one pack per day for one year) and never consumed alcohol were enrolled and classified as smokers. Female subjects who consumed 4–7 drinks/week, and male subjects who had 7–14 drinks/week and never smoked were classified as drinkers. HIV+drinkers with history of no smoking and HIV+smokers with history of no alcohol consumption were recruited. The mean±SD age group of healthy, HIV, drinkers, smokers, HIV+drinkers, and HIV+smokers were 41.4±2.0, 31.6±8.2, 38.8±7.4, 33.5±4.36, 39.0 ±2.6, and 45.7±10.0, respectively. There was no significant difference in age between the study groups. As expected, there was a significant difference in mean±SD for CD4+ T-cell counts of healthy (904±205) when compared with HIV-positive (384±113, p=0.009) and HIV-positive drug abusers (HIV+drinkers, 420±87, p=0.04; HIV+smokers, 387±260, p=0.01) and it didn’t vary when compared with HIV-negative smokers (1231±238, p=0.17) and HIV-negative drinkers (702±91, p=0.65). Individuals suffering from other infectious diseases, such as active TB, malaria, or who were sero-positive for hepatitis A/B/C were excluded in this study. Also, individuals receiving antiretroviral therapy or who took other drugs of abuse and/or with neurological disorders were excluded. Subjects were recruited based on personal interview and clinical screening for HIV, malaria, and hepatitis B.
Exosome isolation and characterization
We isolated exosomes from plasma samples using a modified method from the previously published method (Kumar et al. 2017; Haque et al. 2017) using commercially available precipitation kit (Invitrogen). In brief, samples were run through a 0.22 μm filter to remove any aprticles which are >0.22 μm and centrifuged at 10,000 g for 20 min to remove any large bodies such as microvesicles or cell debris, then mixed with 0.5 volumes of sterile PBS and 0.2 volumes of exosome precipitation reagent. The reaction mixture was then allowed to incubate at room temperature for 10 min. Following incubation, the sample was centrifuged again at 10,000 g for 5 min. to produce a purified exosome pellet. To remove potential contaminants such as soluble albumin and other proteins in exosomes for proteomic work, we performed double isolation procedure as follows. The exosome pellet was resuspended in 1xPBS (volume of 1xPBS =volume of initial plasma+volume of PBS added in the previous step) and mixed with 0.2 volumes of exosome precipitation reagent followed by incubation at room temperature for 10 min and centrifugation at 10,000 g for 5 min to obtain the exosome pellet. To validate the quality of our samples, we characterized the size, shape, and quality of exosomes by using a JEOL 2000EXII transmission electron microscope (The Neuroscience Institute, University of Tennessee Health Science Center), as described previously (Kumar et al. 2017; Haque et al. 2017), and measured the size and zeta potential of exosomes by dynamic light scattering using a Zetasizer Nano-ZS (Malvern Instruments Inc, Malvern, UK) (Kumar et al. 2017; Haque et al. 2017). We quantified and further validated the exosomes by measuring acetylcholinesterase (AchE) activity using the fluorescent Amplex® Red Acetylcholine/Acetylcholinesterase Assay Kit (Molecular Probes, Invitrogen) as described previously (Kodidela et al. 2018) and by testing the samples for expression of the exosomal marker proteins CD63, CD81, and/or Alix by Western blotting (Kumar et al. 2017; Haque et al. 2017).
Protein identification and characterization
A reporter ion quantification approach based on TMT-labeling was employed for differential protein expression analysis as described below.
Sample process: exosome pellets solubilized in 5 volumes of the Lysis Buffer were processed using a commercial kit, (Pierce™ Mass Spec Sample Prep Kit for Cultured Cells 84840, Thermo Fisher) according to the manufacturer’s protocol, in which they underwent nuclease digestion, reduction/alkylation, acetone precipitation, and Lys-C/trypsin digestion. The protein concentration was determined using a Pierce BCA Protein Assay kit (Thermo Fisher) according to the manufacturer’s protocol. The peptide concentration was determined using a Pierce Quantitative Colorimetric Peptide Assay kit (Thermo Fisher) according to the manufacturer’s protocol. TMT-Labeling of peptides was performed using a TMT-10plex commercial kit (Thermo Fisher) according to the manufacturer’s protocol. The labeled set of 10 samples was combined and fractionated using a Pierce High pH Reversed-Phase Peptide Fractionation kit (Thermo Fisher) according to the manufacturer’s protocol - 8 step fractions were collected. The collected peptide fractions were analyzed by LC-MS for peptide/protein identification and quantification. Acquisition of raw MS data was performed on an Orbitrap Fusion Lumos mass spectrometer (Thermo Fisher) operating in line with Ultimate 3000RSLCnano UHPLS system (Thermo Fisher) using MS3 Synchronous Precursor Selection (SPS) method. Post-acquisition analysis of the raw MS data was performed using the mass informatics platform Proteome Discoverer 2.2 (Thermo Fisher) using the Sequest HT search algorithm and human protein database (SwissProt, Homo sapiens, TaxID 9606, v.2017–05-10, 42153 entries). Identification and quantification of peptides/proteins were validated at 0.01 false discovery rate (FDR) and 0.05 p-value corrected for multiple testing, respectively.
Data analysis
a. Protein quantitation
We normalized the protein expression values using a cyclic loess normalization method. We ran a consistency check package for principal component analysis and Pearson’s before and after normalization. Differences in protein expression levels among various groups were compared using Welsh’s t-test and Benjamini Hocherg false discovery methods. Only proteins that passed 0.05 FDR are considered as significant. The heatmap was generated in RStudio (Version 1.0.153, RStudio, Inc.; Boston, MA) using the heatmap3 package. The heatmap includes proteins that had significant fold change in at least one of the study groups compared with control groups. Log (2) fold change of proteins were presented in the heatmap with downregulation depicted in blue and upregulation in red.
b. ExoCarta retrieval
All the significant exosomal proteins were cross checked against the most recent version of the online exosome database ExoCarta(Keerthikumar et al. 2016).
c. Gene Ontology (GO) characterization
To gain broader and more accurate understanding of the significantly altered proteins and their putative role in biological process, we applied Gene Ontology (GO) analysis. The significantly altered proteins in different groups were searched against UniprotKB, EBI, and GO databases via Panther(Thomas et al. 2003). Visualization of GO analysis results was finalized in Excel by importing analyzed data from Panther.
d. GO term enrichment analysis
To identify groups of genes sharing common biology or, alternatively, to identify groups of biological terms sharing common genes, we submitted a list of significantly altered proteins to DAVID(Huang et al. 2009).
e. Examination against HIV-1/host proteins database
To find potential known associations between HIV-1 and the proteins altered between the different groups, we used Human Protein Interaction Database(Ako-Adjei et al. 2015).
Supplementary Material
Supplementary Table S1 online: The list of proteins identified and quantified in all the studied groups
Supplementary Table S2 online: Significantly altered proteins in the study groups
a. Healthy V s HIV -positive subj ects
b. Healthy Vs Drinkers
c. Healthy vs HIV+drinkers
d. Healthy vs Smokers
e. Drinkers vs HIV+drinkers
f. Smokers vs HIV+smokers
g. HIV vs HIV+drinkers
h. HIV vs HIV+smokers
Supplementary Table S3 online. Significantly enriched pathways according to DAVID analysis.
a. Drinkers vs HIV+ Drinkers
b. Smokers vs HIV+ Smokers
c. HIV vs HIV+Drinkers
d. HIV vs HIV+ Smokers
Acknowledgements
We gratefully acknowledge the faculty and staff at the Molecular Research Center at UTHSC for their assistance in performing LC-MS/MS and statistical analysis of proteomics data. We also thank the National Institute on Alcohol Abuse and Alcoholism (AA022063) and National Institute on Drug Abuse (DA042374 and DA031616) for supporting our work.
Footnotes
Competing Interests
The authors declare no competing interests.
Publisher's Disclaimer: This Author Accepted Manuscript is a PDF file of a an unedited peer-reviewed manuscript that has been accepted for publication but has not been copyedited or corrected. The official version of record that is published in the journal is kept up to date and so may therefore differ from this version.
References
- Ako-Adjei D, Fu W, Wallin C, et al. (2015) HIV-1, human interaction database: current status and new features. Nucleic Acids Res 43:D566–D570. doi: 10.1093/nar/gku1126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ande A, McArthur C, Ayuk L, et al. (2015a) Effect of mild-to-moderate smoking on viral load, cytokines, oxidative stress, and cytochrome P450 enzymes in HIV-infected individuals. PLoS ONE 10:e0122402. doi: 10.1371/journal.pone.0122402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ande A, Sinha N, Rao PSS, et al. (2015b) Enhanced oxidative stress by alcohol use in HIV+patients: possible involvement of cytochrome P450 2E1 and antioxidant enzymes. AIDS Res Ther 12:29. doi: 10.1186/s12981-015-0071-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andreu Z, Yáñez-Mó M (2014) Tetraspanins in extracellular vesicle formation and function. Front Immunol 5:442. doi: 10.3389/fimmu.2014.00442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anyanwu SI, Doherty A, Powell MD, et al. (2018) Detection of HIV-1 and Human Proteins in Urinary Extracellular Vesicles from HIV+ Patients. Adv Virol 2018:7863412. doi: 10.1155/2018/7863412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arellano-Orden E, Calero-Acuña C, Cordero JA, et al. (2017) Specific networks of plasma acute phase reactants are associated with the severity of chronic obstructive pulmonary disease: a case-control study. Int J Med Sci 14:67–74. doi: 10.7150/ijms.16907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Armstrong D, Wildman DE (2018) Extracellular Vesicles and the Promise of Continuous Liquid Biopsies. J Pathol Transl Med 52:1–8. doi: 10.4132/jptm.2017.05.21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atienzar-Aroca S, Flores-Bellver M, Serrano-Heras G, et al. (2016) Oxidative stress in retinal pigment epithelium cells increases exosome secretion and promotes angiogenesis in endothelial cells. J Cell Mol Med 20:1457–1466. doi: 10.1111/jcmm.12834 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Awan FM, Naz A, Obaid A, et al. (2015) Identification of Circulating Biomarker Candidates for Hepatocellular Carcinoma (HCC): An Integrated Prioritization Approach. PLoS ONE 10:e0138913. doi: 10.1371/journal.pone.0138913 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barve S, Kapoor R, Moghe A, et al. (2010) Focus on the liver: alcohol use, highly active antiretroviral therapy, and liver disease in HIV-infected patients. Alcohol Res Health 33:229–236 [PMC free article] [PubMed] [Google Scholar]
- Bortner JD, Richie JP, Das A, et al. (2011) Proteomic profiling of human plasma by iTRAQ reveals down-regulation of ITI-HC3 and VDBP by cigarette smoking. J Proteome Res 10:1151–1159. doi: 10.1021/pr100925p [DOI] [PubMed] [Google Scholar]
- Bridges RB, Wyatt RJ, Rehm SR (1986) Effects of smoking on inflammatory mediators and their relationship to pulmonary dysfunction. Eur J Respir Dis Suppl 146:145–152 [PubMed] [Google Scholar]
- Bykov IL, Vákevá A, Járveláinen HA, et al. (2004) Protective function of complement against alcohol-induced rat liver damage. Int Immunopharmacol 4:1445–1454. doi: 10.1016/j.intimp.2004.06.008 [DOI] [PubMed] [Google Scholar]
- Calvo M, Laguno M, Martínez M, Martínez E (2015) Effects of tobacco smoking on HIV-infected individuals. AIDS Rev 17:47–55 [PubMed] [Google Scholar]
- Chen L, Chen R, Kemper S, Brigstock DR (2018) Pathways of production and delivery of hepatocyte exosomes. J Cell Commun Signal 12:343–357. doi: 10.1007/s12079-017-0421-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho Y-E, Song B-J, Akbar M, Baek M-C (2018) Extracellular vesicles as potential biomarkers for alcohol- and drug-induced liver injury and their therapeutic applications. Pharmacol Ther 187:180–194. doi: 10.1016/j.pharmthera.2018.03.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cioe PA (2013) Smoking Cessation Interventions in HIV-Infected Adults in North America: A Literature Review. J Addict Behav Ther Rehabil 2:1000112. doi: 10.4172/2324-9005.1000112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diaz PT, King MA, Pacht ER, et al. (2000) Increased susceptibility to pulmonary emphysema among HIV-seropositive smokers. Ann Intern Med 132:369–372 [DOI] [PubMed] [Google Scholar]
- Dosaka-Akita H, Hommura F, Fujita H, et al. (1998) Frequent loss of gelsolin expression in non-small cell lung cancers of heavy smokers. Cancer Res 58:322–327 [PubMed] [Google Scholar]
- Galvan FH, Bing EG, Fleishman JA, et al. (2002) The prevalence of alcohol consumption and heavy drinking among people with HIV in the United States: results from the HIV Cost and Services Utilization Study. J Stud Alcohol 63:179–186 [DOI] [PubMed] [Google Scholar]
- Gordón-Alonso M, Yañez-Mó M, Barreiro O, et al. (2006) Tetraspanins CD9 and CD81 modulate HIV-1-induced membrane fusion. J Immunol 177:5129–5137 [DOI] [PubMed] [Google Scholar]
- Gould SJ, Booth AM, Hildreth JEK (2003) The Trojan exosome hypothesis. Proc Natl Acad Sci USA 100:10592–10597. doi: 10.1073/pnas.1831413100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutierrez-Vazquez C, Villarroya-Beltri C, Mittelbrunn M, Sanchez-Madrid F (2013) Transfer of extracellular vesicles during immune cell-cell interactions. Immunol Rev 251:125–142. doi: 10.1111/imr.12013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haque S, Sinha N, Ranjit S, et al. (2017) Monocyte-derived exosomes upon exposure to cigarette smoke condensate alter their characteristics and show protective effect against cytotoxicity and HIV-1 replication. Sci Rep 7:16120. doi: 10.1038/s41598-017-16301-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helleberg M, Afzal S, Kronborg G, et al. (2013) Mortality attributable to smoking among HIV-1-infected individuals: a nationwide, population-based cohort study. Clin Infect Dis 56:727–734. doi: 10.1093/cid/cis933 [DOI] [PubMed] [Google Scholar]
- Hicks PL, Mulvey KP, Chander G, et al. (2007) The impact of illicit drug use and substance abuse treatment on adherence to HAART. AIDS Care 19:1134–1140. doi: 10.1080/09540120701351888 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirko KA, Spiegelman D, Willett WC, et al. (2014) Alcohol consumption in relation to plasma sex hormones, prolactin, and sex hormone-binding globulin in premenopausal women. Cancer Epidemiol Biomarkers Prev 23:2943–2953. doi: 10.1158/1055-9965.EPI-14-0982 [DOI] [PMC free article] [PubMed] [Google Scholar]
- HIV Surveillance Report 2016. HIV Surveillance Report 2016. 28:125 [Google Scholar]
- Huang DW, Sherman BT, Lempicki RA (2009) Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc 4:44–57. doi: 10.1038/nprot.2008.211 [DOI] [PubMed] [Google Scholar]
- Jensen TK, Gottschau M, Madsen JOB, et al. (2014) Habitual alcohol consumption associated with reduced semen quality and changes in reproductive hormones; a cross-sectional study among 1221 young Danish men. BMJ Open 4:e005462. doi: 10.1136/bmjopen-2014-005462 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalinin S, Gonzalez-Prieto M, Scheiblich H, et al. (2018) Transcriptome analysis of alcohol-treated microglia reveals downregulation of beta amyloid phagocytosis. J Neuroinflammation 15:141. doi: 10.1186/s12974-018-1184-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keerthikumar S, Chisanga D, Ariyaratne D, et al. (2016) ExoCarta: A Web-Based Compendium of Exosomal Cargo. Journal of Molecular Biology 428:688–692. doi: 10.1016/j.jmb.2015.09.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kodidela S, Ranjit S, Sinha N, et al. (2018) Cytokine profiling of exosomes derived from the plasma of HIV-infected alcohol drinkers and cigarette smokers. PLOS ONE 13:e0201144. doi: 10.1371/journal.pone.0201144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar S, Jin M, Ande A, et al. (2012) Alcohol consumption effect on antiretroviral therapy and HIV-1 pathogenesis: role of cytochrome P450 isozymes. Expert Opin Drug Metab Toxicol 8:1363–1375. doi: 10.1517/17425255.2012.714366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar S, Sinha N, Gerth KA, et al. (2017) Specific packaging and circulation of cytochromes P450, especially 2E1 isozyme, in human plasma exosomes and their implications in cellular communications. Biochem Biophys Res Commun 491:675–680. doi: 10.1016/j.bbrc.2017.07.145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai X, Liangpunsakul S, Crabb DW, et al. (2009) A proteomic workflow for discovery of serum carrier protein-bound biomarker candidates of alcohol abuse using LC-MS/MS. Electrophoresis 30:2207–2214. doi: 10.1002/elps.200800775 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larsen R, Gozzelino R, Jeney V, et al. (2010) A central role for free heme in the pathogenesis of severe sepsis. Sci Transl Med 2:51ra71. doi: 10.1126/scitranslmed.3001118 [DOI] [PubMed] [Google Scholar]
- Lenassi M, Cagney G, Liao M, et al. (2010) HIV Nef is secreted in exosomes and triggers apoptosis in bystander CD4+ T cells. Traffic 11:110–122. doi: 10.1111/j.1600-0854.2009.01006.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang X, Lin T, Sun G, et al. (2009) Hemopexin down-regulates LPS-induced proinflammatory cytokines from macrophages. J Leukoc Biol 86:229–235. doi: 10.1189/jlb.1208742 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma B, Day JP, Phillips H, et al. (2016) Deletion of the hemopexin or heme oxygenase-2 gene aggravates brain injury following stroma-free hemoglobin-induced intracerebral hemorrhage. J Neuroinflammation 13:26. doi: 10.1186/s12974-016-0490-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madison MN, Okeoma CM (2015) Exosomes: Implications in HIV-1 Pathogenesis. Viruses 7:4093–4118. doi: 10.3390/v7072810 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milnerowicz H, Slowinska M (1997) Concentration of metals, ceruloplasmin, metallothionein and the activity of N-acetyl-beta-D-glucosaminidase and gamma-glutamyltransferase in pregnant women who smoke and in those environmentally exposed to tobacco smoke and in their infants. Part I. Int J Occup Med Environ Health 10:187–202 [PubMed] [Google Scholar]
- Momen-Heravi F, Saha B, Kodys K, et al. (2015) Increased number of circulating exosomes and their microRNA cargos are potential novel biomarkers in alcoholic hepatitis. J Transl Med 13:261. doi: 10.1186/s12967-015-0623-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narayanan A, Iordanskiy S, Das R, et al. (2013) Exosomes derived from HIV-1-infected cells contain trans-activation response element RNA. J Biol Chem 288:20014–20033. doi: 10.1074/jbc.M112.438895 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pandrea I, Happel KI, Amedee AM, et al. (2010) Alcohol’s role in HIV transmission and disease progression. Alcohol Res Health 33:203–218 [PMC free article] [PubMed] [Google Scholar]
- Pottiez G, Jagadish T, Yu F, et al. (2012) Plasma proteomic profiling in HIV-1 infected methamphetamine abusers. PLoS ONE 7:e31031. doi: 10.1371/journal.pone.0031031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pretorius A (2014) Fibulin-1 as a marker of cardiovascular disease in HIV-infected black South Africans : a prospective study
- Pritchard MT, McMullen MR, Stavitsky AB, et al. (2007) Differential contributions of C3, C5, and decay-accelerating factor to ethanol-induced fatty liver in mice. Gastroenterology 132:1117–1126. doi: 10.1053/j.gastro.2007.01.053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rahimian P, He JJ (2016) Exosome-associated release, uptake, and neurotoxicity of HIV-1 Tat protein. J Neurovirol 22:774–788. doi: 10.1007/s13365-016-0451-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ranjit S, Kumar S (2018) Recent advances in cancer outcomes in HIV-positive smokers. F1000Res 7:. doi: 10.12688/f1000research.12068.1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren J-C, Zhu Q, Lapaglia N, et al. (2005) Ethanol-induced alterations in Rab proteins: possible implications for pituitary dysfunction. Alcohol 35:103–112. doi: 10.1016/j.alcohol.2005.03.004 [DOI] [PubMed] [Google Scholar]
- Revenfeld ALS, Bæk R, Nielsen MH, et al. (2014) Diagnostic and prognostic potential of extracellular vesicles in peripheral blood. Clin Ther 36:830–846. doi: 10.1016/j.clinthera.2014.05.008 [DOI] [PubMed] [Google Scholar]
- Ritsch A, Scharnagl H, Eller P, et al. (2010) Cholesteryl ester transfer protein and mortality in patients undergoing coronary angiography: the Ludwigshafen Risk and Cardiovascular Health study. Circulation 121:366–374. doi: 10.1161/CIRCULATIONAHA.109.875013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santoro Belangero P, Antônio Figueiredo E, Cohen C, et al. (2018) Changes in the expression of matrix extracellular genes and TGFB family members in rotator cuff tears. J Orthop Res. doi: 10.1002/jor.23907 [DOI] [PubMed] [Google Scholar]
- Shetty V, Jain P, Nickens Z, et al. (2011) Investigation of plasma biomarkers in HIV-1/HCV mono- and coinfected individuals by multiplex iTRAQ quantitative proteomics. OMICS 15:705–717. doi: 10.1089/omi.2011.0004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suriyaprom K, Harnroongroj T, Namjuntra P, et al. (2007) Effects of tobacco smoking on alpha-2-macroglobulin and some biochemical parameters in Thai males. Southeast Asian J Trop Med Public Health 38:918–926 [PubMed] [Google Scholar]
- Süsal C, Kirschfink M, Kröpelin M, et al. (1994) Complement activation by recombinant HIV-1 glycoprotein gp120. J Immunol 152:6028–6034 [PubMed] [Google Scholar]
- Süsal C, Kirschfink M, Kröpelin M, et al. (1996) Identification of complement activation sites in human immunodeficiency virus type-1 glycoprotein gp120. Blood 87:2329–2336 [PubMed] [Google Scholar]
- Takeda Y, He P, Tachibana I, et al. (2008) Double deficiency of tetraspanins CD9 and CD81 alters cell motility and protease production of macrophages and causes chronic obstructive pulmonary disease-like phenotype in mice. J Biol Chem 283:26089–26097. doi: 10.1074/jbc.M801902200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor P, Camp S, Radic Z (2009) Acetylcholinesterase In: Squire LR (ed) Encyclopedia of Neuroscience. Academic Press, Oxford, pp 5–7 [Google Scholar]
- Thomas PD, Campbell MJ, Kejariwal A, et al. (2003) PANTHER: A Library of Protein Families and Subfamilies Indexed by Function. Genome Res 13:2129–2141. doi: 10.1101/gr.772403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tungtrongchitr R, Pongpaew P, Phonrat B, et al. (2002) The effect of cigarette smoking on ceruloplasmin and C3 complement: risk of cardiovascular disease (atherosclerosis). Asian Pac J Allergy Immunol 20:23–28 [PubMed] [Google Scholar]
- Varma VR, Varma S, An Y, et al. (2017) Alpha-2 macroglobulin in Alzheimer’s disease: a marker of neuronal injury through the RCAN1 pathway. Mol Psychiatry 22:13–23. doi: 10.1038/mp.2016.206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vinchi F, Costa da Silva M, Ingoglia G, et al. (2016) Hemopexin therapy reverts heme-induced proinflammatory phenotypic switching of macrophages in a mouse model of sickle cell disease. Blood 127:473–486. doi: 10.1182/blood-2015-08-663245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang AL, Lukas TJ, Yuan M, et al. (2009) Changes in retinal pigment epithelium related to cigarette smoke: possible relevance to smoking as a risk factor for age-related macular degeneration. PLoS ONE 4:e5304. doi: 10.1371/journal.pone.0005304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W, Yang X, Liang J, et al. (2013) Cigarette smoking has a positive and independent effect on testosterone levels. Hormones (Athens) 12:567–577 [DOI] [PubMed] [Google Scholar]
- Wyatt RJ, Bridges RB, Halatek DG (1981) Complement levels in cigarette smokers: elevation of serum concentrations of C5, C9, and C1-inhibitor. J Clin Lab Immunol 6:131–135 [PubMed] [Google Scholar]
- (2014) Extracellular Vesicles in Health and Disease. In: CRC Press; https://www.crcpress.com/Extracellular-Vesicles-in-Health-and-Disease/Harrison-Gardiner-Sargent/p/book/9789814411981. Accessed 30 Jul 2018 [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Table S1 online: The list of proteins identified and quantified in all the studied groups
Supplementary Table S2 online: Significantly altered proteins in the study groups
a. Healthy V s HIV -positive subj ects
b. Healthy Vs Drinkers
c. Healthy vs HIV+drinkers
d. Healthy vs Smokers
e. Drinkers vs HIV+drinkers
f. Smokers vs HIV+smokers
g. HIV vs HIV+drinkers
h. HIV vs HIV+smokers
Supplementary Table S3 online. Significantly enriched pathways according to DAVID analysis.
a. Drinkers vs HIV+ Drinkers
b. Smokers vs HIV+ Smokers
c. HIV vs HIV+Drinkers
d. HIV vs HIV+ Smokers












