Significance
Gene drive can spread beneficial traits through populations, but will never be a one-shot project in which one genetic element provides all desired modifications, for an indefinitely long time. Here, we show that gene drive-mediated population modification in Drosophila can be overwritten with new content while eliminating old, using Cleave and Rescue (ClvR) selfish genetic elements. The ability to carry out cycles of modification that create and then leave behind a modest genetic footprint while entering and exiting a population provides important points of control. It makes possible the replacement of broken elements, upgrades with new elements that better carry out their tasks, and/or provide new functions, all while promoting the removal of modifications no longer needed.
Keywords: gene drive, Cas9, population replacement, selfish genetic element
Abstract
Gene drive-based strategies for modifying populations face the problem that genes encoding cargo and the drive mechanism are subject to separation, mutational inactivation, and loss of efficacy. Resilience, an ability to respond to these eventualities in ways that restore population modification with functional genes, is needed for long-term success. Here, we show that resilience can be achieved through cycles of population modification with “Cleave and Rescue” (ClvR) selfish genetic elements. ClvR comprises a DNA sequence-modifying enzyme such as Cas9/gRNAs that disrupts endogenous versions of an essential gene and a recoded version of the essential gene resistant to cleavage. ClvR spreads by creating conditions in which those lacking ClvR die because they lack functional versions of the essential gene. Cycles of modification can, in principle, be carried out if two ClvR elements targeting different essential genes are located at the same genomic position, and one of them, ClvRn+1, carries a Rescue transgene from an earlier element, ClvRn. ClvRn+1 should spread within a population of ClvRn, while also bringing about a decrease in its frequency. To test this hypothesis, we first show that multiple ClvRs, each targeting a different essential gene, function when located at a common chromosomal position in Drosophila. We then show that when several of these also carry the Rescue from a different ClvR, they spread to transgene fixation in populations fixed for the latter and at its expense. Therefore, genetic modifications of populations can be overwritten with new content, providing an ongoing point of control.
Alleles of genes that confer desired traits are often unlikely to confer an overall fitness benefit on those that carry them (1, 2), particularly if the trait of interest ultimately results in death of carriers (3, 4). In consequence, specific strategies are needed to bring about an increase in the frequency of these genes in wild populations. Gene drive occurs when particular genetic elements—genes, gene complexes, or large chromosomal regions—are transmitted to viable, fertile progeny at rates greater than those of competing allelic variants or other parts of the genome. Transgenes, or alleles of endogenous loci, can be linked with a genetic element conferring drive, and this can promote their spread. A number of approaches to spreading traits through populations (population replacement/alteration/modification) in ways that are self-sustaining, by linking them with genetic elements that mediate drive, have been proposed (5–22). Several of these, Medea (9, 23), UDmel (15), engineered translocations (24), and ClvR (Cleave and Rescue) selfish genetic elements (25), have been implemented and shown to spread to transgene fixation in otherwise wild-type (WT) Drosophila. Sustained modification of a WT mosquito population using a homing-based strategy, resulting in population suppression, has also been reported (26, 27).
Any strategy to modify wild populations must contend with the inevitability of mutation and evolution in response to natural selection. Specifically, genes encoding cargo and constituting the drive mechanism are subject to separation and mutation to inactivity. Such mutations can result in loss of a functional cargo from the population if chromosomes carrying the inactive cargo, an empty drive element, or components of a drive element are more fit than those carrying the full complement of active components. Evolution at other loci in the host, or in a pathogen the cargo is designed to target, can also occur such that the cargo becomes ineffective. Gene drive systems must be made robust—able to withstand forces leading to disruption—in order to delay the breakdown of a functional element. Examples of mechanisms to generate robustness in gene drive for population modification include multiplexing of components required for drive (25, 28, 29); interleaving of drive and Cargo components so as to prevent the creation of recombinant chromosomes that carry an empty drive element or an antidote-only allele (in the case of drive elements that utilize a toxin and antidote) (9, 25); introducing multiple copies of a gene designed to inhibit pathogens; and by using multiple genes that target the pathogen through diverse mechanisms. However, these methods only delay failure, since none of them provides permanent protection against evolution through natural selection in response to all kinds of mutations and genetic diversity. Thus, population modification strategies must also be resilient—able to recover from a breakdown in ways that maintain or restore effective population modification over time. Releases into the wild of first-generation elements may be dependent on the availability—or at least plausibility—of such strategies.
In principle, resilience can be achieved if a new, second-generation drive element can spread within a population fixed for an old element that has failed or lost efficacy. Because second-generation elements are subject to the same evolutionary forces as first-generation elements, it should also be possible to carry out additional cycles of modification. In consequence, a drive mechanism able to achieve resilience will likely use orthogonally acting components such that the presence of old drive elements does not interfere with drive by a newer-generation element. For related reasons, the components that make up a resilient drive mechanism should in some sense be indefinitely extensible in terms of the ability to create orthogonally acting new drive elements. Finally, an ideal system would also minimize genomic clutter from the accumulation of earlier generation nonfunctional elements and/or their components: cargo genes that have lost activity or have undesired effects; drive element components such as guide RNAs (gRNAs) that continue to create new loss-of-function (LOF) alleles at old essential gene loci and which may compete for loading into Cas9 with gRNAs from a current generation element (see below); and dominant markers that serve no purpose and may interfere with monitoring the behavior of newer generation elements. In consequence, drive and population modification with a new element should result in a contemporaneous decrease in frequency of drive elements from earlier generations.
Several strategies for altering the composition of a population following an initial modification by gene drive have been proposed. For gene drive that relies on homing of a transgene cassette into a particular genomic location, second-generation elements can be devised that home themselves into target sequences found in first-generation elements, thereby overwriting old modifications with new ones (16, 30). Populations containing homing-based drive elements can also be altered through the introduction of elements that lack Cas9 but carry gRNAs and potentially other cargo (31, 32). These, depending on whether they are located at the same site as the first element or elsewhere, can lead to loss of Cas9 and/or other components in the first-generation element as they home into the locus and, in some cases, also bring about gRNA-mediated alterations in other regions of the genome. Many other selfish genetic elements found in nature (33), or envisioned as fully synthetic entities (9, 11, 12, 14, 18, 22, 25, 34, 35), can be represented as consisting of a pair of genes sitting at a fixed chromosome position, with one gene encoding a toxin and the other encoding an antidote. Expression of these components in different spatial and temporal patterns results in the death of some or all nonelement-bearing progeny—a killing of the other that can lead to a relative increase in the frequency of element-bearing individuals. Medea and Cleave and Rescue (ClvR) elements are paradigmatic of this type. Because this kind of element does not spread by copying itself, a different kind of approach for achieving cycles of modification is needed (9, 25, 36). In the sections below, we focus on strategies for overwriting ClvR gene drive, but similar principles apply to many chromosome-based toxin-antidote drive systems.
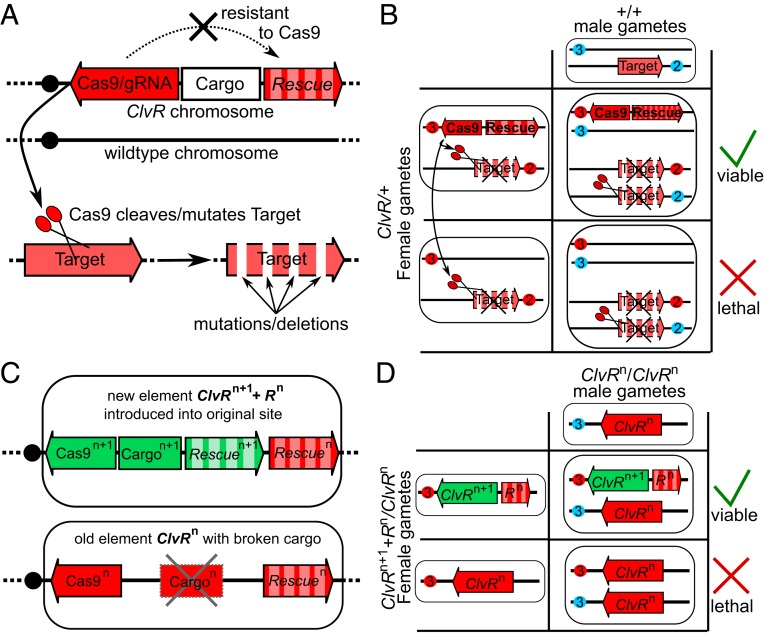
ClvR selfish genetic elements (25), also referred to as Toxin Antidote Recessive Embryo (TARE) in a related implementation (22), comprise two components, a DNA sequence-modifying enzyme such as Cas9 and gRNAs (the toxin/Cleaver) that acts in trans to disrupt the endogenous version of an essential gene through cleavage and inaccurate repair, and a recoded version of the essential gene resistant to cleavage (the antidote/Rescue). When these two components are tightly linked, Cas9 and gRNAs create potentially lethal LOF alleles of the essential gene, wherever it is located (Fig. 1A). However, the lethal LOF phenotype only manifests itself in those who fail to inherit ClvR. In contrast, those who inherit ClvR survive because they always inherit the recoded copy of the essential gene (Fig. 1B). Thus, ClvR spreads by killing those who lack it, creating a population that ultimately becomes dependent on—addicted to—the ClvR-encoded Rescue transgene. ClvR is in principle broadly extensible within a species since any gene that is essential for survival or fertility can be targeted for cleavage and rescue. These components are also orthogonally acting since Cas9/gRNAs only create mutations in the gene to which the gRNAs have homology, and the recoded Rescue transgene only rescues LOF phenotypes due to mutations induced by a gene-specific Cas9/gRNA complex.
Fig. 1.
First- and second-generation ClvR elements and their genetic behavior. (A) Components that make up a ClvR element. (B) A Punnett square highlighting the genetic mechanism by which ClvR elements bias inheritance in their favor. Maternal chromosomes are indicated with a red circle (centromere) and paternal chromosomes in blue. In a heterozygous female, Cas9 (located in the ClvR on chromosome 3 in this example) cleaves and mutates to LOF the endogenous target gene on chromosome 2, in the germline. In addition, active Cas9/gRNA complexes are deposited maternally into all eggs. After mating with a WT male, maternally transmitted Cas9/gRNA cleaves/mutates the target gene on the paternal chromosome. Progeny that do not inherit ClvR, and its recoded Rescue die because they lack essential gene function. (C) A female transheterozygous for ClvRn, which has an inactive Cargon, and ClvRn+1, which carries a new Cargon+1. (D) Gamete and progeny genotypes for a cross between the transheterozygous ClvRn+1/ClvRn female and a homozygous ClvRn/ClvRn male. Chromosomes carrying the two different essential genes being targeted (as in A and B) are not indicated for clarity of presentation.
In earlier work, we briefly outlined a general method by which cycles of population modification with the above characteristics—orthogonally acting components, indefinite extensibility, and contemporaneous removal of earlier generation elements—could in principle be carried out utilizing chromosomally located Medea (9, 36) or ClvR (25) selfish genetic elements, which each utilize a toxin-antidote–based mechanism of action. In brief, these approaches involve creating a series of next-generation drive elements in which each new element competes with and, ultimately, displaces a first or earlier generation element. The basic strategy for carrying out cycles of modification with ClvR is illustrated in Fig. 1 C and D. In this scenario, the original ClvR is known as ClvRn. A second-generation ClvR, ClvRn+1, which is meant to spread and supplant ClvRn, is located at the same position in the genome. Therefore, meiotic recombination cannot bring both ClvRs onto the same chromosome, and they are forced to compete with each other for inheritance in viable progeny through the use of different combinations of Cas9/gRNA toxins and recoded Rescue antidotes. ClvRn targets essential genen for cleavage and rescue. ClvRn+1 targets essential genen+1 for cleavage and rescue, while also carrying the Rescue transgene for essential genen. Because progeny carrying ClvRn or no ClvR element (WT) are sensitive to loss of essential genen+1, those carrying ClvRn+1 have a survival advantage, regardless of their status with respect to ClvRn. Meiosis remains fair and both elements are inherited by progeny in normal Mendelian ratios, but the survival of progeny is biased toward those carrying ClvRn+1. As a result—and provided that the fitness costs associated with carrying ClvRn+1 are less than those experienced by ClvRn and non- ClvR–bearing chromosomes in response to ClvRn+1-dependent killing (25, 34)—the ClvR-bearing chromosome ClvRn+1 is expected to spread into populations of ClvRn (and WT), while bringing about a corresponding decrease in the frequency of ClvRn. A Punnett square example that illustrates this behavior, in which a female transheterozygous for both elements mates with a ClvRn male, is presented in Fig. 1D.
Here, we show that cycles of modification with ClvR elements can be achieved in Drosophila. We first show that when multiple ClvRs, each targeting a different essential gene, are located at a common chromosomal position, they show drive, resulting in rapid spread to transgene fixation in WT populations. We then show that when several of these ClvR elements also carry the Rescue transgene from a different element, a ClvRn element that has failed in some way and needs to be supplanted, the former—now a ClvRn+1 element—spreads to transgene fixation at the expense of ClvRn, and at the expense of WT. These results show that ClvR is extensible with orthogonally acting components and that population modifications can be overwritten with new instructions while eliminating old ones. These features provide important points of control with respect to replacement of broken elements, upgrades with new elements that better carry out their original jobs and/or provide new functions, and removal of old elements whose presence is no longer desired.
Results
Synthesis of Two ClvR Elements at the Same Genomic Position as ClvRtko.
Cycles of population modification with ClvR elements can be carried out in two ways. First, ClvR elements could simply be introduced at new sites in the genome. If new elements freely recombine with old elements, and use orthogonally acting components (different gRNAs and essential genes), a new round of modification should ensue in populations that carry one or more versions of earlier generation elements, at the same rate as for the first-generation element. However, because the components of each ClvR are orthogonally acting, the earlier generation elements and their remnants will remain in the population at frequencies determined by natural selection. This creates the unwanted genomic clutter discussed above. Here, we focus on the alternative strategy in which orthogonally acting ClvR elements are located at the same position in the genome, an arrangement that forces them to compete for survival.
This latter strategy requires that multiple, independently acting ClvR elements show gene drive when located at a common position in the genome. This outcome is not inevitable since local and more global effects of gene location and interaction with near and distant regulatory elements, as well as large-scale chromosomal structure, can influence the expression patterns of genes (37), and the genes required for the essential cell functions that we target (below) must be expressed at sufficient levels in all cells. This potential problem is compounded when one considers that in order for drive of a next-generation element to occur both Rescue transgenes and their regulatory elements must work well, juxtaposed next to each other, in this genomic context.
We previously reported the creation of a single ClvR element, ClvRtko, in Drosophila melanogaster (25). ClvRtko is located on the D. melanogaster third chromosome at map position 68E, spreads rapidly into WT D. melanogaster populations, and is functional in populations from five continents (25). To determine if new, orthogonally acting ClvRs can be created at this same genomic position, we synthesized two ClvR elements using the same approach as for ClvRtko. The targeted essential genes were dribble (dbe) and Transcription-factor-IIA-S (TfIIA-S). Dbe is located at 21E2 on chromosome 2 and encodes a protein required for processing of cytoplasmic preribosomal RNA (38). TfIIA-S is located at 95C8 on chromosome 3 and encodes a small subunit of a basal transcription factor that is a part of the Pol-II transcription machinery (39). Both genes are recessive lethal and expressed ubiquitously in Drosophila. For the Rescue component of tko, we used the tko ortholog from the distantly related species Drosophila virilis. For the new ClvR elements, we used the target gene orthologs from Drosophila suzukii, an agricultural pest of major economic importance. As with ClvRtko, the toxin/Cleaver part of the constructs consisted of Cas9 under the control of the germline–specific nanos promoter, and 5′ and 3′ untranslated regions, and a set of four gRNAs designed to have homology with the D. melanogaster essential gene, but not the antidote/Rescue ortholog from D. suzukii, each expressed from a U6 promoter. The new ClvR elements also carried two dominant markers, ubiquitous opie-td-tomato and eye-specific 3xP3-GFP. A detailed description of construct assembly and fly germ-line transformation is given in Methods and SI Appendix, Figs. S1 and S2.
Synthesis of Second-Generation ClvRn+1 Elements.
A second-generation ClvR, ClvRn+1, consists of a ClvR that utilizes drive components that function orthogonally to those of ClvRn—a different toxin/Cleaver and a different antidote/Rescue—in addition to the antidote/Rescue from ClvRn. The toxin/Cleaver and both antidote/Rescues must work well in order for such an element to spread in populations fixed for ClvRn. To create such elements, we took flies that carried ClvRdbe and inserted into them the Rescue from ClvRtko, thereby creating a second-generation element, ClvRdbe+Rtko, designed to spread into populations of ClvRtko and wild type. We also created the converse second-generation ClvR, ClvRtko+Rdbe, designed to spread into populations of ClvRdbe and wild type. In each case, we first assembled a construct that had the desired Rescuen fragment and a dominant marker consisting of the ubiquitous opie promoter followed by a partial GFP open reading frame (ORF). These elements were flanked by homology arms matching the region surrounding the 3xP3 promoter-driven GFP marker within the first-generation ClvR element, whose components would now become Cas9/gRNAn+1 and Rescuen+1. First-generation ClvR flies were injected with this donor plasmid along with Cas9 protein preloaded with a gRNA that binds between the 3xP3 promoter and the GFP ORF. Once Cas9 creates a double-stranded break (DSB) between the 3xP3 promoter and GFP, the donor template can be used for repair, thereby, inserting the new Rescue. Positive transformants were identified by the change in GFP expression from eye-specific to ubiquitous. Correct insertion of Rescuen was confirmed by sequencing (see SI Appendix, Figs. S1 and S2 for details).
Genetic Behavior of First- and Second-Generation ClvR Elements.
We began our characterization of these four ClvR elements by determining the frequency with which the target essential gene was mutated to LOF in crosses involving female and male ClvR-bearing parents. For females, we crossed heterozygous ClvR/+ (+ for WT locus) virgins to WT males (see the cross with generic elements depicted in Fig. 1B) and scored progeny for the presence of the dominant ClvR marker td-tomato. ClvR frequency was calculated as the number of ClvR-bearing progeny divided by the total number of progeny. The cleavage rate to LOF is the number of ClvR-positive progeny divided by half the total number of progeny, since with Mendelian inheritance 50% of the progeny would be expected to inherit ClvR in the absence of ClvR-dependent killing. These percentages are a function of maternal germ-line cleavage and cleavage in the embryo due to maternal carryover of Cas9/gRNAs. Males carrying ClvR do not show paternal carryover of Cas9 at appreciable frequencies (25). To reveal the LOF mutation status of target loci that are exposed to Cas9/gRNAs in the adult male germline, we crossed heterozygous ClvR/+ males to females that carried a deficiency (Df) for the target gene in trans to a balancer chromosome, which is wild type at the target locus. This allowed us to calculate the male germ-line LOF mutation creation rate as the number of progeny carrying the Df and ClvR divided by half the total number of Df-bearing progeny, since those Df-bearing progeny not carrying ClvR must carry a version of the endogenous essential gene that still retains function and, with Mendelian inheritance, 50% of the Df-bearing progeny would be expected to inherit ClvR in the absence of ClvR-dependent killing. Results of these crosses are summarized in Table 1 and presented in detail in SI Appendix, Tables S1–S3. As with ClvRtko (25), the combined female germline and maternal carryover-dependent cleavage to LOF for all four elements was very high (>99%); the male germ-line cleavage rate to LOF for the two new first-generation ClvR elements was also high (from >94.7 to >99%). In short, the toxin component of the new ClvR elements is very efficient.
Table 1.
Genotype frequencies from crosses to determine the female and male cleavage rates to LOF
| Parental cross | ClvR positive | ClvR negative | ClvR frequency, % | Cleavage to LOF |
| First-generation ClvR elements | ||||
| ♀ClvRdbe/+ XX ♂w1118 | 5,972 | 4 | 99.93 | 99.87 |
| ♀ClvRTfIIA-S/+ XX ♂w1118 | 3,312 | 0 | >99.97 | >99.94 |
| ♂ClvRdbe XX ♀ Df(dbe)/CyO | 776 | 21 | 97.37 | 94.73 |
| ♂ClvRTfIIA-S XX ♀Df(TfIIA-S)/TM6B,Tb | 672 | 2 | 99.70 | 99.40 |
| Second-generation ClvR elements | ||||
| ♀(ClvRdbe+Rtko)/+ XX ♂w1118 | 3,003 | 0 | >99.97 | >99.94 |
| ♀(ClvRtko+Rdbe)/+ XX ♂w1118 | 3,599 | 0 | >99.97 | >99.94 |
Analysis of Target Chromosomes following Exposure to Cas9/gRNAs.
For all flies from Table 1 that did not inherit a LOF allele, we extracted genomic DNA and sequenced the target region. In addition, all male escapers were backcrossed to heterozygous ClvR/+ females, and progeny scored for the absence of ClvR, to determine if the chromosome that escaped LOF allele creation was still sensitive to Cas9 cleavage and LOF allele creation. Details of the characterization are presented in SI Appendix, Tables S4 and S5. To summarize, all of the escaper flies had at least 2 uncleaved WT target sites. Uncleavable target sites resulted mostly from small (3 bp) in-frame deletions or preexisting polymorphisms and one rare 2bp substitution, all of which most likely preserved at least partial gene function. For all of the escapers that we tested in a backcross to ClvR/+ females, cleavage rates to LOF remained high (85% and 94% for two single cases, 100% for all of the rest), indicating that the escaped chromosomes remained sensitive to ClvR-dependent LOF allele creation. Together these results are important because they provide further evidence that the ClvR approach to creation of LOF alleles utilizing four gRNAs is efficient and provides strong protection against the production of alleles at essential gene loci that have mutated target sites but retain essential gene function. Such resistant alleles can slow the rate of spread and decrease the functional lifetime of ClvR elements in the population (25).
We also characterized cleaved target sites that were mutated to LOF following exposure to Cas9/gRNAs. In the case of ClvRtko, all target sites could be cleaved and a variety of indels were created (25). To explore these topics with ClvRdbe and ClvRTfIIA-S, we characterized target sites following one generation of exposure to Cas9 and after 22 generations of a drive experiment (see Fig. 3). The goal in looking at two different timepoints was to gain a sense of which target sites were preferentially cleaved and if all target sites could be cleaved. In each case, we carried out the analysis by taking ClvR-bearing flies and crossing them individually to flies that carried a deficiency for the target gene. From the progeny, we selected one ClvR-bearing fly carrying a chromosome whose target locus had been exposed to Cas9/gRNAs, in trans to a deficiency to the region, and sequenced over the region between the four gRNA target sites. Sequencing results are summarized in SI Appendix, Tables S6–S9. For the dbe locus in ClvRdbe flies, sites 3 and 4 were mutated at a high frequency following a single generation of exposure to Cas9 (site 2 carried a single nucleotide polymorphism), and site 1 was altered in 3 of 16 analyzed flies. After 22 generations, however, all sites were altered. Products of cleavage included small deletions of varying sizes at single gRNA target sites, larger deletions between adjacent target sites, an inversion of the region between the outer target sites, and one fly had the whole locus deleted. In ClvRTfIIA-S, the mutation spectrum was similar after 1 and 22 generations of exposure to Cas9. Most of the sequenced target sites had a deletion between gRNA 1 and 2 and smaller deletions at target sites 3 and 4 (19 and 26 bp). Four flies had small deletions at each of the target sites and one fly had the whole region between gRNA 1 and 4 deleted. In summary, as with ClvRtko, all of the target sites ClvRdbe and ClvRTfIIA-S could be cleaved, and a variety of different LOF indels were observed.
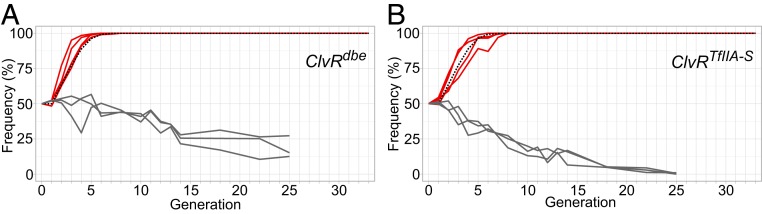
Fig. 3.
Gene drive experiments. Shown are the frequencies of ClvR-bearing flies (ClvR/+ and ClvR/ClvR) in a gene drive experiment with dbe (A) and TfIIA-S (B) as the targeted essential genes. Gene drive replicates are in red, modeled drive behavior for an element with no fitness cost in dotted black (see Methods for model details), and controls in gray.
ClvRdbe and ClvRTfIIA-S Spread to Genotype Fixation in WT Drosophila.
To explore the behavior of the four drive elements, ClvRdbe, ClvRTfIIA-S, ClvRtko+Rdbe, and ClvRdbe+Rtko, we carried out a set of six gene drive experiments, illustrated in Fig. 2, along with controls for each. We first tested the ability of ClvRdbe and ClvRTfIIA-S to spread in a WT population. As shown previously, ClvRtko spread to genotype fixation (>99.5% ClvR-bearing) within seven generations when introduced at a starting allele frequency of 25% (ref. 25 and SI Appendix, Fig. S6). The ability of ClvRdbe and ClvRTfIIA-S to spread was tested under similar conditions. The drive seed generation was set up by crossing male ClvR/+ heterozygous males to WT+/+ females, resulting in 50% of the population carrying one ClvR element in generation 1 (four replicates, with a starting ClvR allele frequency of 25%). Control drive experiments, as with ClvRtko, were performed by crossing males heterozygous for a cassette that includes the ClvR Rescue and td-tomato marker, but not Cas9 and gRNAs, to WT+/+ females (three replicates, with a starting control transgene allele frequency of 25%). Adult flies were allowed to lay eggs for 1 d in a food bottle. After ∼14 d, a large number of the eggs had developed into adults (∼700–1,000). At that time point, we sampled a random selection of the population and scored the frequency of the dominant ClvR (or control) marker. All of the scored flies were transferred to a fresh food bottle to repeat the cycle. Results are plotted in Fig. 3. Both ClvRdbe and ClvRTfIIA-S reached genotype fixation (100% of flies having one or two copies of ClvRdbe) between six and eight generations in all replicates. The control elements slowly decreased in frequency over time, perhaps due to some fitness cost associated with the ubiquitous expression of a dominant marker gene and/or the presence of additional copies (for a total of three to four in transgene-bearing individuals) of the essential target gene present in the Rescue only control element.
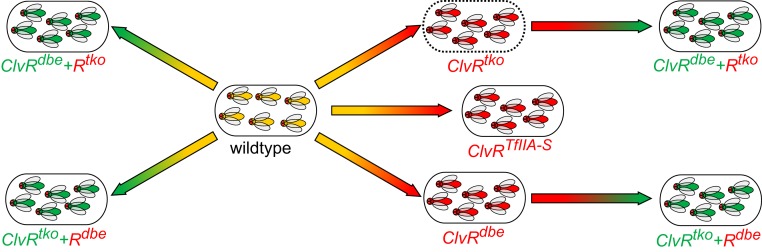
Fig. 2.
Gene drive experiments. Arrows proceed from WT, ClvRtko, or ClvRdbe to some other ClvR-bearing state. The blunt end of the arrow indicates the starting state, and the pointed end the population state (transgene fixation) drive is meant to achieve. Color gradients schematically reflect progress from starting state to end state. Controls for each drive experiment are discussed in the text and in Figs. 3 and 4. Experiments demonstrating drive of ClvRtko into a WT population (dashed outline) were published previously (25). See also SI Appendix, Fig. S6 for further characterization of the ClvRtko experiment over more generations.
After 22 generations (32 generations for ClvRtko), we assayed the allele frequencies in the drive populations (SI Appendix, Table S10). These ranged from 83 to 90% for ClvRdbe, 91–96% for ClvRTfIIA-S, and 94–100% for ClvRtko populations. These observations demonstrate that multiple ClvR elements, each targeting a different gene, can be generated and show gene drive when located at a common site in the genome.
Second-Generation ClvR Elements ClvRtko+Rdbe and ClvRdbe+Rtko Spread to Genotype Fixation.
Second-generation ClvR elements face several potential challenges to spread in WT and ClvRn-bearing populations. First, they carry an additional cargo in the form of the ClvRn Rescue transgene, which may introduce a fitness cost, particularly when drive occurs into a WT background in which the ClvRn Rescue does not function to support drive (it results in ClvR-bearing individuals having three or four functional copies of essential genen). Second, when driving into a population of ClvRn, both Rescue transgenes must work well and be able to rescue LOF phenotypes for two essential genes. Finally, when driving into a population of ClvRn, ClvRn+1 elements will also often find themselves in a transheterozygous state with the earlier generation element. These ClvRn elements include a different set of four gRNAs that do not contribute to drive by ClvRn+1. These could compete with the four gRNAs needed for drive by ClvRn+1 for loading into a complex with Cas9, thereby suppressing drive. To explore the ability of ClvRn+1 elements to thrive in different genetic backgrounds, we carried out drive experiments of ClvRn+1 elements into WT and ClvRn-bearing populations. Drive experiments of ClvRtko+Rdbe and ClvRdbe+Rtko into WT w1118 populations were carried out as with ClvRdbe and ClvRTfIIA-S above, by mating heterozygous ClvR/+ males to w1118; +/+ virgins, for a starting ClvR allele frequency of 25%. As a control for these experiments, we also carried out drive into populations fixed for a ClvR element carrying the same Rescue transgene as that needed for drive by ClvRn+1 (drive of ClvRtko+Rdbe into ClvRtko, and drive of ClvRdbe+Rtko into ClvRdbe). For the experiments involving drive into populations of ClvRdbe and ClvRtko (the controls above, and drive of ClvRtko+Rdbe and ClvRdbe+Rtko into ClvRdbe and ClvRtko, respectively) we crossed heterozygous ClvRn+1/+ males to virgin females taken from the ClvRdbe and ClvRtko drive populations at the generation where we determined allele frequency in the drive populations (generation 22 for ClvRdbe, Fig. 3; generation 32 for ClvRtko, SI Appendix, Fig. S6). These populations consist of mostly ClvR/ClvR homozygotes, with a few ClvR/+ individuals (SI Appendix, Table S10).
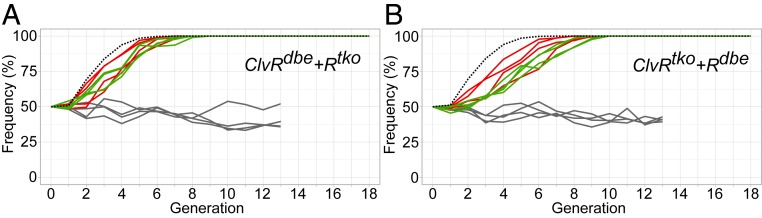
The outcomes of these gene drive experiments are shown in Fig. 4. ClvRdbe+Rtko reached genotype fixation between seven and eight generations when driving into a population of WT w1118; +/+, and between eight and nine generations when driving into a population of ClvRtko. As expected, when driving into a population of ClvRdbe, the second-generation ClvRdbe+Rtko drive element did not increase in frequency (Fig. 4A). ClvRtko+Rdbe performed similarly, although with slightly slower kinetics. ClvRtko+Rdbe reached genotype fixation between 8 and 10 generations when driving into a population of w1118 and between 9 and 10 generations when driving into populations of ClvRdbe. When driving into a population of ClvRtko, ClvRtko+Rdbe did not increase in frequency (Fig. 4B). For the experiments in which a ClvRn+1 was driven into a population of ClvRn (ClvRdbe+Rtko into ClvRtko and ClvRtko+Rdbe into ClvRdbe), we also measured the allele frequency of the ClvRn elements at generation 12. As expected based on the fact that ClvRn and ClvRn+1 share a common genomic location, the high frequency of ClvRn+1 at genotype fixation, with allele frequencies ranging from 84.9 to 91.2%, was associated with a dramatic decrease in the allele frequency of ClvRn, to between 8.2 and 14.5% (details in SI Appendix, Tables S11 and S12). Together these results demonstrate that second-generation ClvR elements can be created; they drive themselves into WT populations; they also drive into populations fixed for an earlier generation element, and they displace the latter as they spread. Thus, cycles of gene drive-mediated population modification can be achieved, while at the same time bringing about a decrease in the frequency of an earlier generation element.
Fig. 4.
ClvRn+1+Rn selfish elements drive into populations carrying ClvRn. Shown are the drive outcomes for ClvRdbe+Rtko driving into a population of w1118 (red lines), ClvRtko (green lines), and ClvRdbe (gray lines, control) (A), and ClvRtko+Rdbe driving into a population of w1118 (red lines), ClvRdbe (green lines), and ClvRtko (gray lines, control) (B). There are four replicates for each drive experiment. Dashed black lines represent model estimated behavior for the spread of ClvRn+1 with no fitness cost into populations consisting primarily of ClvRn population (see Methods for model details). Allele frequencies at generation 12 are presented in SI Appendix, Tables S11 and S12.
Discussion
Genetic modification of a population is unlikely to ever be a one-shot project, with a single genetic element providing all desired modifications, for an indefinitely long time. Mutation, recombination, and natural selection will cause a loss of drive and/or efficacy through one mechanism or another. In addition, as knowledge increases, there are likely to be situations in which one wants to augment (upgrade) a population modification or remove a change whose presence is no longer desired. For all these reasons, it is important that strategies for population modification be extensible for multiple cycles. At the same time, it is also important that introductions of new elements result in the loss (or at least a great decrease in the frequency) of old elements from the population.
Here, we show that a first set of cycles of modification with these properties can be achieved using ClvR selfish genetic elements. While modeling is required to provide a detailed analysis, one way in which it may be possible to carry out cycles of modification indefinitely is as a linear chain of elements, in which the newest element always carries the Rescue of the element from the previous generation, with each new element cleaving and rescuing a new essential gene. Importantly, while each element in the chain must carry the Rescue from the previous generation, it does not need to carry all previous Rescue transgenes from a longer chain. This is because in each new cycle WT alleles at previously targeted loci are brought in along with the new element (since they are not targeted by the current generation Cas9 and gRNAs). As new elements drive out earlier generation elements that carry Cas9 and gRNAs targeting these wildtype alleles, the wildtype alleles spread since they are needed to maintain essential gene function in the absence of Rescue transgenes also present in earlier generation elements. A corollary of this is that the footprint in the genome left by previous cycles of ClvR (LOF alleles at previously targeted loci) fades over time.
In order to achieve multiple cycles of population modification, the drive components need to be orthogonally acting and indefinitely extensible. The components of ClvR are orthogonally acting since Cas9/gRNAs and Rescue transgenes are specific to a particular essential gene. ClvR is also in principle broadly extensible within a species since any gene that is essential for survival or fertility can be targeted for cleavage and rescue. In our earlier work, we created ClvRtko, which is located on the third chromosome and targets an essential gene on the X. Here, we showed that additional ClvRs can be generated at the same site as ClvRtko, targeting essential genes involved in different biological processes, located on chromosomes 2 and 3. Further evidence for the extensibility of the ClvR system comes from recent work in which it was shown that a construct consisting of two gRNAs targeting the gene encoding the essential developmental transcription factor hairy (h) and a recoded rescuing version of h could, when located within the h locus, spread through a population homozygous for Cas9 at an independent locus through a ClvR-like mechanism (22). Spread to transgene fixation occurred rapidly for all four of these elements. Together these observations show that Cleave and Rescue type selfish genetic elements can successfully mutate to LOF and provide rescuing essential gene function for genes involved in a variety of cellular processes, and that no particular spatial relationship in the genome between the drive element and target gene is essential. We also note that while our work herein and in refs. 22 and 25 used Cas9, which generates LOF mutations by creating double-strand DNA breaks that are repaired inaccurately, similar LOF effects can be brought through any mechanism that modifies DNA site-specifically, including methods that do not create DSBs. Examples of other possible mechanisms include a Cas9/gRNA-linked base editor (40), a Cas9/gRNA nickase linked to reverse transcriptase (41), or a pair of site-specific engineered recombinases (42).
For the two ClvRs we generated here, and the one in ref. 25, a variety of indels were created in the essential genes targeted, and all sites were ultimately cleaved. Importantly, in our work and that of Champer et al. (22), alleles of the endogenous essential gene that were completely resistant to cleavage but retained function were not observed. We note that in ref. 25 and the current work, examples were found in which two of the four gRNA target sites were altered in ways that presumably retained function. In some cases, one of these was due to a preexisting polymorphism that was not screened for prior to initiating these experiments. However, other sequence differences were new and due to mutation associated with inaccurate DNA repair. Thus, we recommend multiplexing no less than four gRNAs to maintain functionality in genetically diverse populations. The feasibility of using more than four gRNA to bring about increased robustness in terms of LOF allele creation is suggested by our observation that ClvRn+1 elements showed drive even when in the presence of the four additional gRNAs present in ClvRn, which do not contribute to drive (Fig. 4).
These points notwithstanding, if a first-generation element does fail to spread due to the appearance of multiple resistant target sites in essential genen, this does not prevent the spread of a second-generation element, since such an element will target a different gene, essential genen+1. Mutations in Cas9 itself at significant frequencies have little effect on drive since the activity of remaining active drive elements eliminates WT essential gene alleles, which serves to drive the element lacking Cas9 into the population (25). However, if unlinked suppressor mutations that prevent Cas9 or gRNA expression or function arise and spread in the context of ClvRn, these could block drive of a ClvRn+1 element. It remains to be seen if such suppressor mutations exist. If they do, critical empirical questions will be their frequency and whether the fitness costs associated with specific ClvRs outweigh those associated with the presence of the suppressor, thereby promoting the spread of the latter while slowing or preventing drive. Finally, we note that when two different ClvRs are present in transheterozygous individuals, meiotic recombination could occur between conserved elements such as Cas9. Placing the components of first- and next-generation elements in specific configurations can prevent recombination from creating empty elements that carry Rescuen+1 but not Cargon+1 (SI Appendix, Fig. S9). Inverting the orientation with respect to the centromere of genes that share homology can also be used to ensure that any recombinants formed are on acentric or dicentric chromosomes.
ClvR-type gene drives can modify populations in a number of ways. In one family of approaches, ClvR spreads a LOF allele through the population. This can happen if ClvR is itself located within a gene of interest, thereby disrupting it and driving an increase in frequency of the disrupted allele as it spreads. Alternatively, the ClvR can carry gRNAs or microRNAs that target some other gene to bring about a loss or decrease in function, respectively. In a second family of approaches, ClvR can carry into populations cargo transgenes to which it is linked, or an allele of an endogenous locus to which it is tightly linked through insertion site choice. In all of these scenarios, drive only occurs when the fitness costs to non-ClvR–bearing individuals exceed those associated with being ClvR-bearing, attributes that are frequency-dependent (25, 34). Finally, while more speculative, we note that the mechanism by which ClvR-based population modification occurs provides some unique opportunities for strategies that prevent disease transmission by bringing about the death of host cells and/or hosts in response to infection, or that bring about periodic overall population suppression in response to a cue from the environment. These ideas each take advantage of a key feature of ClvR-dependent drive—that individuals of the modified population are absolutely dependent on the functionality of the Rescue transgene for survival. Given this it is interesting to imagine ways in which the function of the Rescue transgene could be made conditional so as to bring about the death of cells, individuals, or populations under specific circumstances. For example, it may be possible to engineer essential gene function at the level of transcript or protein such that it is sensitive to the presence of viral protease activity (4), small RNAs (43), or other honest markers of infection, resulting in the death of infected host cells or individuals. One can also imagine ways in which entire ClvR-bearing populations could be suppressed in an environmental condition-specific manner. Temperature, to give an example, is often an important seasonal environmental variable. Gene function can be made temperature-sensitive in several ways. A temperature-sensitive intein can be incorporated into the coding region of the essential gene (44). Because self-splicing from the encoded protein is temperature-dependent, survival of a population in which all WT alleles are LOF should be so as well. Alternatively, a temperature-sensitive degron could be linked to the coding region for a similar effect (45). Strategies for engineering essential gene function to be sensitive to the presence of specific chemicals could use a similar strategy, in which protein degradation occurs in response to binding of a small molecule ligand to a specific protein domain (46–48).
One can imagine scenarios in which each of the above approaches to population modification with ClvR is successful and efficacious. However, a similar analysis of each will also identify multiple mechanisms by which drive and efficacy of any cargo can fail over time. This does not mean that population modification should not be attempted. Interventions in any area of biology that involves the forces of mutation and selection typically come with periods of success followed by failure. The evolved resistance of cancers to specific therapies, of bacteria to antibiotics and phage therapy, of plasmodium to antimalarial drugs, of insects and plants to insecticides and herbicides, and the yearly battle of the human immune system and the latest vaccines against the current strain of influenza all reflect the ubiquitous nature of this cycle. The important thing is to have a plan that allows for the continual evolution and implementation of an initially successful strategy. In the case discussed herein, where the goal is to alter the genetic composition of a population toward a specific functional end, the ability to iteratively carry out new modifications while removing old ones provides the essential underpinnings of any plan for long-term success.
Methods
Restriction enzymes, Gibson Assembly enzymes, Q5 and Longamp DNA polymerases were from New England Biolabs. Genomic DNA extraction kit (Quick-DNA 96 Plus Kit), mini plasmid prep (ZymoPURE Plasmid Miniprep), and gel extraction kit (Zymoclean Gel DNA Recovery Kit) were from Zymo Research. DNA maxiprep kit was from Qiagen (EndoFree Plasmid Maxi Kit). All plasmids were cloned with Gibson assembly (49) as described previously (25). All fly embryonic injections were performed by Rainbow Transgenic Flies. Cloning construct design, CRISPR guide design (50), and sequencing alignments with MAFFT (51) were done in the Benchling software suite. All primers, gRNA target sequences, and construct GenBank files are in Dataset S1.
Cloning of ClvR Constructs and Generation of First-Generation ClvR Flies.
First-generation ClvR flies (ClvRdbe and ClvRTfIIA-S) were generated in two steps as described previously (25). We first inserted the Rescue part into the fly genome, followed by integration of the Cleaver (Cas9 and gRNAs) at that same site. The first construct had the Rescue of the target gene, which was amplified from genomic DNA of D. suzukii (Dsuz). The Rescue fragments contained the ORF of the target gene as well as upstream and downstream sequences with potential promoter/enhancer and terminator elements. The Dsuz-dbe Rescue fragment was 2 kb; the Dsuz-TfIIA-S fragment was 3.8 kb (annotated fasta files in SI Appendix, Figs. S7 and S8). In addition, the construct had an opie-td-tomato dominant marker and an attP site. All these elements were flanked by homology arms to facilitate CRISPR-mediated homologous recombination (HR) into the fly genome. Outside the homology arms, the constructs had a U6-driven gRNA that targeted a site at 68E on the third chromosome of Dmel (SI Appendix, Fig. S1A).
The constructs were injected into a stock that had nos-Cas9 on the X chromosome (SI Appendix, Fig. S2A) (52). G0-injected flies were outcrossed to w1118 and the progeny screened for ubiquitous td-tomato expression. Male transformants that came from a male G0 fly were outcrossed again to w1118 to build up a stock. At this point, the Cas9 source on the X chromosome of the injection strain was bred out.
The second part of the ClvR element (the Cleaver) was assembled separately and had Cas9 driven by germ-line–specific nos promoter and UTRs (53) (nos-Cas9 derived from addgene plasmid 62208; ref. 52). A set of four gRNAs were each driven from alternating pairs of U6:3 and U6:1 promoters (29) (similar as in ref. 52). The plasmid further had an attB site to facilitate integration into the genomic location of the first construct and a 3xP3-GFP transformation marker (SI Appendix, Fig. S1B).
This construct was injected into flies that carried the Rescue alongside a helper plasmid as a source of phiC31 integrase. G0-injected flies were outcrossed to w1118 and screened for eye-specific expression of GFP (SI Appendix, Fig. S2B).
Generation of Second-Generation ClvRn+Rn-1 Flies.
We used CRISPR-mediated homologous recombination to modify the ClvR locus of the strain of flies that would become ClvRn+1. A Cas9/gRNA ribonucleoprotein (RNP) complex binding between the 3xP3 promoter and the GFP ORF at the original ClvR locus was injected into ClvR flies alongside a donor plasmid to be inserted via CRISPR HR (SI Appendix, Fig. S2C). The RNP complexes were assembled by mixing Cas9 protein (Alt-R, IDT) and gRNA (sgRNA, IDT) in water and incubating for 5 min at room temperature. Afterward, the donor plasmid was added, and the mixture was stored at −80 °C until injection. Final concentrations in the injection mix were as follows: Cas9 protein 500 ng/μL, gRNA 100 ng/μL, donor plasmid 500 ng/μL. The donor plasmid contained the Rescue of ClvRn and an opie2 promoter (54) with partial GFP sequences that acted as the homology arm. The other homology arm was the 3xP3 promoter and plasmid backbone (SI Appendix, Fig. S1C). We injected a construct carrying the D. suzukii-dbe Rescue into ClvRtko flies to generate second-generation ClvRtko+Rdbe flies. We also injected a construct carrying the D. virilis derived tko Rescue into ClvRdbe flies to create ClvRdbe+Rtko flies. Successful integration of the Rescuen construct was detected by ubiquitous GFP expression. To confirm the integration of the new Rescue at the correct genomic location, we extracted genomic DNA from GFP-positive flies and amplified a fragment with primers binding in the 3xP3 promoter and the nanos 3′UTR downstream of Cas9. The resulting PCR fragments were partially sequenced to confirm that the new Rescue was downstream of 3xP3 and that opie-GFP was downstream of the nos 3′UTR (SI Appendix, Fig. S2C).
Crosses to Determine Male and Female Cleavage Rates to LOF.
We crossed ClvR-bearing males to w1118 virgins to get heterozygous ClvR/+ male and female offspring. To determine the female cleavage rate to LOF, we took ClvR/+ virgins, crossed them to w1118 males, and scored the progeny for the ClvR marker td-tomato. ClvR frequency was calculated as number of td-tomato flies divided by the total number of flies (SI Appendix, Table S1). The cleavage rate to LOF is ClvR-positive progeny divided by half the total progeny, since with Mendelian inheritance 50% of the progeny would be expected to inherit ClvR in the absence of ClvR-dependent killing. The same cross was performed with second-generation ClvRn+1+Rn flies (SI Appendix, Table S3).
For the male ClvR frequency, we crossed heterozygous ClvR/+ males to a stock that carried a deficiency for the target gene. ClvR frequency was calculated by determining the fraction of the total carrying the Df (target essential gene) that were also ClvR-bearing (S2).
Sequencing Analysis of Escapers and Cleavage Events.
Whenever possible we isolated the chromosome that we wanted to sequence over a Df for the essential gene so that there was only one version of the essential gene available. This was done for all sequenced flies except for the four escapers coming from heterozygous ClvRdbe/+ females (SI Appendix, Tables S4 and S5). Genomic DNA was extracted with the Quick-DNA 96 Plus Kit from Zymo. For dbe, we amplified a 2.2-kb genomic region spanning all target sites with primers dbe-genomic-F and dbe-genomic-R and Sanger-sequenced that amplicon with primers dbe-seq-F and dbe-seq-R. For TfIIA-S we amplified a 1.9-kb genomic region with primers tf2-genomic-F and tf2-genomic-R. This amplicon was Sanger-sequenced with primer tf2-genomic-R. See SI Appendix, Fig. S3 for a schematic of the genomic regions and primer binding sites.
Gene Drive Experiments.
All ClvR drive experiments were set up as described previously (25). We crossed heterozygous ClvR/+ males to w1118 virgins in bottles of fly food. After 2 d, the adults were removed and the progeny (seed generation = 0, ClvR-bearing = 50%, allele frequency = 25%) was allowed to eclose in the bottles. After 13–14 d, eclosed flies were anesthetized on a CO2 pad and a random sample of ∼200 flies was scored for the dominant ClvR marker. This sample was then transferred to a bottle with fresh food to continue the next generation. All counts of gene drive experiments are in Dataset S2.
ClvR Computational Model.
Figs. 3 and 4 feature model predicted behavior for a ClvR driving into wild type, or a second-generation ClvRn+1 driving into a population fixed for a first generation, ClvRn, with 0% fitness costs as well as 100% cleavage and maternal carryover rates. We used a deterministic, population proportion model adjusted from a model we have used previously (25), which uses difference equations to track the frequency of each genotype over discrete generations. In this model, we assumed that there is random mating; females produce offspring from a single mating; cleavage occurs during gametogenesis; maternal carryover of Cas9 and gRNAs can cleave any uncleaved allele in the zygote, such as that coming from the father; being heterozygous for a cleaved allele has no fitness effects (the locus is haplosufficient); and two copies of the cleaved target without a Rescue results in death 100% of the time.
Fly Crosses and Husbandry of ClvRtko Flies.
Fly husbandry and crosses were performed under standard conditions at 26 °C. Rainbow Transgenic Flies carried out all of the embryonic injections for germ-line transformation. Containment and handling procedures for ClvR flies were as described previously (29), with G.O and B.A.H. performing all fly handling.
Data Availability.
All data are available in the main text and the supplementary materials. ClvR flies are available on request under the conditions outlined in ref. 25.
Supplementary Material
Acknowledgments
Stocks obtained from the Bloomington Drosophila Stock Center (NIH Grant P40OD018537) were used in this study. This work was carried out with support from the US Department of Agriculture (USDA), National Institute of Food and Agriculture (NIFA) specialty crop initiative under USDA NIFA Award 2012-51181-20086, California University of Technology, and a Beaufort Visiting Fellow award from St. John’s College, Cambridge, UK (to B.A.H.). G.O. was supported by a Baxter Foundation Endowed Senior Postdoctoral Fellowship. T.I. was supported by NIH Training Grant 5T32GM007616-39.
Footnotes
Competing interest statement: The authors have filed patent applications on ClvR and related technologies (U.S. Application No. 15/970,728).
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.1921698117/-/DCSupplemental.
References
- 1.Schmid-Hempel P., Evolutionary ecology of insect immune defenses. Annu. Rev. Entomol. 50, 529–551 (2005). [DOI] [PubMed] [Google Scholar]
- 2.Tripet F., Aboagye-Antwi F., Hurd H., Ecological immunology of mosquito-malaria interactions. Trends Parasitol. 24, 219–227 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Carter J. R., et al. , Suppression of the arboviruses dengue and chikungunya using a dual-acting group-I intron coupled with conditional expression of the bax C-terminal domain. PloS One 10, e0139899 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gao X. J., Chong L. S., Kim M. S., Elowitz M. B., Programmable protein circuits in living cells. Science 361, 1252–1258 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Braig H. R., Yan G., “The spread of genetic constructs in natural insect populations” in Genetically Engineered Organisms: Assessing Environmental and Human Health Effects, Letourneau D. K., Burrows B. E., Eds. (CRC Press, 2001), pp. 251–314. [Google Scholar]
- 6.Davis S., Bax N., Grewe P., Engineered underdominance allows efficient and economical introgression of traits into pest populations. J. Theor. Biol. 212, 83–98 (2001). [DOI] [PubMed] [Google Scholar]
- 7.Burt A., Site-specific selfish genes as tools for the control and genetic engineering of natural populations. Proc. Biol. Sci. 270, 921–928 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gould F., Schliekelman P., Population genetics of autocidal control and strain replacement. Annu. Rev. Entomol. 49, 193–217 (2004). [DOI] [PubMed] [Google Scholar]
- 9.Chen C.-H., et al. , A synthetic maternal-effect selfish genetic element drives population replacement in Drosophila. Science 316, 597–600 (2007). [DOI] [PubMed] [Google Scholar]
- 10.Altrock P. M., Traulsen A., Reeves R. G., Reed F. A., Using underdominance to bi-stably transform local populations. J. Theor. Biol. 267, 62–75 (2010). [DOI] [PubMed] [Google Scholar]
- 11.Marshall J. M., Hay B. A., Inverse Medea as a novel gene drive system for local population replacement: A theoretical analysis. J. Hered. 102, 336–341 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Marshall J. M., Pittman G. W., Buchman A. B., Hay B. A., Semele: A killer-male, rescue-female system for suppression and replacement of insect disease vector populations. Genetics 187, 535–551 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Altrock P. M., Traulsen A., Reed F. A., Stability properties of underdominance in finite subdivided populations. PLoS Comput. Biol. 7, e1002260 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Marshall J. M., Hay B. A., General principles of single-construct chromosomal gene drive. Evolution 66, 2150–2166 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Akbari O. S., et al. , A synthetic gene drive system for local, reversible modification and suppression of insect populations. Curr. Biol. 23, 671–677 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Esvelt K. M., Smidler A. L., Catteruccia F., Church G. M., Concerning RNA-guided gene drives for the alteration of wild populations. eLife 3, e03401 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gokhale C. S., Reeves R. G., Reed F. A., Dynamics of a combined Medea-underdominant population transformation system. BMC Evol. Biol. 14, 98 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Reeves R. G., Bryk J., Altrock P. M., Denton J. A., Reed F. A., First steps towards underdominant genetic transformation of insect populations. PloS One 9, e97557 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sudweeks J., et al. , Locally fixed alleles: A method to localize gene drive to island populations. Sci. Rep. 9, 15821 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nash A., et al. , Integral gene drives for population replacement. Biol. Open 8, bio037762 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.López Del Amo V., et al. , A transcomplementing gene drive provides a flexible platform for laboratory investigation and potential field deployment. Nat. Commun. 11, 352 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Champer J., et al. , A toxin-antidote CRISPR gene drive system for regional population modification. Nat. Commun. 11, 1082 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Akbari O. S., et al. , Novel synthetic Medea selfish genetic elements drive population replacement in Drosophila; a theoretical exploration of medea-dependent population suppression. ACS Synth. Biol. 3, 915–928 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Buchman A. B., Ivy T., Marshall J. M., Akbari O. S., Hay B. A., Engineered reciprocal chromosome translocations drive high threshold, reversible population replacement in Drosophila. ACS Synth. Biol. 7, 1359–1370 (2018). [DOI] [PubMed] [Google Scholar]
- 25.Oberhofer G., Ivy T., Hay B. A., Cleave and Rescue, a novel selfish genetic element and general strategy for gene drive. Proc. Natl. Acad. Sci. U.S.A. 116, 6250–6259 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kyrou K., et al. , A CRISPR-Cas9 gene drive targeting doublesex causes complete population suppression in caged Anopheles gambiae mosquitoes. Nat. Biotechnol. 36, 1062–1066 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pham T. B., et al. , Experimental population modification of the malaria vector mosquito, Anopheles stephensi. PLoS Genet. 15, e1008440 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Champer J., et al. , Reducing resistance allele formation in CRISPR gene drive. Proc. Natl. Acad. Sci. U.S.A. 115, 5522–5527 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Oberhofer G., Ivy T., Hay B. A., Behavior of homing endonuclease gene drives targeting genes required for viability or female fertility with multiplexed guide RNAs. Proc. Natl. Acad. Sci. U.S.A. 115, E9343–E9352 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.DiCarlo J. E., Chavez A., Dietz S. L., Esvelt K. M., Church G. M., Safeguarding CRISPR-Cas9 gene drives in yeast. Nat. Biotechnol. 33, 1250–1255 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wu B., Luo L., Gao X. J., Cas9-triggered chain ablation of cas9 as a gene drive brake. Nat. Biotechnol. 34, 137–138 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gantz V. M., Bier E., The dawn of active genetics. Bioessays 38, 50–63 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Burt A., Trivers R., Genes in Conflict: The Biology of Selfish Genetic Elements (Belknap Press, ed. 1, 2008). [Google Scholar]
- 34.Champer J., Kim I., Champer S. E., Clark A. G., Messer P. W., Performance analysis of novel toxin-antidote CRISPR gene drive systems. bioRxiv: 10.1101/628362 (5 May 2019). [DOI] [PMC free article] [PubMed]
- 35.Maselko M., Heinsch S. C., Chacón J. M., Harcombe W. R., Smanski M. J., Engineering species-like barriers to sexual reproduction. Nat. Commun. 8, 883 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hay B. A., et al. , Engineering the genomes of wild insect populations: Challenges, and opportunities provided by synthetic Medea selfish genetic elements. J. Insect Physiol. 56, 1402–1413 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Schwarzer W., Spitz F., The architecture of gene expression: Integrating dispersed cis-regulatory modules into coherent regulatory domains. Curr. Opin. Genet. Dev. 27, 74–82 (2014). [DOI] [PubMed] [Google Scholar]
- 38.Chan H. Y., Brogna S., O’Kane C. J., Dribble, the Drosophila KRR1p homologue, is involved in rRNA processing. Mol. Biol. Cell 12, 1409–1419 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yokomori K., et al. , Drosophila TFIIA directs cooperative DNA binding with TBP and mediates transcriptional activation. Genes Dev. 8, 2313–2323 (1994). [DOI] [PubMed] [Google Scholar]
- 40.Molla K. A., Yang Y., CRISPR/Cas-Mediated base editing: Technical considerations and practical applications. Trends Biotechnol. 37, 1121–1142 (2019). [DOI] [PubMed] [Google Scholar]
- 41.Anzalone A. V., et al. , Search-and-replace genome editing without double-strand breaks or donor DNA. Nature 576, 149–157 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lansing F., et al. , A heterodimer of evolved designer-recombinases precisely excises a human genomic DNA locus. Nucleic Acids Res. 48, 472–485 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hanewich-Hollatz M. H., Chen Z., Hochrein L. M., Huang J., Pierce N. A., Conditional guide RNAs: Programmable conditional regulation of CRISPR/cas function in bacterial and mammalian cells via dynamic RNA nanotechnology. ACS Cent. Sci. 5, 1241–1249 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zeidler M. P., et al. , Temperature-sensitive control of protein activity by conditionally splicing inteins. Nat. Biotechnol. 22, 871–876 (2004). [DOI] [PubMed] [Google Scholar]
- 45.Dissmeyer N., Conditional modulation of biological processes by low-temperature degrons. Methods Mol. Biol. 1669, 407–416 (2017). [DOI] [PubMed] [Google Scholar]
- 46.Bonger K. M., Chen L.-C., Liu C. W., Wandless T. J., Small-molecule displacement of a cryptic degron causes conditional protein degradation. Nat. Chem. Biol. 7, 531–537 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Nabet B., et al. , The dTAG system for immediate and target-specific protein degradation. Nat. Chem. Biol. 14, 431–441 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Schapira M., Calabrese M. F., Bullock A. N., Crews C. M., Targeted protein degradation: Expanding the toolbox. Nat. Rev. Drug Discov. 18, 949–963 (2019). [DOI] [PubMed] [Google Scholar]
- 49.Gibson D. G., et al. , Enzymatic assembly of DNA molecules up to several hundred kilobases. Nat. Methods 6, 343–345 (2009). [DOI] [PubMed] [Google Scholar]
- 50.Doench J. G., et al. , Optimized sgRNA design to maximize activity and minimize off-target effects of CRISPR-Cas9. Nat. Biotechnol. 34, 184–191 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Katoh K., Standley D. M., MAFFT multiple sequence alignment software version 7: Improvements in performance and usability. Mol. Biol. Evol. 30, 772–780 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Port F., Chen H. M., Lee T., Bullock S. L., Optimized CRISPR/Cas tools for efficient germline and somatic genome engineering in Drosophila. Proc. Natl. Acad. Sci. U.S.A. 111, E2967–E2976 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Van Doren M., Williamson A. L., Lehmann R., Regulation of zygotic gene expression in Drosophila primordial germ cells. Curr. Biol. 8, 243–246 (1998). [DOI] [PubMed] [Google Scholar]
- 54.Theilmann D. A., Stewart S., Molecular analysis of the trans-activating IE-2 gene of Orgyia pseudotsugata multicapsid nuclear polyhedrosis virus. Virology 187, 84–96 (1992). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data are available in the main text and the supplementary materials. ClvR flies are available on request under the conditions outlined in ref. 25.