Significance
Our study reveals the most elaborate opsin expression patterns ever described in any animal eye. In mantis shrimp, a pugnacious crustacean renowned for its visual sophistication, we found unexpected retinal expression patterns highlighting the potential for cryptic photoreceptor functional diversity, including single photoreceptors that coexpress opsins from different spectral clades and a single opsin with a putative nonvisual function important in color vision. This study demonstrates the evolutionary potential for increasing visual system functional diversity through opsin gene duplication and diversification, as well as changes in patterns of gene coexpression among photoreceptors and retinula cells. These results have significant implications for the function of other visual systems, particularly in arthropods where large numbers of retinally expressed opsins have been documented.
Keywords: Stomatopoda, opsin, retinal expression, in situ hybridization, evolution
Abstract
Stomatopod crustaceans possess some of the most complex animal visual systems, including at least 16 spectrally distinct types of photoreceptive units (e.g., assemblages of photoreceptor cells). Here we fully characterize the set of opsin genes expressed in retinal tissues and determine expression patterns of each in the stomatopod Neogonodactylus oerstedii. Using a combination of transcriptome and RACE sequencing, we identified 33 opsin transcripts expressed in each N. oerstedii eye, which are predicted to form 20 long-wavelength–sensitive, 10 middle-wavelength–sensitive, and three UV-sensitive visual pigments. Observed expression patterns of these 33 transcripts were highly unusual in five respects: 1) All long-wavelength and short/middle-wavelength photoreceptive units expressed multiple opsins, while UV photoreceptor cells expressed single opsins; 2) most of the long-wavelength photoreceptive units expressed at least one middle-wavelength–sensitive opsin transcript; 3) the photoreceptors involved in spatial, motion, and polarization vision expressed more transcripts than those involved in color vision; 4) there is a unique opsin transcript that is expressed in all eight of the photoreceptive units devoted to color vision; and 5) expression patterns in the peripheral hemispheres of the eyes suggest visual specializations not previously recognized in stomatopods. Elucidating the expression patterns of all opsin transcripts expressed in the N. oerstedii retina reveals the potential for previously undocumented functional diversity in the already complex stomatopod eye and is a first step toward understanding the functional significance of the unusual abundance of opsins found in many arthropod species’ visual systems.
Stomatopod crustaceans are successful predators in marine habitats. They are famous for their aggressive nature, powerful prey capture, and defense system, and in particular their highly unusual visual system (1). Unlike other arthropods, each stomatopod compound eye consists of three distinct regions: Dorsal and ventral hemispheres separated by an equatorial region of specialized ommatidia referred to as the “midband” (MB). While the hemispheres are typically thought to have anatomically and physiologically similar ommatidia throughout, the MB is most commonly formed from six parallel rows of ommatidia that contain photoreceptors specialized for color (MB rows 1 to 4) and polarization (MB rows 5 and 6) discrimination (Fig. 1) (2–5).
Fig. 1.
The complex photoreceptor arrangement and spectral absorbance in the N. oerstedii compound eye. (A) Image of N. oerstedii. (Scale bar: 5 mm.) (B) The external division of the ommatidia into five regions: The marginal dorsal hemisphere (mDH), dorsal hemisphere (DH), MB composed of six rows of ommatidia, ventral hemisphere (VH), and marginal ventral hemisphere (mVH). (C) A schematic of the arrangement of the dorsal hemisphere, MB (rows 1 to 6), and ventral hemisphere ommatidia, including the R8 cells that sit atop R1–7 cells that form either one photoreceptive unit (hemispheres and MB rows 5 and 6) or two photoreceptive units (MB rows 1 to 4). Photoreceptors are colored to match their spectral absorbance curves (9) as in D.
The Caribbean species Neogonodactylus oerstedii has been particularly well studied and is thought to be representative of many stomatopods with six-row MBs (6–10). Altogether, its retina is composed of 16 spectrally distinct photoreceptive units (10). Similar to other crustaceans, each ommatidium is composed of eight photoreceptor (retinula) cells (R1–8). In the hemispheres and in MB rows 5 and 6, these eight cells are organized into two spectrally distinct layers composed of seven retinula cells (R1–7) situated proximal to a single-celled eighth retinula (R8) photoreceptor. Ommatidia in both hemispheres of N. oerstedii and in the two most ventral ommatidial rows of the MB, MB rows 5 and 6, have two photoreceptive layers: a UV-sensitive (UVS) R8 receptor distal to a middle-wavelength–sensitive (MWS) layer composed of R1–7 cells (6, 7, 9–11).
Most of the spectral diversity found in stomatopod retinas exists in the four dorsal-most ommatidial MB rows (MB rows 1 to 4). In MB rows 1 to 4, the seven proximal photoreceptor cells (R1–7) are subdivided into two separate photoreceptive units, formed by sets of three or four retinula cells. Therefore, in MB rows 1 to 4 each ommatidium is formed from three successive photoreceptive layers comprising the distal UVS R8 photoreceptor, and middle and proximal photoreceptive layers sensitive to the visible spectrum (Fig. 1C). Each of the three photoreceptor layers in each of the first four MB rows is spectrally distinct, adding 12 classes to the overall spectral diversity of the N. oerstedii eye. The spectral diversity of the photoreceptors in these layers is the result of distinct visual pigment expression, filtering through the visual pigments of photoreceptors of successive layers, and additional optical filtering by photostable pigments in the crystalline cones and rhabdoms that further tune their spectral sensitivities (3–7, 10–13).
In all animal visual systems, photoreceptor spectral absorbance is due to the expression of light-absorbing visual pigments. These visual pigments are composed of an opsin protein covalently bound to a vitamin A-derived chromophore. The spectral diversity observed among different photoreceptors within an organism’s visual system is generally based on the expression of different opsin proteins bound to the same chromophore. Crustacean opsin genes have been divided into three distinct phylogenetic groups based on general absorbance properties of the visual pigments: long-wavelength–sensitive (LWS), MWS, and short-wavelength–sensitive or UVS (SWS/UVS) (14). Given the 16 spectrally distinct classes of photoreceptive units in the N. oerstedii retina, a reasonable hypothesis is that there is one visual opsin expressed for each, for a total of 16 opsins. However, previous studies found 15 different LWS opsin transcripts expressed in N. oerstedii retinas, which contain only 6 LWS photoreceptive units, suggesting the potential for rampant coexpression of opsin genes in single photoreceptors (8). Recent studies of N. oerstedii UVS opsins found many fewer transcripts expressed than spectrally distinct R8 photoreceptor types (7). These differences between the number of opsins expressed and number of spectral types of photoreceptors (many more LWS opsins than long-wavelength photoreceptive units and fewer UVS opsins than UV photoreceptors), imply that photoreceptors and opsin proteins tuned to different parts of the visible spectrum may be under different evolutionary pressures.
The aim of this study is to characterize the set of opsin transcripts expressed in N. oerstedii retinal photoreceptors. Furthermore, due to the discrepancy between opsin diversity and photoreceptor diversity within a specific wavelength class, we examine the expression patterns of all identified opsin transcripts. This study provides a fully characterized set of opsins from a stomatopod compound eye and demonstrates that stomatopod visual systems exhibit extreme complexity, both at the level of photoreceptor diversity and at the level of opsin expression patterns.
Results
N. oerstedii Retinal Opsin Transcript Diversity.
In addition to previous results, we identified 15 new N. oerstedii opsins, bringing the expressed retinal visual opsin total to 33 transcripts (7, 8). Of these 33 visual opsins, 24 are full-length transcripts, 8 are partial transcripts missing only the 5′ end of the coding sequence, and 1 is a partial transcript missing coding regions on both the 5′ and 3′ ends. All 33 transcripts contain typical opsin structural motifs, including the predicted seven-transmembrane domains, a lysine residue that forms a protonated Schiff base with the bound retinal chromophore, a glutamic acid residue at the predicted R-type opsin counterion site for the positive charge of the protonated lysine, and a conserved amino acid motif (e.g., glutamic acid/aspartic acid–arginine–tyrosine) in transmembrane-helix 3 that stabilizes the inactive state conformation of class A rhodopsin-like G protein-coupled receptors (15).
In phylogenetic reconstructions including previously published crustacean opsin sequences, all N. oerstedii retinal opsins clustered into the expected three arthropod visual pigment clades (LWS, MWS, and SWS/UVS) (Fig. 2). In total, the 33 N. oerstedii retinal opsins consist of 20 LWS (5 newly identified), 10 MWS (all newly identified), and 3 UVS transcripts (previously identified) (7). Within each spectral clade the opsin transcripts were further divided into well-supported subgroups. The LWS opsins all clustered into the six previously identified stomatopod opsin subgroups A to F (8). The five new LWS opsins identified in this study belong to subgroup B (NoL18), subgroup C (NoL20), and subgroup E (NoL16, NoL17, and NoL19). In particular, the three opsin transcripts in LWS subgroup E are uniquely characterized N. oerstedii sequences from this subgroup. For the MWS clade, the 10 opsins form three subgroups labeled G through I (Fig. 2). We assigned previously described N. oerstedii UVS opsins to new subgroups J (NoUV1 and NoUV3) and K (NoUV2).
Fig. 2.
Maximum-likelihood phylogeny of crustacean opsins, including the 33 opsins characterized from N. oerstedii eyes. N. oerstedii opsins are in bold, and divided into subgroups based on spectral clade (LWS, subgroups A to E; MWS, subgroups G to I; SWS/UVS, subgroups J to K). Clades of opsins from related taxonomic groups have been collapsed for clarity. For stomatopod opsins, clades of opsins from a single species have been collapsed, and the number of opsins indicated in parentheses behind the species name. Maximum-likelihood bootstrap support and Bayesian posterior probabilities are mapped to the visible branches.
N. oerstedii Opsin Transcript Retinal Expression Patterns.
All identified opsins appear to be functional based on conserved structural motifs, so we determined the retinal expression pattern of each transcript (Fig. 3). We will describe the expression patterns of each transcript first by comparing patterns within opsin spectral classes and then by patterns observed based on photoreceptors with similar function or visual task (e.g., spatial and motion vision, color vision, polarization vision, or UV vision).
Fig. 3.
ISH tissue expression patterns for 33 N. oerstedii opsin transcripts. Each 10- to 14-μm retinal tissue section is oriented as in Fig. 1C and has been mapped onto a phylogeny of only N. oerstedii opsins. A schematic showing the expression patterns in the retina for each transcript or set of transcripts is placed side by side with representative ISH images for clarity. (A) LWS opsin clade ISH; (B) MWS opsin clade ISH; (C) SWS/UVS opsin clade ISH. N. oerstedii opsin subgroups are labeled A to K and are colored as in Fig. 2. A condensed tree is shown to depict clade and subgroup relationships. Numbers on branches represent bootstrap support values. mDH = marginal dorsal hemisphere; DH = dorsal hemisphere; MB = midband (rows 1-6); VH = ventral hemisphere; mVH = marginal ventral hemisphere; * = no detected ISH labeling, data not shown. Data for SWS subgroups J and K from ref. 7. (Scale bars in all panels: 200 μm.)
LWS opsin expression patterns.
Nineteen of the 20 LWS opsin transcripts were expressed exclusively in long-wavelength photoreceptors (i.e., the R1–7 photoreceptors of the middle and proximal tiers in MB rows 2, 3, 5, and 6, and both hemispheres). Subgroup A and B opsin transcripts were exclusively expressed in photoreceptive layers composed of R1–7 cells of the hemispheres and MB rows 5 and 6, although there was not a strict division of expression patterns by subgroup (Fig. 3A). The three most similar transcripts in LWS subgroup A (NoL6, -7, -12) were all expressed in MB rows 5 and 6. The fourth transcript in subgroup A (NoL8), however, was expressed in the R1–7 photoreceptive units of both the MB rows 5 and 6 and in a subset of the hemisphere ommatidia, including only those closest to the MB (Fig. 3A). In subgroup B, the single probe designed for five nearly identical transcripts (NoL3–5, NoL10–11) showed that as a set, these were expressed in all of the R1–7 photoreceptive layers of the hemisphere ommatidia; however, we cannot determine if there are expression level differences between individual transcripts. The sixth transcript of this subgroup (NoL18) was expressed only in the R1–7 photoreceptive layer of MB rows 5 and 6 (Fig. 3A).
The remaining transcripts in LWS subgroups D through F were mainly expressed in the long-wavelength middle and proximal layers of MB rows 2 and 3 (Fig. 3A). However, the two subgroup C opsin transcripts (NoL9, NoL20) showed more complex retinal expression patterns. NoL9 had the most unusual expression patterns of all of the LWS opsins, being consistently expressed in the middle and proximal photoreceptive units of all of the MB rows used in color vision (e.g., rows 1 to 4), including photoreceptors in rows 1 and 4 that have short-wavelength sensitivities (Fig. 3A). NoL20 had expression patterns that were difficult to interpret, with strong labeling in the row 3 proximal tier but also weaker labeling in the row 3 middle and row 2 proximal tiers (Fig. 3A). In each of subgroups D, E, and F, the transcripts had comparable expression patterns. In subgroup D, the highly similar transcripts NoL1 and NoL2 were both expressed in the MB row 2 proximal tier (Fig. 3A), while two subgroup E transcripts (NoL16 and NoL17) were expressed in the MB row 2 middle tier (Fig. 3A). The third transcript in subgroup E, NoL19, did not clearly label any retinal photoreceptors, although it had relatively high expression levels based on eye transcriptome estimates (2.4 × 104 transcripts per million [TPM]). Finally, two subgroup F transcripts were expressed in MB row 3 photoreceptive layers, with NoL15 expressed in the proximal tier and NoL13 in the middle tier. Uniquely, the third transcript in subgroup F, NoL14, had a similar expression pattern to subgroup B transcripts and was expressed in all of the R1–7 photoreceptive units in the hemispheres of the eye.
MWS opsin expression patterns.
The 10 N. oerstedii MWS opsin transcripts exhibited two general classes of expression pattern (Fig. 3B). The first was, as expected, expression in single short/middle-wavelength photoreceptive units in the MB (i.e., the middle and proximal tiers of rows 1 and 4). The second, unexpected, pattern of expression was for MWS transcripts found in distinct regionalized sets of photoreceptors in the hemispheres that were previously assumed to be sensitive to long wavelengths of light. Each of the four transcripts in MWS subgroup G had a different expression pattern. Three of the transcripts were expressed in short/middle-wavelength photoreceptive layers: NoM1 in the MB row 1 middle tier, NoM3 in the row 1 proximal tier, and NoM4 in the row 4 middle tier. The final subgroup G transcript, NoM2, was expressed only in the R1–7 photoreceptive unit of ommatidia that line the dorsal border of the eye (marginal dorsal hemisphere) (Fig. 3B).
The various transcripts in subgroup H had very different expression patterns, but all were expressed in photoreceptors previously characterized as LWS. Transcript NoM10 was expressed in the R1–7 layer of MB rows 5 and 6, NoM8 in both dorsal and ventral hemisphere R1–7 photoreceptive units (similar to LWS transcript NoL8), and NoM7 and NoM9 in only the R1–7 layer of the ommatidia that line the dorsal border of the eye (Fig. 3B). Finally, in subgroup I, two similar transcripts shared expression in MB row 4 proximal layers containing SWS/MWS photoreceptors (Fig. 3B).
SWS opsin expression patterns.
The expression patterns of the three transcripts found in the SWS opsin clade were published previously (7). Two of the three SWS transcripts were expressed in the UVS R8 cells of the retina, with a nearly nonoverlapping pattern. Transcript NoUV1 (subgroup J) was expressed in R8 cells of the dorsal hemisphere, MB rows 2 to 6, and the ventral hemisphere ommatidia immediately adjacent to the MB. NoUV2 (subgroup K) was found in the R8 cells of MB row 1 and the ventral hemisphere (Fig. 3C). NoUV3 (subgroup J) was not expressed at significant levels in any retinal photoreceptors, and also had no detectable expression in the transcriptome dataset.
Overall opsin expression in photoreceptor classes.
All of the MWS and LWS opsins, apart from NoL19 (which based on in situ data had no detectable expression in the retina), were expressed in R1–7 cells (Fig. 3). Additionally, all of the LWS and MWS photoreceptive layers throughout the eye (both in hemispheres and in MB rows) expressed from two to nine opsins (Fig. 4A and SI Appendix, Fig. S2). Comparisons of photoreceptive layer spectral sensitivities, spectral absorbances, and opsin expression patterns showed that those with similar spectral properties tend to express similar sets of opsins (Fig. 4B).
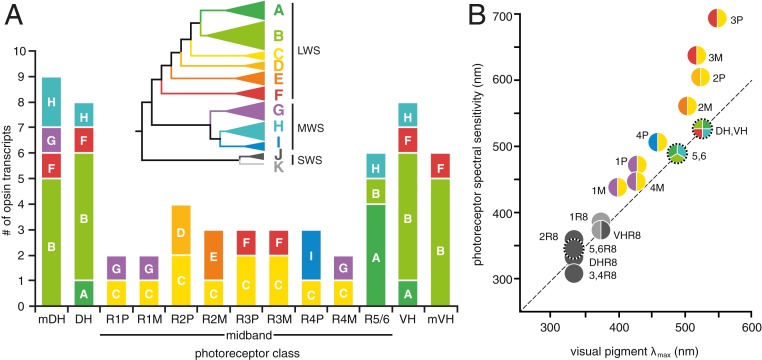
Fig. 4.
(A) Summary of the number of opsin transcripts expressed in each of the LWS and MWS photoreceptor types found in the N. oerstedii retina. Bars are colored by subgroup and labeled A through G as in Fig. 3, and as indicated in the inset collapsed phylogeny of all N. oerstedii opsins. (B) Plot of photoreceptor spectral sensitivity, measured spectral absorbance (visual pigment λmax), and subgroups of opsin transcripts expressed. Each circle represents a photoreceptor, containing a pie chart depicting the number of opsins expressed, colored by subgroup as in A. Photoreceptors specialized for polarization sensitivity are indicated by circles surrounded by tick marks. Photoreceptors in both panels are abbreviated as follows: marginal dorsal hemisphere, mDH; dorsal hemisphere, DH; MB row 1 proximal tier photoreceptor, 1P; MB row 1 middle tier photoreceptor, 1M; MB row 2 proximal tier photoreceptor, 2P; MB row 2 middle tier photoreceptor, 2M; MB row 3 proximal tier photoreceptor, 3P; MB row 3 middle tier photoreceptor, 3M; MB row 4 proximal tier photoreceptor, 4P; MB row 4 middle tier photoreceptor, 4M; MB rows 5 and 6–5/6; ventral hemisphere, VH; marginal ventral hemisphere, mVH; retinula cell 8 photoreceptor, R8. The dashed line represents the margin where the photoreceptor spectral sensitivity and visual pigment spectral absorbance peaks are equivalent. Spectral sensitivity maxima deviating from this line are influenced by tiering and by photostable optical filters.
Differences were also found in expression patterns among photoreceptive units used for different visual tasks. In general, clusters of photoreceptors used for spatial, motion, and polarization vision expressed opsins from multiple subgroups both within and across spectral clades (Fig. 4). For example, the MB row 5 and 6 R1–7 layer, specialized for polarization vision, expressed a total of six transcripts from LWS subgroups A (four transcripts) and B (one transcript), and MWS subgroup H (one transcript). The R1–7 layers in the hemispheres, used for spatial, motion, and polarization vision, express either six transcripts (six LWS) in the most marginal ommatidia of the ventral hemisphere, eight transcripts (seven LWS, one MWS) in the dorsal and ventral hemisphere ommatidia other than those at the margins, or nine transcripts (six LWS and three MWS) in the most marginal ommatidia of the dorsal hemisphere (Figs. 3 and 4). In contrast, the photoreceptive layers in MB ommatidia devoted to color (e.g., rows 1 to 4 middle and proximal tiers) generally expressed fewer opsins consisting of sets of highly similar transcripts from within the same spectral subgroup (e.g., LWS subgroup D transcripts NoL1 and NoL2 were both expressed in the MB row 2 proximal tier, LWS subgroup E transcripts were expressed in the MB row 2 middle tier, and MWS subgroup I opsins in the MB row 4 proximal tier). One notable exception to this pattern is transcript NoL9, which was expressed in all eight of the middle and proximal tiers in MB rows 1 to 4 (Fig. 4A). Finally, in contrast to long-wavelength and short/middle-wavelength photoreceptive units that express multiple opsins, UV R8 photoreceptors all expressed a single opsin transcript (Fig. 3C) (7).
Evidence for Site-Specific Selection.
Amino acid sites near the visual pigment chromophore tend to affect spectral tuning. Through comparison of our N. oerstedii alignment with a recently published jumping spider rhodopsin protein structure (16), we identified 26 sites within 5 Å of the chromophore (Dataset S2). Of these, six sites were invariable, including the chromophore binding site, the counterion site, and one of the cysteine residues involved in disulfide bridge formation. Substitutions in opsins of S-A or F-Y near the chromophore are known to produce spectral shifts in the peak absorbance of visual pigments in many species (17–19). In the identified 26 sites in N. oerstedii opsins, 7 sites contained S-A or F-Y substitutions that varied within or between spectral clades (Table 1). Using tests of selection based on dN/dS ratios (PAML, MEME), we identified a further 10 sites under positive selection across the N. oerstedii opsin phylogeny; 1 site (NoL1#69) was identified by both tests (SI Appendix, Table S4). None of these sites was within 5 Å of the chromophore. Using PAML random sites models, we found there was evidence for positive selection across the N. oerstedii opsin phylogeny (M7 versus M8, P < 0.004; ω = 1.15), with two sites identified as under selection (NoL#69 and NoL#166, P ≤ 0.05). In addition to the potential spectral tuning sites close to the chromophore with S-A or F-Y substitutions, an additional 12 sites from the set of 36 identified from all three methods had variation that corresponded with opsin clade structure (Table 2). The remaining 11 sites from the 36 identified from all three methods had variation both within and across clades, or variation that was not significantly different in amino acid properties (e.g., I-L substitutions).
Table 1.
Amino acid positions identified by proximity to the chromophore that contain S-A or F-Y substitutions (residues in bold)
| Amino acid nos. | LWS | MWS | UVS | ||||||||||
| NoL1 | JSR1 | BVRh | A (4) | B (6) | C (2) | D (2) | E (3) | F (3) | G (2) | H (4) | I (4) | J (2) | K (1) |
| 110 | 107 | 94 | M3 L1 | M6 | M2 | L2 | M3 | L2 V1 | F2 | Y3 F1 | Y2 V1 L1 | F2 | F1 |
| 129 | 126 | 113 | Y4 | Y6 | Y2 | Y2 | Y3 | Y3 | Y2 | Y4 | H2 F1 Y1 | F1 Y1 | F1 |
| 134 | 131 | 118 | N4 | N6 | N2 | N2 | S3 | S3 | G2 | A4 | A2 S1 G1 | A2 | S1 |
| 194 | 191 | 178 | Y4 | Y6 | Y2 | Y2 | Y3 | Y3 | Y2 | Y4 | F3 Y1 | F2 | Y1 |
| 205 | 202 | 189 | T4 | T6 | T2 | T2 | T3 | T3 | F2 | F3 Y1 | F4 | F2 | F1 |
| 226 | 223 | 210 | F4 | F6 | F2 | Y2 | Y3 | F1 Y2 | Y2 | Y4 | F2 Y2 | W2 | Y1 |
| 318 | 317 | 292 | S4 | S5 ?1 | S2 | S2 | S3 | S3 | A2 | A4 | G3 A1 | A2 | A1 |
Amino acid sites are identified based on numbering in the NoL1 opsin of N. oerstedii (NoL1), the crystal structure of rhodopsin from the jumping spider Hasarius adansoni (JSR1; accession no. 6I9K_A), and bovine rhodopsin (BvRh; accession no. NP_001014890.1). Opsins are divided by spectral clades (LWS, MWS, and UVS) and subgroups (A to K). The number of sequences within each subgroup are indicated in the parentheses after each letter. Within each subgroup, all amino acids at that position are indicated, with the number of sequences encoding that amino acid indicated by subscript.
Table 2.
Amino acid positions where residue variation corresponds to clade structure
| Amino acid nos. | Method | LWS | MWS | UVS | ||||||||||
| NoL1 | BVRh | JSR1 | A (4) | B (6) | C (2) | D (2) | E (3) | F (3) | G (2) | H (4) | I (4) | J (2) | K (1) | |
| 106 | 103 | 90 | C | N3, M1 | M6 | M2 | M2 | M3 | M3 | N2 | Q4 | Q4 | K2 | K1 |
| 118 | 115 | 102 | M | T4 | T6 | T2 | T2 | T3 | T3 | V2 | V4 | Y3, V1 | G2 | G1 |
| 138 | 135 | 122 | C | C4 | C6 | C2 | C2 | C3 | C3 | V2 | L4 | D3, V1 | P2 | I1 |
| 204 | 201 | 188 | C | G4 | G6 | G2 | G2 | G3 | G3 | S2 | S4 | S3, T1 | T2 | S1 |
| 218 | 215 | 202 | C | L4 | L6 | L2 | L2 | L3 | L2, V1 | T2 | G4 | G4 | V2 | V1 |
| 221 | 218 | 204 | C | Y4 | Y6 | Y2 | Y2 | Y4 | Y3 | I2 | I3, L1 | L2, M1, V1 | L2 | I1 |
| 222 | 219 | 206 | C | T3, S1 | T6 | S2 | S2 | A2, S1 | T2, C1 | F2 | F4 | F4 | W2 | F1 |
| 225 | 222 | 209 | C | V4 | V6 | V2 | V2 | V3 | C3 | D2 | D3, C1 | C3, Y1 | M1, V1 | S1 |
| 250 | 247 | 236 | M | Q3, A1 | Q5, ?1 | Q2 | Q2 | Q3 | A1, N1, Q1 | S2 | A4 | A4 | A1, Q1 | A1 |
| 296 | 294 | 269 | C | L2, M2 | L5,?1 | L2 | L2 | F2, L1 | A1, C1, L1 | A2 | A4 | A4 | A2 | A1 |
| 299 | 297 | 271 | C | N4 | N5,?1 | N2 | N2 | N3 | N3 | A2 | T3, V1 | V4 | A2 | A1 |
| 321 | 320 | 295 | C | A4 | A5,?1 | A2 | A2 | A4 | A3 | A2 | A4 | L4 | S1 | C2 |
The method of site identification is indicated: C, within 5 Å of the chromophore; M, identified in MEME tests of selection, with a P < 0.05. Amino acid sites are identified based on numbering in the NoL1 opsin of N. oerstedii (NoL1), the crystal structure of rhodopsin from the jumping spider H. adansoni (JSR1; accession no. 6I9K_A), and bovine rhodopsin (BvRh; accession no. NP_001014890.1). Opsins are divided by spectral clades (LWS, MWS, and UVS) and subgroups (A to K). The number of sequences within each subgroup are indicated in the parentheses after each letter. Within each subgroup, all amino acids in that position are indicated, with the number of sequences indicated by subscript.
Discussion
N. oerstedii Retinal Opsin Transcript Numbers.
With 3 UVS, 10 MWS, and 20 LWS transcripts, N. oerstedii expresses an astounding diversity of visual opsins. The high numbers of characterized opsin transcripts are similar to numbers published in transcriptome estimates for other stomatopod species, specifically Hemisquilla californiensis (11 LWS, 2 MWS, and 1 SWS) and Pseudosquilla ciliata (15 LWS, 16 MWS, and 2 SWS) (14). Dragonflies and damselflies (Odonata) also have similar patterns of diversity, generally including 1 UVS, 1 to 8 insect SWS, and 8 to 21 LWS opsin genes (20), with different species expressing anywhere from 4 to 30 visual opsin genes (20–22). Genomic sequencing in many arthropod taxa has revealed unexpectedly large numbers of potential visual opsins, including 12 opsins in the mosquito Anopheles gambia, 9 of which are in visual opsin clades (23); 18 opsins in the horseshoe crab Limulus polyphemus, 13 of which are expressed in eyes (24); 46 opsins in the crustacean Daphnia pulex, 27 of which are likely used for vision (25); and 42 visual opsins (33 LWS, 7 MWS, and 2 SWS) in the penaid shrimp Litopenaeus vannamei (26). Understanding the functional implications of these large numbers of opsin genes in arthropods requires studies that link genomic opsin diversity to retinal expression patterns, as well as research on actual photoreceptor spectral and polarization responses.
In arthropods where an abundance of expressed opsins has been found (i.e., horseshoe crabs, odonates, stomatopods), most gene duplications occur in the LWS opsin clade, with fewer in the MWS clade and very few in the SWS/UVS clade. These differences in frequency of gene duplication suggest that different selection pressures act on visual pigments sensitive to different parts of the spectrum. The stomatopod and odonate visual systems are among the most extreme examples of LWS opsin gene duplication yet characterized, with N. oerstedii expressing 20 LWS and the dragonfly Anax parthenope expressing 21 LWS opsins (20). What drives LWS opsin gene expansion in these taxa? In odonates, Suvorov et al. (22) found that a heterozygote advantage model may explain the opsin gene repertoire expansion, where positive selection acts on alleles that are subsequently duplicated and fixed in the genome. Other possible explanations include genome characteristics and similarities in ecologies (stomatopods and odonates are both highly visual predators).
The high numbers of MWS transcripts found in N. oerstedii (10 opsins) are in line with the numbers of MWS genes revealed in other crustacean studies [D. pulex genome 25 MWS genes (25), L. vannamei genome 7 MWS genes (26)]. Unlike these crustacean examples, one of the few published chelicerate genomes, from L. polyphemus, contains a single MWS opsin (24). Interestingly, among stomatopod species the expressed number of MWS opsin transcripts appears to vary strongly, with only 3 transcripts recovered in H. californiensis, compared to 10 in N. oerstedii and 16 in P. ciliata (this study and ref. 14).
In insects, blue-sensitive opsins are an evolutionarily distinct lineage from the crustacean and horseshoe crab MWS opsins, clustering with what we call here the SWS clade; therefore, the 10 N. oerstedii MWS opsins and the 8 MWS opsins in A. parthenope that form putatively SWS/MWS visual pigments must be an independent set of duplications. Although most orders of insects do not appear to contain sequences closely related to the crustacean MWS clade, recent studies found crustacean-type MWS opsins in early branching hexapod lineages (Sminthurus viridis, Collembola; Machilis hrabei, Archaeognatha) and myriapods (Scutigera coleoptrata) (27).
N. oerstedii Retinal Opsin Expression Patterns.
Only a few genomic and transcriptomic studies of arthropod visual systems have determined the expression pattern of each identified transcript. In this study we demonstrated retinal expression patterns for 31 of the 33 described opsin transcripts. The two opsin transcripts, NoL19 and NoUV3, with no clear retinal expression pattern, may be expressed in parts of the nervous system that we did not include in our study. This idea is supported for NoL19 by two pieces of information. First, NoL19 had expression levels (2.4 × 104 TPM) in the transcriptome similar to other LWS opsin transcripts. Second, a recent study by Donohue et al. (28) found NoL19 (identical to transcript NoT27696g1 in ref. 28) to be expressed in the cerebral ganglion. The case for NoUV3 is less clear, as it had no detectable expression in the transcriptome dataset; this may be due to expression in a very small subset of cells or the possibility that the tissue of expression was not included in the generation of the transcriptome data. Future studies should check the visual neuropils for both of these “orphan” eye transcripts and the CNS for expression of NoUV3.
Determining the retinal expression patterns for all 33 opsins found in the N. oerstedii retina is a major contribution to understanding the function of the complex stomatopod visual system and, more broadly, other arthropod eyes that express a superabundance of opsin transcripts relative to described photoreceptor types. Perhaps the most comprehensive previous study of arthropod opsin expression patterns is that of Battelle et al. (24), which documented 18 opsin genes in the L. polyphemus genome and determined the expression patterns of each across the visual system and CNS. Thirteen of these opsins are expressed in ocular tissues of the median, ventral, or lateral eyes, although only 10 of them are found in what are typically considered visual opsin clades. L. polyphemus eyes all express opsins from multiple spectral clades: The median eyes express seven LWS, one MWS, and one SWS opsins, while the ventral and lateral eyes each express four LWS, one MWS, and one SWS opsins (24). A similar pattern was found in N. oerstedii eyes, where all of the R1–7 photoreceptive layers in MB rows 5 and 6 and the hemispheres (with the exception of the most marginal region of the ventral hemisphere) express numerous LWS opsin transcripts, as well as at least one MWS opsin transcript.
Given that the long-wavelength and short/middle-wavelength photoreceptive units in the N. oerstedii retina express multiple opsin transcripts, an important question for study is how these opsins are distributed among the retinula cells that contribute to each unit. In the ommatidia of the N. oerstedii hemispheres, the R1–7 photoreceptive layer is composed of seven cells that form a fused central rhabdom. Determining which sets of the R1–7 cells express MWS opsin transcripts in the LWS photoreceptive units may provide further clues to the functional significance of these apparently spectrally mismatched visual pigments. Based on LWS and MWS opsin expression patterns across hemisphere ommatidia, there are more opsins expressed (six LWS and three MWS opsins in the marginal dorsal hemisphere ommatidia; seven LWS and one MWS in the dorsal and ventral hemisphere ommatidia) than the number of previously characterized spectrally distinct photoreceptive units (one LWS, zero MWS). Because each of the hemisphere ommatidia contains up to seven photoreceptor cells (R1–7) contributing to LWS sensitivity, this pattern of eight or nine expressed LWS/MWS opsins per ommatidia suggests that some photoreceptor cells in hemisphere ommatidia must express multiple opsin transcripts. However, we cannot predict at this point whether all retinula cells express all opsin transcripts, to what degree subsets of these cells express subsets of opsins, or whether all of the transcripts identified are translated into stable proteins capable of binding chromophore. Coexpression of multiple opsins in arthropod photoreceptor cells has been shown in a number of species, including butterflies (29, 30), mosquitoes (31, 32), horseshoe crabs (24), and fiddler crabs (33). In insects (e.g., butterflies, mosquitoes), opsin coexpression has been shown to broaden the spectral sensitivity of photoreceptors (29) or enhance vision in low-light environments (31). In studies of opsin coexpression in crustaceans, Rajkumar et al. (33) found that the two MWS opsins expressed in the R1–7 layer of the fiddler crab Uca pugilator were coexpressed in only one of the seven retinula cells forming the rhabdom, while each of the remaining six retinula cells expressed only one of the two possible transcripts. Based on this expression pattern involving only two opsin transcripts, the expression patterns at the level of individual retinula cells in stomatopods is likely to be complex, and possibly unique for each opsin. In one of the few published studies on stomatopod opsin expression patterns, at least two LWS opsin transcripts were coexpressed in all retinula cells of the retina of Squilla empusa (34), a species with a relatively simple eye having only two rows of ommatidia in the MB. In the more complicated 6 MB row eye of N. oerstedii, if each retinula cell (R1–7) expressed a unique set of opsins, then photoreceptive units composed of fewer cells (three or four cells in MB photoreceptor tiers versus seven in hemisphere R1–7 photoreceptive units) would be predicted to express fewer transcripts. Indeed, we observe this pattern in the N. oerstedii retina, where tiered middle and proximal photoreceptive layers in the MB express fewer opsin transcripts than the hemisphere R1–7 layer (Fig. 4).
Our data also illustrate how molecular studies can be used to reveal new photoreceptive structures missed by other methods. An example in N. oerstedii is the very marginal region of the dorsal hemisphere, containing ommatidia that line the dorsal border of the eye. The expression of three different MWS opsin transcripts in this region is reminiscent of the “dorsal rim” area of insect eyes typically used to detect shorter-wavelength skylight polarization patterns (35). Histological sections used for in situ hybridization (ISH), although not ideal for anatomical studies, suggest comparable morphological changes in the dorsal-most part of the N. oerstedii eye, where all of the photoreceptors are directed upward. This specialized region is the first identified in a crustacean eye. In insects, photoreceptors in the dorsal rim area have been found to be sensitive to either UV, blue, or green wavelengths of light and may exhibit a suite of morphological changes, including altered or lost lens structures, orthogonally arranged microvilli, loss of screening pigment, an upward directed optical axis, and significantly reduced rhabdom lengths (35). Consistent with what is known from insect eyes, our molecular data suggest the presence of blue-green to blue visual pigments in the dorsal-most eye region of stomatopods. Previous studies of related species suggest this region of the eye is also unique with respect to the presence of UV filters (11). Investigations of ommatidia in the stomatopod eye dorsal rim region are needed, to understand whether or not photoreceptor anatomy and physiology of this region differs from that of the adjacent hemisphere receptors and to learn if N. oerstedii show behavioral evidence of using skylight polarization patterns for navigation underwater.
Why Do Stomatopods Have So Many Opsins in Their Retinas?
The retinal expression patterns of identified opsin transcripts were far more complex than we predicted. Photoreceptive layers formed from retinula cells R1–7 in N. oerstedii express as many as nine different opsin transcripts (Fig. 4). This is particularly surprising because absorbance spectra previously measured by microspectrophotometry (MSP) suggest that each photoreceptive unit, with the possible exception of those of the hemispheres, possesses only a single spectral class of visual pigment (10). However, absorbance spectral data are unsuitable for detecting the presence of multiple visual pigments if they have similar absorption spectra or if one or more components are present in relatively low concentration.
When multiple opsins are expressed in single photoreceptors, the mixture’s presence is often obvious from the broadening of the spectral absorption or sensitivity of the receptor. In vertebrates, for example cichlid fish or nocturnal rodents, the absorption effects of visual pigment coexpression have generally been interpreted as adaptations to the spectral or luminosity features of the environment (36–38). In arthropods the situation is often more puzzling. The best data available are from butterflies, where opsin coexpression is common in subsets of retinal photoreceptors that have also been physiologically characterized. In some cases, the mixture produces spectrally obvious effects, forming receptors with extremely broad, or even double, spectral peaks (29, 30, 39). In others, however, photoreceptors with mixed opsins have spectral sensitivities similar to receptors with only one rhodopsin class (30, 40). Similar to our MSP findings in N. oerstedii, previous crustacean studies on opsin coexpression in an intertidal grapsid crab (41) and a deep-sea lophogastrid (42) found that spectral sensitivity of photoreceptors was explainable by a single visual pigment in the retina despite the mix of expressed opsins. Electrophysiological recordings from single photoreceptor cells in the color receptors (i.e., from photoreceptive units in the dorsal four MB rows) of N. oerstedii indicate narrow-band sensitivity (43), essentially identical to predictions made based on models using single visual pigments plus the overlying filters (10). This is not particularly surprising, given that these receptors express few opsins and are heavily filtered. On the other hand, physiological measurements from receptors in the hemispheres, which are not filtered and which express up to nine opsin transcripts, also find spectral sensitivities explainable by the presence of a single visual pigment (44). Given the consistency of these results from multiple crustaceans, it appears that the multiple opsin expression we report here rarely (or never) leads to spectral sensitivity broadening in these animals.
One possible explanation of the seeming contradiction between the complex opsin expression results and the MSP measurements is that some transcribed opsins, for example the LWS-subgroup C opsins expressed in all of the visible light photoreceptors in MB rows 1 to 4, are either untranslated, nonfunctional as proteins, or have a yet unidentified nonvisual function (“functionality” hypothesis). Alternatively, only one (or at least a very few) of the expressed opsins dominates the visual pigments in the rhabdom of a given photoreceptive layer (“dominance” hypothesis). A third possibility is that opsins belonging to the same subgroup (i.e., A, B, C, and so forth) that are expressed in the same cluster of photoreceptors may have similar or identical absorption spectra (“similarity” hypothesis). A final possibility is that some of the expressed visual opsins have nonvisual functions within the retina (“nonvisual” hypothesis).
Although these hypotheses are not mutually exclusive, and the expression patterns of different transcripts may be explained by alternate hypotheses, our data allow us to investigate these alternatives using several approaches: 1) Analyzing transcript sequences for sites important to visual pigment function (functionality hypothesis); 2) investigating expression levels of selected transcripts expressed in similar sets of photoreceptors (dominance hypothesis); and 3) inspecting the residue identities for sites under positive selection and potential spectral tuning sites within groups of related sequences expressed in the same sets of photoreceptors (similarity hypothesis). For hypotheses of some transcripts having nonvisual functions, we can only speculate as to the potential roles based on previous studies in other arthropods.
Functionality hypothesis.
All 33 transcripts characterized here contained typical opsin structural motifs, including the chromophore binding and counterion sites, and can therefore be translated into functional proteins. Within the N. oerstedii opsins a set of 10 sets of sites were identified that were under positive selection (SI Appendix, Table S4). Because most of these sites are far from the chromophore binding pocket, it is likely that variations at these sites affect overall opsin function and G protein-coupled receptor signaling, rather than spectral tuning (34). Although many of these sites have amino acid substitutions that vary across or within clades (Table 2), at this point it is difficult to determine how this amino acid variation may affect overall visual pigment function and thus the veracity of the functionality hypothesis. Future studies of protein localization will help identify transcripts translated into functional proteins, while the sites under positive selection may serve as candidates for in vitro studies of arthropod opsin functional diversity unrelated to spectral tuning.
Dominance hypothesis.
As a first approach to investigating whether single transcripts dominate in a particular photoreceptive unit, we considered both the relative level of labeling in ISH sections and the calculated retinal expression levels of coexpressed sets of opsin transcripts. Although this is a poor measure of abundance, some sets of transcripts expressed in the same photoreceptive layer all had strong in situ labeling, suggesting significant expression levels for multiple transcripts (e.g., the expression patterns of transcripts in LWS subgroups A and B in MB rows 5 and 6, or NoL1 and NoL2 expression in the MB row 2 proximal tier) (Fig. 3A). Due to complex and overlapping expression patterns in multiple types of photoreceptive units, we have chosen not to present estimated expression levels of each transcript here because they are difficult to interpret. However, there are several sets of LWS transcripts with similar expression patterns, making estimated expression levels comparable. For example, transcripts from two different subgroups (e.g., NoL6-7 and NoL12 from group A, and NoL18 from group B) that have the same expression pattern in only the R1–7 layer of MB rows 5 and 6 all had similar levels of expression (NoL6 = 1.2 × 105 TPM; NoL7 = 1.4 × 105 TPM; NoL12 = 1.5 × 105 TPM; NoL18 = 1.0 × 105 TPM), illustrating that at least for some photoreceptive units there is not a single dominant expressed opsin. Similarly, the closely related sets of transcripts expressed in the MB row 2 proximal tier (NoL1 = 2.8 × 104 TPM; NoL2 = 2.5 × 104 TPM) and the row 2 middle tier (NoL16 = 1.3 × 104 TPM; NoL17 = 4.0 × 103 TPM) all had similar levels of expression (Fig. 3A). Together, although neither of these methods can specifically test this question, results suggest that in most stomatopod retinal photoreceptive layers there is probably not a single dominant visual pigment responsible for the previously measured spectral absorbances, and therefore we consider the dominance hypothesis an unlikely explanation for stomatopod retinas. However, to establish this conclusion definitively, methods for determining translation efficiency as well as visual pigment functionality are needed.
Similarity hypothesis.
The similarity hypothesis, at its core, addresses the fate of opsin gene duplicates in the stomatopod genome. There is a large literature on evolutionary models for the maintenance of gene duplicates (45), distilling down to three outcomes: 1) The evolution of a new function (“neofunctionalization”) (46); 2) the division of ancestral functions among duplicates (“subfunctionalization”); 3) the conservation of function in both duplicates (“gene conservation”). In the context of stomatopod opsin genes that encode visual pigments, gene duplication followed by subfunctionalization and neofunctionalization has led to the large set of visual pigments with diverse peak spectral absorbances expressed in distinct photoreceptor types. However, what processes are at play for duplicated genes expressed in the same photoreceptor is still an open question. There is evidence from S. empusa that coexpressed, closely related gene variants may interact differently with the phototransduction machinery (34) and thus may tune photoreceptor cell temporal responses.
The only potential evolutionary model that we can address here, given our data for opsin gene duplicates expressed within a single photoreceptive structure, is gene conservation. Mechanisms for maintaining a gene duplicate include dosage models, where producing more of a gene product provides a functional advantage. To address whether related sets of opsin transcripts may form visual pigments with similar peak absorbances, we compared the residues at predicted spectral tuning sites across transcript sequences that were closely related and also expressed in the same set of photoreceptor cells. For example, the MB row 2 proximal and middle tiers both express two opsin transcripts that are evolutionarily closely related (Fig. 3A), and that do not vary in amino acid at any of the potential spectral tuning sites (e.g., subgroup E NoL16 and NoL17 in the MB row 2 middle tier and subgroup D NoL1 and NoL2 in the MB row 2 proximal tier) (Table 1). This suggests that spectral absorbance of the encoded visual pigments does not vary, at least for these sets of transcripts, and they therefore may adhere to the dosage model of gene duplicate conservation. Another possible factor in gene conservation is heterodimerization, particularly in the LWS polarization-sensitive photoreceptive layers in the hemispheres and MB rows 5 and 6, which all express six or more opsin transcripts (Fig. 4). Large numbers of expressed transcripts with similar spectral properties may play a role in increased dimerization and heterodimerization and improved chromophore orientation along the microvillar axis, improving the polarization sensitivity of these photoreceptors (47–49).
Nonvisual hypothesis.
Although we have no way to test this hypothesis, given the data presented here, previous studies of Drosophila have shown that visual opsins can have light-independent functions, including thermal preference (50), mechanoreception (51, 52), and larval locomotion. Therefore, we cannot rule out the possibility that some of the opsin transcripts expressed in the N. oerstedii retina have nonvisual functions. Further studies are needed to begin to sort out these potential functions.
Summary.
Elucidating the expression patterns of all 33 opsin transcripts found in N. oerstedii retinas is a major step toward understanding the functional significance of the large abundance of opsins described from many arthropod species’ visual systems. This study demonstrates that the already complex photoreceptor diversity of the stomatopod visual system is even more complex at the molecular level of phototransduction. The observed expression patterns, particularly for the MWS opsins, were unexpected and revealed the potential for previously unsuspected functional diversity in the photoreceptors of the already complex stomatopod eye. In the retina of N. oerstedii, we show that all LWS and SWS/MWS photoreceptive layers express multiple opsins, that the R1–7 photoreceptive units of the hemispheres and MB rows 5 and 6 express more transcripts than any of the middle or proximal tiers involved in color vision in MB rows 1 to 4, and that all of the hemisphere and MB row 5 and 6 LWS photoreceptive units express at least one MWS opsin transcript. In addition, there is a single opsin transcript that is uniquely expressed in all eight of the MB rows 1 to 4 middle and proximal tiers devoted to color vision.
Materials and Methods
Data Availability.
The generated N. oerstedii transcriptome and opsin transcript sequences are available at GenBank (SRA PRJNA609025, accession nos. MT112859–MT112888). The full details of our methods, including sequence alignments, are presented in the SI Appendix and Datasets S1–S6.
RNA-Sequencing and Opsin Transcript Identification.
Total RNA was isolated using TRIzol (Life Technologies) and purified using a RNeasy kit (Qiagen). A transcriptome generated and assembled by Cofactor Genomics was queried using local BLAST+ (v2.2.28) to identify opsin contigs. Identified opsin contigs, as well as previously published N. oerstedii opsin sequences (7, 8), were confirmed using transcript-specific primers in RT-PCR, while degenerate primers were used to search for possible transcripts missed by all other methods. RACE methods were used to sequence the ends of incomplete transcripts (8).
Opsin Phylogenetic Analysis.
Each opsin transcript was translated and aligned with other opsin sequences using MUSCLE (53, 54) as implemented by MEGA 6.06 (55) to create two datasets: 1) N. oerstedii transcripts with other published arthropod opsin sequences; and 2) N. oerstedii opsins only to use for mapping ISH results. Phylogenetic analyses were carried out using maximum-likelihood methods executed in RAxML 8.1.5 (56–58), with the best-fit model as selected by ProtTest3 (59, 60). Additionally, posterior probabilities using Bayesian inference methods coupled with a Markov chain Monte Carlo algorithm executed in MrBayes 3.2.6 (61, 62) were generated on the CIPRES Science Gateway v3.3 (63, 64).
Analyses of Spectral Tuning Sites, Gene Conversion, and Selection.
SWISS-PDBVIEWER (65–67) was used to fit a N. oersterdii opsin (NoL1) to the jumping spider rhodopsin protein structure (PDB ID code 619K) (16), and to identify all residues predicted to be within 5 Å of the chromophore. These candidate sites were screened based on how residue identities mapped to spectral clades, specific amino acid properties, and correspondence with sites previously identified to cause shifts in spectral absorbance of the visual pigment. Tests for selection across branches and sites were accomplished using the CodeML program in the PAML software package (68, 69) as implemented in EasycodeML (70), and the HyPhy software package on the Datamonkey webserver (71–74). For residue identification, amino acids were numbered based on their position in the N. oerstedii reference sequence (NoL1#), the Hasarius adansoni crystal structure (JSR1#), or bovine rhodopsin (BvRh#).
In Situ Hybridization.
Dissected eyes were fixed in 4% paraformaldehyde in 10 mM phosphate buffer (pH 7.4), cryoprotected with sucrose, permeated with Tissue-Tek OCT Compound (Sakura Finetek), quick-frozen, and sectioned at a thickness of 12 µm in a cryostat cooled to −20 °C. With the exception of five transcripts (NoL3 to -5, -10, and -11) with similar sequences, unique riboprobes were designed for each transcript based on the 3′ UTR of each opsin mRNA. Riboprobes were generated as described in ref. 75. Hybridizations were performed on cryosections as described in ref. 75 and 76. Individual hybridization temperatures were calculated by an equation for RNA duplex formation derived by marginal dorsal hemisphere in and Knudson (77).
Supplementary Material
Acknowledgments
Thanks to Mireille Steck for help with in situ hybridization and expression data. This research was funded by grants from the National Science Foundation (IOS 0721608) and the Air Force Office of Scientific Research through Grants FA9550-06-1-0117, FA9550-09-1-0149, and FA9550-12-0321.
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
Data deposition: The sequences reported in this paper have been deposited in the GenBank database (SRA PRJNA609025, accession nos. MT112859–MT112888).
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.1917303117/-/DCSupplemental.
References
- 1.Schram F. R., et al. , “Subclass Hoplocarida Calman, 1904: Order Stomatopoda Latreille, 1817” in Treatise on Zoology—Anatomy, Taxonomy, Biology; The Crustacea; Revised and Updated, as Well as Extended from the Traité de Zoologie, Vaupel Klein J. C., Charmantier-Daures M., Schram F. R., Eds. (Brill, Leiden-Boston, 2013), Vol. 4, pp. 179–356. [Google Scholar]
- 2.Marshall N. J., A unique colour and polarization vision system in mantis shrimps. Nature 333, 557–560 (1988). [DOI] [PubMed] [Google Scholar]
- 3.Marshall N. J., Land M. F., King C. A., Cronin T. W., The compound eyes of mantis shrimps (Crustacea, Hoplocarida, Stomatopoda). I. Compound eye structure: The detection of polarized light. Philos. Trans. R. Soc. Lond. B Biol. Sci. 334, 33–56 (1991). [Google Scholar]
- 4.Marshall N. J., Land M. F., King C. A., Cronin T. W., The compound eyes of mantis shrimps (Crustacea, Hoplocarida, Stomatopoda). II. Colour pigments in the eyes of stomatopod crustaceans: Polychromatic vision by serial and lateral filtering. Philos. Trans. R. Soc. Lond. B Biol. Sci. 334, 57–84 (1991). [Google Scholar]
- 5.Marshall J., Cronin T. W., Kleinlogel S., Stomatopod eye structure and function: A review. Arthropod Struct. Dev. 36, 420–448 (2007). [DOI] [PubMed] [Google Scholar]
- 6.Marshall J., Oberwinkler J., The colourful world of the mantis shrimp. Nature 401, 873–874 (1999). [DOI] [PubMed] [Google Scholar]
- 7.Bok M. J., Porter M. L., Place A. R., Cronin T. W., Biological sunscreens tune polychromatic ultraviolet vision in mantis shrimp. Curr. Biol. 24, 1636–1642 (2014). [DOI] [PubMed] [Google Scholar]
- 8.Porter M. L., Bok M. J., Robinson P. R., Cronin T. W., Molecular diversity of visual pigments in Stomatopoda (Crustacea). Vis. Neurosci. 26, 255–265 (2009). [DOI] [PubMed] [Google Scholar]
- 9.Cronin T. W., Marshall N., Caldwell R., Visual pigment diversity in two genera of mantis shrimps implies rapid evolution (Crustacea; Stomatopoda). J. Comp. Physiol. A Neuroethol. Sens. Neural Behav. Physiol. 179, 371–384 (1996). [Google Scholar]
- 10.Cronin T. W., Marshall N. J., Multiple spectral classes of photoreceptors in the retinas of gonodactyloid stomatopod crustaceans. J. Comp. Physiol. A Neuroethol. Sens. Neural Behav. Physiol. 166, 261–275 (1989). [Google Scholar]
- 11.Bok M. J., Porter M. L., Cronin T. W., Ultraviolet filters in stomatopod crustaceans: Diversity, ecology and evolution. J. Exp. Biol. 218, 2055–2066 (2015). [DOI] [PubMed] [Google Scholar]
- 12.Cronin T. W., Bok M. J., Marshall N. J., Caldwell R. L., Filtering and polychromatic vision in mantis shrimps: Themes in visible and ultraviolet vision. Philos. Trans. R. Soc. Lond. B Biol. Sci. 369, 20130032 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cronin T. W., Marshall N. J., A retina with at least ten spectral types of photoreceptors in a mantis shrimp. Nature 339, 137–140 (1989). [Google Scholar]
- 14.Porter M. L., et al. , The evolution of complexity in the visual systems of stomatopods: Insights from transcriptomics. Integr. Comp. Biol. 53, 39–49 (2013). [DOI] [PubMed] [Google Scholar]
- 15.Vogel R., et al. , Functional role of the “ionic lock”—An interhelical hydrogen-bond network in family A heptahelical receptors. J. Mol. Biol. 380, 648–655 (2008). [DOI] [PubMed] [Google Scholar]
- 16.Varma N., et al. , Crystal structure of jumping spider rhodopsin-1 as a light sensitive GPCR. Proc. Natl. Acad. Sci. U.S.A. 116, 14547–14556 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Salcedo E., Farrell D., Zheng L., Phistry M., Bagg E., The green-absorbing Drosophila Rh6 visual pigment contains a blue-shifting. J. Biol. Chem. 284, 5717–5722 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hope A. J., Partridge J. C., Dulai K. S., Hunt D. M., Mechanisms of wavelength tuning in the rod opsins of deep-sea fishes. Proc. Biol. Sci. 264, 155–163 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Takahashi Y., Ebrey T. G., Molecular basis of spectral tuning in the newt short wavelength sensitive visual pigment. Biochemistry 42, 6025–6034 (2003). [DOI] [PubMed] [Google Scholar]
- 20.Futahashi R., et al. , Extraordinary diversity of visual opsin genes in dragonflies. Proc. Natl. Acad. Sci. U.S.A. 112, E1247–E1256 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chauhan P., et al. , De novo transcriptome of Ischnura elegans provides insights into sensory biology, colour and vision genes. BMC Genom. 15, 808 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Suvorov A., et al. , Opsins have evolved under the permanent heterozygote model: Insights from phylotranscriptomics of Odonata. Mol. Ecol. 26, 1306–1322 (2017). [DOI] [PubMed] [Google Scholar]
- 23.Hill C. A., et al. , G protein-coupled receptors in Anopheles gambiae. Science 298, 176–178 (2002). [DOI] [PubMed] [Google Scholar]
- 24.Battelle B. A., et al. , Opsin repertoire and expression patterns in horseshoe crabs: Evidence from the genome of Limulus polyphemus (Arthropoda: Chelicerata). Genome Biol. Evol. 8, 1571–1589 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Colbourne J. K., et al. , The ecoresponsive genome of Daphnia pulex. Science 331, 555–561 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhang X., et al. , Penaeid shrimp genome provides insights into benthic adaptation and frequent molting. Nat. Commun. 10, 356 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Henze M. J., Oakley T. H., The dynamic evolutionary history of pancrustacean eyes and opsins. Integr. Comp. Biol. 55, 830–842 (2015). [DOI] [PubMed] [Google Scholar]
- 28.Donohue M. W., Carleton K. L., Cronin T. W., Opsin expression in the central nervous system of the mantis shrimp Neogonodactylus oerstedii. Biol. Bull. 233, 58–69 (2017). [DOI] [PubMed] [Google Scholar]
- 29.Arikawa K., Mizuno S., Kinoshita M., Stavenga D. G., Coexpression of two visual pigments in a photoreceptor causes an abnormally broad spectral sensitivity in the eye of the butterfly Papilio xuthus. J. Neurosci. 23, 4527–4532 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ogawa Y., et al. , Coexpression of three middle wavelength-absorbing visual pigments in sexually dimorphic photoreceptors of the butterfly Colias erate. J. Comp. Physiol. A Neuroethol. Sens. Neural Behav. Physiol. 198, 857–867 (2012). [DOI] [PubMed] [Google Scholar]
- 31.Hu X., Whaley M. A., Stein M. M., Mitchell B. E., O’Tousa J. E., Coexpression of spectrally distinct rhodopsins in Aedes aegypti R7 photoreceptors. PLoS One 6, e23121 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hu X., Leming M. T., Whaley M. A., O’Tousa J. E., Rhodopsin coexpression in UV photoreceptors of Aedes aegypti and Anopheles gambiae mosquitoes. J. Exp. Biol. 217, 1003–1008 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rajkumar P., Rollmann S. M., Cook T. A., Layne J. E., Molecular evidence for color discrimination in the Atlantic sand fiddler crab, Uca pugilator. J. Exp. Biol. 213, 4240–4248 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Valdez-Lopez J. C., et al. , Sequence, structure, and expression of opsins in the monochromatic stomatopod Squilla empusa. Integr. Comp. Biol. 58, 386–397 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Labhart T., Meyer E. P., Detectors for polarized skylight in insects: A survey of ommatidial specializations in the dorsal rim area of the compound eye. Microsc. Res. Tech. 47, 368–379 (1999). [DOI] [PubMed] [Google Scholar]
- 36.Peichl L., Diversity of mammalian photoreceptor properties: Adaptations to habitat and lifestyle? Anat. Rec. A Discov. Mol. Cell. Evol. Biol. 287, 1001–1012 (2005). [DOI] [PubMed] [Google Scholar]
- 37.Dalton B. E., Loew E. R., Cronin T. W., Carleton K. L., Spectral tuning by opsin coexpression in retinal regions that view different parts of the visual field. Proc. Biol. Sci. 281, 20141980 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Dalton B. E., Lu J., Leips J., Cronin T. W., Carleton K. L., Variable light environments induce plastic spectral tuning by regional opsin coexpression in the African cichlid fish, Metriaclima zebra. Mol. Ecol. 24, 4193–4204 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chen P.-J., Awata H., Matsushita A., Yang E.-C., Arikawa K., Extreme spectral richness in the eye of the common bluebottle Graphium sarpedon. Front. Ecol. Evol. 4, 1–18 (2016). [Google Scholar]
- 40.Kitamoto J., Sakamoto K., Ozaki K., Mishina Y., Arikawa K., Two visual pigments in a single photoreceptor cell: Identification and histological localization of three mRNAs encoding visual pigment opsins in the retina of the butterfly Papilio xuthus. J. Exp. Biol. 201, 1255–1261 (1998). [DOI] [PubMed] [Google Scholar]
- 41.Sakamoto K., Hisatomi O., Tokunaga F., Eguchi E., Two opsins from the compound eye of the crab Hemigrapsus sanguineus. J. Exp. Biol. 199, 441–450 (1996). [DOI] [PubMed] [Google Scholar]
- 42.Frank T. M., Porter M., Cronin T. W., Spectral sensitivity, visual pigments and screening pigments in two life history stages of the ontogenetic migrator Gnathophausia ingens. J. Mar. Biol. Assoc. U. K. 89, 119–129 (2009). [Google Scholar]
- 43.Cronin T. W., Marshall J., “The unique visual world of mantis shrimps” in Complex Worlds from Simpler Nervous Systems, Prete F. R., Ed. (MIT Press, Cambridge, MA, 2004), pp. 239–268. [Google Scholar]
- 44.Cronin T. W., King C. A., Spectral sensitivity of vision in the mantis shrimp, Gonodactylus oerstedii, determined using noninvasive optical techniques. Biol. Bull. 176, 308–316 (1989). [DOI] [PubMed] [Google Scholar]
- 45.Hahn M. W., Distinguishing among evolutionary models for the maintenance of gene duplicates. J. Hered. 100, 605–617 (2009). [DOI] [PubMed] [Google Scholar]
- 46.Force A., et al. , Preservation of duplicate genes by complementary, degenerative mutations. Genetics 151, 1531–1545 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Roberts N. W., Porter M. L., Cronin T. W., The molecular basis of mechanisms underlying polarization vision. Philos. Trans. R. Soc. Lond. B Biol. Sci. 366, 627–637 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Prinster S. C., Hague C., Hall R. A., Heterodimerization of G protein-coupled receptors: Specificity and functional significance. Pharmacol. Rev. 57, 289–298 (2005). [DOI] [PubMed] [Google Scholar]
- 49.Awata H., Wakakuwa M., Arikawa K., Evolution of color vision in pierid butterflies: Blue opsin duplication, ommatidial heterogeneity and eye regionalization in Colias erate. J. Comp. Physiol. A Neuroethol. Sens. Neural Behav. Physiol. 195, 401–408 (2009). [DOI] [PubMed] [Google Scholar]
- 50.Shen W. L., et al. , Function of rhodopsin in temperature discrimination in Drosophila. Science 331, 1333–1336 (2011). [DOI] [PubMed] [Google Scholar]
- 51.Katana R., et al. , Chromophore-independent roles of opsin apoproteins in Drosophila mechanoreceptors. Curr. Biol. 29, 2961–2969.e4 (2019). [DOI] [PubMed] [Google Scholar]
- 52.Zanini D., et al. , Proprioceptive opsin functions in Drosophila larval locomotion. Neuron 98, 67–74.e4 (2018). [DOI] [PubMed] [Google Scholar]
- 53.Edgar R. C., MUSCLE: Multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 32, 1792–1797 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Edgar R. C., MUSCLE: A multiple sequence alignment method with reduced time and space complexity. BMC Bioinf. 5, 113 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Tamura K., Stecher G., Peterson D., Filipski A., Kumar S., MEGA6: Molecular evolutionary genetics analysis version 6.0. Mol. Biol. Evol. 30, 2725–2729 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Stamatakis A., RAxML-VI-HPC: Maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics 22, 2688–2690 (2006). [DOI] [PubMed] [Google Scholar]
- 57.Stamatakis A., RAxML version 8: A tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 30, 1312–1313 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Stamatakis A., Hoover P., Rougemont J., A rapid bootstrap algorithm for the RAxML Web servers. Syst. Biol. 57, 758–771 (2008). [DOI] [PubMed] [Google Scholar]
- 59.Abascal F., Zardoya R., Posada D., ProtTest: Selection of best-fit models of protein evolution. Bioinformatics 21, 2104–2105 (2005). [DOI] [PubMed] [Google Scholar]
- 60.Darriba D., Taboada G. L., Doallo R., Posada D., ProtTest 3: Fast selection of best-fit models of protein evolution. Bioinformatics 27, 1164–1165 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Huelsenbeck J. P., Ronquist F., MRBAYES: Bayesian inference of phylogenetic trees. Bioinformatics 17, 754–755 (2001). [DOI] [PubMed] [Google Scholar]
- 62.Ronquist F., et al. , MrBayes 3.2: Efficient Bayesian phylogenetic inference and model choice across a large model space. Syst. Biol. 61, 539–542 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Miller M., Pfeiffer W., Schwartz T., “Creating the CIPRES Science Gateway for inference of large phylogenetic trees” in Gateway Computing Environments Workshop (GCE), 2010 (IEEE, 2010), pp. 1–8. [Google Scholar]
- 64.Miller M. A., et al. , A RESTful API for access to phylogenetic tools via the CIPRES Science Gateway. Evol. Bioinform. Online 11, 43–48 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Guex N., Peitsch M. C., SWISS-MODEL and the Swiss-PdbViewer: An environment for comparative protein modeling. Electrophoresis 18, 2714–2723 (1997). [DOI] [PubMed] [Google Scholar]
- 66.Guex N., Peitsch M. C., Schwede T., Automated comparative protein structure modeling with SWISS-MODEL and Swiss-PdbViewer: A historical perspective. Electrophoresis 30, S162–S173 (2009). [DOI] [PubMed] [Google Scholar]
- 67.Johansson M. U., Zoete V., Michielin O., Guex N., Defining and searching for structural motifs using DeepView/Swiss-PdbViewer. BMC Bioinf. 13, 173 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Yang Z., PAML 4: Phylogenetic analysis by maximum likelihood. Mol. Biol. Evol. 24, 1586–1591 (2007). [DOI] [PubMed] [Google Scholar]
- 69.Yang Z., PAML: A program package for phylogenetic analysis by maximum likelihood. Comput. Appl. Biosci. 13, 555–556 (1997). [DOI] [PubMed] [Google Scholar]
- 70.Gao F., et al. , EasyCodeML: A visual tool for analysis of selection using CodeML. Ecol. Evol. 9, 3891–3898 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Pond S. L., Frost S. D., Datamonkey: Rapid detection of selective pressure on individual sites of codon alignments. Bioinformatics 21, 2531–2533 (2005). [DOI] [PubMed] [Google Scholar]
- 72.Poon A. F., Frost S. D., Pond S. L., Detecting signatures of selection from DNA sequences using Datamonkey. Methods Mol. Biol. 537, 163–183 (2009). [DOI] [PubMed] [Google Scholar]
- 73.Pond S. L. K., Muse S. V., “HyPhy: Hypothesis testing using phylogenies” in Statistical Methods in Molecular Evolution, Nielsen R., Ed. (Springer, 2005), pp. 125–181. [Google Scholar]
- 74.Weaver S., et al. , Datamonkey 2.0: A modern web application for characterizing selective and other evolutionary processes. Mol. Biol. Evol. 35, 773–777 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Cronin T. W., Porter M. L., Bok M. J., Wolf J. B., Robinson P. R., The molecular genetics and evolution of colour and polarization vision in stomatopod crustaceans. Ophthalmic Physiol. Opt. 30, 460–469 (2010). [DOI] [PubMed] [Google Scholar]
- 76.Ishii T., Hirota J., Mombaerts P., Combinatorial coexpression of neural and immune multigene families in mouse vomeronasal sensory neurons. Curr. Biol. 13, 394–400 (2003). [DOI] [PubMed] [Google Scholar]
- 77.Bodkin D. K., Knudson D. L., Assessment of sequence relatedness of double-stranded RNA genes by RNA-RNA blot hybridization. J. Virol. Methods 10, 45–52 (1985). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The generated N. oerstedii transcriptome and opsin transcript sequences are available at GenBank (SRA PRJNA609025, accession nos. MT112859–MT112888). The full details of our methods, including sequence alignments, are presented in the SI Appendix and Datasets S1–S6.