Polyketide synthases (PKSs) assemble activated carboxylic acids to elaborate chemical compounds (1). The key synthetic step is the C-C bond-forming condensation of an acyl moiety (e.g., acetyl-coenzyme A [CoA]) with an α-carboxyacyl moiety (e.g., malonyl-CoA) on release of CO2. The emerging β-ketoacyl compound can optionally be further modified by three accessory catalytic functions. Since this reaction sequence can be repeated, with each elongation varying in accessory catalytic functions, polyketides can be rich in chemistry (Fig. 1A). Many polyketides are of pharmaceutical relevance, among them several top-selling small molecule drugs (2): for example, antibiotics (e.g., erythromycin and tetracycline), antineoplastics (e.g., daunorubicin), and immunosuppressants (e.g., rapamycin) (3).
Fig. 1.
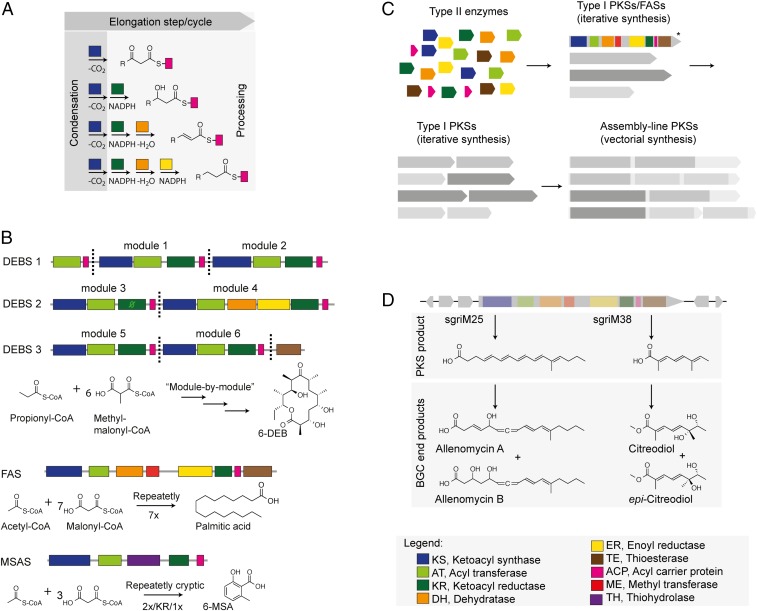
Iterative PKS. (A) Concept of FAS/PKS-mediated synthesis. Processing varies, resulting in different chemistries as indicated. Domain coloring is as indicated in the legend. (B) Biosynthesis of the erythromycin precursor 6-DEB, MSASs, and palmitic acid. DEBS, deoxyerythronolide B synthase. (C) Model for the PKS evolution specifically including iterative type I PKSs. Iterative PKSs predate PKS modules that comprise assembly lines. *For clarity, one multidomain module is shown with domains in color code. (D) Bacterial monomodular PKS gene clusters discovered by Wang et al. (7). Example on two biosynthetic gene clusters (BGCs) indicating PKS products and BGC end products. The scheme of BGC is for illustration only and does not reflect gene organization in BGCs sgriM25 or sgriM38.
PKSs are subdivided into three categories: type I PKSs are large multifunctional proteins composed of several catalytic and functional domains, type II PKSs are characterized by discrete monofunctional enzymes, and type III PKSs catalyze polyketide formation within a single active site (1). Type I PKSs are mainly responsible for the wealth of pharmacologically important activities found in this compound class.
Type I PKSs further divide in iterative and modular systems. Modular type I PKSs occur as large multimodular assembly-line complexes in which acyl substrates are successively condensed module by module. The modules assemble in a vectorial manner, and the sequence of the modules essentially defines the sequence of functional groups in the final compound. The 6-deoxyerythronolide B (6-DEB) synthase from the bacterium Saccharopolyspora erythraea is the prototypical modular PKS producing the 6-DEB as precursor for the antibiotic erythromycin (4) (Fig. 1B). The high degree of organization of modular PKSs, forming assembly lines of up to several megadaltons, is unique among proteins. On the other hand, iterative type I PKSs perform synthesis in a recursive manner, similar to fatty acid synthases (FASs), with the catalytic domains of a single multidomain protein repeatedly condensing precursor units until the specific length of the compound is reached (Fig. 1B). Probably, the best-studied representatives are PksA from the fungal genus Aspergillus, initiating biosynthesis of the environmental carcinogen aflatoxin B1 (5), and the 6-methylsalicylic acid synthase (MSAS) from the fungus Penicillium patulum (6) (Fig. 1B).
Since the synthesis is performed within single multidomain proteins in recursive manner, the chemical complexity of compounds derived from the iterative systems is usually less sophisticated compared to compounds from the more versatile modular systems. This is the reason why iterative PKSs usually receive far less comment than the modular ones. In their PNAS article, Wang et al. (7) extend the relevance of iterative PKSs in several aspects: as efficient producers of natural compounds, as targets for protein engineering, and as an inherent part in the evolution of the PKS protein family.
Understanding the evolution of PKSs, including FASs and the related nonribosomal peptide synthetases, is fundamental for disclosing basic processes that drive protein evolution in general (8, 9). Modular PKSs also bear high potential for the custom synthesis of polyketides, and approaches inspired by evolution can increase success rates in custom polyketide assembly (10, 11). In the generally accepted model of PKS evolution, the type II proteins are considered as the precursors that gave rise to the type I multidomain modules. Gene duplication of modules followed by more subtle adaptions played the fundamental role for the emergence of the multimodular systems capable of performing elaborate assembly-line–like synthesis. Details of this model still remain elusive. For example, the role of iterative PKSs in the evolution of the modular PKSs is not understood. Based on current knowledge, iterative PKSs are primarily found in fungi (and just occasionally in bacteria), while the modular assembly-line PKSs are widespread in bacteria. Can monomodular iterative PKSs play a role in the evolution of multimodular PKS assembly lines when hardly present in bacteria?
The work of Wang et al. (7) is essentially built on the question of whether iterative PKSs are more broadly distributed in bacteria than currently assumed. Their answers revise the current view on their relevance in bacteria and shed light onto their role in PKS evolution. In an array of experiments, Wang et al. (7) first identify monomodular PKSs from the bacterial genus Streptomyces by genome mining using strict selection criteria for a PKS module to be standalone in biological gene clusters. The separation of genes encoding PKSs is a valid argument for the PKSs to act iteratively similar as fungal iterative PKSs. From this pool, several hundred hits were selected and further classified by a ketoacyl synthase-based phylogenetic analysis. PKSs from several classes of bacterial monomodular PKSs were then selected for experimental validation of their iterative working mode. This study shows convincingly that iterative PKSs are indeed more broadly distributed in bacteria than predicted so far. Tight phylogenetic relationships among KS domains suggest that bacterial PKSs (iterative and modular) and fungal iterative PKSs are evolutionarily related.
Based on the broad distribution of iterative PKSs in bacteria, Wang et al. (7) posit that the multimodular PKSs occurring in bacteria are the direct descendants of monomodular (iterative) PKSs (Fig. 1C). Since duplicated modules inherit catalytic properties of their descendants, a monomodular (iterative) PKS could be particularly competent for generating multimodular PKSs because of its high substrate specificity and chain length tolerance. Wang et al. (7) see evidence for this scenario in the mycolactone-producing PKS assembly line in which KS domains with >97% sequence identity (12), reminiscent of the bacterial monomodular (iterative) precursor, accept substrates of remarkably different chemistry and chain length.
In the following, the biosynthetic aspects of this work are highlighted before revisiting PKS evolution again. Type I FASs are prototypical iterative megasynthases. Fatty acids are fundamental building blocks of essential cellular compounds but chemically primitive owing to FASs’ low versatility in the number of elongations performed per product and their narrow substrate tolerance (13). Although iterative PKSs are similar to FASs, they can increase the chemical complexity of products by accepting a broad spectrum of acyl substrates and by varying the catalytic steps used per elongation cycle. For analysis of product spectra of monomodular PKSs and their gene clusters, Wang et al. (7) transformed encoding genes under control of a strong promoter in the heterologous expression host Streptomyces lividans. The products that they identified are remarkably complex, which raises the relevance of iterative PKSs as a rich source for complex polyketides. Not least, Wang et al. (7) determined allenic polyketides and citreodiols as end products of two gene clusters in the native host of which the former may be involved in chemical communication (Fig. 1D). The product spectra unambiguously reveal the monomodular PKSs, identified in this study, as iterative PKSs. They perform repeated elongations, accept “unusual” acyl moieties, and mostly produce products via cryptic pathways. Specifically, PKSs characterized by Wang et al. (7) seem to rely on cryptic use of the enoyl reductase and/or the methyl transferase domain (Fig. 1D).
To outline the nature of cryptic coding in more detail and to return to PKS evolution again, the synthetic pathway of MSAS is described exemplarily in the following (Fig. 1C). MSAS first condenses the acetyl moiety with two moieties of malonyl. The triketide is then restricted from further KS-mediated condensation unless reduced by the ketoacyl reductase domain. After another elongation, the compound is released on hydrolysis. Product synthesis by MSAS relies on well-coordinated condensation and reduction reactions, calling for fine-tuned kinetic properties of the catalytic domains. Despite the remarkable synthetic capacity of MSAS, this example also shows the dilemma of iterative PKSs. Accessible product spectra are constrained by the number of catalytic processes that can be coordinated on the single multidomain protein. As mentioned above, Wang et al. (7) suggest that monomodular (iterative) PKSs are the direct ancestors of multimodular PKSs. A glimpse of the synthetic capacity of bacterial iterative PKSs, the already known ones (14) and the newly discovered (7), offers an interesting additional perspective on this hypothesis. A driver for evolving multimodular PKSs with the iterative proteins could have been the increased synthetic capacity of duplicated iterative PKSs. When collaborating in polyketide synthesis, a bimodular offspring can directly reward gene duplication by providing a more elaborate polyketide core structure (Fig. 1C). The specific features for vectorial synthesis in multimodular PKSs (15) appear later in evolution at multimodular level. The hidden iterative function of some modules of multimodular PKS assembly lines may reflect their origin as monomodular iterative PKSs. In contrast, a direct benefit of an initial gene duplication of a monomodular noniterative PKS is less obvious. Since operating noniteratively, compounds of low-modular assembly lines would remain of low complexity while produced at high metabolic costs.
Recently, Nivina et al. (16) have summarized current insights in the evolution of PKSs and suggested an “alternative model” for PKS evolution that is able to broadly include variants of PKSs. Key to the model is the emergence of modular PKS gene clusters by recombination events from a pool of “PKS gene material” but not by extensive gene duplication of modules within the same gene cluster. In this model, gene clusters initially appeared as a patchwork of sequences from different sources, and gene conversion, a process homogenizing homologous sequences, played a key role in shaping the modular PKSs as they occur today. A body of evidence is supporting this alternative model, and some of the arguments for the “conventional model” outlined above are also valid here. For example, the high conservation of the KS domain in mycolactone synthesis can also be regarded as the fingerprint of extensive gene conversion. Also, in the alterative model, iterative PKSs can play an important role as part of a pool of genes and fragments that assembled to modular PKS assembly lines.
Likely, the role of bacterial iterative PKS in the evolution of the intricate PKS assembly lines has been prominent. The article of Wang et al. (7), demonstrating the broad distribution of iterative PKSs in bacteria and their complex product spectra, gives even more reason to think so.
Acknowledgments
Research of M.G. is supported by a Lichtenberg Grant of the Volkswagen Foundation and the Landes-Offensive zur Entwicklung wissenschaftlich-ökonomischer Exzellenz Program of the state of Hesse conducted within the framework of the MegaSyn Research Cluster. M.G. thanks Aleksandra Nivina for discussions about PKS evolution.
Footnotes
The author declares no competing interest.
See companion article, “Unraveling the iterative type I polyketide synthases hidden in Streptomyces,” 10.1073/pnas.1917664117.
References
- 1.Hertweck C., The biosynthetic logic of polyketide diversity. Angew. Chem. Int. Ed. Engl. 48, 4688–4716 (2009). [DOI] [PubMed] [Google Scholar]
- 2.Newman D. J., Cragg G. M., Natural products as sources of new drugs over the 30 years from 1981 to 2010. J. Nat. Prod. 75, 311–335 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Staunton J., Weissman K. J., Polyketide biosynthesis: A millennium review. Nat. Prod. Rep. 18, 380–416 (2001). [DOI] [PubMed] [Google Scholar]
- 4.Cortes J., Haydock S. F., Roberts G. A., Bevitt D. J., Leadlay P. F., An unusually large multifunctional polypeptide in the erythromycin-producing polyketide synthase of Saccharopolyspora erythraea. Nature 348, 176–178 (1990). [DOI] [PubMed] [Google Scholar]
- 5.Crawford J. M., Dancy B. C., Hill E. A., Udwary D. W., Townsend C. A., Identification of a starter unit acyl-carrier protein transacylase domain in an iterative type I polyketide synthase. Proc. Natl. Acad. Sci. U.S.A. 103, 16728–16733 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Parascandolo J. S., et al. , Insights into 6-methylsalicylic acid bio-assembly by using chemical probes. Angew. Chem. Int. Ed. Engl. 55, 3463–3467 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wang B., Guo F., Huang C., Zhao H., Unraveling the iterative type I polyketide synthases hidden in Streptomyces. Proc. Natl. Acad. Sci. U.S.A., 10.1073/pnas.1917664117 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jenke-Kodama H., Sandmann A., Müller R., Dittmann E., Evolutionary implications of bacterial polyketide synthases. Mol. Biol. Evol. 22, 2027–2039 (2005). [DOI] [PubMed] [Google Scholar]
- 9.Sherman D. H., Smith J. L., Clearing the skies over modular polyketide synthases. ACS Chem. Biol. 1, 505–509 (2006). [DOI] [PubMed] [Google Scholar]
- 10.Peng H., Ishida K., Sugimoto Y., Jenke-Kodama H., Hertweck C., Emulating evolutionary processes to morph aureothin-type modular polyketide synthases and associated oxygenases. Nat. Commun. 10, 3918 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wlodek A., et al. , Diversity oriented biosynthesis via accelerated evolution of modular gene clusters. Nat. Commun. 8, 1206 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Stinear T. P., et al. , Giant plasmid-encoded polyketide synthases produce the macrolide toxin of Mycobacterium ulcerans. Proc. Natl. Acad. Sci. U.S.A. 101, 1345–1349 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Heil C. S., Wehrheim S. S., Paithankar K. S., Grininger M., Fatty acid biosynthesis: Chain-length regulation and control. ChemBioChem 20, 2298–2321 (2019). [DOI] [PubMed] [Google Scholar]
- 14.Zhang Q., Pang B., Ding W., Liu W., Aromatic polyketides produced by bacterial iterative type I polyketide synthases. ACS Catal. 3, 1439–1447 (2013). [Google Scholar]
- 15.Lowry B., Li X., Robbins T., Cane D. E., Khosla C., A turnstile mechanism for the controlled growth of biosynthetic intermediates on assembly line polyketide synthases. ACS Cent. Sci. 2, 14–20 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nivina A., Yuet K. P., Hsu J., Khosla C., Evolution and diversity of assembly-line polyketide synthases. Chem. Rev. 119, 12524–12547 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]