Significance
Eastern equine encephalitis virus (EEEV) is an important pathogenic alphavirus that causes severe encephalitis and neuron damage with a high mortality rate of ∼50%. Currently, no approved antiviral agents are available for treatments of EEEV infection. Heparan sulfate (HS) proteoglycan on host cell surfaces has been identified as a host attachment factor/receptor for EEEV infection. This study demonstrates the first cryo-EM structure of the wild-type EEEV that has been examined when bound to an HS-mimicking polysaccharide, heparin. The identified axial and peripheral HS binding sites on each spike show structural similarity. The discovered HS binding sites on EEEV may also serve as potential targets for the development of antiviral agents against this highly infectious alphavirus.
Keywords: alphavirus, cryoelectron microscopy, heparan sulfate proteoglycan, infection, structure
Abstract
Eastern equine encephalitis virus (EEEV), a mosquito-borne icosahedral alphavirus found mainly in North America, causes human and equine neurotropic infections. EEEV neurovirulence is influenced by the interaction of the viral envelope protein E2 with heparan sulfate (HS) proteoglycans from the host’s plasma membrane during virus entry. Here, we present a 5.8-Å cryoelectron microscopy (cryo-EM) structure of EEEV complexed with the HS analog heparin. “Peripheral” HS binding sites were found to be associated with the base of each of the E2 glycoproteins that form the 60 quasi-threefold spikes (q3) and the 20 sites associated with the icosahedral threefold axes (i3). In addition, there is one HS site at the vertex of each q3 and i3 spike (the “axial” sites). Both the axial and peripheral sites are surrounded by basic residues, suggesting an electrostatic mechanism for HS binding. These residues are highly conserved among EEEV strains, and therefore a change in these residues might be linked to EEEV neurovirulence.
Eastern equine encephalitis virus (EEEV) is a mosquito-borne virus prevalent in North America that can cause severe encephalitis. A total of 121 human EEEV infections were reported in the United States between 2003 and 2016. Of these, 119 cases were classified as neuroinvasive and 110 were diagnosed as causing encephalitis or meningoencephalitis, resulting in 50 deaths (1). Although EEEV vaccination is available for horses, human vaccines against EEEV are not yet available. As symptomatic infections of EEEV in humans have been associated with high mortality rates (2, 3), vaccines or antiviral agents are urgently needed to prevent and cure EEEV infections.
EEEV is a member of the genus Alphavirus, belonging to the Togaviridae family. Pathogenic alphaviruses are transmitted by mosquitoes or other arthropods. Alphaviruses infect vertebrates including humans and have been broadly classified as Old World or New World viruses (4). In general, Old World alphaviruses such as Chikungunya (CHIKV), Ross River (RRV), Sindbis (SINV), Semliki Forest, and Mayaro viruses can cause fevers, rashes, and arthritis. In contrast, New World alphaviruses such as Venezuelan (VEEV), Eastern (EEEV), and Western (WEEV) equine encephalitis viruses can cause severe encephalitis in humans or domestic animals (4).
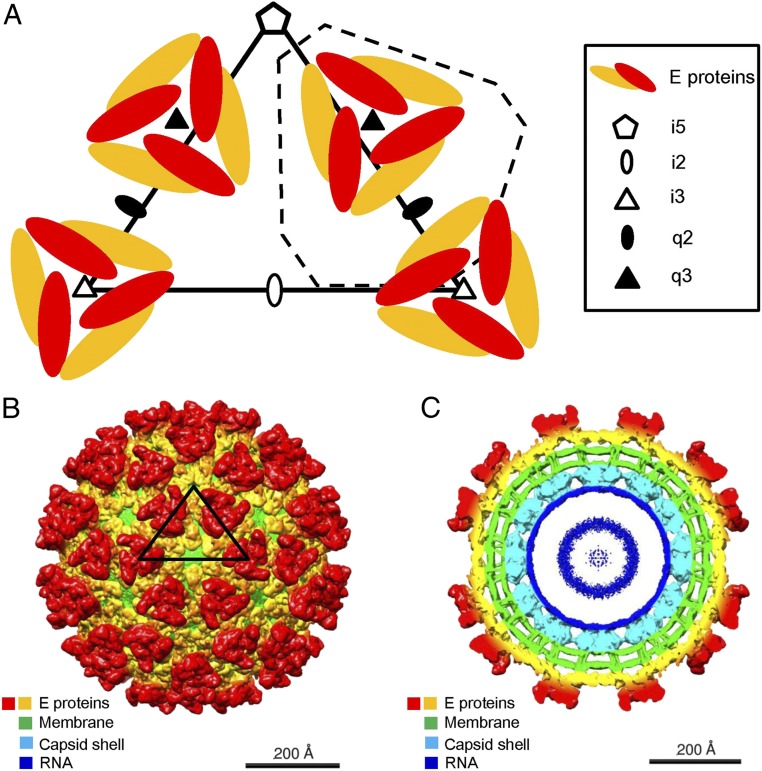
Alphaviruses are single-stranded, positive-sense RNA viruses that contain a host-derived internal lipid bilayer membrane surrounding the RNA genome, with a diameter of about 650 Å (5–13). The RNA genome encodes four nonstructural proteins (nsP1, nsP2, nsP3, and nsP4) and five structural proteins (E1, E2, E3, 6k, and capsid) (4, 14). Heterodimers of the transmembrane envelope proteins pE2 and E1 form trimeric spikes in the endoplasmic reticulum membrane of the host. The pE2 polypeptide is cleaved into E2 and E3 by a furin-like enzyme during maturation in the Golgi network (15). The assembled virions contain 240 copies of the two transmembrane envelope glycoproteins, E1 and E2, arranged into a T = 4 icosahedral lattice (16) (Fig. 1A). The virus structure consists of 60 quasi-threefold spikes (“q3” spikes) in general positions and 20 spikes located on the icosahedral threefold axes (“i3” spikes). Each spike contains three copies of the E1–E2 heterodimer (17, 18).
Fig. 1.
Cryo-EM reconstruction of EEEV–heparin (Hp). (A) Diagrammatic representation of the E1–E2 glycoprotein spikes (E proteins) arranged with T = 4 quasi-icosahedral symmetry. One asymmetric unit is outlined by a black triangle. An alternative asymmetric unit is outlined by a dashed line. (B) Three-dimensional cryo-EM map of the EEEV–Hp complex, viewed down an icosahedral twofold (i2) axis. An asymmetric unit is outlined by a black triangle. (C) A central cross section of the EEEV–Hp cryo-EM map viewed down an i2 axis. The coloring in B and C is by radius according to the sequence of colors in a rainbow with the largest radius colored red and the smallest colored blue. Much of the red to orange regions are occupied by the E1 and E2 proteins.
The E1 ectodomain is divided into three domains: DI, DII, and DIII (SI Appendix, Fig. S1) (19). The “fusion loop” on DII can form a tight trimer at the tip of each spike that can insert itself into the endosomal membrane of the host cell endosome under acidic conditions to trigger fusion of the viral and host membranes. The E2 ectodomain consists of domain A (implicated in receptor binding), domain B (implicated in protecting the E1 fusion loop from premature exposure), domain C (an Ig fold probably involved in host recognition), and a β-ribbon connector (stabilizing domain B at neutral pH) (SI Appendix, Fig. S1) (19). The C-terminal portions of the E2 proteins penetrate the viral membrane and interact with the internal capsid proteins (20). The latter form a shell around the ∼11-kb viral RNA genome. Previous investigations have shown that the E2 protein binds to host cell receptors when initiating cell entry, whereas the E1 protein is required for pH-triggered membrane fusion within acidified endosomes (21). Following the fusion of viral and endosomal membranes, the nucleocapsid is delivered into the cytosol where it dissociates to release the viral RNA genome to initiate replication and subsequent assembly of progeny particles.
Alphaviruses infect a wide variety of tissues in arthropod vectors and mammalian hosts. Tissues associated with lymphoid, nervous, musculoskeletal, and gastrointestinal-related systems have been suggested to be involved in virus entry and propagation (22). Host membrane proteins such as DC-SIGN, L-SIGN (23), NRAMP2 (24), Mxra8 (25), and HS (22) have been suggested to function as receptors and attachment factors for alphaviruses. HS polysaccharides are composed of repeating disaccharide units of glucuronic acid and acetyl-glucosamine (GlcAβ1–4GlcNAcα1–4)n. Heparin (Hp) polysaccharides are similar to highly sulfated HS and composed of repeating disaccharide units of sulfated iduronic acid and acetyl-glucosamine (IdoAβ1–4GlcNAcα1–4)n. Incubation of an EEEV strain representative of naturally circulating viruses with Hp polymers or infection of HS-nonexpressing cells significantly reduces infectivity as demonstrated by reduction of plaque formation (22), implying that HS can act as an attachment receptor for wild-type EEEV (22). Hence, identification of the heparan sulfate (HS) binding sites on the EEEV surface should provide information on the mechanism of viral entry and neurovirulence.
Here, we determine and present the cryo-electron microscopic (cryo-EM) reconstruction of the wild-type EEEV in complex with Hp. A difference map generated by subtracting the EEEV virus density map from the EEEV–Hp complex map showed four Hp peaks on each q3 and i3 spike. All six “peripheral” sites are at chemically equivalent sites. Similarly, both axial sites are chemically equivalent. We further show that these HS binding sites on the E2 glycoprotein involve positively charged residues that are conserved in EEEV strains but not in other alphaviruses. Chondroitin sulfate (CS), a sulfated glycosaminoglycan (GAG), was unable to effectively inhibit EEEV viral infection, probably because it has only one negatively charged sulfate per disaccharide as opposed to about an average of two for Hp.
Results
The Cryo-EM Structure of the EEEV–Hp Complex.
Purified EEEV particles were mixed with low–molecular-weight Hp (6 kDa) at pH ∼8.0 and flash-frozen in liquid ethane on lacey carbon grids for cryo-EM analysis. About 1,200 micrographs were recorded. After motion correction (26) and contrast transfer function (CTF) estimation (27), ∼28,000 EEEV–Hp complexed particles were selected for further processing. After reference-free two-dimensional (2D) classification using the RELION software (28), ∼17,000 particles with consistent features were used to reconstruct the structure of EEEV complexed with Hp to 5.8-Å resolution (Fig. 1 B and C) using the jspr software (29). Similarly, ∼12,000 uncomplexed native EEEV particles were used as a control experiment to produce a 5.2-Å resolution structure of EEEV (Table 1 and SI Appendix, Fig. S2 A and B).
Table 1.
Statistics of cryo-EM data collection, processing, and model fitting
| EEEV | EEEV–Hp | |
| Data collection and processing | ||
| Nominal magnification | 38,000 | 81,000 |
| Voltage, kV | 300 | 300 |
| Total electron dose, e−/Å2 | 32.05 | 32.07 |
| No. of frames per movie | 40 | 60 |
| Exposure time, s | 10 | 12 |
| Frame exposure time, ms | 250 | 200 |
| Dose rate, e−⋅Å−2⋅s−1 | 3.20 | 2.67 |
| Defocus range, μm | 0.5–3.5 | 0.5–3.5 |
| Pixel size, Å | 1.58 | 1.73 |
| Number of micrographs collected | 696 | 1,581 |
| Final particle images used for single particle reconstruction, no. | 11,867 | 17,022 |
| Symmetry imposed | Icosahedral | Icosahedral |
| Map resolution, Å (FSC = 0.143) | ||
| Unmasked | 5.17 | 5.79 |
| Masked | 4.76 | 5.56 |
| Sharpening B factor, Å2 | −174 | −305 |
| Model fitting | ||
| Model used | — | E1–E2 heterodimer (PDB: 6MX4) |
| Fitting method | — | Rigid-body fitting using EMfit |
| Sumf* (%) value (EMfit) | — | 27.58 |
| Model-map correlation coefficient (Phenix)† | — | 0.7440 |
Difference between the EEEV–Hp and the Native EEEV Structures.
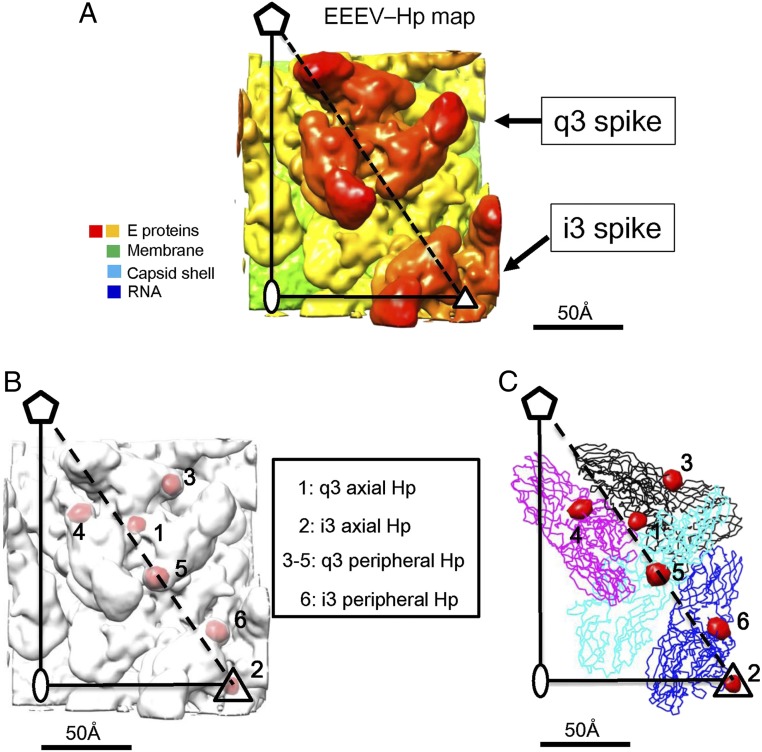
The EEEV–Hp cryo-EM map (Fig. 2A) was compared with the native EEEV map to identify the HS binding sites on the viral surface (Fig. 2B). Although the external diameter of the native EEEV cryo-EM reconstruction was ∼630 Å, similar to the size of the 4.4-Å resolution VEEV cryo-EM structure reported earlier (5), it was smaller than the external diameter of the EEEV–Hp complex by ∼30 Å as determined from their radial density distributions (SI Appendix, Fig. S2 A and B). Independent calibrations of the microscope magnification did not show any dramatic change. Thus, the difference in diameter of the native and complexed EEEV structures was assumed to be real. Possibly the additional negative charge of Hp on neighboring q3 and i3 spikes might have caused the virus to expand slightly to relieve the interspike electrostatic forces or the binding of Hp caused a change in the conformation of the E1–E2 heterodimers. Thus, for calculating a difference map between the native and Hp-bound EEEV complexed reconstructions, the pixel size of the cryo-EM map of native EEEV particles was modified from 1.58 to 1.66 Å to match the radius of the EEEV and EEEV–Hp reconstructions. The native and complexed EEEV structures were each Fourier transformed to allow least-squares scaling of the electron density from separate resolution ranges. The scaled difference densities were then Fourier transformed back into real space for calculation of a 5.8-Å resolution difference density map. This difference map (Fig. 2C and SI Appendix, Fig. S3) showed four roughly spherical difference peaks associated with each q3 and each i3 spikes. These peaks had heights varying between 22σ and 23σ, where σ is the SD of the electron density in the map. The SD of the highest noise peak was 7σ. Presumably, these peaks were Hp molecules bound to the virus. The four Hp molecules were similarly associated with each i3 and each q3 trimeric spike. Three of these peaks (“peripheral”) were related by the icosahedral or quasi-threefold symmetry of each spike. These were separated by ∼57 Å from each other (SI Appendix, Fig. S3). The fourth peak (“axial”) was located on the threefold axis and at the vertex of each spike. On each spike, the axial site was 33 Å distant from the three surrounding peripheral sites (SI Appendix, Fig. S3). These results suggest that there were four HS binding sites on each glycoprotein spike of EEEV.
Fig. 2.
Identification of HS binding sites in an asymmetric unit of the EEEV viral particle. (A) The cryo-EM map of EEEV–Hp is shown in radial coloring from yellow (∼250 Å) to red (∼350 Å). (B) The superposition of the difference map (red) and the EEEV–Hp map (white). The larger difference densities were located at the axial sites (labeled 1 and 2) and peripheral sites (labeled 3 to 5) around the q3 spike and 6 at the i3 spike. (C) The superposition of the difference map (red blobs) and the structures of the E1–E2 glycoproteins represented as ribbon drawings. The four E1–E2 heterodimers are colored black, magenta, cyan, and blue. These were fitted into the EEEV–Hp cryo-EM map using the EMfit program (31, 32).
Analysis of the HS-Binding Sites.
The EEEV–Hp map was interpreted by fitting the E1 and E2 structures of EEEV from the earlier cryo-EM determination (30) into the present cryo-EM map (Fig. 2C) while imposing T = 4 quasi-icosahedral symmetry using the EMfit program (31, 32). The peripheral peaks were located close to the E2 β-connector (SI Appendix, Fig. S1). The spherical shape of the difference peaks (SI Appendix, Fig. S3) is consistent with an average of multiple orientations and conformations of the Hp molecules at each site (33).
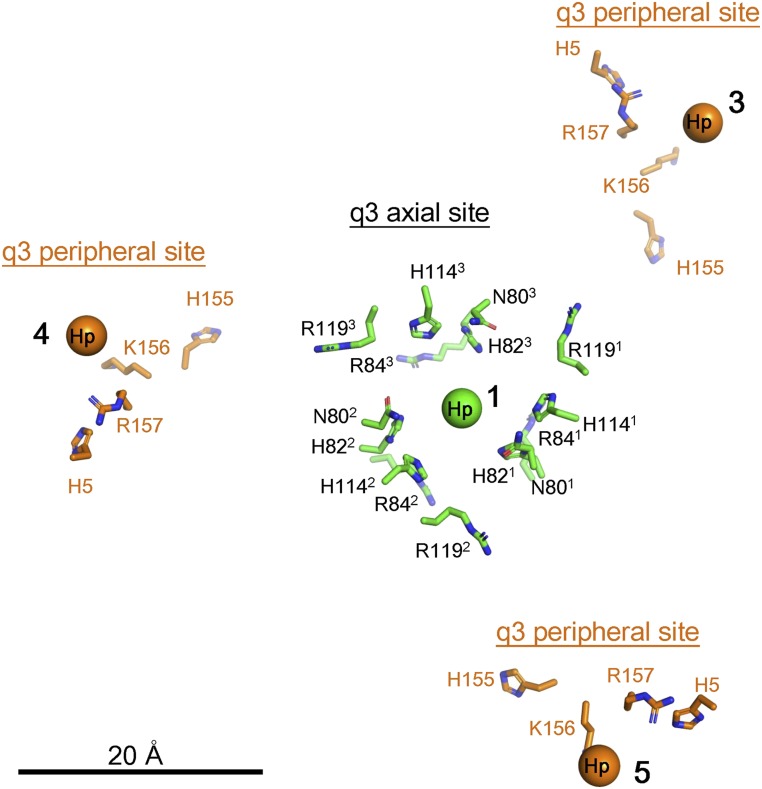
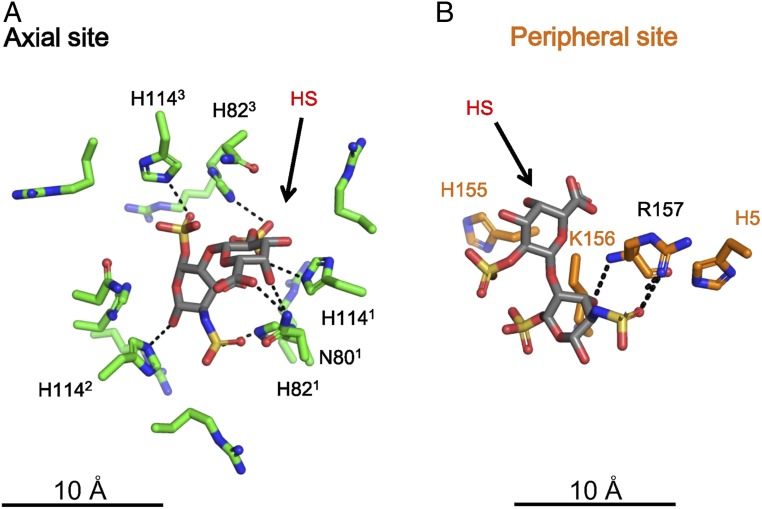
The axial HS sites were surrounded by sets of threefold-related His82, Arg84, His114, and Arg119 residues on E2 (Fig. 3), whereas the peripheral HS sites were located near the basic residues His5, His155, Arg156, and Lys157 of E2 (Fig. 3), suggesting that the negatively charged HS molecules (SI Appendix, Fig. S4) were bound to the virus by the positive charges surrounding each HS binding site (Fig. 3). These basic residues were found to be highly conserved in EEEV strains (Fig. 4A) but poorly conserved in other alphaviruses (Fig. 4B), indicating that alphaviruses may interact with cellular receptors at different sites to infect other types of host cells.
Fig. 3.
HS binding environment at the axial and peripheral HS sites. The q3 axial sites are labeled in black, and the q3 peripheral sites are labeled in orange. The center of the axial HS density (labeled 1) is shown as a green sphere. The side chains of the residues in the axial site are shown in green. The center of the peripheral Hp densities (labeled 3, 4, and 5) are shown as orange spheres, and the side chains of the residues, H5, H155, K156, and R157 of E2 are shown in orange.
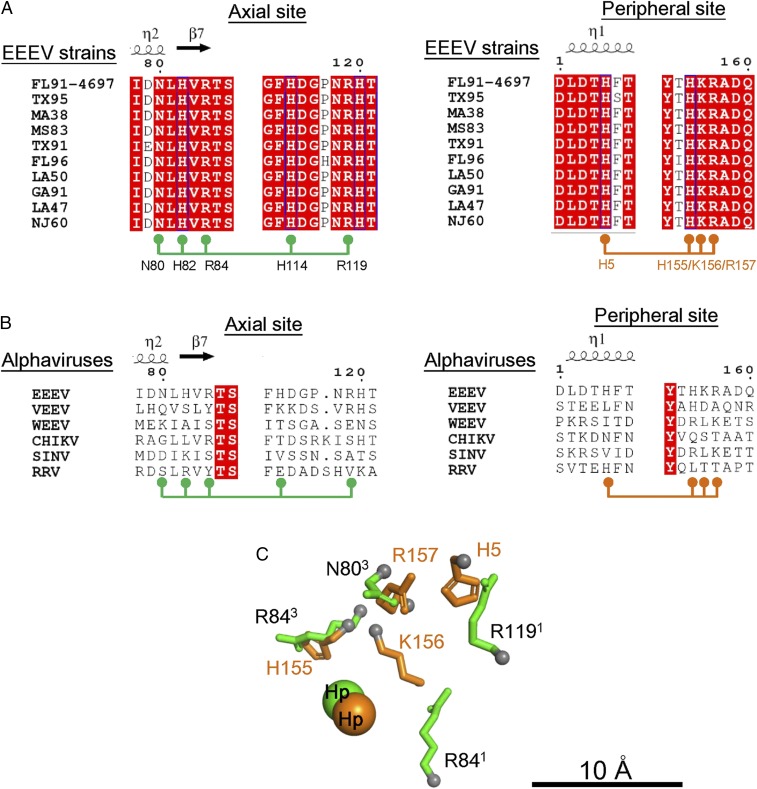
Fig. 4.
Chemically equivalent residues in the axial and peripheral HS binding sites. (A) Sequence alignment of E2 of different EEEV strains (FL91-4679, TX95, MA38, MS83, TX91, FL96, LA50, GA91, LA47, and NJ60) of the E2 glycoproteins. (B) Sequence alignment of E2 of the different alphaviruses (VEEV, EEEV, WEEV, CHIKV, SINV, and RRV) of the E2 glycoproteins. In A and B, residues in the axial (N80, H82, R84, H114, and R119) and peripheral (H5, H155, K156, and R157) sites are indicated below the extracted sequence regions. Residues highlighted in red are completely conserved, in blue are highly conserved, and in white are not conserved. (C) Structural alignment of residues in the axial (labeled black) and peripheral (labeled orange) sites. The Cα atoms of residues in the axial and peripheral sites are shown as gray spheres. The “best” structural alignment is shown with the lowest RMSD between the axial and peripheral sites as listed in Table 2.
In EEEV, both the axial and peripheral binding sites contain arginine and histidine residues. To compare these sites, the atoms on the side chains of the peripheral sites (Nε2 of H5, Nε2 of H155, Nζ of K156, and Cζ of R157 on E2) were aligned with the atoms of the side chains of the axial site (Nε2 of H82, Cζ of R84, Nδ2 of N80, Nε2 of H114, and Cζ of R119 on E2). In addition, the centers of the axial and peripheral Hp densities were also considered. There were numerous ways in which the peripheral and axial sites could be aligned. However, they could be differentiated by the root-mean-square deviation (RMSD) of the aligned atoms (Table 2). The “best” alignment gave a RMSD difference between the aligned atoms of ∼2.3 Å (Fig. 4C). Possible HS binding configurations from docking experiments also suggested that the sulfate groups in HS can interact with basic residues at the axial (Fig. 5A) and peripheral HS sites (Fig. 5B).
Table 2.
Structure alignment of HS binding residues at the peripheral and axial sites
| HS site | Alignment | RMSD, Å | |||
| Peripheral | H5 | R157 | K156 | H155 | 2.36 |
| Axial | R1191 | N803 | R841 | R843 | |
| Peripheral | H5 | R157 | K156 | H155 | 2.44 |
| Axial | H821 | H1141 | R842 | R843 | |
| Peripheral | H5 | R157 | K156 | H155 | 2.73 |
| Axial | N801 | H1141 | R1192 | R842 | |
| Peripheral | H5 | R157 | K156 | H155 | 2.84 |
| Axial | H1141 | R841 | N801 | R1192 | |
| Peripheral | H5 | R157 | K156 | H155 | 2.84 |
| Axial | H821 | N801 | R841 | R1191 | |
| Peripheral | H5 | R157 | K156 | H155 | 2.97 |
| Axial | N801 | H821 | H1142 | R842 | |
The E2 residues H5, H155, K156 and R157 surrounding the peripheral HS site, and H82, R84, N80, H114 and R119 surrounding the axial HS site were selected for structural comparison between the two HS sites. The atoms, Nε2 of H5, Nε2 of H155, Nζ of K156 from the peripheral sites and Cζ of R157, Nε2 of H82, Cζ of R84, Nδ2 of N80, Nε2 of H114, and Cζ of R119 from the axial sites and the centers of the axial and peripheral HS densities included in the structural alignment and RMSD calculations. In the axial HS site, trimeric copies of five residues are surrounding the axial HS binding sites. The first (1), second (2), or third (3) sets of axial residues are labeled in a clockwise direction when looking down the E1–E2 glycoprotein trimeric spike. These residues are used for the structural alignment.
Fig. 5.
Modeling of HS into the axial and peripheral HS binding sites. (A) The E2 residues in the axial site are labeled in black, and the HS molecule from one of the docking results is labeled in red. Residues that make polar contacts with HS are indicated with black dashed lines. (B) The positively charged residues of E2 in one of the peripheral sites are shown in orange, and the HS molecule from one of the docking results is shown in red. The residues that make polar contacts with HS are indicated with black dashed lines.
Specificity of EEEV–HS Interactions.
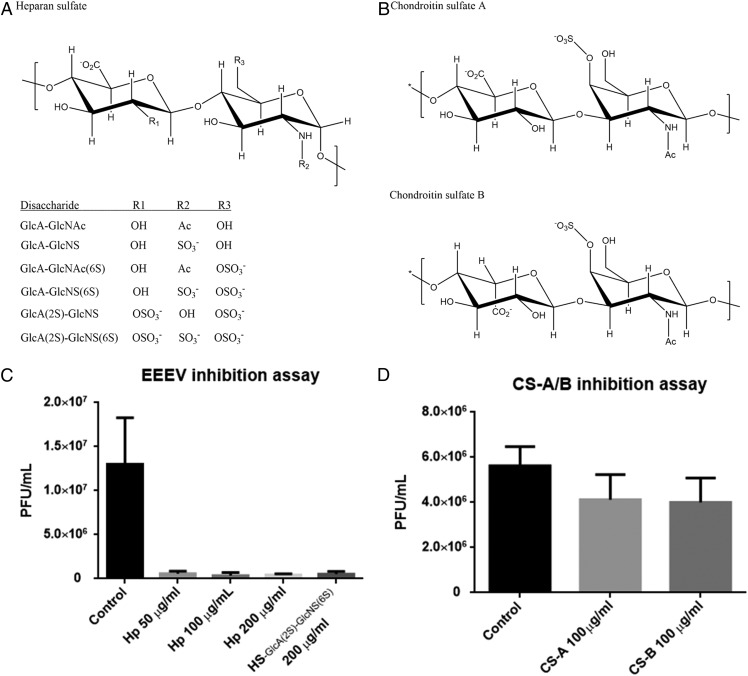
GAGs had also been reported previously as attachment factors for viral entry in alphaviruses, although it is unclear which of these phenotypes occur in naturally circulating viruses versus those resulting from adaptation of laboratory strains to cultured cells (34, 35). In brain tissues, CS and HS constitute 30% and 50% of the total GAGs, respectively (36). Similar to HS (Fig. 6A), CS is a sulfated GAG with repeating disaccharide units of glucuronic/iduronic acid and acetylgalactosamine (GlcAβ1–3GalNAcβ1–4)n (Fig. 6B). To identify whether HS or CS function as the major cellular attachment factor for EEEV infection, cell-based inhibition assays were performed. When low–molecular-weight Hp (6 kDa) at a concentration of 100 μg/mL was added during EEEV infection of baby hamster kidney (BHK) cells, infectivity of the virus (as measured by plaque counts) was reduced by ∼90% (Fig. 6C), whereas CS at the same concentration showed a reduction of only ∼30% (Fig. 6D). Therefore, EEEV binds preferably to HS as opposed to CS, implying that EEEV initiates virus infection mostly but not exclusively by binding to HS.
Fig. 6.
Inhibition of EEEV by GAGs. (A) Chemical structure of HS. The scaffold of HS contains repeating units of glucuronic acid and N-glucosamine (GlcAβ1–4GlcNAcα1–4)n. The R1, R2, and R3 groups represent different moieties in the six different disaccharide units. (B) Chemical structures of CS-A and CS-B. CS-A/B contains repeating disaccharide units of glucuronic/iduronic acid and acetylgalactosamine (GlcAβ1/IdoAβ1–3GalNAcβ1–4)n, respectively. (C) Inhibition of EEEV by Hp treatment. Hp (6 kDa) concentrations of 50, 100, or 200 μg/mL were used to determine the EEEV inhibition. A HS disaccharide [GlcA(2S)–GlcNS(6S)] was tested at a concentration of 200 μg/mL. (D) EEEV inhibition assays by CS treatment. Chondroitin sulfate A (CS-A) and B (CS-B) were tested at a concentration of 100 μg/mL.
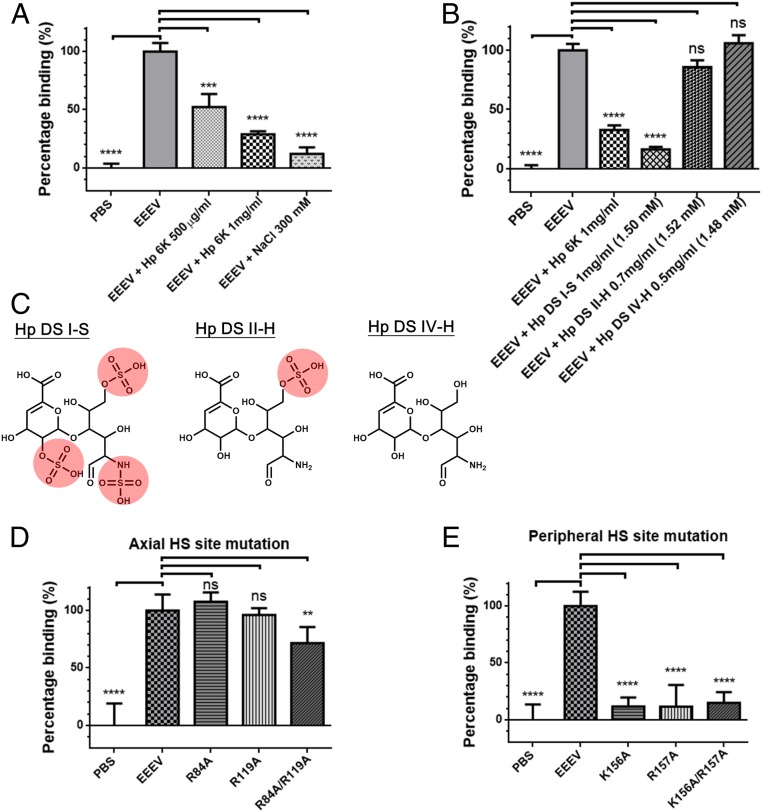
To test whether the binding of EEEV to HS is mediated by electrostatic interactions between the positively charged sulfate groups of HS and the negatively charged basic residues of EEEV, in vitro binding assays were conducted. This included varying the ionic strength with sodium chloride (Fig. 7A) and the addition of Hp as well as Hp disaccharides with varying degree of sulfations (Fig. 7 A–C). The results showed that increased ionic strength and highly sulfated Hp effectively disrupted the binding of EEEV onto the cell surface. Furthermore, EEEV mutants with mutations of E2 glycoprotein residues located either around the axial HS site (R84A/R119A) (Fig. 7D) or the peripheral HS site (K156A, R157A, K156A/R157A) (Fig. 7E) show significant decrease of binding to BHK cells. These results suggest that the electrostatic attraction between EEEV and the host cell HS is mediated by highly sulfated HS and the basic residues on the surface of EEEV.
Fig. 7.
In vitro binding assays of EEEV and mutants. (A) Treatments of EEEV with heparin (Hp) and sodium chloride (NaCl). EEEV, EEEV without treatment; Hp 1 mg/mL, EEEV incubated with 1 mg/mL Hp; Hp 500 μg/mL, EEEV incubated with 500 μg/mL Hp; NaCl 300 mM, EEEV incubated with 300 mM NaCl; PBS, controls without EEEV viral particles. (B) Treatment of EEEV with different Hp molecules. The 6-kDa Hp and three Hp disaccharide molecules, I-S, II-H, and IV-H, were incubated with EEEV and the compound concentrations of Hp molecules were indicated. (C) The chemical structures of three Hp disaccharide molecules (DS I-S, II-H, and IV-H). Sulfated groups were highlighted in red. (D) Examination of binding of wild-type EEEV and the three axial-HS site mutants, R84A, R119A, and R84A/R119A, to BHK cells. Virus concentrations of the mutant viruses were adjusted to the equal amount of wild-type EEEV before virus attachment. (E) Examination of binding of wild-type EEEV and the three peripheral-HS site mutants, K156A, R157A, and K156A/R157A, to BHK cells. Virus concentrations of the mutant viruses were adjusted to the equal amount of wild-type EEEV before virus attachment. For each set of experiments, five repeats were conducted, and the error bar represents the SEM. Data were normalized according to the results of PBS controls and EEEV (Materials and Methods). ns, not significant, P > 0.05; **P ≤ 0.01; ***P ≤ 0.001; ****P ≤ 0.0001.
Discussion
As the side-chain positions in the current structure of EEEV complexed with Hp were identified by fitting the E1–E2 structure derived from the recently published 4.4-Å native EEEV reconstruction (19, 30), it is possible to compare the amino acid side chains that comprise the chemically equivalent axial and peripheral HS sites on the i3 and q3 spikes (Table 2 and Fig. 4B). Both the axial and peripheral sites are formed by Arg and His residues, providing a positively charged environment for HS binding, as is also the case in other alphavirus strains (35, 37–39), in which mutations of residues to Arg within these regions as well as other sites resulted in increased HS binding ability and virus infectivity in vitro. These basic residues interact electrostatically with the HS sulfate groups, while other residues in the binding site make hydrogen bonds with HS (40).
In general, HS on cell surfaces is ∼80% less sulfated (less than one sulfate group per disaccharide unit on average) and ∼20% highly sulfated (more than one sulfate group per disaccharide unit on average) (41). Hp used in the current studies is more equivalent to the highly sulfated HS form. The EEEV axial and peripheral HS sites are both composed of polar and positively charged residues, suggesting that EEEV preferably binds to highly sulfated HS. A recent study on the porcine circovirus 2 (PCV2) in complex with HS (42) showed that PCV2 has three HS binding sites (SI Appendix, Fig. S5A) that are similar to the EEEV peripheral HS sites (SI Appendix, Fig. S5 B–D), implying that the Lys–Arg residues (or positive charge distribution) in a HS binding site might have a specific 3D arrangement that favors HS binding.
In previous studies, the 71KXX74KXXK77 sequence in the E2 glycoprotein of EEEV was shown to be involved in HS binding (22, 39, 43, 44). Furthermore, positively charged cell culture-adapted mutations that increase HS binding affinity occur at positions 70 (SINV) (43), 76 (VEEV) (44), and 79 (CHIKV) (39). However, the EEEV–Hp reconstruction showed that there were no Hp molecules bound at the 71KXX74KXXK77 sequence. It is possible that the 71KXX74KXX77K site does not directly interact with the low–molecular-weight Hp used in this study. It is also possible that the binding of low–molecular-weight Hp to the 71KXX74KXX77K site does not follow icosahedral symmetry, and thus we could not detect binding due to the imposed icosahedral symmetry during the reconstruction. By structural comparison, the axial and peripheral HS binding sites are composed of clustered positively charged residues, and these two sites share similarities. However, the 71KXX74KXXK77 sequence has Lys71, Lys74, and Lys77 separated by ∼10 Å and is less similar to the peripheral site of EEEV (SI Appendix, Fig. S6). On each i3 and q3 glycoprotein spike, there are threefold-related 71KXX74KXXK77 sequences which were ∼34 Å distant from each other. Thus, the 71KXX74KXXK77 sequences may form a larger positively charged area for EEEV to bind higher molecular-weight HS molecules on the cell surface (>100 kDa), consistent with mutational experiments in which one Lys at a time was changed to Ala (45), resulting in a reduced EEEV HS-dependent viral infectivity. Based on the identified binding sites, it is unlikely that HS induces fusion or interferes with the fusion process. Instead, the binding sites are located primarily on domain A and are responsible for initial attachment events. It is likely that larger HS molecules that are present on cells cross-link the axial and peripheral sites in an asymmetric unit.
Currently, there are no effective treatments for EEEV infections. The identified axial and peripheral HS binding sites, together with the previously identified 71KXX74KXXK77 sequence, should serve as promising targets for developing antiviral agents against EEEV by interfering with virus–host interactions.
Methods
Purification of SINV–EEEV Viral Particles.
Cell culture and virus infection experiments were conducted under biological safety level 2 (BSL-2) conditions using a chimeric Sindbis–EEEV virus (44). BHK-15 cells were cultured in minimum essential medium (MEM) supplemented with 1% nonessential amino acids (NEAA) and 10% heat-inactivated FBS and grown to ∼90% confluency. During virus infection, the infection medium (5 mL) containing MEM and 1% NEAA and EEEV virus (multiplicity of infection of ∼1 to 5) were added to obtain virus attachment at room temperature. The modified MEM (10 mL) was added for ∼17-h incubation at 37 °C. The culture medium containing viruses was passed through a 0.22-μm filter to remove cell debris. The viral particles were precipitated by 14% (wt/vol) PEG 6000 and 4.6% (wt/vol) NaCl. The precipitated viral particles were pelleted at 2,500 × g for 30 min at 4 °C and resuspended in 1 mL of TNE buffer (25 mM Tris, pH 8, 100 mM NaCl, and 0.1% EDTA). Further purification of EEEV viral particles was performed using a continuous Opti-prep density gradient (0∼90%) at 247,000 × g for 1.5 h at 4 °C (SW-41 swing rotor). The purified EEEV particles were buffer-exchanged with TNE buffer and concentrated to 1 mg/mL E2 glycoprotein for cryo-EM sample preparation.
Plaque Assay.
BHK-15 cells were grown in six-well plates to nearly 100% confluency. Before virus attachment, the culture medium was removed and the cells were rinsed with PBS. The virus attachment procedure was conducted by adding 0.5 mL of infection medium containing viruses to each well for 1 h of incubation at room temperature. The infection medium was removed from the wells, and the BHK cells were overlaid with 3 mL of MEM-containing soft agar (1% melting agar, 1× MEM, 1% NEAA, and 5% FBS). Plates were incubated at 37 °C for 17 h, and plaques were stained with neutral red (0.1% neutral red in PBS) for 4 h of incubation at 37 °C. The dead cells after virus infection could not be stained by neutral red and thus visualized in the plaques.
Cryo-EM Sample Preparation and Data Collection.
Cryo-EM samples of EEEV and EEEV–Hp were prepared under BSL-2 conditions. To prepare EEEV–Hp samples, EEEV (∼1 mg/mL of E2 glycoproteins) and 16 mg/mL Hp were mixed and incubated for 1 d at 4 °C. Cryo-EM samples were prepared using the Cryo-plunge 3 system (Gatan). Aliquots of 3 μL of virus solutions with or without Hp were applied onto glow-discharged Lacey carbon EM grids (400 mesh; Ted Pella). After blotting for 3.3 to 6 s, grids were plunge-frozen in liquid ethane, and virus samples were embedded in vitreous ice. The samples were visualized on a Titan Krios transmission electron microscope (ThermoFisher Scientific). Forty or 60 consecutive frames were recorded on a Gatan K2 Summit direct electron detector with 3,838 × 3,710 pixels. All of the datasets were collected in counting mode using Leginon (46). Experimental parameters include a defocus range of 0.5 to 3.5 μm with a pixel size of 1.73 Å. The total electron dose for data collection was ∼32 e−/Å2.
Image Processing.
For the datasets of EEEV and EEEV–Hp, beam-induced motion correction was performed using the MotionCor2 program (26). The CTF value of each frame was estimated using CTFFIND4 (27). Particles from the corrected frames were boxed using the FindEM template picking method in the Appion (47) software with templates generated from the manually picked particles. Particle images were initially binned eightfold and subjected to nonreference 2D classification in RELION (28) to filter out particles that failed to generate good average projections. The remaining 17,022 EEEV–Hp particles were used for the 3D reconstruction. Similarly, ∼12,000 particles were extracted for the uncomplexed control dataset.
Single-Particle Reconstruction.
The 3D icosahedral reconstruction for native EEEV and EEEV–Hp were conducted using the jspr program (29). In each case, the gold-standard 3D reconstruction was used to estimate the effective resolution. A total of 500 particles was randomly selected to generate an initial model for each half dataset. The final electron density map was reconstructed, and the resolution was estimated to be 5.8 Å for the EEEV–Hp map based on the Fourier shell correlation criterion of 0.143 (48). The same approach was taken for the control dataset which produced a 5.2-Å structure of native EEEV.
Model Fitting, Difference Map Calculation, and Docking Experiments.
The published structure of the EEEV E1–E2 glycoproteins derived from the 4.4-Å native EEEV cryo-EM reconstruction (30) was fitted to the EEEV–Hp map using the EMfit program (32). As the native and complexed structures showed significantly different diameters, both maps were scaled to the same size by modifying the pixel size of the native EEEV map from 1.58 to 1.66 Å. Subsequently, a difference map was calculated in reciprocal space using programs in CCP4, sfall to convert the EEEV and EEEV–Hp maps to structure factors, rstats for nonlinear least-squares map scaling, and FFT for difference map calculation. A HS disaccharide with three sulfate groups were generated in the PRODRG server (49). The “Vina” autodocking program (50), as implemented in the University of California San Francisco Chimera software (51), was used to prepare ligands (HS) and receptors (E1–E2 glycoprotein), to define docking areas in the axial and the peripheral HS sites [a box volume of 15 Å × 15 Å × 15 Å around the axial HS site (center: 90.6, 55.0, 290.0 from the origin) and a box volume of 15 Å × 15 Å × 15 Å around the peripheral HS site (center: 67.0, 78.0, 297.0 from the origin)].
Sequence Alignment.
The sequences of E2 glycoproteins from six alphaviruses (VEEV, AAB02517.1; EEEV, ADW86015.2; WEEV, ACN87273.1; CHIKV, AAU43881.1; SINV, AAA96976.1; Ross River Virus, AAA47405.1) and different strains of EEEV (FL91-4679, ADW86015.2; TX95, AAF04797.1; MA38, AAF04792.1; MS83, AHL83695.1; TX91, AAF04795.1; FL96, AAF04798.1; LA50, AAF04793.1; GA91, AAF04799.1; LA47, ANB41591.1; NJ60, ABQ63086.1) were aligned using EBI Clustal Omega (52). The secondary structure elements were drawn using ESPript3.0 (53, 54) based on the structural model of the published EEEV E2 glycoprotein.
Structural Alignment.
The coordinates of HS binding residues in the axial and peripheral HS binding sites were extracted from the native EEEV E2 glycoprotein structure (30). The centers of the axial and peripheral HS densities, the atoms (Nε2 of H5, Nε2 of H155, Nζ of K156, and Cζ of R157) from the residues in the peripheral site, and the atoms (Nε2 of H82, Cζ of R84, Nδ2 of N80, Nε2 of H114, and Cζ of R119) from the axial site were used for structural alignment and RMSD calculations by the Pair-fit program in Pymol (55).
In Vitro EEEV Binding Assay.
The binding assay was designed based on the combined cell culture ELISA (56). A 96-well plate with BHK cells was precooled at 4 °C for 1 h. The culture medium was removed from each well, and cells were rinsed with chilled PBS three times. A concentrated EEEV virus stock (∼3 mg/mL of structural proteins) was diluted in chilled PBS (1:5,000 dilution) and then 100 μL of the diluted virus solution with different treatments (500 μg/mL and 1 mg/mL of Hp 6 kDa, 300 mM NaCl, Hp disaccharide I-S, II-H, and IV-H [purchased from Sigma]) were added to each well for virus attachment at 4 °C for 2 h. Similarly, purified mutant viruses (R84A, R119A, R84/R119A, K156A, R157A, and K156A/R157A) were diluted to an equal amount of EEEV before binding assays. Unbound virus particles were removed, and each well was rinsed by chilled PBS two times, followed by addition of the diluted primary antibodies (EEEV-22, 12 mg/mL; 1:5,000 dilution), which recognizes E2 domain B (57) for 1 h at 4 °C. Free antibodies were removed and each well was rinsed by chilled PBS for two times, followed by addition of the goat anti-mouse antibodies conjugated with horseradish peroxidase (GAM-HRP) (Abcam) for 1 h at 4 °C. After removal of GAM-HRP, each well was rinsed by chilled PBS for two times before adding 100 μL of the TMB substrate solution (Thermo Fisher). Within 30 min, the reactions were then terminated by adding 100 μL of 1 M H2SO4 and end-point absorbance at 450 nm was measured for each experiment. For plotting, data points were adjusted by subtracting the mean of the negative control from all of the absorbance values in the data and normalizing each data point with the PBS control set as 0% and EEEV alone as 100%.
Reagent and Resource Sharing.
Further information and requests for resources and reagents should be directed to and will be fulfilled by the corresponding author. An approved Material Transfer Agreement may be required for resource sharing.
Data Availability.
The atomic coordinates for the E1–E2 glycoproteins bound with HS disaccharides, and the cryo-EM density maps of the EEEV–Hp complex structures and native EEEV have been submitted to the Protein Data Bank and Electron Microscopy Data Bank with ID codes 6ODF, EMD-20019, and EMD-20025, respectively. The authors declare that all other data supporting the findings of this study are available within the article and its SI Appendix.
Supplementary Material
Acknowledgments
We thank Valorie D. Bowman (Purdue University) for technical help in cryo-EM data collection. Cryo-EM data were collected at the Purdue University Cryo-EM Facility. This work was supported by NIH Grants AI095366 (M.G.R.) and AI095436 (W.B.K.); and Defense Threat Reduction Agency Award HDTRA1-15-1-0047 (W.B.K.).
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
Data deposition: The atomic coordinates for the E1–E2 glycoproteins bound with heparan sulfate disaccharides, and the cryo-EM density maps of the EEEV–heparin complex structures and native EEEV reported in this paper have been deposited to the Protein Data Bank and Electron Microscopy Data Bank (ID codes 6ODF, EMD-20019, and EMD-20025, respectively).
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.1910670117/-/DCSupplemental.
References
- 1.Lindsey N. P., Staples J. E., Fischer M., Eastern equine encephalitis virus in the United States, 2003–2016. Am. J. Trop. Med. Hyg. 98, 1472–1477 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Armstrong P. M., Andreadis T. G., Eastern equine encephalitis virus—old enemy, new threat. N. Engl. J. Med. 368, 1670–1673 (2013). [DOI] [PubMed] [Google Scholar]
- 3.Villari P., Spielman A., Komar N., McDowell M., Timperi R. J., The economic burden imposed by a residual case of eastern encephalitis. Am. J. Trop. Med. Hyg. 52, 8–13 (1995). [DOI] [PubMed] [Google Scholar]
- 4.Strauss J. H., Strauss E. G., The alphaviruses: Gene expression, replication, and evolution. Microbiol. Rev. 58, 491–562 (1994). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhang R., et al. , 4.4 Å cryo-EM structure of an enveloped alphavirus Venezuelan equine encephalitis virus. EMBO J. 30, 3854–3863 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhang W., et al. , Placement of the structural proteins in Sindbis virus. J. Virol. 76, 11645–11658 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mukhopadhyay S., et al. , Mapping the structure and function of the E1 and E2 glycoproteins in alphaviruses. Structure 14, 63–73 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sun S., et al. , Structural analyses at pseudo atomic resolution of Chikungunya virus and antibodies show mechanisms of neutralization. eLife 2, e00435 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sherman M. B., Weaver S. C., Structure of the recombinant alphavirus Western equine encephalitis virus revealed by cryoelectron microscopy. J. Virol. 84, 9775–9782 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kostyuchenko V. A., et al. , The structure of Barmah Forest virus as revealed by cryo-electron microscopy at a 6-angstrom resolution has detailed transmembrane protein architecture and interactions. J. Virol. 85, 9327–9333 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhang W., Heil M., Kuhn R. J., Baker T. S., Heparin binding sites on Ross River virus revealed by electron cryo-microscopy. Virology 332, 511–518 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mancini E. J., Clarke M., Gowen B. E., Rutten T., Fuller S. D., Cryo-electron microscopy reveals the functional organization of an enveloped virus, Semliki Forest virus. Mol. Cell 5, 255–266 (2000). [DOI] [PubMed] [Google Scholar]
- 13.Cheng R. H., et al. , Nucleocapsid and glycoprotein organization in an enveloped virus. Cell 80, 621–630 (1995). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gaedigk-Nitschko K., Schlesinger M. J., The Sindbis virus 6K protein can be detected in virions and is acylated with fatty acids. Virology 175, 274–281 (1990). [DOI] [PubMed] [Google Scholar]
- 15.Heidner H. W., Johnston R. E., The amino-terminal residue of Sindbis virus glycoprotein E2 influences virus maturation, specific infectivity for BHK cells, and virulence in mice. J. Virol. 68, 8064–8070 (1994). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Caspar D. L., Klug A., Physical principles in the construction of regular viruses. Cold Spring Harb. Symp. Quant. Biol. 27, 1–24 (1962). [DOI] [PubMed] [Google Scholar]
- 17.Melancon P., Garoff H., Processing of the Semliki Forest virus structural polyprotein: Role of the capsid protease. J. Virol. 61, 1301–1309 (1987). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Vogel R. H., Provencher S. W., von Bonsdorff C. H., Adrian M., Dubochet J., Envelope structure of Semliki Forest virus reconstructed from cryo-electron micrographs. Nature 320, 533–535 (1986). [DOI] [PubMed] [Google Scholar]
- 19.Voss J. E., et al. , Glycoprotein organization of Chikungunya virus particles revealed by X-ray crystallography. Nature 468, 709–712 (2010). [DOI] [PubMed] [Google Scholar]
- 20.Lee S., et al. , Identification of a protein binding site on the surface of the alphavirus nucleocapsid and its implication in virus assembly. Structure 4, 531–541 (1996). [DOI] [PubMed] [Google Scholar]
- 21.Kondor-Koch C., Burke B., Garoff H., Expression of Semliki Forest virus proteins from cloned complementary DNA. I. The fusion activity of the spike glycoprotein. J. Cell Biol. 97, 644–651 (1983). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gardner C. L., Ebel G. D., Ryman K. D., Klimstra W. B., Heparan sulfate binding by natural eastern equine encephalitis viruses promotes neurovirulence. Proc. Natl. Acad. Sci. U.S.A. 108, 16026–16031 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Klimstra W. B., Nangle E. M., Smith M. S., Yurochko A. D., Ryman K. D., DC-SIGN and L-SIGN can act as attachment receptors for alphaviruses and distinguish between mosquito cell- and mammalian cell-derived viruses. J. Virol. 77, 12022–12032 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rose P. P., et al. , Natural resistance-associated macrophage protein is a cellular receptor for sindbis virus in both insect and mammalian hosts. Cell Host Microbe 10, 97–104 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhang R., et al. , Mxra8 is a receptor for multiple arthritogenic alphaviruses. Nature 557, 570–574 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zheng S. Q., et al. , MotionCor2: Anisotropic correction of beam-induced motion for improved cryo-electron microscopy. Nat. Methods 14, 331–332 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rohou A., Grigorieff N., CTFFIND4: Fast and accurate defocus estimation from electron micrographs. J. Struct. Biol. 192, 216–221 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Scheres S. H., RELION: Implementation of a Bayesian approach to cryo-EM structure determination. J. Struct. Biol. 180, 519–530 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Guo F., Jiang W., Single particle cryo-electron microscopy and 3-D reconstruction of viruses. Methods Mol. Biol. 1117, 401–443 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hasan S. S., et al. , Cryo-EM structures of eastern equine encephalitis virus reveal mechanisms of virus disassembly and antibody neutralization. Cell Rep. 25, 3136–3147.e5 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rossmann M. G., Fitting atomic models into electron-microscopy maps. Acta Crystallogr. D Biol. Crystallogr. 56, 1341–1349 (2000). [DOI] [PubMed] [Google Scholar]
- 32.Rossmann M. G., Bernal R., Pletnev S. V., Combining electron microscopic with x-ray crystallographic structures. J. Struct. Biol. 136, 190–200 (2001). [DOI] [PubMed] [Google Scholar]
- 33.Xie Q., et al. , The 2.8 Å electron microscopy structure of adeno-associated virus-DJ bound by a heparinoid pentasaccharide. Mol. Ther. Methods Clin. Dev. 5, 1–12 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Byrnes A. P., Griffin D. E., Binding of Sindbis virus to cell surface heparan sulfate. J. Virol. 72, 7349–7356 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Klimstra W. B., Ryman K. D., Johnston R. E., Adaptation of Sindbis virus to BHK cells selects for use of heparan sulfate as an attachment receptor. J. Virol. 72, 7357–7366 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Margolis R. U., Margolis R. K., Chang L. B., Preti C., Glycosaminoglycans of brain during development. Biochemistry 14, 85–88 (1975). [DOI] [PubMed] [Google Scholar]
- 37.Gorchakov R., et al. , Attenuation of Chikungunya virus vaccine strain 181/clone 25 is determined by two amino acid substitutions in the E2 envelope glycoprotein. J. Virol. 86, 6084–6096 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bear J. S., Byrnes A. P., Griffin D. E., Heparin-binding and patterns of virulence for two recombinant strains of Sindbis virus. Virology 347, 183–190 (2006). [DOI] [PubMed] [Google Scholar]
- 39.Gardner C. L., et al. , Deliberate attenuation of Chikungunya virus by adaptation to heparan sulfate-dependent infectivity: A model for rational arboviral vaccine design. PLoS Negl. Trop. Dis. 8, e2719 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ancsin J. B., Kisilevsky R., The heparin/heparan sulfate-binding site on apo-serum amyloid A. Implications for the therapeutic intervention of amyloidosis. J. Biol. Chem. 274, 7172–7181 (1999). [DOI] [PubMed] [Google Scholar]
- 41.Mochizuki H., Yoshida K., Shibata Y., Kimata K., Tetrasulfated disaccharide unit in heparan sulfate: Enzymatic formation and tissue distribution. J. Biol. Chem. 283, 31237–31245 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Dhindwal S., Avila B., Feng S., Khayat R., Porcine circovirus 2 uses a multitude of weak binding sites to interact with heparan sulfate, and the interactions do not follow the symmetry of the capsid. J. Virol. 93, e02222–e02218 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ryman K. D., et al. , Heparan sulfate binding can contribute to the neurovirulence of neuroadapted and nonneuroadapted Sindbis viruses. J. Virol. 81, 3563–3573 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bernard K. A., Klimstra W. B., Johnston R. E., Mutations in the E2 glycoprotein of Venezuelan equine encephalitis virus confer heparan sulfate interaction, low morbidity, and rapid clearance from blood of mice. Virology 276, 93–103 (2000). [DOI] [PubMed] [Google Scholar]
- 45.Gardner C. L., et al. , Natural variation in the heparan sulfate binding domain of the eastern equine encephalitis virus E2 glycoprotein alters interactions with cell surfaces and virulence in mice. J. Virol. 87, 8582–8590 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Suloway C., et al. , Automated molecular microscopy: The new Leginon system. J. Struct. Biol. 151, 41–60 (2005). [DOI] [PubMed] [Google Scholar]
- 47.Lander G. C., et al. , Appion: An integrated, database-driven pipeline to facilitate EM image processing. J. Struct. Biol. 166, 95–102 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Scheres S. H., Chen S., Prevention of overfitting in cryo-EM structure determination. Nat. Methods 9, 853–854 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Schüttelkopf A. W., van Aalten D. M., PRODRG: A tool for high-throughput crystallography of protein-ligand complexes. Acta Crystallogr. D Biol. Crystallogr. 60, 1355–1363 (2004). [DOI] [PubMed] [Google Scholar]
- 50.Trott O., Olson A. J., AutoDock Vina: Improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J. Comput. Chem. 31, 455–461 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Pettersen E. F., et al. , UCSF Chimera—a visualization system for exploratory research and analysis. J. Comput. Chem. 25, 1605–1612 (2004). [DOI] [PubMed] [Google Scholar]
- 52.Sievers F., et al. , Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol. Syst. Biol. 7, 539 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Robert X., Gouet P., Deciphering key features in protein structures with the new ENDscript server. Nucleic Acids Res. 42, W320-4 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Gouet P., Robert X., Courcelle E., ESPript/ENDscript: Extracting and rendering sequence and 3D information from atomic structures of proteins. Nucleic Acids Res. 31, 3320–3323 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Schrödinger LLC, The PyMOL Molecular Graphics System (Version 1.5.0.4, Schrödinger LLC, 2015).
- 56.Wahby A. F., Combined cell culture enzyme-linked immunosorbent assay for quantification of poliovirus neutralization- relevant antibodies. Clin. Diagn. Lab. Immunol. 7, 915–919 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kim A. S., et al. , Protective antibodies against eastern equine encephalitis virus bind to epitopes in domains A and B of the E2 glycoprotein. Nat. Microbiol. 4, 187–197 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Adams P. D., et al. , PHENIX: A comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr. 66, 213–221 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The atomic coordinates for the E1–E2 glycoproteins bound with HS disaccharides, and the cryo-EM density maps of the EEEV–Hp complex structures and native EEEV have been submitted to the Protein Data Bank and Electron Microscopy Data Bank with ID codes 6ODF, EMD-20019, and EMD-20025, respectively. The authors declare that all other data supporting the findings of this study are available within the article and its SI Appendix.