Significance
Noncoding RNAs (ncRNAs) constitute critical components of major cellular protein complexes. However, the vast majority of human ncRNAs, known to date, have unknown functions and their protein interactors are poorly characterized. Here, we introduce an approach that charts the cosedimentation of human ncRNAs with diverse cellular protein machineries. This approach uncovered ncRNA MaIL1 as an integral component of the Toll-like receptor 4 (TLR4) pathway, which induces innate antibacterial defense responses. MaIL1 stabilizes the TLR4 signaling-protein OPTN and is required for TBK1-kinase–dependent IRF3 transcription factor phosphorylation and immune gene activation. These results revisit our understanding of ncRNA functions in cytosolic protein networks and predict further ncRNAs to constitute integral signaling pathway components in different cell types and organisms.
Keywords: lncRNA, glycerol gradient, interferon, macrophage, immunity
Abstract
RNA has been proposed as an important scaffolding factor in the nucleus, aiding protein complex assembly in the dense intracellular milieu. Architectural contributions of RNA to cytosolic signaling pathways, however, remain largely unknown. Here, we devised a multidimensional gradient approach, which systematically locates RNA components within cellular protein networks. Among a subset of noncoding RNAs (ncRNAs) cosedimenting with the ubiquitin–proteasome system, our approach unveiled ncRNA MaIL1 as a critical structural component of the Toll-like receptor 4 (TLR4) immune signal transduction pathway. RNA affinity antisense purification–mass spectrometry (RAP-MS) revealed MaIL1 binding to optineurin (OPTN), a ubiquitin-adapter platforming TBK1 kinase. MaIL1 binding stabilized OPTN, and consequently, loss of MaIL1 blunted OPTN aggregation, TBK1-dependent IRF3 phosphorylation, and type I interferon (IFN) gene transcription downstream of TLR4. MaIL1 expression was elevated in patients with active pulmonary infection and was highly correlated with IFN levels in bronchoalveolar lavage fluid. Our study uncovers MaIL1 as an integral RNA component of the TLR4–TRIF pathway and predicts further RNAs to be required for assembly and progression of cytosolic signaling networks in mammalian cells.
Since the discovery of mRNA as an intermediate between DNA and protein, RNA has been recognized as a key component of many cellular machineries, including the spliceosome (small nuclear RNAs [snRNAs]), the ribosome (ribosomal RNA [rRNA] and transfer RNA [tRNA]), or the microprocessor complex (micro-RNA [miRNA]). Recently, systematic genome annotations by the ENCODE and FANTOM projects have unveiled ∼20,000 mostly uncharacterized long noncoding RNAs (lncRNAs) (1–3)—a heterogeneous group of transcripts ≥200 nucleotides (nt) without protein-coding capacity. Mechanistically, lncRNAs may act in cis or trans by functioning as protein scaffolds, decoys, or guides (4). Pioneering studies have revealed critical lncRNA functions, e.g., in X-inactivation (5), control of pattern formation (6), or regulation of cellular life span (7). Unlike other RNA classes, however, the global association of lncRNAs with cellular protein complexes has remained unknown, hampering the dissection of lncRNA mechanisms in human disease trajectories, including exaggerated or failed antibacterial immunity.
Recognition of infectious agents by the immune system relies on germline-encoded receptors, which sense conserved pathogen structures. A prototypical example is the plasma membrane-bound Toll-like receptor 4 (TLR4), which recognizes bacterial lipopolysaccharide (LPS). TLR4 activation triggers dichotomous signaling cascades. Signaling through TLR4-adapter MyD88 induces proinflammatory cytokines such as IL1β through the NFκB and AP-1 transcription factors, while TRIF-dependent signaling activates the IRF3 transcription factor and type I interferon (IFN) expression. To prevent from inflammation-induced tissue damage, an intricate network of regulators counterbalances TLR signaling. Both protein regulators (e.g., A20 or IRAK-M) and miRNAs (e.g., miR-146, miR-155, let-7) provide TLR4 feedback and feedforward control (8–11). Most recently, lncRNAs, such as Linc-Cox2, LincRNA-EPS, or Lnc13 have been implicated in immune modulation downstream of pathogen sensors (12–17). Thus, TLR signaling is tightly balanced by a network of protein, miRNA, and lncRNA regulators, preventing from exaggerated or failed immunity. While control mechanisms regulating TLR activity are well documented, the spatial coordination and assembly of TLR signaling components in the crowded intracellular milieu are still not fully understood. Recently, scaffolding proteins were found to contribute to the intracellular coordination of TLR signaling. OPTN, for example, binds ubiquitin chains, to establish a proteasome-sensitive platform, which mediates TBK1-kinase–dependent IRF3 phosphorylation in the final step of the TLR4–TRIF cascade (18–21). Besides proteins, noncoding RNAs (ncRNAs) have been known for decades to contribute to cellular protein complex organization (e.g., rRNAs and tRNAs). Recently, several lncRNAs were found to adopt scaffolding functions to maintain nuclear protein complex and chromatin architecture (22, 23). Little is known, however, about possible architectural roles of lncRNAs in cytosolic signal transduction networks.
Here, we devised a multipronged approach, using gradient ultracentrifugation and multiomics, to chart the global cosedimentation of mammalian RNA with cellular protein machineries. In stark contrast to other RNA classes, lncRNAs were heterogeneously distributed over the gradient and cosedimented with diverse machineries, including the ribosome, the proteasomal network, or mitochondrial proteins. Notably, among a subgroup of lncRNAs cosedimenting with ubiquitin–proteasome-associated proteins, we discovered MaIL1 as an architectural RNA component of the TLR4 pathway. Combination of our approach with RNA antisense purification (RNA antisense purification–mass spectrometry [RAP-MS]) revealed MaIL1 to bind the TLR4 signal transduction protein and ubiquitin-adapter OPTN. MaIL1 stabilized OPTN and was essential for OPTN aggregation and OPTN-TBK1 kinase-dependent IRF3 transcription factor phosphorylation. Consequently, knockout or knockdown of MaIL1 blunted TLR4–TRIF–IRF3-mediated IFN expression and antibacterial defense. In line with its requirement for TLR–TRIF pathway progression, MaIL1 levels were predictive of IFN expression in patients with bronchopulmonary infection. Our approach thus uncovers MaIL1 as a critical RNA element within the TLR–TRIF pathway and predicts further integral RNA components to contribute to the functionality of cytosolic protein networks in mammalian cells.
Results
lncRNA Profiles of Resting and Activated Primary Human Macrophages.
To systematically interrogate lncRNA elements within cellular protein complexes, we devised a multipronged approach, charting the global cosedimentation of RNAs with proteins, using glycerol gradient centrifugation, RNA-seq, and protein MS. As a cell model, we used human blood-derived macrophages, an abundant primary cell type with key roles in the immune system. In a first step, we characterized the baseline and TLR-stimulus–dependent RNA expression landscape of this cell type and charted the nuclear and cytosolic distribution of transcripts by RNA-seq (Fig. 1A) (25). These data aided the interpretation of gradient-based RNA–protein cosedimentation data and the prioritization of candidate ncRNAs with potential architectural roles in immune-relevant protein networks (Fig. 1A).
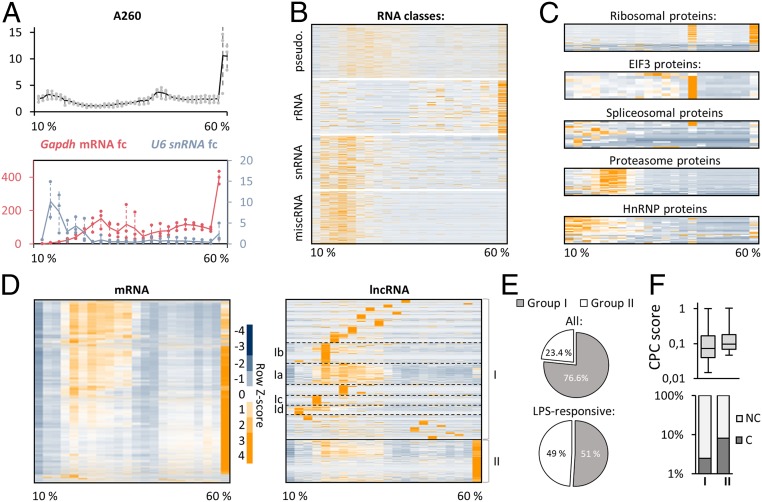
Fig. 1.
RNA-seq charts lncRNA profiles of resting and activated human blood-derived macrophages. (A) Experimental outline: lncRNA profiling in resting and immune-activated macrophages is followed by investigation of lncRNA distribution in subcellular fractions and RNA/protein cosedimentation analysis using glycerol gradient centrifugation. RAP-MS is used to reveal specific lncRNA interactors. (B) qRT-PCR analysis of IL1β and CD80 mRNA induction (8-h LPS versus mock, greater than three independent experiments). (C) RNA class distribution in LPS-treated macrophages from B (RNA-seq; averaged over three replicates). (D) Fold changes and numbers of regulated mRNAs and lncRNAs in samples from B (Padj ≤ 0.05, fold change ≥ 2 or ≤ 0.5). (E) LncRNA fold changes and adjusted P values (RNA-seq data from D). (F) Top 10 up- and down-regulated lncRNAs from D (C, mock treatment). (G) qRT-PCR validations. Macrophages were exposed to indicated agonists (L.pn. = Legionella pneumophila) for 8 h and compared to mock treatment (eight independent experiments). (H) RNA-seq analysis of mRNA and lncRNA localization to nucleus and cytoplasm (in percent, two averaged replicates; 8-h mock or LPS treatment). (I) Comparison of lncRNA cytoplasmic localization in mock and LPS-stimulated cells (datasets from H).
For the characterization of lncRNA expression profiles of blood-derived macrophages, cells were challenged with Salmonella enterica LPS or mock treated, respectively. Successful macrophage activation by LPS was verified by qRT-PCR analysis of IL1β and CD80 mRNA induction (Fig. 1B). RNA-seq revealed lncRNAs to account for ∼4.4% of the cellular poly(A)-RNA in LPS-stimulated macrophages (Fig. 1C). Differential gene expression analysis revealed 773 mRNAs to be up- and 460 to be down-regulated in response to LPS (fold change ≥ 2, adjusted P ≤ 0.05; Fig. 1D). Notably, using the same strict cutoffs, we identified 43 up- and 14 down-regulated lncRNAs, suggesting their involvement in macrophage immunity (Fig. 1 D and E, SI Appendix, Fig. S1A, and Dataset S1). Underpinning the current gap in the understanding of lncRNA functions in human cells, except for LINC01268 (ROCKI) (24) none of the top 10 LPS-induced lncRNAs (Fig. 1F) has been mechanistically characterized so far. Validation by qRT-PCR revealed even increased up-regulation of selected lncRNAs AP003354.2 (henceforth MaIL1, for macrophage interferon-regulatory lncRNA 1), LINC01215, and NRIR, upon coactivation with LPS and IFN-γ (Fig. 1G). MaIL1 and LINC01215 induction was also observed upon macrophage infection with the Gram-negative bacterial pathogen Legionella pneumophila (Fig. 1G). Of note, NRIR was only mildly induced following Legionella infection, indicative of differential sensitivity of macrophage lncRNAs to microbial infection (Fig. 1G).
Irrespective of TLR sensitivity, a total of 201 expressed lncRNAs was detected in blood-derived macrophages (reads per kilobase per million mapped reads [RPKM] cutoff of 0.5). Analysis of cytoplasmic and nuclear distribution of these lncRNAs by RNA-seq confirmed the nuclear enrichment of well-characterized lncRNAs NEAT1 and MALAT1 (26) and cytosolic enrichment of lncRNA ZFAS1 (27) (Fig. 1H). Overall, lncRNAs localized predominantly to the cytoplasm, irrespective of LPS treatment and similar to mRNAs (Fig. 1H and Dataset S1). Only few LPS-induced changes in subcellular lncRNA distribution were detected. Potential shifts in subcellular distribution of Hotairm1 and BIC, however, did not reach significance at the P ≤ 0.05 level (Fig. 1I and SI Appendix, Fig. S1B). Taken together, human blood-derived macrophage lncRNAs are predominantly cytoplasmic, indicative of their implication in chromatin-independent processes. TLR activation substantially impacts the macrophage lncRNA landscape (∼28% being up- or down-regulated ≥2-fold) at the expression but not the subcellular localization level. Presently, most of these lncRNAs remain of unknown function.
Gradient Profiling Charts the Cosedimentation of Functional RNA and Protein Classes.
Upon characterization of the expression dynamics and subcellular distribution of lncRNAs, we sought to illustrate their association with cellular protein complexes, to infer potential functions of the predominantly cytoplasmic lncRNAs. To this end, we established a 10 to 60% glycerol gradient ultracentrifugation protocol, to chart the global RNA cosedimentation with cellular machineries lighter than ribosomes. We chose an early time point, at which lncRNA induction was already visible (2-h LPS, SI Appendix, Fig. S1C), to account for immune-regulatory RNA–protein interactions, preceding the fully developed inflammatory response. Gradient-separated macrophage lysates were divided into 22 consecutive fractions, which were subjected to RNA-seq and protein MS. Quantification of RNA concentration by A260 analysis verified the expected sharp increase toward the last fractions (Fig. 2A), which contain the heavy ribosomal complexes (see below). qRT-PCR confirmed the expected peak of U6 snRNA in the early gradient fractions (light-weight spliceosome complexes), while GAPDH mRNA spiked in the late (ribosomal) fractions (Fig. 2A). Further verifying successful separation of RNA–protein complexes, RNA-seq analysis confirmed rRNA to peak in the last fraction, whereas snRNA was enriched in the first fractions (Fig. 2B). Of note, miscRNA, a category of poorly investigated RNA, exhibited a distinct sedimentation profile, reminiscent of snRNA (Fig. 2B). Pseudogene-RNA primarily occupied fractions succeeding snRNAs and miscRNAs and unlike rRNA displayed a markedly reduced abundance in the last (ribosomal) fraction (Fig. 2B).
Fig. 2.
Glycerol gradient profiling reveals the cosedimentation of RNA classes with cellular machineries. (A, Top) A260 absorbance in 44 successive 10 to 60% glycerol gradient fractions (lysates from 2-h LPS-treated macrophages). (A, Bottom) qRT-PCR analysis of Gapdh mRNA and U6 snRNA levels, compared to fraction 1, in samples from A, reduced to 22 successive fractions. Three independent experiments. (B) Sedimentation profiles of major RNA classes, determined by RNA-seq analysis of samples from A (row Z-score heatmap). (C) Same as B but for proteins, based on MS. (D) mRNA and lncRNA sedimentation profiles (gradient RNA-seq data). Major lncRNA subclusters are highlighted. (E) Percentages of lncRNAs in cluster I and II. (Top) All detected lncRNAs; (Bottom) LPS-responsive lncRNAs from Fig. 1D. (F, Top) CPC2 coding-scores (box plots) of cluster I and II lncRNAs. (F, Bottom) Percentages of lncRNAs deemed “protein coding” in CPC2 analysis for cluster I and II.
Separation of protein complexes along the gradient was confirmed by silver staining (SI Appendix, Fig. S1D). MS verified the accumulation of ribosomal components in the last fraction, with a secondary peak in fraction 15 (Fig. 2C). This pattern is explained by the localization of EIF3 proteins to fraction 15, which are involved in ribosomal recycling (28) (Fig. 2C). Combined with the RNA-seq profiles, these data confirmed the aspired migration of active ribosomes, containing rRNA, and ribosomal proteins, into the last gradient fractions. Spliceosomal proteins showed a more heterogeneous distribution, but predominantly occupied the first seven gradient fractions, matching the pattern observed for snRNA. Similarly, heterogeneous nuclear ribonucleoproteins (hnRNPs), which are involved in mRNA processing, were primarily found in the first fractions (Fig. 2C). The proteasomal proteins, establishing another abundant cellular machinery, accumulated in fractions 5 to 7 (Fig. 2C). Thus, our approach accurately discriminates functional protein and RNA classes. This allowed us to portray the global cosedimentation of lncRNAs, coding transcripts, and proteins and pinpoint lncRNA components within cytosolic protein networks.
lncRNAs Form Subgroups Cosedimenting with Different Protein Machineries.
lncRNAs are considered a functionally heterogeneous type of RNA, and recent evidence even suggests a minor fraction of annotated lncRNAs to be translated (29–31). In line with this notion, principal-component analysis of gradient RNA-seq data revealed a subset of lncRNAs to cocluster with rRNAs and mRNAs (SI Appendix, Fig. S2A). The majority of lncRNAs, however, cosegregated with other types of ncRNA (snRNA, miscRNA). Complementary results were obtained in hierarchical clustering analysis. More than 90% of all mRNAs followed a single, uniform migration pattern along the gradient, with the expected peak toward the last (ribosomal) fraction. lncRNA clustering yielded a markedly different result, revealing ∼22 lncRNA subgroups with distinct sedimentation profiles (Fig. 2D and Dataset S2). Generally, lncRNAs that were detected in fractions other than the ribosome-containing fraction 22, accounted for 76.6% of all macrophage lncRNAs. We refer to these as group I lncRNAs. In total, 23.4% of lncRNAs, which we refer to as group II lncRNAs, cosedimented with ribosomes, similar to mRNA (Fig. 2 D and E, Upper). When restricting the analysis to LPS-responsive lncRNAs, group II lncRNAs increased to 49% (Fig. 2 E, Lower). Both the percentage of lncRNAs predicted as “coding” by the CPC2 algorithm (32) and the average CPC2 coding score were elevated in group II lncRNAs, in agreement with their ribosomal localization (Fig. 2F). For the remainder of this study, we focused on the highly diverse group I lncRNAs, which are absent from the ribosomal fraction and therefore likely to be truly noncoding.
lncRNA group I could be further divided into ∼21 subgroups, based on their sedimentation profiles, representing categories of lncRNAs associating with different cellular machineries lighter than ribosomes (Fig. 2D and SI Appendix, Fig. S2B). A subgroup, which we refer to as group Ib, for instance, cosedimented with proteins involved in mitochondrial metabolism and contained known mitochondrial regulatory lncRNA RMRP (33), (SI Appendix, Fig. S2 B–D). Other subgroups cosedimented with proteins involved, e.g., in vesicle trafficking and neutrophil degranulation (group Ic) or innate immunity and infectious diseases (group Id) (SI Appendix, Fig. S2B). A prominent subgroup, which we refer to as group Ia, contained several well-described lncRNAs such as PACER, NORAD, or NEAT1 (Fig. 2D and Dataset S2) and several of the above-identified LPS-responsive lncRNAs (SI Appendix, Fig. S2E). Reactome pathway analysis of proteins cosedimenting with group Ia lncRNAs (SI Appendix, Fig. S2B and Fig. 3 A and B) predicted their involvement in innate immunity and the ubiquitin–proteasome system. Protein ubiquitination critically controls macrophage innate immune signaling at multiple levels (34). To evaluate these gradient-based predictions, we exemplarily selected a group Ia lncRNA of presently unknown function for in-depth characterization.
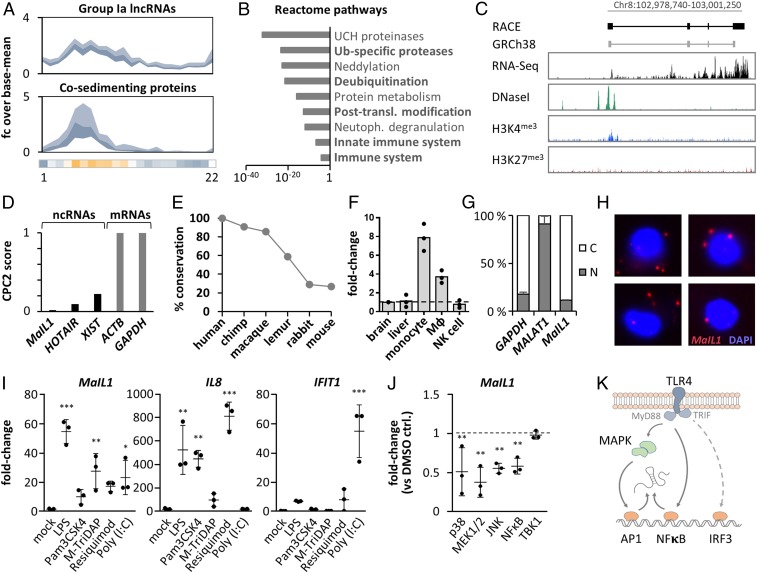
Fig. 3.
Characterization of MaIL1 as a TLR-responsive noncoding RNA. (A) Sedimentation profile (fold-change plot, inner quartiles, relative to base mean) of LPS-responsive group Ia lncRNAs (Top) and cosedimenting proteins (Bottom). Horizontal bar: MaIL1 Z scores, color-coded according to Fig. 2. (B) Reactome pathway analysis (proteins from A). (C, Top) RACE PCR-determined MaIL1 architecture, compared to GRh38 annotation. (C, Bottom) CD14+ monocyte RNA-seq, DNaseI-seq, H3K4me3 or H3K27me3 ChIP-seq signals. (D) CPC2 score of MaIL1 and GRCh38 reference RNAs. (E) MaIL1 sequence conservation in reference genomes of indicated species. (F) qRT-PCR analysis of MaIL1 abundance in indicated tissues and cell types, relative to brain (three replicates). (G) Localization of indicated RNAs to cytoplasm (white) and nucleus (gray) in RNA-seq replicates from Fig. 1H (LPS). (H) RNA-FISH visualization of MaIL1 (8-h LPS). Nucleus counterstained with DAPI. Representative pictures. (I) qRT-PCR-analysis of MaIL1, IL8, and IFIT1 induction upon immune-agonist stimulation for 8 h (versus mock treatment, three replicates). (J) qRT-PCR analysis of MaIL1 levels in 8-h LPS-stimulated macrophages pretreated with pathway inhibitors (fold change compared to LPS + DMSO, three replicates). (K) Model of MaIL1 activation by TLR4-triggered MAPK/NFκB signaling. Where applicable, SDs and data points from three experiments are shown. P values (***P ≤ 0.0001, **P ≤ 0.01, *P ≤ 0.05) determined by one-way ANOVA.
MaIL1 lncRNA Is a TLR-Response Gene.
Among group Ia lncRNAs (Fig. 3 A and B), we recognized MaIL1 as one of the most highly induced lncRNAs during macrophage activation with LPS (Fig. 1G). The latest GENCODE release recently removed MaIL1 (AP003354.2) from the list of intergenic lncRNAs and associated it with a neighboring antisense RNA. We therefore reassessed the MaIL1 cDNA ends and splice-structure by rapid amplification of cDNA ends–PCR (RACE-PCR), which verified the previously predicted four-exon architecture and confirmed MaIL1 as an independent lincRNA with a length of 1,543 nt (Fig. 3C and SI Appendix, Fig. S3 A–D). Analysis of ENCODE monocyte DNase-, RNA-, and ChIP-seq data revealed a DNaseI hypersensitive element and an H3K4 trimethylation site (hallmarks of transcriptional start sites) at the RACE-PCR determined MaIL1 5′ end, as well as specific RNA-seq coverage across the gene body (Fig. 3C). Thus, MaIL1 is a four-exon intergenic lncRNA, primed from a unique transcriptional start site. Reanalysis of the RACE-refined RNA sequence by the CPC2 algorithm confirmed a low coding score, similar to well-characterized noncoding RNAs, and distinct from mRNAs (Fig. 3D), which is also in agreement with our classification as a group I lncRNA (Fig. 2). Human MaIL1 cDNA sequence conservation was high (>85%) in catarrhine primates, but moderate to low in more distant species (Fig. 3E). Indicative of a potential function in myeloid cell immunity, MaIL1 expression was elevated in macrophages and monocytes compared to brain and liver or NK cells (Fig. 3F). RNA-seq analysis of subcellular fractions from LPS-treated macrophages suggested that MaIL1 is a cytoplasmic RNA (Fig. 3G). This could be confirmed by RNA–fluorescence in situ hybridization (RNA-FISH) analysis (Fig. 3H and SI Appendix, Fig. S3E). Control-treated macrophages were found to contain ∼1 copy per cell, whereas LPS-stimulated cells contained ∼20 to 60 copies of MaIL1 per cell (SI Appendix, Fig. S3F). Similar copy numbers have been described for other functional lncRNAs (35). Besides with LPS, MaIL1 was also strongly induced upon stimulation with other pattern recognition receptor (PRR) ligands, such as agonists of TLR2 (Pam3csk4), TLR3/RIG-I [poly(I:C)], TLR7/8 (Resiquimod), and NOD1/NOD2 (M-TriDAP) (Fig. 3I). Overall, LPS conferred the strongest MaIL1 induction (Fig. 3I). The TLR4-mediated response to LPS is relayed by transcription factors such as NFκB and AP-1 (via MyD88) and IRF3 (via TRIF) (36). Inhibition of MEK/ERK, p38, and JNK kinases upstream of AP-1 and of NFκB significantly attenuated MaIL1 expression upon LPS stimulation (Fig. 3J). TANK-binding kinase-1 (TBK1) inhibition upstream of IRF3 did not impact on MaIL1 expression (Fig. 3J). Temporally, MaIL1 induction preceded induction of TRIF-target IFNβ1, but not of MyD88-target IL8 (SI Appendix, Fig. S3G). Thus, MaIL1 is a PRR-induced, cytoplasmic noncoding RNA, up-regulated via MyD88- but not TRIF-signaling downstream of TLRs (Fig. 3K).
MaIL1 Binds Ubiquitin Adapter OPTN in the TLR4 Pathway.
Our gradient data predicted MaIL1 to operate within a proteasome-ubiquitin–dependent innate immune network (SI Appendix, Figs. S2B and S3B). To pinpoint the specific interactors of MaIL1 within this network, we performed coimmunoprecipitations (co-IPs) and RNA affinity chromatographies. Among the RNAs cosedimenting with MaIL1 on glycerol gradients was NEAT1 (Fig. 4A), which has been implicated in innate immune responses through association with RNA binding protein SFPQ (37). SFPQ was also identified as a potential interactor of NORAD, another group Ia lncRNA (38). Co-IP confirmed the interaction of NEAT1 with SFPQ and revealed an additional interaction with MaIL1 (SI Appendix, Fig. S4A). However, gradient analysis detected SFPQ only within the first, but not the second, and major MaIL1 peak (Fig. 4A), suggesting further MaIL1 interactors in macrophages. We therefore performed endogenous RAP using UV–cross-linking and biotinylated antisense oligonucleotide tiling, followed by MS (39) (Fig. 4B). Success and specificity of endogenous MaIL1 RAP from macrophage lysates, compared to control RAP with random oligonucleotides, was confirmed by RNA-seq analysis of RAP eluates (RAP-seq) and qRT-PCR (Fig. 4C and SI Appendix, Fig. S4B).
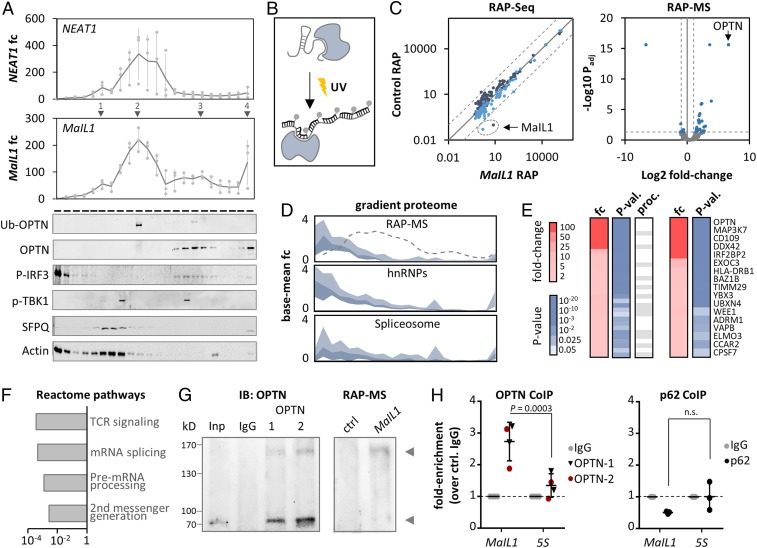
Fig. 4.
MaIL1 binds ubiquitin adapter OPTN. (A, Top) NEAT1 and MaIL1 profiles on 10 to 60% glycerol gradients (qRT-PCR, 2-h LPS, relative to fraction 1, three replicates). (A, Bottom) Representative Western blots (Ub, ubiquitin-associated form). Proteins detected on independent blots, with the same protein samples (full scans in SI Appendix, Fig. S7). (B) RNA affinity purification (RAP) procedure: UV–cross-linking is followed by hybridization of biotinylated oligos to target RNA and streptavidin-based purification. (C, Left) RNA-seq analysis of eluates from control and MaIL1 RAP (RPKMs, two color-coded replicates). The dashed lines indicate 10-fold enrichment over perfect-correlation line. (C, Right) Protein MS results (eluate fractions from three control and MaIL1 RAPs). Dashed lines: twofold (MaIL1 vs. control RAP) and Padj ≤ 0.05 cutoffs. (D) Top: Fold-change plot (inner two quartiles, relative to base mean), depicting sedimentation profiles of RAP-MS copurified proteins from C (fold enrichment ≥ 2, P ≤ 0.05; data from Fig. 2C). Dashed line: MaIL1. (Middle and Bottom) Same as Top, but with hnRNP and spliceosomal proteins from Fig. 2C. (E, Left) Fold change (red) and P value (blue) for RAP-MS purified proteins (fold enrichment ≥ 2, P ≤ 0.05). Gray cells: RNA processing (proc.) factors. (E, Right) Same as Left but excluding processing factors. (F) Reactome pathway analysis of RAP-MS purified proteins. (G, Left) OPTN co-IP Western blot (input: Inp; control co-IP: IgG; OPTN co-IP with two different antibodies: 1, 2). (G, Right) OPTN Western blot with RAP-MS eluates. The arrows indicate ubiquitin-associated and nonassociated OPTN. (H) MaIL1 and 5S rRNA levels in OPTN (two antibodies) and p62 (control) co-IP eluates, compared to control IgG (qRT-PCR). Significances determined by one-way ANOVA (n.s., not significant). Results of three or more experiments are shown.
MS analysis of RAP eluates (RAP-MS) uncovered multiple MaIL1 coenriched proteins (Padj ≤ 0.05, fold change ≥ 2; Fig. 4C). Expectedly, among the MaIL1-interacting proteins were many RNA processing factors, which cosedimented with hnRNP and spliceosomal proteins in the gradient datasets (Fig. 4 D–F). As these proteins did not overlap with the MaIL1 main gradient peak (Fig. 4D), we considered them to represent factors transiently involved in MaIL1 maturation rather than stable interactors. Irrespective of their involvement in RNA processing, among all MaIL1 copurified proteins the critical TLR4–TRIF signal transduction component and ubiquitin-adapter optineurin (OPTN) was most highly and significantly enriched (Fig. 4 C and E). Ubiquitin-associated OPTN establishes a platform for TBK1-kinase–mediated phosphorylation of the IRF3 transcription factor downstream of TLR4, which mediates type I IFN expression (18, 19). Like most signaling proteins, OPTN was not detected in the gradient datasets. Western blot, however, revealed enrichment of ubiquitin-associated OPTN in MaIL1 RAP-MS eluates (Fig. 4G and SI Appendix, Fig. S4 C and D) and localization to the MaIL1 main peak on glycerol gradients (fraction 10, Fig. 4A and SI Appendix, Fig. S4 C and D). Ubiquitin-free OPTN peaked in later gradient fractions and cosedimented with the MaIL1 minor peaks 3 and 4 (around fraction 16 and 22, Fig. 4A). TLR4–TRIF signaling proteins TBK1 and IRF3 did not colocalize with the MaIL1 main peak (in line with their absence from RAP-MS data), but their phosphorylated forms overlapped with the MaIL1 minor peak 3, which also contains ubiquitin-free and ubiquitin-associated OPTN (Fig. 4A and SI Appendix, Fig. S4E). Co-IP qRT-PCR confirmed the interaction of MaIL1 with OPTN, but not with the OPTN-regulatory protein p62, as a control (Fig. 4H). Thus, MaIL1 is a TLR-MyD88–sensitive ncRNA interacting with the ubiquitin-binding signaling protein OPTN.
MaIL1 Sequesters OPTN to Promote TLR-TBK1–Dependent IRF3 Phosphorylation.
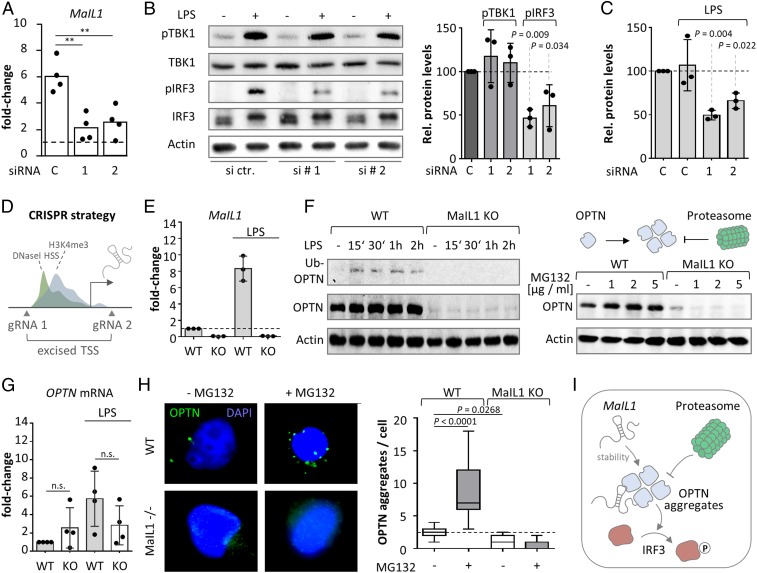
OPTN is a multifunctional ubiquitin-adapter, platforming TBK1 kinase. OPTN aggregate structures, forming around ubiquitin chains, are eventually cleared by the proteasome (40). Prior to their proteasomal degradation, higher-order ubiquitin–OPTN–TBK1 structures mediate phosphorylation of IRF3 and thereby type I IFN transcription (18, 19, 41). We therefore predicted OPTN-bound MaIL1 to be implicated in the TBK1–IRF3 signaling step. Accordingly, MaIL1 silencing by RNA interference (RNAi) significantly reduced phospho-IRF3 (but not phospho-TBK1) levels in LPS-activated macrophages (Figs. 5 A and B and 6A). MaIL1 silencing also reduced ubiquitin-associated OPTN levels (Fig. 5C and SI Appendix, Fig. S4F), suggesting that the interaction with MaIL1 affects OPTN stability and subsequently TLR–TBK1–IRF3 signaling. Non–ubiquitin-bound OPTN levels were also reduced upon MaIL1 knockdown, reaching significance for one out of two siRNAs (SI Appendix, Fig. S4G). Since transient knockdown may not reveal the full regulatory capacity of MaIL1, we generated MaIL1-deficient THP1 macrophage lines by CRISPR/Cas9-based genome editing. We knocked out the epigenetically marked transcriptional start site (DNaseI HSS, H3K4me3, Fig. 3C), spanning the proximal promoter region and first exon, which resulted in loss of MaIL1 expression (Fig. 5 D and E). In line with the RNAi-based results, MaIL1 knockout resulted in reduced OPTN protein, but not mRNA levels, compared to wild-type cells (Fig. 5 F, Left and Fig. 5G). IRF3 phosphorylation was impaired in MaIL1-deficient cells, similar to the results with siRNAs (SI Appendix, Fig. S4H), and IFNB1 expression was reduced (SI Appendix, Fig. S4I). OPTN forms higher-order ubiquitin-bound, proteasome-sensitive structures, required for TBK1 signaling (Fig. 5 F, Right) (40). Proteasome inhibition using increasing doses of proteasome inhibitor MG132 elevated OPTN levels in wild-type but not in MaIL1-deficient cells (Fig. 5 F, Right). Immunofluorescence confirmed OPTN to form cytosolic aggregates, which increased in number upon MG132 treatment (Fig. 5H). In MaIL1-deficient cells, residual OPTN levels were sufficient for the formation of OPTN aggregates, which were fewer in number compared to wild-type cells (Fig. 5H). The accumulation of OPTN aggregates upon proteasome inhibition, however, was lost in MaIL1-deficient cells (Fig. 5H). Of note, FISH and immunofluorescence visualized MaIL1 in vicinity to OPTN and MG132-induced ubiquitin foci (SI Appendix, Fig. S5 A and B). These results suggest that MaIL1 stabilizes OPTN in a proteasome-independent manner and promotes formation of OPTN signaling aggregates, which are required for TLR4–IRF3 signaling and eventually cleared by the ubiquitin–proteasome (Fig. 5I). These findings support our cosedimentation-based prediction that group Ia lncRNAs are involved in innate immune-relevant ubiquitin-proteasome–associated circuitries (SI Appendix, Fig. S2B and Fig. 3B).
Fig. 5.
MaIL1 sequesters OPTN to promote TLR4-TBK1–dependent IRF3 phosphorylation. (A) qRT-PCR validation of MaIL1 knockdown (8-h LPS, two siRNAs, four replicates). (B, Left) Representative Western blot showing total- and phospho-TBK1 and -IRF3 levels (mock- and LPS-treated macrophages, control and MaIL1 knockdown with siRNAs 1 and 2). (B, Right) Quantification of phospho-TBK1 and -IRF3 (three independent Western blots, relative to actin). (C) Quantification of OPTN signal (ubiquitin-associated form, relative to actin, three independent Western blots) upon control or MaIL1 siRNA transfection and LPS treatment. (D) CRISPR/cas9 strategy for MaIL1 knockout. (E) MaIL1 expression in 8-h mock- or LPS-treated wild-type (WT) or MaIL1-deficient (KO) THP-1 macrophages (qRT-PCR, three replicates). (F, Left) Western blot analysis of OPTN expression in WT and MaIL1 KO THP1 cells, mock- or LPS treated as indicated. (F, Top Right) Scheme of OPTN aggregation and proteasomal degradation. (F, Bottom Right) Western blot analysis of OPTN expression in THP1 cells following 4-h mock or MG132 treatment of WT and MaIL1 KO THP1 cells. (G) Same as E, but for OPTN mRNA (four replicates). (H, Left) Representative OPTN immunofluorescence pictures (WT and MaIL1 KO THP1 cells, 4-h MG132 or mock treatment). (H, Right) Quantification of OPTN aggregates per cell (23 pictures per condition). (I) Model showing OPTN stabilization and aggregation by MaIL1, which promotes IRF3 phosphorylation. OPTN aggregates are subsequently resolved by the proteasome. P values determined by one-way ANOVA (**P ≤ 0.01; n.s., not significant).
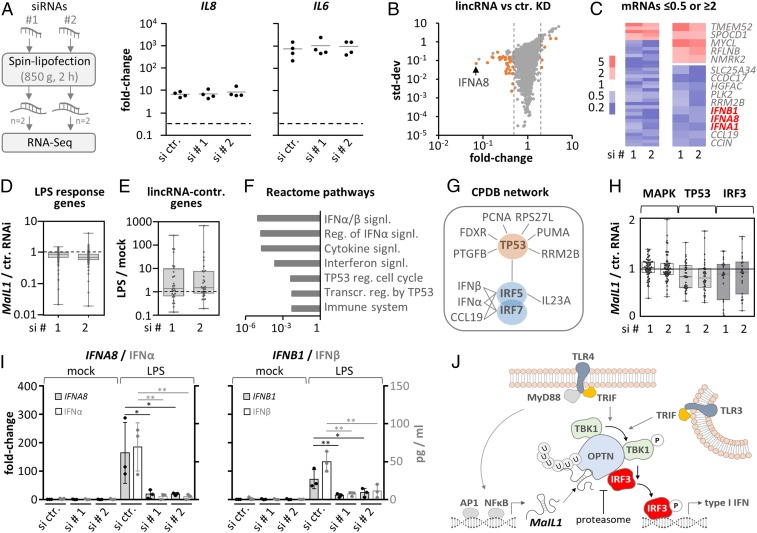
Fig. 6.
MaIL1 controls type I IFN expression. (A, Left) Knockdown strategy using independent siRNA designs. (A, Right) qRT-PCR analysis of IL8 and IL6 induction (8-h LPS versus mock, four replicates), in MaIL1-silenced macrophages. (B) RNA-seq plot showing mean fold changes and SD (MaIL1 siRNA 1 or 2 vs. control siRNA knockdown). Genes with fold changes ≥2 or ≤0.5 in both runs are highlighted (orange). (C) Fold change heatmap for orange-labeled genes from B. Five up- and top 10 down-regulated genes are shown to the Right. (D) Box plot showing regulation of LPS-response genes (RNA-seq determined, fold change ≥ 2, LPS vs. mock) upon MaIL1 compared to control knockdown. (E) Box plot showing RNA-seq–determined regulation of MaIL1-controlled genes from C in response to LPS. (F) Reactome pathway analysis using down-regulated genes from C. (G) ConsensusPathDB network analysis using down-regulated genes from C. (H) Regulation of MAPK-, TP53-, and IRF3-dependent genes (MaIL1 versus control knockdown, data from B). (I) qRT-PCR and ELISA validation of diminished type I IFN mRNA (primary axis) and protein (secondary axis) induction upon MaIL1 knockdown (mRNA fold changes compared to control siRNA and mock stimulation, three replicates). (J) Model: MaIL1 fosters OPTN aggregation to promote TLR-TRIF-TBK1–dependent IRF3 phosphorylation and type I IFN expression. MaIL1 expression and thereby TLR–TRIF activity is induced by TLR–Myd88 signaling. The proteasome eventually resolves MaIL1-dependent OPTN aggregates. *P ≤ 0.05, **P ≤ 0.01 (one-way ANOVA).
MaIL1 Deficiency Blunts TLR-IRF3–Induced Type I IFN Production.
Phosphorylation of IRF3 represents the final step in the TLR4-TRIF–dependent type I IFN induction pathway. The requirement of MaIL1 for IRF3 phosphorylation therefore predicts it to be essential for TLR4-triggered type I IFN expression. We addressed this hypothesis by RNA-seq analysis of control and MaIL1 siRNA-treated blood-derived macrophages (Fig. 6A, samples from Fig. 5A). Whereas MaIL1 silencing (Fig. 5A) did not impact LPS-triggered induction of TLR-MyD88–dependent IL8 or IL6 (Fig. 6A), a marked reduction of type I IFN expression was observed (Fig. 6 B and C). When relaxing the regulation cutoff below twofold, this effect extended to other LPS-responsive genes (Fig. 6D). Vice versa, genes sensitive to MaIL1 knockdown were mostly LPS inducible (Fig. 6E). Reactome pathway analysis of MaIL1-controlled genes revealed an overrepresentation of type I IFN and TP53-related pathways (Fig. 6F). Consequently, induced network analysis predicted an underlying IRF and TP53 transcription factor network (Fig. 6G). Furthermore, genes belonging to the IRF3 or TP53 but not to the MAPK regulon, as a control, were down-regulated upon MaIL1 knockdown (Fig. 6H), in line with the autocrine activation of TP53 genes by type I IFN (42). Enzyme-linked immunosorbent assay (ELISA) and qRT-PCR confirmed a marked suppression of TLR4-mediated type I IFN but not IL6 induction upon MaIL1 knockdown, which extended to a milder suppression of other TLR-response genes (Fig. 6I and SI Appendix, Fig. S6 A and B). Attenuated type I IFN induction was also observed in MaIL1-silenced cells activated with TLR3 agonist poly(I:C) (SI Appendix, Fig. S6C). As expected, no type I IFN induction was observed in macrophages treated with TLR2 agonist Pam3csk4, and MaIL1 knockdown did not affect the induction of TLR-MyD88–dependent cytokines IL6 and IL8 (SI Appendix, Fig. S6D). Taken together, these results suggest a transistor model, in which TLR–TRIF pathway activity is a function of TLR-MyD88–dependent MaIL1 expression, which regulates OPTN platform assembly, and thus TBK1-dependent IRF3 phosphorylation and IFN expression (Fig. 6J).
MaIL1 Levels Are Elevated in Pulmonary Infections and Correlate with IFN Expression.
To determine the contribution of TLR4–TRIF–IFN pathway component MaIL1 to host defense, we performed bacterial infection studies (Fig. 7 A, Left). Classically regarded as antiviral mediators, type I IFNs are increasingly recognized as regulators of antibacterial immunity (43). Legionella pneumophila, for instance, which preferentially replicates inside macrophages, is highly sensitive to type I IFN (44). To assess the contribution of MaIL1 to cell-autonomous antibacterial defense, macrophages were infected with GFP-expressing L. pneumophila at a physiological multiplicity of infection (MOI), which is normally cleared by macrophages (44). MaIL1 knockdown resulted in a 1.7- to 3.1-fold increase in the percentage of Legionella-infected (GFP-positive) macrophages (Fig. 7 A, Right and SI Appendix, Fig. S8B). This increased susceptibility to Legionella was reduced by exogenous type I IFN supplementation (Fig. 7 A, Right and SI Appendix, Fig. S8B). In line with the increased infection rates, MaIL1 silencing diminished IFNα and IFNβ production by Legionella-infected macrophage cultures (SI Appendix, Fig. S8C). Thus, MaIL1 is required for cell-autonomous IFN-dependent antibacterial defense.
Fig. 7.
MaIL1 impacts bacterial replication and is elevated in BALF from infected patients. (A, Left) siRNA-transfected macrophages are infected with GFP-Legionella, followed by FACS quantification. (A, Right) Quantification of infection rates in percent (MOI 0.1; 24 h), upon control or MaIL1 knockdown (with or without IFN treatment, MaIL1 siRNA 1 and 2). Three replicates and one-way ANOVA. (B) qRT-PCR analysis of MaIL1 (Left) and IFNβ1 (Right) expression, relative to U6 snRNA and compared against median CT value in BALF pellets of 10 control patients compared to 12 patients with bacterial or fungal pulmonary infection (two-tailed Student’s t test, assuming equal variances). (C) Correlation of MaIL1 and IFNβ1 mRNA expression in the same BALF samples.
Given the important role of MaIL1 in TLR-TRIF–dependent defense, we predicted MaIL1 expression to impact IFN immunity in vivo. We therefore determined MaIL1 levels in bronchoalveolar lavage fluid (BALF) of human patients with bacterial or fungal bronchopulmonary infection, and control patients without apparent lung disease, and investigated the correlation with IFN expression. MaIL1 levels were significantly increased in patients with pulmonary infection, compared to the control group (Fig. 7 B, Left). Similarly, IFNβ1 levels were significantly increased in BALF from patients with pulmonary infection (Fig. 7 B, Right), and IFNβ1 expression correlated linearly with MaIL1 (R2 = 0.92, Fig. 7C), supporting a critical role of MaIL1 in type I IFN induction in vivo.
In summary, our approach successfully identifies critical RNA components within cytosolic protein networks through global cosedimentation analysis of lncRNAs and proteins. Among lncRNAs cosedimenting with the ubiquitin–proteasome system, MaIL1 constitutes an architectural RNA component of the TLR4–TRIF pathway, stabilizing the OPTN–TBK1 platform to promote IRF3 phosphorylation and type I IFN production. Our cosedimentation data predict many other ncRNA components to be involved in cytoplasmic protein complex dynamics and organization in mammalian organisms.
Discussion
Already in the 1950s to 1960s, RNA was recognized as an essential component of the translational machinery, when rRNAs and tRNAs were discovered. The protein interactors of less abundant ncRNA species, including lncRNAs, however, have remained poorly characterized. Hampering their investigation, lncRNAs are vastly heterogeneous in function. Attempts have been made to subclassify lncRNAs, i.e., into cis- or trans-acting, genic or intergenic transcripts to facilitate their investigation. Moreover, our own and other recently published data suggest that lncRNAs need to be subclassified according to their association with ribosomes. Our glycerol gradient data revealed 23.4% of all lncRNAs in human macrophages to cosediment with ribosomes. Among the LPS-inducible lncRNAs, even every second lncRNA was found in the ribosomal fraction (Fig. 2). Jackson et al. (29) recently reported >35% of lncRNAs to bind to ribosomes in murine cells, matching the numbers determined here for human lncRNAs. In line with these observations, polysome profiling has revealed large numbers of lncRNAs to associate with ribosomal monosomes and polysomes (30, 31). Although Jackson et al. (29) found evidence for noncanonical open reading frames in a number of ribosome-associated lncRNAs, it is also conceivable that some cytoplasmic lncRNAs associate with ribosomes to regulate translation rather than being translated. While the classic narrative assumes lncRNAs to function mostly in the nucleus, recent literature supports our notion that lncRNAs may locate to the cytoplasm in large quantities (45, 46). Our observation that >50% of these lncRNAs associate with complexes lighter than ribosomes, suggests that lncRNAs, besides their well-documented functions in nuclear architecture, are part of many cytoplasmic protein complexes, other than the translational machinery.
Recently, glycerol gradient profiling was introduced as a novel tool to illustrate the differential association of bacterial regulatory RNA with RNA-binding proteins (47). Adopting a similar strategy for eukaryotic cells, the present study charts the global cosedimentation of lncRNAs with cellular protein machineries. These data revealed lncRNAs, unlike other RNA classes, to display a vastly heterogeneous sedimentation behavior, indicative of their association with various different protein complexes. We recognized several subclusters of cosedimenting lncRNAs and could confirm known lncRNAs such as RMRP, NORAD, or NEAT1 to cosediment with nonribosomal complexes (SI Appendix, Fig. S2B). Exemplarily, we investigated MaIL1 as an uncharacterized member of lncRNA subgroup Ia, which cosediments with proteins involved in the ubiquitin–proteasome system and innate immunity (Figs. 2 and 3B). RAP-MS revealed MaIL1 as an integral component of the TLR–TRIF signaling network, binding and sequestering ubiquitin-adapter OPTN. OPTN forms a platform for TBK1 and is required for TBK1-dependent IRF3 phosphorylation. OPTN colocalizes with ubiquitin foci (48), and mutations in the polyubiquitin binding domain impair IRF3 phosphorylation and IFN induction (18). Interestingly, TBK1 phosphorylates OPTN to promote its binding to ubiquitin chains (49), which likely results in the formation of covalent oligomers (50). Thus, ubiquitin-associated OPTN structures are promoted by TBK1 and critically contribute to TBK1-dependent IRF3 activation. Notably, our study reveals ncRNA MaIL1 to bind ubiquitin-associated OPTN, to promote formation of subcellular OPTN foci and IRF3 phosphorylation. Our observations suggest MaIL1 to constitute an integral component of the TLR4–TRIF pathway, fostering OPTN aggregate formation and induction of IFN immunity. Eventually, the MaIL1-dependent OPTN foci are resolved and turned over by the proteasome, as indicated by the accumulation of OPTN aggregates upon proteasome inhibition in wild-type but not MaIL1-deficient cells (Fig. 5). An open question is how MaIL1 impacts on basal expression of non–ubiquitin-associated OPTN, as reduction of basal OPTN protein levels upon MaIL1 knockdown or knockout was not rescued by proteasome inhibition. Alternatively, autophagy or mitophagy-dependent processes could mediate increased OPTN turnover in MaIL1-deficient cells, as OPTN serves as an autophagy adapter, accumulating in inclusion bodies and at mitochondrial surfaces (49, 51). MaIL1 might also impact on OPTN levels through one of the still poorly characterized proteasome-independent processes. Regardless of the exact mechanism of OPTN stabilization, the versatile regulatory functions of OPTN suggest that its interaction partner MaIL1 may have additional cellular functions, beyond the critical role in TLR–TRIF signaling.
With MaIL1, our study provides evidence for an integral RNA component within the TLR–TRIF–IFN pathway. Recently, several regulatory RNAs (e.g., Lnczc3h7a, Lnc-Lsmb3, lnc-ITPRIP-1, or NEAT1) were reported to stabilize cellular type I IFN expression programs (35, 37, 52–54). This suggests a model in which type I IFN immunity is controlled by RNA at multiple levels—while architectural RNAs such as MaIL1 are essential for signaling pathway assembly and progression, a network of regulatory RNAs provides positive and negative feedback to confer optimal IFN levels. In the TLR4–TRIF–IFN induction pathway, another regulatory instance is established by the TLR-MyD88–activated transcription factor NFκB. In the nucleus, NFκB coregulates IFN expression through cooperative binding to IFN gene promoters, in proximity to IRF3 binding sites (55). Notably, MaIL1 expression may be elevated in an NFκB-dependent manner (Fig. 3 I–K). This suggests a transistor model, in which TLR–TRIF activity is tweaked by TLR–MyD88 activity in the nucleus via NFκB binding to IFN gene promoters and in the cytosol through TLR4-MyD88–dependent elevation of MaIL1 levels and thus enhanced OPTN–TBK1 signaling. This model is also supported by the early induction of MaIL1 upon TLR4 activation, preceding type I IFN induction (SI Appendix, Fig. S3G). Taken together, our present study and previously published data suggest that IFN activation relies on an intricate network of RNA and protein components, acting together to confer efficient pathogen eradication without excessive inflammatory damage. The MaIL1–OPTN complex likely represents a central hub within this complex circuitry.
Besides the OPTN–MaIL1 complex, our RNA–protein cosedimentation data predict numerous other lncRNAs to act within the ubiquitin–proteasome system and other protein machineries, including the respiratory chain, the membrane trafficking network, and even the translational machinery (Fig. 2 and SI Appendix, Fig. S2). Beyond lncRNAs, our datasets also chart the cosedimentation of smaller ncRNA species with proteins. For instance, miscRNAs, a poorly studied group of RNAs, displayed an unexpectedly uniform sedimentation behavior and cosedimented with metabolic enzymes and proteins implicated in immunity and vesicle trafficking (SI Appendix, Fig. S9). These results are in support of an ongoing dogma change and match the expected pleiotropic molecular implications and diverse subcellular localization patterns of ncRNAs, recently reviewed by Carlevaro-Fita and Johnson (45). This view is also supported by the RNA-world and RNP-world theories, which assume life to have evolved from early self-replicating RNA or RNP constellations (56). Instead of having been replaced by the more versatile proteins, nonmessenger functions of RNA might have, throughout evolution of complex organisms, continued to play important roles in subcellular organization. Strong evidence for this view was delivered by the discovery of the abundant rRNAs and tRNAs as essential structural components within the translational machinery (57, 58), using similar techniques as the current study. The advent of unbiased and sensitive high-throughput sequencing methods has uncovered further, less abundant types of RNA, such as lncRNA, but also vault- or y-RNA, which are now emerging as critical components within diverse cellular circuitries. Our study provides a global snapshot of the cosedimentation of these RNAs with cellular machineries in eukaryotic cells. With MaIL1, we provide an important example of an integral RNA component within a major cellular signaling pathway. We expect our study to facilitate the discovery of further critical RNA elements within the protein networks of mammalian cells.
Materials and Methods
Cell Culture and Infection Assays.
Buffy coats were obtained through the Transfusion Medicine department, University Clinics Giessen and Marburg, and deidentified prior to use. Leukocyte populations were isolated by Miltenyi MACS purification. Cells were cultivated in X-Vivo 15 medium (Lonza) or RPMI (RNA-FISH experiments). Macrophages were generated with 15 ng/mL GM-CSF or 20 nM PMA (THP1). Cells were treated with LPS, Pam3csk4, or IFNγ (all 100 ng/mL), M-Tridap or Resiquimod (both 1 µg/mL), or poly(I:C) (20 µg/mL). For bacterial infections, GFP-expressing Legionella pneumophila strain Corby (59) was used. Pathway inhibitors were used at 50 µM. Recombinant IFNβ was used at 1 ng/mL. Cell culture conditions are further specified in SI Appendix, Supplementary Methods. Throughout this study, all replicates shown (e.g., data points and SD for qRT-PCR or ELISA measurements) represent independent experiments performed with cells obtained from different donors (primary cells) or different passages (cell lines).
RNAi.
The 50 nM Silencer Select siRNAs (Thermo Fisher; http://sidirect2.rnai.jp/) per 5 × 105 cells (12-well format) were complexed with Lipofectamine 2000 according to the manufacturer’s instructions. Complexed siRNAs were added to macrophage cultures, followed by centrifugation at 850 × g for 2 h. Stimulations were carried out 16 h posttransfection, and knockdown was verified by qRT-PCR. siRNAs were as follows: Silencer Select control siRNA 1, MaIL1 siRNA 1 (guide, AAAUACAUGGCUUUCAUGCUA; passenger, GCAUGAAAGCCAUGUAUUUAA); MaIL1 siRNA 2 (guide, UUUACAUUUCUAUUAUGUGUG; passenger, CACAUAAUAGAAAUGUAAAAG).
CRIPSR/Cas9.
Cells were transfected with pX458 vector constructs containing control or MaIL1 gene locus targeting guide RNAs, using Lipofectamine 2000, according to the manufacturer’s instructions. Constructs were designed, and knockout cells were generated as previously described (10). MaIL1 guide RNA pair sequences were as follows: TTCTTGTGTAACTTACGACA; GTGCAGTGGCCAATCATTAG. Cell clones were tested for MaIL1 TSS deletion by genomic PCR (SI Appendix, Fig. S4L).
qRT-PCR.
RNA was isolated using TRIzol (Ambion)/chloroform extraction. Purified RNA was treated with DNase I (Ambion) in the presence of recombinant RNase inhibitor (Promega), followed by phenol/chloroform/isoamyl alcohol (PCI) (Roth) extraction. Concentrations were determined using a Nanodrop 2000 instrument. Expression changes in all experiments except those listed below were measured using a two-step qRT-PCR approach: reverse transcription (High-Capacity cDNA Reverse Transcription Kit; Thermo Fisher) and qPCR (PowerUP SYBR Green Master Mix; Thermo Fisher) were done according to the manufacturer’s instructions. For cytoplasm–nucleus fractionation, glycerol gradient, and co-IP samples, relative target RNA abundance was determined by one-step qRT-PCR (Power SYBR RNA-to-Ct 1-Step Kit; Thermo Fisher). A QuantStudio 3 instrument was used (Applied Biosystems). Fold changes, where applicable normalized to U6 snRNA, were calculated using the 2−ΔΔCT method (60). Primers are listed in SI Appendix, Table S1.
RACE-PCR.
RACE-PCR was done using the SMARTer 5′/3′ RACE kit (Takara) and primers listed in SI Appendix, Table S1 according to the manufacturer’s instructions with the following modifications: For PCR amplification, Advantage2 Polymerase (Takara) was used, and PCR products were cloned for downstream Sanger sequencing analysis (Seqlab) into the Strataclone U/A PCR Cloning vector (Agilent).
Copy Number Determination.
RACE-PCR determined MaIL1 cDNA was subcloned (Stratagene PCR Cloning kit; Agilent) and verified by Sanger sequencing (Seqlab). A T7 promoter was fused to the cDNA 5′ end by PCR using Taq polymerase and primers listed in SI Appendix, Table S1. Two hundred nanograms of PCR product were used for in vitro transcription (Megascript T7 kit; Thermo Fisher), according to the manufacturer’s instructions. Upon template degradation with Turbo DNase (Thermo Fisher), RNA was purified (Qiagen RNeasy mini columns) and RNA integrity was determined (Bioanalyzer RNA Nano chips; Agilent). RNA concentration was validated by Bioanalyzer and Nanodrop 2000 quantification. MaIL1 copy number was determined by qRT-PCR using RNA extracted from a defined number of cells (manual counting in triplicates) and serial dilutions of in vitro transcribed MaIL1 RNA as a reference, assuming 1.63 × 109 MaIL1 copies per nanogram:
Protein Staining and Western Blot.
BCA assay was performed according to manufacturer’s instructions (Pierce BCA Protein Assay kit; Thermo Fisher). For SDS/PAGE, 10% polyacrylamide gels were prepared. For silver staining, gels were fixed with 50% methanol, 12% acetic acid, and 0.5 mL/L formaldehyde (37%). After washing with 50% and 30% ethanol, gels were pretreated with 0.2 mg/mL Na2S2O3 and impregnated in silver solution (2 g/L). Gels were developed with 60 g/L Na2CO3, 4 mg/L Na2S2O3 × 5H2O, and 0.5 mL/L formaldehyde (>37%) solution, and development was stopped with 1% glycine. For Western blotting, proteins were transferred onto nitrocellulose (Amersham Protran; Sigma-Aldrich; GE10600003) by semidry blotting. For blot development, a Chemostar Imager (INTAS Science Imaging) was used. Western blot full scans are shown in SI Appendix, Fig. S7. Antibodies and dilutions are specified in SI Appendix, Supplementary Methods.
Immunofluorescence.
Cells were seeded on 18-mm round cover glass and differentiated with PMA in 12-well plates. Cells were optionally incubated with 10 μM MG132 for 12 h, washed with PBS, and fixed for 10 min at room temperature (RT) with 3.7% formaldehyde. One milliliter of PBS containing 0.1% Triton X-100 was used for permeabilization (5 min; RT). Cells were blocked with PBS, 2% BSA for 1 h. OPTN and ubiquitin antibody, respectively, was diluted 1:500 in PBS and added to the cells, followed by incubation for 2 h at RT. Cells were washed three times (1 mL of PBS; 10-min incubations) and exposed to secondary goat anti-mouse IgG cross-adsorbed Alexa Fluor 488 (Thermo Fisher; A11001), diluted 1:2,000 in PBS, for 1 h at RT. After one wash, cells were fixed again for 10 min at RT with 3.7% formaldehyde. For nuclear staining, cells were incubated in 1 mL of DAPI solution (AAT Bioquest; diluted 1:1,000 in PBS) for 30 min at RT. MOWIOL4-88 (Carl-Roth) was used for mounting. Slides were dried overnight. Images were acquired on a Zeiss AXIO Vert.A1 microscope, and image analysis was performed using ImageJ (NIH).
RNA-FISH.
Macrophages were grown in Nunc Lab-Tek II CC2 Chamber Slides and fixed with 4% paraformaldehyde. RNA-FISH was performed using the QuantiGene ViewRNA ISH Cell Assay (Affymetrix). Probes were designed by Affymetrix (Homo sapiens EEF1A1 catalog no. VA1-10418; Homo sapiens ENSG00000254281 catalog no. VPRWEK4) (sequence provided by L.N.S.). EEF1A1 served as positive control, whereas probe set diluent served as negative control (SI Appendix, Fig. S3E). Nuclei were counterstained with Roti-Mount FluorCare DAPI (Carl Roth). Images were acquired at an Olympus DP 80 microscope (600× magnification). The detailed RNA-FISH procedure is described in SI Appendix, Supplementary Methods.
ELISA.
Plates were coated overnight at 4 °C with 50 µL/well 1:1,000 dilution of α-human IFNα (21100-2-10; PBL; 100 µg/mL). Blocking was done with PBS, 0.05% Tween 20, 1% BSA, for 1 h at RT. Protein standard (human IFNα; 300-02A; Preprotech) was prepared in blocking buffer. Detection was performed with 50 µL/well of 1:1,000 antibody dilution (α-human–IFNα HRP-conjugate; BMS216MSTK; eBioscience) and 50 µL/well substrate solution (one OPD-tablet [20 mg]; P7288; Sigma; in 20 mL of substrate buffer; A0626,2500; AppliChem). Reaction was stopped with 25 µL/well 2 M H2SO4. Plates were read at 490 nm, with reference wavelength 650 nm, using an Emax plate reader and SoftMaxPro5 software. For IFNβ and IL6 detection, the DuoSet IFNβ ELISA kit (DY814-05; R&D Systems) and the IL6 Ready-SET-Go kit (88-7060-88; Thermo Fisher) were used according to the manufacturer’s instructions. Plates were read at 450 nm, with reference wavelength 540 nm (IFNβ) or 570 nm (IL6), using a Tecan Infinite 200 PRO plate reader.
Flow Cytometry.
Data were acquired using a Guava easyCyte (Millipore) or FACSCalibur (BD) cytometer. Intact cells were gated through size (forward scatter) and granularity (side scatter) and analyzed for GFP (510- to 540-nm channel) versus background (652- to 671-nm channel) fluorescence using FlowJo. Cells in the GFP-positive gate were considered infected. The gating strategy is detailed in SI Appendix, Fig. S8.
Subcellular Fractionation.
A total of 3 × 106 cells was washed twice with PBS, resuspended in 200 µL of lysis buffer (10 mM Tris, pH 8; 140 mM NaCl; 1.5 mM MgCl2; 0.5% Igepal; 2 mM vanadyl ribonucleoside complex) and incubated on ice for 5 min. On centrifugation at 1,000 × g for 3 min, the supernatant was transferred to a new tube, centrifuged for 3 min at maximum speed, and transferred to a new tube for RNA extraction. The nuclear pellet was washed twice with lysis buffer without and once with 0.5% deoxycholic acid (at 1,000 × g), followed by RNA extraction (see RNA-Seq).
Glycerol Gradient Ultracentrifugation.
Gradients were poured in 5% increments, starting at 10% and ending at 60% glycerol in buffer (10 mM Tris [pH 8], 150 mM NaCl, 10 mM KCl, 1.5 mM MgCl2, 0.5% Triton, 0.5 mM EDTA, 1 mM DTT). A total of 108 macrophages was passed through a 26 G needle (five times) followed by disruption of nuclei using a Dounce homogenizer (10 strokes). Lysate was supplemented with RNase inhibitor (Promega) and layered onto gradient. Gradients were centrifuged at 50,200 rcf (Servall S-34 rotor), acceleration level 1, brakes off, for 20 h at 4 °C using a Servall Evolution ultracentrifuge (Hitachi). Protein and RNA were recovered by PCI extraction. A detailed protocol (including a step-by-step protocol) is provided in SI Appendix, Supplementary Methods.
RNA Affinity Chromatography.
RAP-MS was performed as previously described by the Guttman laboratory (https://www.guttmanlab.caltech.edu/RAP_MS_Protocol_April2015.pdf) (39), using the whole-cell lysate procedure, with 8 × 107 UV–cross-linked cells per capture, grown in X-Vivo 15 medium (Lonza), without SILAC isotopes. RAP-MS oligonucleotide antisense pools were generated by segmenting RACE-PCR refined MaIL1 cDNA sequence or a random sequence into 80-mers with 13- to 18-nt spacing. Sequences (SI Appendix, Table S2) were synthesized at Metabion AG and 3′ monobiotinylated using terminal transferase (New England Biolabs) and Biotin-11-ddUTP (Jena Bioscience) according to the manufacturer’s instructions. RAP-MS conditions and probe design are further specified in SI Appendix, Supplementary Methods.
Co-IP.
Co-IP was performed as described (61) using 107 cells per sample, with minor modifications: Cells were UV–cross-linked with 300 mJ/cm2 and proteins were purified with protein A or protein G Dynabeads (Thermo Fisher). Five micrograms of antibodies listed above (Protein Staining and Western Blot) and control IgGs were used per co-IP. Protein and RNA were extracted using PCI and precipitated with acetone and 30:1 ethanol/3M sodium acetate, respectively (Glycerol Gradient Ultracentrifugation).
RNA-Seq.
RNA was purified using the miRVana miRNA isolation kit (Thermo Fisher; AM1560; total RNA isolation procedure). RNA was DNase I digested and transferred to the in-house transcriptomics facility (Philipps-University Marburg, medical faculty) for generation of Illumina stranded mRNA libraries or to Vertis Biotech (Freising, Germany) for total RNA libraries. RNA from glycerol gradient fractions was supplemented with Escherichia coli K12 MG1655 spike-in RNA for normalization. After RNA quality control (Experion RNA analysis kit; Bio-Rad), barcoded libraries were generated using the TruSeq Stranded mRNA Kit (Illumina) or Vertis Biotech in-house kits. Libraries were sequenced (single ends) on a HiSeq1500 machine in rapid mode with 50-bp read length or a NextSeq500 device with 75-bp read length.
Proteomics.
Samples bound to (magnetic) beads were washed three times with 100 µL of 0.1 M ammonium bicarbonate solution. Upon “on-bead” digestion by addition of Sequencing Grade Modified Trypsin (Serva) and incubation at 37 °C for 45 min, the supernatant was transferred to fresh tubes. Peptides were desalted and concentrated using Chromabond C18WP spin columns (Macherey-Nagel; Part No. 730522). Finally, Peptides were dissolved in 25 µL of water with 5% acetonitrile and 0.1% formic acid. The mass spectrometric analysis was performed using an Orbitrap Velos Pro mass spectrometer (Thermo Scientific). MS equipment and conditions are further detailed in SI Appendix, Supplementary Methods.
Bioinformatics.
Reads were mapped to the GRCh38 annotation using the CLC genomics workbench (Qiagen) with standard settings. Differential expression analysis was done using DeSeq2 (62). For subcellular localization analysis, RPKM values were corrected to account for the different RNA content of nucleus and cytoplasm, as described in SI Appendix, Supplementary Methods. RNA-seq data from glycerol gradient fractions were normalized to Escherichia coli spike-in RNA. RNA-seq data can be accessed through the National Center for Biotechnology Information Gene Expression Omnibus (GEO) pipeline (accession no. GSE101409).
MS data analysis was performed using Proteome Discoverer 2.2 (Thermo Scientific). For gradient samples, abundance values were normalized to the relative protein content in each fraction. Gradient proteomics data are provided in Dataset S3.
For data visualization, Cluster (Eisen laboratory), JAVA TreeView (63), and ConsensusPathDB (64) and Cytoscape version 3.7.2 and the R-script prcomp (stats) were used. Bioinformatics analysis are further detailed in SI Appendix, Supplementary Methods.
Bronchoalveolar Lavage and Patient Selection.
BALF was obtained at the Department of Infectious Diseases and Respiratory Medicine (Charité University Hospital Berlin). All patients underwent bronchoalveolar lavage (BAL) on clinical indication and had provided oral and written informed consent. The study was approved by the ‘Ethics committee of Charité–Universitätsmedizin Berlin’, 10117 Berlin, Germany (EA2/086/16). Only patients with nonmycobacterial infection were selected for the infection group. Control patients underwent BAL because of idiopathic coughing or for exclusion of pulmonary tuberculosis or pulmonary involvement of systemic diseases. For patient characterization, see SI Appendix, Table S3. For liver tissue, cirrhosis was an exclusion criterion. All patients had provided oral and written informed consent, and the study was approved by the local ethics committee (Marburg FB20 Ethikkomission Az.: Studie 14/17). Sample processing and patient selection are further detailed in SI Appendix, Supplementary Methods.
Data Availability.
The datasets produced in this study are available in the following database: RNA-seq data: GEO, https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE101409 (accession no. GSE101409).
Supplementary Material
Acknowledgments
This work was supported by the SFB-TR84 (to L.N.S., B.S., L.E.S., and S.B.) and von-Behring-Röntgen-Stiftung (Project 63-0036, to L.N.S.). Part of this work was funded by Bundesministerium für Bildung und Forschung (JPI-AMR Restrict-Pneumo, Project 01Kl1702, to B.S.; ERACoSysMed2–SysMed-COPD, Project 031L0140, to B.S.; e:Med CAPSYS, Project 01ZX1304E, to B.S.). S.M.V. was supported by Jürgen Manchot Foundation (Doctoral Research Fellowship).
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
Data deposition: The RNA-seq data reported in this paper have been deposited in the Gene Expression Omnibus (GEO) database, https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE101409 (accession no. GSE101409).
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.1920393117/-/DCSupplemental.
References
- 1.Derrien T., et al. , The GENCODE v7 catalog of human long noncoding RNAs: Analysis of their gene structure, evolution, and expression. Genome Res. 22, 1775–1789 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Djebali S., et al. , Landscape of transcription in human cells. Nature 489, 101–108 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hon C. C., et al. , An atlas of human long non-coding RNAs with accurate 5′ ends. Nature 543, 199–204 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rinn J. L., Chang H. Y., Genome regulation by long noncoding RNAs. Annu. Rev. Biochem. 81, 145–166 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Penny G. D., Kay G. F., Sheardown S. A., Rastan S., Brockdorff N., Requirement for Xist in X chromosome inactivation. Nature 379, 131–137 (1996). [DOI] [PubMed] [Google Scholar]
- 6.Rinn J. L., et al. , Functional demarcation of active and silent chromatin domains in human HOX loci by noncoding RNAs. Cell 129, 1311–1323 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kotzin J. J., et al. , The long non-coding RNA Morrbid regulates Bim and short-lived myeloid cell lifespan. Nature 537, 239–243 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Schulte L. N., Eulalio A., Mollenkopf H. J., Reinhardt R., Vogel J., Analysis of the host microRNA response to Salmonella uncovers the control of major cytokines by the let-7 family. EMBO J. 30, 1977–1989 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Schulte L. N., Westermann A. J., Vogel J., Differential activation and functional specialization of miR-146 and miR-155 in innate immune sensing. Nucleic Acids Res. 41, 542–553 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Janga H., et al. , Cas9-mediated excision of proximal DNaseI/H3K4me3 signatures confers robust silencing of microRNA and long non-coding RNA genes. PLoS One 13, e0193066 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Taganov K. D., Boldin M. P., Chang K. J., Baltimore D., NF-kappaB-dependent induction of microRNA miR-146, an inhibitor targeted to signaling proteins of innate immune responses. Proc. Natl. Acad. Sci. U.S.A. 103, 12481–12486 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Atianand M. K., Caffrey D. R., Fitzgerald K. A., Immunobiology of long noncoding RNAs. Annu. Rev. Immunol. 35, 177–198 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Carpenter S., et al. , A long noncoding RNA mediates both activation and repression of immune response genes. Science 341, 789–792 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tong Q., et al. , LincRNA-Cox2 modulates TNF-α-induced transcription of Il12b gene in intestinal epithelial cells through regulation of Mi-2/NuRD-mediated epigenetic histone modifications. FASEB J. 30, 1187–1197 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hu G., et al. , LincRNA-Cox2 promotes late inflammatory gene transcription in macrophages through modulating SWI/SNF-mediated chromatin remodeling. J. Immunol. 196, 2799–2808 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Atianand M. K., et al. , A long noncoding RNA lincRNA-EPS acts as a transcriptional brake to restrain inflammation. Cell 165, 1672–1685 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Castellanos-Rubio A., et al. , A long noncoding RNA associated with susceptibility to celiac disease. Science 352, 91–95 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gleason C. E., Ordureau A., Gourlay R., Arthur J. S., Cohen P., Polyubiquitin binding to optineurin is required for optimal activation of TANK-binding kinase 1 and production of interferon β. J. Biol. Chem. 286, 35663–35674 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Munitic I., et al. , Optineurin insufficiency impairs IRF3 but not NF-κB activation in immune cells. J. Immunol. 191, 6231–6240 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Slowicka K., et al. , Optineurin deficiency in mice is associated with increased sensitivity to Salmonella but does not affect proinflammatory NF-κB signaling. Eur. J. Immunol. 46, 971–980 (2016). [DOI] [PubMed] [Google Scholar]
- 21.Meena N. P., et al. , The TBK1-binding domain of optineurin promotes type I interferon responses. FEBS Lett. 590, 1498–1508 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chujo T., Yamazaki T., Hirose T., Architectural RNAs (arcRNAs): A class of long noncoding RNAs that function as the scaffold of nuclear bodies. Biochim. Biophys. Acta 1859, 139–146 (2016). [DOI] [PubMed] [Google Scholar]
- 23.Quinodoz S., Guttman M., Long noncoding RNAs: An emerging link between gene regulation and nuclear organization. Trends Cell Biol. 24, 651–663 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhang Q., et al. , The long noncoding RNA ROCKI regulates inflammatory gene expression. EMBO J. 38, e100041 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Schulte L. N., Subcellular profiling of macrophage long non-coding RNAs. Gene Expression Omnibus. https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE101409. Deposited 13 July 2017.
- 26.Bhatt D. M., et al. , Transcript dynamics of proinflammatory genes revealed by sequence analysis of subcellular RNA fractions. Cell 150, 279–290 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hansji H., et al. , ZFAS1: A long noncoding RNA associated with ribosomes in breast cancer cells. Biol. Direct 11, 62 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pisarev A. V., Hellen C. U. T., Pestova T. V., Recycling of eukaryotic posttermination ribosomal complexes. Cell 131, 286–299 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jackson R., et al. , The translation of non-canonical open reading frames controls mucosal immunity. Nature 564, 434–438 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.van Heesch S., et al. , Extensive localization of long noncoding RNAs to the cytosol and mono- and polyribosomal complexes. Genome Biol. 15, R6 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Carlevaro-Fita J., Rahim A., Guigó R., Vardy L. A., Johnson R., Cytoplasmic long noncoding RNAs are frequently bound to and degraded at ribosomes in human cells. RNA 22, 867–882 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kang Y. J., et al. , CPC2: A fast and accurate coding potential calculator based on sequence intrinsic features. Nucleic Acids Res. 45, W12–W16 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hsieh C. L., et al. , The gene for the RNA component of the mitochondrial RNA-processing endoribonuclease is located on human chromosome 9p and on mouse chromosome 4. Genomics 6, 540–544 (1990). [DOI] [PubMed] [Google Scholar]
- 34.Hu H., Sun S. C., Ubiquitin signaling in immune responses. Cell Res. 26, 457–483 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lin H., et al. , The long noncoding RNA Lnczc3h7a promotes a TRIM25-mediated RIG-I antiviral innate immune response. Nat. Immunol. 20, 812–823 (2019). [DOI] [PubMed] [Google Scholar]
- 36.Lu Y. C., Yeh W. C., Ohashi P. S., LPS/TLR4 signal transduction pathway. Cytokine 42, 145–151 (2008). [DOI] [PubMed] [Google Scholar]
- 37.Morchikh M., et al. , HEXIM1 and NEAT1 long non-coding RNA form a multi-subunit complex that regulates DNA-mediated innate immune response. Mol. Cell 67, 387–399.e5 (2017). [DOI] [PubMed] [Google Scholar]
- 38.Munschauer M., et al. , The NORAD lncRNA assembles a topoisomerase complex critical for genome stability. Nature 561, 132–136 (2018). [DOI] [PubMed] [Google Scholar]
- 39.McHugh C. A., et al. , The Xist lncRNA interacts directly with SHARP to silence transcription through HDAC3. Nature 521, 232–236 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mao J., et al. , A critical role of Hrd1 in the regulation of optineurin degradation and aggresome formation. Hum. Mol. Genet. 26, 1877–1889 (2017). [DOI] [PubMed] [Google Scholar]
- 41.Ryan T. A., Tumbarello D. A., Optineurin: A coordinator of membrane-associated cargo trafficking and autophagy. Front. Immunol. 9, 1024 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rivas C., Aaronson S. A., Munoz-Fontela C., Dual role of p53 in innate antiviral immunity. Viruses 2, 298–313 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Boxx G. M., Cheng G., The roles of type I interferon in bacterial infection. Cell Host Microbe 19, 760–769 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lippmann J., et al. , Dissection of a type I interferon pathway in controlling bacterial intracellular infection in mice. Cell. Microbiol. 13, 1668–1682 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Carlevaro-Fita J., Johnson R., Global positioning system: Understanding long noncoding RNAs through subcellular localization. Mol. Cell 73, 869–883 (2019). [DOI] [PubMed] [Google Scholar]
- 46.Benoit Bouvrette L. P., et al. , CeFra-seq reveals broad asymmetric mRNA and noncoding RNA distribution profiles in Drosophila and human cells. RNA 24, 98–113 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Smirnov A., et al. , Grad-seq guides the discovery of ProQ as a major small RNA-binding protein. Proc. Natl. Acad. Sci. U.S.A. 113, 11591–11596 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Shen W. C., Li H. Y., Chen G. C., Chern Y., Tu P. H., Mutations in the ubiquitin-binding domain of OPTN/optineurin interfere with autophagy-mediated degradation of misfolded proteins by a dominant-negative mechanism. Autophagy 11, 685–700 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Richter B., et al. , Phosphorylation of OPTN by TBK1 enhances its binding to Ub chains and promotes selective autophagy of damaged mitochondria. Proc. Natl. Acad. Sci. U.S.A. 113, 4039–4044 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Gao J., Ohtsubo M., Hotta Y., Minoshima S., Oligomerization of optineurin and its oxidative stress- or E50K mutation-driven covalent cross-linking: Possible relationship with glaucoma pathology. PLoS One 9, e101206 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Slowicka K., Vereecke L., van Loo G., Cellular functions of optineurin in health and disease. Trends Immunol. 37, 621–633 (2016). [DOI] [PubMed] [Google Scholar]
- 52.Jiang M., et al. , Self-recognition of an inducible host lncRNA by RIG-I feedback restricts innate immune response. Cell 173, 906–919.e13 (2018). [DOI] [PubMed] [Google Scholar]
- 53.Xie Q., et al. , Long noncoding RNA ITPRIP-1 positively regulates the innate immune response through promotion of oligomerization and activation of MDA5. J. Virol. 92, e00507-18 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ma H., et al. , The long noncoding RNA NEAT1 exerts antihantaviral effects by acting as positive feedback for RIG-I signaling. J. Virol. 91, e02250-16 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Honda K., Takaoka A., Taniguchi T., Type I interferon [corrected] gene induction by the interferon regulatory factor family of transcription factors. Immunity 25, 349–360 (2006). Erratum in: Immunity 25, 849 (2006). [DOI] [PubMed] [Google Scholar]
- 56.Lehman N., The RNA world: 4,000,000,050 years old. Life (Basel) 5, 1583–1586 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Hoagland M. B., Stephenson M. L., Scott J. F., Hecht L. I., Zamecnik P. C., A soluble ribonucleic acid intermediate in protein synthesis. J. Biol. Chem. 231, 241–257 (1958). [PubMed] [Google Scholar]
- 58.Scherrer K., Darnell J. E., Sedimentation characteristics of rapidly labelled RNA from HeLa cells. Biochem. Biophys. Res. Commun. 7, 486–490 (1962). [DOI] [PubMed] [Google Scholar]
- 59.Köhler R., et al. , Expression and use of the green fluorescent protein as a reporter system in Legionella pneumophila. Mol. Gen. Genet. 262, 1060–1069 (2000). [DOI] [PubMed] [Google Scholar]
- 60.Livak K. J., Schmittgen T. D., Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔC(T) method. Methods 25, 402–408 (2001). [DOI] [PubMed] [Google Scholar]
- 61.Tawk C., Sharan M., Eulalio A., Vogel J., A systematic analysis of the RNA-targeting potential of secreted bacterial effector proteins. Sci. Rep. 7, 9328 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Love M. I., Huber W., Anders S., Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15, 550 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Saldanha A. J., Java Treeview—extensible visualization of microarray data. Bioinformatics 20, 3246–3248 (2004). [DOI] [PubMed] [Google Scholar]
- 64.Kamburov A., Wierling C., Lehrach H., Herwig R., ConsensusPathDB—a database for integrating human functional interaction networks. Nucleic Acids Res. 37, D623–D628 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets produced in this study are available in the following database: RNA-seq data: GEO, https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE101409 (accession no. GSE101409).