Significance
Auxin signaling plays essential roles in almost every aspect of plant growth and development. Auxin response factors (ARFs) are key transcriptional regulators of auxin signaling. However, it is not clear what roles ARF transcription factors may play in plant–pathogen, and specifically plant–virus, interactions. This study reveals that an ARF transcription factor is targeted by several independently evolved viral proteins of very different plant RNA viruses. These viral proteins impede the ARF activity in different ways, but in each case, these interactions benefit viral infection. These findings demonstrate that manipulation of the auxin signaling by viral proteins is a common pathogenicity strategy in plant RNA viruses.
Keywords: auxin signaling, auxin response factor, OsARF17, rice virus, viral proteins
Abstract
Plant auxin response factor (ARF) transcription factors are an important class of key transcriptional modulators in auxin signaling. Despite the well-studied roles of ARF transcription factors in plant growth and development, it is largely unknown whether, and how, ARF transcription factors may be involved in plant resistance to pathogens. We show here that two fijiviruses (double-stranded RNA viruses) utilize their proteins to disturb the dimerization of OsARF17 and repress its transcriptional activation ability, while a tenuivirus (negative-sense single-stranded RNA virus) directly interferes with the DNA binding activity of OsARF17. These interactions impair OsARF17-mediated antiviral defense. OsARF17 also confers resistance to a cytorhabdovirus and was directly targeted by one of the viral proteins. Thus, OsARF17 is the common target of several very different viruses. This suggests that OsARF17 plays a crucial role in plant defense against different types of plant viruses, and that these viruses use independently evolved viral proteins to target this key component of auxin signaling and facilitate infection.
Virus infection often causes abnormal host plant development. For instance, infection of rice by fijiviruses (rice black streaked dwarf virus, RBSDV, and the closely related Southern rice black streaked dwarf virus, SRBSDV) results in the stunting of plants and leaves that are dark green and stiff (1). Plants infected by rice stripe virus (RSV) have leaves with necrotic stripes, and the plants show wilting and stunting. These symptoms are associated with changes in concentrations of plant hormones (2), and viral infections have been widely reported to interfere with plant hormone homeostasis (3–5). For example, the P2 protein of rice dwarf virus (RDV) interacts with ent-kaurene oxidase and affects GA biosynthesis, resulting in a dwarf phenotype (6). Our previous studies also indicated that many plant hormone pathways were dramatically changed following RBSDV invasion (7–9). For example, the resistance response to RBSDV includes up-regulation of the jasmonate (JA) pathways and down-regulation of the brassinosteroid and abscisic acid pathways.
Auxin, the first discovered plant growth hormone, regulates many aspects of plant growth and development (10). Three indispensable protein families are known to be involved in the auxin signaling pathway: auxin receptors TIR1/AFB, Aux/IAA repressor proteins, and auxin response factors (ARFs) (11, 12). Auxin is first perceived by the coreceptor complex (TIR1/AFB protein and Aux/IAA repressor protein). When auxin concentrations are low, Aux/IAA proteins are more stable and repress ARF transcription factors by direct interaction with their domains III and IV (13). At higher concentrations, auxin acts as a molecular glue directly binding to domain II of Aux/IAA repressors and TIR1/AFB proteins, thereby triggering Aux/IAA ubiquitination by SCFTIR1/AFB E3 ligase and proteolysis mediated by the 26S proteasome (14). This degradation causes de-repression of the ARFs and allows transcription of auxin response genes. In plants, ARF transcription factors regulate auxin gene transcription. A typical ARF contains three components: a conserved N-terminal DNA-binding domain (DBD), a nonconserved middle region (MR), and a conserved C-terminal dimerization domain (CTD) (15). The DBD domain of ARF contains a B3 DNA binding motif that binds specifically to AuxREs (TGTCH, where H is A, C, or T) to regulate the transcription of auxin-responsive genes (16). In rice, the ARF protein family contains 25 members that can be divided into two groups on the basis of the variable MR domain. Nine (including OsARF17) are transcriptional activators, whereas the remaining 16 are transcriptional repressors (17). The CTD regions of ARFs are similar to Aux/IAA protein domains III-IV, and are protein–protein interaction domains (18, 19). The specific interactions ARF–ARF, ARF–Aux/IAA, and Aux/IAA–ARF may affect the auxin-responsive gene expression. ARF–ARF interactions via the DNA-binding domain and/or through the CTD domain are critical for ARF function (13, 20, 21). The oligomerization of Aux/IAA proteins through the CTD domain may be required for ARF repression (20, 22).
Auxin not only regulates plant growth and development but also plays key roles in plant defense against pathogen invasion (23). The positive or negative roles of auxin in pathogen infection depend on the host and pathogen combination. For example, the auxin pathway enhanced plant defense against necrotrophic pathogens by interacting with the JA pathway in Arabidopsis plants (24), but elevated auxin levels promoted disease symptoms in plants infected by the bacterial pathogen Pseudomonas syringae (25). Several studies have demonstrated that plant viruses regulate the auxin pathway to affect virus accumulation and spread (26–28). As key regulators of auxin signaling, ARFs play essential roles in various auxin signaling-mediated biological processes. However, it has rarely been reported that ARFs play a role in plant resistance to pathogens.
This study used several very different viruses of rice that each cause major destructive diseases in Asia (1, 29). SRBSDV is a fijivirus with a double-stranded RNA genome of 10 segments (S1-S10) (30, 31) and 13 ORFs. Not all of the viral proteins have been clearly identified, but SP8 and SP10 are, respectively, core capsid and outer capsid proteins (32). RSV is a tenuivirus with a single-stranded RNA genome of four segments and has some proteins translated directly from the virion strand, while others are translated from the complementary strand. RNA2 of RSV encodes P2 (a weak RNA silencing suppressor) and Pc2 (a glycoprotein) (33, 34). Rice stripe mosaic virus (RSMV) is a negative-sense single-stranded RNA virus with a monopartite genome that encodes seven proteins and is classified in the genus Cytorhabdovirus (35). We show here that both the SP8 of SRBSDV and the P2 of RSV interact with OsARF17 to facilitate virus infection, but in different ways. SRBSDV P8 disturbed the auxin pathway by repressing the transcriptional activation activity of OsARF17 and interfering with OsARF-OsARF dimerization. RSV P2 interacted with OsARF17 through the DBD domain and impeded the DNA binding ability of OsARF17. Moreover, OsARF17 was also shown to specifically interact with RSMV M protein via its MR domain. Thus, our findings suggest that these different viruses use independently evolved viral proteins to target this key component of auxin signaling for infection by distinct strategies.
Results
The SRBSDV SP8 Protein Interacts with Auxin Response Transcription Factor OsARF17.
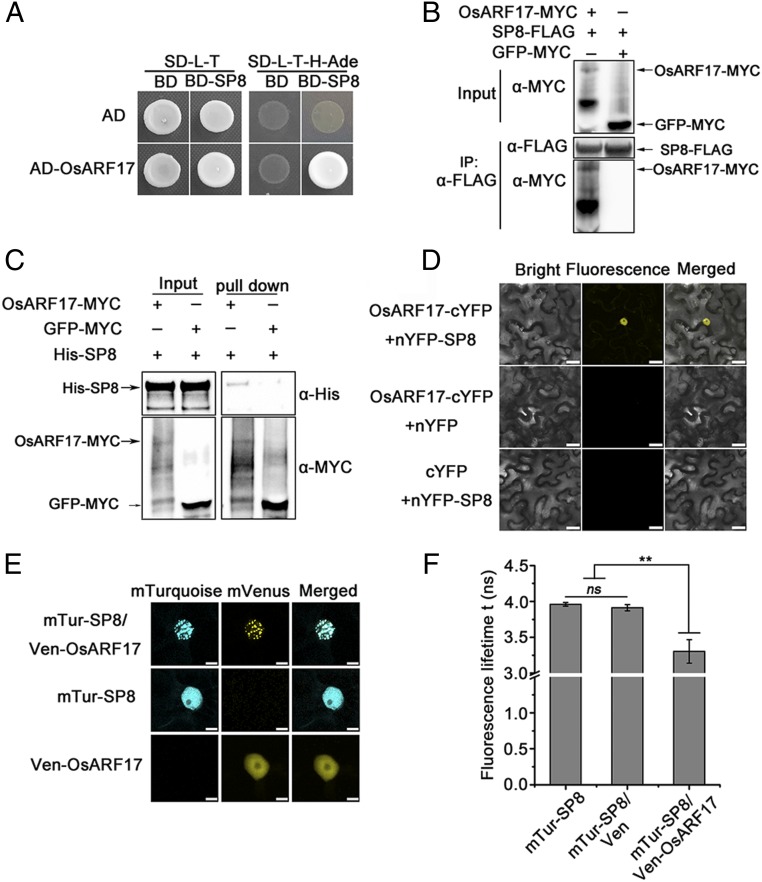
Our previous results showed that auxin signaling played vital roles in defending rice against virus infection (36). To investigate the interaction between auxin signaling and SRBSDV, the full-length viral proteins of SRBSDV were used as baits to screen a rice cDNA library in a yeast two-hybrid (Y2H) assay, and a rice auxin transcription factor OsARF17 was found to interact with SP8 (Fig. 1A). The full-length OsARF17 interacted only with SP8 and not with any other SRBSDV protein (SI Appendix, Fig. S1A). Coimmunoprecipitation (Co-IP) assays also demonstrated that OsARF17 interacted with SP8 in vivo (Fig. 1B), and this was further confirmed using an in vitro pull-down assay of overexpressed OsARF17–MYC in Nicotiana benthamiana leaves and a recombinant His–SP8 fusion protein (Fig. 1C). Bimolecular fluorescence complementation (BiFC) assays were also performed in tobacco cells. When OsARF17-cYFP and nYFP-SP8 were transiently coexpressed in N. benthamiana leaves, there were strong YFP fluorescence signals in the nucleus, but there were no detectable signals in the negative control combinations OsARF17-cYFP and nYFP or cYFP and YFP-SP8 (Fig. 1D). Collectively, these results confirm that SP8 directly associates with OsARF17 in planta.
Fig. 1.
SRBSDV P8 protein interacts with OsARF17. (A) Y2H assays illustrating the interaction between SP8 and OsARF17 proteins. The different combinations of constructs transformed into yeast cells were grown on selective media SD/-Trp/-Leu (SD-L-T), and interactions were tested with SD/-Trp/-Leu/-His/-Ade (SD-L-T-H-Ade). Pictures were taken after 3 d of incubation at 30 °C. (B) Co-IP assays confirm that SP8 interacts with OsARF17 in N. benthamiana leaves. Total proteins were extracted and immunoprecipitated by anti-FLAG magnetic beads. The coimmunoprecipitated proteins were detected by anti-MYC antibody. (C) In vitro pull-down assays demonstrating the interaction of SP8 with OsARF17. His–SP8 proteins were used to pull-down with OsARF17–MYC and GFP–MYC, and further detected with anti-His antibody. (D) BiFC assays of the interaction between SP8 and OsARF17 in N. benthamiana cells. (Scale bars, 25 μm.) (E) The subcellular localization of SP8 and OsARF17, shown by expressing mTur-SP8 (CFP) or Ven-OsARF17 (YFP) alone or by coexpressing them. (Scale bars, 10 μm.) (F) Mean lifetime measurements of mTur-SP8 expressed alone or in the presence of Ven or Ven-OsARF17 in a fluorescence lifetime imaging assay to monitor the interaction between CFP-(donor) and YFP-(acceptor) tagged SP8 and OsARF17, respectively. Values shown are the means ± SD of 3 biological replicates. Significant differences were identified using Fisher’s least significant difference tests. **At the top of columns indicates significant difference at P ≤ 0.01. ns, no significant difference.
The interaction between OsARF17 and SP8 suggested that OsARF17 would be colocalized with SP8 in the nucleus. To test this, either OsARF17 fused to the Yellow Fluorescent Protein mVenus (Ven-OsARF17) or SRBSDV SP8 fused to the Cyan Fluorescent Protein mTurquoise (mTur-SP8) was transiently expressed in N. benthamiana epidermal cells. In each case, there was a diffuse fluorescent signal in the nucleus (Fig. 1E). When Ven-OsARF17 and mTur-SP8 were coexpressed, they were clearly colocalized in the nucleus (Fig. 1E), but appeared to be in distinct punctate structures. A fluorescence lifetime imaging assay was then used to monitor the interaction between the two tagged proteins (Fig. 1F and SI Appendix, Table S2). The lifetime of CFP in the nucleus coexpressing mTur-SP8 and Ven-OsARF17 was 3.2 ± 0.15 ns, which is significantly less than when mTur-SP8 was expressed alone (3.9 ± 0.03 ns) or when mTur-SP8 and mVenus were coexpressed (3.9 ± 0.04). This reflects a direct physical association between the two proteins in the nucleus.
The CTD Domain of OsARF17 Is Required for Its Interaction with SP8 and for Dimerization.
OsARF proteins contain three important domains: a DBD, a MR, and a CTD (37). Using a series of truncated mutants of OsARF17 in Y2H assays, we showed that SP8 strongly interacted with both the MR and CTD domains, but not with the DBD domain (SI Appendix, Fig. S2 A and B). In CoIP assays, SP8-FLAG was coimmunoprecipitated with CTD–MYC, but not with DBD–MYC (SI Appendix, Fig. S2C). The results indicate that the CTD domain is required for the SP8–OsARF17 interaction.
The seven other transcriptional activator OsARFs (OsARF6, OsARF11, OsARF12, OsARF16, OsARF19, OsARF21, and OsARF25) were cloned and tested for any interaction with SP8 by Y2H. Only OsARF6, OsARF12, and OsARF25 interacted with SP8 (SI Appendix, Fig. S3B), and these are the ones that have the closest similarity to OsARF17 (SI Appendix, Fig. S3A). Previous research showed that ARFs can interact with one another through their DBD or CTD domains to form self-dimers or heterodimers (12, 17, 19). Y2H assays were next performed to test whether OsARF17 could form a self-dimer or a heterodimer with OsARF6, OsARF12, and OsARF25 via either the DBD or CTD domains. As shown in SI Appendix, Fig. S4A, OsARF17 formed both self-dimers and heterodimers through its CTD domain, but not via the DBD domain. These results therefore show that the CTD domain is required for OsARF17 dimerization.
Extensive research suggests that ARFs regulate the expression of auxin response genes by interacting with Aux/IAA proteins via their CTD domains (13, 38). When the Aux/IAA protein OsIAA20 is overexpressed in transgenic rice, the plants are more susceptible to RBSDV, and we were able to confirm that OsARF17 interacted with OsIAA20 through its CTD domain (SI Appendix, Fig. S4B), consistent with previous studies (17). Overall, these results indicate that the CTD domain is indispensable for OsARF17 dimerization with itself, other OsARFs, and OsIAA20 (17, 39, 40).
SP8 Attenuates Auxin Signaling by Interfering with OsARF17 Dimerization.
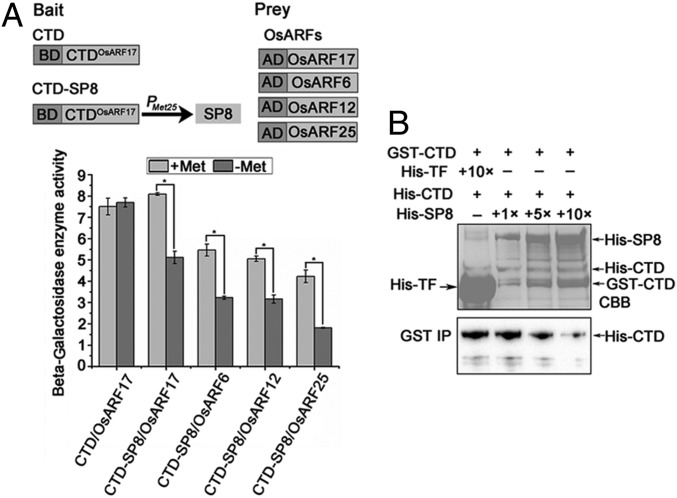
Since the CTD domain of OsARF17 is essential for its interactions with SP8, other OsARFs, and OsIAA20, we speculated that SP8 might act by competing for CTD domain binding sites with these other proteins. We first examined whether SP8 interfered with OsARF17 dimerization using yeast three-hybrid (Y3H) assays with the CTD domain of OsARF17 as the bait and driving the expression of SP8 by the methionine-inducible Met25 promoter (Fig. 2 A, Upper). In this assay, the expression of SP8 was inhibited in the presence of methionine and induced by its absence. β-Galactosidase enzyme activity assays showed that SP8 could affect the CTD-OsARF17 interaction (Fig. 2 A, Lower). In further experiments, dose-dependent pull-down assays were used to assess whether SP8 interfered with the CTD–CTD interaction in vitro. The same amounts of GST–CTD and His–CTD were mixed with different amounts of His–SP8 or His–TF (negative control) and immobilized onto anti-GST beads. The amounts of GST–CTD bound to His–CTD decreased with increasing amounts of His–SP8 (Fig. 2B), indicating that SP8 disrupted CTD dimerization in vitro. Y3H assays were also used to determine whether SP8 could interfere with the OsARF17–OsIAA20 interaction. β-Galactosidase enzyme activity measurements showed that SP8 affected the OsARF17–OsIAA20 interaction (SI Appendix, Fig. S5). Together, these results indicate that SP8 disturbs OsARF17 dimerization by competing with the CTD domain.
Fig. 2.
SP8 interferes with OsARF17 dimerization. (A) Y3H assays demonstrating the influence of SP8 on OsARF17 dimerization. (Upper) Schematic diagrams of the bait and the prey constructs used in Y3H assays. The CTD domain of OsARF17 ligated into pBridge vector was the bait, and the expression of SP8 was driven by the methionine-inducible Met25 promoter. The OsARFs (OsARF17, OsARF6, OsARF12, OsARF25) were cloned into pGADT7 vectors as prey. (Lower) Yeast cells containing bait and prey vectors were grown on selective media SD/-Trp/-Leu (SD-L-T) for 3 d. The yeast cells were shaken with SD-L-T-H-Ade or SD/-Trp/-Leu/-His/-Met liquid selective media. β-Galactosidase enzyme activity assays with or without SP8. When methionine (20 mM) was present, the expression of SP8 was inhibited. Values shown are the means ± SD of 3 biological replicates. Significant differences were identified using Fisher’s least significant difference tests. *At the top of columns indicates significant difference at P ≤ 0.05. (B) Dose-dependent pull-down assays to assess SP8 interference with the CTD-CTD interaction in vitro. Equal amounts of purified GST–CTD and His–CTD mixed with proteins were incubated with increasing amounts of His–SP8 (0, 2, 10, or 20 μg) or His–TF (negative control) in vitro. The interaction between GST–CTD and His–CTD was weakened by His–SP8. His–CTD proteins were used to pull-down with GST–CTD, and further detected with anti-His antibody. (Upper) The loading of His–SP8, GST–CTD, His–CTD, and His–TF is shown by Coomassie brilliant blue staining.
SP8 Suppresses the Transcriptional Activation Activity of OsARF17 and Restrains Auxin-Induced Root Growth Inhibition.
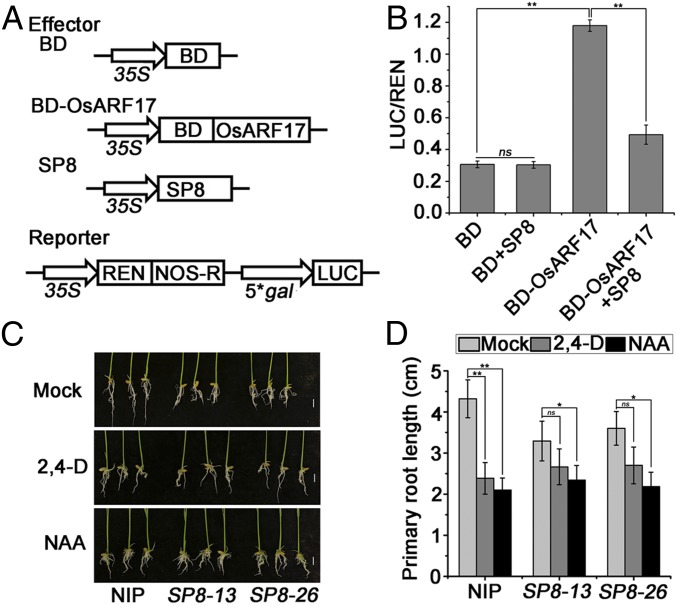
Several reports have shown that OsARF17 belongs to the class of transcriptional activators (17, 41). It has previously been reported that the RBSDV P8 protein, a homolog of SRBSDV SP8, acts as a transcriptional repressor (42). We next confirmed that SP8 also has transcriptional repressor activity using a GAL4 DNA-BD binding element (5*gal)-luciferase (LUC) reporter assay in N. benthamiana leaves. The promoter of 5*gal was used to drive the firefly luciferase gene (LUC) as a reporter, and the Renillia luciferase (REN) reporter gene controlled by Cauliflower mosaic virus promoter (35S) was used as the reference. The BD domain, which can bind the 5*gal promoter, was fused with SP8 protein (SI Appendix, Fig. S6A). There was a significant decrease in LUC activity in the presence of BD–SP8 compared with the empty effector BD, and this was not affected by coexpression with OsARF17 (SI Appendix, Fig. S6B).
To examine whether SP8 affected the transcriptional activation of OsARF17, we performed dual-LUC reporter assays in N. benthamiana leaves, using BD, BD-OsARF17, and SP8 as the effectors (Fig. 3A). The LUC reporter activity was significantly enhanced in the presence of BD-OsARF17 compared with the BD control, but this effect was inhibited when BD-OsARF17 and SP8 were coexpressed (Fig. 3B). Together, these results suggest that OsARF17 is indeed a transcriptional activator and that SP8 can repress its effects.
Fig. 3.
SP8 reduced the transcriptional activation activity of OsARF17. (A) Schematic diagrams of the effectors and reporters used in the dual-LUC experiments. The effectors BD, OsARF17-BD, and SP8 fused FLAG-tag. The reporters 35S: REN-5*gal pro:LUC plasmids. The BD domain can bind the 5*gal promoter. (B) The relative LUC activities were measured in N. benthamiana cells, using the combinations shown in A. The empty BD effector was used as a negative control. The LUC/REN ratio represents the relative LUC activity. (C) The effect of synthetic auxin (2,4-D or NAA) treatments on transgenic rice plants expressing the SP8 gene (SP8-13 and SP8-26) driven by the CaMV 35S promoter. The seedling shoots of SP8-13 and SP8-26 were immersed in 0.1 μm 2, 4-D, and 0.1 μm NAA culture solution in the dark and compared with 0.1% Triton X-100 treated rice controls for 10 d at 37 °C. Each treatment used at least 25 to 30 plants. Pictures were taken after 10 d of treatment. (Scale bar, 1 cm.) (D) Statistical analyses of primary root length. Values shown are the means ± SD of 3 biological replicates. Significant differences were identified using Fisher's least significant difference tests. At the top of columns, significant difference at *P ≤ 0.05 and **P ≤ 0.01, respectively. ns, no significant difference.
To explore the biological significance of this inhibition by the SP8 protein of the transcriptional activity of OsARF17, transgenic rice plants were generated that expressed the SP8 gene driven by the CaMV 35S promoter. Two genetically stable homozygous lines, SP8-13 and SP8-26, were used in these experiments; the relative transcript levels of SP8 are shown in SI Appendix, Fig. S7A. To evaluate the auxin responsiveness of these plants, they were treated with the auxin analogs 2, 4-D, and NAA at 37 °C for 7 d, and the inhibition of root growth was compared with that in NIP control plants. Compared with untreated transgenic seedlings 2, 4-D treatment caused about 46% inhibition in the control NIP primary roots, but less than 25% in SP8-13 and SP8-26 plants, and there was a similar effect from NAA treatment (Fig. 3 C and D). These results suggest that expression of SP8 made plants less sensitive to the auxin analogs. Together, these results show that SP8 represses the auxin response by disrupting the function of OsARF17.
OsARF17 Enhances Rice Defense Against SRBSDV Infection.
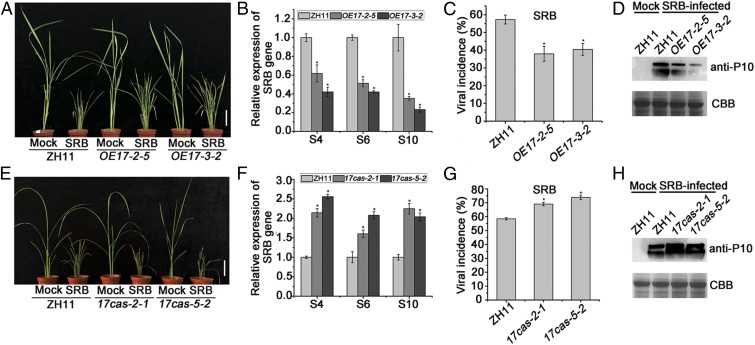
The role of OsARF17 in plant immunity against SRBSDV infection was next investigated. Transgenic rice plants overexpressing OsARF17 were first constructed (lines OE17-2-5 and OE17-3-2). The expression levels of OsARF17 in the T3 generation transgenic rice seedlings were more than 180 to 250 times higher than those in the wild-type ZH11 (SI Appendix, Fig. S7B). The primary roots of transgenic plants overexpressing OsARF17 were significantly longer than those of control ZH11 plants (SI Appendix, Fig. S7 C and D). This is consistent with previous studies showing that OsARFs participate in regulating root elongation and auxin response (43, 44). Transgenic or control plants were then infested with SRBSDV-infected or virus-free insect vector Sogatella furcifera, and subsequent symptoms were monitored. The SRBSDV-infected transgenic lines were less dwarfed than the infected controls (Fig. 4A), and quantitative real-time PCR (qRT-PCR) indicated that the levels of SRBSDV S4, S6, and S10 RNAs in transgenic lines were less than 40% of those in control rice plants (Fig. 4B). The viral incidence in both transgenic lines (<40%) was significantly less than in ZH11 plants (57%; Fig. 4C), and there were similar differences in the amounts of viral coat protein P10 detected (Fig. 4D). Thus, rice overexpressing OsARF17 was less susceptible to viral infection.
Fig. 4.
OsARF17 positively modulates rice resistance to SRBSDV. (A and E) The symptoms on SRBSDV-infected WT (ZH11), transgenic, and mutant plants. The phenotypes were observed and photos taken at 30 dpi. (Scale bars, 5 cm.) (B and F) qRT-PCR results showing the relative expression levels of SRBSDV (S4, S6, and S10) in SRBSDV-infected OE17 (OE17-2-5 and OE17-3-–2) and 17cas (17cas-2-1 and 17cas-5-2) plants compared with SRBSDV-infected control plants. UBQ5 was used as the internal reference gene to normalize the relative expression. Values shown are the means ± SD of 3 biological replicates. Significant differences were identified using Fisher's least significant difference tests. *At the top of columns indicates significant difference at P ≤ 0.05. (C and G) Disease incidence in ZH11 (control) and OE17 lines (OE17-2-5 and OE17-3-2) or 17cas (17cas-2-1 and 17cas-5-2) lines, respectively. The percentage of plants infected by SRBSDV was determined by RT-PCR at 30 dpi. (D and H) The accumulation of SRBSDV P10 protein in SRBSDV-infected plants determined by Western blotting.
To further shed light on the role of OsARF17 in SRBSDV infection, the CRISPR/Cas9 system was used to generate mutants of the OsARF17 transgenic plants in the background of ZH11 (45). Two homozygous mutants called 17cas-2-1 and 17cas-5-2 were identified and characterized (SI Appendix, Fig. S8). The primary roots of 17cas-2-1 and 17cas-5-2 plants were significantly shorter than those of ZH11 (SI Appendix, Fig. S7 E and F). After challenge with SRBSDV, the mutant plants were more dwarfed and had more severe symptoms than the ZH11 controls (Fig. 4E). The mutants also had a greater accumulation of SRBSDV genomic RNAs (S4, S6, S10; Fig. 4F), a higher concentration of viral coat protein (Fig. 4H), and a higher incidence of infected plants (Fig. 4G). Overall, these results suggest a critical role for OsARF17 in enhancing host defense against SRBSDV.
OsARF17-Mediated Rice Defense Against RBSDV Infection.
SRBSDV and RBSDV are closely related members of the genus Fijivirus. We therefore tested whether RBSDV P8, the homolog of SRBSDV SP8, also interacted with OsARF17, using Y2H. The results showed that RBSDV P8 protein could also associate with OsARF17 via the CTD domain in a similar manner to SRBSDV SP8 (SI Appendix, Fig. S9). The OsARF17 transgenic plants were resistant to RBSDV, as well as SRBSDV: following inoculation using the insect vector, qRT-PCR showed that there were lower levels of transcripts of RBSDV genes (S4, S6, S10) in the transgenic lines compared with the controls. There were also higher virus RNA levels in RBSDV-infected OsARF17 mutants than in ZH11 plants (SI Appendix, Fig. S10 A and B). Thus, OsARF17 functions in antiviral defense against both these pathogens of rice.
To eliminate the possibility that OsARF17 transgenic plants differed from wild-type plants in their resistance to the insect vectors, the survival rates of these insects were tested during a 5-d period, as described previously (7). Insect mortality rates did not differ between the transgenic and control plants (SI Appendix, Fig. S11A).
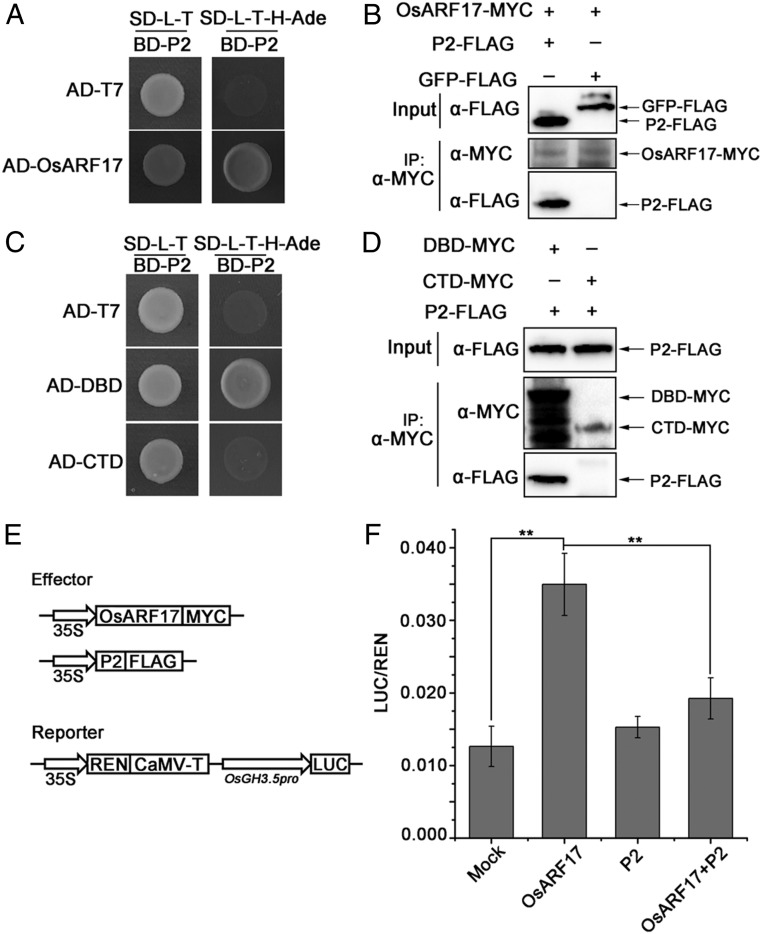
The RSV P2 Protein Interacts with OsARF17 and Impedes Its DNA Binding Ability.
To determine whether a different rice virus interacts with OsARF17 and perturbs the auxin signaling pathway, we used a Y2H assay to test for any interaction between OsARF17 and the proteins of a single-stranded RNA virus, RSV. Interestingly, OsARF17 specifically interacted with RSV P2 in yeast cells (Fig. 5A), but not with other viral proteins (SI Appendix, Fig. S12). Co-IP assays confirmed this interaction in N. benthamiana leaves when OsARF17–MYC and P2-FLAG were coexpressed (Fig. 5B). While SRBSDV SP8 specifically binds to the CTD domain of OsARF17, the RSV P2 interacted with the DBD domain and not the CTD domain in a Y2H assay (Fig. 5C). This result was confirmed by Co-IP assays (Fig. 5D). The DBD domain of ARF specifically binds to AuxREs to regulate the expression of auxin response genes (GH3 and SAUR) (7, 16, 46, 47). In rice, OsARF19 affects the leaf angles by positively regulating the expression of the OsGH3.5 gene (48). Interestingly, we found that auxin response elements (AuxREs) were present in the OsGH3.5 promoter. To determine whether OsARF17 directly binds to the promoter of OsGH3.5, we conducted electromobility shift assays with a recombinant His–DBD fusion protein and His–TF purified from E. coli. The DIG-labeled DNA probes contained the AuxREs motif (SI Appendix, Fig. S13A). The His–DBD fusion protein specifically bound the DIG-labeled OsGH3.5 probes in competition with unlabeled probes (SI Appendix, Fig. S13A), and no band was observed when His–TF was used in place of His–DBD. These results show that OsARF17 can directly bind the AuxRE element in the OsGH3.5 promoter. Next, to evaluate whether OsARF17-P2 association affects the ability of OsARF17 to bind to the promoter of OsGH3.5, we performed electromobility shift assays by increasing the amount of MBP-P2 fusion protein purified from E. coli (SI Appendix, Fig. S13B). With increasing concentrations of MBP-P2, the intensity of the shifted band significantly decreased, whereas the control MBP had no influence on the DNA binding activity of OsARF17. We further analyzed the effect of P2 on the function of OsARF17 in N. benthamiana leaf cells by dual-LUC assays. OsARF17 bound to the OsGH3.5 promoter and activated the expression of the LUC reporter (Fig. 5 E and F), but this expression was significantly decreased when both P2 and OsARF17 were coexpressed. P2 did not affect the transcriptional activation of OsARF17 when it was fused to the BD domain and tested in 5*gal-LUC reporter assays (SI Appendix, Fig. S14 A and B). Together, these data showed that P2 mainly interacted with and influenced the DNA binding ability of OsARF17, but did not interfere with its transcriptional activation activity.
Fig. 5.
RSV P2 protein interacts with OsARF17. (A) Y2H assays showing the interaction between RSV P2 and OsARF17 proteins. The different combinations of constructs transformed into yeast cells were grown on selective media SD/-Trp/-Leu (SD-L-T), and interactions were tested with SD/-Trp/-Leu/-His/-Ade (SD-L-T-H-Ade). Pictures were taken after 3 d of incubation at 30 °C. (B) Co-IP assays confirm that P2 interacts with OsARF17 in N. benthamiana leaves. Total proteins were extracted and immunoprecipitated by anti-MYC magnetic beads. The coimmunoprecipitated proteins were probed with an anti-FLAG antibody. (C) Y2H assays show that P2 interacts with the DBD domain and not with the CTD domain of OsARF17. (D) Co-IP assays confirm that P2 interacts with the DBD domain in N. benthamiana leaves. Total proteins were extracted and immunoprecipitated by anti-MYC magnetic beads. The coimmunoprecipitated proteins were probed with an anti-FLAG antibody. (E and F) P2 affects the transcriptional activation activity of OsARF17 in N. benthamiana leaf cells by dual-LUC assays. (E) Schematic diagrams of the dual-LUC assays. The OsGH3.5 promoter driving the firefly luciferase (LUC) was used as the reporter. The Renilla luciferase (REN) is the internal control. OsARF17 and P2 are the effectors. (F) The OsGH3.5 promoter was activated by OsARF17 proteins, but this activation was significantly suppressed by coexpressing P2 in N. benthamiana cells. The reporters expressed alone or coexpressed with P2 were used as negative controls. The LUC/REN ratio represents the relative LUC activity. Values shown are the means ± SD of 3 biological replicates. Significant differences were identified using Fisher’s least significant difference tests. **At the top of columns indicates significant difference at P ≤ 0.01.
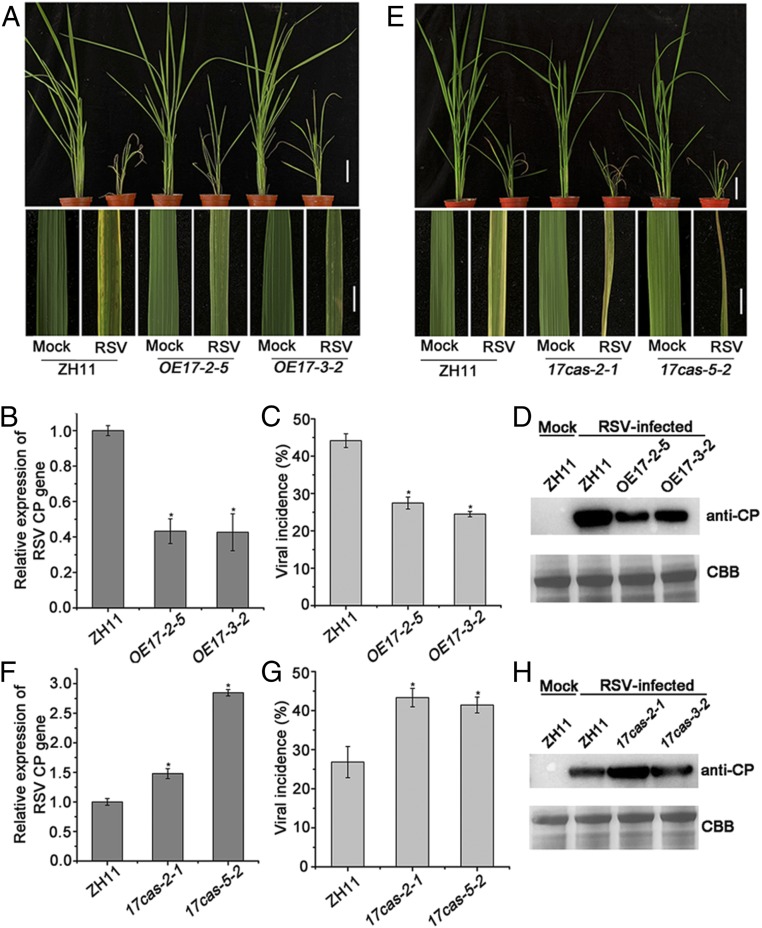
OsARF17 Enhances Rice Resistance to RSV.
To investigate the effect of OsARF17 on RSV resistance, OsARF17 transgenic lines were challenged with RSV. RSV-infected plants exhibited necrotic stripes and stunting, but symptoms on the transgenic lines overexpressing OsARF17 were milder than in the control ZH11 plants (Fig. 6A). qRT-PCR results showed that the transgenic lines had substantially (two- to threefold) less viral CP RNA than the controls (Fig. 6B), and viral incidence (the proportion of infected plants) was also significantly lower in the two transgenic lines (28% and 25%) than in ZH11 plants (45%; Fig. 6C). Similar trends were observed in RSV CP protein expression levels (Fig. 6D). These results confirm that overexpressing the OsARF17 protein in rice enhanced resistance to RSV. RSV-infected mutants 17cas-2-1 and 17cas-3-2 had much more severe disease symptoms than the controls (Fig. 6E); the levels of RSV CP RNA and protein were correspondingly much higher (Fig. 6 F and H), and viral incidence (44% and 42%) was also greater than in ZH11 (27%; Fig. 6G). Together, these data suggest that OsARF17 plays important roles in rice defense against RSV infection.
Fig. 6.
OsARF17 enhanced the resistance of rice to RSV. (A and E) The phenotypes of RSV-infected plants at 30 dpi. (Scale bars, 5 cm.) (Upper) and 1 cm (Lower) (B and F) qRT-PCR results showing the relative expression levels of the RSV CP gene in RSV-infected OE17 (OE17-2-5 and OE17-3-2) and 17cas (17cas-2-1 and 17cas-5-2) plants compared with RSV-infected control plants. UBQ5 was used as the internal reference gene to normalize the relative expression. Values shown are the means ± SD of 3 biological replicates. Significant differences were identified using Fisher's least significant difference tests. *At the top of columns indicates significant difference at P ≤ 0.05. (C and G) Disease incidence in ZH11 (control) and OE17 lines (OE17-2-5 and OE17-3-2) or 17cas (17cas-2-1 and 17cas-5-2) lines, respectively. The percentage of plants infected by RSV plants was determined by RT-PCR at 30 dpi. Each experiment used at least three biological replicates. (D and H) The accumulation of RSV CP protein in RSV-infected plants determined by Western blotting.
OsARF17 Is Also Targeted by a Cytorhabdovirus.
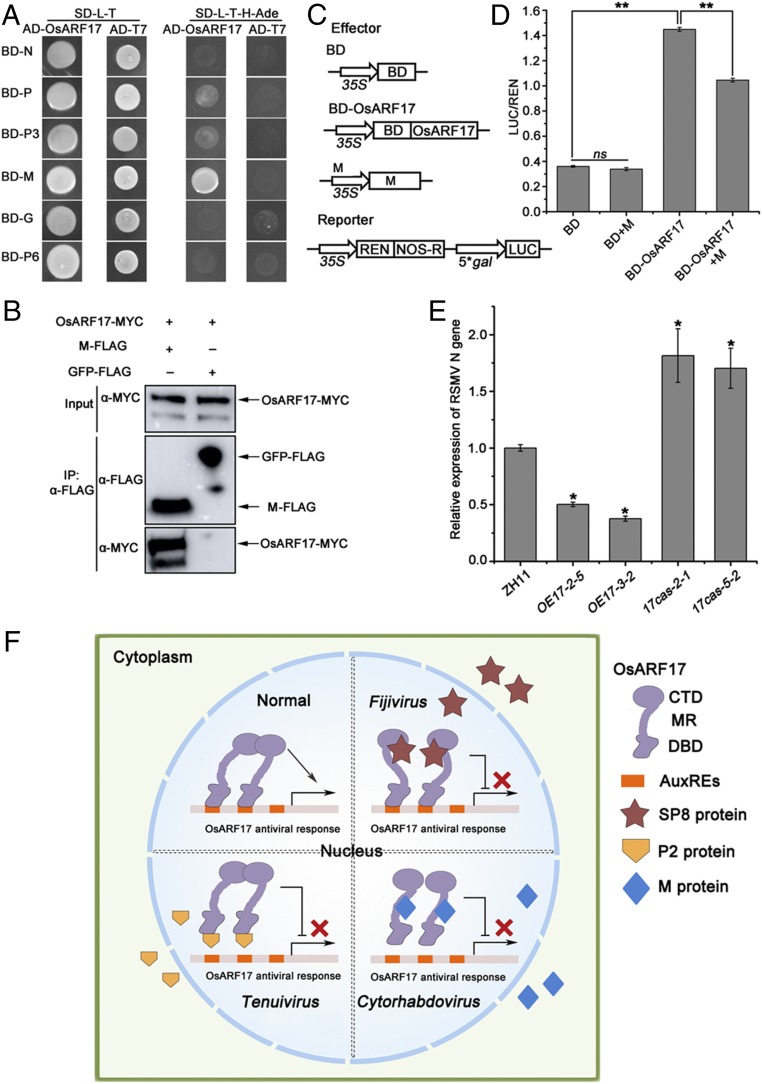
Following the finding that OsARF17 is targeted by viruses of the different genera Fijivirus and Tenuivirus, we finally investigated whether another distinct type of rice virus also modulated its function. RSMV, a negative-sense single-stranded RNA virus with a monopartite genome belonging to the genus Cytorhabdovirus, is not closely related to either fijiviruses or tenuiviruses and has recently been found in southern China (35). Its genome is predicted to have seven ORFs that are believed to encode the following proteins: nucleocapsid protein (N), phosphoprotein (P), two nonstructural proteins (P3 and P6), M protein, glycoprotein (G), and a large polymerase protein (L) (35). In Y2H assays, OsARF17 was able to specifically interact with the M protein of RSMV, but not with the other viral proteins (Fig. 7A). Further experiments showed that only the MR-CTD domain of OsARF17 was responsible for interacting with M protein, indicating that the MR domain might be involved in the M–OsARF17 interaction (SI Appendix, Figs. S2A and S15). Co-IP assays confirmed that OsARF17 interacted with M protein in vivo (Fig. 7B). In addition, we found the M protein inhibited the transcriptional activation of OsARF17 by dual-LUC reporter assays (Fig. 7 C and D). Together, these results suggest that RSMV also modulates the transcriptional activation activity of OsARF17, in this case by means of its M protein.
Fig. 7.
RSMV M protein interacts with OsARF17. (A) Y2H experiments showing the interaction between M protein and OsARF17. (B) Co-IP assays confirm that M protein interacts with OsARF17 in N. benthamiana leaves. Total proteins were extracted and immunoprecipitated by anti-MYC magnetic beads. The coimmunoprecipitated proteins were probed with an anti-FLAG antibody. (C) Schematic diagrams of the effectors and reporters used in the dual-LUC experiments. The effectors were BD, OsARF17-BD, and M. The reporters were 35S: REN-5*gal pro:LUC plasmids. The BD domain can bind the 5*gal promoter. (D) The relative LUC activities were measured in N. benthamiana cells, using the combinations shown in C. The empty BD effector was used as a negative control. The LUC/REN ratio represents the relative LUC activity. Values shown are the means ± SD of 3 biological replicates. Significant differences were identified using Fisher’s least significant difference tests. **At the top of columns indicates significant difference at P ≤ 0.01. ns, no significant difference. (E) qRT-PCR results showing the relative expression levels of the RSMV N gene in RSMV-infected OE17 (OE17-2-5 and OE17-3-2) and 17cas (17cas-2-1 and 17cas-5-2) plants compared with RSMV-infected control plants. UBQ5 was used as the internal reference gene to normalize the relative expression. Values shown are the means ± SD of 5 biological replicates. Significant differences were identified using Fisher’s least significant difference tests. *At the top of columns indicates significant difference at P ≤ 0.05. (F) Model of OsARF17-mediated antiviral defense response. In plant–virus interaction, different types of virus target the same key component of auxin signaling to inhibit OsARF17-mediated antiviral response by different strategies. Lines ending with arrows show activation; the solid line ending with perpendicular lines and multiplication signs display suppression.
To further investigate the effect of OsARF17 on RSMV infection, the virus was inoculated to plants, as previously described (35), by infesting them with either virus-free or viruliferous leafhoppers. qRT-PCR analysis showed that RSMV N gene RNA was much less in two transgenic lines overexpressing OsARF17, but dramatically more in 17cas-2-1 and 17cas-3-2 mutants than in the control Zh11 plants (Fig. 7E). There was no difference between the transgenic and control plants in resistance to the leafhopper vectors (SI Appendix, Fig. S11C). Thus, overexpression of OsARF17 in rice also enhanced resistance to RSMV.
Discussion
As one of the earliest found phytohormones, auxin plays essential roles in almost every aspect of plant growth and development. More recently, there have increasing reports that auxin plays a vital role in plant defense to diverse pathogens (23). For example, disruption of auxin signaling enhanced resistance of Arabidopsis to the bacterial pathogen P. syringae, but increased susceptibility to the fungus Alternaria brassicicola (24, 25). In addition, auxin signaling was reported to increase the susceptibility of Arabidopsis to the fungus Fusarium oxysporum (49). Hence, the roles of the auxin pathway in plant defense appear complicated and may depend on the particular pathogen. We recently reported that repression of auxin signaling by overexpression of Aux/IAA proteins increased rice susceptibility to the virus RBSDV (36). However, the effect of rice viruses on auxin signaling and the mechanism or mechanisms by which viruses regulate the auxin pathway remain largely unknown. In this study, we found that both the dsRNA viruses SRBSDV/RBSDV and the ssRNA virus RSV have viral proteins that suppress OsARF17-mediated antiviral defense. Since the cytorhabdovirus RSMV also targeted OsARF17, it appears that this protein is a common target for various rice viruses.
There have been a number of reports that the Aux/IAA proteins are targeted in pathogen virulence strategies (50). In Arabidopsis, SA stabilized Aux/IAA to inhibit auxin signaling as a part of the SA-mediated defense mechanism. The Pseudomonas syringae type III effector AvrRpt2 stabilized the interaction between TIR1 and Aux/IAA proteins to promote Aux/IAA degradation, making plants susceptible to pathogen invasion (25). RDV P2 protein specifically interacted with and stabilized OsIAA11 protein to facilitate virus infection (28). TMV replicase p126 protein altered and prevented the nuclear localization of AtIAA26 and AtIAA27, leading to a reprogramming of auxin signaling (4) that enhanced virus phloem loading and systemic movement (27). In plants, Aux/IAA proteins act as the master repression regulators in auxin signaling by interacting with and controlling ARF transcription factors. However, the direct involvement of ARF transcription factors in plant–pathogen interactions has been little reported.
ARFs are a class of transcriptional activators and repressors that bind with specificity to the AuxRE element in the promoters of early auxin response genes (13, 51). In rice, the 25 OsARF proteins are divided into two groups, transcriptional activators and transcriptional repressors, based on the MR amino acid sequence (17). In this study, we found that the transcriptional activator OsARF17 and its homologs could interact with SRBDV SP8 protein. The results indicated that SP8 modulated auxin signaling by binding to the CTD domain of OsARF17, thus inhibiting the transcriptional activation and disturbing the dimerization of OsARF17. In contrast, RSV P2 interacted with the DBD domain of OsARF17. This domain binds to AuxREs elements in promoters of auxin response genes to regulate their expression. We confirmed that OsARF17 could bind to the AuxREs element in the promoter of OsGH3.5, and our results therefore indicate that P2 modulated the function of OsARF17 by inhibiting its DNA binding activity. Since the M protein of another ssRNA virus, RSMV, interacted with OsARF17 protein in Y2H and Co-IP assays, it therefore appears that OsARF17 is a key target for multiple viral proteins.
Previous reports have shown that ARFs are critical for normal growth and development in plants (41). Thus, in rice, OsARF1 regulates the crown root by regulating the expression of OsCRL1 (52). OsARF12 regulates root elongation and iron accumulation (43). OsARF19 controls rice leaf angles by binding to the promoters of OsGH3-5 and OsBRI1. However, the role of ARFs in plant defense remains unclear. In this study, we found that overexpression of OsARF17 increased rice resistance to several very different rice viruses, whereas OsARF17 CRISPR/Cas9 mutant plants had more severe symptoms than WT plants. Our recent results showed that the induction of JA signaling in response to viral infection was suppressed in transgenic plants overexpressing OsIAA20 and OsIAA31. OsRboh-mediated ROS levels also participated in auxin signaling-mediated rice defense (36). In future work, we aim to determine whether OsARF17 directly modulates JA signaling or regulates ROS levels, and to investigate how the roles of OsARF17 in plant growth development and antiviral defense are balanced.
In conclusion, the results presented here show how proteins of different viruses bind in different ways to OsARF17, resulting in either a repression of its transcriptional activation or inhibition of its DNA binding activity. This interference with auxin signaling makes the plants more susceptible to viruses (Fig. 7F). In this previously unreported plant–virus interaction, different types of virus deploy independently evolved proteins to target the same key component of auxin signaling and thus facilitate infection.
Materials and Methods
The SP8 transgenic plants expressing the SRBSDV SP8 protein were in the Nipponbare (Oryza sativa L. cv. Japonica, NIP) cultivar background, whereas plants overexpressing OsARF17 and the OsARF17 mutant plants used Zhonghua 11 (ZH11) seedlings as the background. Details of experimental methods including plant materials treatments, qRT-PCR, plasmid construction, virus inoculation, agroinfiltration, Y2H and Y3H, Co-IP, pull-down, electrophoretic mobility shift, Western blot analysis, fluorescence resonance energy transfer (FRET)-fluorescence lifetime imaging measurements (FLIM), dual-LUC assays are provided in SI Appendix, Materials and Methods. Primers used in this study are listed in SI Appendix, Table S1.
Data Availability.
All of the materials and data that were used or generated in this study are described and available in the manuscript and SI Appendix.
Supplementary Material
Acknowledgments
We are indebted to Prof. Jianxiang Wu (Zhejiang University) for providing viral proteins antibody. We thank Prof. Guohui Zhou and Dr. Tong Zhang for providing RSMV-infected plants. We thank Mike Adams for critically reading and improving the manuscript. This work was funded by the National Key Research and Development Plan (2016YFD0200804), Ningbo Science and Technology Innovation 2025 Major Project (2019B10004) and the Major Project of New Varieties of Genetically Modified Organism of China (2016ZX08001-002). This work was sponsored by the K.C. Wong Magna Fund in Ningbo University.
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.1918254117/-/DCSupplemental.
References
- 1.Zhou G., Xu D., Xu D., Zhang M., Southern rice black-streaked dwarf virus: A white-backed planthopper-transmitted fijivirus threatening rice production in Asia. Front. Microbiol. 4, 270 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Huang R., et al. , Dynamic phytohormone profiling of rice upon rice black-streaked dwarf virus invasion. J. Plant Physiol. 228, 92–100 (2018). [DOI] [PubMed] [Google Scholar]
- 3.Alazem M., Lin N.-S., Roles of plant hormones in the regulation of host-virus interactions. Mol. Plant Pathol. 16, 529–540 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Padmanabhan M. S., Shiferaw H., Culver J. N., The Tobacco mosaic virus replicase protein disrupts the localization and function of interacting Aux/IAA proteins. Mol. Plant Microbe Interact. 19, 864–873 (2006). [DOI] [PubMed] [Google Scholar]
- 5.Collum T. D., Culver J. N., The impact of phytohormones on virus infection and disease. Curr. Opin. Virol. 17, 25–31 (2016). [DOI] [PubMed] [Google Scholar]
- 6.Zhu S., et al. , The rice dwarf virus P2 protein interacts with ent-kaurene oxidases in vivo, leading to reduced biosynthesis of gibberellins and rice dwarf symptoms. Plant Physiol. 139, 1935–1945 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.He Y., et al. , Jasmonic acid-mediated defense suppresses brassinosteroid-mediated susceptibility to Rice black streaked dwarf virus infection in rice. New Phytol. 214, 388–399 (2017). [DOI] [PubMed] [Google Scholar]
- 8.Xie K., et al. , Abscisic acid negatively modulates plant defence against rice black-streaked dwarf virus infection by suppressing the jasmonate pathway and regulating reactive oxygen species levels in rice. Plant Cell Environ. 41, 2504–2514 (2018). [DOI] [PubMed] [Google Scholar]
- 9.Zhang H., et al. , The dual effect of the brassinosteroid pathway on rice black streaked dwarf virus infection by modulating the peroxidase-mediated oxidative burst and plant defense. Mol. Plant Microbe Interact. 32, 685–696 (2019). [DOI] [PubMed] [Google Scholar]
- 10.Teale W. D., Paponov I. A., Palme K., Auxin in action: Signalling, transport and the control of plant growth and development. Nat. Rev. Mol. Cell Biol. 7, 847–859 (2006). [DOI] [PubMed] [Google Scholar]
- 11.Weijers D., Wagner D., Transcriptional responses to the auxin hormone. Annu. Rev. Plant Biol. 67, 539–574 (2016). [DOI] [PubMed] [Google Scholar]
- 12.Korasick D. A., et al. , Molecular basis for AUXIN RESPONSE FACTOR protein interaction and the control of auxin response repression. Proc. Natl. Acad. Sci. U.S.A. 111, 5427–5432 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Guilfoyle T. J., Hagen G., Auxin response factors. Curr. Opin. Plant Biol. 10, 453–460 (2007). [DOI] [PubMed] [Google Scholar]
- 14.Gray W. M., Muskett P. R., Chuang H. W., Parker J. E., Arabidopsis SGT1b is required for SCF(TIR1)-mediated auxin response. Plant Cell 15, 1310–1319 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tatematsu K., et al. , MASSUGU2 encodes Aux/IAA19, an auxin-regulated protein that functions together with the transcriptional activator NPH4/ARF7 to regulate differential growth responses of hypocotyl and formation of lateral roots in Arabidopsis thaliana. Plant Cell 16, 379–393 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ulmasov T., Hagen G., Guilfoyle T. J., ARF1, a transcription factor that binds to auxin response elements. Science 276, 1865–1868 (1997). [DOI] [PubMed] [Google Scholar]
- 17.Shen C., et al. , Functional analysis of the structural domain of ARF proteins in rice (Oryza sativa L.). J. Exp. Bot. 61, 3971–3981 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sumimoto H., Kamakura S., Ito T., Structure and function of the PB1 domain, a protein interaction module conserved in animals, fungi, amoebas, and plants. Sci. STKE 2007, re6 (2007). [DOI] [PubMed] [Google Scholar]
- 19.Nanao M. H., et al. , Structural basis for oligomerization of auxin transcriptional regulators. Nat. Commun. 5, 3617 (2014). [DOI] [PubMed] [Google Scholar]
- 20.Salehin M., Bagchi R., Estelle M., SCFTIR1/AFB-based auxin perception: Mechanism and role in plant growth and development. Plant Cell 27, 9–19 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Boer D. R., et al. , Structural basis for DNA binding specificity by the auxin-dependent ARF transcription factors. Cell 156, 577–589 (2014). [DOI] [PubMed] [Google Scholar]
- 22.Tiwari S. B., Wang X.-J., Hagen G., Guilfoyle T. J., AUX/IAA proteins are active repressors, and their stability and activity are modulated by auxin. Plant Cell 13, 2809–2822 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Spaepen S., Vanderleyden J., Auxin and plant-microbe interactions. Cold Spring Harb. Perspect. Biol. 3, a001438 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Qi L., et al. , Arabidopsis thaliana plants differentially modulate auxin biosynthesis and transport during defense responses to the necrotrophic pathogen Alternaria brassicicola. New Phytol. 195, 872–882 (2012). [DOI] [PubMed] [Google Scholar]
- 25.Chen Z., et al. , Pseudomonas syringae type III effector AvrRpt2 alters Arabidopsis thaliana auxin physiology. Proc. Natl. Acad. Sci. U.S.A. 104, 20131–20136 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Padmanabhan M. S., Goregaoker S. P., Golem S., Shiferaw H., Culver J. N., Interaction of the tobacco mosaic virus replicase protein with the Aux/IAA protein PAP1/IAA26 is associated with disease development. J. Virol. 79, 2549–2558 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Collum T. D., Padmanabhan M. S., Hsieh Y. C., Culver J. N., Tobacco mosaic virus-directed reprogramming of auxin/indole acetic acid protein transcriptional responses enhances virus phloem loading. Proc. Natl. Acad. Sci. U.S.A. 113, E2740–E2749 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jin L., et al. , Rice dwarf virus P2 protein hijacks auxin signaling by directly targeting the rice OsIAA10 protein, enhancing viral infection and disease development. PLoS Pathog. 12, e1005847 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wei T., Li Y., Rice reoviruses in insect vectors. Annu. Rev. Phytopathol. 54, 99–120 (2016). [DOI] [PubMed] [Google Scholar]
- 30.Zhou G. H., et al. , Southern rice black-streaked dwarf virus: A new proposed Fijivirus species in the family Reoviridae. Chin. Sci. Bull. 53, 3677–3685 (2008). [Google Scholar]
- 31.Qiang W., et al. , The complete genome sequence of two isolates of southern rice black-streaked dwarf virus, a new member of the genus fijivirus. J. Phytopathol. 158, 733–737 (2010). [Google Scholar]
- 32.Qin F., et al. , Invasion of midgut epithelial cells by a persistently transmitted virus is mediated by sugar transporter 6 in its insect vector. PLoS Pathog. 14, e1007201 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Du Z., et al. , p2 of rice stripe virus (RSV) interacts with OsSGS3 and is a silencing suppressor. Mol. Plant Pathol. 12, 808–814 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lu G., et al. , Tenuivirus utilizes its glycoprotein as a helper component to overcome insect midgut barriers for its circulative and propagative transmission. PLoS Pathog. 15, e1007655 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yang X., et al. , Rice Stripe Mosaic Virus, a novel cytorhabdovirus infecting rice via leafhopper transmission. Front. Microbiol. 7, 2140 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhang H., et al. , Suppression of auxin signalling promotes rice susceptibility to Rice black streaked dwarf virus infection. Mol. Plant Pathol. 20, 1093–1104 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tiwari S. B., Hagen G., Guilfoyle T., The roles of auxin response factor domains in auxin-responsive transcription. Plant Cell 15, 533–543 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Dinesh D. C., et al. , Solution structure of the PsIAA4 oligomerization domain reveals interaction modes for transcription factors in early auxin response. Proc. Natl. Acad. Sci. U.S.A. 112, 6230–6235 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Liscum E., Reed J. W., Genetics of Aux/IAA and ARF action in plant growth and development. Plant Mol. Biol. 49, 387–400 (2002). [PubMed] [Google Scholar]
- 40.Kim J., Harter K., Theologis A., Protein-protein interactions among the Aux/IAA proteins. Proc. Natl. Acad. Sci. U.S.A. 94, 11786–11791 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ulmasov T., Hagen G., Guilfoyle T. J., Activation and repression of transcription by auxin-response factors. Proc. Natl. Acad. Sci. U.S.A. 96, 5844–5849 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Liu H., Wei C., Zhong Y., Li Y., Rice black-streaked dwarf virus minor core protein P8 is a nuclear dimeric protein and represses transcription in tobacco protoplasts. FEBS Lett. 581, 2534–2540 (2007). [DOI] [PubMed] [Google Scholar]
- 43.Qi Y., et al. , OsARF12, a transcription activator on auxin response gene, regulates root elongation and affects iron accumulation in rice (Oryza sativa). New Phytol. 193, 109–120 (2012). [DOI] [PubMed] [Google Scholar]
- 44.Ding Z., Friml J., Auxin regulates distal stem cell differentiation in Arabidopsis roots. Proc. Natl. Acad. Sci. U.S.A. 107, 12046–12051 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lu Y., et al. , Genome-wide targeted mutagenesis in rice using CRISPR/Cas9 system. Mol. Plants 10, 1242–1245 (2017). [DOI] [PubMed] [Google Scholar]
- 46.Liu Z. B., Ulmasov T., Shi X., Hagen G., Guilfoyle T. J., Soybean GH3 promoter contains multiple auxin-inducible elements. Plant Cell 6, 645–657 (1994). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Li Y., Liu Z. B., Shi X., Hagen G., Guilfoyle T. J., An auxin-inducible element in soybean SAUR promoters. Plant Physiol. 106, 37–43 (1994). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zhang S., et al. , The auxin response factor, OsARF19, controls rice leaf angles through positively regulating OsGH3-5 and OsBRI1. Plant Cell Environ. 38, 638–654 (2015). [DOI] [PubMed] [Google Scholar]
- 49.Kidd B. N., et al. , Auxin signaling and transport promote susceptibility to the root-infecting fungal pathogen Fusarium oxysporum in Arabidopsis. Mol. Plant Microbe Interact. 24, 733–748 (2011). [DOI] [PubMed] [Google Scholar]
- 50.Cui F., et al. , The Pseudomonas syringae type III effector AvrRpt2 promotes pathogen virulence via stimulating Arabidopsis auxin/indole acetic acid protein turnover. Plant Physiol. 162, 1018–1029 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Li S. B., Xie Z. Z., Hu C. G., Zhang J. Z., A review of auxin response factors (ARFs) in plants. Front Plant Sci. 7, 47 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Waller F., Furuya M., Nick P., OsARF1, an auxin response factor from rice, is auxin-regulated and classifies as a primary auxin responsive gene. Plant Mol. Biol. 50, 415–425 (2002). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All of the materials and data that were used or generated in this study are described and available in the manuscript and SI Appendix.