Abstract
The value of optical redox imaging (ORI) of cells/tissues based on the intrinsic fluorescences of NADH (nicotinamide adenine dinucleotide) and oxidized flavoproteins (containing flavin adenine dinucleotide, i.e., FAD) has been demonstrated for potential biomedical applications including diagnosis, prognosis, and determining treatment response. However, the Chance redox scanner (a 3D cryogenic tissue imager) is limited by spatial resolution (~50 μm), and tissue ORI using fluorescence microscopy (single or multi-photon) is limited by the light penetration depth. Furthermore, viable or snap-frozen tissues are usually required. In this project, we aimed to study whether ORI may be achieved for unstained fixed tissue using a state-of-the-art modern Serial Two-Photon (STP) Tomography scanner that can rapidly acquire multi-plane images at micron resolution. Tissue specimens of mouse muscle, liver, and tumor xenografts were harvested and fixed in 4% parafor-maldehyde (PFA) for 24 h. Tissue blocks were scanned by STP Tomography under room temperature to acquire the autofluorescence signals (NADH channel: excitation 750 nm, blue emission filter; FAD channel: excitation 860 nm, green emission filter). We observed remarkable signals with significant intra-tissue heterogeneity in images of NADH, FAD and redox ratio (FAD/(NADH+FAD)), which are worthy of further investigation for extracting biological information.
Keywords: Autofluorescence, NADH, FAD, Two photon imaging, Fixed tissue
1. Introduction
Optical redox imaging (ORI) or optical metabolic imaging (OMI) based on endogenous fluorescence signal sources including NADH and oxidized flavoproteins containing FAD have been increasingly employed for probing the metabolic/redox status of tissues and cells in order to understand disease pathology and develop tools for diagnosis and monitoring and predicting treatment response [1–6]. Optical redox ratio FAD/(NADH+FAD) is commonly used as an index sensitive to metabolic and redox status, and has been shown to correlate linearly with NAD+/(NAD++NADH) measured biochemically [7]. However, ORI is limited by the light penetration depth for tissue imaging with conventional fluorescence, confocal, or two-photon microscopes.
The 3D metabolic/redox indices (FAD, NADH, and redox ratio FAD/(NADH+FAD)) of deep tissues can be imaged by a cryogenic fluorescence imager (e.g. the Chance redox scanner [8, 9]) that is coupled to a cryostat or microtome with alternating tissue sectioning and imaging. Since NADH and FAD signals are labile with time in resected fresh tissue, tissue snap-freezing and imaging under cryogenic condition are used as necessary means to preserve or minimize the change of metabolism in tissue during sample harvesting, post-processing and imaging. Nevertheless, these procedures pose technical or operational challenges to applying ORI to clinical specimens.
In the clinic, paraffin embedded fixed tissue blocks and slides are widely available to provide pathophysiological evaluation. The long-term goal of this study is to evaluate whether ORI can be applicable to unstained fixed tissues so as to obtain metabolic/redox information that may potentially be clinically useful. We have reported some preliminary ORI results of formalin-fixed-paraffin-embedded unstained tissue slides using a conventional fluorescence microscope [10]. Here we present some ORI results for tissue blocks using two-photon microscopy.
2. Methods
Various fresh or frozen tissue blocks (size ~4 mm or less) including mouse organs and xenograft tumors were fixed in PFA (4%) for 24 h and then imaged on a STP Tomography scanner (TissueCyte® 1000, TissueVision Inc., Somerville, MA, USA) [11–13]. Specifically, the formalin-fixed tissues were embedded in a 4.5% oxidized agarose block and was subsequently placed in a solution of 0.5% sodium borohydride in a 0.5 M sodium borate buffer to covalently link the tissue to the agarose. Tissue blocks were sectioned by a microtome to expose a surface plane for imaging. 2D or 3D imaging was achieved with the TissueCyte® 1000 coupled to a titanium sapphire laser (Spectra Physics InSight DeepSee™, Santa Clara, CA, USA) operating at 750 nm and 860 nm. A Nikon 16× 0.8 NA objective (Nikon, Melville, NY, USA) was used and the sampling resolution was 1.2 μm in the XY plane. The emitted fluorescent light was separated into three spectral channels: red (≥560 nm), green (500–560 nm), and blue (≤500 nm). Images generated from 750 nm excitation and blue emission corresponded to the NADH channel, whereas images with 860 nm excitation and green emission corresponded to the FAD channel. These images were post-processed with a customized Matlab® program to generate pseudocolor images of NADH, FAD and redox ratio FAD/(FAD+NADH).
3. Results and Discussion
Figure 1 shows the white light photo and 2D ORI images of a typical mouse muscle sample obtained by STP imaging. The muscle fiber structures are clearly seen. The ORI images of NADH and FAD demonstrate significant signal heterogeneities within the whole muscle. This is even clearer in the blown-up images such as a field of interest shown in Fig. 1d–f, indicating fibers with either high or low redox indices (FAD, NADH and redox ratio). Similar phenomena are observed in the ORI images of three additional muscle samples.
Fig. 1.
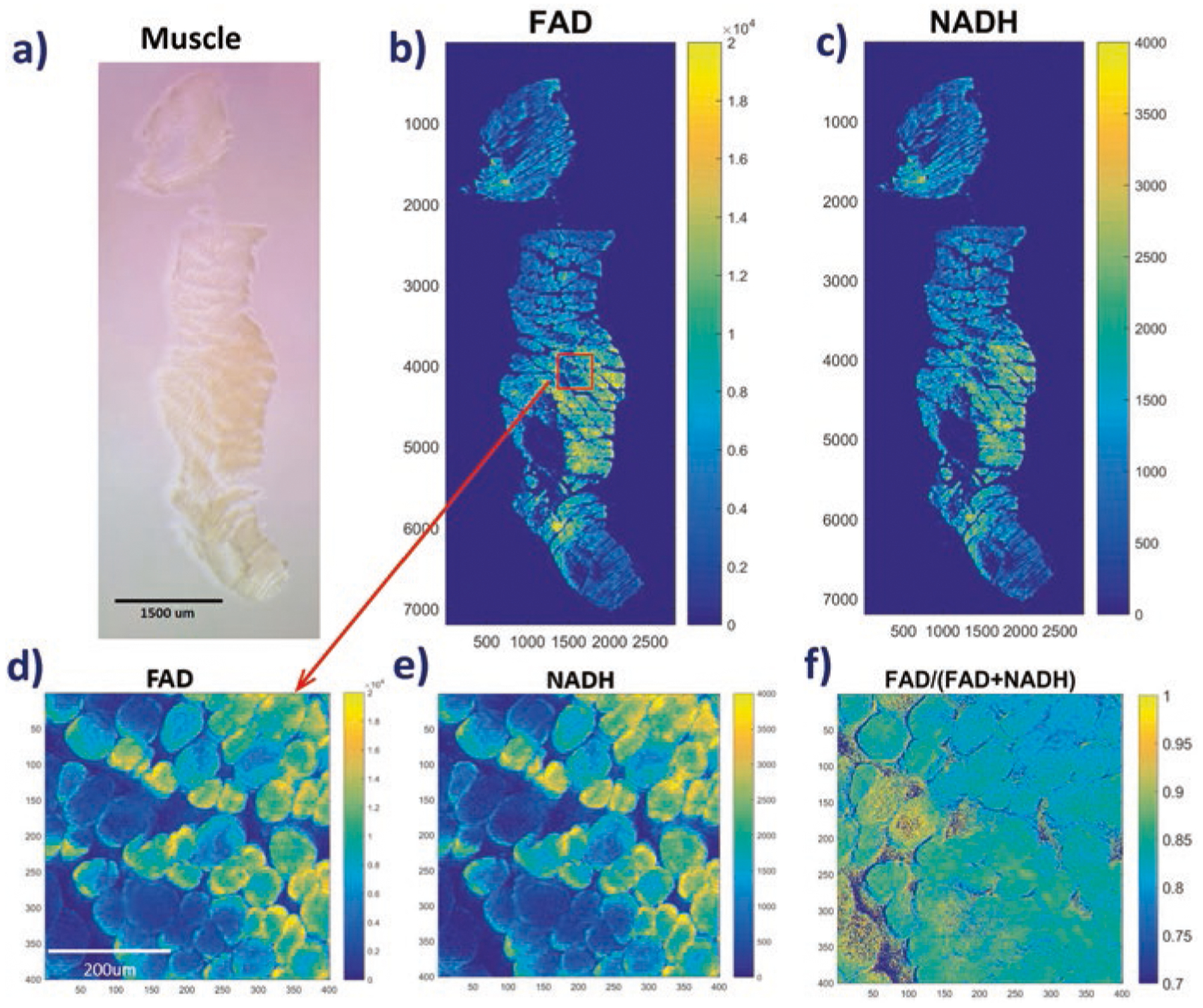
ORI of a typical fixed mouse muscle block. (a) White light photo of the sample; (b) FAD and (c) NADH images for the sample; (d–f) FAD, NADH and redox ratios images, respectively, for a FOV blown-up from (b). The numbers below and on the left sides of individual images represent x and y coordinates of the image matrices. The color bars on the right sides of images indicate the ranges of displayed parameters
It is known that muscle fibers exhibit two major metabolic subtypes, i.e., glycolytic and oxidative with different mitochondrial functional phenotypes [14]. Since redox indices correlate with the metabolic state and oxygen consumption rate [1, 7, 15], it would be interesting to investigate whether and how the redox subtypes of the fixed muscle relate to the known metabolic subtypes.
Figure 2 shows the white light photo and ORI images of a liver sample from a mouse of 19 weeks old. In addition to the general intra-tissue heterogeneity across the whole sample, a new but very interesting phenomenon was observed as shown in the blown-up images of FOVs (Fig. 2 d–f). Both the FAD and NADH images exhibit scattered patterns indicating localized areas of high signals surrounded by areas of low signals. The size of these localized high-signal areas range from ~5–20 microns in NADH images and relatively less in FAD images. The distribution of these high-signal areas in FAD and NADH channels appears to be highly correlated but not fully matched in locations. This can be shown more clearly by the redox ratio image in Fig. 2e, in which a number of blue areas correlate spatially with the high intensity areas in FAD and NADH images. For such blue areas in the redox ratio image, we observed the dark blue regions indicating low FAD and high NADH and the light blue regions indicating high FAD and high NADH. Thus, there is a spatial mismatch between the high signal areas of FAD and NADH images.
Fig. 2.
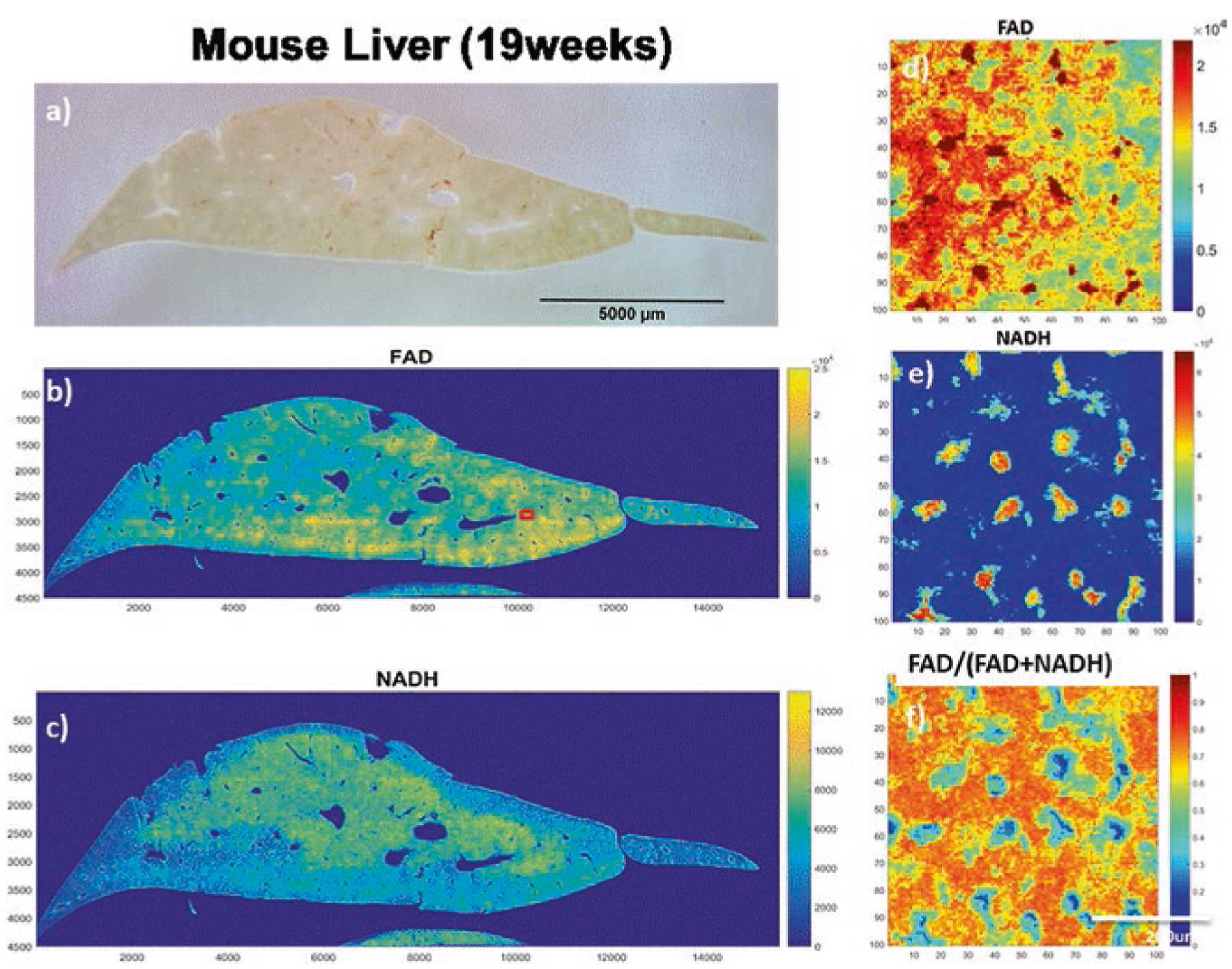
Optical redox images of a fixed mouse liver sample from the two-photon scanner. (a) White light photo of the liver sample (19 weeks); (b, c) FAD and NADH images, respectively; (d–f) Blown-up FAD, NADH and redox ratios images, respectively, for an area of interest (100 × 100 pixel) marked as a red square in (b)
The ORI images from a mouse xenograft of human breast cancer MDA-MB-231 cell line are shown in Fig. 3. Again we can see significant intratumor heterogeneity for all three indices, i.e., FAD, NADH and redox ratio.
Fig. 3.
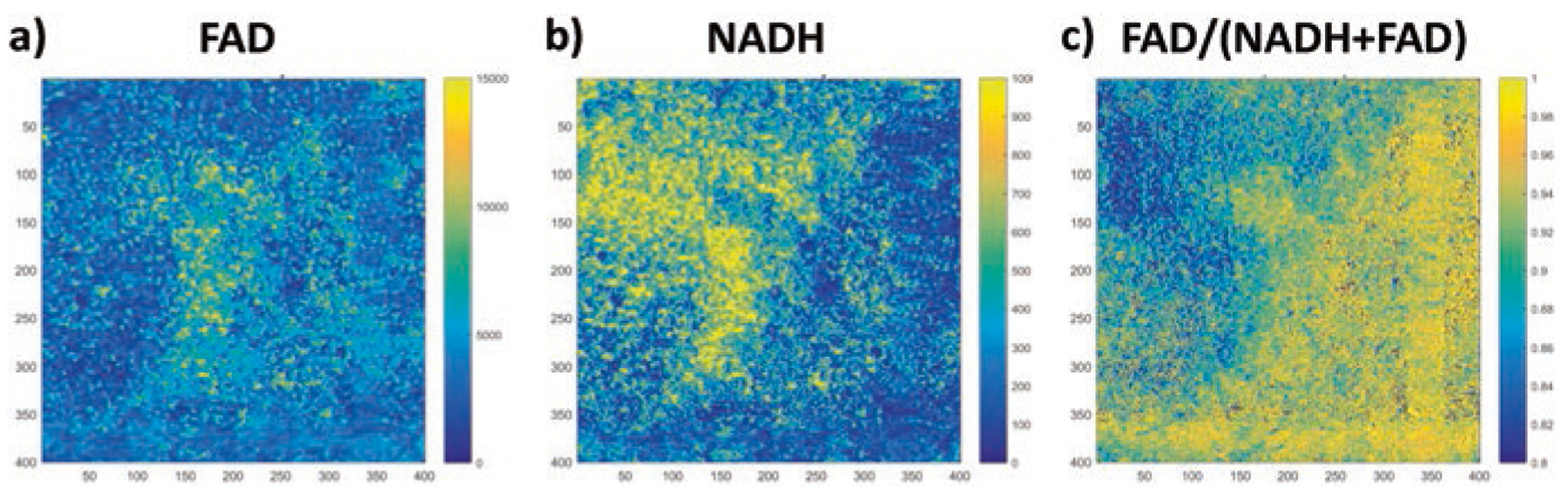
FAD, NADH, and redox ratio images of a region of interest in a fixed mouse xenograft of human breast cancer MDA-MB-231 cells
In summary, we have obtained high resolution autofluorescence images of fixed unstained tissue blocks and observed significant intra-tissue signal variations. It is known that mitochondria in cytosol account for the most of autofluorescence signals in the NADH and FAD channels [16], whereas nuclei should be relatively dark. The intratissue spatial heterogeneity of NADH and FAD signals have been reported in normal and cancer tissues imaged by cryogenic redox scanner [16, 17]. However, the interpretation of these spatial heterogeneity remains open. Other biological sources may contribute to these autofluorescence signals including NADPH, keratin, elastin, collagen, and lipofuscin [7, 18, 19]. More sophisticated imaging and data analysis approaches can be used to resolve specifically the NADH and FAD signals and exclude contributions from other sources [7, 18, 20]. Co-registration of autofluorescence imaging of tissue slides with H&E staining may also help to understand the histological basis of autofluorescence [21].
Moreover, tissue NADH and FAD signals are expected to change due to the deprivation of oxygen during the period after death and before fixation. Fixation may further change the tissue micro- and ultra-structure, and autofluorescence intensity may depend on the tissue penetration speed and depth of fixatives. More studies are needed to understand the effects of tissue processing and PFA fixation on tissue autofluorescence [10], and to investigate whether these signals from fixed tissues may contain any biological or pathological information that could be useful for the clinic.
Acknowledgments
We would like to thank Ms. Lily Moon, Ms. Min Feng and Dr. Karthikeyani Chellappa for preparation and handling of tissue samples. Note that Tim Ragan is an employee and shareholder of TissueVision Inc and Marissa Masek was an employee of TissueVision. TissueVision manufactures and sells the TissueCyte system on which some of the data taken in the paper was taken. We appreciate the support from NIH R01 CA191207 (Li LZ) and a pilot grant (Li LZ and Baur J) from the Institute On Aging of the University of Pennsylvania
Contributor Information
Lin Z. Li, Department of Radiology & Britton Chance Laboratory of Redox Imaging, Johnson Research Foundation, Perelman School of Medicine, University of Pennsylvania, Philadelphia, PA, USA; Abramson Cancer Center, University of Pennsylvania, Philadelphia, PA, USA; Institute of Translational Medicine and Therapeutics, Perelman School of Medicine, University of Pennsylvania, Philadelphia, PA, USA.
Marissa Masek, TissueVision, Inc, Somerville, MA, USA.
Ting Wang, Department of Radiology & Britton Chance Laboratory of Redox Imaging, Johnson Research Foundation, Perelman School of Medicine, University of Pennsylvania, Philadelphia, PA, USA.
He N. Xu, Department of Radiology & Britton Chance Laboratory of Redox Imaging, Johnson Research Foundation, Perelman School of Medicine, University of Pennsylvania, Philadelphia, PA, USA
Shoko Nioka, Department of Radiology & Britton Chance Laboratory of Redox Imaging, Johnson Research Foundation, Perelman School of Medicine, University of Pennsylvania, Philadelphia, PA, USA.
Joseph A. Baur, Department of Physiology, Institute for Diabetes, Obesity and Metabolism, University of Pennsylvania, Philadelphia, PA, USA
Timothy M. Ragan, TissueVision, Inc, Somerville, MA, USA.
References
- 1.Chance B, Schoener B, Oshino R et al. (1979) Oxidation-reduction ratio studies of mitochondria in freeze-trapped samples. NADH and flavoprotein fluorescence signals. Journal of Biological Chemistry 254:4764–4771 [PubMed] [Google Scholar]
- 2.Li LZ, Zhou R, Xu HN et al. (2009) Quantitative magnetic resonance and optical imaging biomarkers of melanoma metastatic potential. Proc Natl Acad Sci U S A 106:6608–6613 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Li LZ, Xu HN, Ranji M et al. (2009) Mitochondrial redox imaging for cancer diagnostic and therapeutic studies. J Innov Opt Health Sci 2:325–341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Georgakoudi I, Quinn KP (2012) Optical imaging using endogenous contrast to assess metabolic state. Annu Rev Biomed Eng 14:351–367 [DOI] [PubMed] [Google Scholar]
- 5.Walsh AJ, Cook RS, Sanders ME et al. (2014) Quantitative optical imaging of primary tumor organoid metabolism predicts drug response in breast cancer. Cancer Res 74:5184–5194 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Xu HN, Tchou J, Feng M et al. (2016) Optical redox imaging indices discriminate human breast cancer from normal tissues. J Biomed Opt 21:114003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Varone A, Xylas J, Quinn KP et al. (2014) Endogenous two-photon fluorescence imaging elucidates metabolic changes related to enhanced glycolysis and glutamine consumption in precancerous epithelial tissues. Cancer Res 74:3067–3075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Quistorff B, Haselgrove JC, Chance B (1985) High spatial resolution readout of 3-D metabolic organ structure: an automated, low-temperature redox ratio-scanning instrument. Anal Biochem 148:389–400 [DOI] [PubMed] [Google Scholar]
- 9.Gu Y, Qian Z, Chen J et al. (2002) High-resolution three-dimensional scanning optical image system for intrinsic and extrinsic contrast agents in tissue. Rev Sci Instrum 73:172–178 [Google Scholar]
- 10.Xu HN, Tchou J, Li Y et al. (2018) Optical redox imaging of fixed unstained tissue slides to identify biomarkers for breast cancer diagnosis/prognosis: feasibility study. Proc SPIE 10472:104720U1-6 [Google Scholar]
- 11.Ragan T, Kadiri LR, Venkataraju KU et al. (2012) Serial two-photon tomography for automated ex vivo mouse brain imaging. Nat Methods 9:255–258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Amato SP, Pan F, Schwartz J et al. (2016) Whole brain imaging with serial two-photon tomography. Front Neuroanat 10:31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ragan T, Sylvan JD, Kim KH et al. (2007) High-resolution whole organ imaging using two-photon tissue cytometry. J Biomed Opt 12:014015. [DOI] [PubMed] [Google Scholar]
- 14.Picard M, Hepple RT, Burelle Y (2012) Mitochondrial functional specialization in glycolytic and oxidative muscle fibers: tailoring the organelle for optimal function. Am J Physiol Cell Physiol 302:26. [DOI] [PubMed] [Google Scholar]
- 15.Hou J, Wright HJ, Chan N et al. (2016) Correlating two-photon excited fluorescence imaging of breast cancer cellular redox state with seahorse flux analysis of normalized cellular oxygen consumption. J Biomed Opt 21:060503–060503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Xu HN, Li LZ (2014) Quantitative redox imaging biomarkers for studying tissue metabolic state and its heterogeneity. Journal of Innovative Optical Health Sciences 7:1430002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Xu HN, Zhou R, Moon L et al. (2014) 3D imaging of the mitochondrial redox state of rat hearts under normal and fasting conditions. Journal of Innovative Optical Health Sciences 7:1350045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Xu Z, Reilley M, Li R et al. (2017) Mapping absolute tissue endogenous fluorophore concentrations with chemometric wide-field fluorescence microscopy. J Biomed Opt 22:66009. [DOI] [PubMed] [Google Scholar]
- 19.Alzbeta C, Dusan C (2014) Tissue fluorophores and their spectroscopic characteristics In: Fluorescence lifetime spectroscopy and imaging. CRC Press, pp 47–84 [Google Scholar]
- 20.Blacker TS, Duchen MR (2016) Investigating mitochondrial redox state using NADH and NADPH autofluorescence. Free Radic Biol Med 100:53–65 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Xu HN, Zhou R, Nioka S et al. (2009) Histological basis of MR/optical imaging of human melanoma mouse xenografts spanning a range of metastatic potentials. Adv Exp Med Biol 645:247–253 [DOI] [PMC free article] [PubMed] [Google Scholar]


