Significance
Nonribosomal peptides (NRPs) are a vast class of natural products and an important source of therapeutics. Typically, these secondary metabolites are assembled by NRP synthetases (NRPSs) that function on substrates covalently linked to the enzyme by a thioester, in a process known as thiotemplated biosynthesis. Although NRPS-independent assembly pathways are known, all are nonthiotemplated. Here we report an NRPS-independent yet thiotemplated pathway for NRP biosynthesis and demonstrate that members of the ATP-grasp and cysteine protease families form the β-peptide backbone of an antibiotic. Armed with this knowledge, we provide genomic evidence that this noncanonical assembly pathway is widespread in bacteria. Our results will inspire future genome mining efforts for the discovery of potential therapeutics that otherwise would be overlooked.
Keywords: biosynthesis, natural products, nonribosomal peptide, thiotemplated assembly
Abstract
Closthioamide (CTA) is a rare example of a thioamide-containing nonribosomal peptide and is one of only a handful of secondary metabolites described from obligately anaerobic bacteria. Although the biosynthetic gene cluster responsible for CTA production and the thioamide synthetase that catalyzes sulfur incorporation were recently discovered, the logic for peptide backbone assembly has remained a mystery. Here, through the use of in vitro biochemical assays, we demonstrate that the amide backbone of CTA is assembled in an unusual thiotemplated pathway involving the cooperation of a transacylating member of the papain-like cysteine protease family and an iteratively acting ATP-grasp protein. Using the ATP-grasp protein as a bioinformatic handle, we identified hundreds of such thiotemplated yet nonribosomal peptide synthetase (NRPS)-independent biosynthetic gene clusters across diverse bacterial phyla. The data presented herein not only clarify the pathway for the biosynthesis of CTA, but also provide a foundation for the discovery of additional secondary metabolites produced by noncanonical biosynthetic pathways.
Nonribosomal peptides (NRPs) compose a structurally diverse class of secondary metabolites displaying myriad biological activities. Despite the large chemical space occupied by NRPs, the vast majority share a common biosynthetic origin. Typically, such compounds are biosynthesized by multimodular, thiotemplated NRP synthetases (NRPSs) (1). However, over the last decade, it has become apparent that NRPs may also be assembled by NRPS-independent pathways. Two prominent examples are the amide synthases involved in siderophore (2) and diketopiperazine (3) biosyntheses. In both pathways, the peptide ligases function on nonthiotemplated substrates, but the pathways differ in their strategy for carboxylate activation. The siderophore amide synthetases use ATP to activate the carboxylic acids of their substrates as adenylates (2), while the diketopiperazine amide synthases circumvent the requirement for ATP by using preactivated amino acids in the form of acyl-tRNAs (3). Apart from these widespread NRPS-independent pathways, a handful of additional nonthiotemplated peptide ligases have also been identified (4).
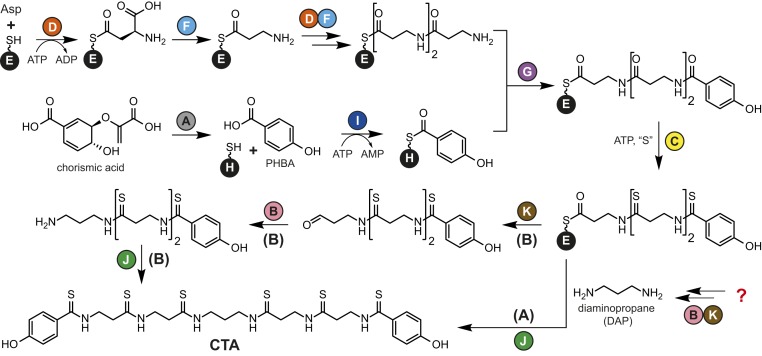
With the increasing number of characterized NRP biosynthetic pathways, a pattern became apparent: thiotemplated pathways are NRPS-dependent, while those that use free-standing peptide ligases are nonthiotemplated. However, a notable exception came with the recent discovery of a thiotemplated yet NRPS-independent pathway for the biosynthesis of closthioamide (CTA; Fig. 1 A and B) (5, 6).
Fig. 1.
CTA is biosynthesized by an NRPS-independent thiotemplated pathway. (A) Structure of CTA. (B) CTA biosynthetic gene cluster. (C) Proposed biosynthetic pathway for CTA maturation. Proteins with proven functions are in red.
CTA is a DNA gyrase-targeting antibiotic with potent inhibitory activity against clinically important bacterial pathogens, including drug-resistant strains (7–10), and is a rare example of a thioamidated NRP (11, 12). This unusual copper chelating NRP (13) is produced by Ruminiclostridium cellulolyticum DSM 5812 (7, 14) and is one of only a handful of natural products isolated from anaerobic bacteria (15–17). Knockout studies and in vitro biochemical assays led to the identification of the enzymes responsible for sulfur insertion and late-stage intermediate dimerization (Fig. 1C) (5, 6). The biosynthetic strategy for assembly of the amide backbone of CTA has remained a mystery, however.
Here we used a combination of bioinformatics and in vitro biochemical assays to unravel the atypical amide backbone biosynthetic pathway in CTA maturation. We demonstrate that amide formation is thiotemplated and is catalyzed by novel members of the ATP-grasp and cysteine protease protein families. Using the ATP-grasp protein as a bioinformatic handle, we provide evidence that similar thiotemplated, NRPS-independent pathways are widespread in Actinobacteria and Firmicutes. In addition to deciphering the early steps in CTA maturation, the data presented herein provide a paradigm for NRPS-independent amide assembly and should inform future genome mining efforts for such noncanonical thiotemplate pathways.
Results and Discussion
Identification of the CtaH Loading Enzyme as an Entry Point to Backbone Assembly.
Setting a starting point to elucidate the backbone biosynthetic pathway proved very challenging. Based on knockout studies and reconstitution of the CTA thioamide synthetase, we knew that the pathway was thiotemplated and that sulfur incorporation occurs on p-hydroxybenzoic acid (PHBA) end-capped β-alanine (βAla) polymers attached to the peptidyl carrier protein (PCP) CtaE (5, 6). However, as all of the biosynthetic intermediates identified to date from both wild-type and knockout strains contain thioamides (5–7, 14), the origin of this PHBA-βAla conjugate was a mystery. Further complicating matters, the biosynthetic enzymes expected to be responsible for forming this CtaE-linked intermediate—CtaD, CtaG, and CtaI—belong to protein families more commonly associated with primary metabolism than with NRP assembly, making the a priori prediction of their substrates difficult.
The CTA biosynthetic locus also encodes a second PCP (Fig. 1B), CtaH, with no proven role in CTA assembly (5, 6). Given the conservation of this second PCP in similar biosynthetic gene clusters, we predicted that CtaH would be critical for CTA maturation. Taking this into account, we envisioned a biosynthetic pathway in which CtaH and CtaE would serve as carrier proteins for PHBA and the poly-β-Ala backbone of CTA, respectively (Fig. 1C). Subsequently, CtaE would receive PHBA from CtaH to afford the PHBA end-capped conjugate. Therefore, we selected CtaH as our entry point for studying CTA backbone formation.
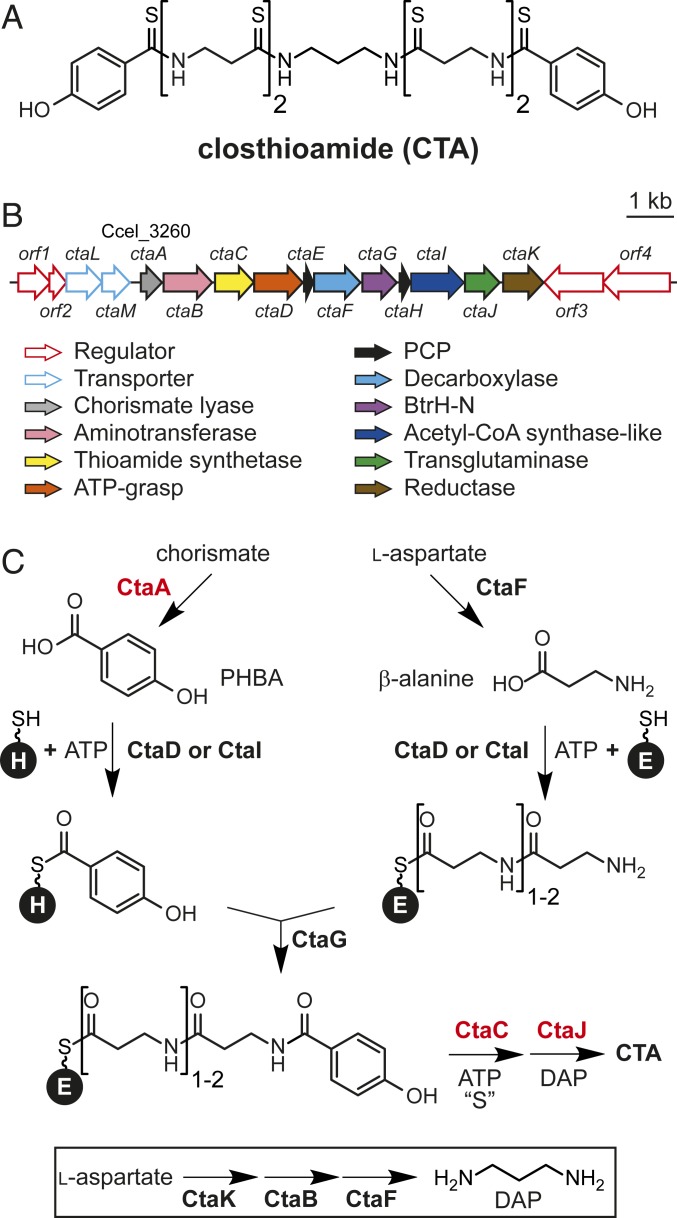
PCP-based biosynthetic pathways begin with the ATP-dependent attachment of a precursor or intermediate to the 4′-phosphopantetheine arm of the carrier protein (1). Among the proteins encoded in the CTA gene cluster are members of the ATP-grasp (CtaD) and the acetyl-CoA synthetase-like (CtaI) protein families (Fig. 1B). Characterized members of both protein families catalyze the ATP-dependent coupling of carboxylic acids to diverse nucleophiles (18–20), making them likely candidates for the PCP-loading enzymes in CTA biosynthesis. Therefore, both proteins were isolated as N-terminal His-tagged proteins from Escherichia coli (SI Appendix, Fig. S1) and tested for holo-CtaH loading activity. Reactions were performed with PHBA, and progress was monitored by matrix-assisted laser desorption/ionization-time-of-flight-mass spectrometry (MALDI-TOF-MS). Whereas reactions performed with CtaD did not show any processing of holo-CtaH, a new signal with a mass shift consistent with PHBA loading (+120 Da; PHBA-S-CtaH) was observed in CtaI reactions (Fig. 2A). Reactions were also performed with β-alanine (βAla) and with holo-CtaE. In neither instance was there any processing of the corresponding PCP, demonstrating that CtaI is specific for holo-CtaH and PHBA (SI Appendix, Fig. S2).
Fig. 2.
CtaI loads PHBA onto holo-CtaH. (A) MALDI-TOF spectral overlay of holo-CtaH loading reactions performed with PHBA and either CtaI or CtaD. The expected average masses of holo-CtaH and PHBA-S-CtaH are indicated. (B) HPLC absorbance profiles (250 nm) of reactions performed with NAC, PHBA, and CtaI. (C) Quantification of ATP hydrolysis products using a malachite green assay. Error bars represent the SD from the mean (n = 3). PPase, inorganic pyrophosphatase. (D) HPLC profiles of CtaI reactions with NAC. Red strikethrough indicates heat-inactivated enzyme; red asterisk, NAC-specific peak.
Characterized members of the acetyl-CoA synthetase-like protein superfamily activate substrates by adenylation and thereby produce AMP and pyrophosphate (PPi) as byproducts of the reaction (19). Consistent with the predicted ATP-dependence of CtaI, no processing of holo-CtaH was observed in reactions lacking ATP (Fig. 2A). However, attempts to ascertain the ATP hydrolysis products of the reaction were stymied owing to the relatively poor solubility of holo-CtaH. To overcome this challenge, CtaI was tested for the ability to accept N-acetylcysteamine (NAC) as a surrogate for holo-CtaH. Liquid chromatography high-resolution mass spectrometry (LC-HR-MS) profiles of the reactions showed that this 4′-phosphopantetheine mimic was indeed processed by CtaI to form the PHBA-NAC conjugate (Fig. 2B and SI Appendix, Fig. S3). The ATP hydrolysis products formed during processing were monitored by both a malachite green assay (21) (phosphate and PPi detection) and high-performance liquid chromatography (HPLC; AMP and ADP detection). Consistent with the characterized members of the acetyl-CoA synthetase family, reactions containing CtaI, PHBA, and NAC produced both PPi and AMP (Fig. 2 C and D), suggesting that PHBA transfer proceeds through an adenylated intermediate.
Reconstitution of CtaD Activity.
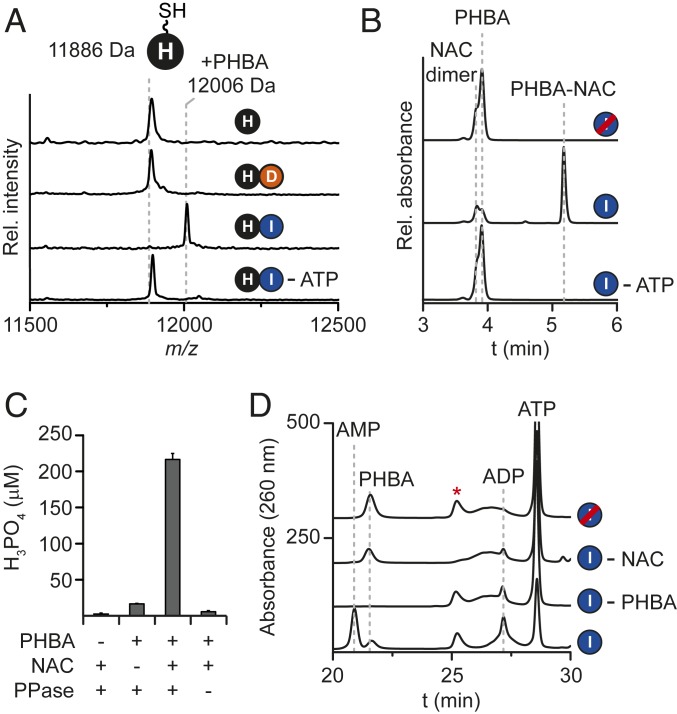
Based on the structure of CTA, we reasoned that holo-CtaE would be iteratively loaded with βAla to form the backbone of CTA. With the function of CtaI assigned, CtaD—which was previously shown to be necessary for CTA maturation (5)—was the logical candidate to catalyze this reaction. We performed holo-CtaE loading reactions with CtaD, βAla, and ATP and monitored reaction progress by MALDI-TOF-MS. No modification of holo-CtaE was observed irrespective of the concentration of CtaD or the reaction time (Fig. 3A), suggesting that this biosynthetic proposal was incorrect.
Fig. 3.
CtaD and CtaF work together to polymerize β-alanine onto holo-CtaE. (A) MALDI-TOF-MS spectral overlay of holo-CtaE loading reactions. The expected average masses of holo-CtaE, Asp-S-CtaE, and 3βAla-S-CtaE are indicated. (B) LC-HR-MS profiles of NAC loading assays. Traces correspond to the extracted ion chromatography data of the [M+H]+ ionic species for Asp-NAC and are displayed with m/z values ±5 ppm from the calculated exact mass. (C) Quantification of ATP hydrolysis products using a malachite green assay. Error bars represent the SD from the mean (n = 3). (D) HPLC profiles of CtaD reactions with NAC. (E) MALDI-TOF-MS spectral overlay of a CtaD-F reaction time course. The expected masses of holo-CtaE, 3βAla-S-CtaE, and all predicted intermediates are indicated. Red strikethrough indicates heat-inactivated enzyme; red asterisk, NAC-specific peak.
To help guide the development of a new hypothesis for the function of CtaD, we performed a bioinformatic search for characterized biosynthetic gene clusters encoding CtaD and CtaE homologs. Among the resulting hits, the genetic locus responsible for butirosin biosynthesis (22, 23) stood out as it also encodes a homolog of the putative pyridoxal phosphate (PLP)-dependent decarboxylase CtaF. Butirosin is an unusual amino acid-functionalized aminoglycoside antibiotic produced by Bacillus circulans (24). During butirosin maturation, an ATP-grasp protein loads l-glutamate onto a PCP and the conjugate is subsequently decarboxylated to 4-aminobutyrate by a PLP-dependent decarboxylase (25). The decarboxylated product is further modified by the ATP-grasp protein to afford a PCP-linked 4-aminobutyrate-glutamate dipeptide.
Inspired by the butirosin system, we repeated the CtaD-CtaE loading assays with l-aspartate (Asp) rather than βAla and monitored reaction progress by MALDI-TOF-MS. Under these conditions, holo-CtaE was partially converted to a new species ∼115 Da heavier than the unmodified PCP (Fig. 3A), indicating successful Asp loading. Attempts to improve holo-CtaE loading by increasing the concentration of CtaD and extending the reaction time were unsuccessful (SI Appendix, Fig. S4). In fact, we noted that the Asp-holo-CtaE (Asp-S-CtaE) was hydrolyzed to holo-CtaE with longer reaction times (SI Appendix, Fig. S4). Importantly, control experiments performed with holo-CtaH and in the absence of ATP established that Asp-loading was both specific to holo-CtaE and dependent on ATP (Fig. 3A and SI Appendix, Fig. S5).
Characterized members of the ATP-grasp protein superfamily activate substrates by phosphorylation and thus produce ADP and phosphate (Pi) as byproducts of the reaction (18). To circumvent issues with CtaE solubility, we checked CtaD for the ability to accept NAC as a holo-CtaE mimic. Indeed, LC-HR-MS profiles of NAC supplemented reactions showed that CtaD can produce Asp-NAC (Fig. 3B and SI Appendix, Fig. S6). Next, we monitored the ATP-hydrolysis products in the NAC reactions by both a malachite green assay and HPLC. Consistent with the characterized members of the ATP-grasp protein family, ADP and Pi were detected in reactions containing all components (Fig. 3 C and D), suggesting that Asp transfer proceeds through a phosphorylated intermediate.
CtaD Works with the Decarboxylase CtaF to Generate the Amide Backbone of CTA.
The CTA biosynthetic gene cluster encodes a putative member of the ornithine/diaminopimelate/arginine PLP-dependent decarboxylase family, CtaF (Fig. 1B), which we anticipated would catalyze the conversion of l-aspartate to βAla (Fig. 1C) (5, 6). Although we originally predicted that CtaF would function on free acids, the selectivity of CtaD for Asp suggested that the decarboxylation activity would be thiotemplated.
To test this proposal, we isolated CtaF as an N-terminal His-tagged protein from E. coli (SI Appendix, Fig. S1) and repeated holo-CtaE loading assays with CtaF supplementation. Although CtaF solutions were slightly yellow and had the characteristic UV-visible absorption spectrum of PLP cofactor-containing enzymes (SI Appendix, Fig. S7), PLP was also added to reaction mixtures to ensure maximal activity from the putative decarboxylase. While control reactions containing only CtaF and Asp did not show any processing of holo-CtaE, a new peak with a mass increase of 213 Da relative to holo-CtaE was observed in CtaD-containing reactions (Fig. 3A). This mass is consistent with the attachment of three βAla (3βAla) residues to holo-CtaE (3βAla-S-CtaE). To confirm the identity of the product, the product was cleaved from the PCP under basic conditions, derivatized with o-phthaldialdehyde (OPA) (26) and analyzed by LC-HR-MS. By comparison with a synthetic reference (SI Appendix, Fig. S8) and by MS/MS fragmentation (SI Appendix, Fig. S9), we confirmed that the released product was 3βAla. This suggests that CtaF not only decarboxylates Asp-S-CtaE, but also that the resulting product is further processed by CtaD and CtaF.
In analogy to the biosynthesis of the amino acid side chain of butirosin (25), we expected that 3βAla-S-CtaE was formed by the sequential and iterative action of CtaD and CtaF on holo-CtaE. However, this CtaE conjugate could also be formed by the transfer of a fully polymerized βAla chain to holo-CtaE. In this case, no biosynthetic intermediates would be expected to be formed on CtaE. To differentiate between these two possibilities, we performed a reaction time course and monitored the production of holo-CtaE conjugates by MALDI-TOF-MS. At the earliest time point, four new peaks were observed with mass shifts consistent with the additions of Asp (+115 Da), βAla (+71 Da), Asp-βAla (+186 Da), and di-βAla (2βAla; +142 Da) (Fig. 3E). At longer reaction times, the intensity of these peaks decreased, and new peaks emerged with mass shifts consistent with the additions of Asp-2βAla (+257 Da) and 3βAla (+213 Da) (Fig. 3E). Together with the observation that free l-aspartate is not a substrate for CtaF (SI Appendix, Fig. S10), these data demonstrate that formation of the β-alanine backbone of CTA is thiotemplated and catalyzed by the cooperative action of an ATP-grasp protein and a decarboxylase.
While ATP-grasp proteins are commonly known as peptide ligases in both primary and secondary metabolism, to the best of our knowledge, the butirosin pathway is the only other example in which the activity of an ATP-grasp protein is thiotemplated (4, 20, 25). However, in butirosin maturation, the ATP-grasp protein is involved in the biosynthesis of the amino acid side chain of this unusual aminoglycoside (25) rather than in the biosynthesis of an NRP backbone.
CtaG Is a Transacylase Responsible for PHBA-3βAla Synthesis.
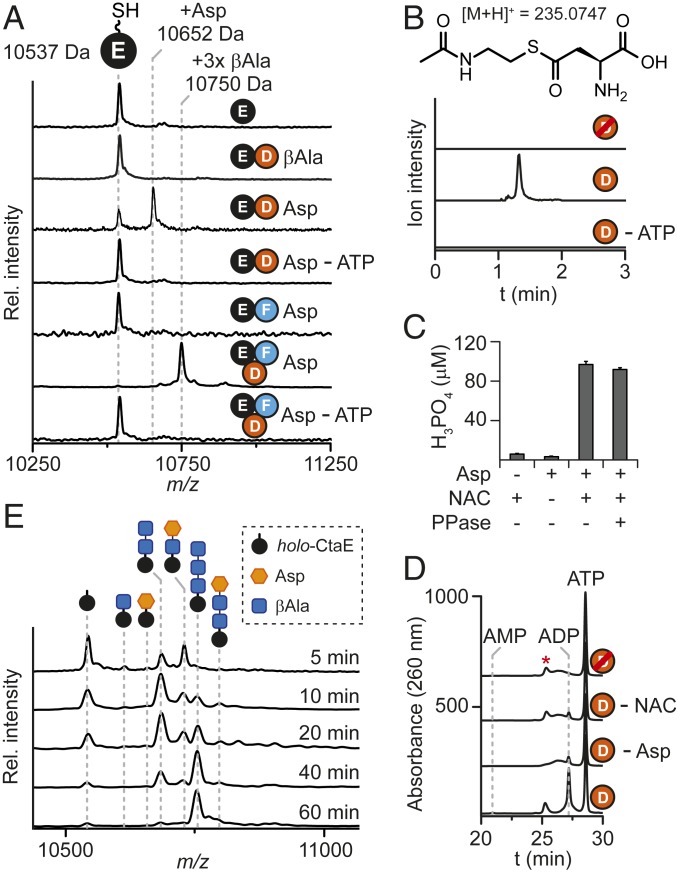
With the PCP loading enzymes and pathway to β-alanine backbone formation uncovered, we focused on the identification and characterization of the enzyme responsible for transferring PHBA from PHBA-S-CtaH to 3βAla-S-CtaE. Of the enzymes encoded in the CTA gene cluster, CtaG was the likely candidate to catalyze this reaction. Although CtaG is annotated as a hypothetical protein, bioinformatic analysis using HHpred (27, 28) identified it as a member of the C39 protease (PF11814) and BtrH (PF14399) protein families (5). While the former is composed of proteases involved in ribosomally synthesized and posttranslationally modified peptide biosynthesis (29), the latter is a PCP-dependent amide bond-forming transacylase involved in butirosin maturation (30). Despite this difference in activity, members of both protein families share a papain-like protease fold and a conserved active-site Cys-His-Asp/Asn catalytic triad and fall within the CA clan of proteases (Pfam clan CL0125) (31, 32). Notably, this protease superfamily also includes other transacylating enzymes (33, 34), including a thiotemplated peptide synthase involved in the biosynthesis of the acylated NRP andrimid (35).
Owing to solubility issues with His6-tagged CtaG, we isolated CtaG from E. coli as a maltose binding protein (MBP)-tagged fusion protein (SI Appendix, Fig. S1). We performed reactions with CtaD, CtaF, CtaI, holo-CtaE/CtaH, and MBP-CtaG and monitored reaction progress by MALDI-TOF-MS. A new peak with a mass consistent with a PHBA-3βAla-holo-CtaE conjugate (PHBA-3βAla-S-CtaE) was formed in reactions containing MBP-CtaG but was absent in reactions performed with heat-denatured MBP-CtaG (Fig. 4A). Moreover, this species was not formed in reactions lacking holo-CtaH and CtaI, demonstrating that CtaG activity is dependent on the presence of PHBA-S-CtaH (Fig. 4A). The product was cleaved from the PCP under basic conditions and analyzed by LC-HR-MS to confirm its identity. By comparison to a synthetic reference (Fig. 4B) and by MS/MS fragmentation (SI Appendix, Fig. S11), we verified that the released product was PHBA-3βAla.
Fig. 4.
CtaG is a novel transacylase. (A) MALDI-TOF-MS spectral overlay of CtaD–I reactions. The expected average masses of 3βAla-S-CtaE and PHBA-3βAla-S-CtaE are indicated. (B) LC-HR-MS profiles of CtaD–I reactions following thioester cleavage. Traces correspond to the extracted ion chromatogram of the [M-H]− ionic species for PHBA-3βAla and are displayed with m/z values ±5 ppm from the calculated exact mass. (C) MALDI-TOF-MS spectral overlay of CtaG reactions performed with 2βAla-S-CtaE. The expected average masses of 2βAla-S-CtaE and PHBA-2βAla-S-CtaE are indicated. (D) MALDI-TOF-MS spectral overlay of CtaG reactions performed with 3βAla-S-CtaE. The expected average masses of 3βAla-S-CtaE and PHBA-3βAla-S-CtaE are indicated. (E and F) MALDI-TOF-MS spectral overlays of CtaG (E) and CtaGC11A (F) reactions performed with holo-CtaH and CtaI but without CtaD-F. The expected [M+2H]2+ for MBP-CtaG/MBP-CtaGC11A and the corresponding PHBA conjugate are displayed. Red strikethrough indicates heat-inactivated enzyme; red asterisk, apo-CtaE. Con., control.
With the activity of CtaG assigned, we next turned our attention to determining its substrate specificity. The inability of free PHBA to substitute for PHBA-S-CtaH in CtaG reactions indicates that CtaG uses this acyl-PCP as the PHBA source for the transacylation reaction. To test this hypothesis, we performed reactions with increased concentrations of holo-CtaH and monitored the modification state of holo-CtaH over the course of the reaction by MALDI-TOF-MS. Indeed, on the addition of MBP-CtaG, the peak corresponding to PHBA-S-CtaH disappeared, and a new peak with a mass consistent with holo-CtaH appeared (SI Appendix, Fig. S12).
The thioamide synthetase CtaC can accept both PHBA-3βAla-S-CtaE and PHBA-2βAla-S-CtaE as substrates (6), but since the preceding and ensuing steps in CTA maturation remained unclear, the biosynthetically relevant substrate could not be definitively determined. Given the promiscuity of CtaC, we speculated that an upstream biosynthetic enzyme likely serves as a gatekeeper to ensure that only a substrate harboring the correct number of βAla units is presented to CtaC. As CtaG catalyzes the transfer of PHBA to the CtaE-linked βAla chain, we reasoned that CtaG could serve as this gatekeeper. While the sole production of the PHBA-3βAla conjugate in assays performed with CtaD-I (Fig. 4A) suggested that the longer βAla chain is the correct biosynthetic intermediate, we could not exclude the possibility that a nonnative ratio of the enzymes caused a shift in pathway specificity. Therefore, to interrogate the βAla chain length selectivity of CtaG more rigorously, we synthesized synthetic CoA-linked substrates bearing two (2βAla-CoA) or three (3βAla-CoA) β-alanine residues. The putative substrates were then transferred to apo-CtaE using the promiscuous phosphopantetheinyl transferase Sfp from Bacillus subtilis (36) to afford the corresponding peptidyl-holo-CtaE. Following the successful generation of these CtaE-linked substrates, we performed reactions with holo-CtaH, CtaI, and MBP-CtaG and monitored CtaE processing by MALDI-TOF-MS. While only trace PHBA transfer occurred in reactions containing 2βAla-S-CtaE after 18 h (Fig. 4C), 3βAla-S-CtaE processing was complete in 1 h (Fig. 4D). These data support a CTA biosynthetic pathway in which PHBA-3βAla is a key intermediate and the biosynthetically relevant substrate for the thioamide synthetase.
Characterized transacylating members of the CA protease clan form an enzyme-bound thioester intermediate with the cysteine residue of the catalytic triad during their catalytic cycle (34, 35, 37). Despite the large sequence divergence between CtaG and characterized CA clan members, a putative catalytic triad—C11, H128, and D144—is also present in CtaG (SI Appendix, Fig. S13). To determine whether CtaG-mediated PHBA transfer proceeds through a similar mechanism, we generated mutant versions of the transacylase bearing alanine substitutions to the putative catalytic residues CtaGC11A, CtaGH128A, and CtaGD144A (SI Appendix, Fig. S1) and tested them for catalytic competency. Consistent with the bioinformatic prediction, all mutants were devoid of activity (Fig. 4 A and B).
Next, we repeated CtaG reactions with the omission of CtaD-F and monitored the modification state of CtaG by MALDI-TOF-MS to check for the accumulation of an acylated species. Compared with control reactions lacking holo-CtaH, a new signal consistent with the attachment of PHBA was detected in reactions containing all components (Fig. 4E). Following the digestion of CtaG with trypsin, this modification was localized to the peptide fragment containing the catalytic cysteine residue (SI Appendix, Fig. S14). Reactions performed with the CtaG mutant lacking the catalytic cysteine residue, CtaGC11A, did not show this modification (Fig. 4F and SI Appendix, Fig. S14).
Taken together, these data confirm that CtaG uses the canonical catalytic triad of the papain-like cysteine proteases to catalyze a thiotemplated transacylation reaction in CTA biosynthesis. The thiotemplated nature of CtaG is notable. Apart from CtaG, the only other examples of thiotemplated transacylating papain-like proteases are found in the biosynthesis of the aminoglycoside butirosin (30) and the NRP andrimid (35).
Updated Model for CTA Biosynthesis.
In light of the data presented herein, we propose the following updated model for the early steps in CTA biosynthesis (Scheme 1). These early steps in CTA maturation proceed through two parallel-acting PCP-loading pathways. In the first pathway, PHBA is formed from chorismate by CtaA (5) and is loaded onto holo-CtaH by CtaI in an ATP-dependent reaction. In the second pathway, CtaD catalyzes the ATP-dependent coupling of the γ-carboxylate of l-aspartate and holo-CtaE. The resultant conjugate is decarboxylated by CtaF to afford βAla-S-CtaE. Two additional cycles of l-aspartate addition and decarboxylation by CtaD and CtaF, respectively, result in the formation of 3βAla-S-CtaE. At this stage, the CTA biosynthetic pathway converges, and CtaG catalyzes the transfer of PHBA from CtaH to 3βAla-S-CtaE. The resultant product, PHBA-3βAla-S-CtaE, is iteratively thioamidated by CtaC to afford thioamidated-3βAla-S-CtaE (6).
Scheme 1.
Updated CTA biosynthetic pathway. Two potential pathways (A and B) for DAP biosynthesis and incorporation are displayed.
Although the remaining steps in CTA biosynthesis are not fully elucidated, based on previous knockout studies (5) and the detection of biosynthetic intermediates (5, 14), we foresee two possible pathways for diaminopropane (DAP) incorporation and thioamidated PHBA-3βAla dimerization. In our original proposal, we suggested that DAP would be biosynthesized from l-aspartate by the sequential action of a reductase (CtaK), an aminotransferase (CtaB), and the decarboxylase CtaF (5) (Fig. 1C). DAP would then be used by the transglutaminase CtaJ to convert two molecules of thioamidated-PHBA-3βAla-S-CtaE into CTA. The third β-alanine residue would remain on holo-CtaE and serve as a priming residue for another round of β-alanine polymerization by CtaD/CtaF. However, the inability of CtaF to decarboxylate free substrates requires the use of an alternative, unknown metabolite as the source of DAP. Alternatively, CtaK could catalyze the reductive release of thioamidated PHBA-3βAla from holo-CtaE, and the resultant ketone could be converted to the corresponding primary amine by CtaB. Finally, CtaJ would couple two molecules of the DAP-containing product to afford CTA.
Genomic Prospecting for Noncanonical Thiotemplate Pathways.
Bioinformatic tools are proficient in the identification of canonical thiotemplated biosynthetic gene clusters encoding a NRPS or a polyketide synthase (PKS). However, biosynthetic pathways lacking these megasynthases, as is the case for the CTA assembly, are recalcitrant to genome mining efforts, as the biosynthetic enzymes resemble proteins involved in primary metabolism. As such, elucidation of the CTA amide backbone assembly pathway provided a rare opportunity to mine genomes for additional noncanonical thiotemplated (NCT) biosynthetic pathways.
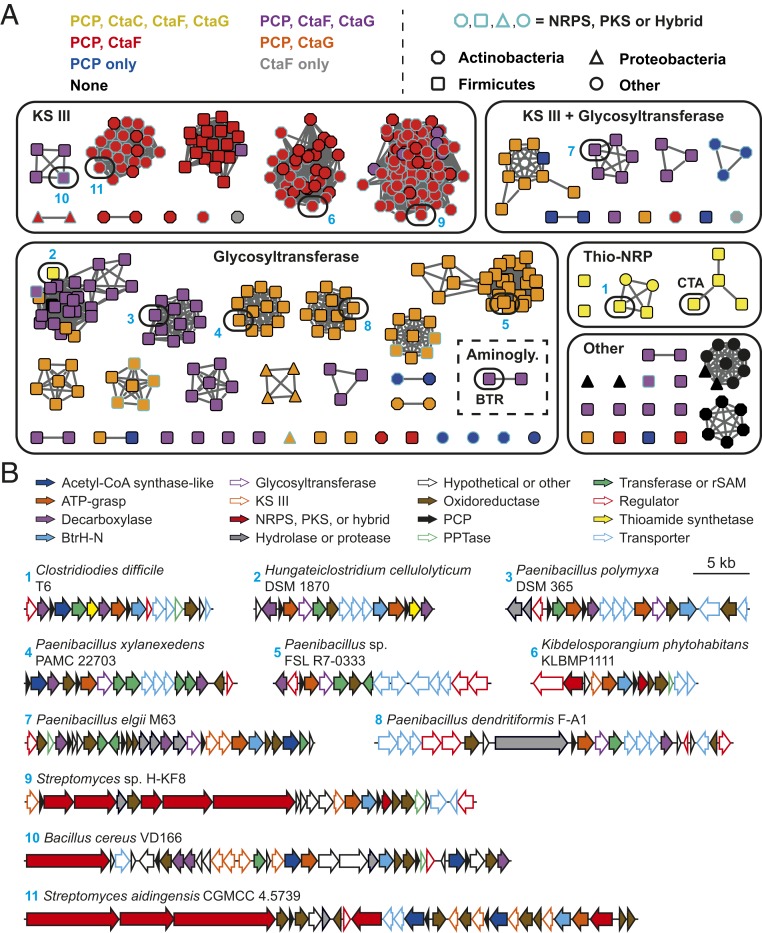
To find candidate NCT gene clusters, BLAST was used to identify 660 homologs of CtaD in GenBank, and these proteins were used to generate a sequence similarity network using the Enzyme Function Initiative-Enzyme Similarity Tool (EFI-EST) (38, 39) (Fig. 5A). Collapsing proteins with ≥95% sequence identity into a single node resulted in a network composed of 402 sequences. Next, we analyzed the local genome neighborhood for each CtaD homolog in the network for the presence of genes encoding CtaF and CtaG homologs, as well as genes encoding PCPs. Although the gene neighborhoods surrounding each homolog were highly diverse (Fig. 5B), 95% were encoded in a genetic locus containing at least one PCP (Fig. 5A and SI Appendix, Table S4). Approximately 65% and 45% of these PCP-associated CtaD homologs were found in loci encoding a decarboxylase or CtaG-like transacylase, respectively. Furthermore, more than 20% of the putative thiotemplated biosynthetic gene clusters encoded both a CtaF homolog and a CtaG homolog (Fig. 5A and SI Appendix, Table S4).
Fig. 5.
Genome mining for additional noncanonical thiotemplate pathways. (A) Sequence similarity network for CtaD homologs (95% identical sequences are conflated; alignment score 115). Nodes are color-coded based on the presence of genes encoding PCPs and homologs of the indicated CTA biosynthetic enzymes in the genomic region surrounding the CtaD homolog. Node shapes correspond to the phylum of the organism harboring the biosynthetic gene cluster, while the color of the node outline indicates the presence of genes encoding an NRPS or PKS in the gene neighborhood. The ATP-grasp proteins from the CTA and butirosin (BTR) biosynthetic pathways are indicated. SI Appendix, Table S4 lists all the sequences in the network. Aminogly., aminoglycoside; Thio-NRP, thioamidated NRP. (B) Representative biosynthetic gene clusters that encode a CtaD homolog (selected based on architecture diversity). Numbers correspond to the node numbering in A. Genes encoding methyltransferases, acetyltransferases, nucleotidyltransferases, aminotransferases, and radical S-adenosyl-l-methionine enzymes are grouped under the designation “Transferase or rSAM”.
Notably, only one of the CtaD homologs was found in a biosynthetic gene cluster with a known metabolite (butirosin). To gain insight into the biosynthetic landscape of the uncharacterized putative NCT assembly lines identified in our bioinformatic analysis, we next searched each gene neighborhood for the presence of secondary metabolite biosynthetic genes. From this analysis, we were able to group the majority of the biosynthetic loci into two large families: loci encoding a ketoacylsynthase III-like protein (PF08541) and loci encoding a glycosyltransferase family 1 protein (PF13528) (Fig. 5A and SI Appendix, Table S4), suggesting that the corresponding products will be acylated (40) or glycosylated (41), respectively. Moreover, canonical thiotemplated biosynthetic machinery (NRPS, PKS, or hybrid) is encoded near the CtaD homolog in approximately one-third of instances (Fig. 5A and SI Appendix, Table S4), hinting that such NCT cassettes may work in concert with the well-characterized megasynthases.
Perhaps most notably, we found that CTA-like and butirosin-like biosynthetic gene clusters are very rare. Apart from a handful of clusters that closely resemble the CTA biosynthetic locus, only one other CtaD homolog was found in a neighborhood containing a CtaC-like thioamide synthetase (Fig. 5 A and B). The results are even more striking for butirosin. Although many neighborhoods encoded putative glycosyltransferases, only one other cluster contained the requisite machinery for the assembly of an aminoglycoside (42, 43) (Fig. 5A). Thus, while the shared biosynthetic logic of thiotemplated amide bond formation typified by butirosin and closthioamide maturation is widespread, these compounds represent biosynthetic outliers for such NCT pathways rather than the norm. Although the co-occurrence of an ATP-grasp protein and a PCP in a putative biosynthetic gene cluster does not necessarily mean that the resulting product will be an NRP, the ∼400 uncharacterized gene clusters identified serve as a starting point for the discovery of additional NCT assembly lines and their corresponding products.
Conclusions
In summary, we have characterized four CTA biosynthetic enzymes responsible for the thiotemplated assembly of the CTA amide backbone. In doing so, we have demonstrated that amide bond formation is catalyzed by novel members of the ATP-grasp and cysteine protease families. Although members of both protein families are known to be NRP amide synthases (4, 20, 30, 35), most either work in concert with an NRPS or are carrier protein-independent. Indeed, the only other example of such a thiotemplated yet NRPS-independent pathway for amide bond formation is found in the biosynthesis of the amino acid side chain of the aminoglycoside butirosin (25, 30). Remarkably, the biosynthetic logic for the biosynthesis and transfer of this amino acid to the butirosin glycone is mirrored in the assembly of the peptide backbone of CTA. Thus, the backbone biosynthesis strategy for CTA maturation uncovered herein represents a rare example of a thiotemplated yet NRPS-independent peptide assembly pathway. However, based on the widespread co-occurrence of ATP-grasp proteins and PCPs in diverse organisms, we predict that such NRPS-independent, thiotemplated secondary metabolite biosynthetic pathways are present in diverse bacterial phyla. In this way, our results not only address the mystery of amide backbone formation in CTA maturation, but also serve as a starting point for the characterization of previously bioinformatically undetectable gene clusters.
Materials and Methods
Expanded descriptions of the experiments are provided in SI Appendix, Materials and Methods.
CtaD and CtaI Reconstitution Assays.
Standard assays were performed with 0.1 μM enzyme (CtaD or CtaI), 30 μM holo-PCP (CtaE or CtaH), 1 mM substrate (PHBA, l-aspartate, or β-alanine), and 1 mM ATP in reaction buffer (50 mM Hepes pH 7.5, 125 mM NaCl, 20 mM MgCl2, and 1 mM TCEP) at 30 °C. Unless indicated otherwise, reactions were allowed to proceed for either 1 h (CtaD) or 2 h (CtaI) before PCP loading was determined by MALDI-TOF-MS. In an attempt to improve holo-CtaE processing, CtaD reactions were also performed for longer times (3 h and 16 h) and with 1 μM CtaD.
NAC assays were performed with 1 μM enzyme (CtaD or CtaI), 400 mM NAC, 1 mM substrate (l-aspartate or PHBA), and 1 mM ATP in modified reaction buffer (50 mM Hepes pH 8.0, 125 mM NaCl, 20 mM MgCl2, and 1 mM DTT). Reactions were allowed to proceed for 18 h at 25 °C. CtaD and CtaI reactions were quenched by the addition of 6 volumes of 0.4 M KH2PO4 or 1 volume of methanol, respectively. NAC processing was then determined by LC-HR-MS. Control reactions were performed with heat-inactivated enzyme.
CtaD–F Reconstitution Assays.
Standard assays were performed with 0.1 μM CtaD, 0.1 μM CtaF, 30 μM holo-CtaE, 1 mM l-aspartate, 10 μM PLP, and 1 mM ATP in reaction buffer. Reactions were allowed to proceed for 2 h at 30 °C before PCP loading was determined by MALDI-TOF-MS. Control reactions were performed with heat-inactivated enzyme.
For identification of the CtaD–F reaction product by LC-HR-MS, reactions were performed as described above. After a 2-h reaction, potassium hydroxide was added to a final concentration of 100 mM, and samples were heated to 55 °C for 1.5 h. Samples were then cooled to room temperature, and amine-containing compounds were derivatized by the addition of 2 volumes of OPA complete reagent (Sigma-Aldrich; P0532), according to a previously established protocol (26). After a 1- to 2-min reaction at room temperature, KH2PO4 was added to a final concentration of 0.225 M, and samples were incubated for another 3 min at room temperature. Derivatized samples were then immediately subjected to analysis by LC-HR-MS. Enzymatically produced 3βAla was compared with an authentic synthetic standard.
For the detection of early-stage intermediates, reactions were performed for the indicated times with either 0.05 μM (5-min time point) or 0.1 μM (10-, 20-, 40-, and 60-min time points) CtaD and CtaF. PCP loading was determined by MALDI-TOF-MS.
CtaG Reconstitution Assays with CtaD–F and CtaH–I.
Standard reactions were performed with 1 μM CtaG (wild-type and mutants), 0.1 μM CtaD, 0.1 μM CtaF, 0.1 μM CtaI, 30 μM holo-CtaE, 1 μM holo-CtaH, 1 mM l-aspartate, 1 mM PHBA, 10 μM PLP, and 1 mM ATP in reaction buffer at 25 °C. To generate 3βAla-S-CtaE and PHBA-S-CtaH, reaction mixtures were incubated for 1 h at 25 °C, followed by the addition of CtaG. At the indicated time points, reactions were quenched and analyzed by either MALDI-TOF-MS or LC-HR-MS. Control reactions were performed with heat-inactivated CtaG. For reactions performed with unactivated PHBA, holo-CtaH and CtaI were omitted.
For LC-HR-MS detection of CtaE-bound products from CtaG reactions, thioester cleavage was performed as described above. Following product release and methanol addition, samples were analyzed by LC-HR-MS. Enzymatically produced PHBA-3βAla was compared with an authentic synthetic standard.
CtaG Selectivity Assays.
Reactions were performed with 1 μM CtaG, 0.1 μM CtaI, 30 μM peptidyl-holo-CtaE (2βAla-S-CtaE or 3βAla-S-CtaE), 1 μM holo-CtaH, 1 mM PHBA, and 1 mM ATP in reaction buffer at 25 °C. To generate PHBA-S-CtaH, reaction mixtures were incubated for 1 h at 25 °C, followed by the addition of CtaG. At the indicated time points, reactions were quenched and analyzed by MALDI-TOF-MS. Control reactions were performed with heat-inactivated CtaG.
CtaH Offloading by CtaG.
Reactions were performed with 1 μM CtaG (wild-type or CtaGC11A), 10 μM 3βAla-S-CtaE, and 7.5 μM PHBA-S-CtaH in reaction buffer at 25 °C. After 30 or 60 min, reactions were analyzed by MALDI-TOF-MS. Control reactions were performed with heat-denatured CtaG.
To generate PHBA-S-CtaH, 50 μM holo-CtaH was mixed with 0.1 μM CtaI, 1 mM PHBA, and 1 mM ATP in reaction buffer. To generate 3βAla-S-CtaE, 30 μM holo-CtaE was mixed with 0.1 μM CtaD, 0.1 μM CtaF, 10 μM PLP, 1 mM l-aspartate, and 1 mM ATP in reaction buffer. Reactions were performed for 2 h at 25 °C, then stopped by transferring the loaded PCPs into fresh reaction buffer lacking substrate and ATP using a Zeba Spin desalting column (Thermo Fisher Scientific) according to the manufacturer’s instructions. The desalted, loaded PCPs were used in CtaG assays as detailed above.
MALDI-TOF-MS.
For detection of PCPs, samples were acidified by the addition of trifluoroacetic acid (TFA) to a final concentration of 0.2% and desalted using a C18 ZipTip (Merck Millipore) according to the manufacturer’s instructions. Protein mixtures were eluted from the ZipTip in 1.5 μL of 50% acetonitrile supplemented with 0.1% TFA. Eluates were diluted with 1.5 μL of 2% TFA and 1.5 µL of 100 mM 2′-5′-dihydroxyacetophenone (DHAP) and spotted onto a MALDI plate.
For detection of MBP-CtaG, 1 μL of sample was mixed with 1 μL of DHAP and 1 μL of 2% TFA. The resultant mixture (0.5 μL) was spotted onto a MALDI plate and allowed to dry. The dried spot was then washed three times with 1.5 μL of 2% TFA.
Samples were analyzed using a Bruker Daltonics UltrafleXtreme MALDI-TOF mass spectrometer. Spectra were obtained in linear positive mode, and the instrument was calibrated to a commercially available standard (PCPs, Protein Calibration Standard I; MBP-CtaG, Protein Calibration Standard II; Bruker) before each measurement. Data analysis was performed with flexAnalysis 3.3 (Bruker).
LC-HR-MS.
HPLC-HR-MS and HPLC-HR-MS/MS measurements were performed with an Accela HPLC system coupled to a QExactive Hybrid Quadrupole Orbitrap (Thermo Fisher Scientific) mass spectrometer equipped with an electrospray ion source. Separation was performed with an Accucore C18 column (2.1 × 100 mm, 2.6 µm; Thermo Fisher Scientific) operating at a flow rate of 200 µL min−1, with 0.1% formic acid (solvent A) and acetonitrile +0.1% formic acid (solvent B) and the following gradient: 5% solvent B for 0.1 min, 5% to 98% solvent B over 10 min, and hold at 98% solvent B for 12 min.
CtaG Covalent Intermediate Detection.
Reactions were performed with 15 μM CtaG (wild-type or CtaGC11A), 1 μM CtaI, 2 μM holo-CtaH, 1 mM PHBA, and 1 mM ATP in reaction buffer at 25 °C. To generate PHBA-S-CtaH, reaction mixtures were incubated for 30 min at 25 °C, followed by the addition of CtaG. After an 18- h reaction at 25 °C, samples were either analyzed directly by MALDI-TOF-MS or trypsin-digested before MALDI-TOF-MS analysis.
For trypsin digestion reactions, trypsin (CtaG:trypsin, 1:1,000), 100 mM CaCl2, and acetonitrile (15% final concentration) were added to samples, and digestions were allowed to proceed for 18 h at 37 °C.
Detection of ADP and AMP.
Assays were performed with 400 mM NAC, 1 mM substrate (PHBA or l-aspartate), 1 mM ATP, and either 1 μM (CtaD) or 2 μM (CtaI) enzyme in NAC reaction buffer. Reactions were allowed to proceed for 5 h at 25 °C before being quenched by the addition of 1 volume of methanol. Samples were analyzed by HPLC according to an established method (44). The HPLC profiles were compared with those of reference compounds (Jena Bioscience).
Phosphate and Pyrophosphate Detection.
The concentrations of phosphate and pyrophosphate were determined using a previously established malachite green method (21). The malachite green working reagent was made fresh each day by mixing 1.5 mL of malachite green (1.22 g/L in 3 M sulfuric acid), 0.375 mL of 7.5% (vol/vol) ammonium molybdate, and 30 μL of 11% (vol/vol) Tween 20. Enzyme reactions were diluted either 5-fold or 10-fold with deionized water, and 40 μL of this diluted sample was transferred to a 384-well plate. The malachite green reaction was initiated by the addition of 10 μL of the malachite green working reagent. Reactions were allowed to develop for 10 to 15 min at 25 °C, after which the absorbance at 620 nm was measured on a Varioskan Lux plate reader (Thermo Fisher Scientific). A standard curve was created from a known concentration of phosphate in appropriately diluted reaction mixture. The absorbance for each of the samples was corrected for any background signal originating from the buffers and substrates (n = 3).
Assays were performed with 0.5 μM enzyme (CtaI or CtaD), 400 mM NAC, 1 mM substrate (PHBA or l-aspartate), and either 1 mM (CtaD reactions) or 2 mM ATP (CtaI reactions) in NAC reaction buffer. CtaI and CtaD reactions were performed at 25 °C for 30 min and 60 min, respectively. Where indicated, 0.01 U of inorganic pyrophosphatase (New England BioLabs) was added to the reaction mixture. All reactions were initiated by the addition of enzyme.
Materials and Data Availability.
All data are included in the manuscript and SI Appendix.
Supplementary Material
Acknowledgments
We thank A. Perner, T. Kindel, and M. García-Altares Peréz for MS measurements, H. Heinecke for NMR measurements, and E. Molloy for critical feedback on the manuscript. K.L.D. was supported by the Humboldt Research Fellowship for Postdoctoral Researchers. Financial support from the German Research Foundation (Leibniz Award, to C.H.) is gratefully acknowledged.
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.1918759117/-/DCSupplemental.
References
- 1.Finking R., Marahiel M. A., Biosynthesis of nonribosomal peptides 1. Annu. Rev. Microbiol. 58, 453–488 (2004). [DOI] [PubMed] [Google Scholar]
- 2.Carroll C. S., Moore M. M., Ironing out siderophore biosynthesis: A review of non-ribosomal peptide synthetase (NRPS)-independent siderophore synthetases. Crit. Rev. Biochem. Mol. Biol. 53, 356–381 (2018). [DOI] [PubMed] [Google Scholar]
- 3.Giessen T. W., Marahiel M. A., The tRNA-dependent biosynthesis of modified cyclic dipeptides. Int. J. Mol. Sci. 15, 14610–14631 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Goswami A., Van Lanen S. G., Enzymatic strategies and biocatalysts for amide bond formation: Tricks of the trade outside of the ribosome. Mol. Biosyst. 11, 338–353 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dunbar K. L., et al. , Genome editing reveals novel thiotemplated assembly of polythioamide antibiotics in anaerobic bacteria. Angew. Chem. Int. Ed. Engl. 57, 14080–14084 (2018). [DOI] [PubMed] [Google Scholar]
- 6.Dunbar K. L., Dell M., Molloy E. M., Kloss F., Hertweck C., Reconstitution of iterative thioamidation in closthioamide biosynthesis reveals tailoring strategy for nonribosomal peptide backbones. Angew. Chem. Int. Ed. Engl. 58, 13014–13018 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lincke T., Behnken S., Ishida K., Roth M., Hertweck C., Closthioamide: An unprecedented polythioamide antibiotic from the strictly anaerobic bacterium Clostridium cellulolyticum. Angew. Chem. Int. Ed. Engl. 49, 2011–2013 (2010). [DOI] [PubMed] [Google Scholar]
- 8.Chiriac A. I., et al. , Mode of action of closthioamide: The first member of the polythioamide class of bacterial DNA gyrase inhibitors. J. Antimicrob. Chemother. 70, 2576–2588 (2015). [DOI] [PubMed] [Google Scholar]
- 9.Miari V. F., Solanki P., Hleba Y., Stabler R. A., Heap J. T., In vitro susceptibility to closthioamide among clinical and reference strains of Neisseria gonorrhoeae. Antimicrob. Agents Chemother. 61, e00929-17 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kloss F., Lincke T., Hertweck C., Highly efficient total synthesis of the Clostridium-derived anti-MRSA antibiotic closthioamide. Eur. J. Org. Chem. 2011, 1429–1431 (2011). [Google Scholar]
- 11.Mahanta N., Szantai-Kis D. M., Petersson E. J., Mitchell D. A., Biosynthesis and chemical applications of thioamides. ACS Chem. Biol. 14, 142–163 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dunbar K. L., Scharf D. H., Litomska A., Hertweck C., Enzymatic carbon-sulfur bond formation in natural product biosynthesis. Chem. Rev. 117, 5521–5577 (2017). [DOI] [PubMed] [Google Scholar]
- 13.Kloss F., Pidot S., Goerls H., Friedrich T., Hertweck C., Formation of a dinuclear copper (I) complex from the Clostridium-derived antibiotic closthioamide. Angew. Chem. Int. Ed. Engl. 52, 10745–10748 (2013). [DOI] [PubMed] [Google Scholar]
- 14.Behnken S., Lincke T., Kloss F., Ishida K., Hertweck C., Antiterminator-mediated unveiling of cryptic polythioamides in an anaerobic bacterium. Angew. Chem. Int. Ed. Engl. 51, 2425–2428 (2012). [DOI] [PubMed] [Google Scholar]
- 15.Li J. S., Barber C. C., Zhang W., Natural products from anaerobes. J. Ind. Microbiol. Biotechnol. 46, 375–383 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Schieferdecker S., et al. , Biosynthesis of diverse antimicrobial and antiproliferative acyloins in anaerobic bacteria. ACS Chem. Biol. 14, 1490–1497 (2019). [DOI] [PubMed] [Google Scholar]
- 17.Schieferdecker S., Shabuer G., Knuepfer U., Hertweck C., Clostrindolin is an antimycobacterial pyrone alkaloid from Clostridium beijerinckii. Org. Biomol. Chem. 17, 6119–6121 (2019). [DOI] [PubMed] [Google Scholar]
- 18.Fawaz M. V., Topper M. E., Firestine S. M., The ATP-grasp enzymes. Bioorg. Chem. 39, 185–191 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gulick A. M., Conformational dynamics in the Acyl-CoA synthetases, adenylation domains of non-ribosomal peptide synthetases, and firefly luciferase. ACS Chem. Biol. 4, 811–827 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ogasawara Y., Dairi T., Biosynthesis of oligopeptides using ATP-grasp enzymes. Chemistry 23, 10714–10724 (2017). [DOI] [PubMed] [Google Scholar]
- 21.Baykov A. A., Evtushenko O. A., Avaeva S. M., A malachite green procedure for orthophosphate determination and its use in alkaline phosphatase-based enzyme immunoassay. Anal. Biochem. 171, 266–270 (1988). [DOI] [PubMed] [Google Scholar]
- 22.Ota Y., et al. , Butirosin-biosynthetic gene cluster from Bacillus circulans. J. Antibiot. (Tokyo) 53, 1158–1167 (2000). [DOI] [PubMed] [Google Scholar]
- 23.Kudo F., et al. , Extended sequence and functional analysis of the butirosin biosynthetic gene cluster in Bacillus circulans SANK 72073. J. Antibiot. (Tokyo) 58, 373–379 (2005). [DOI] [PubMed] [Google Scholar]
- 24.Dion H. W., et al. , Butirosin, a new aminoglycosidic antibiotic complex: Isolation and characterization. Antimicrob. Agents Chemother. 2, 84–88 (1972). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Li Y., Llewellyn N. M., Giri R., Huang F., Spencer J. B., Biosynthesis of the unique amino acid side chain of butirosin: Possible protective-group chemistry in an acyl carrier protein-mediated pathway. Chem. Biol. 12, 665–675 (2005). [DOI] [PubMed] [Google Scholar]
- 26.Jarrett H. W., Cooksy K. D., Ellis B., Anderson J. M., The separation of o-phthalaldehyde derivatives of amino acids by reversed-phase chromatography on octylsilica columns. Anal. Biochem. 153, 189–198 (1986). [DOI] [PubMed] [Google Scholar]
- 27.Söding J., Biegert A., Lupas A. N., The HHpred interactive server for protein homology detection and structure prediction. Nucleic Acids Res. 33, W244-8 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zimmermann L., et al. , A completely reimplemented MPI bioinformatics toolkit with a new HHpred server at its core. J. Mol. Biol. 430, 2237–2243 (2018). [DOI] [PubMed] [Google Scholar]
- 29.Bobeica S. C., et al. , Insights into AMS/PCAT transporters from biochemical and structural characterization of a double glycine motif protease. eLife 8, e42305 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Llewellyn N. M., Li Y., Spencer J. B., Biosynthesis of butirosin: Transfer and deprotection of the unique amino acid side chain. Chem. Biol. 14, 379–386 (2007). [DOI] [PubMed] [Google Scholar]
- 31.Rawlings N. D., et al. , The MEROPS database of proteolytic enzymes, their substrates and inhibitors in 2017 and a comparison with peptidases in the PANTHER database. Nucleic Acids Res. 46, D624–D632 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Barrett A. J., Rawlings N. D., Evolutionary lines of cysteine peptidases. Biol. Chem. 382, 727–733 (2001). [DOI] [PubMed] [Google Scholar]
- 33.Blum R., et al. , Function of phytochelatin synthase in catabolism of glutathione-conjugates. Plant J. 49, 740–749 (2007). [DOI] [PubMed] [Google Scholar]
- 34.Rea P. A., Vatamaniuk O. K., Rigden D. J., Weeds, worms, and more. Papain’s long-lost cousin, phytochelatin synthase. Plant Physiol. 136, 2463–2474 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Fortin P. D., Walsh C. T., Magarvey N. A., A transglutaminase homologue as a condensation catalyst in antibiotic assembly lines. Nature 448, 824–827 (2007). [DOI] [PubMed] [Google Scholar]
- 36.Beld J., Sonnenschein E. C., Vickery C. R., Noel J. P., Burkart M. D., The phosphopantetheinyl transferases: Catalysis of a post-translational modification crucial for life. Nat. Prod. Rep. 31, 61–108 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Griffin M., Casadio R., Bergamini C. M., Transglutaminases: Nature’s biological glues. Biochem. J. 368, 377–396 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gerlt J. A., et al. , Enzyme function initiative-enzyme similarity tool (EFI-EST): A web tool for generating protein sequence similarity networks. Biochim. Biophys. Acta 1854, 1019–1037 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zallot R., Oberg N. O., Gerlt J. A., “Democratized” genomic enzymology web tools for functional assignment. Curr. Opin. Chem. Biol. 47, 77–85 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Nofiani R., Philmus B., Nindita Y., Mahmud T., 3-Ketoacyl-ACP synthase (KAS) III homologues and their roles in natural product biosynthesis. MedChemComm 10, 1517–1530 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Coutinho P. M., Deleury E., Davies G. J., Henrissat B., An evolving hierarchical family classification for glycosyltransferases. J. Mol. Biol. 328, 307–317 (2003). [DOI] [PubMed] [Google Scholar]
- 42.Kudo F., Eguchi T., Biosynthetic genes for aminoglycoside antibiotics. J. Antibiot. (Tokyo) 62, 471–481 (2009). [DOI] [PubMed] [Google Scholar]
- 43.Kudo F., Eguchi T., Aminoglycoside antibiotics: New insights into the biosynthetic machinery of old drugs. Chem. Rec. 16, 4–18 (2016). [DOI] [PubMed] [Google Scholar]
- 44.Litomska A., et al. , Enzymatic thioamide formation in a bacterial antimetabolite pathway. Angew. Chem. Int. Ed. Engl. 57, 11574–11578 (2018). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.