Significance
Glioblastoma has an extremely poor prognosis, driven by its invasive and angiogenic potential. The development of effective therapies for glioblastoma will require the identification of molecular factors that promote these hallmarks. Our results show that PHIP drives glioblastoma motility, invasion, and angiogenesis. These functions are enabled by localization of PHIP to focal adhesions. PHIP regulates expression of focal adhesion proteins and physically interacts with vinculin, mechanisms by which it promotes tumor cell motility and invasion. The presence of elevated PHIP copy number in distinct molecular glioblastoma subtypes provides additional support for its importance to glioblastoma biology. These studies also identify PHIP as a compelling target for glioblastoma therapy.
Keywords: PHIP, glioblastoma, invasion, motility, angiogenesis
Abstract
The invasive behavior of glioblastoma is essential to its aggressive potential. Here, we show that pleckstrin homology domain interacting protein (PHIP), acting through effects on the force transduction layer of the focal adhesion complex, drives glioblastoma motility and invasion. Immunofluorescence analysis localized PHIP to the leading edge of glioblastoma cells, together with several focal adhesion proteins: vinculin (VCL), talin 1 (TLN1), integrin beta 1 (ITGB1), as well as phosphorylated forms of paxillin (pPXN) and focal adhesion kinase (pFAK). Confocal microscopy specifically localized PHIP to the force transduction layer, together with TLN1 and VCL. Immunoprecipitation revealed a physical interaction between PHIP and VCL. Targeted suppression of PHIP resulted in significant down-regulation of these focal adhesion proteins, along with zyxin (ZYX), and produced profoundly disorganized stress fibers. Live-cell imaging of glioblastoma cells overexpressing a ZYX-GFP construct demonstrated a role for PHIP in regulating focal adhesion dynamics. PHIP silencing significantly suppressed the migratory and invasive capacity of glioblastoma cells, partially restored following TLN1 or ZYX cDNA overexpression. PHIP knockdown produced substantial suppression of tumor growth upon intracranial implantation, as well as significantly reduced microvessel density and secreted VEGF levels. PHIP copy number was elevated in the classical glioblastoma subtype and correlated with elevated EGFR levels. These results demonstrate PHIP’s role in regulating the actin cytoskeleton, focal adhesion dynamics, and tumor cell motility, and identify PHIP as a key driver of glioblastoma migration and invasion.
Gliomas represent one of the most lethal tumor types, with a median survival of 15–16 mo for patients with high-grade (grade IV) gliomas (glioblastoma), and account for over 15,000 deaths each year in the United States (1). The marked histopathological heterogeneity of glioblastomas has led to efforts to classify them according to various molecular alterations (2, 3). Glioblastomas have been classified at the molecular level using defined somatic mutations or copy number alterations. Thus, a significant proportion of gliomas harbor mutations in TP53, IDH1, or NF1, or amplification of EGFR (4–6). The integration of gene expression profiling with mutational analysis has identified three major molecular subtypes of IDH-WT glioblastoma: classical, mesenchymal, and proneural (7). The proneural subtype is typified by alterations in PDGFRA. The mesenchymal subtype is associated with NF1 mutations, and alterations in the PI3K-AKT pathway (8). The classical subtype is characterized by EGFR amplifications (3). While this scheme has proved extremely useful in glioblastoma classification, to date, identification of novel targets for therapy has proved elusive.
The aggressive behavior of glioblastomas is driven in part by their migratory and invasive characteristics, capable of infiltrating diffusely into normal brain parenchyma (9). During invasion, glioblastoma cells become polarized, and their leading edge is characterized by dynamic cytoskeletal rearrangements, regulated by integrins, FAK, and AKT. In addition to a highly motile phenotype, glioblastomas are highly angiogenic, which further enhances glioma cell survival (9). Development of successful targeted interventions for glioblastoma will require the identification of factors that mediate these essential features of its aggressiveness. Recently, we demonstrated a role for PHIP in the progression of human breast and lung cancer (10) as well as of human melanoma, by virtue of its effects on tumor cell invasion and angiogenesis (11, 12). PHIP was initially localized to the nucleus and shown to mediate proliferative responses following activation of the type I insulin-like growth factor receptor (IGF1R) in pancreatic islet cells (13). In the current study, we examine the role of PHIP in the progression of human glioblastoma, and describe previously uncharacterized cellular localization and functions that enable key roles for PHIP in driving glioblastoma progression, identifying it as a rational target for therapy.
Results
To evaluate the role of PHIP in glioblastoma progression, we utilized two effective shRNAs targeting PHIP in human melanoma, breast and lung cancer cells (10). We initially generated U-251 human glioblastoma cells stably expressing either a control shRNA targeting firefly luciferase (luc) or anti-PHIP shRNA#1. Stable shRNA expression resulted in 89.8% reduction in PHIP expression by qRT-PCR (SI Appendix, Fig. S1A) and 55% decrease by Western blot analysis (SI Appendix, Fig. S1B). We used a proteome profiler human phospho-kinase array to identify signaling pathways altered following stable PHIP knockdown, and detected marked reduction in the phosphorylation level of several kinases that promote tumor progression, including pAKT (∼70% inhibition) (SI Appendix, Fig. S1C). Levels of pPXN (Y118) and pFAK (Y925) were also suppressed (by 35% and 32%, respectively) in U-251 cells expressing anti-PHIP shRNA (SI Appendix, Fig. S1C). AKT and PXN promote tumor cell motility and invasion following IGF1R pathway activation (14), and pAKT was down-regulated following PHIP knockdown in melanoma (11), breast and lung cancer cells (10). Decreased expression of pAKT and pPXN was confirmed using Western analysis (SI Appendix, Fig. S1B).
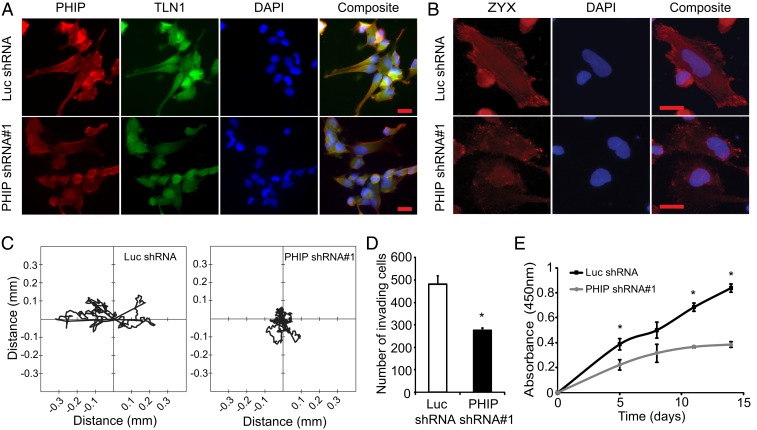
Given the down-regulation of pFAK and pPXN following PHIP silencing, we analyzed the expression and localization of PHIP, together with that of several proteins involved in cell motility and cytoskeletal organization using fluorescence microscopy. Surprisingly, the pattern of immunopositivity and quantitative analysis of pixel intensity indicated prominent staining for PHIP at the leading edge of motile U-251 cells, suggesting its localization with focal adhesion proteins (SI Appendix, Figs. S2 and S3). In addition, PHIP silencing suppressed the expression levels of pFAK (Y925) and ITGB1, as shown by quantitative immunofluorescence (SI Appendix, Figs. S2 and S3), as well as expression of TLN1, VCL, pPXN (Y118), and ZYX (Fig. 1 A–D and SI Appendix, Fig. S3). The PHIP protein has previously been localized to the nucleus of normal pancreatic β cells (13). Its localization at the leading edge of cancer cells, together with its downstream regulation of focal adhesion proteins, including ZYX, has not been previously reported. Visualization of stress fibers using rhodamine phalloidin revealed a strikingly disorganized F-actin pattern in U-251 cells stably expressing anti-PHIP shRNA (Fig. 1E), indicating a role for PHIP in regulating assembly of the cytoskeletal apparatus.
Fig. 1.
Qualitative immunofluorescence analysis of PHIP and focal adhesion proteins in U-251 stable cells. Images representing immunofluorescent detection of PHIP and pPXN (Y118) (A), TLN1 (B), ZYX (C), VCL (D), and F-actin (E) in U-251 transformants stably expressing anti-luc shRNA or anti-PHIP shRNA#1. DAPI staining was used to counterstain the nuclei (Scale bars: 20 µm.) White arrows point to the leading edge of cells. Quantification of the immunofluorescence results, including statistical analysis, is provided in SI Appendix, Fig. S3.
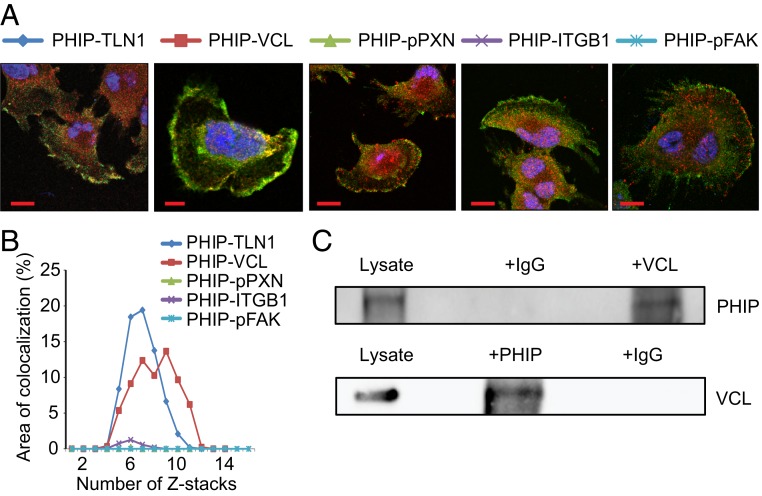
We then determined the precise localization of PHIP within the focal adhesion complex. Recent studies have indicated that focal adhesions are organized into three distinct domains with unique protein compositions (reviewed by ref. 15): 1) a distal tip (the integrin signaling layer) facing the leading edge, composed of integrins, FAK, and PXN; 2) a proximal tip (the actin regulatory layer) that interacts with the stress fiber, composed of α-actinin, ZYX, and vasodilator-stimulated phosphoprotein (VASP); and 3) a force transduction layer spanning these two layers, composed of talin and VCL. Confocal microscopy of U-251 cells, together with analysis of the colocalization coefficient of PHIP immunopositivity against TLN1, VCL, pPXN, pFAK, and ITGB1, demonstrated that PHIP specifically colocalizes with TLN1 and VCL in the force transduction layer (Fig. 2 A and B). Given this colocalization, we assessed whether PHIP interacted with either of the proteins in the force transduction layer. Coimmunoprecipation analysis of cytoplasmic extracts of U-251 cells revealed a physical interaction between PHIP and VCL. Both immunoprecipitated fractions were positive by Western blot analysis, for their counterpart, indicating that PHIP binds to VCL (Fig. 2C).
Fig. 2.
Colocalization of PHIP, TLN1, and VCL at the leading edge of U-251 cells. (A) Representative confocal images (single Z-stack) of U-251 cells after performing immunostaining for PHIP, TLN1, VCL, pPXN (Y118), ITGB1, and pFAK (Y925). (Scale bars: 20 µm.) (B) Graph showing percentage of area that PHIP colocalizes with the covisualized proteins in consecutive Z-stacks. (C) Cytoplasmic extracts from U-251 cells were subjected to the immunoprecipitation of endogenous VCL using an anti-VCL antibody or IgG control antibody followed by Western blot analysis of endogenous PHIP using an anti-PHIP antibody (Upper); cytoplasmic extracts from U-251 cells were subjected to the immunoprecipitation of endogenous PHIP using an anti-PHIP antibody or IgG control antibody followed by Western analysis of endogenous VCL using an anti-VCL antibody (Lower).
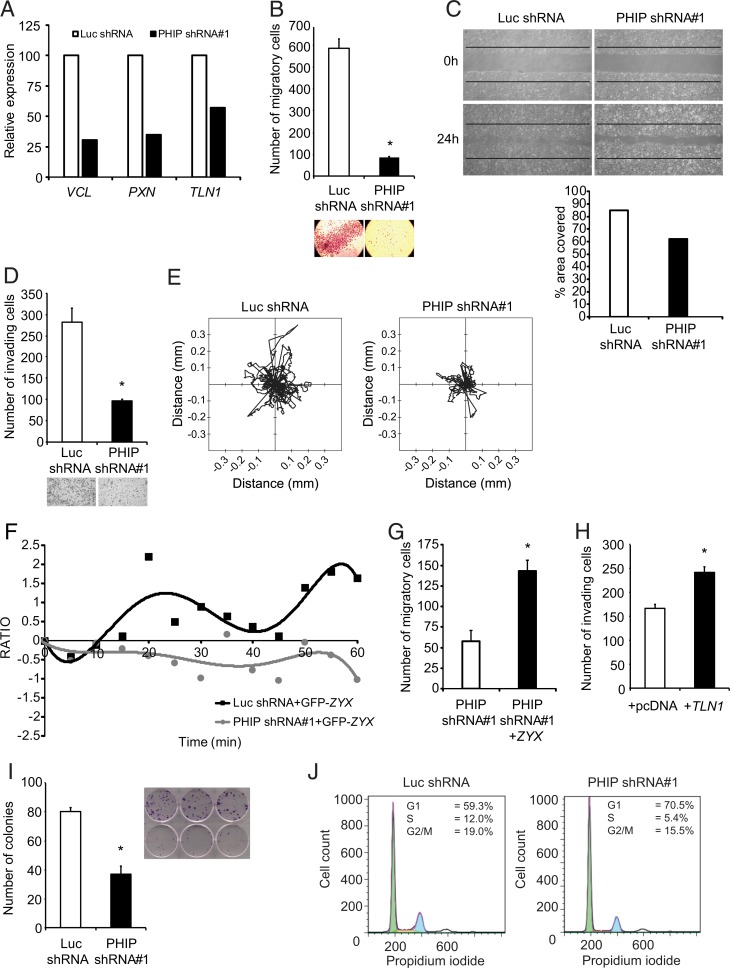
We then assessed the functional effects of suppressing PHIP expression on glioblastoma progression. Initially, qRT-PCR analysis indicated down-regulation of several focal adhesion genes at the RNA level (Fig. 3A). This was accompanied by significantly reduced U-251 cell migration in the Transwell Boyden chamber assay (∼86% reduction, P < 0.05, Fig. 3B), in a wound-healing assay (Fig. 3C), and significantly decreased U-251 cell invasion into Matrigel (66% reduction, P < 0.05, Fig. 3D). Detailed analysis of U-251 cell motility revealed that PHIP down-regulation significantly suppressed the migratory capacity of these cells, as evidenced by the distance traveled within the in vitro 2D environment (P < 0.0001, Fig. 3E and Movies S1 and S2), suggestive of contact inhibition of locomotion (16).
Fig. 3.
Effects of stable suppression of PHIP in U-251 transformants stably expressing anti-luc shRNA or anti-PHIP shRNA#1 in vitro. (A) Relative expression of focal adhesion genes in U-251 transformants. (B) Migration assay of U-251 transformants in a Boyden chamber with representative images. (C) Phase-contrast images of scratch assay at 0 h and 24 h after wound and quantification of the area covered. (D) Invasion into Matrigel of U-251 transformants with representative images. (E) Two-dimensional projections of cell tracks from U-251 transformants stably expressing anti-PHIP shRNA#1 (n = 100 cells) or anti-luc shRNA (n = 93 cells) plotted normalized to their starting positions. (F) Plot presenting the fluorescence intensity of GFP-tagged ZYX at the leading edge of U-251 transformants as measured in consecutive frames of live-cell imaging. (G) Effects of ZYX cDNA overexpression on migration in Boyden chamber of U-251 transformants. (H) Effects of TLN1 cDNA overexpression on invasion into Matrigel of U-251 transformants. (I) Colony formation ability of U-251 transformants, with representative images. (J) Cell cycle analysis of U-251 transformants (Dean-Jett-Fox quantification by FlowJo). All graphs represent mean ± SEM, *P < 0.05 versus control.
Since the polarization of motile cells is tightly controlled by the organization of stress fibers, including recruitment of ZYX, we used GFP-tagged ZYX to visualize the role of PHIP in regulating focal adhesion dynamics in glioblastoma cells. Initially, conventional immunofluorescence showed that upon PHIP down-regulation, the pattern of immunopositivity for ZYX was drastically affected, wherein anti-PHIP shRNA-expressing U-251 cells presented a disorganized and weak signal when compared to control cells. Live-cell imaging of GFP-tagged ZYX provided further support for the static imaging provided by immunofluorescence. At the leading edge of motile cells, areas of positivity from GFP-tagged ZYX (the site of ZYX recruitment along the actin filaments) showed a specific pattern of movement along the axis perpendicular to the direction of motile cells (Movies S3 and S4). Plotting these areas as points of positivity (based on pixel intensity for GFP fluorescence) against time revealed a periodic pattern, analogous to assembly and disassembly of the focal complex, that was absent in cells expressing anti-PHIP shRNA (Fig. 3F). Because ZYX is a marker of focal adhesion maturation (17), these results provide support for PHIP as an important regulator of focal adhesion assembly, which is vital for the formation of stable focal adhesion points and for maintaining the invasive phenotype of glioblastoma cells.
We then assessed whether any of the proteins down-regulated following targeted suppression of PHIP are involved in mediating its effects on glioblastoma migration and invasion. Lentiviral infection of GFP-ZYX cDNA or transfection of a plasmid encoding TLN1 cDNA in U-251 cells stably expressing anti-PHIP shRNA significantly restored their migratory and invasive capacity, respectively (Fig. 3 G and H). Thus, the proinvasive and promigratory roles of PHIP in glioblastoma are mediated, at least in part, by TLN1 and ZYX.
In addition, shRNA-mediated targeting of PHIP resulted in significant inhibition of U-251 colony formation (P < 0.05, Fig. 3I). This was accompanied by a reduction in the number of glioblastoma cells in S phase (Fig. 3J), as well as by decreased expression of CCND1 (SI Appendix, Fig. S1B), which operates downstream of AKT to promote tumor cell proliferation (18). Several of the effects observed in U-251 cells were confirmed using a second anti-PHIP shRNA (shRNA#2, SI Appendix, Fig. S1 D–H).
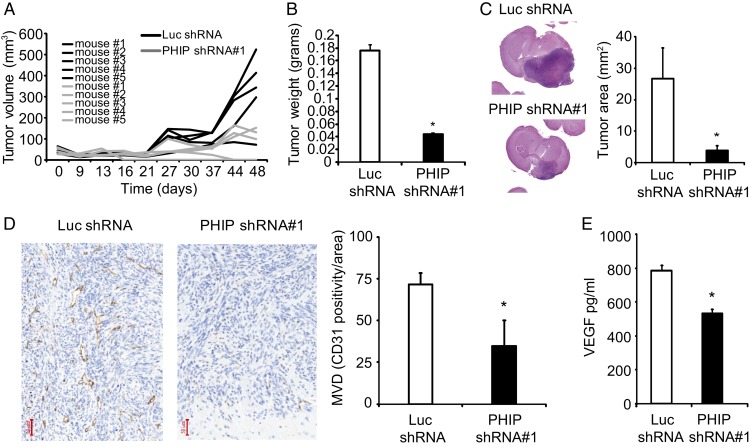
Having shown a role for PHIP in the progression of U-251 cells in culture, we next assessed its function in vivo, using a murine xenograft model. First, subcutaneous (s.c.) injection of U-251 transformants stably expressing anti-PHIP shRNA#1 in the flanks of nude mice resulted in a significant decrease in tumor growth (P < 0.05, Fig. 4A). Importantly, two of the mice (40%) injected with the U-251 transformants expressing anti-PHIP shRNA showed complete regression of their tumor (Fig. 4A). This was accompanied by a significant reduction in weights of anti-PHIP shRNA-expressing tumors (P < 0.05, Fig. 4B). Next, we evaluated the in vivo growth potential of U-251 transformants when injected intracranially in nude mice. There was a dramatic (85%) reduction in tumor size (as determined by the cross-sectional area occupied by the tumor) observed in the brains of mice injected with U-251 cells expressing the anti-PHIP shRNA when compared to the brains of control animals (P < 0.05, Fig. 4C).
Fig. 4.
Effects of stable suppression of PHIP in U-251 transformants stably expressing anti-luc shRNA or anti-PHIP shRNA#1 in vivo. (A) Individual tumor volume measurements following s.c. injection of U-251 transformants. (B) Weight of s.c. U-251 tumors in the flanks of nude mice. (C) Tumor area following intracranial injection of U-251 transformants, with representative hematoxylin-eosin staining of intracranial tumors. (D) Microvessel density in intracranial tumors from U-251 transformants, with representative CD31 immunostaining in the intracranial tumors. (E) Quantification of VEGF levels by ELISA in U-251 transformants. All graphs represent mean ± SEM, *P < 0.05 versus control.
As glioblastomas are highly angiogenic (9), CD31 immunostaining was used to evaluate intratumoral microvessel density following PHIP knockdown. Tumors expressing anti-PHIP shRNA showed a significant (51.6%) reduction in the number of CD31-positive vessels (P < 0.05, Fig. 4D), accompanied by a significant reduction in secreted VEGF levels, as measured by enzyme-linked immunosorbent assay (ELISA) (P < 0.05, Fig. 4E).
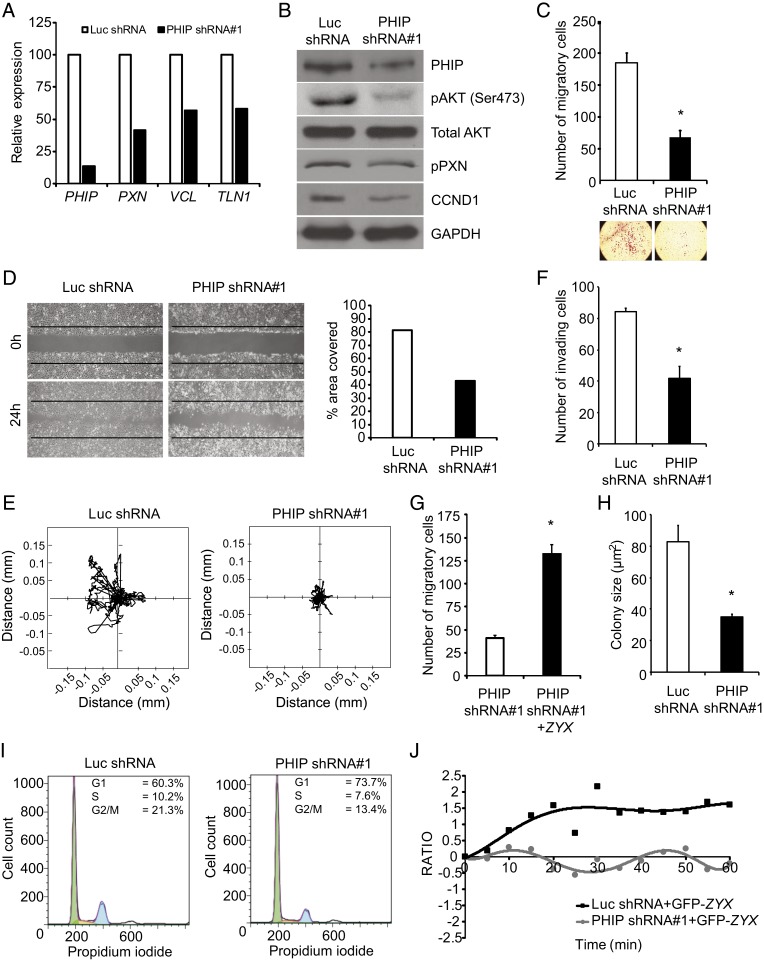
To further analyze the impact of PHIP in glioblastoma progression, we assessed the consequences of PHIP silencing in a second cell line. Stable expression of anti-PHIP shRNA#1 in LN-18 cells resulted in a pronounced decrease in PHIP expression by qRT-PCR (Fig. 5A) and Western blot analysis (Fig. 5B), accompanied by suppression of RNA expression of several focal adhesion genes (Fig. 5A). Qualitative and quantitative immunofluorescence analyses confirmed the localization of PHIP at the leading edge of LN-18 cells and demonstrated significantly reduced expression of ITGB1, pFAK (Y925), VCL, TLN1, pPXN (Y118), and ZYX following PHIP silencing (SI Appendix, Figs. S4–S6). Qualitative analysis of stress fibers showed a disorganized F-actin pattern in LN-18 cells following stable PHIP knockdown (SI Appendix, Fig. S5C). The decreased expression of pAKT and pPXN was confirmed using Western blot analysis (Fig. 5B). PHIP silencing significantly reduced LN-18 cell migration in the Transwell Boyden chamber (62.6% reduction, P < 0.05, Fig. 5C), in a wound assay (Fig. 5D), and in time-lapse (P < 0.0001, Fig. 5E), as well as significantly reduced LN-18 cell invasion into Matrigel (50.4% reduction, P < 0.05, Fig. 5F). Lentiviral infection of GFP-ZYX cDNA in LN-18 cells stably expressing anti-PHIP shRNA significantly restored their migration capacity (Fig. 5G). PHIP targeting resulted in significant inhibition of LN-18 colony formation (P < 0.05, Fig. 5H) and was accompanied by a reduction in the number of cells in S-phase (Fig. 5I) and by decreased CCND1 expression (Fig. 5B). Again, live-cell imaging of GFP-tagged ZYX at the leading edge of LN-18 cells showed a specific and periodic pattern of movement along the axis perpendicular to the direction of motile cells that was not apparent in cells expressing anti-PHIP shRNA (Fig. 5J and Movies S5 and S6). Confocal microscopy of LN-18 cells confirmed the colocalization of PHIP with TLN1 and VCL in the force transduction layer (SI Appendix, Fig. S5 D and E). Several of these effects were confirmed following PHIP silencing with shRNA#2 (SI Appendix, Fig. S7 A–E).
Fig. 5.
Effects of stable suppression of PHIP in LN-18 transformants stably expressing anti-luc shRNA or anti-PHIP shRNA#1 in vitro. (A) Relative expression of PHIP and focal adhesion genes in LN-18 transformants. (B) Western blot analysis of PHIP and other proteins in LN-18 transformants. (C) Migration assay of LN-18 transformants in a Boyden chamber with representative images. (D) Phase-contrast images of scratch assay at 0 h and 24 h after wound and quantification of the area covered. (E) Two-dimensional projections of cell tracks from LN-18 transformants stably expressing anti-PHIP shRNA#1 (n = 87 cells) or anti-luc shRNA (n = 99 cells) plotted normalized to their starting positions. (F) Invasion into Matrigel of LN-18 transformants. (G) Effects of ZYX cDNA overexpression on migration in Boyden chamber of LN-18 transformants stably expressing anti-PHIP shRNA#1. (H) Colony formation ability of LN-18 transformants. (I) Cell cycle analysis of LN-18 transformants (Dean-Jett-Fox quantification by FlowJo). (J) Plot presenting the fluorescence intensity of GFP-tagged ZYX at the leading edge of LN-18 transformants as measured in consecutive frames of live-cell imaging. All graphs represent mean ± SEM, *P < 0.05 versus control.
In addition, several of PHIP’s functional properties were confirmed in U-87 human glioblastoma cells. Stable shRNA expression resulted in decreased PHIP expression by qRT-PCR (SI Appendix, Fig. S7F) and immunofluorescence analysis (SI Appendix, Fig. S7 G and H), resulting in reduced pAKT expression (54.7% reduction, SI Appendix, Fig. S7I), invasion (50.4% reduction, P < 0.05, SI Appendix, Fig. S7J), and colony formation (P < 0.05, SI Appendix, Fig. S7K) when compared to control U-87 cells expressing the anti-luc shRNA. Subsequently, the impact of shRNA-mediated PHIP knockdown on U-87 cell progression was demonstrated using shRNA#2 (SI Appendix, Fig. S7 L–N).
Having shown a role for PHIP in the progression of three established human glioblastoma cell lines, we next assessed its function in a patient-derived primary glioblastoma culture. We generated short-term transformants of human primary patient-derived 3832 cells expressing anti-luc or anti-PHIP shRNA. Expression of anti-PHIP shRNA in 3832 cells resulted in a significant decrease in PHIP expression by immunofluorescence analysis (Fig. 6A and SI Appendix, Fig. S8A), accompanied by significantly reduced expression of TLN1 (Fig. 6A and SI Appendix, Fig. S8B) and ZYX (Fig. 6B and SI Appendix, Fig. S8C), and associated with significantly reduced tumor cell motility (P < 0.05, Fig. 6C and Movies S7 and S8), invasion (P < 0.05, Fig. 6D), and proliferation (P < 0.05, Fig. 6E).
Fig. 6.
Effects of stable suppression of PHIP in 3832 primary glioblastoma transformants stably expressing anti-luc shRNA or anti-PHIP shRNA#1. (A) Qualitative immunofluorescence analysis of PHIP and TLN1 expression in primary 3832 transformants. DAPI staining was used to counterstain the nuclei. Quantification of the immunofluorescence results, including statistical analysis, is provided in SI Appendix, Fig. S8. (B) Qualitative immunofluorescence analysis of ZYX expression in primary 3832 transformants. DAPI staining was used to counterstain the nuclei. Quantification of the immunofluorescence results, including statistical analysis, is provided in SI Appendix, Fig. S8. (C) Two-dimensional projections of cell tracks from primary 3832 transformants stably expressing anti-PHIP shRNA#1 (n = 9 cells) or anti-luc shRNA (n = 11 cells) plotted normalized to their starting positions. (D) Invasion into Matrigel of primary 3832 transformants. (E) Cell survival analysis of primary 3832 transformants. All graphs represent mean ± SEM, *P < 0.05 versus control. (Scale bars: 20 µm.)
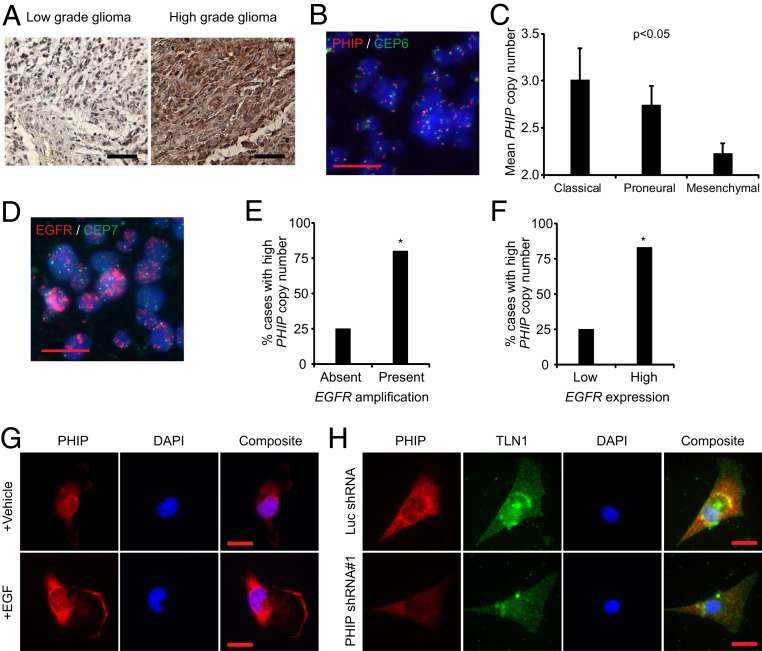
We next examined PHIP expression in glioma progression. We performed immunohistochemical analysis of PHIP protein expression in a tissue microarray containing 10 normal human brain tissue specimens, 58 low-grade gliomas, and 50 glioblastomas and scored the specimens for intensity of PHIP immunostaining on a 0–3 scale. High PHIP expression (defined as a score of 3) was not observed in either normal brain tissue or in low-grade gliomas but was present in a significantly higher proportion (8/50, or 16%) of glioblastomas (P < 0.001, Fig. 7A). These results suggest PHIP as a potential marker of glioma progression.
Fig. 7.
Analysis of PHIP levels in human glioma specimens and glioblastoma cell lines. (A) Representative PHIP immunostaining in low- and high-grade glioma. (B) Representative image of glioblastoma 180 with elevated PHIP copy number as detected using probes for PHIP (red) and centromere of chromosome 6 (green) together with DAPI as nuclear counterstain (blue). (C) Mean PHIP copy number analysis by FISH in different molecular subtypes of glioblastoma. (D) Representative image of glioblastoma 180 with EGFR amplification as detected by probes for EGFR (red) and centromere of chromosome 7 (green) together with DAPI as nuclear counterstain (blue). (E) Bar graphs showing the percentage of glioblastoma cases with high PHIP copy number versus EGFR amplification. (F) Bar graphs showing the percentage of glioblastoma cases with high PHIP copy number versus the level of EGFR expression. (G) Qualitative immunofluorescence analysis of PHIP in U-251 cells upon stimulation with EGF versus vehicle. (H) Qualitative immunofluorescence analysis of PHIP and TLN1 in 218 classical low-passage glioblastoma cells following expression of anti-PHIP shRNA#1 versus anti-luc shRNA. Quantification of the immunofluorescence results, including statistical analysis, is provided in SI Appendix, Fig. S8. All graphs represent mean ± SEM, *P < 0.05 versus control. (Scale bars: A, 100 µm; B, D, G, and H, 20 µm.)
In addition, we assessed PHIP levels in distinct molecular subtypes of glioblastoma (3, 6–8). Recent studies have demonstrated PHIP copy number elevations in melanoma, including its association with and enrichment in distinct molecular subtypes (19). We thus assessed PHIP copy number in glioblastoma using fluorescence in situ hybridization (FISH). As aldehyde-based fixatives (formalin, paraformaldehyde, and glutaraldehyde) may alter tissues and induce autofluorescence and given that brain tissue contains a variety of autofluorescent proteins such as lipofuscin and lipofuscin-like granules (20), FISH analysis of PHIP copy number could not be reliably performed in formalin-fixed and paraffin-embedded glioblastoma samples. Thus, FISH analysis was performed on a cohort of 25 fresh frozen glioblastoma cases in which transcriptomic analysis was utilized to classify the tumors into the three major molecular subtypes of glioblastoma (i.e., classical, proneural, and mesenchymal, SI Appendix, Table S1). Elevated PHIP copy number (defined as mean copy number of ≥ 3.0, Fig. 7B) was present in 20% of the glioblastomas analyzed and was observed in both classical and proneural subtypes. PHIP copy number was significantly different in the three glioblastoma subtypes (P < 0.05, ANOVA), with the highest levels observed in the classical subtype (Fig. 7C and SI Appendix, Table S1). Given that classical glioblastomas are characterized by enhanced EGFR levels (whether through amplification or overexpression), we assessed the potential association between PHIP copy number and 1) EGFR copy number (Fig. 7D) and 2) EGFR expression, separately, in this cohort. There was a significant association between mean PHIP and mean EGFR copy number. Eighty percent of cases with EGFR amplification had elevated PHIP copy number compared with 25% of cases without EGFR amplification (P = 0.04, Fig. 7E). Similarly, there was a significant association between mean PHIP copy number and mean EGFR RNA expression level (Fig. 7F), as 83.3% of cases with high EGFR expression had elevated PHIP copy number compared with 25% of cases with low PHIP copy number (P = 0.03). In addition, PHIP expression increased significantly following EGF stimulation of serum-starved U-251 and LN-18 cells (Fig. 7G and SI Appendix, Fig. S8 D–F). Finally, shRNA-mediated suppression of PHIP (Fig. 7H and SI Appendix, Fig. S8G) in 218, a low-passage primary classical glioblastoma line, resulted in reduced expression of TLN1 (Fig. 7H and SI Appendix, Fig. S8H) and ZYX (SI Appendix, Fig. S8 I and J), and was accompanied by significantly reduced invasion into Matrigel (SI Appendix, Fig. S8K).
Discussion
In this work, we explored the potential role of PHIP in mediating glioblastoma progression. Previously, PHIP was shown to exert a proinvasive role in melanoma, another neuroectodermal tumor (11). Immunofluorescence analysis of glioblastoma cells revealed immunopositivity for PHIP specifically at the leading edge of motile tumor cells. Prior studies had demonstrated that the PHIP protein localizes to the nucleus of pancreatic islet cells (13). The expression of PHIP at the leading edge of glioblastoma cells suggested its colocalization with various focal adhesion proteins, each with a demonstrated role in tumor progression (21–24). Beyond its colocalization with focal adhesion proteins, PHIP silencing resulted in significantly reduced expression of several focal adhesion proteins, suggesting an important role for PHIP in regulating assembly of the cytoskeletal apparatus. This hypothesis was initially suggested by a proteome profiler array, which showed a direct effect of PHIP suppression on pPXN and pFAK expression. Confocal microscopy indicated a precise colocalization of PHIP with TLN1 and VCL in the force transduction layer. Coimmunoprecipitation analysis revealed a direct physical interaction between PHIP and VCL, demonstrating a function for the PHIP protein in supporting the actin cytoskeleton, as well as providing a mechanistic basis for its regulation of glioblastoma motility and invasion.
The force transduction at the leading edge of migrating cells is regulated by an organized and conserved nanoscale architecture of a “molecular clutch,” consisting of the structural proteins TLN and VCL, as well as other mechanosensitive protein–protein interactions (15). TLN-mediated integrin activation is well understood, as is TLN’s role in the formation of focal adhesions with PXN and VCL (25). In addition, TLN1 protects β1-integrins from proteasomal degradation (26). Integrin immobilization in focal adhesions requires simultaneous binding to both TLN and the ECM. As TLN binds F-actin flowing away from the leading edge, it activates integrins and triggers the assembly of dynamic nascent focal adhesions. TLN plays vital roles in linking activated integrins to the actin cytoskeleton, sensing and reinforcing the response to mechanical force and regulating focal adhesion formation and turnover. Depletion of TLN in glioblastoma cells resulted in decreased traction forces and migratory capability (27). In prostate cancer cells, TLN1 regulated adhesion, migration, and invasion (28). Previously, TLN1 expression was significantly suppressed following PHIP silencing in melanoma, breast, and lung cancer cells (10) and has now been extended to glioblastoma cells. In addition, overexpression of TLN1 cDNA alone rescued the invasive capability of U-251 cells following PHIP knockdown.
VCL is a highly conserved cytoskeletal scaffold protein that localizes to focal adhesions and is essential to regulating focal adhesion assembly and disassembly. VCL interacts with many focal adhesion proteins. Its interaction with F-actin regulates motility, mechanotransduction, and adhesion dynamics at the leading edge of motile cells. VCL recruitment stabilizes focal adhesions by simultaneously binding to TLN and F-actin and helps to mechanically reinforce the talin–actin linkage. Expression of a mutant form of VCL resulted in reduced motile forces (29). Our demonstration of colocalization as well as a physical interaction between PHIP and VCL suggest a docking function for PHIP in the force transduction layer, providing a mechanism by which it supports the cytoskeletal apparatus.
FAK and PXN are also important components of focal adhesions, as focal adhesions containing ITGB1, FAK, and PXN form in the protruding lamellipodia. FAK-mediated phosphorylation of PXN is critical for focal adhesion turnover at the leading edge of migrating cells (30). PXN proteolysis has been shown to negatively regulate focal adhesion dynamics and cell migration (31). An important feature of this study was PHIP’s regulation of expression of several focal adhesion proteins, both at the RNA and protein levels. The PHIP protein contains two bromodomains, implicating it in chromatin remodeling (10, 32–35), thereby providing a potential mechanism by which PHIP can regulate expression of these focal adhesion proteins.
A strong link between PHIP and adhesion dynamics was demonstrated by the effects of the anti-PHIP shRNA on ZYX and stress fibers. ZYX is a zinc-binding phosphoprotein that concentrates at focal adhesions along the actin cytoskeleton, thereby functioning as a key messenger in the signal transduction pathway that mediates adhesion-stimulated changes in gene expression, resulting in the cytoskeletal organization of actin bundles (36, 37). PHIP’s regulation of ZYX suggests not only that the recruitment of ZYX along the actin filaments can be orchestrated by PHIP, but also indicates a more broad-based role for PHIP in modulating the mechanotransduction signaling cascade required for glioblastoma cell motility. Previously, it has been shown that suppression of ZYX expression can result in tumor regression by affecting cytoarchitecture and motility in oral squamous cell carcinoma cells (38). The effects on stress fiber organization observed here following PHIP targeting are analogous to those described following ZYX targeting. Further, live-cell imaging of ZYX-GFP–expressing glioblastoma cells showed a profound disorganization of ZYX spikes and stress fibers following PHIP silencing, demonstrating an important role for PHIP in regulating focal adhesion dynamics in glioblastoma cells. As rapid actin polymerization is the major force that regulates cell shape and migration (39), the changes in the actin cytoskeleton following PHIP silencing identify PHIP as a cell adhesion effector protein that, by regulating the expression and recruitment of focal adhesion proteins, can drive glioblastoma cell migration and invasion. Intriguingly, PHIP’s ortholog in Drosophila has also been implicated in migration (40), suggesting the evolutionary conservation of PHIP’s role in regulation of cell motility.
Beyond its impact on glioblastoma cell motility and invasion, targeted suppression of PHIP also resulted in significant suppression of glioblastoma proliferation. This was accompanied by reduced expression of pAKT and CCND1, providing a pathway by which PHIP promotes tumor cell proliferation. Further evidence for a role for PHIP in glioblastoma progression was provided by the dramatic reductions observed in growth of U-251 cells following both s.c. and intracranial tumor cell implantation. The in vivo suppression of glioblastoma growth was associated with significantly reduced microvessel density, further supported by reduced VEGF levels. Ample evidence exists to link IGF1R/PI3K pathway activation to promotion of tumor angiogenesis (41). Thus, PHIP promotes multiple hallmarks of glioblastoma aggressiveness, including tumor cell motility and invasion, as well as proliferative and angiogenic potential.
Taken together, our observations identify PHIP as a key mediator of glioblastoma progression, as well as a rational therapeutic target, given the identification of small molecule inhibitors of PHIP (42). Our results were further supported by analysis of PHIP levels in brain tissues, demonstrating high PHIP expression only in glioblastomas, as opposed to normal brain tissue or low-grade gliomas. Moreover, PHIP copy number was significantly different in the three molecular subtypes of glioblastoma and was enriched in the classical subtype (3, 7). Accordingly, there was a significant increase in PHIP copy number in tumors with either elevated EGFR copy number or expression. These studies using fresh frozen primary tumor samples further demonstrate PHIP’s importance to glioblastoma pathophysiology and were supported by functional studies assessing regulation of PHIP expression in a low-passage classical glioblastoma line. Furthermore, EGF stimulation of glioblastoma cells resulted in increased PHIP expression. Since targeting of EGFR in glioblastoma patients has not yet demonstrated a clinical benefit (43), our studies suggest the potential therapeutic utility of cotargeting PHIP and EGFR. Ultimately, whether PHIP targeting prolongs survival associated with glioblastoma awaits the further development of pharmacological inhibitors of PHIP.
In conclusion, our results show important mechanistic roles for PHIP in regulating tumor cell motility and invasion, processes fundamental to the aggressive phenotype of glioblastoma. These functions are facilitated by PHIP copy number elevations, as well as PHIP’s localization to the leading edge of tumor cells, enabling its interaction with VCL and its activation of the focal adhesion apparatus. These studies assign an important role to PHIP in regulating focal adhesion dynamics and identify PHIP as a rational therapeutic target to block the lethal invasive progression of glioblastoma.
Materials and Methods
Experimental details and methods can be found in SI Appendix, including sources of cell lines and cell culture conditions, assays of cell cycle, proliferation, migration and invasion, transfections, construction of lentiviral vectors and procedures for virus production, infection and generation of stable transformants, and animal studies. Details of microscopy and image processing for FISH, live-cell imaging and image analysis using AxioVision software or ImageJ, and fixed-cell imaging and image analysis are described in SI Appendix. Methods for Western blot, cytoplasmic and nuclear fractionation, coimmunoprecipitation, qRT-PCR, ELISA, immunostaining of tissue arrays, human phospho-kinase profiling, and statistics are also described in SI Appendix.
Data Availability Statement.
All data generated in this study are included in the paper and SI Appendix.
Supplementary Material
Acknowledgments
This work was supported by National Institutes of Health Grants CA114337, CA122947, CA215755 (to M.K.-S.), and CA109171 (to R.J.D.); the Cancer Avatar Project at the CPMC Research Institute by the CPMC Charitable Foundation; and The Edward A. Dickson Emeritus Professorship Award of University California, San Francisco (to J.E.C.). We thank Dr. Jeremy Rich (School of Medicine at the University of California San Diego) for providing the 3832 primary glioblastoma cells.
Footnotes
Competing interest statement: C.C. and J.D.L. are coauthors on a 2018 research article.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.1914505117/-/DCSupplemental.
References
- 1.Siegel R. L., Miller K. D., Jemal A., Cancer statistics, 2019. CA Cancer J. Clin. 69, 7–34 (2019). [DOI] [PubMed] [Google Scholar]
- 2.Phillips H. S., et al. , Molecular subclasses of high-grade glioma predict prognosis, delineate a pattern of disease progression, and resemble stages in neurogenesis. Cancer Cell 9, 157–173 (2006). [DOI] [PubMed] [Google Scholar]
- 3.Verhaak R. G., et al. ; Cancer Genome Atlas Research Network , Integrated genomic analysis identifies clinically relevant subtypes of glioblastoma characterized by abnormalities in PDGFRA, IDH1, EGFR, and NF1. Cancer Cell 17, 98–110 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Parsons D. W., et al. , An integrated genomic analysis of human glioblastoma multiforme. Science 321, 1807–1812 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Westphal M., Lamszus K., The neurobiology of gliomas: From cell biology to the development of therapeutic approaches. Nat. Rev. Neurosci. 12, 495–508 (2011). [DOI] [PubMed] [Google Scholar]
- 6.Brennan C. W., et al. ; TCGA Research Network , The somatic genomic landscape of glioblastoma. Cell 155, 462–477 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wang Q., et al. , Tumor evolution of glioma-intrinsic gene expression subtypes associates with immunological changes in the microenvironment. Cancer Cell 32, 42–56.e6 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Network T. C. G. A. R.; Cancer Genome Atlas Research Network , Comprehensive genomic characterization defines human glioblastoma genes and core pathways. Nature 455, 1061–1068 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Onishi M., Ichikawa T., Kurozumi K., Date I., Angiogenesis and invasion in glioma. Brain Tumor Pathol. 28, 13–24 (2011). [DOI] [PubMed] [Google Scholar]
- 10.de Semir D., et al. , PHIP as a therapeutic target for driver-negative subtypes of melanoma, breast, and lung cancer. Proc. Natl. Acad. Sci. U.S.A. 115, E5766–E5775 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.De Semir D., et al. , Pleckstrin homology domain-interacting protein (PHIP) as a marker and mediator of melanoma metastasis. Proc. Natl. Acad. Sci. U.S.A. 109, 7067–7072 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bezrookove V., et al. , Prognostic impact of PHIP copy number in melanoma: Linkage to ulceration. J. Invest. Dermatol. 134, 783–790 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Podcheko A., et al. , Identification of a WD40 repeat-containing isoform of PHIP as a novel regulator of beta-cell growth and survival. Mol. Cell. Biol. 27, 6484–6496 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Metalli D., et al. , The insulin-like growth factor receptor I promotes motility and invasion of bladder cancer cells through Akt- and mitogen-activated protein kinase-dependent activation of paxillin. Am. J. Pathol. 176, 2997–3006 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Case L. B., Waterman C. M., Integration of actin dynamics and cell adhesion by a three-dimensional, mechanosensitive molecular clutch. Nat. Cell Biol. 17, 955–963 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Roycroft A., Mayor R., Forcing contact inhibition of locomotion. Trends Cell Biol. 25, 373–375 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zaidel-Bar R., Ballestrem C., Kam Z., Geiger B., Early molecular events in the assembly of matrix adhesions at the leading edge of migrating cells. J. Cell Sci. 116, 4605–4613 (2003). [DOI] [PubMed] [Google Scholar]
- 18.Liang J., Slingerland J. M., Multiple roles of the PI3K/PKB (Akt) pathway in cell cycle progression. Cell Cycle 2, 339–345 (2003). [PubMed] [Google Scholar]
- 19.Bezrookove V., et al. , Role of elevated PHIP copy number as a prognostic and progression marker for cutaneous melanoma. Clin. Cancer Res. 24, 4119–4125 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Duong H., Han M., A multispectral LED array for the reduction of background autofluorescence in brain tissue. J. Neurosci. Methods 220, 46–54 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sulzmaier F. J., Jean C., Schlaepfer D. D., FAK in cancer: Mechanistic findings and clinical applications. Nat. Rev. Cancer 14, 598–610 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Desgrosellier J. S., Cheresh D. A., Integrins in cancer: Biological implications and therapeutic opportunities. Nat. Rev. Cancer 10, 9–22 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Desiniotis A., Kyprianou N., Significance of talin in cancer progression and metastasis. Int. Rev. Cell Mol. Biol. 289, 117–147 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kato H., et al. , The primacy of β1 integrin activation in the metastatic cascade. PLoS One 7, e46576 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Calderwood D. A., Campbell I. D., Critchley D. R., Talins and kindlins: Partners in integrin-mediated adhesion. Nat. Rev. Mol. Cell Biol. 14, 503–517 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Liu J., et al. , Talin1 regulates integrin turnover to promote embryonic epithelial morphogenesis. Mol. Cell. Biol. 31, 3366–3377 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sen S., Ng W. P., Kumar S., Contributions of talin-1 to glioma cell-matrix tensional homeostasis. J. R. Soc. Interface 9, 1311–1317 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sakamoto S., McCann R. O., Dhir R., Kyprianou N., Talin1 promotes tumor invasion and metastasis via focal adhesion signaling and anoikis resistance. Cancer Res. 70, 1885–1895 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jannie K. M., et al. , Vinculin-dependent actin bundling regulates cell migration and traction forces. Biochem. J. 465, 383–393 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Webb D. J., et al. , FAK-Src signalling through paxillin, ERK and MLCK regulates adhesion disassembly. Nat. Cell Biol. 6, 154–161 (2004). [DOI] [PubMed] [Google Scholar]
- 31.Cortesio C. L., Boateng L. R., Piazza T. M., Bennin D. A., Huttenlocher A., Calpain-mediated proteolysis of paxillin negatively regulates focal adhesion dynamics and cell migration. J. Biol. Chem. 286, 9998–10006 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jang S. M., et al. , The replication initiation determinant protein (RepID) modulates replication by recruiting CUL4 to chromatin. Nat. Commun. 9, 2782 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ji X., et al. , Chromatin proteomic profiling reveals novel proteins associated with histone-marked genomic regions. Proc. Natl. Acad. Sci. U.S.A. 112, 3841–3846 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Filippakopoulos P., et al. , Histone recognition and large-scale structural analysis of the human bromodomain family. Cell 149, 214–231 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Morgan M. A. J., et al. , A cryptic Tudor domain links BRWD2/PHIP to COMPASS-mediated histone H3K4 methylation. Genes Dev. 31, 2003–2014 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yoshigi M., Hoffman L. M., Jensen C. C., Yost H. J., Beckerle M. C., Mechanical force mobilizes zyxin from focal adhesions to actin filaments and regulates cytoskeletal reinforcement. J. Cell Biol. 171, 209–215 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hirata H., Tatsumi H., Sokabe M., Zyxin emerges as a key player in the mechanotransduction at cell adhesive structures. Commun. Integr. Biol. 1, 192–195 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yamamura M., et al. , Functional analysis of Zyxin in cell migration and invasive potential of oral squamous cell carcinoma cells. Int. J. Oncol. 42, 873–880 (2013). [DOI] [PubMed] [Google Scholar]
- 39.Pollard T. D., Cooper J. A., Actin, a central player in cell shape and movement. Science 326, 1208–1212 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bai S. W., et al. , Identification and characterization of a set of conserved and new regulators of cytoskeletal organization, cell morphology and migration. BMC Biol. 9, 54 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Karar J., Maity A., PI3K/AKT/mTOR pathway in angiogenesis. Front. Mol. Neurosci. 4, 51 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Cox O. B., et al. , A poised fragment library enables rapid synthetic expansion yielding the first reported inhibitors of PHIP(2), an atypical bromodomain. Chem. Sci. 7, 2322–2330 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Westphal M., Maire C. L., Lamszus K., EGFR as a target for glioblastoma treatment: An unfulfilled promise. CNS Drugs 31, 723–735 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data generated in this study are included in the paper and SI Appendix.