Significance
The mechanisms by which autoimmunity is triggered/amplified in T1D remain to be clarified, and the lack of this basic understanding hampers efforts to prevent or arrest the disease. Testing the hypothesis that T1D-associated genetic variants in lncRNAs affect important pathways that modify inflammation and pancreatic β-cell survival will increase our understanding of T1D pathogenesis. In the present paper, we describe a molecular mechanism by which the T1D-associated lncRNA Lnc13 regulates pancreatic β-cell inflammation. These findings provide information on the molecular mechanisms by which disease-associated SNPs in lncRNAs influence disease pathogenesis and open the door for the development of novel diagnostic and therapeutic approaches based on lncRNA targeting.
Keywords: type 1 diabetes, polymorphism, lncRNA, pancreatic β-cell, inflammation
Abstract
The vast majority of type 1 diabetes (T1D) genetic association signals lie in noncoding regions of the human genome. Many have been predicted to affect the expression and secondary structure of long noncoding RNAs (lncRNAs), but the contribution of these lncRNAs to the pathogenesis of T1D remains to be clarified. Here, we performed a complete functional characterization of a lncRNA that harbors a single nucleotide polymorphism (SNP) associated with T1D, namely, Lnc13. Human pancreatic islets harboring the T1D-associated SNP risk genotype in Lnc13 (rs917997*CC) showed higher STAT1 expression than islets harboring the heterozygous genotype (rs917997*CT). Up-regulation of Lnc13 in pancreatic β-cells increased activation of the proinflammatory STAT1 pathway, which correlated with increased production of chemokines in an allele-specific manner. In a mirror image, Lnc13 gene disruption in β-cells partially counteracts polyinosinic-polycytidylic acid (PIC)-induced STAT1 and proinflammatory chemokine expression. Furthermore, we observed that PIC, a viral mimetic, induces Lnc13translocation from the nucleus to the cytoplasm promoting the interaction of STAT1 mRNA with (poly[rC] binding protein 2) (PCBP2). Interestingly, Lnc13-PCBP2 interaction regulates the stability of the STAT1 mRNA, sustaining inflammation in β-cells in an allele-specific manner. Our results show that the T1D-associated Lnc13 may contribute to the pathogenesis of T1D by increasing pancreatic β-cell inflammation. These findings provide information on the molecular mechanisms by which disease-associated SNPs in lncRNAs influence disease pathogenesis and open the door to the development of diagnostic and therapeutic approaches based on lncRNA targeting.
T1D is a chronic autoimmune disease in which insulin-producing pancreatic β-cells are specifically destroyed by the immune system. During the initial stages of the disease, pancreatic islets are infiltrated by immune cells leading to the generation of an inflammatory environment that promotes pancreatic β-cell destruction (1). Local islet inflammation (insulitis) is partially driven by a “dialog” between β-cells and the infiltrating immune cells, through the release of proinflammatory chemokines and cytokines by both β-cells and immune cells. Increasing evidence suggests that the triggering of insulitis depends on the interaction between T1D candidate genes (2) and environmental factors, such as viral infections (3, 4). The molecular mechanisms by which viruses induce an autoimmune response targeting β-cells remain unclear, however, genetically regulated host-immune response could play an important role in β-cell demise through the stimulation of immune autoreactivity, first triggered and then maintained by potential persistent infections in the pancreas (3–5). Although virus-induced activation of specific transcription factors (IRF7, NFκB, STAT1, and STAT2, among others) is certainly a contributory factor to gene expression changes associated with β-cell failure (3, 5), recent studies indicate that a new class of gene-regulatory molecules named lncRNAs actively participate in the regulation of innate immune responses in different cell types (6, 7).
LncRNAs are noncoding RNAs that are 200 nucleotides or longer in transcript length and whose structure is similar to the one of protein-coding genes (8). Several studies have implicated lncRNAs in a wide range of biological and cellular processes, including the modulation of the innate antiviral immune response through the activation of pattern recognition receptor‐related signal transduction (9), the regulation of innate immune‐associated chemokines (10), and other inflammatory genes (11). LncRNAs regulate a variety of normal immune responses, but there is limited information on their implication in the development of autoimmunity, and only few lncRNAs have been identified in this context (12).
Interestingly, the majority of immune disease-related genome-wide association signals map to noncoding regions of the genome, including lncRNAs. This suggests that risk variants located in lncRNAs could play regulatory roles, altering their function and leading to dysregulated expression of genes or gene networks potentially important for the development of autoimmunity.
Lnc13 is a lncRNA located in chromosome 2q12 that harbors a polymorphism (rs917997) associated with several autoimmune diseases, including celiac disease (CeD) (13), inflammatory bowel disease (14), and T1D (15). The expression level of this lncRNA is significantly decreased in small intestinal biopsy samples from patients with CeD, and a recent study described that Lnc13 regulates the expression of a subset of CeD-associated inflammatory genes through interaction with heterogeneous nuclear ribonucleoprotein D (16). Interestingly, while the risk signal in CeD corresponds to the rs917997*T allele, the opposite allele (rs917997*C) is the risk allele in T1D, suggesting that the single nucleotide polymorphism (SNP) may alter the function of this lncRNA in a tissue- or disease-specific mode.
In the current study, we describe a molecular mechanism by which T1D-associated Lnc13 regulates virus-induced pancreatic β-cell inflammation. We observed that Lnc13 acts as a linker between the RNA binding protein PCBP2 and the 3′UTR of STAT1 to stabilize the mRNA of STAT1. This stabilization leads to increased activation of the STAT1 proinflammatory pathway, contributing to local inflammation and eventual β-cell destruction.
In conclusion, our results show that Lnc13 participates in pancreatic β-cell inflammation via regulation of the STAT1 signaling pathway. This pathway plays a crucial role in T1D-related β-cell dysfunction and death, therefore, the ability of Lnc13 to influence STAT1 activation supports its role in the pathogenesis of the disease. These results serve as a proof of concept of the role of T1D-associated lncRNAs in the progressive loss of pancreatic β-cells in T1D and open an avenue for the development of therapeutic approaches based on lncRNA expression modification.
Results
Lnc13 Is Expressed in Human Pancreatic β-Cells, Is Up-Regulated by Viral Infections, and Correlates with STAT1 Expression in Human Pancreatic Islets.
We first evaluated the expression of Lnc13 in the human pancreatic β-cell line EndoC-βH1 compared with a set of human tissues. As previously described (16), Lnc13 is ubiquitously expressed, albeit at various levels in several human tissues (Fig. 1A). The expression of Lnc13 in the human EndoC-βH1 is around 20 times higher than the most Lnc13-expressing human tissues (heart, liver, and muscle), likely due to the embryonic nature of the cell line. In contrast, expression of Lnc13 in pancreatic human islets in which the typical proportion of β-cells is ∼50–60% (17) is similar to that of the thymus, colon, and lung (Fig. 1A). Mining of publicly available RNA sequencing data in a set of human primary cells confirmed that Lnc13 is expressed in most of the cell types, although its expression level is very low (less than two transcripts per million) (SI Appendix, Fig. S1A, upper graph). In terms of Lnc13 copies per cell, EndoC-βH1 cells harbor more Lnc13 molecules per cell (mean ± SEM; 2.78 ± 0.31 copies/cell) than other cell lines, such as HEK293 (kidney; mean ± SEM; 0.98 ± 0.49 copies/cell) and SHSY5Y (brain; mean ± SEM; 0.87 ± 0.29 copies/cell), among others (SI Appendix, Fig. S1B). These results are in concordance with previous studies showing that the majority of lncRNAs are of very low abundance, having a copy number even lower than one per cell (18–20).
Fig. 1.
Lnc13 is expressed in pancreatic β-cells, up-regulated by viral infections, and correlated with STAT1 expression in human pancreatic islets. (A) Lnc13 expression was analyzed in the human β-cell line EndoC-βH1, in human pancreatic islets, and in a set of human tissues (heart, liver, muscle, brain, spleen, kidney, intestine, colon, thymus, lung, and stomach). Lnc13 expression was determined by Q-PCR and normalized by the housekeeping gene β-actin. Results are means ± SEM of three experimental replicates. (B) Human EndoC-βH1 cells were left untransfected (—white bars) or transfected with PIC (1 µg/mL; gray bars) for 24 h. Expression of Lnc13, STAT2, and IRF7 (positive controls), INS and PAX6 (negative controls) was assayed by Q-PCR and normalized by the housekeeping gene β-actin. Results are means ± SEM of four independent experiments; ***P < 0.001 vs. untreated cells; Student’s t test. (C) Human EndoC-βH1 cells were left uninfected (—) or infected with the CVB5 (CVB5; MOI 5) for 24 h. Lnc13 expression was determined by Q-PCR and normalized by the housekeeping gene β-actin. Results are means ± SEM of five independent experiments; **P < 0.01; Student’s t test. (D) EndoC-βH1s were kept untreated (i.e., 0 h) or treated with PIC for 8 or 24 h. Relative Lnc13 and STAT1 expressions were determined by Q-PCR and normalized by the housekeeping gene β-actin. Results are means ± SEM of six independent experiments; ***P < 0.001 vs. time 0 h; Student’s t test. (E) Human EndoC-βH1 cells were left uninfected (—) or infected with the CVB5 (CVB5) for 24 h. STAT1 expression was determined by Q-PCR and normalized by the housekeeping gene β-actin. Results are means ± SEM of five independent experiments. **P < 0.01; Student’s t test. (F) Lnc13 and STAT1 expressions were determined in 43 human pancreatic islets of nondiabetic individuals. Expression values were normalized by the housekeeping gene β-actin. Spearman’s correlation analysis was performed to check for correlation between Lnc13 and STAT1 expression values; Spearman’s R = 0.51 (0.24–0.0.71); P < 0.001.
Proinflammatory stimuli, such as proinflammatory cytokines and viral infections have been shown to play a crucial role in pancreatic β-cell demise through activation of several inflammation- and apoptosis-related pathways (21–23). The proinflammatory cytokines IL-1β+IFNγ did not modify Lnc13 expression in human pancreatic β-cells (SI Appendix, Fig. S2A). However, intracellular PIC, a synthetic mimic of viral double-stranded RNA (dsRNA), up-regulated Lnc13 expression by around 42% (Fig. 1B). Intracellular PIC-induced Lnc13 up-regulation was similar to that observed in other known PIC-inducible genes (STAT2 and IRF7) (Fig. 1B). In addition, we observed that infection of pancreatic human islets with the diabetogenic Coxsackie Virus B5 ([CVB5]; multiplicity of infection [MOI] of 5) for 24 h led to a 2.5-fold increase in Lnc13 expression (Fig. 1C). Previous studies have demonstrated that interaction of the mRNA decapping protein DCP2 with Lnc13 under inflamed conditions contributes to the degradation of Lnc13. Indeed, in bone marrow derived macrophages from DCP2 knockout mice, the Lnc13 transcript was up-regulated after lipopolysaccharide stimulation, strongly suggesting that DCP2 recognized Lnc13 and caused its degradation (16). In accordance with the increase in Lnc13 expression, we observed that intracellular PIC exposure for 8 and 24 h led to a significant decrease in the expression of DCP2 (SI Appendix, Fig. S2B), suggesting that PIC-induced DCP2 decay facilitates Lnc13 up-regulation as previously shown in other models (16). However, infection with CVB5 in human pancreatic islets did not alter DCP2 expression (SI Appendix, Fig. S2C), probably due to the ability of some RNA viruses to sequester the cellular decay machinery to increase their infectivity, which will, in turn, reduce DCP2 function (24). In addition, we observed a very close correlation between the PIC-induced Lnc13 up-regulation and the increased mRNA expression of the proinflammatory transcription factor STAT1 (r2 = 0.99; P < 0.05) (Fig. 1D). Moreover, CVB5-induced Lnc13 up-regulation in human islets was also accompanied with a 2.8-fold increase in STAT1 mRNA expression when compared to noninfected islets (Fig. 1E). Interestingly, STAT1 expression levels in primary human cells (SI Appendix, Fig. S1A, lower graph) were correlated with Lnc13 expression (SI Appendix, Fig. S1C). In line with these results, determination of Lnc13 and STAT1 expression levels in human pancreatic islets from 43 individuals revealed that, in general, islets expressing higher amounts of Lnc13 also had higher expression levels of STAT1 (Spearman’s R = 0.51 [0.24–0.71]; P < 0.001) (Fig. 1F).
The T1D-Associated SNP Risk Genotype in the Lnc13 Gene Correlates with Increased STAT1 Expression in Human Pancreatic Islets.
To determine whether the genotype of the T1D-associated SNP in Lnc13 affects STAT1 expression in human pancreatic islets, we next genotyped rs917997 in complementary DNA (cDNA) samples from human pancreatic islets. Out of these 43 samples, 15 were homozygous for the risk allele rs917997*C, two samples were homozygous for the protective allele rs917997*T and 26 samples were heterozygous (CT). As shown in Fig. 2A, human islets from individuals homozygous for the T1D risk allele rs917997*C showed higher STAT1 mRNA expression levels as compared to individuals heterozygous (CT) or homozygous for the T1D protective allele rs917997*T.
Fig. 2.
The T1D-associated SNP genotype in the Lnc13 gene correlates with STAT1 expression in human pancreatic islets, and overexpression of Lnc13 in β-cells leads to an up-regulation of STAT1 mRNA expression in an allele-specific manner. (A) Expression of STAT1 in human pancreatic islets stratified by the genotype of the T1D-associated SNP rs917997 in the Lnc13 gene. Results are means ± SEM of 15 samples with the homozygous risk genotype (CC), 26 samples with the heterozygous genotype (CT), and two samples with the homozygous protective genotype (TT) genotype. **P < 0.01; Student’s t test. (B and C) EndoC-βH1 cells were left untransfected (NT) or transfected with pCMV6, pLnc13-C, or pLnc13-T. After 48 h, expression levels of Lnc13 (B) and STAT1 (C) were determined by Q-PCR and normalized by the housekeeping gene β-actin. Results are means ± SEM of four independent experiments; ***P < 0.001 and *P < 0.05 vs. pCMV6-transfected cells; †††P < 0.001 vs. pLnc13-C-transfected cells; Student’s t test. (D) STAT1 mRNA expression in pLnc13-C- and pLnc13-T-transfected β-cells corrected by Lnc13 expression values to control for differences in Lnc13 allele stability. Results are means ± SEM of four independent experiments; **P < 0.01; Student’s t test.
Lnc13 Overexpression Activates the STAT1 Signaling Pathway and Increases Production of Proinflammatory Chemokines in an Allele-Specific Manner.
In order to clarify the role of Lnc13 variants in STAT1 up-regulation, we overexpressed either the T1D risk allele rs917997*C (pLnc13-C) or the nonrisk allele rs917997*T (pLnc13-T) in EndoC-βH1 cells. Both, the Lnc13-C and the Lnc13-T, increased Lnc13 mRNA expression by 783- and 2,000-fold, respectively (Fig. 2B) which were paralleled by a drastic up-regulation of STAT1 mRNA expression when compared to nontransfected or pCMV6-transfected pancreatic β-cells (Fig. 2C). The increase in transcript levels correlated with increased levels of total STAT1 and phosphorylated STAT1 (SI Appendix, Fig. S3), suggesting an increased activation of the STAT1 signaling pathway even in the absence of proinflammatory stimuli.
The plasmid harboring the nonrisk allele T (pLnc13-T) increased Lnc13 mRNA expression more efficiently than the plasmid containing the risk allele C (pLnc13-C) (Fig. 2B). Thus, to control for differences in Lnc13-overexpression levels, we next normalized STAT1 expression by Lnc13 expression values. Interestingly, this normalization revealed that STAT1 mRNA expression was exacerbated in pLnc13-C-transfected β-cells when compared to pLnc13-T-transfected ones (Fig. 2D). These results suggest that Lnc13 harboring the risk allele for T1D (C allele) induces a more pronounced increase in STAT1 expression than Lnc13 containing the nonrisk allele.
To evaluate the effect of Lnc13 overexpression on STAT1-regulated proinflammatory chemokine expression, we next determined the expression of CXCL10, CXCL9, CCL5, and CXCL1 in Lnc13-overexpressing EndoC-βH1 cells. Lnc13 overexpression (T and C) led to an increase in all proinflammatory chemokine transcripts (SI Appendix, Fig. S4 A–D). Similar to STAT1, transcript levels for CXCL10, CXCL9, CCL5, and CXCL1 were higher in pLnc13-C-transfected cells compared to pLnc13-T-transfected ones (Fig. 3 A–D), suggesting that Lnc13 harboring the T1D-associated rs917997*C allele promotes the expression of proinflammatory chemokines more efficiently than the Lnc13 containing the nonrisk allele (rs917997*T).
Fig. 3.
Overexpression of Lnc13 up-regulates proinflammatory chemokine production in an allele-specific manner in pancreatic β-cells through STAT1 signaling activation. (A–D) EndoC-βH1 cells were transfected with pLnc13-C or pLnc13-T, and mRNA levels of CXCL10, CXCL9, CCL5, and CXCL1 were determined by Q-PCR and normalized by the housekeeping gene β-actin and corrected by Lnc13 expression values to control for differences in Lnc13 allele stability. Results are means ± SEM of four independent experiments; **P < 0.01 and *P < 0.05 vs. pLnc13-C-transfected cells; Student’s t test. (E and F) EndoC-βH1 cells were transfected with pLnc13-C or pLnc13-T, and CXCL10 (E), and CCL5 (F) protein release to the medium was determined by ELISA in cell supernatants. CXCL10 and CCL5 values (pg/mL) were corrected by Lnc13 expression values to control for differences in Lnc13 allele stability. Results are means ± SEM of six and three independent experiments, respectively. *P < 0.05 vs. pLnc13-C-transfected cells; Student’s t test. (G) Human EndoC-βH1 cells were transfected with pCMV6 or with pLnc13-C and, subsequently, left untreated (NT), treated with intracellular PIC (1 µg/mL) for 24 h (PIC), or treated with PIC and ruxolitinib for 24 h (PIC + Inhib). CXCL10 mRNA expression was measured by Q-PCR and normalized by the housekeeping gene β-actin. The results are means ± SEM of three independent experiments; **P < 0.01 and *P < 0.05 vs. NT and transfected with the same plasmid; †††P < 0.001, ††P < 0.01, and †P < 0.05 as indicated; ANOVA followed by Student’s t test followed with Bonferroni correction. (H) EndoC-βH1s were transfected with pCMV6 or pLnc13-C, chromatin was fragmented and precipitated with anti-STAT1 or anti-IgG (as the negative control), and CXCL10 promoter or a control region (Oct4 gene body) was amplified by Q-PCR. Results are means ± SEM of four independent experiments; *P < 0.05 as indicated; ANOVA followed by Student’s t test followed with Bonferroni correction. (I) Supernatants of pCMV6- or pLnc13-C-transfected EndoC-βH1 cells were used to determine chemotactic migration of Jurkat cells using a transwell system and a fluorescence-based assay. Supernatant of PIC-transfected cells was used as the positive control. The results are means ± SEM of three independent experiments. *P < 0.05 vs. pCMV6-transfected cells; Student’s t test.
The up-regulation of CXCL10 and CCL5 mRNA expression in Lnc13-overexpressing β-cells was confirmed at the protein level (SI Appendix, Fig. S4 E and F). Similar to the results at the mRNA level, Lnc13-C-overexpressing cells secreted higher amounts of CXCL10 and CCL5 per Lnc13 molecule than the Lnc13-T-overexpressing cells (Fig. 3 E and F).
To determine whether Lnc13 affects PIC-induced proinflammatory chemokine expression through modulation of the STAT signaling pathway, we exposed Lnc13-C-overexpressing EndoC-βH1 cells to PIC in the absence or presence of ruxolitinib, a Janus kinase inhibitor (Inhib.). As shown in Fig. 3G, Lnc13-C overexpression enhanced PIC-induced CXCL10 mRNA expression, and this effect was counteracted by the presence of ruxolitinib. In addition, we observed that, in Lnc13-C-overexpressing β-cells, binding of the STAT1 transcription factor to the CXCL10 promoter was increased when compared to pCMV6-transfected control cells (Fig. 3H), confirming that the up-regulation of chemokine expression in Lnc13-overexpressing β-cells is driven by increased activation of the STAT1 signaling pathway.
To determine the biological relevance of Lnc13 up-regulation-induced chemokine production, we performed a chemotactic migration assay by incubating activated Jurkat T cells with supernatants of pCMV6- or pLnc13-C-transfected β-cells. As shown in Fig. 3I, supernatants of Lnc13-C-overexpressing β-cells induce a higher migration of Jurkat T cells than the control supernatants.
We next disrupted the Lnc13 gene in the EndoC-βH1 cell line by generating a deletion of 1698 bp in the Lnc13 gene using the CRISPR-Cas9 technique (Fig. 4A). Due to the low proliferation rate and fragility of these cells, we did not obtain a homogenous Lnc13-disrupted β-cell line. Nevertheless, the mix population of Lnc13-disrupted cells (CRISPR-Lnc13) showed a reduced expression of Lnc13 of about 60% (Fig. 4B). In a mirror image of the overexpression experiments, Lnc13 disruption partially counteracted the effect of intracellular PIC in STAT1, CXCL10, and CCL5 up-regulation (Fig. 4 C–E).
Fig. 4.
Lnc13 gene disruption in pancreatic β-cells partially counteracts the effect of PIC in STAT1 and proinflammatory chemokine expression up-regulation. (A) Lnc13 disruption was performed by generating a deletion of 1,698 bp using the CRISPR-Cas9 technique and single guide RNAs (sgRNAs) targeting the Lnc13 gene. The presence of the deletion was confirmed by PCR using a pair of primers located inside the deleted region (for detection of unedited cells; wild type forward (WTF) and wild type reverse (WTR)) and a pair of primers located outside the deleted region (for detection of edited cells; mutF and mutR). (B) EndoC-βH1 cells were transfected with an empty px459 vector (CRISPR-Ctrl) or with a vector harboring the Lnc13 targeting sgRNAs (CRISPR-Lnc13). Lnc13 expression was determined by Q-PCR and normalized by the housekeeping gene β-actin. The results are means ± SEM of three independent cell populations. **P < 0.01; Student’s t test. (C–E) Control and Lnc13-disrupted mixed cell populations were exposed to intracellular PIC for 24 h, STAT1 (C), CXCL10 (D), and CCL5 (E) expressions were determined by Q-PCR and normalized by the housekeeping gene β-actin. The results are represented as fold induction and are means ± SEM of three independent cell populations. ***P < 0.001, **P < 0.01 and *P < 0.05 vs. nontreated cells; †††P < 0.0001, and †P < 0.05 as indicated; Student’s t test.
Lnc13 Does Not Directly Regulate Transcription of the STAT1 Gene but Participates in Its mRNA Stabilization in an Allele-Specific Manner.
To determine the molecular mechanisms by which Lnc13 activates the STAT1 signaling pathway in pancreatic β-cells, we first analyzed the potential effect of Lnc13 on the transcriptional regulation of STAT1. Transcription of STAT1 is regulated by several transcription factors, including homodimers of phosphorylated STAT1 and heterodimers of phosphorylated STAT1/STAT2 or STAT1/STAT3, and, thus, increased phosphorylated STAT1 may lead to increased STAT1 transcription through an autoregulatory mechanism (25, 26). Taking into account that Lnc13 overexpression led to increased pSTAT1 (SI Appendix, Fig. S3), we first inhibited protein translation using cycloheximide to inhibit the synthesis of the STAT1 protein and determine whether Lnc13 by itself was promoting STAT1 expression up-regulation. To this end, after overexpressing Lnc13-C, we exposed β-cells to cycloheximide for 24 h and determined the expression levels of STAT1 total mRNA and the STAT1 primary transcript using specific primers. Cycloheximide treatment for 24 h efficiently decreased STAT1 protein amounts (SI Appendix, Fig. S5A). As shown in SI Appendix, Fig. S5B, cycloheximide treatment led to a fivefold decrease in STAT1 primary transcript expression but did not affect the expression level of STAT1 total mRNA. These data suggested that, in the absence of Lnc13-induced pSTAT1 protein synthesis, Lnc13 was not up-regulating STAT1 expression through a direct effect on its transcription and suggested an effect on the stabilization of already produced STAT1 mRNA molecules.
To analyze whether Lnc13 contributes to the stabilization of STAT1 mRNA molecules, we next analyzed the amount of STAT1 mRNA in pLnc13-C- or pLnc13-T-overexpressing β-cells in the absence or presence of actinomycin D, an inhibitor of transcription. Actinomycin D treatment revealed that Lnc13-C was less stable than Lnc13-T as exposure to actinomycin D for 6 h down-regulated Lnc13 expression by 40% in pLnc13-C-transfected β-cells but did not have any effect in pLnc13-T-transfected cells (SI Appendix, Fig. S5C). Despite being less stable, Lnc13-C showed an increased capacity to stabilize STAT1 mRNA when compared to Lnc13-T. Indeed, actinomycin D treatment did not affect STAT1 mRNA levels in pLnc13-C-transfected β-cells while pLnc13-T-transfected cells showed a 1.9-fold decrease in Lnc13-normalyzed STAT1 mRNA expression after 6 h of actinomycin D exposure (SI Appendix, Fig. S5D).
Lnc13 Interacts with PCBP2 in the Cytoplasm Leading to Enhanced STAT1 Expression in Response to Viral dsRNA.
Cellular location often determines the functional role of lncRNAs (27), so we next examined the subcellular localization of Lnc13 in nontreated and PIC-transfected pancreatic β-cells. Consistent with previous studies, we found that, in untreated cells, Lnc13 is preferentially expressed in the nucleus, while PIC transfection induces its translocation to the cytoplasm (Fig. 5A).
Fig. 5.
Upon exposure to PIC, Lnc13 translocates from the nucleus and facilitates the interaction between the PCBP2 and the 3′-UTR of STAT1 mRNA. (A) EndoC-βH1 cells were exposed to PIC (1 µg/mL) for 24 h, and relative Lnc13 expression was determined in nucleus and whole extracts. Expression of MALAT1 and RPLPO was used as a control for nucleus and whole cell fractions, respectively. Amounts of specific nuclear RNA were measured by Q-PCR and compared to the total amount of RNA in the whole cell. Results are represented as a logarithm (nucleus/whole) and are means ± SEM of three independent experiments; *P < 0.05; Student’s t test. (B) EndoC-βH1s were exposed to PIC (1 µg/mL) for 24 h and cytoplasmic (Cyt), nuclear (Nuc), and chromatin (Chro) fractions were purified. PCBP2 protein expression was determined in all cellular compartments by Western blot and protein expression of Hsp90, HDAC1, and H3 was used as a control for cytoplasmic, nuclear, and chromatin fractions, respectively. The results are representative of three independent experiments. (C) EndoC-βH1 cells were left nontransfected (NT) or transfected with PIC for 24 h. RNA immunoprecipitation was performed using a specific antibody for PCBP2 and Lnc13, and STAT1 expression was determined in PCBP2-bound RNA by Q-PCR. Results are means ± SEM of three independent experiments, and the amounts of Lnc13 and STAT1 are expressed as relative to the input. IgG was used as the negative control; ***P < 0.001 and **P < 0.01 as indicated; ANOVA followed by Student’s t test with Bonferroni correction. (D) In vitro transcribed biotinylated 3′-UTR region of STAT1 was incubated with cellular extracts overexpressing Lnc13-C+PCBP2, Lnc13-T+PCBP2, or PCBP2. Afterward, 3′-UTR-STAT1-bound proteins were purified using streptavidin beads, and PCBP2 and Hsp90 (as the negative control) were detected by Western blot. Incubation with streptavidin beads alone was used as the negative control. The results are representative of three independent experiments. (E) Densitometry results for purified PCBP2 amounts in RNA pull down experiments are represented as means ± SEM of three independent experiments. ANOVA followed by Student’s t test with Bonferroni correction; ***P < 0.001 and *P < 0.01 vs. negative control; †††P < 0.001 and †P < 0.05 as indicated. (F) Lnc13 antisense purification was performed in nontreated (NT) and PIC-treated nuclear and cytoplasmic fractions of EndoC-βH1 cells. Lnc13-bound 3′-UTR-STAT1 amounts were determined by Q-PCR using specific primers. A nonrelated similar lncRNA was used as the negative control (Ctrl). Results are represented as fold enrichment and are means ± SEM of four independent experiments. *P < 0.05 as indicated; ANOVA followed by Student’s t test with Bonferroni correction. (G) RNA-protein interaction assay. Cells were transfected with pPCBP2 alone, pPCBP2+pLnc13-C, pPCBP2+pLnc13-T, or pPCBP2+pLnc13-delSNP. Cell lysates were incubated with in vitro transcribed 3′-UTR-STAT1 molecules, and a native agarose gel electrophoretic mobility shift assay was performed (Lower). After electrophoresis, proteins were transferred to a nitrocellulose membrane for PCBP2 visualization (Upper). The 3′-UTR-STAT1 RNA molecule alone was loaded as the Ctrl. The results are representative of three independent experiments. (H) EndoC-βH1 cells were transfected with a plasmid encoding Lnc13-C or a mutant Lnc13 in which the region containing the T1D-associated SNP was deleted (pLnc13-delSNP). STAT1, CXCL10, and CCL5 expressions were determined by Q-PCR and normalized by the housekeeping gene β-actin and corrected by Lnc13 expression values. Results are means ± SEM of four independent experiments. P < 0.05 as indicated; Student’s t test.
Interestingly, previous data (16) showed that, in mouse macrophages, Lnc13 can bind PCBP2, a RNA-binding protein that participates in antiviral cellular responses through 3′-UTR stabilization of the STAT1 transcript (28). Exposure to intracellular PIC led to increased PCBP2 protein expression both in the nucleus and in the cytoplasm of EndoC-βH1, mirroring Lnc13 kinetics (Fig. 5B and SI Appendix, Fig. S6).
To test whether Lnc13 interacts with PCBP2 in human pancreatic β-cells, we performed a RNA immunoprecipitation assay using cellular extracts of nontreated and PIC-transfected EndoC-βH1 cells. Using a specific antibody against PCBP2, we isolated PCBP2-bound RNA molecules and determined Lnc13 levels by Q-PCR. As shown in Fig. 5C, Lnc13 exhibited modest binding to PCBP2 in untreated cells, and the amount of PCBP2-bound Lnc13 RNA increased by threefold after PIC treatment. Similarly, we observed that STAT1 mRNA was poorly bound to PCBP2 under basal conditions, and this interaction was enhanced 17-fold after intracellular PIC exposure (Fig. 5C).
To examine whether Lnc13 is needed to facilitate the binding between PCBP2 and STAT1, we next performed a RNA pulldown experiment. As shown in Fig. 5D, the interaction between the PCBP2 and the 3′-UTR of STAT1 was only detectable in the presence of Lnc13. Moreover, when the Lnc13 harboring the risk allele for T1D (rs917997*C) was overexpressed, there was a stronger interaction between the PCBP2 and the 3′-UTR-STAT1 than when the Lnc13 harboring the protective allele for T1D (rs917997*T) was overexpressed (Fig. 5E). These results suggest that Lnc13 acts as a linker between PCBP2 and STAT1, and that the presence of the risk allele in Lnc13 promotes a stronger interaction between these two molecules, inducing a more efficient stabilization of STAT1 mRNA.
To further confirm the interaction between the Lnc13 and the 3′-UTR of STAT1, we next performed a RNA antisense purification (RAP) experiment to purify Lnc13-bound RNA molecules in nontreated and PIC-treated nuclear and cytoplasmic fractions of pancreatic β-cells. Lnc13 was purified using biotinylated antisense complementary oligos (SI Appendix, Fig. S7A), and antisense oligos complementary to a similar lncRNA were used as the control (SI Appendix, Fig. S7B). As shown in Fig. 5F, both in nontreated and in PIC-treated nuclear fractions, the 3′-UTR of STAT1 was not bound to purified Lnc13 molecules. In contrast, in PIC-treated cytoplasmic fractions, the 3′-UTR of STAT1 was significantly bound to purified Lnc13 when compared to the control, suggesting that the interaction between the Lnc13 and the 3′-UTR of STAT1 occurs in the cytoplasm after PIC exposure.
Finally, to clarify whether the region containing the T1D-associated SNP in the Lnc13 molecule was implicated in the binding between the PCBP2 and the 3′-UTR-STAT1 RNA molecule, we generated a mutant Lnc13 (Lnc13-delSNP) that lacked a 507 bp region (1,771–2,278 bp) harboring the SNP (rs917997; 2058 bp) and performed a RNA-protein interaction assay using a native agarose gel electrophoretic mobility shift assay. As shown in Fig. 5G, we observed changes in the migration pattern of the in vitro transcribed 3′-UTR-STAT1 RNA molecule upon mixing with protein lysates from PCBP2+Lnc13-C-, PCBP2+Lnc13-T-, or PCBP2+Lnc13-delSNP-overexpressing cells. While the migration pattern was similar between the PCBP2+Lnc13-C- and the PCBP2+Lnc13-T-overexpressing cells, cells expressing the mutant Lnc13 (Lnc13-delSNP) presented a different migration profile, suggesting that the T1D-associated SNP region was implicated in the configuration of the Lnc13-STAT1-PCBP2 complex. Moreover, these results confirmed that, in the absence of Lnc13, PCBP2 did not bind to the 3′-UTR of STAT1 as the 3′UTR-STAT1 band was not detectable in cells in which only PCBP2 was overexpressed (Fig. 5G).
The implication of the T1D-associated SNP region in Lnc13 was further confirmed by analyzing the expression of STAT1, CXCL10, and CCL5 in Lnc13-C- and Lnc13-delSNP-overexpressing EndoC-βH1 cells. As shown in Fig. 5H, cells overexpressing the mutant Lnc13 expressed less STAT1, CXCL10, and CCL5 than cells overexpressing the wild-type Lnc13-C. Taking together, these results indicate that Lnc13 enhances STAT1 expression levels by stabilizing its mRNA in the cytoplasm through interaction with the RNA-binding protein PCBP2. Moreover, these results suggest that the region harboring the T1D-associated SNP in Lnc13 is important for the interaction between PCBP2 and 3′-UTR of STAT1, and, thus, for the up-regulation of STAT1 and proinflammatory chemokine expression in pancreatic β-cells.
Discussion
Most of the genetic variants associated with T1D detected through genome-wide association studies are located in noncoding regions of the genome, making the characterization of their functional impact on the pathogenesis of the disease challenging (29). Advances in the annotation of the human genome have revealed that many disease-associated SNPs are located within lncRNAs, affecting their function through the disruption of their secondary structure (30). In the context of T1D, previous studies have pointed out the potential role of lncRNAs in pancreatic islet biology and T1D pathogenesis (31, 32), but the precise mechanisms involved remain to be characterized.
Herein we highlight a molecular mechanism by which the T1D-associated Lnc13 modulates virus-induced inflammation in pancreatic β-cells, namely, by sustaining the activation of the STAT1 proinflammatory pathway in an allele-specific manner. We observed that viral dsRNA (a by-product of viral replication) and infection by CVB5 induces parallel up-regulation of Lnc13 and STAT1 expression in pancreatic β-cells. In line with this, human pancreatic islets expressing higher amounts of Lnc13 generally have increased mRNA expression of STAT1. Previous studies have demonstrated a link between activation of the STAT1 signaling pathway and T1D pathogenesis. Indeed, STAT1 and its downstream transcription factor IRF1 are aberrantly expressed in pancreatic islets of T1D patients (33, 34), and deletion of STAT1 in nonobese diabetic mice prevents T1D development (35), suggesting a key role for this proinflammatory pathway in the pathogenesis of the disease.
Overexpression of Lnc13 in pancreatic β-cells led to increased STAT1 activation and up-regulated proinflammatory chemokine expression, suggesting that Lnc13 plays a regulatory role in STAT1-driven inflammation at the pancreatic β-cell level. Indeed, Lnc13 disruption in pancreatic β-cells partially prevented viral dsRNA-induced inflammation as revealed by reduced STAT1 and chemokine up-regulation in Lnc13-disrupted β-cells.
Interestingly, we observed that pancreatic islets harboring the risk T1D genotype in Lnc13 (rs917997*CC) displayed higher STAT1 expression levels than islets carrying the heterozygous (rs917997*CT) or protective genotype (rs917997*TT). These data suggest that individuals with the risk genotype in Lnc13 express higher amounts of STAT1 in their islets, and this may contribute to increased β-cell inflammation and apoptosis observed in T1D pathogenesis (36). Indeed, Lnc13-C-overexpressing β-cells showed higher activation of STAT1 in comparison with Lnc13-T-overexpressing cells. In line with this increase in STAT1 signaling, Lnc13-C-overexpressing cells expressed and released higher amounts of proinflammatory chemokines. Expression of chemokines has been shown to be altered in several autoimmune diseases, including T1D (37, 38). Indeed, during early stages of T1D, proinflammatory chemokines released by pancreatic β-cells play a crucial role in attracting immune cells to the islets and generating a local inflammatory environment that eventually leads to pancreatic β-cell loss (1). In this context, the fact that Lnc13 up-regulation leads to increased STAT1 activation and chemokine production potentially rising T cell migration indicates its functional implication in the pathogenesis of the disease.
In contrast to T1D, in CeD where the disease-associated risk allele in Lnc13 is the T allele, down-regulation of Lnc13 in intestinal biopsies of active CeD patients correlated with increased expression of STAT1 and other proinflammatory genes (16). The functional characterization of Lnc13 in this model revealed that Lnc13 represses the expression of CeD-related proinflammatory genes via its interaction with two chromatin-associated proteins (hnRNPD and HDAC1) that negatively regulate transcription. The CeD risk allele in Lnc13 (allele T) decreased its affinity to bind hnRNPD and chromatin, resulting in higher expression of proinflammatory genes and thereby increasing the predisposition to develop CeD (16).
The results presented here demonstrate that the function of Lnc13 is cell and stimuli specific, but it is equally affected by the disease-associated SNP, impacting in the regulation of T1D-associated pathways at the pancreatic β-cell level. Indeed, the molecular mechanism by which Lnc13 regulates the STAT1 signaling pathway and inflammation in pancreatic β-cells is linked to a PCBP2-driven mRNA-stabilization mechanism in the cytoplasm. We found that, in the presence of a viral infection (a potential environmental trigger in T1D) (3, 39, 40), Lnc13 translocates from the nucleus to the cytoplasm acting as a linker between the 3′-UTR region of STAT1 and PCBP2, forming a complex that stabilizes the STAT1 mRNA molecule. It has been demonstrated that PCBP2 facilitates the antiviral activity of IFNα against the Hepatitis C virus by stabilizing the mRNA of STAT1 and STAT2 in hepatocyte cell lines (28). Stabilization of mRNA molecules usually implies the binding of PCBP2 to the 3′-UTRs (28, 41), but some studies have revealed that PCBP2 can also bind 5′-UTRs of mRNA molecules, inhibiting their translation (42).
We observed that Lnc13 harboring the risk allele for T1D (rs917797*C) promoted a stronger interaction between the PCBP2 and the 3′-UTR of the STAT1 mRNA molecule than Lnc13 carrying the protective allele (rs917797*T). As previously described, this autoimmune disease-associated SNP (rs917997) induces a change in the secondary structure of Lnc13 that most likely disrupts its function and capacity to bind specific proteins (16). Indeed, the function of lncRNAs relies in their ability to form different structures and induce molecular interactions with proteins and nucleic acids (43). In this sense, we have observed that deletion of the T1D-associated SNP region in the Lnc13 molecule alters the configuration of the Lnc13-STAT1-PCBP2 complex, affecting its capacity to promote STAT1 activation and β-cell inflammation.
As links between structure and function continue to emerge, it is increasingly apparent that polymorphisms or mutations located within lncRNAs contribute to disease pathogenesis through the dysregulation of disease-associated pathways (16, 44, 45). For example, lncRNA CCAT2, which harbors a cancer risk-associated SNP, regulates the metabolism of cancer cells through an interaction with the CFIm complex in an allele-specific manner (44). Furthermore, arteriosclerotic vascular disease-associated SNPs modifying the structure and splicing of the lncRNA ANRIL have been suggested to change its function and alter the expression of downstream inflammatory genes (45).
In conclusion, our results demonstrate that Lnc13 is implicated in the regulation of virus-induced inflammation in pancreatic β-cells through a molecular mechanism that implies allele-specific modulation of the STAT1 signaling pathway activation (Fig. 6). These findings provide information on the molecular mechanisms by which disease-associated SNPs in lncRNAs influence disease pathogenesis and open the door to the development of diagnostic and therapeutic approaches based on lncRNA targeting.
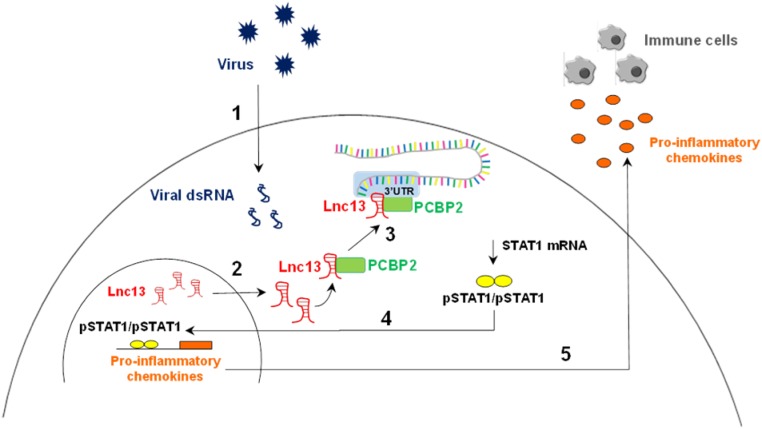
Fig. 6.
Lnc13 participates in virus-induced pancreatic β-cell inflammation by stabilizing STAT1 mRNA through interaction with the PCBP2 protein. Viral infections in pancreatic β-cells lead to the generation of viral dsRNA that is recognized by viral dsRNA cytoplasmic receptors (1). Viral dsRNA induces a decrease in DCP2 expression that leads to an increase in Lnc13 RNA levels. In addition, viral dsRNA provokes the translocation of Lnc13 from the nucleus to the cytoplasm (2). Once in the cytoplasm, Lnc13 interacts with PCBP2 to bind the 3′UTR of STAT1 mRNA and induce its stabilization (3). Increased STAT1 mRNA levels lead to increased protein and provide the substrate for generation of activated STAT1 (phosphorylated STAT1; pSTAT1). pSTAT1 forms homodimers and translocates to the nucleus, promoting the expression of several proinflammatory chemokines (4). Chemokines are then released by pancreatic β-cells, attracting immune cells into the islets (5). In genetically susceptible individuals harboring the T1D risk genotype in Lnc13 (rs917997*CC), the function of Lnc13 is affected, leading to an excessive antiviral inflammatory response that contributes to β-cell destruction in T1D.
Materials and Methods
For this study, the EndoC-βH1 human pancreatic β-cell line (Univercell Biosolutions, Paris, France), HEK-293 cell line (ATCC, CRL-1573), and JURKAT (ATCC, TIB-152) were used. cDNA samples from human pancreatic islets were obtained from two centers, namely, Cisanello University Hospital, Pisa, Italy (directly or via the ULB Center for Diabetes Research, Brussels, Belgium) and the Andalusian Center for Molecular Biology and Regenerative Medicine, CABIMER, Seville, Spain. Experiments using human islets were approved by the corresponding Ethical Committee.
The EndoC-βH1 human pancreatic β-cell line was transfected using synthetic viral dsRNA mimic PIC (InvivoGen, San Diego, CA). Lnc13 and coding gene expression levels were quantified by Q-PCR, starting from DNaseI (Qiagen) treated RNA using TaqMan expression assays (Applied Biosystems) or specific primers. Plasmids overexpressing Lnc13 were generated by cDNA cloning in a modified pCMV6 vector. For confirmation of Lnc13 binding to PCPB2 and/or 3′-UTR-STAT1, we used the RNA immunoprecipitation, RNA pulldown and RAP techniques. Mixed mutant cell populations with disrupted Lnc13 were generated using CRISPR-Cas9 system.
Data Availability.
All of the data, associated protocols, and materials for this study are available in the SI Appendix.
Supplementary Material
Acknowledgments
This work was supported by a Research Project Grant from the Basque Department of Health (2015111068) and a Research Grant from Fundación de la Sociedad Española de Diabetes (FSED) to I.S. I.G.-M. and A.O.-G. were supported by Predoctoral Fellowship Grants from the UPV/EHU (Universidad del Pais Vasco/Euskal Herriko Unibertsitatea) and the Basque Department of Education, respectively. A.C.-R. was funded by a Grant from the Spanish Ministry of Science, Innovation and Universities (PGC2018-097573-A-I00). D.L.E. received support from the Fonds National de la Recherche 420 Scientifique (FNRS), Welbio Grant CR-2015A-06. B.R.G. was funded by the Spanish Ministry of Science, Innovation and Universities; the European Fund for Regional Development (FEDER) (Grant BFU2017-83588-P); and the Juvenile Diabetes Research Foundation (Grants 17-2013-372 and 2-SRA-2019-837-S-B).
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission. K.A.F. is a guest editor invited by the Editorial Board.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.1914353117/-/DCSupplemental.
References
- 1.Eizirik D. L., Colli M. L., Ortis F., The role of inflammation in insulitis and beta-cell loss in type 1 diabetes. Nat. Rev. Endocrinol. 5, 219–226 (2009). [DOI] [PubMed] [Google Scholar]
- 2.Santin I., Eizirik D. L., Candidate genes for type 1 diabetes modulate pancreatic islet inflammation and β-cell apoptosis. Diabetes Obes. Metab. 15 (suppl. 3), 71–81 (2013). [DOI] [PubMed] [Google Scholar]
- 3.Dotta F., et al. , Coxsackie B4 virus infection of beta cells and natural killer cell insulitis in recent-onset type 1 diabetic patients. Proc. Natl. Acad. Sci. U.S.A. 104, 5115–5120 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Op de Beeck A., Eizirik D. L., Viral infections in type 1 diabetes mellitus–Why the β cells? Nat. Rev. Endocrinol. 12, 263–273 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rodriguez-Calvo T., Enterovirus infection and type 1 diabetes: Unraveling the crime scene. Clin. Exp. Immunol. 195, 15–24 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wang P., The opening of Pandora’s Box: An emerging role of long noncoding RNA in viral infections. Front. Immunol. 9, 3138 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lin H., et al. , The long noncoding RNA Lnczc3h7a promotes a TRIM25-mediated RIG-I antiviral innate immune response. Nat. Immunol. 20, 812–823 (2019). [DOI] [PubMed] [Google Scholar]
- 8.Ponting C. P., Oliver P. L., Reik W., Evolution and functions of long noncoding RNAs. Cell 136, 629–641 (2009). [DOI] [PubMed] [Google Scholar]
- 9.Jiang M., et al. , Self-recognition of an inducible host lncRNA by RIG-I feedback restricts innate immune response. Cell 173, 906–919.e13 (2018). [DOI] [PubMed] [Google Scholar]
- 10.Carpenter S., et al. , A long noncoding RNA mediates both activation and repression of immune response genes. Science 341, 789–792 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhang Q., et al. , The long noncoding RNA ROCKI regulates inflammatory gene expression. EMBO J. 38, e100041 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mirza A. H., Kaur S., Pociot F., Long non-coding RNAs as novel players in β cell function and type 1 diabetes. Hum. Genomics 11, 17 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hunt K. A., et al. , Newly identified genetic risk variants for celiac disease related to the immune response. Nat. Genet. 40, 395–402 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhernakova A., et al. , Genetic analysis of innate immunity in Crohn’s disease and ulcerative colitis identifies two susceptibility loci harboring CARD9 and IL18RAP. Am. J. Hum. Genet. 82, 1202–1210 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Smyth D. J., et al. , Shared and distinct genetic variants in type 1 diabetes and celiac disease. N. Engl. J. Med. 359, 2767–2777 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Castellanos-Rubio A., et al. , A long noncoding RNA associated with susceptibility to celiac disease. Science 352, 91–95 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cabrera O., et al. , The unique cytoarchitecture of human pancreatic islets has implications for islet cell function. Proc. Natl. Acad. Sci. U.S.A. 103, 2334–2339 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mercer T. R., et al. , Targeted RNA sequencing reveals the deep complexity of the human transcriptome. Nat. Biotechnol. 30, 99–104 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Derrien T., et al. , The GENCODE v7 catalog of human long noncoding RNAs: Analysis of their gene structure, evolution, and expression. Genome Res. 22, 1775–1789 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Seiler J., et al. , The lncRNA VELUCT strongly regulates viability of lung cancer cells despite its extremely low abundance. Nucleic Acids Res. 45, 5458–5469 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cardozo A. K., et al. , IL-1beta and IFN-gamma induce the expression of diverse chemokines and IL-15 in human and rat pancreatic islet cells, and in islets from pre-diabetic NOD mice. Diabetologia 46, 255–266 (2003). [DOI] [PubMed] [Google Scholar]
- 22.Colli M. L., et al. , Exposure to the viral by-product dsRNA or Coxsackievirus B5 triggers pancreatic beta cell apoptosis via a Bim/Mcl-1 imbalance. PLoS Pathog. 7, e1002267 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Santin I., et al. , USP18 is a key regulator of the interferon-driven gene network modulating pancreatic beta cell inflammation and apoptosis. Cell Death Dis. 3, e419 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hogg J. R., Viral evasion and manipulation of host RNA quality control pathways. J. Virol. 90, 7010–7018 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.He F., et al. , A positive autoregulatory loop of Jak-STAT signaling controls the onset of astrogliogenesis. Nat. Neurosci. 8, 616–625 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yuasa K., Hijikata T., Distal regulatory element of the STAT1 gene potentially mediates positive feedback control of STAT1 expression. Genes Cells 21, 25–40 (2016). [DOI] [PubMed] [Google Scholar]
- 27.Ransohoff J. D., Wei Y., Khavari P. A., The functions and unique features of long intergenic non-coding RNA. Nat. Rev. Mol. Cell Biol. 19, 143–157 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Xin Z., et al. , PCBP2 enhances the antiviral activity of IFN-α against HCV by stabilizing the mRNA of STAT1 and STAT2. PLoS One 6, e25419 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tak Y. G., Farnham P. J., Making sense of GWAS: Using epigenomics and genome engineering to understand the functional relevance of SNPs in non-coding regions of the human genome. Epigenetics Chromatin 8, 57 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Castellanos-Rubio A., Ghosh S., Disease-associated SNPs in inflammation-related lncRNAs. Front. Immunol. 10, 420 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Eliasson L., Esguerra J. L. S., Role of non-coding RNAs in pancreatic beta-cell development and physiology. Acta Physiol. 211, 273–284 (2014). [DOI] [PubMed] [Google Scholar]
- 32.Motterle A., Gattesco S., Caille D., Meda P., Regazzi R., Involvement of long non-coding RNAs in beta cell failure at the onset of type 1 diabetes in NOD mice. Diabetologia 58, 1827–1835 (2015). [DOI] [PubMed] [Google Scholar]
- 33.Colli M. L., et al. , PDL1 is expressed in the islets of people with type 1 diabetes and is up-regulated by interferons-α and-γ via IRF1 induction. EBioMedicine 36, 367–375 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Richardson S. J., et al. , Islet cell hyperexpression of HLA class I antigens: A defining feature in type 1 diabetes. Diabetologia 59, 2448–2458 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kim S., et al. , Essential role for signal transducer and activator of transcription-1 in pancreatic β-cell death and autoimmune type 1 diabetes of nonobese diabetic mice. Diabetes 56, 2561–2568 (2007). [DOI] [PubMed] [Google Scholar]
- 36.Moore F., et al. , STAT1 is a master regulator of pancreatic beta-cell apoptosis and islet inflammation. J. Biol. Chem. 286, 929–941 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Atkinson M. A., Wilson S. B., Fatal attraction: Chemokines and type 1 diabetes. J. Clin. Invest. 110, 1611–1613 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Collier J. J., Sparer T. E., Karlstad M. D., Burke S. J., Pancreatic islet inflammation: An emerging role for chemokines. J. Mol. Endocrinol. 59, R33–R46 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Krogvold L., et al. , Pancreatic biopsy by minimal tail resection in live adult patients at the onset of type 1 diabetes: Experiences from the DiViD study. Diabetologia 57, 841–843 (2014). [DOI] [PubMed] [Google Scholar]
- 40.Krogvold L., et al. , Detection of a low-grade enteroviral infection in the islets of langerhans of living patients newly diagnosed with type 1 diabetes. Diabetes 64, 1682–1687 (2015). [DOI] [PubMed] [Google Scholar]
- 41.Chen C., et al. , Poly(rC) binding protein 2 (PCBP2) promotes the viability of human gastric cancer cells by regulating CDK2. FEBS Open Bio 8, 764–773 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Smirnova V. V., et al. , eIF4G2 balances its own mRNA translation via a PCBP2-based feedback loop. RNA 25, 757–767 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Guttman M., Rinn J. L., Modular regulatory principles of large non-coding RNAs. Nature 482, 339–346 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Redis R. S. S., et al. , Allele-specific reprogramming of cancer metabolism by the long non-coding RNA CCAT2. Mol. Cell 61, 520–534 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Aguilo F., Di Cecilia S., Walsh M. J., Long non-coding RNA ANRIL and Polycomb in human cancers and cardiovascular disease. Curr. Top. Microbiol. Immunol. 394, 29–39 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All of the data, associated protocols, and materials for this study are available in the SI Appendix.