Significance
Homologous recombination is initiated by a two-step nucleolytic processing of DNA breaks to generate overhangs required for the downstream repair steps. The first short-range resection step is slow but versatile because it can remove noncanonical structures from the break end. The second long-range resection step is fast but sensitive to protein adducts or secondary DNA structures. Our results support a model where CtIP is a cofactor that stimulates not only the short-range pathway as defined previously, but also the long-range nucleolytic processing. Specifically, we define here the interplay of CtIP and the DNA2 nuclease-helicase that functions in long-range resection. The involvement of CtIP in both resection steps might help coordinate DNA end processing and initiation of homologous recombination.
Keywords: DNA, homologous recombination, nuclease, helicase, DNA end resection
Abstract
To repair a DNA double-strand break by homologous recombination, 5′-terminated DNA strands must first be resected to reveal 3′-overhangs. This process is initiated by a short-range resection catalyzed by MRE11-RAD50-NBS1 (MRN) stimulated by CtIP, which is followed by a long-range step involving EXO1 or DNA2 nuclease. DNA2 is a bifunctional enzyme that contains both single-stranded DNA (ssDNA)-specific nuclease and motor activities. Upon DNA unwinding by Bloom (BLM) or Werner (WRN) helicase, RPA directs the DNA2 nuclease to degrade the 5′-strand. RPA bound to ssDNA also represents a barrier, explaining the need for the motor activity of DNA2 to displace RPA prior to resection. Using ensemble and single-molecule biochemistry, we show that CtIP also dramatically stimulates the adenosine 5′-triphosphate (ATP) hydrolysis-driven motor activity of DNA2 involved in the long-range resection step. This activation in turn strongly promotes the degradation of RPA-coated ssDNA by DNA2. Accordingly, the stimulatory effect of CtIP is only observed with wild-type DNA2, but not the helicase-deficient variant. Similarly to the function of CtIP to promote MRN, also the DNA2 stimulatory effect is facilitated by CtIP phosphorylation. The domain of CtIP required to promote DNA2 is located in the central region lacking in lower eukaryotes and is fully separable from domains involved in the stimulation of MRN. These results establish how CtIP couples both MRE11-dependent short-range and DNA2-dependent long-range resection and define the involvement of the motor activity of DNA2 in this process. Our data might help explain the less severe resection defects of MRE11 nuclease-deficient cells compared to those lacking CtIP.
DNA double-strand breaks (DSBs) in eukaryotes are repaired by end-joining or homology-directed pathways (1). End-joining, including both nonhomologous and microhomology-mediated subpathways, requires only minimal processing of the DNA ends and may involve limited microhomology at the break sites to facilitate ligation. Homologous recombination instead utilizes genetic information stored in an intact DNA copy, usually the sister chromatid in vegetative cells, to serve as a template for mostly accurate repair. There are several distinct subpathways of homology-directed repair, including single-strand annealing (SSA) that leads to large deletions, synthesis-dependent strand annealing, break-induced replication, and canonical DNA double-strand break repair (1). While these pathways differ in mechanisms and resulting genetic outcomes, their common denominator is the first step, termed DNA end resection. All recombination pathways are initiated by nucleolytic degradation of the 5′-terminated DNA strand at DSB sites, leading to extended stretches of 3′-terminated single-stranded DNA (ssDNA), which are required for the downstream steps in all of the respective pathways (2, 3). Extensive DNA end resection commits the DSB repair to recombination as resected ends are no longer ligatable by end-joining pathways. Cyclin-dependent kinases (CDKs) regulate DNA end resection to be allowed only in the S and G2 phases of the cell cycle, when sister chromatids are available (4–9).
It has been established that DNA end resection generally consists of two subsequent steps. In human cells, resection is initiated by the MRE11–RAD50–NBS1 (MRN) complex, which functions in conjunction with CtIP (10, 11). CtIP activates the MRE11 endonuclease within the MRN complex (12, 13). The activated MRN complex then preferentially cleaves the 5′-terminated DNA strand past protein blocks. Therefore, this step is particularly important to process DNA ends with noncanonical structures, including bound proteins like Ku and topoisomerase-DNA cleavage complexes, as well as secondary DNA structures (14–18). CtIP is phosphorylated by multiple kinases including CDK, ATM, and ATR. In particular, the T847 CDK site of CtIP must be phosphorylated to promote MRN, which represents a mechanism that allows cell cycle-dependent control of DNA end resection (5, 13, 19–21).
Downstream of this initial short-range processing, either of two partially redundant nucleases catalyze long-range 5′-end resection that can extend up to several kilobases in length (22–27). The Exonuclease 1 (EXO1) degrades the 5′-terminated DNA strand within double-stranded DNA (dsDNA). The nuclease-helicase DNA2 instead degrades ssDNA and thus requires a helicase partner to act upstream to unwind dsDNA, which can be either the Bloom (BLM) or Werner (WRN) RecQ family helicase (28–30). Both BLM/WRN and DNA2 function in conjunction with the single-strand DNA binding protein RPA, which promotes DNA unwinding by BLM/WRN and directs DNA2 to degrade the 5′-terminated ssDNA strand (31–33). The structure of murine DNA2 revealed that the polypeptide possesses a central cavity through which ssDNA needs to thread before it can be cleaved (33). As the size of the cavity only accommodates ssDNA, bound proteins, including RPA, need to be displaced prior to degradation (33, 34). The nuclease activity of DNA2 is essential for its function in resection and cell viability (24). However, DNA2 also has a conserved helicase domain (34, 35). Although the helicase-deficient DNA2 mutants are equally lethal, the function of this activity has remained enigmatic (36). The helicase of DNA2 in resection cannot replace the helicase of BLM/WRN (26). The unwinding capacity of wild-type (WT) DNA2 is very weak, and processive dsDNA unwinding is only observed upon inactivation of the nuclease activity (29, 35, 37). The motor activity of DNA2 was thus proposed to promote resection, not as a helicase to unwind dsDNA, but rather as an efficient ssDNA translocase to act downstream of the RecQ family helicase partner (38, 39). In this model, BLM and WRN function as the lead helicases that unwind dsDNA. This provides long stretches of RPA coated 5′-terminated ssDNA to DNA2, which uses its motor activity to translocate along the ssDNA to displace RPA and feed the ssDNA strand to the nuclease domain (38).
The phenotypes of human CtIP-depleted cells or corresponding Saccharomyces cerevisiae sae2Δ mutants were reported to be more severe than those of MRE11/Mre11 nuclease point mutants, indicating that CtIP/Sae2 have additional functions that go beyond stimulating the MRE11 nuclease. To this point, both Sae2 and CtIP were described to possess an intrinsic nuclease activity with functions in DNA end resection and beyond, which could explain the phenotypic differences (40–42). However, the Sae2/CtIP nuclease function in resection remains controversial (12, 18, 43). Sae2/CtIP may also have a structural role in DNA break repair to bridge DNA (18, 44). In yeast, sae2Δ mutants were found to hyperactivate checkpoint signaling, and Sae2 was shown to have a structural function to dampen checkpoint signaling (45). Checkpoint-defective sae2Δ mutants exhibited identical DNA damage sensitivity and resection defects as mre11 nuclease-dead mutants, showing that checkpoint hyperactivation accounted for the differential phenotypes of the respective mutants (45–47). In higher eukaryotes, however, the situation is more complex. While some studies reported similar sensitivities of MRE11 nuclease-deficient (resulting from point mutations or small-molecule inhibitors) and CtIP-deficient cells (11), other reports point out more severe defects upon CtIP depletion. Specifically, in Xenopus egg extracts, complete inhibition of resection was observed upon depletion of EXO1 and CtIP, implying that DNA2 requires CtIP to perform long-range resection (48). Likewise, epistatic relationships between CtIP and DNA2 were found in chicken DT40 cells (49). Disruption of the MRE11 nuclease active site did not dramatically reduce resection, in contrast to CtIP-deficient cells, which exhibited a severe defect (50). Similarly, inactivation of the MRE11 nuclease was less severe than disruption of CtIP in human TK6 cells (50). Finally, single-molecule analysis of DNA resection tracks showed that CtIP contributes to fast resection at long distances from the DNA end (51), which disagrees with the expected limited range of the MRN-dependent short-range resection. Together, these reports suggest that CtIP may promote long-range DNA end resection by an unknown mechanism. In accord, CtIP was shown to physically interact with and promote DNA unwinding by the BLM helicase, as well as to modestly stimulate DNA degradation by the DNA2 nuclease (52). Using ensemble and single-molecule biochemistry, we show here that phosphorylated CtIP (pCtIP) dramatically stimulates the motor activity of DNA2. This accelerates degradation of RPA-coated ssDNA by DNA2, showing the need for the motor activity of DNA2 to facilitate resection. Our results show that CtIP is thus a cofactor not only of MRE11, but also of DNA2, and demonstrate that the domains of CtIP required for the stimulation of MRN and DNA2 are physically separate. Our data support a model where CtIP first activates the MRE11 nuclease and then helps couple short-range resection with the downstream long-range step by promoting DNA2. These results might help explain the dramatic DNA end resection defect observed in CtIP-depleted human cells.
Results
CtIP Promotes DNA2-Dependent Long-Range DNA End Resection Pathway.
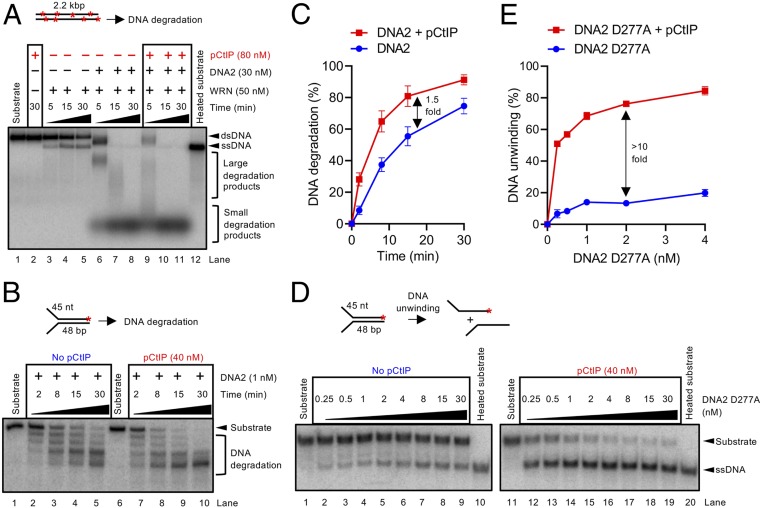
pCtIP functions as a cofactor of the MRN endonuclease acting in the initial short-range DNA end resection pathway (13) (SI Appendix, Fig. S1A). Multiple studies, however, indicated that deficiency of CtIP has a stronger impact on resection than mutations or inhibition of the MRE11 nuclease (48–50) (SI Appendix, Fig. S1 B and C), suggesting that CtIP has additional functions in resection beyond promoting MRE11. To study the effect of CtIP on long-range DNA end resection pathways in a defined system, we expressed and purified phosphorylated CtIP (pCtIP) in the presence of phosphatase inhibitors, as well as DNA2, BLM, WRN, and EXO1 (SI Appendix, Fig. S1 D–H) in Sf9 cells. Recombinant pCtIP did not promote DNA degradation by EXO1 (SI Appendix, Fig. S2 A and B) but stimulated DNA end resection by BLM-DNA2-RPA (SI Appendix, Fig. S2 C and D), as well as by WRN-DNA2-RPA (Fig. 1A, compare lanes 6 and 9, SI Appendix, Fig. S2E). To understand the stimulation of resection by pCtIP in detail, we next investigated its effect on the activities of BLM/WRN and DNA2 individually. pCtIP stimulated DNA unwinding by BLM approximately twofold (SI Appendix, Fig. S2 F and G), as well as to a similar extent the nuclease of DNA2 on oligonucleotide-based substrates (Fig. 1 B and C), as noted previously (52). In contrast, pCtIP had only a very minor effect on DNA unwinding by WRN (SI Appendix, Fig. S2 H and I). The strongest stimulatory effect by pCtIP was observed when we assayed DNA unwinding by nuclease-dead DNA2 D277A (Fig. 1 D and E). With pCtIP, 0.25 nM DNA2 D277A unwound more than 50% of the Y-structured DNA substrate, which was more than that unwound by 30 nM DNA2 D277A without pCtIP, corresponding to >10-fold stimulation (Fig. 1 D and E). Recombinant pCtIP thus predominantly promotes the motor activity of DNA2. These results indicate that CtIP functions as a cofactor in both short- and long-range resection pathways. To confirm the physiological relevance of our observations, we reasoned that the depletion of CtIP, which affects both resection pathways simultaneously, should result in the same DNA end resection defect as the depletion of DNA2 in combination with chemical inhibition of MRE11 nuclease, which affects both pathways individually. To this point, we used an SSA reporter assay in U2OS cells, which requires resection of several thousand nucleotides (nt) in length and which is often used as a physiological readout of DNA end resection efficacy (53, 54). As shown in SI Appendix, Fig. S2J, the depletion of CtIP almost completely abrogated SSA while the depletion of DNA2 and the inhibition of the MRE11 nuclease by small molecules had moderate effects. However, the combination of DNA2 depletion and MRE11 inhibitors resulted in resection defects comparable to the depletion of CtiP (SI Appendix, Fig. S2J), in agreement with the model that CtIP is involved in both resection pathways.
Fig. 1.
pCtIP promotes DNA2-dependent long-range DNA end resection pathway. (A) DNA end resection by WRN, DNA2, and human RPA (176 nM) using 2.2-kilobase pair (kbp)-long randomly labeled dsDNA substrate in the presence or absence of pCtIP. The reaction buffer contained 50 mM NaCl. Reaction products were separated by 1% agarose gel electrophoresis. Panel shows a representative experiment. Red asterisks indicate random labeling. (B) Representative 15% denaturing polyacrylamide gel showing the degradation kinetics of a Y-structured (45 nt/48 bp) DNA by DNA2 with or without pCtIP, in the presence of human RPA (15 nM) and 100 mM NaCl. The red asterisk indicates the position of the labeling. (C) Quantitation of overall substrate utilization from experiments such as shown in B. n = 3; error bars, SEM. (D) A representative experiment showing DNA unwinding by nuclease-deficient DNA2 D277A with or without pCtIP on oligonucleotide-based Y-structured (45 nt/48 bp) DNA. Reactions were supplemented with human RPA (7.5 nM) and 50 mM NaCl and analyzed on 10% native acrylamide gel electrophoresis. A red asterisk indicates the position of the labeling. (E) Quantitation of overall substrate unwinding from experiments such as shown in D. n = 3; error bars, SEM.
CtIP Dramatically Promotes Long-Range ssDNA Degradation by DNA2.
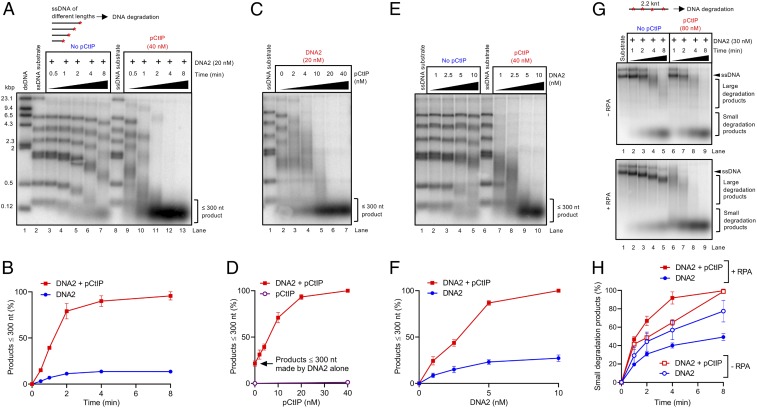
To define the function of pCtIP in regulating DNA2 in a simple system, we next used an assay that monitors 5′→3′ degradation of 3′ end-labeled fragments of ssDNA by WT DNA2. The use of ssDNA bypasses the requirement for BLM or WRN helicase. Previously, we demonstrated that the motor of DNA2 particularly promoted the degradation of RPA-coated ssDNA (38) so we next tested for a function of CtIP. Without pCtIP, the degradation of ssDNA by DNA2 was slow (Fig. 2A, lanes 3–7) and was, strikingly, ∼10-fold stimulated when pCtIP was included in the reactions (Fig. 2A, lanes 9–13, Fig. 2B). While pCtIP appears to accelerate ssDNA degradation by DNA2, it did not change the 5′→3′ polarity of DNA degradation in the presence of RPA (SI Appendix, Fig. S3 A and B). pCtIP also did not allow DNA2 to cleave ssDNA endonucleolytically when DNA ends were blocked (SI Appendix, Fig. S3 A and B). Likewise, pCtIP did not facilitate extensive dsDNA degradation by DNA2 (SI Appendix, Fig. S3C), showing that DNA2 is very unlikely to function independently of BLM/WRN in DNA end resection, in agreement with cellular data (28). Low nanomolar concentrations of both pCtIP and DNA2 were required for maximal DNA degradation under our conditions (Fig. 2 C–F). We note that 1 nM DNA2 in the presence of pCtIP was more efficient in DNA degradation than 20 nM DNA2 without pCtIP (compare Fig. 2E, lane 7 with Fig. 2A, lane 7), highlighting the dramatic stimulation of the ssDNA degradative capacity of DNA2 by pCtIP.
Fig. 2.
pCtIP dramatically promotes degradation of long stretches of ssDNA by DNA2. (A) Representative 1% agarose gel showing degradation kinetics of 3′ 32P-labeled ssDNA fragments (derived from λDNA) of various lengths by DNA2 without or with pCtIP in the presence of 864 nM human RPA. The sizes of the corresponding dsDNA fragments are indicated on the left. A red asterisk indicates the position of the labeling. (B) Quantitation of products smaller than ∼300 nt from experiments such as shown in A. n = 3; error bars, SEM. (C) Representative experiment as in A with various concentrations of pCtIP incubated for 8 min. (D) Quantitation of data such as shown in C; n = 3; error bars, SEM. The degradation activity of pCtIP alone is the same as in Fig. 3A (lane 7). (E) Experiment such as in A with various concentrations of DNA2 incubated for 8 min in the presence or absence of pCtIP. (F) Quantitation of data such as shown in E; n = 3; error bars, SEM. (G) Degradation of 2.2-kilonucleotide (knt)-long randomly labeled ssDNA substrate by DNA2 in the presence or absence of pCtIP. The reaction buffer contained 50 mM NaCl and, where indicated, 352 nM RPA. Reaction products were separated by 1% agarose gel electrophoresis. The panel shows a representative experiment. Red asterisks indicate random labeling of substrate DNA. (H) Quantitation of data such as shown in G; n = 3; error bars, SEM.
RPA protects the 3′-terminated ssDNA strand from degradation by DNA2 (29, 31, 32). To explain the role of RPA in the interplay of pCtIP and DNA2 and carry out reactions without RPA, we next prepared an internally labeled substrate. Using ssDNA randomly labeled at internal sites, we observed that RPA inhibited ssDNA degradation, in agreement with our previous data (38). pCtIP stimulated ssDNA degradation also without RPA, but much greater stimulatory effect was observed in the presence of RPA (Fig. 2 G and H). We conclude that pCtIP promotes ssDNA degradation by DNA2, which becomes particularly important in the presence of RPA.
The Motor Activity of DNA2 Mediates the Accelerated ssDNA Degradation with pCtIP.
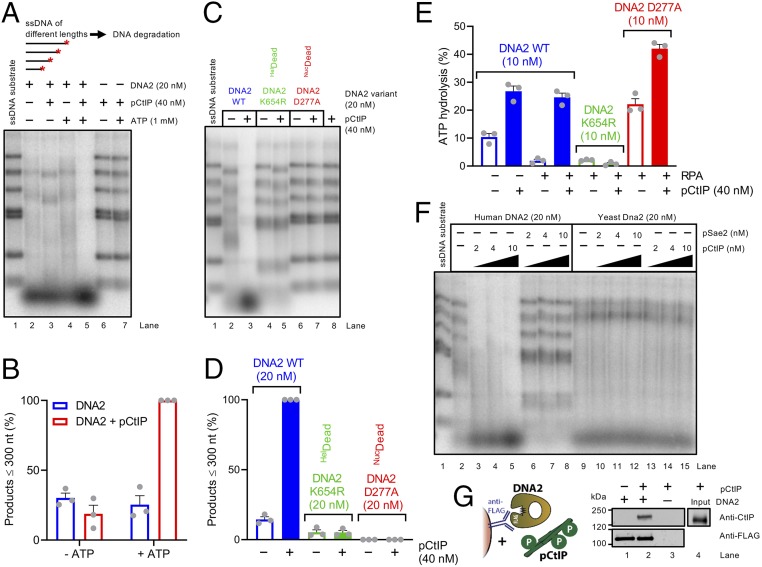
To define the mechanism of the stimulatory effect of pCtIP on the DNA2 nuclease, we set out to investigate the role of the ATPase-driven motor activity of DNA2 in this process. Strikingly, pCtIP strongly stimulated ssDNA degradation by DNA2 only in the presence of adenosine 5′-triphosphate (ATP) (Fig. 3 A and B). Importantly, pCtIP had no activity on its own with or without ATP (Fig. 3A, lanes 6 and 7). Furthermore, pCtIP did not notably promote ssDNA degradation by the helicase-deficient DNA2 K654R variant with disrupted ATPase active site (Fig. 3 C and D).
Fig. 3.
The motor activity of DNA2 mediates the accelerated ssDNA degradation with pCtIP. (A) Degradation of 3′ end-labeled ssDNA fragments (derived from λDNA) of various lengths by DNA2 in the presence or absence of pCtIP without or with ATP, as indicated. All reactions contained human RPA (864 nM). The experiment was incubated at 37 °C for 8 min. ATP is required for the stimulatory effect of pCtIP on DNA2. A red asterisk indicates the position of the labeling. (B) Quantitation of data such as shown in A. n = 3; error bars, SEM. The grey circles represent data points from indepedent experiments. (C) Degradation of ssDNA fragments by WT, helicase-deficient K654R, or nuclease-deficient D277A DNA2 variants without or with pCtIP. All reactions contained human RPA (864 nM). The experiment was incubated at 37 °C for 8 min. HelDead, helicase-dead; NucDead, nuclease-dead. (D) Quantitation of small degradation products from experiments such as shown in C. n = 3; error bars, SEM. (E) ATP hydrolysis by WT, helicase-deficient K654R, or nuclease-deficient D277A DNA2 alone (10 nM) or with pCtIP (40 nM). Reactions contained 10.3-kbp-long substrate denatured at 95 °C for 5 min, 395.5 nM human RPA where indicated and 20 mM NaCl. (F) Degradation of 3′ end-labeled ssDNA fragments derived from λDNA by human or yeast DNA2/Dna2 without or with human pCtIP or yeast pSae2. Reactions with human DNA2 were carried out at 37 °C for 8 min with human RPA (864 nM). Reactions with yeast Dna2 were performed at 25 °C for 1 min with yeast RPA (1.09 µM). (G) Analysis of DNA2 interaction with pCtIP. DNA2-FLAG was immobilized on M2 anti-FLAG affinity resin and incubated with purified recombinant pCtIP (phosphorylation sites depicted as P in green circles). The Western blot was performed with anti-FLAG and anti-CtIP antibodies.
Using thin-layer chromatography (TLC), we found that pCtIP strongly promoted the ATPase activity of WT DNA2 in the presence of ssDNA as cofactor, both without and with RPA (Fig. 3E and SI Appendix, Fig. S4A). pCtIP also stimulated the ATPase of nuclease-dead DNA2 D277A while no ATP hydrolysis was observed using the ATPase-deficient DNA2 K654R variant, as expected (Fig. 3E). Using a kinetic ATPase assay where ATP hydrolysis is linked to reduced nicotinamide-adenine dinucleotide oxidation monitored spectrophotometrically, we estimate the apparent catalytic constant for DNA2 without pCtIP of 3.4 s−1 and 17.3 s−1 in the presence of pCtIP, which corresponds to approximately fivefold stimulation by pCtIP, in agreement with the TLC assays (Fig. 3E). These experiments together demonstrate the importance of the ATPase-driven translocase activity of DNA2 in extended ssDNA degradation.
No DNA degradation was observed when using the nuclease-deficient DNA2 D277A (Fig. 3 C and D), even in the presence of pCtIP, further demonstrating that the observed nuclease activity was intrinsic to DNA2 and not a result of nonspecific contamination in our assays. Human pCtIP did not promote ssDNA degradation by yeast Dna2, and, vice versa, phosphorylated yeast Sae2 (pSae2) did not promote the nuclease of human DNA2 (Fig. 3F and SI Appendix, Fig. S4 B and C), implicating direct species-specific interactions between the human polypeptides. Furthermore, pSae2 did not notably stimulate yeast Dna2, showing that the interplay described here is specific to higher eukaryotes. The species-specific interplay of cognate human pCtIP and DNA2 suggested that the proteins might directly physically interact. Indeed, FLAG-tagged DNA2 could readily pull down pCtIP, demonstrating a direct physical interaction (Fig. 3G). Together, these results establish that, by forming a complex, pCtIP stimulates the ATPase-driven translocase activity of DNA2, which in turn accelerates its ssDNA degradative capacity.
Single-Molecule Experiments Reveal the Acceleration of DNA Unwinding Rate of DNA2 by pCtIP.
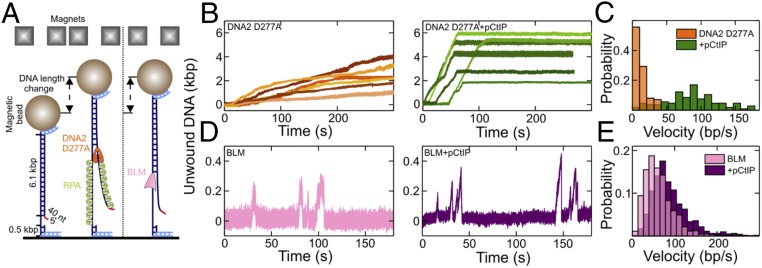
To further mechanistically define the stimulation of DNA2 motor by pCtIP, we employed magnetic tweezers, which monitor the position of a magnetic bead linked by a single DNA molecule to a fixed surface. As ssDNA is extended compared to dsDNA, DNA unwinding can be inferred from the relative position of the bead and DNA extension curve. We could not directly monitor the DNA2 nuclease in our setup because DNA must be attached on both ends. Instead, we analyzed the effect of pCtIP on DNA unwinding by the nuclease-dead DNA2 D277A (Fig. 4A). The mean velocity of DNA2 D277A in the absence of pCtIP was 13 ± 1 base pairs (bp)/s (SEM), which was increased approximately sevenfold in the presence of pCtIP to 86 ± 4 bp/s (SEM) (Fig. 4 B and C and SI Appendix, Fig. S5A). In contrast, pCtIP had no effect on the processivity of DNA2 D277A (SI Appendix, Fig. S5B). While the motor activity of human DNA2 alone is slower than that of the yeast ortholog (29, 37), we note that, in the presence of pCtIP, the unwinding activity of human DNA2 is faster than that by yeast Dna2, making human DNA2 one of the most efficient known motor proteins in eukaryotes.
Fig. 4.
Single-molecule experiments demonstrate accelerated DNA2 motor activity in the presence of pCtIP. (A) Sketch of the employed magnetic tweezers assay and the DNA construct carrying a 40-nt 5′ flap to allow loading of either helicase. (B) Representative DNA unwinding events of the DNA2 nuclease-dead mutant (D277A, 25 nM) in the absence (orange) and presence (green) of pCtIP (25 nM). Both reactions were also supplemented with 25 nM human RPA. (C) Histograms of the observed unwinding velocities for nuclease-dead DNA2 (n = 40 traces for each case). (D) Representative DNA unwinding events of BLM (25 nM) in the absence (magenta) and presence (purple) of pCtIP (25 nM). (E) Histogram of the observed unwinding velocities for BLM (n = 852 events). The force in all experiments was F = 19 ± 3 pN (SD).
As pCtIP also modestly promotes DNA unwinding by BLM (SI Appendix, Fig. S2 F and G) (52), we also assayed this stimulatory effect by magnetic tweezers. The DNA unwinding by BLM, as by other helicases from the RecQ family such as Sgs1, is characterized by bursts of DNA unwinding, followed by rewinding (Fig. 4D) (55). We observed ∼1.5-fold stimulation of DNA unwinding by BLM when pCtIP was included in the reaction (Fig. 4 D and E). The mean BLM velocity in the absence of pCtIP was 61 ± 4 bp/s (SEM), and 91 ± 4 bp/s (SEM) in the presence of pCtIP. As with DNA2 D277A, pCtIP did not affect the processivity of BLM (SI Appendix, Fig. S5C). Additionally, pCtIP did not stimulate DNA unwinding by WRN (SI Appendix, Fig. S5 D–F), in agreement with the biochemical data (SI Appendix, Fig. S2 H and I). No DNA unwinding was observed without ATP, or by CtIP alone (SI Appendix, Fig. S5 G–K). In summary, pCtIP promotes the velocity of DNA unwinding by both BLM and DNA2 without affecting their processivity. The effect of pCtIP on DNA2 motor is much greater than that on BLM (compare Fig. 4 C and E).
Domains of pCtIP That Promote MRN and DNA2 Are Fully Separable.
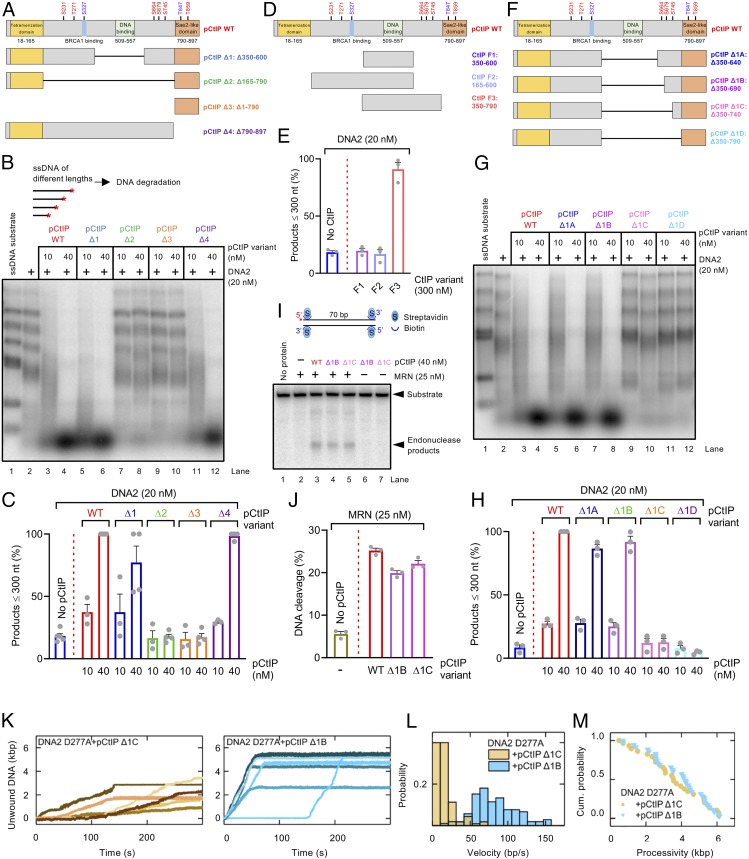
The pCtIP protein is composed of an N-terminal tetramerization domain, an unstructured central region—largely lacking in S. cerevisiae—that binds DNA and contains a number of phosphorylation sites, and finally a conserved C-terminal domain that is required to activate MRN, has a secondary DNA binding site, and is likewise subject to phosphorylation (Fig. 5A) (10, 13, 18, 56). A key cyclin-dependent kinase site, T847, which enables cell cycle-regulated control of DNA end resection by licensing stimulation of MRN, is located within the C-terminal domain of pCtIP (Fig. 5A) (5). Only the N-terminal tetramerization and the C-terminal domains bear limited similarity to yeast Sae2. To identify pCtIP regions that are required for the stimulation of DNA2, we designed pCtIP variants lacking stretches of amino acids from the internal region, such as pCtIP Δ1 (lacking residues 350 to 600 including the DNA-binding region) and pCtIP Δ2 (lacking residues 165 to 790 comprising the entire region between the tetramerization domain and the Sae2-like C-terminal domain). As WT full-length pCtIP, these internally truncated variants were expressed in Sf9 cells and purified in the presence of phosphatase inhibitors to preserve phosphorylation. We observed that pCtIP Δ1, but not pCtIP Δ2, was fully proficient in stimulating DNA2 (Fig. 5 B and C and SI Appendix, Fig. S6A). This indicated that the region in pCtIP between residues 350 and 600 containing the DNA binding domain is dispensable for the stimulation of DNA2, and the stimulatory activity is dependent on a region upstream or downstream of these residues. We also prepared the C-terminal domain of pCtIP alone (pCtIP Δ3, containing residues 790 to 897), and a mutant lacking this C-terminal domain (pCtIP Δ4) (Fig. 5A). Remarkably, the pCtIP Δ4 variant, lacking the C-terminal Sae2-like region, while incapable to stimulate MRN (SI Appendix, Fig. S6B), as expected (13), was fully proficient in stimulating DNA2 (Fig. 5 B and C and SI Appendix, Fig. S6A). In accord with these data, pCtIP Δ1 and Δ4, but not the pCtIP Δ2 variant, were capable to promote the ATPase of DNA2 (SI Appendix, Fig. S6C). Also, pCtIP Δ1 physically interacted with DNA2 similarly as full-length pCtIP while the interaction was abolished when using pCtIP Δ2 (SI Appendix, Fig. S6D).
Fig. 5.
Separate domains of pCtIP promote the MRN and DNA2 nucleases. (A) A schematic representation of the primary structure of WT pCtIP and internal deletion variants (pCtIP Δ1 to Δ4) purified from Sf9 cells in the presence of phosphatase inhibitors. Main ATM phosphorylation sites are indicated in red, and CDK phosphorylation sites are indicated in blue. (B) Degradation ssDNA fragments of various lengths by DNA2 without or with pCtIP variants, as indicated. All reactions contained human RPA (864 nM). A red asterisk indicates the position of the radioactive label. (C) Quantitation of data such as shown in B. n = 3–4; error bars, SEM. (D) A schematic representation of purified recombinant CtIP fragments (F1 to F3) expressed in E. coli. Full-length pCtIP is again shown as a reference. (E) Quantitation of ssDNA fragment degradation into products smaller than ∼300 nt by DNA2 without or with F1 to F3 CtIP fragments. n = 3; error bars, SEM. (F) A schematic representation of internal deletion variants (pCtIP Δ1A to Δ1D) purified from Sf9 cells in the presence of phosphatase inhibitors. Full-length pCtIP is shown as a reference. (G) Degradation of ssDNA fragments of various lengths by DNA2 without or with various concentrations of WT pCtIP or Δ1A to Δ1D mutants, in the presence of human RPA (864 nM). (H) Quantitation of data such as shown in G. n = 3; error bars, SEM. (I) Endonuclease assay with MRN (25 nM) and WT full-length or pCtIP Δ1B and Δ1C variants. A red asterisk indicates the position of the radioactive label. (J) Quantitation of data such as shown in I. n = 3; error bars, SEM. (K) Representative DNA unwinding events using the DNA2 nuclease-dead mutant (D277A, 25 nM) in the presence of pCtIP Δ1C (25 nM) (yellow) and pCtIP Δ1B (25 nM) (cyan). Both reactions were also supplemented with 25 nM RPA. (L) Histograms of the observed unwinding velocities for nuclease-dead DNA2 (n > 25 traces for each case) in the presence of pCtIP Δ1C (yellow) and pCtIP Δ1B (cyan) with mean values of 16 ± 1 bp/s (SEM) and 85 ± 3 bp/s (SEM), respectively. (M) Cumulative probability distributions (shown as survival probability) of the processivity of the individual DNA unwinding events by nuclease-dead DNA2 D277A in the presence of pCtIP Δ1C (Left) and pCtIP Δ1B (Right) with mean values of 3.6 ± 0.3 kbp (SEM) and 3.8 ± 0.3 kbp (SEM), respectively.
To further map the CtIP region required to stimulate DNA2, we expressed various portions of the central CtIP domain in Escherichia coli (Fig. 5D). We prepared the central region corresponding to residues 350 to 600, as well as fragments containing additionally the upstream and downstream regions from the wider central domain (Fig. 5D). We observed that the CtIP F3 fragment, containing residues 350 to 790, was capable to promote DNA2 (Fig. 5E). This indicated that the CtIP region downstream of residue 600 was required for DNA2 stimulation. We note that this domain is lacking in yeast Sae2, and our result is thus in agreement with the observation that yeast Sae2 does not promote yeast or human DNA2 (Fig. 3F). We observed that a high concentration of the short F3 fragment compared to the longer pCtIP variants expressed in insect cells was needed to observe DNA2 stimulation (compare Fig. 5 C and E).
To narrow down the stimulatory region in pCtIP even further, we expressed in Sf9 cells and purified an additional series of internal pCtIP truncations lacking also residues downstream of position 600 (Fig. 5F). We observed that pCtIP Δ1B fragment (lacking residues 350 to 690) was fully proficient in stimulating DNA2, but additional shortening of this mutant by 50 residues (pCtIP Δ1C, lacking residues 350 to 740) entirely eliminated this capacity (Fig. 5 G and H and SI Appendix, Fig. S7A). In contrast, the pCtIP Δ1C construct was fully proficient in stimulating the endonuclease of MRN (Fig. 5 I and J). As above, the capacity of the pCtIP internal truncation variants to promote ssDNA degradation by DNA2 correlated with their ability to stimulate DNA2’s ATP hydrolysis activity (SI Appendix, Fig. S7B), as well as to promote DNA unwinding by DNA2 D277A as established by magnetic tweezers (Fig. 5 K and M). Accordingly, the CtIP Δ1C and Δ1D variants that did not promote DNA degradation and ATPase of DNA2 lost their capacity to physically interact with DNA2 (SI Appendix, Fig. S7C). We also note that, while the CtIP region 690 to 740 is necessary to stimulate DNA2, it is not sufficient because no DNA2 stimulation was observed using this CtIP fragment alone (SI Appendix, Fig. S7 D–F).
To confirm the physiological relevance of our in vitro data, we carried out a knockdown-rescue experiment in U2OS cells using the SSA reporter system. Upon down-regulation of CtIP by small interfering RNA (siRNA), SSA efficacy was rescued by siRNA-resistant CtIP Δ1C to a lesser extent than by WT full-length CtIP (SI Appendix, Fig. S7G). As CtIP Δ1C is proficient in stimulating MRE11 and deficient in stimulating DNA2, our results indicate that the interplay between CtIP and DNA2 described here might be important for the efficacy of extended DNA end resection required for SSA. These experiments together establish that the pCtIP region between residues 690 and 740 is required for the stimulation of the DNA2 motor activity while it is dispensable for the stimulation of MRN. Therefore, pCtIP promotes MRN and DNA2 via distinct domains that are fully separable.
pCtIP Phosphorylation Facilitates Its Capacity to Promote DNA2.
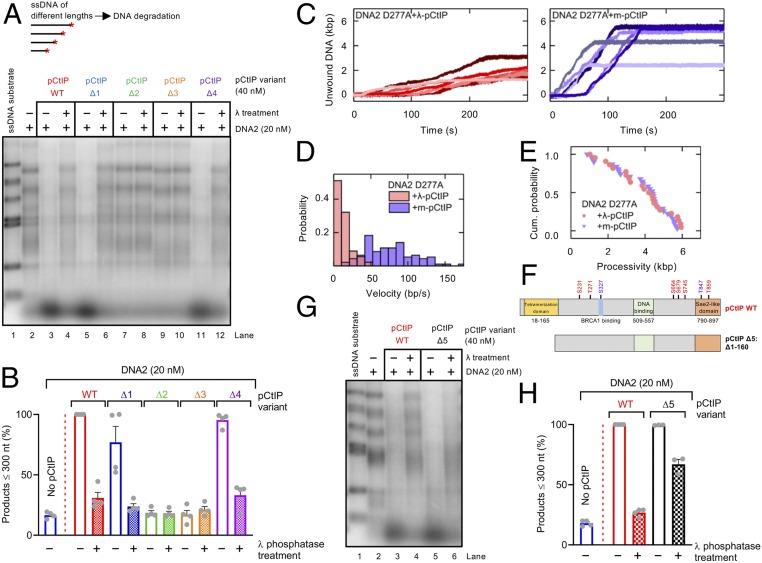
Due to the established requirement for pCtIP phosphorylation in the stimulation of MRN in vitro (13, 57) and in vivo (5, 6), we set out to define the function of pCtIP phosphorylation in the regulation of DNA2. Dephosphorylation of full-length Δ1 and Δ4 pCtIP variants expressed in Sf9 cells dramatically reduced their capacity to stimulate DNA2 (Fig. 6 A and B and SI Appendix, Fig. S8 A–C). Likewise, lambda phosphatase treatment of pCtIP reduced its capacity to promote DNA unwinding by DNA2 D277A in gel-based assays (SI Appendix, Fig. S8 D and E), as well as in single-molecule experiments (Fig. 6 C–E). Dephosphorylated CtIP could also no longer bind DNA2 (SI Appendix, Fig. S8F). These results are in agreement with data from Fig. 5E where we observed only limited stimulatory activity of the CtIP fragment expressed in E. coli, which was not expected to be phosphorylated.
Fig. 6.
pCtIP phosphorylation facilitates its capacity to promote DNA2. (A) Degradation of ssDNA fragments of various lengths with DNA2 alone (20 nM) or with pCtIP WT, Δ1, Δ2, Δ3, or Δ4, mock-treated or λ-treated, in the presence of human RPA (864 nM). A red asterisk indicates the position of the labeling. (B) Quantitation of small degradation products from experiments such as shown in A. n = 4; error bars, SEM. (C) Representative DNA unwinding events of the DNA2 nuclease-dead mutant (D277A, 25 nM) in the presence of pCtIP (25 nM) treated with λ phosphatase (Left) and mock buffer (Right). Both reactions were also supplemented with 25 nM RPA. (D) Histograms of the observed unwinding velocities for nuclease-dead DNA2 (n > 25 traces for each case) in the presence of pCtIP treated with λ phosphatase (pink) and mock buffer (violet) with mean values of 16 ± 2 bp/s (SEM) and 74 ± 3 bp/s (SEM), respectively. (E) Cumulative (Cum.) probability distributions (such as survival probability) of the processivity of the individual unwinding events of nuclease-dead DNA2 D277A in the presence of pCtIP treated with λ phosphatase (pink) and mock buffer (violet) with mean values of 3.8 ± 0.3 kbp (SEM) and 3.9 ± 0.3 kbp (SEM), respectively. (F) A schematic representation of purified recombinant pCtIP Δ5 lacking the first 160 amino acids. pCtIP WT is again shown as a reference. (G) Degradation of ssDNA fragments of various lengths with DNA2 alone (20 nM) or with pCtIP WT or Δ5, mock-treated or λ-treated, in the presence of human RPA (864 nM). (H) Quantitation of data such as shown in G. n = 3–4; error bars, SEM.
Dephosphorylated CtIP and their homologs are thought to aggregate so the apparent stimulatory function of phosphorylation may stem from abrogated CtIP aggregation (4, 58). To address whether phosphorylation enables CtIP to promote DNA2 by preventing its aggregation, we employed pCtIP Δ5, which lacks the N-terminal tetramerization domain (residues 1 to 160) (56, 59) (Fig. 6F). Lack of the tetramerization domain is expected to prevent aggregation, even in the dephosphorylated state (4). This pCtIP Δ5 variant was proficient in stimulating DNA2, which was largely but not completely resistant to lambda phosphatase treatment (Fig. 6 G and H and SI Appendix, Fig. S8 G and H). Accordingly, pCtIP Δ5 could stimulate the ATPase of DNA2 and physically interact with DNA2 irrespective of its phosphorylation state (SI Appendix, Fig. S8I). We also analyzed several point mutants nonphosphorylatable on some of the previously established phosphorylation sites in pCtIP within or close to the 690-to-740 region identified above, including S664A, S679A, and S754A forming pCtIP 3A (SI Appendix, Fig. S9A). The pCtIP 3A stimulated DNA2 equally as WT pCtIP (SI Appendix, Fig. S9 B and C). In contrast, a pCtIP mutant S723A, nonphosphorylatable at a recently identified PLK1 site (60), was impaired in DNA2 stimulation (SI Appendix, Fig. S9 A–C). This indicated that the region at S723 of pCtIP is important for its interplay with DNA2, but detailed understanding of this relationship will require further experimentation. We conclude that pCtIP phosphorylation clearly facilitates the stimulation of DNA2. This is likely caused, at least in part, indirectly by preventing pCtIP aggregation. Furthermore, while CtIP tetramerization is essential to stimulate MRN (13, 56), it may be dispensable to stimulate DNA2.
Discussion
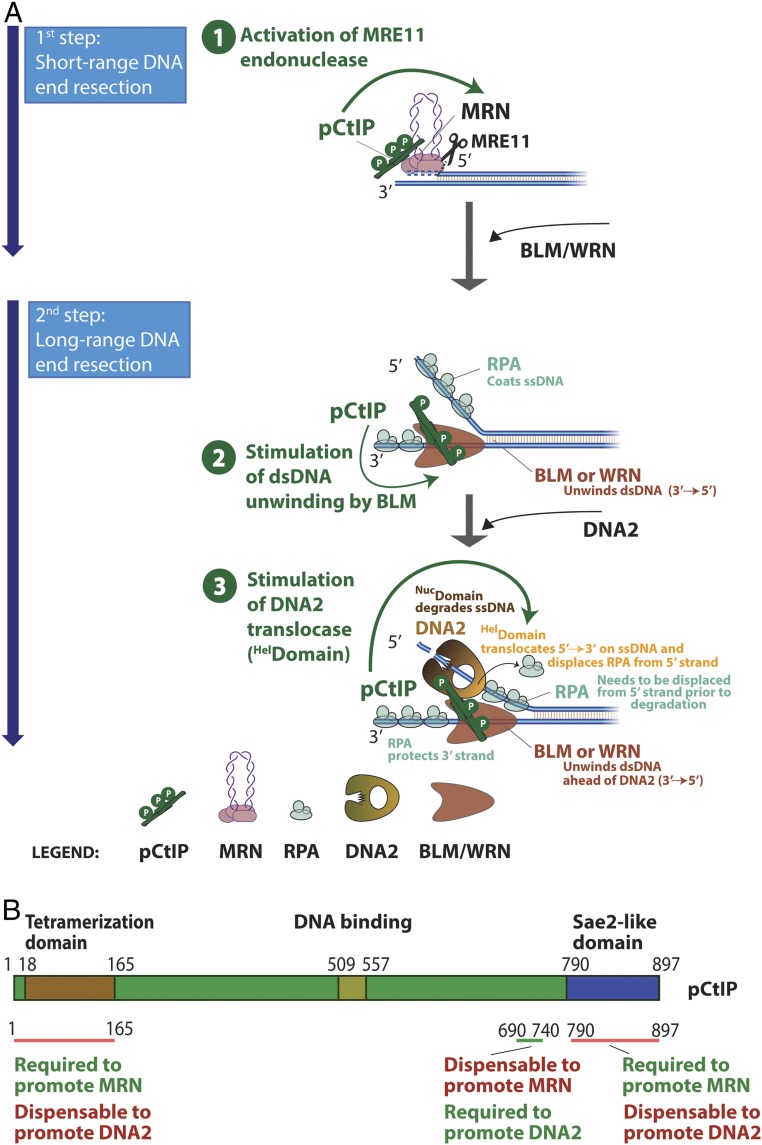
CtIP has a well-established role as an activator of the MRE11 nuclease activity within the MRN complex, which initiates DNA end resection. This function is conserved in evolution from yeast to human cells (12, 13, 15, 61). The results presented here demonstrate that CtIP also controls DNA2, a long-range DNA end resection nuclease that functions downstream of MRE11. Therefore, similarly to SLX4 that controls a number of structure-specific nucleases acting late in the recombination pathway (62, 63), CtIP controls both short- and long-range resection nucleases (Fig. 7A). We propose that coupling of both resection steps by CtIP allows to better coordinate short- and long-range DNA end resection.
Fig. 7.
Model for pCtIP functions in DNA end resection. (A) A schematic depicting the role of pCtIP in short-range resection by MRE11 within the MRN complex and in long-range resection by DNA2-BLM. In long-range resection, pCtIP stimulates both the helicase activity of BLM to unwind dsDNA and the translocase activity of DNA2 downstream to facilitate degradation of unwound, RPA-coated ssDNA. Unwinding of dsDNA and ssDNA degradation in long-range resection is likely coordinated and was depicted as separate for easy visualization. (B) A schematic representation of the primary structure of pCtIP and the domains required for the stimulation of MRN or DNA2 nucleases.
The CtIP domains required to promote MRE11 and DNA2 are at least partially separate. We found that the very N-terminal and C-terminal domains of CtIP, while being required to promote MRE11, are largely dispensable to regulate DNA2 (Fig. 7B). Instead, we identified a region in the central domain of pCtIP between residues 690 and 740 that is essential to stimulate DNA2, but dispensable for stimulating MRN (Fig. 7B). The central domain of CtIP is lacking in homologs of low eukaryotes, such as in S. cerevisiae Sae2. In accord, we failed to observe stimulation of yeast Dna2 by yeast Sae2. Therefore, while the regulation of MRE11 by CtIP is conserved in evolution, the regulation of DNA2 appears to be restricted to higher eukaryotes.
Deletion of yeast SAE2 results in profound cellular sensitivity to DNA-damaging drugs and a resection defect. While Sae2 and the Mre11 nuclease are critical to remove Spo11, stalled topoisomerases, or secondary DNA structures, the MRX-Sae2 resection pathway, per se, is partially dispensable for the processing of clean DSBs in yeast, as shown by a less pronounced resection defect of mre11 nuclease-deficient mutants compared to sae2Δ (64). What is then causing the more severe resection defect of sae2Δ cells? The phenotypic difference between mre11 nuclease-deficient and sae2Δ has been linked to checkpoint hyperactivation in sae2Δ (46); Sae2 was shown to have a structural function to dampen Tel1-dependent checkpoint signaling that leads to a permanent checkpoint arrest in its absence (45). Upon DNA damage without Sae2, the activated Rad9 checkpoint protein limits resection by Sgs1-Dna2 and causes cellular lethality (46, 47, 65). Importantly, in the rad9Δ checkpoint-defective background, the phenotypes of mre11 nuclease-deficient and sae2Δ mutants are indistinguishable, and the phenotypic differences between mre11 nuclease-deficient and sae2Δ can thus be fully explained by differential checkpoint activation (45). These results support our observation that yeast Sae2 does not directly stimulate long-range resection by Dna2.
In contrast to yeast, defects in CtIP almost completely inhibit DNA end resection in human cells, including that of nuclease-generated clean breaks (10). Accordingly, depletion of CtIP is used to abrogate resection in many experimental systems in human cells. While a signaling loop similar to that established in yeast has not been uncovered in human cells yet, our results presented here demonstrate a direct function of CtIP to promote long-range resection by DNA2, which could help explain the differential requirement for Sae2 and CtIP in resection in high and low eukaryotes. At the same time, it is likely that the short-range resection is generally more important in human cells. The critical function of CtIP in resection could thus also result from an upstream placement of the MRN-CtIP–dependent DNA clipping. Abrogation of a critical upstream step would then mask an involvement of CtIP downstream of MRE11, explaining similar phenotypes resulting from the down-regulation of CtIP and chemical inhibition of the MRE11 nuclease observed in some studies (11). Other studies, including ours (SI Appendix, Fig. S1 B and C) (49, 50), in contrast, noted a more severe resection defect associated with CtIP depletion compared to inhibition or mutagenesis of the MRE11 nuclease active site, implicating other proresection functions for CtIP beyond the regulation of MRE11. Mre11 nuclease- and Sae2-deficiency both result in embryonic lethality in mice; however, Sae2-deficient embryos succumb earlier (20, 66). Additionally, studies in human and murine cells, as well as Xenopus egg extracts, noted that CtIP may promote the long-range dependent pathway by DNA2 (48–50). The results presented here strongly support these data and provide a mechanistic explanation for CtIP and DNA2 functions in long-range resection.
Our data indicate that CtIP primarily functions to stimulate the ATPase-dependent ssDNA translocase activity of DNA2. While DNA2 is primarily active as a nuclease, the function of its helicase domain remained somewhat elusive. We previously reported that the helicase activity of DNA2 becomes only apparent after inactivation of the nuclease (29, 37). Therefore, the DNA2 motor does not likely function as a helicase to unwind dsDNA, but rather as an ssDNA translocase, which is readily observed with the WT protein (38, 39). According to this model, the ssDNA translocase activity helps feed ssDNA to the nuclease domain of DNA2 and facilitates thus its degradation. Moreover, DNA unwound by the cognate RecQ family helicase partner is coated by RPA, which directs the DNA2 nuclease to degrade the 5′-terminated ssDNA strand (31, 32). However, as only naked ssDNA can pass through the channel within the DNA2 structure to reach the nuclease domain (33), RPA needs to be displaced, requiring an active mechanism. The motor activity of DNA2 plays an important function during this process as it accelerates DNA degradation primarily when RPA is present (38). The results presented here demonstrate that pCtIP further accelerates the motor activity of DNA2 and thus indirectly stimulates ssDNA degradation by its nuclease domain (Fig. 7A). The stimulation of the DNA2 motor is not the only way how CtIP promotes the BLM-DNA2 long-range resection pathway. Sung and colleagues previously reported that CtIP also stimulates DNA unwinding by BLM (52), which functions upstream of DNA2 (Fig. 7A). We could confirm this stimulatory effect. In single-molecule experiments, we observed that pCtIP accelerated DNA unwinding by BLM by ∼1.5-fold while it stimulated DNA2 by ∼10-fold under the same experimental conditions in single-molecule assays, demonstrating a comparatively larger effect of pCtIP on DNA2 compared to BLM. Finally, we demonstrate that phosphorylation of pCtIP facilitates its capacity to promote DNA2, possibly rather nonspecifically by preventing CtIP aggregation. Therefore, similarly to the interplay with MRE11, phosphorylation may represent a key regulatory mechanism that keeps DNA2 activity in check although more research is needed to better define this regulatory function. Control of DNA end resection by phosphorylation allows resection only in the S–G2 phases of the cell cycle and upon DNA damage, which limits illegitimate recombination.
Materials and Methods
Data Availability Statement.
All primary data and DNA constructs used for protein expression are available upon request.
Cloning, Expression, and Purification of Recombinant Proteins.
WT DNA2, nuclease-dead DNA2 D277A, helicase-dead DNA2 K654R, and double-dead DNA2 D277A K654R (DNA2 DA/KR) were expressed in Spodoptera frugiperda 9 (Sf9) insect cells and purified by affinity chromatography exploiting the N-terminal 6×his-tag and the C-terminal FLAG-tag (29, 67). BLM, WRN, phosphorylated Sae2, and phosphorylated WT CtIP and variants were obtained by taking advantage of the maltose-binding protein (MBP) tag at the N terminus and 10×his-tag at the C terminus. The MBP tag was cleaved off during purification (4, 12, 13, 29). The cloning, expression, and purification of other proteins and their variants are detailed in SI Appendix.
Biochemical Assays.
DNA end resection assays with PCR-based dsDNA substrate were performed in a 15-µL volume in 25 mM Tris-acetate, pH 7.5, 2 mM magnesium acetate, 1 mM ATP, 1 mM dithiothreitol (DTT), 0.1 mg/mL bovine serum albumin (BSA) (New England Biolabs), 1 mM phosphoenolpyruvate (PEP), 80 U/mL pyruvate kinase (Sigma), 50 mM NaCl, and 1 nM substrate (in molecules). When randomly labeled ssDNA was used, 2 nM substrate (in molecules) was added instead. Human RPA was included as indicated to saturate all ssDNA. Additional recombinant proteins were then added on ice, and the reactions were incubated at 37 °C as indicated to perform kinetic experiments. Reactions were stopped by adding 5 µL of 2% stop solution (150 mM ethylenediaminetetraacetic acid [EDTA], 2% sodium dodecyl sulfate, 30% glycerol, bromophenol blue) and 1 µL of proteinase K (18 mg/mL; Roche) and incubated at 37 °C for 10 min. Samples were analyzed by 1% agarose gel electrophoresis. Gels were dried on DE81 chromatography paper (Whatman), exposed to storage phosphor screens (GE Healthcare), and scanned by a Typhoon 9500 phosphorimager (GE Healthcare). Analysis of ssDNA degradation was performed with 0.15 nM (in molecules) 3′-labeled λ DNA/HindIII fragments that were heat-denatured for 5 min at 95 °C before adding to the reaction mixture. Differently from the resection assays above, the reaction buffer contained 3 mM magnesium acetate and 20 mM NaCl, and, unless indicated otherwise, reactions were incubated at 37 °C for 8 min. When yeast proteins were used, the experiment was performed at 25 °C for 1 min, and yeast RPA was added in each reaction. Nuclease assays with 32P-labeled Y-shaped DNA substrate (0.1 nM in molecules) were carried out in a similar buffer containing 100 mM NaCl at 37 °C, and the reactions were stopped by adding 0.5 µL of 0.5 M EDTA and 1 μL of proteinase K (18 mg/mL; Roche) and incubated at 50 °C for 30 min. An equal amount of formamide dye (95% [vol/vol] formamide, 20 mM EDTA, and bromophenol blue) was added, and samples were heated at 95 °C for 4 min and separated on 15% denaturing polyacrylamide gels (ratio acrylamide:bisacrylamide 19:1; Bio-Rad). After fixing in a solution containing 40% methanol, 10% acetic acid, and 5% glycerol for 30 min, the gels were dried on 3MM paper (Whatman) and analyzed as described above. Endonuclease assays with MRN and pCtIP (15-µL volume) were performed in nuclease buffer containing 25 mM Tris⋅HCl, pH 7.5, 5 mM magnesium acetate, 1 mM manganese acetate, 1 mM ATP, 1 mM DTT, 0.25 mg/mL BSA, 1 mM PEP, 80 U/mL pyruvate kinase, and 1 nM oligonucleotide-based DNA substrate (in molecules). Biotinylated DNA ends were blocked by adding 15 nM streptavidin and incubating the samples 5 min at room temperature. Samples were then processed and analyzed as described above with 15% denaturing gels (13, 68). Where unlabeled pAttP-S–based DNA substrates were used, the reaction buffer was prepared with 20 mM NaCl, but with 3 mM magnesium acetate and 30 nM streptavidin to block biotinylated DNA ends as indicated. Reactions were incubated at 37 °C for 30 min, and DNA was visualized by staining with GelRed (Biotium).
Helicase, ATPase, magnetic tweezer, protein interaction, and SSA assays are described in SI Appendix. Additional primary data underlying graphs and uncropped gel images are shown in SI Appendix, Fig. S10.
Supplementary Material
Acknowledgments
We thank Dr. Davide Moiani and Dr. John Tainer (MD Anderson Cancer Center) for providing MRE11 inhibitors. We thank members of the P.C. laboratory (A. Acharya, G. Reginato, E. Cannavo, A. Sanchez, and S. Halder) for critical comments on the manuscript. This work was supported by the European Research Council (ERC) (Grant HRMECH, 681630) and the Swiss National Science Foundation (Grant 31003A_175444).
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2001165117/-/DCSupplemental.
References
- 1.Ranjha L., Howard S. M., Cejka P., Main steps in DNA double-strand break repair: An introduction to homologous recombination and related processes. Chromosoma 127, 187–214 (2018). [DOI] [PubMed] [Google Scholar]
- 2.Cejka P., DNA end resection: Nucleases team up with the right partners to initiate homologous recombination. J. Biol. Chem. 290, 22931–22938 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bonetti D., Colombo C. V., Clerici M., Longhese M. P., Processing of DNA ends in the maintenance of genome stability. Front. Genet. 9, 390 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cannavo E., et al. , Regulatory control of DNA end resection by Sae2 phosphorylation. Nat. Commun. 9, 4016 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Huertas P., Jackson S. P., Human CtIP mediates cell cycle control of DNA end resection and double strand break repair. J. Biol. Chem. 284, 9558–9565 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Huertas P., Cortés-Ledesma F., Sartori A. A., Aguilera A., Jackson S. P., CDK targets Sae2 to control DNA-end resection and homologous recombination. Nature 455, 689–692 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ira G., et al. , DNA end resection, homologous recombination and DNA damage checkpoint activation require CDK1. Nature 431, 1011–1017 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Aylon Y., Liefshitz B., Kupiec M., The CDK regulates repair of double-strand breaks by homologous recombination during the cell cycle. EMBO J. 23, 4868–4875 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chen X., et al. , Cell cycle regulation of DNA double-strand break end resection by Cdk1-dependent Dna2 phosphorylation. Nat. Struct. Mol. Biol. 18, 1015–1019 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sartori A. A., et al. , Human CtIP promotes DNA end resection. Nature 450, 509–514 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Shibata A., et al. , DNA double-strand break repair pathway choice is directed by distinct MRE11 nuclease activities. Mol. Cell 53, 7–18 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cannavo E., Cejka P., Sae2 promotes dsDNA endonuclease activity within Mre11-Rad50-Xrs2 to resect DNA breaks. Nature 514, 122–125 (2014). [DOI] [PubMed] [Google Scholar]
- 13.Anand R., Ranjha L., Cannavo E., Cejka P., Phosphorylated CtIP functions as a co-factor of the MRE11-RAD50-NBS1 endonuclease in DNA end resection. Mol. Cell 64, 940–950 (2016). [DOI] [PubMed] [Google Scholar]
- 14.Reginato G., Cannavo E., Cejka P., Physiological protein blocks direct the Mre11-Rad50-Xrs2 and Sae2 nuclease complex to initiate DNA end resection. Genes Dev. 31, 2325–2330 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wang W., Daley J. M., Kwon Y., Krasner D. S., Sung P., Plasticity of the Mre11-Rad50-Xrs2-Sae2 nuclease ensemble in the processing of DNA-bound obstacles. Genes Dev. 31, 2331–2336 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Liao S., Tammaro M., Yan H., The structure of ends determines the pathway choice and Mre11 nuclease dependency of DNA double-strand break repair. Nucleic Acids Res. 44, 5689–5701 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hoa N. N., et al. , Mre11 is essential for the removal of lethal topoisomerase 2 covalent cleavage complexes. Mol. Cell 64, 580–592 (2016). [DOI] [PubMed] [Google Scholar]
- 18.Wilkinson O. J., et al. , CtIP forms a tetrameric dumbbell-shaped particle which bridges complex DNA end structures for double-strand break repair. eLife 8, e42129 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Orthwein A., et al. , A mechanism for the suppression of homologous recombination in G1 cells. Nature 528, 422–426 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 20.Polato F., et al. , CtIP-mediated resection is essential for viability and can operate independently of BRCA1. J. Exp. Med. 211, 1027–1036 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Liu X., et al. , CtIP is essential for early B cell proliferation and development in mice. J. Exp. Med. 216, 1648–1663 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cannavo E., Cejka P., Kowalczykowski S. C., Relationship of DNA degradation by Saccharomyces cerevisiae exonuclease 1 and its stimulation by RPA and Mre11-Rad50-Xrs2 to DNA end resection. Proc. Natl. Acad. Sci. U.S.A. 110, E1661–E1668 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gravel S., Chapman J. R., Magill C., Jackson S. P., DNA helicases Sgs1 and BLM promote DNA double-strand break resection. Genes Dev. 22, 2767–2772 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhu Z., Chung W. H., Shim E. Y., Lee S. E., Ira G., Sgs1 helicase and two nucleases Dna2 and Exo1 resect DNA double-strand break ends. Cell 134, 981–994 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mimitou E. P., Symington L. S., Sae2, Exo1 and Sgs1 collaborate in DNA double-strand break processing. Nature 455, 770–774 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nimonkar A. V., et al. , BLM-DNA2-RPA-MRN and EXO1-BLM-RPA-MRN constitute two DNA end resection machineries for human DNA break repair. Genes Dev. 25, 350–362 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Liao S., Toczylowski T., Yan H., Mechanistic analysis of Xenopus EXO1‘s function in 5’-strand resection at DNA double-strand breaks. Nucleic Acids Res. 39, 5967–5977 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sturzenegger A., et al. , DNA2 cooperates with the WRN and BLM RecQ helicases to mediate long-range DNA end resection in human cells. J. Biol. Chem. 289, 27314–27326 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pinto C., Kasaciunaite K., Seidel R., Cejka P., Human DNA2 possesses a cryptic DNA unwinding activity that functionally integrates with BLM or WRN helicases. eLife 5, e18574 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yan H., Toczylowski T., McCane J., Chen C., Liao S., Replication protein A promotes 5′–>3′ end processing during homology-dependent DNA double-strand break repair. J. Cell Biol. 192, 251–261 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cejka P., et al. , DNA end resection by Dna2-Sgs1-RPA and its stimulation by Top3-Rmi1 and Mre11-Rad50-Xrs2. Nature 467, 112–116 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Niu H., et al. , Mechanism of the ATP-dependent DNA end-resection machinery from Saccharomyces cerevisiae. Nature 467, 108–111 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhou C., Pourmal S., Pavletich N. P., Dna2 nuclease-helicase structure, mechanism and regulation by Rpa. eLife 4, e09832 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Balakrishnan L., Polaczek P., Pokharel S., Campbell J. L., Bambara R. A., Dna2 exhibits a unique strand end-dependent helicase function. J. Biol. Chem. 285, 38861–38868 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Masuda-Sasa T., Imamura O., Campbell J. L., Biochemical analysis of human Dna2. Nucleic Acids Res. 34, 1865–1875 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Duxin J. P., et al. , Okazaki fragment processing-independent role for human Dna2 enzyme during DNA replication. J. Biol. Chem. 287, 21980–21991 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Levikova M., Klaue D., Seidel R., Cejka P., Nuclease activity of Saccharomyces cerevisiae Dna2 inhibits its potent DNA helicase activity. Proc. Natl. Acad. Sci. U.S.A. 110, E1992–E2001 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Levikova M., Pinto C., Cejka P., The motor activity of DNA2 functions as an ssDNA translocase to promote DNA end resection. Genes Dev. 31, 493–502 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Miller A. S., et al. , A novel role of the Dna2 translocase function in DNA break resection. Genes Dev. 31, 503–510 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lengsfeld B. M., Rattray A. J., Bhaskara V., Ghirlando R., Paull T. T., Sae2 is an endonuclease that processes hairpin DNA cooperatively with the Mre11/Rad50/Xrs2 complex. Mol. Cell 28, 638–651 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Makharashvili N., et al. , Catalytic and noncatalytic roles of the CtIP endonuclease in double-strand break end resection. Mol. Cell 54, 1022–1033 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wang H., et al. , CtIP maintains stability at common fragile sites and inverted repeats by end resection-independent endonuclease activity. Mol. Cell 54, 1012–1021 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Andres S. N., Williams R. S., CtIP/Ctp1/Sae2, molecular form fit for function. DNA Repair (Amst.) 56, 109–117 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Andres S. N., Li Z. M., Erie D. A., Williams R. S., Ctp1 protein-DNA filaments promote DNA bridging and DNA double-strand break repair. J. Biol. Chem. 294, 3312–3320 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yu T. Y., Kimble M. T., Symington L. S., Sae2 antagonizes Rad9 accumulation at DNA double-strand breaks to attenuate checkpoint signaling and facilitate end resection. Proc. Natl. Acad. Sci. U.S.A. 115, E11961–E11969 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Gobbini E., et al. , Sae2 function at DNA double-strand breaks is bypassed by dampening Tel1 or Rad53 activity. PLoS Genet. 11, e1005685 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Colombo C. V., et al. , Uncoupling Sae2 functions in downregulation of Tel1 and Rad53 signaling activities. Genetics 211, 515–530 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Peterson S. E., et al. , Activation of DSB processing requires phosphorylation of CtIP by ATR. Mol. Cell 49, 657–667 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hoa N. N., et al. , BRCA1 and CtIP are both required to recruit Dna2 at double-strand breaks in homologous recombination. PLoS One 10, e0124495 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hoa N. N., et al. , Relative contribution of four nucleases, CtIP, Dna2, Exo1 and Mre11, to the initial step of DNA double-strand break repair by homologous recombination in both the chicken DT40 and human TK6 cell lines. Genes Cells 20, 1059–1076 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Cruz-García A., López-Saavedra A., Huertas P., BRCA1 accelerates CtIP-mediated DNA-end resection. Cell Rep. 9, 451–459 (2014). [DOI] [PubMed] [Google Scholar]
- 52.Daley J. M., et al. , Enhancement of BLM-DNA2-mediated long-range DNA end resection by CtIP. Cell Rep. 21, 324–332 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Bhargava R., Onyango D. O., Stark J. M., Regulation of single-strand annealing and its role in genome maintenance. Trends Genet. 32, 566–575 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Gunn A., Stark J. M., I-SceI-based assays to examine distinct repair outcomes of mammalian chromosomal double strand breaks. Methods Mol. Biol. 920, 379–391 (2012). [DOI] [PubMed] [Google Scholar]
- 55.Kasaciunaite K., et al. , Competing interaction partners modulate the activity of Sgs1 helicase during DNA end resection. EMBO J. 38, e101516 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Davies O. R., et al. , CtIP tetramer assembly is required for DNA-end resection and repair. Nat. Struct. Mol. Biol. 22, 150–157 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Anand R., et al. , NBS1 promotes the endonuclease activity of the MRE11-RAD50 complex by sensing CtIP phosphorylation. EMBO J. 38, e101005 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Fu Q., et al. , Phosphorylation-regulated transitions in an oligomeric state control the activity of the Sae2 DNA repair enzyme. Mol. Cell. Biol. 34, 778–793 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Andres S. N., et al. , Tetrameric Ctp1 coordinates DNA binding and DNA bridging in DNA double-strand-break repair. Nat. Struct. Mol. Biol. 22, 158–166 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wang H., et al. , PLK1 targets CtIP to promote microhomology-mediated end joining. Nucleic Acids Res. 46, 10724–10739 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Deshpande R. A., Lee J. H., Arora S., Paull T. T., Nbs1 converts the human mre11/rad50 nuclease complex into an endo/exonuclease machine specific for protein-DNA adducts. Mol. Cell 64, 593–606 (2016). [DOI] [PubMed] [Google Scholar]
- 62.Fekairi S., et al. , Human SLX4 is a Holliday junction resolvase subunit that binds multiple DNA repair/recombination endonucleases. Cell 138, 78–89 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Wyatt H. D., Laister R. C., Martin S. R., Arrowsmith C. H., West S. C., The SMX DNA repair tri-nuclease. Mol. Cell 65 , 848–860.e11 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Llorente B., Symington L. S., The Mre11 nuclease is not required for 5′ to 3′ resection at multiple HO-induced double-strand breaks. Mol. Cell. Biol. 24, 9682–9694 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Bonetti D., et al. , Escape of Sgs1 from Rad9 inhibition reduces the requirement for Sae2 and functional MRX in DNA end resection. EMBO Rep. 16, 351–361 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Buis J., et al. , Mre11 nuclease activity has essential roles in DNA repair and genomic stability distinct from ATM activation. Cell 135, 85–96 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Anand R., Pinto C., Cejka P., Methods to study DNA end resection I: Recombinant protein purification. Methods Enzymol. 600, 25–66 (2018). [DOI] [PubMed] [Google Scholar]
- 68.Pinto C., Anand R., Cejka P., Methods to study DNA end resection II: Biochemical reconstitution assays. Methods Enzymol. 600, 67–106 (2018). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All primary data and DNA constructs used for protein expression are available upon request.