Significance
We previously identified the cellular target of halofuginone (HF), a drug that suppresses tissue damage in a variety of disease settings, but its mechanism of tissue action remains unclear. HF inhibits a mammalian enzyme that fuses proline to its cognate tRNA during protein synthesis, thereby mimicking amino acid limitation. Mammalian cellular amino acid levels are understood to be monitored via two signaling pathways: The amino acid response and the mammalian target of rapamycin complex 1. We find that both HF treatment and cellular amino acid restriction can suppress inflammatory responses in cytokine-stimulated synoviocytes, without utilizing either of these pathways. Thus, we clarify the tissue action of HF and identify a previously unrecognized amino acid sensing-pathway that mediates inflammatory suppression.
Keywords: halofuginone (HF), aminoacyl-tRNA synthetase (aaRS) inhibition, GCN2, GCN1, amino acid catabolism
Abstract
Signaling pathways that sense amino acid abundance are integral to tissue homeostasis and cellular defense. Our laboratory has previously shown that halofuginone (HF) inhibits the prolyl-tRNA synthetase catalytic activity of glutamyl-prolyl-tRNA synthetase (EPRS), thereby activating the amino acid response (AAR). We now show that HF treatment selectively inhibits inflammatory responses in diverse cell types and that these therapeutic benefits occur in cells that lack GCN2, the signature effector of the AAR. Depletion of arginine, histidine, or lysine from cultured fibroblast-like synoviocytes recapitulates key aspects of HF treatment, without utilizing GCN2 or mammalian target of rapamycin complex 1 pathway signaling. Like HF, the threonyl-tRNA synthetase inhibitor borrelidin suppresses the induction of tissue remodeling and inflammatory mediators in cytokine-stimulated fibroblast-like synoviocytes without GCN2, but both aminoacyl-tRNA synthetase (aaRS) inhibitors are sensitive to the removal of GCN1. GCN1, an upstream component of the AAR pathway, binds to ribosomes and is required for GCN2 activation. These observations indicate that aaRS inhibitors, like HF, can modulate inflammatory response without the AAR/GCN2 signaling cassette, and that GCN1 has a role that is distinct from its activation of GCN2. We propose that GCN1 participates in a previously unrecognized amino acid sensor pathway that branches from the canonical AAR.
The ability to successfully respond to environmental stimuli, such as changing nutrient conditions, is essential to an organism’s survival. Reprogramming of the transcriptome and proteome is an evolutionarily conserved means of homeostatic adjustment to stress (1–5). The protein synthetic apparatus senses and responds to organismal and cellular stress by multiple means (6–10). Stringent control, first described in bacteria, is a programmatic adaption to nutrient limitation that utilizes the protein synthetic machinery for signal generation (1, 11). The stringent response pathway senses amino acid stress by detecting nonaminoacylated (uncharged) tRNA and responds by inducing comprehensive transcriptional reprogramming that elicits growth arrest, up-regulates survival genes, and increases virulence in pathogenic bacteria (10, 11). In both prokaryotic and eukaryotic cells, amino acid insufficiency leads to depletion of available aminoacylated tRNA for addition to nascent peptide chains, resulting in paused ribosomes that have an uncharged tRNA occupying the ribosomal A site (10, 12, 13). Mammalian cells, like bacteria, sense amino acid limitation through the accumulation of uncharged tRNA, but the mammalian cell relays this signal through an arm of the integrated stress response (ISR), called the amino acid response (AAR) (2, 14–16). Our laboratory has shown that the natural product derivative halofuginone (HF) targets human glutamyl-prolyl-tRNA synthetase (EPRS) and inhibits its proly-tRNA synthetase (PRS) activity to confer therapeutic benefits (17). Aminoacyl-tRNA synthetases (aaRSs), such as EPRS, are ubiquitous, essential protein synthetic enzymes that fuse an amino acid to its cognate tRNA. Both HF inhibition of EPRS and amino acid insufficiency reduce the cell’s capacity to aminoacylate tRNA and, therefore, trigger the accumulation of uncharged tRNA (5, 8, 17). HF prevents tissue damage in a wide variety of disease settings (18–23). In our effort to understand how HF’s inhibition of EPRS mediates a programmatic change in diverse inflamed tissues, we considered the evolutionarily conserved utilization of the protein synthetic apparatus for adaptive signaling, as well as the immune motif of regulated amino acid catabolism for inflammatory suppression.
Metabolic stress pathways and nutrient availability instruct immunity (24–31). To maintain tissue homeostasis and safeguard organ integrity, the mammalian immune system balances protective wound healing and inflammatory responses with their timely resolution. It utilizes the induced breakdown of amino acids to temper hyperinflammatory innate responses, foster tolerance to self, and promote tissue integrity (26, 28, 29, 32). Immune cells operate in both nutrient-rich and -restricted niches, such as the gut and tumor microenvironment, which impact their function (33). Although competition for resources is intense in both inflamed and cancerous tissues, in each of these settings multiple types of immune cells up-regulate amino acid catabolizing enzymes, such as indoleamine 2,3-dioxygenase (IDO1) or arginase 1 (Arg1), to regulate their environment (29, 34–36). Amino acid catabolism at sites of inflammation and in the tumor microenvironment suppresses tissue inflammation, but impedes antitumor immunity (34, 35, 37, 38). Amino acid availability is monitored by two recognized sensors: The AAR and the mammalian target of rapamycin complex 1 (mTORC1) pathway (26, 39). The AAR and mTORC1 pathways each have important regulatory roles in immune response and healthspan (32, 40–45). Both pathways sense and respond to cellular amino acid restriction, but they are differentially activated and elicit distinct sets of transcriptional responses and biological effects. Whereas the mTORC1 pathway can sense amino acids directly (39) and is inhibited by amino acid limitation, the AAR pathway is activated by insufficiency of any amino acid (46, 47) and by the proximal trigger of uncharged tRNA accumulation (48–50). Uncharged tRNA binds and activates the protein kinase GCN2 (50–52), resulting in GCN2 autophosphorylation and the phosphorylation of eIF2α, the shared component of the ISR (2, 5, 14). Cellular treatment with an aaRS-inhibitor activates the AAR (17, 53), but does not directly affect mTORC1 signaling (22, 54), making it a useful means to distinguish between cellular responses that are initiated by uncharged tRNA accumulation/ribosomal pausing and those that result from inhibition of the mTORC1 pathway.
Cross-talk between nonimmune cells and tissue resident or recruited immune cells within organs or at barrier sites is essential for response to injury and infection (55, 56). This communication involves diverse cell types, both parenchymal and stromal constituents of tissue structures (hereafter structural cells), and underpins both response to damage, inflammatory resolution, and the development of chronic disease (55, 57, 58). The language of this cross-talk is incompletely understood, but amino acid levels in the tissue microenvironment are central to the maintenance of tissue and immune homeostasis (29), and amino acid sensor pathways have crucial immunoregulatory functions (28, 42, 59). HF mimics amino acid restriction by inhibiting EPRS, and acts directly upon fibroblastic and immune cells to suppress tissue remodeling and inflammatory responses (17, 21, 60–63). Because HF competes with proline for binding to the active site of PRS, the in vitro addition of excess proline reverses HF-mediated cellular responses that are due to PRS inhibition (17). Using proline rescue of HF-treated cells, we’ve shown that HF’s suppression of both collagen transcription in fibroblasts and differentiation CD4+ T helper 17 (TH17) cells (22) is due to PRS inhibition (17). In addition to in vitro studies, HF has been used to treat a wide range of fibrotic manifestations in animal models (61, 63–66) and human disease. HF also has been used for the effective treatment of autoimmune disease, consistent with its activity as an inhibitor of TH17 cell differentiation and proinflammatory function (18). Because HF has been studied extensively as an antifibrotic drug (21, 67), much has been made of its ability to inhibit collagen deposition. Others have proposed that HF’s primary mechanism of action in this setting is the direct inhibition of TGF-β–stimulated Smad signaling (21, 62, 63, 68). The observation that HF acts directly upon both nonimmune and immune cells via PRS inhibition, regulating tissue homeostasis and immunity (69), strongly suggests to us that mimicking amino acid stress is the basis of HF’s broad tissue benefit.
To better understand how HF might mediate a programmatic change in inflamed structural tissues, our laboratory utilized an in vitro culture system of TNF-α–stimulated fibroblast-like synoviocytes (FLS). FLS are the resident mesenchymal cells in the lining of synovial joints, which play a key role in the pathogenesis of rheumatoid arthritis (RA) (70–72). In the RA synovium, FLS become activated, expressing a well-characterized program of matrix metalloproteinases (MMPs), inflammatory cytokines, and chemokines that attract and activate immune cells, promoting ongoing inflammation and tissue destruction (71–74). Reflecting key differences in the disease pathologies of RA and fibrosis, activated FLS induce net collagen destruction, rather than collagen deposition in inflamed tissues. This difference allows us to study our proposed therapeutic mechanism of HF action—that HF acts to suppress inflammatory tissue remodeling in structural cells—apart from the more narrow, proposed mechanism, that HF acts as an inhibitor of TGF-β–induced collagen deposition. For these reasons, FLS are an ideal complement to stromal fibroblasts for a broad study of HF’s therapeutic tissue mechanism.
Using either primary RA-FLS or established FLS cultures, we find that HF suppresses a subset of TNF-α–stimulated responses, mirroring HF’s suppression of proinflammatory functions in mature TH17 memory cells (18) and its suppression of TGF-β–stimulated fibrotic tissue remodeling. Importantly, we show here that HF-mediated suppression of diverse inflammatory programs can occur in cells that lack GCN2, the signature effector of the AAR. We further show that amino acid deprivation and treatment with borrelidin, a catalytic site inhibitor of threonyl-tRNA synthetase (TRS) (75), each recapitulate significant aspects of HF treatment in activated FLS, without utilizing GCN2 or mTORC1 signaling. The capacity of amino acid stress and aaRS inhibitors to signal in cells that lack GCN2 indicates the presence of a previously unrecognized cellular pathway that responds to a reduction in aminoacyl-tRNA levels (SI Appendix, Fig. S1). We further find that when FLS are depleted of the upstream AAR pathway component GCN1 (76), HF- and borrelidin-mediated suppression of the inflammatory response is impaired.
Results
HF Suppresses a Distinct Subset of the TNF-α–Induced Program in FLS.
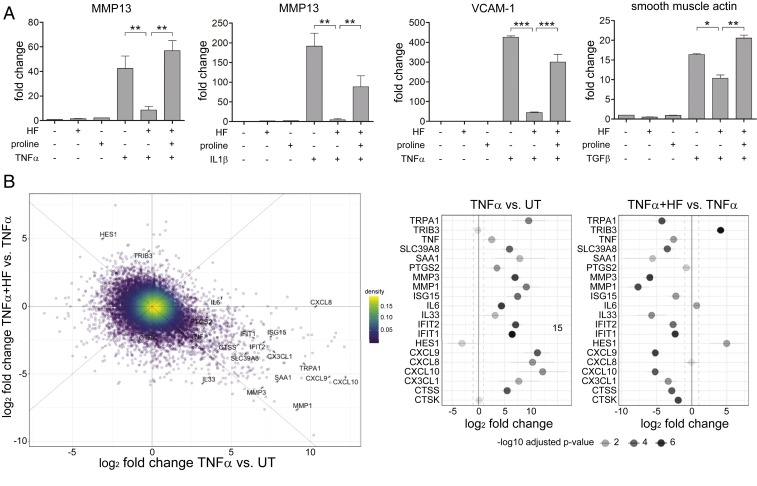
To study the effects of HF treatment on cytokine-stimulated structural cells, we first treated primary RA-FLS with either TNF-α or IL-1β, in the presence or absence of HF, and assayed for MMP13 expression using qRT-PCR. In RA-FLS, both TNF-α and IL-1β strongly induced the expression of MMP13, a collagenase involved in the pathophysiology of arthritis (77) (Fig. 1A), and HF potently suppressed this induction. The suppression of cytokine-induced MMP13 expression was reversed by the addition of excess proline to culture medium (Fig. 1A), confirming the role of PRS inhibition in this treatment model. HF treatment of RA-FLS at therapeutic doses did not affect global protein synthesis over 24 h (SI Appendix, Fig. S2), and cells that were pretreated with HF remained refractory to TNF-α–induced MMP13 up-regulation for up at least 72 h (SI Appendix, Fig. S3). These results indicate that EPRS-mediated HF treatment results in durable, selective reprogramming of RA-FLS that suppresses a key participant in the TNF-α– and IL-1β–driven, tissue-destructive response in RA.
Fig. 1.
HF inhibits multiple cytokine responses in a range of primary cell types. (A, Left two panels) HF inhibition of MMP13 induction by TNF-α or IL1-β in human FLS (RA-FLS). Human RA-FLS were pretreated with 200 nM HF and/or proline (2 mM) overnight and treated with 10 ng/mL TNF-α for 6 h and mRNA analyzed for MMP13 expression by qRT-PCR. (Right two panels) HUVEC were treated and analyzed for gene expression as described for FLS, above; human LL29 lung fibroblasts were pretreated with HF and/or proline for 6 h and treated with TGF-β for 24 h and analyzed for gene expression, as above. Results are representative of three independent experiments (means ± SD, n = 3). *P < 0.05, **P < 0.01, ***P < 0.001. (B) RNA-seq analysis of HF effects on TNF-α responses. (B, Left) Selective regulation of TNF-α–inducible proinflammatory gene expression in FLS, determined by RNA-seq after 24-h culture in media alone, TNF-α, or TNF-α plus HF. Limma-voom derived log2-fold transformed gene-expression changes for two comparisons are shown; treatment with TNF-α (x axis) as compared to untreated (x axis) or combined TNF-α and HF treatment as compared to TNF-α treatment (y axis). Selected genes are highlighted in the figure with lines connecting to their respective data points. Only genes with real reads per kilobase of transcript per million mapped reads (RPKM) values (>0) are shown (n = 12,973). (Right) Representation of fold-changes following treatment with TNF-α or TNF-α and HF in specific genes referred to in the text. Log2-fold transformed gene-expression changes and their respective 95% confidence intervals are shown for TNF-α versus untreated (UT, Left) and TNF-α and HF treatment versus TNF-α alone treatment (Right) for genes of interest. The respective false-discovery rate-adjusted P values for the gene-expression changes are shown via shading of the plotted values. The vertical black line represents no change (log2 fold-change = 0, or fold-change = 1) with the dotted lines representing twofold expression change in either direction of the comparison.
To expand our observations to the cytokine stimulation of other structural cells, we treated TNF-α– or IL-1β–stimulated primary human endothelial cells (HUVEC) with HF, and saw HF inhibition of the cytokine-induction of VCAM-1 and E-selectin (Fig. 1A and SI Appendix, Fig. S4), two critical mediators of vascular inflammation (78–80). Similarly, in lung fibroblasts HF inhibited TGF-β–driven induction of smooth muscle actin and CTGF (Fig. 1A and SI Appendix, Fig. S4), two key profibrotic factors (81, 82). In each of these cases, HF’s suppression of proinflammatory and profibrotic structural cell responses was opposed by the presence of excess proline (Fig. 1A and SI Appendix, Fig. S4). These data demonstrate that HF acts via PRS inhibition to suppress the cytokine induction of key inflammatory and tissue remodeling responses in diverse cell types.
To provide an unbiased look at the effects of HF on a program of inflammatory response, we performed RNA sequencing (RNA-seq) and differential expression analysis on RA-FLS treated with TNF-α, HF, or both. More than 80% (10 of 12) of genes induced ≥1,000-fold by TNF-α stimulation were suppressed at least 5-fold in HF-treated FLS, whereas only half (83 of 171) of the genes induced between 16- and 64-fold by TNF-α stimulation, and only 5% of the total FLS transcriptome were similarly sensitive to HF treatment (Fig. 1B). Examples of TNF-α–inducible genes in FLS that were inhibited by HF included: 1) Several matrix degrading proteases (MMP1, MMP3, MMP9, MMP13, CTSS, CTSK); 2) a series of proinflammatory chemokines and cytokines (e.g., TNF-α, IL-33, CXCL9, CXCL10); 3) SLC39A8/ZIP8, a zinc transporter implicated in RA-associated joint damage (83); and 4) the serum amyloids SAA1 and SAA2, secreted factors known to promote TH17 cell activation (84, 85). Because cytokine induced SLC39A8/ZIP8 has a key role in the regulation of tissue damage in arthritis (83), we directly confirmed that HF treatment dramatically reduces SLC39A8/ZIP8 protein levels in TNF-α–treated cells in parallel with mRNA inhibition (SI Appendix, Fig. S5). Conversely, HF treatment of RA-FLS induced the expression of a smaller number of genes that were repressed by TNF-α (e.g., 65 genes inhibited by TNF-α ≥ 10-fold) (Fig. 1B and Dataset S1) (86). HES1, a repressor of transcriptional elongation that suppresses CXCL1 production and thus neutrophil recruitment (87), is one notable example from this group. Overall, HF showed a broad pattern of inhibition of TNF-α–induced genes or enhancement of TNF-α–inhibited genes, having less effect on genes that were neither induced or inhibited by TNF-α (SI Appendix, Fig. S6). Notably, these effects were selective in that HF somewhat enhanced or had no effect on the expression a distinct subset of genes that were potently induced by TNF-α, including PTGS2, IL6, and CXCL8 (Fig. 1B). These HF-resistant genes, like most of the HF-sensitive TNF-α–induced genes, are well established transcriptional targets of NF-κB-activation downstream of both TNF-α and IL-1β signaling (88, 89), indicating that HF’s inhibition of cytokine signaling is not associated with a global inhibition of NF-κB activation by cytokines. Taken together, these observations establish that HF can suppress a cytokine-mediated program of inflammatory responses in nonimmune cells, and that this mechanism of inhibition is selective for a subset of transcriptional responses to cytokine treatment.
Both HF treatment and amino acid catabolism instigate the accumulation of uncharged tRNA and resultant ribosomal stalling, which activates GCN2 signaling (8, 52, 90). Once considered to be the mechanistic core of IDO1 immunosuppression (40), the role of GCN2 signaling in this process has been challenged (32, 91, 92). Because HF treatment overlaps with IDO1-induction, both in mechanism and therapeutic tissue response, we next asked: Do TNF-α–stimulated FLS respond to HF treatment in the absence of GCN2 signaling?
HF Suppression of TNF-α–Induced Responses Can Occur Without GCN2 Signaling.
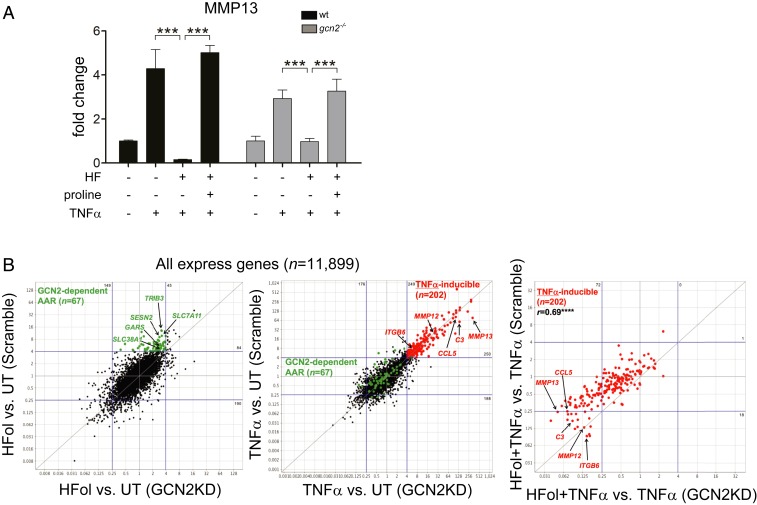
To address this point, we examined the effect of HF treatment on TNF-α–induced responses in GCN2−/− or GCN2-depleted FLS. Phosphorylation of eIF2α by GCN2 kinase leads to transient reduction in cap-dependent mRNA translation, and a concomitant increase in translation of a subset of mRNAs, notably the transcription factor ATF4 (93, 94). Inhibition of mass protein translation conserves cellular amino acids, while up-regulation of ATF4 translation initiates an adaptive response to amino acid stress that includes the expression of genes involved in amino acid transport and biosynthesis, such as TRIB3, ASNS, and SESN2 (53, 95, 96). To assess the presence or absence of intact AAR signaling, we assayed several AAR pathway responses. We show here that, as expected, HF does not induce eif2α phosphorylation or the translation of ATF4 in Gcn2−/− FLS (SI Appendix, Fig. S7). Surprisingly, the ability of HF to suppress TNF-α–induction of Mmp13 expression was not diminished in primary FLS from Gcn2−/− (also known as Eif2ak4−/−) mice (Fig. 2A). Furthermore, HF suppression of MMP13 expression in TNF-α–stimulated Gcn2−/− FLS was reversed by the addition of excess proline (Fig. 2), confirming PRS inhibition as the mechanism of action. To further assess whether the GCN2-eif2α-ATF4 signaling cassette might be dispensable for many of HF’s effects on cytokine-treated cells, we used shRNA-mediated depletion to knock down GCN2 in primary human RA-FLS and in dermal fibroblasts. HF effects on TNF-α–induced responses were unaffected by shRNA-mediated depletion of GCN2 in primary human RA-FLS (SI Appendix, Fig. S8), and depletion of GCN2 did not affect HF suppression of TGF-β–stimulated Col1A1 induction in dermal fibroblasts (SI Appendix, Fig. S9).
Fig. 2.
HF regulates cytokine responses in cells lacking GCN2. (A) FLS from wild-type and gcn2−/− mice were pretreated with HF and/or proline and treated with TNF-α for 6 h. Transcript levels of target genes were quantified by qPCR. Results are representative of three independent experiments (means ± SD, n = 3); ***P < 0.001. (B) Summary of results from RNA-seq transcriptomics comparing HFol and TNF-α action in wild-type and GCN2-depleted K4 FLS. (Left) GCN2 depletion reduces HFol induction of canonical AAR genes (highlighted in green). (Center) GCN2 depletion has no significant effect on TNF-α–induced genes (highlighted in red). (Right) GCN2 depletion either has no effect, or enhances, HFol effects on TNFα induced genes. ****P < 0.0001, two-tailed Pearson correlation test.
We wanted to further examine, in an unbiased manner, the effect of GCN2 on a PRS-inhibitor’s ability to suppress a TNF-α program in the immortalized human FLS cell line K4. As such, we performed, and compared, transcriptomic analysis of wild-type, TNF-α–stimulated K4 cells with or without Halofuginol (HFol) treatment to their GCN2-depleted, TNF-α–stimulated, ±HFol, K4 companions (Fig. 2B and Dataset S2) (86). The PRS-inhibitor HFol is closely chemically related to HF (17), with favorable treatment parameters. GCN2-depletion in K4s markedly reduced HFol induction of AAR pathway response genes (Fig. 2 B, Left), such as TRIB3, SESN2, and the amino acid transporter SLC7A11 (97), confirming an effective functional knockdown of GCN2 signaling in these cells. In contrast, HFol-mediated suppression of TNF-α–responsive genes was either unaffected or slightly enhanced following GCN2 knockdown (Fig. 2 B, Right). Furthermore, GCN2-depletion had no systematic impact on TNF-α–induced gene expression in the absence of HFol treatment (Fig. 2 B, Center). Together, these findings indicate that GCN2 is broadly dispensable for HF/HFol action on TNF-α–induced responses.
TRS Inhibition Recapitulates HF Treatment in GCN2-Depleted Cells.
Inhibition of the catalytic activity of any aaRS, such as PRS, leads to the accumulation of nonaminoacylated tRNA, activating the AAR via GCN2 phosphorylation (17, 22, 53). We demonstrate here, however, that HF inhibition of PRS, as confirmed by proline rescue, suppresses a TNF-α–mediated program of inflammatory responses in the absence of GCN2 signaling, making the tissue mechanism of HF action unclear. We, therefore, considered whether the observed therapeutic effects were unique to EPRS, possibly stemming from a noncanonical EPRS activity (98, 99) incidental to catalytic PRS inhibition, or a general feature of aaRS inhibition and, thus, elicited by uncharged tRNA accumulation. Previously, we showed that selective amino acid depletion and treatment with the TRS inhibitor borrelidin (100) inhibits TH17 cell differentiation comparably to HF (22). Recently, others have shown that borrelidin or leucinol treatment mimics the antifibrotic effects of HF in human cardiac fibroblasts, inhibiting type I collagen deposition, as well as Col1A1 gene and protein expression (61). Therefore, we tested the effects of borrelidin on cytokine responses in primary and established FLS. In primary human RA-FLS, both HF and borrelidin inhibited TNF-α induction of MMP13 and CXCL10 (SI Appendix, Fig. S10) at doses matched to reflect similar activation of the AAR pathway (SI Appendix, Fig. S10). As in the case of HF treatment, borrelidin did not inhibit TNF-α induction of IL-6 or IL-8 (SI Appendix, Fig. S10), confirming that the distinctive specificity of HF action to inhibit some TNF-α–stimulated responses, but not others is shared by another aaRS inhibitor. We then compared the effects of borrelidin and HF on GCN2-depleted K4 FLS; depletion of GCN2 was confirmed by a substantial reduction in borrelidin-induced TRIB3 compared to control K4 cells. Like HF treatment, borrelidin treatment suppressed TNF-α induction of MMP13 and CXCL10 in these cells, as in non-GCN2–depleted control K4s (SI Appendix, Fig. S10), indicating that the effects of HF are a general property of aaRS inhibition and not specific to EPRS inhibition.
Single Amino Acid Depletion Mimics HF Treatment, without Utilizing GCN2 or mTORC1 Signaling.
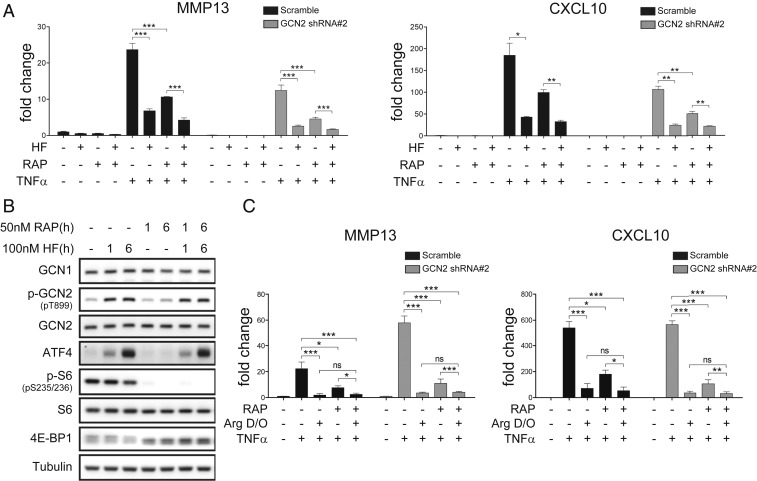
Next, we examined whether single amino acid depletion can suppress inflammatory responses without utilizing GCN2 or mTORC1 signaling. Consistent with prior work (17), HF did not inhibit mTORC1-signaling (measured using S6 phosphorylation and EIF4E-BP1 migration as readouts) in K4 cells, and mTORC1 signaling was completely blocked by 50 nM rapamycin (Fig. 3). Although rapamycin did reduce MMP13 and CXCL10 activation by TNF-α, HF still clearly inhibited MMP13 and CXCL10 induction even when mTORC1 signaling was completely blocked by rapamycin, indicating that HF does not require mTORC1 signaling to exert its effects (Fig. 3A). This rapamycin-insensitive effect of HF on inflammatory transcription was unaffected by knockdown of GCN2 (Fig. 3A), indicating that neither of the known amino acid-sensing pathways is necessary for the action of HF on inflammatory transcription. To test whether the same was true for amino acid depletion, which can be directly coupled to mTORC1 regulation, we used DMEM lacking arginine (Arg dropout [D/O] medium). Incubation of cells in Arg D/O medium activated GCN2 comparably to HF, and consistent with prior studies (39), substantially reduced mTORC1 signaling (SI Appendix, Fig. S11). As in the case of HF treatment, incubation in Arg D/O medium potently inhibited TNF-α induction of MMP13 and CXCL10. This inhibition was unaffected by GCN2 knockdown, and was still evident in the presence of 50 nM rapamycin (Fig. 3C). Incubation of cells in histidine (His) or lysine (Lys) D/O medium had effects identical to Arg D/O on TNF-α responses (SI Appendix, Fig. S11), and these effects were still clear in rapamycin-treated GCN2 knockdown cells (SI Appendix, Fig. S11). In contrast to Arg D/O treatment, His D/O treatment had only a weak effect on mTORC1 activity, but depletion of either amino acid inhibited inflammatory responses to a similar extent (SI Appendix, Fig. S11). These findings further indicate that mTORC1 regulation does not account for the effects of amino acid depletion on cytokine-induced transcriptional responses. Collectively, these observations establish that depletion of any of several amino acids can, like aaRS inhibitors, suppress TNF-α–stimulated responses in FLS, and that this signal is transduced without requiring GCN2 or mTORC1 signaling pathways.
Fig. 3.
HF and amino acid limitation each inhibit inflammatory responses in the absence of mTORC1 signaling. (A) HF inhibits TNF-α induction of MMP13 in the absence of detectable mTORC1 pathway activity. Wild-type or shRNA GCN2-depleted K4 fibroblasts were pretreated with 100 nM HF for 16 h, and then treated with 10 ng/mL TNF-α for 6 h. Transcript levels of target genes (MMP13, CXCL10) were quantified by qPCR. (B) Effects of HF and rapamycin on AAR (p-GCN2, ATF4) and mTORC1 (p-S6, 4E-BP1) pathway activation. (C) Arginine deprivation inhibits TNF-α induction of MMP13 or CXCL10 in the absence of detectable mTORC1 signaling, in both wild-type and GCN2 KD K4 cells. Wild-type or shRNA GCN2 depleted K4 fibroblasts were switched to arginine D/O DMEM for 16 h, and then treated with 10 ng/mL TNF-α for 6 h. Results are representative of three independent experiments (means ± SD, n = 3), *P < 0.05, **P < 0.01, ***P < 0.001.
HF Effects on Proinflammatory TH17 Cells Occur in Gcn2−/− Cells.
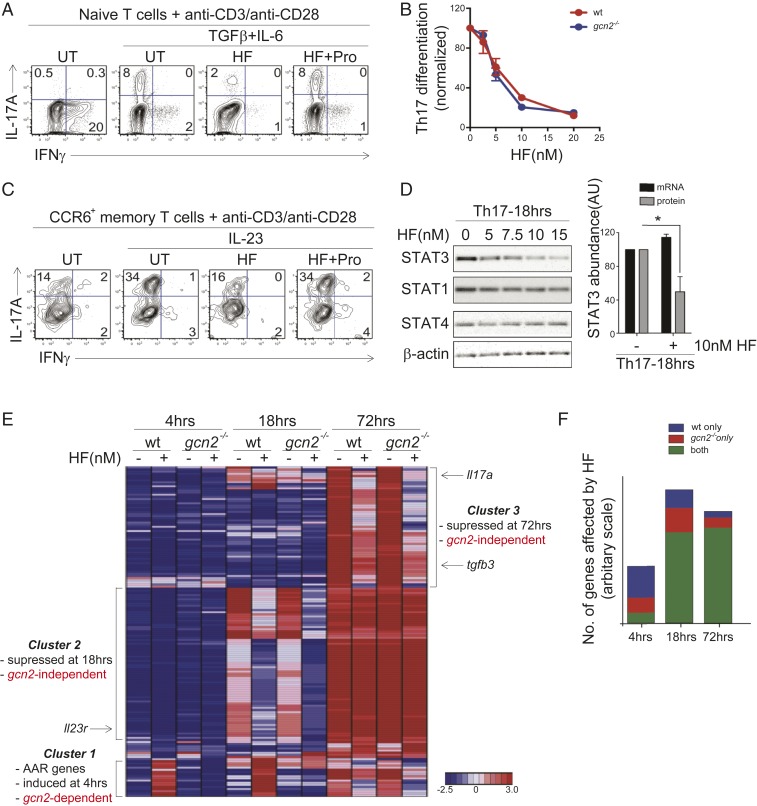
Next, we sought to establish whether GCN2 signaling is required for each of our previously reported, HF-mediated observations in T cells: 1) Inhibition of cytokine-directed TH17 differentiation and 2) inhibition of proinflammatory functions in mature TH17 memory cells. Using T cells obtained from Gcn2−/− mice (40), we confirmed that, in these cells, HF neither induces eif2α phosphorylation (SI Appendix, Fig. S12A), nor does HF induce of a cluster of adaptive AAR-associated responses, including expression amino acid transport and biosynthesis genes. (SI Appendix, Fig. S12B). Strikingly, HF treatment inhibits TH17 differentiation with similar potency in wild type and Gcn2−/− cells (Fig. 4 A and C); this inhibition is rescued by addition of proline (Fig. 4B). Naïve CD4+ T cells from Gcn2−/− mice show no overt functional defects in vitro (SI Appendix, Fig. S12), indicating that, consistent with prior work, Gcn2 is dispensable for most basic T cell functions in vitro (40).
Fig. 4.
HF regulates TH17 differentiation and effector function in the absence of GCN2. (A) FACS analysis of IL-17A and IFN-γ expression in gcn2-deficient CD4+ T cells 4 d after activation in the absence (no cytokines) or presence of TH17-polarizing cytokines (TGF-β + IL-6). TH17-polarized cells were treated with vehicle (UT), 10 nM HF, or 10 nM HF plus 50 mM l-proline (HF+Pro), as indicated. Representative of three experiments. (B) Dose–response of HF on wild-type (red) or gcn2-deficient (blue) TH17 differentiation. Percentages of IL-17A+ cells (+SEM; n = 3) were determined by intracellular staining and FACS analysis as in A and are normalized UT cells. (C) FACS analysis of cytokine (IL-17A, IFN-γ) expression in gcn2-deficient CCR6+ memory TH17 cells 2 d after in vitro stimulation (anti-CD3/anti-CD28) in the presence or absence of IL-23. Cells were treated with vehicle (UT), 10 nM HF, or 10 nM HF plus 50 mM l-proline (HF+Pro), as indicated. Representative of three experiments. (D, Left) STAT protein levels, determined by Western blot, in gcn2-deficient CD4+ T cells stimulated in TH17-polarizing conditions as in A for 18 h. Cells were treated with titrating concentrations of HF. Representative of three experiments. (Right) Relative abundance (+SEM; n = 3) of STAT3 protein or Stat3 mRNA in gcn2-deficient CD4+ T cells stimulated in TH17-polarizing conditions for 18 h ± 10 nM HF. STAT3 protein levels determined by Western blot as above; Stat3 mRNA levels were determined by microarray. Abundance shown as fold-change in HF- vs. DMSO-treated samples, *P < 0.05; paired two-tailed Student's t test. (E) Hierarchical clustering of differentially-expressed genes (>2.5-fold change) in wild-type and gcn2-deficient CD4+ T cells cultured for the indicated times in TH17-polarizing conditions ±10 nM HF. Gene clusters (1–3) are indicated by text; examples of genes within clusters are indicated by text and arrows; figure based on mean gene expression values obtained from biological duplicates. (F) Proportion of genes affected by HF (10 nM) in wild-type CD4+ T cells, gcn2-deficient T cells, or both, determined by microarray as in E.
In addition to blocking the differentiation of naïve T cell precursors into TH17 cells, we previously have shown that HF suppresses the proinflammatory response of already-differentiated TH17 “memory” cells to IL-23 (18). In vivo, TH17 memory cells are distinguished from other memory T cell subsets by the expression of the mucosal chemokine receptor, CCR6, and unlike in naive T cells, T cell receptor stimulation is sufficient to induce basal IL-17A expression in TH17 memory cells (18, 101, 102). IL-23 stimulation further enhances proinflammatory function of TH17 memory cells, and is required for TH17 cell-driven autoimmunity in vivo (103, 104). As in wild-type TH17 memory cells (18), those lacking GCN2 expressed IL-17A following T cell receptor stimulation, and up-regulated IL-17A further in the presence of IL-23 (Fig. 4C). HF suppressed IL-23–induced IL-17A expression in Gcn2−/− TH17 memory cells, leaving intact basal IL-17A expression, and this suppression was reversed by provision of excess proline. (Fig. 4C).
Both IL-6–mediated TH17 differentiation and IL-23–dependent memory TH17 cell function proceeds through the transcription factor Stat3 (105). We previously have shown that HF inhibits Stat3 activation (i.e., Tyr705 phosphorylation) downstream of multiple cytokines in wild-type TH cells, including IL-6, IL-23, and IL-27, and that this inhibition is associated with decreased Stat3 protein but not mRNA levels (18). Indeed, HF also decreased Stat3 protein levels in Gcn2−/− TH17 cells in a dose-dependent (Fig. 4 D, Left). Consistent with posttranscriptional regulation, HF reduced Stat3 protein abundance in Gcn2−/− TH17 cells without impacting Stat3 mRNA levels (Fig. 4 D, Right). Thus, all aspects of HF-dependent TH17 regulation that we previously have described in wild-type cells occur in cells lacking GCN2.
To better understand the relative contribution of GCN2-dependent and GCN2-independent pathways in the response of TH17 cells to HF, we performed microarray time-course experiments, in which wild-type or Gcn2−/− naïve CD4+ T cells were stimulated in TH17-polarizing conditions ±HF, and RNA was isolated at various time points postactivation (e.g., 4, 18, 72 h). Whereas more than half of all genes acutely induced by HF-treatment in wild-type cells at 4 h (39 of 71) represented GCN2-dependent, AAR-associated genes, most HF-regulated genes evident at 18 h (120 of 177) and 72 h (122 of 143) were suppressed to comparable levels in wild-type and Gcn2−/− TH17 cells (Fig. 4 E and F and Dataset S3) (106). Key molecules involved in TH17 cell differentiation and proinflammatory function—Il23r, Tgfb3, Il17a—were among the genes suppressed by HF in the absence of GCN2 (Fig. 4 E and F) (104). These findings indicate that, in the absence of GCN2 signaling, HF-mediated inhibition of EPRS suppresses key aspects of proinflammatory cytokine programs responsible for the differentiation of TH17 cells.
Effects of aaRS Inhibition on Cytokine Responses Are Sensitive to Depletion of GCN1.
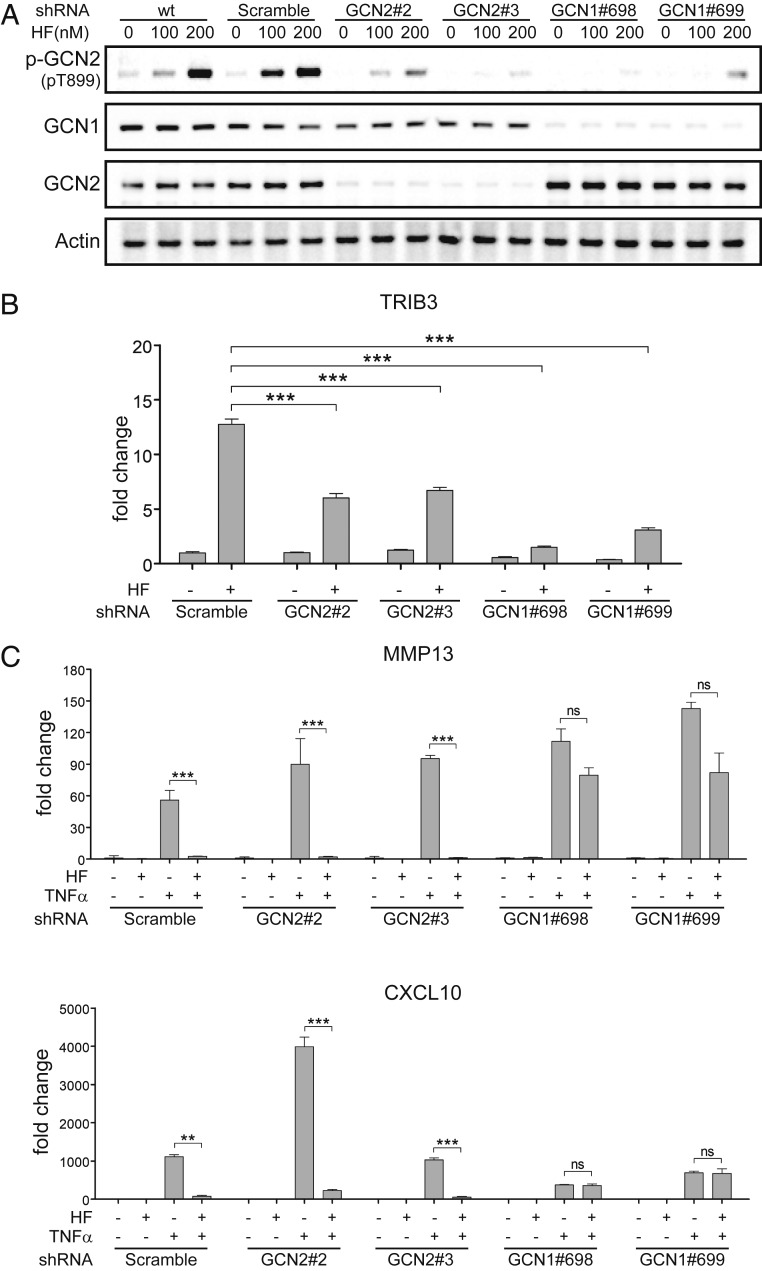
GCN1 was originally identified in yeast as an AAR pathway gene required for activation of GCN2 (76). We, therefore, examined whether mammalian GCN1 (GCN1L1) might have a role in mediating the effects of HF inhibition on TNF-α–stimulated inflammatory responses, distinct from its recognized role in GCN2 activation. To test this possibility, we used shRNA-mediated knockdowns of GCN1 or GCN2, matched with respect to their effects on AAR pathway readouts, to compare the effects of each depletion on HF suppression of inflammatory responses (Fig. 5). Depletion of GCN1 or GCN2, each with two independently targeted shRNAs, reduced HF-induced GCN2 phosphorylation (Fig. 5A), and reduced HF induction of the AAR-responsive transcription factor TRIB3 (Fig. 5B). GCN1-depletion, but not GCN2-depletion, largely prevented the ability of HF to suppress its characteristic subset TNF-α–responsive genes (Fig. 5C), using either of two independently targeted shRNAs. Similarly, GCN1 depletion impaired the capacity of borrelidin treatment to inhibit key TNF-α–responsive genes (SI Appendix, Fig. S13). These knockdown data indicate that GCN1 mediates an inflammatory-suppressive response to aaRS inhibition, distinct from its role in the activation of GCN2, and points to an upstream branch-point in the canonical AAR pathway (SI Appendix, Fig. S1).
Fig. 5.
HF Inhibition of cytokine responses is sensitive to depletion of GCN1, but not GCN2. (A) Characterization of protein expression and GCN2 phosphorylation in GCN1- and GCN2-depleted K4 cells. K4 were transfected with a lentiviral vector carrying two independently targeted shRNAs against GCN2 (#2 or #3) or GCN1 (#698 or #699), and protein expression and GCN2 phosphorylation examined by Western blot 15 min after HF treatment. Scrambled shRNA (SC) infected cells and uninfected cells were used as negative controls. (B) Evaluation of knockdown effects on induction of the AAR marker Trib3. GCN2 or GCN1 knockdown cells were treated with HF (100 nM) and/or TNF-α (10 ng/mL). For transcript levels of TRIB3, cells treated with HF for 6 h. (C) Evaluation of knockdown effects on HF inhibition of TNF-α–induced genes. For transcript levels of MMP13 (Upper) or CXCL10 (Lower), cells were pretreated with HF and treated with TNF-α for 6 h. Transcript levels of target genes were quantified by qPCR, Results are representative of three independent experiments (means ± SD, n = 3). **P < 0.01, ***P < 0.001; ns, not significant.
Discussion
In our effort to understand how the EPRS inhibitor HF mediates programmatic change in diverse, inflamed tissues, we discovered a nutrient stress pathway that senses an amino acid restriction signal via the cell’s protein synthetic apparatus to induce a program of inflammatory suppression in cultured FLS. aaRS inhibitors, like HF, act as amino acid restriction mimetics by inducing the accumulation of uncharged tRNA (5) and consequent ribosomal pausing (8, 90, 107). This pathway, which we call the ribosome-induced inflammatory suppressive pathway, branches from the canonical AAR, as demonstrated by its capacity to signal in cells that lack GCN2 and by its sensitivity to removal of the AAR pathway component GCN1 (SI Appendix, Fig. S1). The AAR is the oldest arm of the ISR, conserved to plants (108), worms (109), and yeast (110), and GCN1 has orthologs that are conserved across all eukaryotic phyla. Extensive work from the Hinnebusch laboratory (5, 76, 111), and others, on general amino acid control in Saccharomyces cerevisiae shows that GCN1 is bound to the elongating ribosome, and is necessary for the adaptive response to amino acid starvation and GCN2 activation. In mammalian cells, as in yeast, GCN1 is required for activation of GCN2 (112). Interestingly, studies in Caenorhabditis elegans and Arabidopsis thaliana each point to roles for GCN1 that are distinct from GCN2 activation (109, 113). In A. thaliana, mutants in AtGCN1/ILITHYIA (ILA) that impact plant defense and adaptation to abiotic stresses (114–116) are not phenocopied by loss-of-function mutations in AtGCN2 (113), suggesting that the signaling apparatus that allows amino acid deprivation to modulate cellular defense through GCN1, independently of GCN2, arose early in the evolution of immunity in multicellular organisms. In mammalian cells, amino acid stress is sensed both at the translating ribosome (5) and at the lysosome via amino acid sensors upstream of mTORC1 (39). We show here that HF treatment can suppress a TNF-α–driven tissue remodeling program in cultured FLS, and that restriction of any of several amino acids can mimic this inflammatory suppression even when both GCN2 and mTORC1 signaling have been abrogated. This clearly demonstrates that, in mammals, an amino acid stress signal can be sensed and transduced without GCN2 or mTORC1 signaling, and suggests that HF may owe its therapeutic value to regulation of stress pathways activated endogenously by amino acid catabolism.
HF treatment and amino acid catabolizing enzymes, such as IDO1 and Arg1, have overlapping mechanisms of action and shared tissue consequences. Previously, we showed that HF treatment potently inhibits cytokine-directed TH17 differentiation (17) and suppresses IL-23–stimulated proinflammatory functions in mature TH17 memory cells (18). We now show that these HF-mediated T cell effects can occur in cells that lack GCN2. HF and other aaRS inhibitors are powerful tools with which to parse the amino acid catabolic signal; they act as amino acid restriction mimetics by causing the accumulation of uncharged tRNA but, unlike amino acid catabolism, they neither generate bioactive catabolites nor do they directly inhibit the mTORC1 pathway. Although GCN2 signaling plays distinct roles in the immune system (26, 91, 117–120), GCN2-independent responses, both to IDO1-induced tryptophan catabolism (32, 91, 121) and to general amino acid restriction, have been reported (122–125). Moreover, evidence that IDO-induced tryptophan depletion has a central role in local immunosuppression (40, 126), and reinforcement of peripheral tolerance is at apparent odds with the absence of a prominent immunologic phenotype in either IDO-null or GCN2-null mice (29, 32). It has been noted that this discrepancy suggests redundancy in the signaling networks that govern these processes (32). One example of such redundancy is the observation that TREGS stimulate the expression of multiple amino acid-consuming enzymes within dendritic cells and successful skin grafts (32). Whereas GCN2 kinase signaling was once thought to be the core mechanism that mediates proliferative arrest and anergy in T cells (40), several studies now directly challenge the notion that immunoregulated amino acid catabolism acts through AAR pathway activation (32, 92), or that GCN2 is required for sensing amino acid depletion in T cells (32, 127). Cobbold et al. (32) showed that the in vitro restriction of a single amino acid was sufficient to suppress the proliferation of T cells in response to antigen, and that this response could occur in cells that lack GCN2. Similarly, Van de Velde et al. (127) demonstrated that GCN2-deficient T cells retain the capacity to sense and respond to tryptophan limitation. To date, studies of amino acid depletion or IDO1-induced immunosuppression in the GCN2-null context have focused on the mechanism of mTORC1 pathway inhibition (29, 32, 127) and on tissue responses to kynurenine metabolites (92). Our discovery of an amino acid sensor pathway that doesn’t signal through GCN2 activation provides an additional potential explanation for observed immunoregulation in the GCN2-null context, and prompts further study of the IDO1 mechanism in pathological contexts, such as the chronic inflammatory or tumor microenvironment.
Type 2 (TH2) immune responses provide host protection against parasites, such as helminth infection, control inflammation, and promote wound repair (128). In wild-type mice, infection with Schistosoma mansoni results in a protective granuloma formation, and a TH2 response sufficient to kill schistosome eggs (129). This TH2 response is associated with a strong activation of the arginine-degrading enzyme Arg1 in M2 macrophages by TH2 cytokines (130). This potent induction of Arg1, combined with up-regulation of arginine import, make M2 macrophages a big sink for the tissue-depletion of arginine. In mice that are deficient in macrophage-specific Arg1 (Arg1ΔM); however, schistosomiasis results in a nonresolving granulomatous pathology, unrestricted Th2 cell proliferation, and marked liver fibrosis (131). These data demonstrate that Arg1-expressing macrophages function as suppressors of fibrosis, and governors of TH2-dependent inflammation (29). We surmise that aaRS inhibitors mimic the endogenous effects of up-regulated amino acid catabolism, suppressing T cell proliferation and fibrosis, by inducing amino acid stress pathway signals, including the pathway described here.
Over the last 25 years, HF has been studied extensively as an antifibrotic drug (21, 67) and, more recently, as a treatment in animal models of autoimmune disease (18, 20, 22). The overarching assumption has been that HF’s efficacy is due to action upon a key cell or tissue, such as dermal fibroblasts or TH17 cells. The breadth of conditions for which HF has demonstrated therapeutic value, however, coupled with the fact that HF targets a ubiquitous enzyme, suggested to us that HF could act directly upon multiple cooperating cell types in vivo. The convergence of findings in T cells and FLS reported here points strongly to a common pathway linking HF inhibition of EPRS to the suppression of inflammatory programs in immune and structural cells. In HF-treated T cells, cultured under TH17-polarizing conditions, transcriptomic analyses demonstrate that HF inhibits a broad program of genes in these cells, even in the absence of GCN2. Similarly, in human FLS, HF and HFol potently suppress a TNF-α–stimulated transcriptional program, irrespective of GCN2. HF further suppresses key proinflammatory responses, in IL-23–stimulated TH17 memory cells and IL-1β–stimulated FLS, also in cells that lack GCN2. These findings make HF, and its chemical derivatives, attractive drug candidates for disease settings in which anti-TNF biologics have been successful, such as RA and Crohn’s disease (132, 133). Small-molecule inhibitors of TNF-α tissue programs could be much needed adjuncts, or alternatives to biologic therapies in instances of immunogenicity (133), while providing added tissue benefit via inhibition of both TH17-driven inflammation and TGF-β–stimulated fibrosis.
Our work, and an important body of work by others, demonstrate that perturbations that trigger the cellular accumulation of uncharged tRNA, such as inhibition of aaRSs and amino acid restriction, can reproduce key features of HF’s tissue action (17, 22, 23, 61). Borrelidin treatment inhibits the differentiation of TH17 cells (17) and suppresses inflammatory responses in TNF-α–stimulated FLS (SI Appendix, Figs. S11 and S13). Qin et al. (61) recently showed that HF, borrelidin, and leucinol inhibit collagen deposition in cardiac fibroblasts, and that these effects can be rescued by the addition of proline, threonine, or leucine, respectively. We additionally show that restriction of histidine, arginine, or lysine can mimic key aspects of HF’s inflammatory suppression program in FLS, and that these effects occur without GCN2 or mTORC1 signaling. These data indicate that the tissue action of HF, whether inhibition of extracellular matrix deposition, inhibition of cartilage destruction, or inhibition of differentiation and function of a proinflammatory T helper cell subset, is part of broader homeostatic signal that is a shared feature of aaRS inhibition, and of amino acid restriction, in general. The dose for onset of HF-induced tissue benefit tracks reliably with AAR pathway activation, but can occur in cells that lack GCN2. Taken together, these data suggest the presence of a signaling pathway that shares the AAR’s activation strategy, but branches from the AAR upstream from GCN2. Thus, we considered AAR pathway components upstream from GCN2 as potential candidates to transduce the observed inflammatory-suppressive signal.
The requirement of GCN1 for GCN2 activation in mammalian cells (112) places it immediately genetically upstream from GCN2 in the AAR pathway. This makes GCN1 an attractive candidate for involvement in detection and transduction of an HF-induced, GCN2-independent, inflammatory-suppressive signal. Significantly, we find that HF suppression of TNF-α–induced FLS inflammatory mediators is sensitive to depletion of GCN1. Furthermore, we find that borrelidin mimics HF in that borrelidin suppression of TNF-α–stimulated inflammatory responses is intact in GCN2 knockdown FLS, but impaired in GCN1 knockdown FLS cells. Currently, GCN2 is the only protein in mammalian cells that is known to sense and transduce an uncharged tRNA-based signal (134). The AAR is believed to detect amino acid stress through the build-up of uncharged tRNA that activates GCN2, by binding to a histidyl-tRNA synthetase-like domain in its C terminus (50). GCN2 then responds by autophosphorylation and phosphorylation of eIF2α (14, 53). In contrast, our findings indicate that treatment with aaRS inhibitors or amino acid restriction, perturbations that generate the accumulation of uncharged tRNA, can be sensed and transduced without GCN2, but require the presence of GCN1.
In yeast, as in Escherichia coli (10), uncharged tRNA is detected in the context of the stalled ribosome (13, 135). Classic work in yeast shows that GCN1 is necessary for coupling amino acid availability to GCN2 activation (76, 111). This work indicates that GCN1 binds to elongating ribosomes and to GCN2 and facilitates the activation of GCN2 by uncharged tRNA (111). Although GCN1 is dispensable for GCN2 kinase activity per se (76), physical interaction between GCN1 and GCN2 is necessary for GCN2 activation and for general amino acid control in yeast (111). More recent work shows that GCN2 can be potently stimulated by direct interaction with the ribosomal P-stalk (90), supporting the notion that ribosomes are an important link between translational stress and GCN2 activation. The Hinnebusch model places GCN1 at the ribosome and in proximity to sense and resolve uncharged tRNA on a stalled ribosome. Consistent with this model, we propose that GCN1 acts downstream of aaRS inhibition to link the sensing of uncharged tRNA in the ribosomal A site to signal transduction by non-GCN2 effectors. GCN1 is large protein that lacks any recognized enzymatic activity or tRNA-binding motif, but instead contains a series of evolutionarily conserved HEAT motif repeats (76, 136), through which it forms complexes with other proteins (137). In addition to scaffolding the ribosome to GCN2, we propose that GCN1 bridges the ribosome to an unknown effector, or effectors, that couple amino acid stress to downstream regulation of inflammatory and tissue remodeling programs.
The ribosome is a highly evolutionarily conserved signaling hub that mediates responses to a wide range of cellular stressors (7, 8, 10). Immunoregulated amino acid catabolism and the amino acid stress pathways that sense and transduce these signals have important emerging roles in immunity and tissue homeostasis (26, 29, 35). We identify here a mammalian pathway that senses amino acid limitation to regulate a program of immune and inflammatory responses in FLS. Like stringent control and the AAR, this pathway utilizes the cell’s protein synthetic machinery to signal via the accumulation of uncharged tRNA to induce programmatic cellular change. We previously showed that HF’s inhibition of PRS catalytic activity is sufficient to explain the therapeutic action of this compound (17), and now we show that HF’s tissue effects cannot be attributed solely to the activation of the canonical AAR pathway. We establish that HF inhibition of PRS mediates suppression of distinct inflammatory programs in immune and structural cells, notably, even in cells that lack the signature AAR effector GCN2. Furthermore, cellular treatment with other aaRS inhibitors or restriction of any of several amino acids reproduce inflammatory suppression, also in the absence of GCN2 and mTORC1 signaling. We provide insight into the tissue mechanism of aaRS inhibitors for the treatment of human disease, both by characterizing HF’s suppression of disease-relevant inflammatory and tissue remodeling programs, and by demonstrating the involvement of a pathway that we call the ribosome-induced inflammatory suppressive pathway (SI Appendix, Fig. S1). Future work will be required to establish the tissue effects of GCN1 knockdown in other cells types, and to understand how signaling pathway components that are highly conserved to all cells can transduce an amino acid stress signal to regulate tissue remodeling and inflammatory programs.
Materials and Methods
Cell Culture and Media.
Primary human synovial fibroblasts (RA-FLS).
Human synovial tissues from patients with RA were obtained as discarded tissue, de-identified, and FLS-isolated, as previously described (74). For gene-expression analysis, 80% subconfluent cells were left in serum reduced media (0.2% FBS) for 24 to 48 h, treated with HF (300 nM) and/or proline (2 mM) for 16 h, and treated with TNF-α (10 ng/mL) for 4 h. Both TNF-α responsiveness and HF sensitivity were confirmed in FLS isolates from multiple patients, with some variability in TNF-α responsiveness between individual isolates. TNF-α–sensitivity also declined slowly over passage in culture; primary FLS were used before passage 10 to 12.
Immortalized K4 FLS.
K4 FLS were a kind gift from Evelyn Murphy, University College Dublin, Dublin, Ireland. For TNF-α–stimulated experiments, cells were shifted into K4 cell media with 0.2% FBS instead of 10% FBS 24 h before the HF treatment, pretreated with HF for 16 h, and then with TNF-α for times indicated in the figure legends.
Human lung fibroblasts (LL29, AnHa).
LL29 and AnHa were purchased from the American Type Culture Collection.
HUVEC (Lonza EGM [CC-2517]).
HUVECs were cultured in EGM-2 (CC-3162, Lonza). For experiments, cells were serum-starved in 0.2% FBS for 24 h, treated with 200 nM HF and 2 mM proline for 16 h, and then treated 10 ng/mL TNF-α for 4 h.
Normal human dermal fibroblasts.
Normal human dermal fibroblasts were purchased from Lonza (CC-2511) and used in accordance with the manufacturer’s instructions.
GCN2−/− FLS isolation and culture.
Primary cell cultures were obtained from wild-type and general control nonderepressible 2 GCN2−/− (eif2ak4−/−) mice purchased from Jackson Laboratories (stock no. 008240). Primary murine FLS were isolated as previously described (79). Murine FLS were cultured in the same media used for human primary FLS. For treatment, murine FLS were cultured in serum-reduced media (2%) for 72 h.
Primary CD4+ T Cells Analysis.
Primary CD4+ T cells from wild-type (C57BL/6J; stock no. 000664) or Gcn2−/− (eif2ak4−/−) mice were isolated, treated ±HF ±l-proline, and cultured as previously described (18). For TH17 polarization, cells were treated as described in detailed methods in SI Appendix.
Cell Lysates and Western Blot.
Primary or established FLS were harvested in RIPA buffer supplemented with protease inhibitor tablets and phosphatase inhibitors (Roche). Cultured T cells were harvested at the indicated time points, washed once in PBS, and lysed on ice at 5 to 10 × 107 cells/mL in Laemmli sample buffer (Bio-Rad) supplemented with protease inhibitor tablets (Roche) and phosphatase inhibitors. Cell lysates were cleared by centrifugation at 12,000 × g for 10 min at 4 °C, and stored at −80 °C. Additional details are provided in SI Appendix.
Quantitative Reverse-Transcription PCR.
RNA was extracted using TRIzol reagent in accordance with the manufacturer’s instructions (Invitrogen), and retro-transcribed to cDNA using the High Capacity cDNA Reverse Transcription kit (Applied Biosystems). Transcriptional analysis was performed using the Universal Probe Library System (UPL; Roche). Details and probes are provided in SI Appendix.
Stable Knockdown of GCN1 and GCN2 in K4 FLS and Human RA-FLS.
Lentiviral-transduction was performed using the pLKO.1 lentiviral constructs encoding shRNA against human GCN2 or GCN1 in K4 FLS; details are provided in SI Appendix.
RNA-Seq.
RNA-seq was performed on primary human FLS (Dataset S1) or K4 FLS (Dataset S2). Primary FLS were left in serum-reduced media (0.2% FBS) for 24 h, pretreated for 16 h with 300 nM HF or vehicle, and then treated for 8 or 24 h with 10 ng/mL TNF-α or vehicle. K4 FLS stably transformed with lentiviral shRNAs (scrambled or GCN2 shRNA #2), as described above, were cultured in serum-reduced media (0.2% FBS) for 24 h, then treated for 24 h with 250 nM HFol, an HF derivative with similar activity to HF as an EPRS inhibitor (17), or vehicle, and then with 10 ng/mL TNF-α for 8 h. Biological duplicates were collected for each condition. Details of data analysis are provided in SI Appendix.
T Cell Microarrays and Data Analyses.
Total RNA was isolated from Primary CD4+ T cells from wild-type or Gcn2−/− (eif2ak4−/−) mice treated ±HF, as described in SI Appendix. RNA was sent to the Boston University Microarray Resource Facility and analyzed, as described in SI Appendix.
Statistical Analysis.
For qRT-PCR results, values were obtained from three independent experiments and expressed as means ± SD. Significant differences were assessed using one-way ANOVA followed by Tukey’s honest significant difference post hoc test or Welch’s ANOVA followed by Games–Howell post hoc test. *P < 0.05, **P < 0.01, ***P < 0.001. All statistical data were calculated using SPSS software.
Data Availability.
All protocols and data supporting the findings of this study are available within the main text and SI Appendix. The primary sequencing data and microarray information that were generated for this publication have been deposited in the National Center for Biotechnology Information’s Gene Expression Omnibus (GEO). Information for SI Appendix, Tables S1 and S2 are accessible through GEO Series accession no. GSE145205, https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE145205. Information for SI Appendix, Table S3 are accessible through GEO accession no. GSE47478, https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE47478.
Supplementary Material
Acknowledgments
We thank Thaddeus Carlson, Ivana Djuretic, and Radha Ramesh (Tempero Pharmaceuticals) for technical support for T cell experiments; Kamil Slowikowski (Broad Institute) for valuable advice and assistance with the analysis of RNA sequencing data; John Hutchinson (Harvard School of Public Health) for data analysis and figure preparation of transcriptomics in Fig. 1; and Fred Goldberg (Harvard Medical School) and Vicki Rosen (Harvard School of Dental Medicine) for critical reading of the manuscript. M.W. and T.L.K. were supported by Department of Defense Award W81XWH-15-1-0396, National Institute of Allergy and Infectious Diseases Grant AI142343, and an unrestricted research grant from Allied Bristol Life Sciences. M.S.S. was supported by The Scripps Research Institute Florida via the state of Florida. A.R. was supported by National Institute of Allergy and Infectious Diseases Grant AI128589. C.-Y.Y. was supported by the National Research Foundation of Korea grant funded by the Korea government (Grants 2012R1A5A1048236 and 2015R1A2A2A05027993).
Footnotes
The authors declare no competing interest.
Data deposition: The data reported in this paper have been deposited and are publicly available in the Gene Expression Omnibus (GEO) database, https://www.ncbi.nlm.nih.gov/geo (accession no. GSE145205; microarray data accession no. GSE47478).
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.1913788117/-/DCSupplemental.
References
- 1.Hauryliuk V., Atkinson G. C., Murakami K. S., Tenson T., Gerdes K., Recent functional insights into the role of (p)ppGpp in bacterial physiology. Nat. Rev. Microbiol. 13, 298–309 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wek R. C., Role of eIF2α kinases in translational control and adaptation to cellular stress. Cold Spring Harb. Perspect. Biol. 10, a032870 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Walter P., Ron D., The unfolded protein response: From stress pathway to homeostatic regulation. Science 334, 1081–1086 (2011). [DOI] [PubMed] [Google Scholar]
- 4.Boniecka J., Prusińska J., Dąbrowska G. B., Goc A., Within and beyond the stringent response-RSH and (p)ppGpp in plants. Planta 246, 817–842 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hinnebusch A. G., Translational regulation of GCN4 and the general amino acid control of yeast. Annu. Rev. Microbiol. 59, 407–450 (2005). [DOI] [PubMed] [Google Scholar]
- 6.Sonenberg N., Hinnebusch A. G., Regulation of translation initiation in eukaryotes: Mechanisms and biological targets. Cell 136, 731–745 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Crawford R. A., Pavitt G. D., Translational regulation in response to stress in Saccharomyces cerevisiae. Yeast 36, 5–21 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Darnell A. M., Subramaniam A. R., O’Shea E. K., Translational control through differential ribosome pausing during amino acid limitation in mammalian cells. Mol. Cell 71, 229–243.e11 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Juszkiewicz S., et al. , ZNF598 is a quality control sensor of collided ribosomes. Mol. Cell 72, 469–481.e7 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Brown A., Fernández I. S., Gordiyenko Y., Ramakrishnan V., Ribosome-dependent activation of stringent control. Nature 534, 277–280 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Liu K., Bittner A. N., Wang J. D., Diversity in (p)ppGpp metabolism and effectors. Curr. Opin. Microbiol. 24, 72–79 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Srivatsan A., Wang J. D., Control of bacterial transcription, translation and replication by (p)ppGpp. Curr. Opin. Microbiol. 11, 100–105 (2008). [DOI] [PubMed] [Google Scholar]
- 13.Sattlegger E., Hinnebusch A. G., Polyribosome binding by GCN1 is required for full activation of eukaryotic translation initiation factor 2alpha kinase GCN2 during amino acid starvation. J. Biol. Chem. 280, 16514–16521 (2005). [DOI] [PubMed] [Google Scholar]
- 14.Harding H. P., et al. , An integrated stress response regulates amino acid metabolism and resistance to oxidative stress. Mol. Cell 11, 619–633 (2003). [DOI] [PubMed] [Google Scholar]
- 15.Baird T. D., Wek R. C., Eukaryotic initiation factor 2 phosphorylation and translational control in metabolism. Adv. Nutr. 3, 307–321 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Brown M. V., Reader J. S., Tzima E., Mammalian aminoacyl-tRNA synthetases: Cell signaling functions of the protein translation machinery. Vascul. Pharmacol. 52, 21–26 (2010). [DOI] [PubMed] [Google Scholar]
- 17.Keller T. L., et al. , Halofuginone and other febrifugine derivatives inhibit prolyl-tRNA synthetase. Nat. Chem. Biol. 8, 311–317 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Carlson T. J., et al. , Halofuginone-induced amino acid starvation regulates Stat3-dependent Th17 effector function and reduces established autoimmune inflammation. J. Immunol. 192, 2167–2176 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Guo L.-W., et al. , Halofuginone stimulates adaptive remodeling and preserves re-endothelialization in balloon-injured rat carotid arteries. Circ. Cardiovasc. Interv. 7, 594–601 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Park M.-K., et al. , Halofuginone ameliorates autoimmune arthritis in mice by regulating the balance between Th17 and Treg cells and inhibiting osteoclastogenesis. Arthritis Rheumatol. 66, 1195–1207 (2014). [DOI] [PubMed] [Google Scholar]
- 21.Pines M., Halofuginone for fibrosis, regeneration and cancer in the gastrointestinal tract. World J. Gastroenterol. 20, 14778–14786 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sundrud M. S., et al. , Halofuginone inhibits TH17 cell differentiation by activating the amino acid starvation response. Science 324, 1334–1338 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Peng W., et al. , Surgical stress resistance induced by single amino acid deprivation requires Gcn2 in mice. Sci. Transl. Med. 4, 118ra111 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wang A., et al. , Opposing effects of fasting metabolism on tissue tolerance in bacterial and viral inflammation. Cell 166, 1512–1525.e12 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wang A., Luan H. H., Medzhitov R., An evolutionary perspective on immunometabolism. Science 363, eaar3932 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Walls J., Sinclair L., Finlay D., Nutrient sensing, signal transduction and immune responses. Semin. Immunol. 28, 396–407 (2016). [DOI] [PubMed] [Google Scholar]
- 27.Chang C. H., Pearce E. L., Emerging concepts of T cell metabolism as a target of immunotherapy. Nat. Immunol. 17, 364–368 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Grohmann U., et al. , Amino-acid sensing and degrading pathways in immune regulation. Cytokine Growth Factor Rev. 35, 37–45 (2017). [DOI] [PubMed] [Google Scholar]
- 29.Murray P. J., Amino acid auxotrophy as a system of immunological control nodes. Nat. Immunol. 17, 132–139 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hotamisligil G. S., Inflammation, metaflammation and immunometabolic disorders. Nature 542, 177–185 (2017). [DOI] [PubMed] [Google Scholar]
- 31.Tsalikis J., Croitoru D. O., Philpott D. J., Girardin S. E., Nutrient sensing and metabolic stress pathways in innate immunity. Cell. Microbiol. 15, 1632–1641 (2013). [DOI] [PubMed] [Google Scholar]
- 32.Cobbold S. P., et al. , Infectious tolerance via the consumption of essential amino acids and mTOR signaling. Proc. Natl. Acad. Sci. U.S.A. 106, 12055–12060 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Buck M. D., Sowell R. T., Kaech S. M., Pearce E. L., Metabolic instruction of immunity. Cell 169, 570–586 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Munn D. H., Mellor A. L., IDO in the tumor microenvironment: Inflammation, counter-regulation, and tolerance. Trends Immunol. 37, 193–207 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lemos H., Huang L., Prendergast G. C., Mellor A. L., Immune control by amino acid catabolism during tumorigenesis and therapy. Nat. Rev. Cancer 19, 162–175 (2019). [DOI] [PubMed] [Google Scholar]
- 36.Ravishankar B., et al. , Tolerance to apoptotic cells is regulated by indoleamine 2,3-dioxygenase. Proc Natl Acad Sci U S A. 109, 3909–3914. (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sharma M. D., et al. , Plasmacytoid dendritic cells from mouse tumor-draining lymph nodes directly activate mature Tregs via indoleamine 2,3-dioxygenase. J. Clin. Invest. 117, 2570–2582 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mondanelli G., et al. , A relay pathway between arginine and tryptophan metabolism confers immunosuppressive properties on dendritic cells. Immunity 46, 233–244 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wolfson R. L., Sabatini D. M., The dawn of the age of amino acid sensors for the mTORC1 pathway. Cell Metab. 26, 301–309 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Munn D. H., et al. , GCN2 kinase in T cells mediates proliferative arrest and anergy induction in response to indoleamine 2,3-dioxygenase. Immunity 22, 633–642 (2005). [DOI] [PubMed] [Google Scholar]
- 41.Jones R. G., Pearce E. J., MenTORing immunity: mTOR signaling in the development and function of tissue-resident immune cells. Immunity 46, 730–742 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gallinetti J., Harputlugil E., Mitchell J. R., Amino acid sensing in dietary-restriction-mediated longevity: Roles of signal-transducing kinases GCN2 and TOR. Biochem. J. 449, 1–10 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Saxton R. A., Sabatini D. M., mTOR signaling in growth, metabolism, and disease. Cell 168, 960–976 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Haigis M. C., Yankner B. A., The aging stress response. Mol. Cell 40, 333–344 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kapahi P., et al. , With TOR, less is more: A key role for the conserved nutrient-sensing TOR pathway in aging. Cell Metab. 11, 453–465 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Palii S. S., et al. , Specificity of amino acid regulated gene expression: Analysis of genes subjected to either complete or single amino acid deprivation. Amino Acids 37, 79–88 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tang X., et al. , Comprehensive profiling of amino acid response uncovers unique methionine-deprived response dependent on intact creatine biosynthesis. PLoS Genet. 11, e1005158 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Krupitza G., Thireos G., Translational activation of GCN4 mRNA in a cell-free system is triggered by uncharged tRNAs. Mol. Cell. Biol. 10, 4375–4378 (1990). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zaborske J. M., et al. , Genome-wide analysis of tRNA charging and activation of the eIF2 kinase Gcn2p. J. Biol. Chem. 284, 25254–25267 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wek R. C., Jackson B. M., Hinnebusch A. G., Juxtaposition of domains homologous to protein kinases and histidyl-tRNA synthetases in GCN2 protein suggests a mechanism for coupling GCN4 expression to amino acid availability. Proc. Natl. Acad. Sci. U.S.A. 86, 4579–4583 (1989). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zhu S., Sobolev A. Y., Wek R. C., Histidyl-tRNA synthetase-related sequences in GCN2 protein kinase regulate in vitro phosphorylation of eIF-2. J. Biol. Chem. 271, 24989–24994 (1996). [DOI] [PubMed] [Google Scholar]
- 52.Dong J., Qiu H., Garcia-Barrio M., Anderson J., Hinnebusch A. G., Uncharged tRNA activates GCN2 by displacing the protein kinase moiety from a bipartite tRNA-binding domain. Mol. Cell 6, 269–279 (2000). [DOI] [PubMed] [Google Scholar]
- 53.Kilberg M. S., Pan Y.-X., Chen H., Leung-Pineda V., Nutritional control of gene expression: How mammalian cells respond to amino acid limitation. Annu. Rev. Nutr. 25, 59–85 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wang X., et al. , Re-evaluating the roles of proposed modulators of mammalian target of rapamycin complex 1 (mTORC1) signaling. J. Biol. Chem. 283, 30482–30492 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Medzhitov R., Origin and physiological roles of inflammation. Nature 454, 428–435 (2008). [DOI] [PubMed] [Google Scholar]
- 56.Nowarski R., Jackson R., Flavell R. A., The stromal intervention: Regulation of immunity and inflammation at the epithelial-mesenchymal barrier. Cell 168, 362–375 (2017). [DOI] [PubMed] [Google Scholar]
- 57.Serhan C. N., Levy B. D., Resolvins in inflammation: Emergence of the pro-resolving superfamily of mediators. J. Clin. Invest. 128, 2657–2669 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Richards C. D., Innate immune cytokines, fibroblast phenotypes, and regulation of extracellular matrix in lung. J. Interferon Cytokine Res. 37, 52–61 (2017). [DOI] [PubMed] [Google Scholar]
- 59.Munn D. H., Mellor A. L., Indoleamine 2,3 dioxygenase and metabolic control of immune responses. Trends Immunol. 34, 137–143 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.O’Connell D. J., et al. , Simultaneous pathway activity inference and gene expression analysis using RNA sequencing. Cell Syst. 2, 323–334 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Qin P., et al. , Activation of the amino acid response pathway blunts the effects of cardiac stress. J. Am. Heart Assoc. 6, e004453 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.McGaha T. L., Phelps R. G., Spiera H., Bona C., Halofuginone, an inhibitor of type-I collagen synthesis and skin sclerosis, blocks transforming-growth-factor-beta-mediated Smad3 activation in fibroblasts. J. Invest. Dermatol. 118, 461–470 (2002). [DOI] [PubMed] [Google Scholar]
- 63.Xavier S., et al. , Amelioration of radiation-induced fibrosis: Inhibition of transforming growth factor-beta signaling by halofuginone. J. Biol. Chem. 279, 15167–15176 (2004). [DOI] [PubMed] [Google Scholar]
- 64.Oishi H., et al. , Halofuginone treatment reduces interleukin-17A and ameliorates features of chronic lung allograft dysfunction in a mouse orthotopic lung transplant model. J. Heart Lung Transplant. 35, 518–527 (2016). [DOI] [PubMed] [Google Scholar]
- 65.Pines M., et al. , Reduction in dermal fibrosis in the tight-skin (Tsk) mouse after local application of halofuginone. Biochem. Pharmacol. 62, 1221–1227 (2001). [DOI] [PubMed] [Google Scholar]
- 66.Turgeman T., et al. , Prevention of muscle fibrosis and improvement in muscle performance in the mdx mouse by halofuginone. Neuromuscul. Disord. 18, 857–868 (2008). [DOI] [PubMed] [Google Scholar]
- 67.Pines M., Nagler A., Halofuginone: A novel antifibrotic therapy. Gen. Pharmacol. 30, 445–450 (1998). [DOI] [PubMed] [Google Scholar]
- 68.Song D. G., et al. , Glutamyl-prolyl-tRNA synthetase induces fibrotic extracellular matrix via both transcriptional and translational mechanisms. FASEB J. 33, 4341–4354 (2019). [DOI] [PubMed] [Google Scholar]
- 69.Afroz S., et al. , Amino acid starvation enhances vaccine efficacy by augmenting neutralizing antibody production. Sci. Signal. 12, eaav4717 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Zhang F., et al. ; Accelerating Medicines Partnership Rheumatoid Arthritis and Systemic Lupus Erythematosus (AMP RA/SLE) Consortium , Defining inflammatory cell states in rheumatoid arthritis joint synovial tissues by integrating single-cell transcriptomics and mass cytometry. Nat. Immunol. 20, 928–942 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Noss E. H., Brenner M. B., The role and therapeutic implications of fibroblast-like synoviocytes in inflammation and cartilage erosion in rheumatoid arthritis. Immunol. Rev. 223, 252–270 (2008). [DOI] [PubMed] [Google Scholar]
- 72.Bottini N., Firestein G. S., Duality of fibroblast-like synoviocytes in RA: Passive responders and imprinted aggressors. Nat. Rev. Rheumatol. 9, 24–33 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Chang S. K., Gu Z., Brenner M. B., Fibroblast-like synoviocytes in inflammatory arthritis pathology: The emerging role of cadherin-11. Immunol. Rev. 233, 256–266 (2010). [DOI] [PubMed] [Google Scholar]
- 74.Nguyen H. N., et al. , Autocrine loop involving IL-6 family member LIF, LIF receptor, and STAT4 drives sustained fibroblast production of inflammatory mediators. Immunity 46, 220–232 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Ruan B., A unique hydrophobic cluster near the active site contributes to differences in borrelidin inhibition among threonyl-tRNA synthetases. J. Biol. Chem. 280, 571–577 (2005). [DOI] [PubMed] [Google Scholar]
- 76.Marton M. J., Crouch D., Hinnebusch A. G., GCN1, a translational activator of GCN4 in Saccharomyces cerevisiae, is required for phosphorylation of eukaryotic translation initiation factor 2 by protein kinase GCN2. Mol. Cell. Biol. 13, 3541–3556 (1993). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Goldring M. B., Otero M., Inflammation in osteoarthritis. Curr. Opin. Rheumatol. 23, 471–478 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Kakkar A. K., Lefer D. J., Leukocyte and endothelial adhesion molecule studies in knockout mice. Curr. Opin. Pharmacol. 4, 154–158 (2004). [DOI] [PubMed] [Google Scholar]
- 79.Agarwal S. K., Brenner M. B., Role of adhesion molecules in synovial inflammation. Curr. Opin. Rheumatol. 18, 268–276 (2006). [DOI] [PubMed] [Google Scholar]
- 80.Marcos-Ramiro B., García-Weber D., Millán J., TNF-induced endothelial barrier disruption: Beyond actin and Rho. Thromb. Haemost. 112, 1088–1102 (2014). [DOI] [PubMed] [Google Scholar]
- 81.Leask A., Targeting the TGFbeta, endothelin-1 and CCN2 axis to combat fibrosis in scleroderma. Cell. Signal. 20, 1409–1414 (2008). [DOI] [PubMed] [Google Scholar]
- 82.Krieg T., Abraham D., Lafyatis R., Fibrosis in connective tissue disease: The role of the myofibroblast and fibroblast-epithelial cell interactions. Arthritis Res. Ther. 9 (suppl. 2), S4 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Kim J.-H., et al. , Regulation of the catabolic cascade in osteoarthritis by the zinc-ZIP8-MTF1 axis. Cell 156, 730–743 (2014). [DOI] [PubMed] [Google Scholar]
- 84.Sano T., et al. , An IL-23R/IL-22 circuit regulates epithelial serum amyloid A to promote local effector Th17 responses. Cell 164, 324 (2016). [DOI] [PubMed] [Google Scholar]
- 85.Atarashi K., et al. , Th17 cell induction by adhesion of microbes to intestinal epithelial cells. Cell 163, 367–380 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Kim Y., Edenius M., Whitman M., Keller T.. Transcriptional responses of fibroblast like synoviocytes to TNFα in the presence or absence of Halofuginone (HF). Gene Expression Omnibus. https://www.ncbi.nlm.nih.gov/geo/query/acc.cgiacc=GSE145205. Deposited 10 February 2020.
- 87.Shang Y., et al. , The transcriptional repressor Hes1 attenuates inflammation by regulating transcription elongation. Nat. Immunol. 17, 930–937 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Mestre J. R., et al. , Redundancy in the signaling pathways and promoter elements regulating cyclooxygenase-2 gene expression in endotoxin-treated macrophage/monocytic cells. J. Biol. Chem. 276, 3977–3982 (2001). [DOI] [PubMed] [Google Scholar]
- 89.Vanden Berghe W., et al. , p38 and extracellular signal-regulated kinase mitogen-activated protein kinase pathways are required for nuclear factor-kappaB p65 transactivation mediated by tumor necrosis factor. J. Biol. Chem. 273, 3285–3290 (1998). [DOI] [PubMed] [Google Scholar]
- 90.Inglis A. J., et al. , Activation of GCN2 by the ribosomal P-stalk. Proc. Natl. Acad. Sci. U.S.A. 116, 4946–4954 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Van de Velde L. A., et al. , Stress kinase GCN2 controls the proliferative fitness and trafficking of cytotoxic T cells independent of environmental amino acid sensing. Cell Rep. 17, 2247–2258 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Jasperson L. K., et al. , Inducing the tryptophan catabolic pathway, indoleamine 2,3-dioxygenase (IDO), for suppression of graft-versus-host disease (GVHD) lethality. Blood 114, 5062–5070 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Donnelly N., Gorman A. M., Gupta S., Samali A., The eIF2α kinases: Their structures and functions. Cell. Mol. Life Sci. 70, 3493–3511 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Harding H. P., et al. , Regulated translation initiation controls stress-induced gene expression in mammalian cells. Mol. Cell 6, 1099–1108 (2000). [DOI] [PubMed] [Google Scholar]
- 95.Ye J., et al. , GCN2 sustains mTORC1 suppression upon amino acid deprivation by inducing Sestrin2. Genes Dev. 29, 2331–2336 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Ohoka N., Yoshii S., Hattori T., Onozaki K., Hayashi H., TRB3, a novel ER stress-inducible gene, is induced via ATF4-CHOP pathway and is involved in cell death. EMBO J. 24, 1243–1255 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Lee J. I., et al. , HepG2/C3A cells respond to cysteine deprivation by induction of the amino acid deprivation/integrated stress response pathway. Physiol. Genomics 33, 218–229 (2008). [DOI] [PubMed] [Google Scholar]
- 98.Mukhopadhyay R., Jia J., Arif A., Ray P. S., Fox P. L., The GAIT system: A gatekeeper of inflammatory gene expression. Trends Biochem. Sci. 34, 324–331 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Guo M., Schimmel P., Essential nontranslational functions of tRNA synthetases. Nat. Chem. Biol. 9, 145–153 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Fang P., et al. , Structural basis for full-spectrum inhibition of translational functions on a tRNA synthetase. Nat. Commun. 6, 6402 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Ramesh R., et al. , Pro-inflammatory human Th17 cells selectively express P-glycoprotein and are refractory to glucocorticoids. J. Exp. Med. 211, 89–104 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Acosta-Rodriguez E. V., et al. , Surface phenotype and antigenic specificity of human interleukin 17-producing T helper memory cells. Nat. Immunol. 8, 639–646 (2007). [DOI] [PubMed] [Google Scholar]
- 103.Cua D. J., et al. , Interleukin-23 rather than interleukin-12 is the critical cytokine for autoimmune inflammation of the brain. Nature 421, 744–748 (2003). [DOI] [PubMed] [Google Scholar]
- 104.Lee Y., et al. , Induction and molecular signature of pathogenic TH17 cells. Nat. Immunol. 13, 991–999 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Durant L., et al. , Diverse targets of the transcription factor STAT3 contribute to T cell pathogenicity and homeostasis. Immunity 32, 605–615 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Sundrud M. S., Carlson T. J.. Transcriptional responses of wild-type and Gcn2-/- Th17 cells to halofuginone and rapamycin. Gene Expression Omnibus. https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE47478. Deposited 29 May 2013.
- 107.Ishimura R., Nagy G., Dotu I., Chuang J. H., Ackerman S. L., Activation of GCN2 kinase by ribosome stalling links translation elongation with translation initiation. eLife 5, e14295 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Zhang Y., Dickinson J. R., Paul M. J., Halford N. G., Molecular cloning of an arabidopsis homologue of GCN2, a protein kinase involved in co-ordinated response to amino acid starvation. Planta 217, 668–675 (2003). [DOI] [PubMed] [Google Scholar]
- 109.Hirose T., Horvitz H. R., The translational regulators GCN-1 and ABCF-3 act together to promote apoptosis in C. elegans. PLoS Genet. 10, e1004512 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Hinnebusch A. G., Gene-specific translational control of the yeast GCN4 gene by phosphorylation of eukaryotic initiation factor 2. Mol. Microbiol. 10, 215–223 (1993). [DOI] [PubMed] [Google Scholar]
- 111.Marton M. J., Vazquez de Aldana C. R., Qiu H., Chakraburtty K., Hinnebusch A. G., Evidence that GCN1 and GCN20, translational regulators of GCN4, function on elongating ribosomes in activation of eIF2alpha kinase GCN2. Mol. Cell. Biol. 17, 4474–4489 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Silva R. C., Sattlegger E., Castilho B. A., Perturbations in actin dynamics reconfigure protein complexes that modulate GCN2 activity and promote an eIF2 response. J. Cell Sci. 129, 4521–4533 (2016). [DOI] [PubMed] [Google Scholar]
- 113.Luna E., et al. , Plant perception of β-aminobutyric acid is mediated by an aspartyl-tRNA synthetase. Nat. Chem. Biol. 10, 450–456 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Izquierdo Y., et al. , Arabidopsis nonresponding to oxylipins locus NOXY7 encodes a yeast GCN1 homolog that mediates noncanonical translation regulation and stress adaptation. Plant Cell Environ. 41, 1438–1452 (2018). [DOI] [PubMed] [Google Scholar]
- 115.Faus I., et al. , Arabidopsis ILITHYIA protein is necessary for proper chloroplast biogenesis and root development independent of eIF2α phosphorylation. J. Plant Physiol. 224-225, 173–182 (2018). [DOI] [PubMed] [Google Scholar]
- 116.Monaghan J., Li X., The HEAT repeat protein ILITYHIA is required for plant immunity. Plant Cell Physiol. 51, 742–753 (2010). [DOI] [PubMed] [Google Scholar]
- 117.Ravindran R., et al. , The amino acid sensor GCN2 controls gut inflammation by inhibiting inflammasome activation. Nature 531, 523–527 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Ravindran R., et al. , Vaccine activation of the nutrient sensor GCN2 in dendritic cells enhances antigen presentation. Science 343, 313–317 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.McGaha T. L., IDO-GCN2 and autophagy in inflammation. Oncotarget 6, 21771–21772 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Rodriguez P. C., Quiceno D. G., Ochoa A. C., L-arginine availability regulates T-lymphocyte cell-cycle progression. Blood 109, 1568–1573 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Jasperson L. K., et al. , Indoleamine 2,3-dioxygenase is a critical regulator of acute graft-versus-host disease lethality. Blood 111, 3257–3265 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Deval C., et al. , Amino acid limitation regulates the expression of genes involved in several specific biological processes through GCN2-dependent and GCN2-independent pathways. FEBS J. 276, 707–718 (2009). [DOI] [PubMed] [Google Scholar]
- 123.Shan J., et al. , A mitogen-activated protein kinase/extracellular signal-regulated kinase kinase (MEK)-dependent transcriptional program controls activation of the early growth response 1 (EGR1) gene during amino acid limitation. J. Biol. Chem. 289, 24665–24679 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Chaveroux C., et al. , Identification of a novel amino acid response pathway triggering ATF2 phosphorylation in mammals. Mol. Cell. Biol. 29, 6515–6526 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.De Vito A., et al. , Amino acid deprivation triggers a novel GCN2-independent response leading to the transcriptional reactivation of non-native DNA sequences. PLoS One 13, e0200783 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Muller A. J., et al. , Chronic inflammation that facilitates tumor progression creates local immune suppression by inducing indoleamine 2,3 dioxygenase. Proc. Natl. Acad. Sci. U.S.A. 105, 17073–17078 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Van de Velde L. A., et al. , T cells encountering myeloid cells programmed for amino acid-dependent immunosuppression use rictor/mTORC2 protein for proliferative checkpoint decisions. J. Biol. Chem. 292, 15–30 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Gause W. C., Wynn T. A., Allen J. E., Type 2 immunity and wound healing: Evolutionary refinement of adaptive immunity by helminths. Nat. Rev. Immunol. 13, 607–614 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Allen J. E., Sutherland T. E., Host protective roles of type 2 immunity: Parasite killing and tissue repair, flip sides of the same coin. Semin. Immunol. 26, 329–340 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Herbert D. R., et al. , Arginase I suppresses IL-12/IL-23p40-driven intestinal inflammation during acute schistosomiasis. J. Immunol. 184, 6438–6446 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Pesce J. T., et al. , Arginase-1-expressing macrophages suppress Th2 cytokine-driven inflammation and fibrosis. PLoS Pathog. 5, e1000371 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Jani M., Barton A., Warren R. B., Griffiths C. E., Chinoy H., The role of DMARDs in reducing the immunogenicity of TNF inhibitors in chronic inflammatory diseases. Rheumatology (Oxford) 53, 213–222 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Pagnini C., Pizarro T. T., Cominelli F., Novel pharmacological therapy in inflammatory bowel diseases: Beyond anti-tumor necrosis factor. Front. Pharmacol. 10, 671 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Castilho B. A., et al. , Keeping the eIF2 alpha kinase Gcn2 in check. Biochim. Biophys. Acta 1843, 1948–1968 (2014). [DOI] [PubMed] [Google Scholar]
- 135.Sattlegger E., Hinnebusch A. G., Separate domains in GCN1 for binding protein kinase GCN2 and ribosomes are required for GCN2 activation in amino acid-starved cells. EMBO J. 19, 6622–6633 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Andrade M. A., Petosa C., O’Donoghue S. I., Müller C. W., Bork P., Comparison of ARM and HEAT protein repeats. J. Mol. Biol. 309, 1–18 (2001). [DOI] [PubMed] [Google Scholar]
- 137.Vazquez de Aldana C. R., Marton M. J., Hinnebusch A. G., GCN20, a novel ATP binding cassette protein, and GCN1 reside in a complex that mediates activation of the eIF-2 alpha kinase GCN2 in amino acid-starved cells. EMBO J. 14, 3184–3199 (1995). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All protocols and data supporting the findings of this study are available within the main text and SI Appendix. The primary sequencing data and microarray information that were generated for this publication have been deposited in the National Center for Biotechnology Information’s Gene Expression Omnibus (GEO). Information for SI Appendix, Tables S1 and S2 are accessible through GEO Series accession no. GSE145205, https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE145205. Information for SI Appendix, Table S3 are accessible through GEO accession no. GSE47478, https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE47478.