Abstract
Environmental, genetic and epigenetic risk factors have been closely related to the development of type-2 diabetes (T2D). It has been reported that the expression in H19 and MALAT1 are related to metabolic diseases. To analyze the relationship between the expression of H19 and MALAT1 lncRNAs with diabetic patients. A study was conducted in subjects with T2D and nondiabetic controls, residents of Mexico City. Anthropometric measurements were made, and serum concentrations of glucose, glycosylated hemoglobin, total cholesterol, triglycerides, high- and low-density lipoprotein cholesterol were analyzed. Total RNA was extracted from serum and serum exosomes. The H19 and MALAT1 expression levels were quantified by RT-qPCR. A significant reduction in the expression of MALAT1 from serum or serum exosomes were found in patients with T2D, metabolic syndrome and low levels of HDL-c. Significant increase in H19 levels was found in diabetic subjects with poor glycemic control. Additionally, the principal component analyzes showed that serum MALAT1 expression was associated with total cholesterol and HDL-c levels, and the exosomes H19 expression was associated with waist circumference. The results obtained suggest that MALAT1 expression levels could be an epigenetic biomarker of diabetes risk or of its comorbidities.
Keywords: Type-2 diabetes, Exosomes, Metabolic syndrome, lncRNA-MALAT1, lncRNA-H19
1. Introduction
Diabetes is one of the most important pathologies worldwide, it is considered one of the main causes of morbidity and mortality among the adult population. The International Diabetes Federation for the year 2015 estimated 415 million cases of diabetes among adults aged 20–79 years. For 2040, it has been estimated that 642 million people aged 20–79, could have diabetes [1]. The chronic hyperglycemia of diabetes is associated with long-term damage and therefore, dysfunction of various organs, especially the eyes, kidneys, nerves and blood vessels. The relationship between type-2 diabetes (T2D) and environmental factors, diet and level of physical activity is well known [1,2]. However, the identification of new genetic and epigenetic factors involved in these pathologies is also relevant.
The transport of molecules contained within extracellular vesicles (EVs), such as exosomes, have emerged as important biomarkers in the intercellular communication in physiological and pathophysiological states [3]. Exosomes can be detected in a variety of fluids, including serum, plasma, urine, saliva, and cerebrospinal fluid, among other. At present, several investigations have focused on studying the relationship of exosomes and their content with complex diseases and cancer [4,5]. In particular, it has been shown that in T2D, exosomes secreted from skeletal muscle, visceral adipose tissue and hepatocytes can transfer both functional proteins and RNA species that regulate the metabolic function of both remote tissues and of adjacent cells. Individuals with varying complications due to diabetes showed diverse patterns of exosome molecules [6], suggesting that exosomes may contribute to the stage-specific pathogenic mechanisms of T2D progression and the complications of diabetes. It has been noted that when EVs derived from adipose tissue are transferred from obese mice to mice without obesity, an inflammatory phenotype and insulin resistance are favored [7]. Recent evidence indicates that the expression of various lncRNAs in diabetic animal models and in clinical studies can serve as new biomarkers for the prognosis of T2D [8].
The long non-coding RNAs (lncRNAs) are involved in the expression of genes that regulate several functions. It has been observed that lncRNAs act as activators or repressors of transcription, enhancers of gene expression through the recruitment of transcriptional complexes. In addition, lncRNAs are molecular scaffolds or decoy lncRNAs for the recruitment of chromatin-modifying complexes, inhibiting splicing or translation [[9], [10], [11], [12]]. It has been reported that mutations and dysregulations of lncRNAs are associated with the development and progression of several diseases, including cancer, diabetes, metabolic syndrome (MetS), obesity, and Alzheimer, among others [[13], [14], [15], [16]]. It has been identified that some lncRNAs such as H19, showed a reduced expression in skeletal muscle from diabetic subjects and mice with insulin resistance [17]. On the other hand, the levels of expression of MALAT1 have been associated with processes related to inflammation and hypoxia within the context of diabetes, as well as with diabetic retinopathy and nephropathy, and in the dysregulation of glucose metabolism [18,19].
Clinical studies that have reported association of lncRNAs and metabolic diseases have been performed primarily in peripheral blood mononuclear cells (PBMCs). Likewise, the primary evidences of these relationships have been carried out mainly in cell lines and animal models. It has been reported that H19 may regulate muscle glucose metabolism by acting as a novel upstream regulator of let-7 family miRNAs whose role in glucose metabolism has been established. The decrease in H19 leads to increased bioavailability of let-7 display insulin resistance and impaired glucose tolerance, a phenotype that occurs in part through let-7-mediated repression of multiple components of the insulin-PI3K-mTOR pathway, including insulin receptor, insulin receptor substrate 2 and insulin-like growth factor receptor 1 [17]. On the other hand, it has been reported that acute H19 elevation leads to decreased promoter methylation and increased expression of hepatocyte nuclear factor 4-alpha (HNF4-α) and subsequent gluconeogenic gene expression, and therefore, increased glucose production [20].
On the other hand, it has been shown that MALAT1 regulates inflammatory cytokine production in a state of diabetes, such as TNFα, IL-6 and IL-1β, through the activation of serum amyloid antigen 3 (SAA3), which is a regulator of glucose-induced inflammatory changes and oxidative stress. The low-grade inflammatory process is a key mechanism in the development of diabetes comorbidities [19,21]. Another study, using STZ‐induced diabetic mice and immortalized mouse podocytes exposed to high glucose, demonstrated increased MALAT1 levels, accompanied by β‐catenin nuclear translocation, with increased MMP‐2 activity and albumin permeability, which was attenuated by MALAT1 knock‐down. The researchers have proposed that β‐catenin expression is modulated at post‐transcriptional level by MALAT1, potentially via recognition of β‐catenin pre‐mRNAs, and thereby altered at protein levels. Additionally, the researchers report that MALAT1 levels were affected by β-catenin as well through binding to the promotor region of MALAT1. β‐catenin shRNA transduction led to decreased mutual binding and thus diminished MALAT1 expression [22]. The purpose of this study was to analyze the relationship between the levels of H19 and MALAT1 expression from serum and serum exosomes in patients with T2D.
2. Materials and methods
2.1. Subjects
Patients with a diagnosis of less than 5 years of T2D and subjects without T2D were analyzed. The participants were residents of Mexico City served by the medical services from the Mexican Institute of Social Security (IMSS). The research was approved by the National Ethical Committee of the Mexican Institute of Social Security in accordance with the Helsinki Declaration. All participants signed an informed consent. Each participant was measured for weight, height, waist circumference (WC), and blood pressure (BP). Venous blood samples were also obtained after a 12-h fast, for RNA extraction and biochemical studies. Concentrations of total cholesterol (c-total), triglycerides, high-density lipoprotein cholesterol (HDL-c), low-density lipoprotein cholesterol (LDL-c) and glycosylated hemoglobin (HbA1c) were performed. From each participant serum samples were frozen at −70 °C until use.
Additionally, according to International Diabetes Federation (IDF) criteria, a MetS diagnosis of the study participants was performed. These criteria define that a person has MetS when they have central obesity (for the population of the Americas, the criterion of a waist circumference ≥88 cm in females and ≥102 cm in males continues to be used), plus any two of the following four factors: Raised triglycerides: ≥150 mg/dL; reduced HDL cholesterol: < 40 mg/dL (1.03 mmol/L) in males and < 50 mg/dL (1.29 mmol/L) in females, or specific treatment for these lipid abnormalities; raised blood pressure: systolic BP ≥ 130 or diastolic BP ≥ 85 mmHg, or treatment of previously diagnosed hypertension; raised fasting plasma glucose: ≥ 100 mg/dL (5.6 mmol/L), or previously diagnosed type 2 diabetes [23].
2.2. Isolation of exosomes and RNA extraction
Serum samples were thawed on ice and subsequently centrifuged at 3000 g for 15 min. The exosomes and RNA contained were isolated using the exoRNeasy Serum/Plasma Midi Kit (cat# 77044, QIAGEN, Hilden, Germany), following manufacturer instructions. Briefly, serum samples were mixed with 2X binding buffer XBP, after which the mixture was then passed through the exoEasy membrane affinity column to bind the EVs to the membrane and centrifuged at 500 g for 1 min. The flow-through was discarded and the wash buffer XWP was added to the column. After another centrifugation and discarding of the flow-through, the vesicles were lysed by adding of 700 μL QIAzol to the spin column, and the lysate was collected by centrifugation at 3000 g for 5 min. Subsequently, 90 μL of chloroform was added and centrifuged at 12,000 g by 15 min at 4 °C. The aqueous phase was recovered and mixed with ethanol, the sample-ethanol mixture was added to a RNeasy MinElute spin column and centrifuged and the column was washed once with buffer RWT after it was washed twice with buffer RPE followed by elution of RNA into water.
2.3. Extraction of serum RNA
Total RNA was extracted of serum samples using TRizol™ reagent (Thermo Fisher Scientific, Waltham, MA, USA). Briefly, 500 μL of serum was placed 1.5 mL of trizol reagent and incubated for 5 min, then 40 μL of chloroform was added. It was mixed and incubated for 15 min and centrifugated at 12,000 g for 25 min at 4 °C. The aqueous phase was transferred to a new tube and adding 1 mL of isopropanol and incubate for 10 min. It was then centrifuged at 21,100 g for 15 min at 4 °C, after which the supernatant was carefully removed. The pellet was mixed with 1.5 mL of ethanol for resuspending the RNA.
2.4. Obtaining cDNA from serum and exosomes
Prior to the synthesis of the cDNA, the DNase I RNase free kit (Thermo Fisher Scientific, Waltham, MA, USA) was used to eliminate the gDNA and to avoid interference in the results. Subsequently, 10 μL was taken for cDNA synthesis using the High-Capacity cDNA Reverse Transcription Kit (Thermo Fisher Scientific, Waltham, MA, USA), following the manufacturer's instructions. Briefly, the master mix 2X RT was prepared on ice, adding for each reaction 2 μL of 10X RT buffer, 0.8 μL of 25X dNTP, 2 μL of RT 10X primers, 1 μL of reverse transcriptase (RT), 1 μL of RNase inhibitor, and 3.2 μL of nuclease-free water. Subsequently, 10 μL of master mix 2X RT was placed in each reaction well, adding 10 μL of RNA (2 μg). The reaction volume was adjusted to 20 mL, and the amplification was performed in the MAXYGEN II Thermal Cycler (Axygen Scientific, Union City, CA, USA).
2.5. Expression of long non-coding RNAs H19 and MALAT1
The expression of lncRNAs was quantified using TaqMan Gene Expression Assays (Thermo Fisher Scientific, Waltham, MA, USA), H19 (cat# Hs00399294_g1) and MALAT1 (cat# Hs00273907_s1) by RT-qPCR using the 7900HT detection system (Applied Biosystems, Foster City, CA, USA). Gene expression data was normalized to the expression levels of the GAPDH housekeeping gene (cat# Hs02786624_g1) and analyzed using the method [24].
2.6. Statistical analysis
The data of continuous variables are represented as means ± standard deviation or as medians and percentiles (p25th - p75th) based on their distribution, and in frequencies for qualitative variables. Comparisons between groups were performed by Mann-Whitney test for variables without normal distribution, or t-test for variables with normal distribution, and Chi-square test by qualitative data. Generalized linear models were performed to evaluate the association between lncRNAs with T2D, MetS and biochemical measurements. Principal Component Analysis (PCA) was performed to examine the correlation between lncRNAs expression and anthropometrics and biochemicals variables. We reported only components that showed eigenvalues greater than 1. Two-tailed statistical tests were conducted with a significance level of 5% using STATA v.15 software (Stata Corp, College Station, TX, USA).
3. Results
Sixty patients with T2D and sixty subjects without T2D were studied, 68.3% of which were women and 31.7% were men. Diabetic patients had significantly higher BMI and WC, as well as elevated glucose and triglycerides levels and reduced HDL-c levels, compared to individuals without T2D. In addition, 58.3% of diabetic individuals had MetS and 10% of subjects without T2D (Table 1). Serum levels of lncRNA-H19 were 14% lower in T2D patients than control subjects. For exosome-derived levels of H19, T2D patients showed a reduction of 24% in comparison with the control group. However, these differences were not significant. Relative lncRNA-MALAT1 expression levels were 45% or 44% lower in the T2D group than in the control group for serum and serum exosomes respectively. However, only the reduction in MALAT1 expression from serum of T2D group was significant. In addition, serum or exosomes-derived levels of MALAT1 were similar (Fig. 1).
Table 1.
Clinic characteristics of the study participants.
| Characteristic | Total n = 120 | T2D n = 60 | Without T2D n = 60 | P |
|---|---|---|---|---|
| Age (years) | 49.9 ± 6.3 | 50.2 ± 6.1 | 49.6 ± 6.5 | 0.630a |
| Years of diagnosis | 2.8 ± 1.7 | – | ||
| Gender, n (%) | ||||
| Male | 38 (31.7) | 19 (31.7) | 19 (31.7) | 1.0b |
| Female | 82 (68.3) | 41 (68.3) | 41 (68.3) | |
| BMI (kg/m2) | 28.9 ± 5.4 | 30.2 ± 5.4 | 27.7 ± 5.1 | 0.012a |
| WC (cm) | 93 ± 12.5 | 96.8 ± 13.7 | 89.3 ± 10.0 | <0.001a |
| < 88, n (%) | 60 (50) | 23 (38.3) | 37 (61.7) | 0.011b |
| ≥ 88, n (%) | 60 (50) | 37 (61.7) | 23 (38.3) | |
| Systolic BP (mmHg) | 113 ± 14 | 113 ± 16 | 113 ± 13 | 0.975a |
| Diastolic BP (mmHg) | 78 ± 11 | 80 ± 12 | 76 ± 10 | 0.067a |
| BP < 135 or 85, n (%) | 92 (76.7) | 40 (66.7) | 52 (86.7) | 0.010b |
| BP ≥ 135 or 85, n (%) | 28 (23.3) | 20 (33.3) | 8 (13.3) | |
| Glucose (mg/dL) | 94.5 (87–120) | 120 (102–158) | 87.5 (83–92) | <0.001c |
| < 100, n (%) | 73 (60.8) | 13 (21.7) | 60 (100) | <0.001b |
| ≥ 100, n (%) | 47 (78.2) | 47 (78.3) | 0 | |
| Triglycerides (mg/dL) | 168 (125.5–229) | 188 (140–257) | 142 (101–196.5) | <0.001c |
| < 150, n (%) | 52 (43.3) | 18 (30) | 34 (56.7) | 0.003 |
| ≥ 150, n (%) | 68 (56.7) | 42 (70) | 26 (43.3) | |
| HDL-c (mg/dL) | 45.5 (39–55) | 41 (35.5–49) | 50.5 (41.5–57) | <0.001c |
| ≥ 40 M or 50 F, n (%) | 62 (51.7) | 22 (36.7) | 40 (66.7) | 0.001 |
| < 40 M or 50 F, n (%) | 58 (48.3) | 38 (63.3) | 20 (33.3) | |
| LDL-c (mg/dL) | 140 ± 36 | 140 ± 34 | 142 ± 38 | 0.731a |
| Cholesterol (mg/dL) | 201 ± 45 | 200 ± 41 | 202 ± 48 | 0.873a |
| Metabolic syndrome, n (%) | 41 (34.2) | 35 (58.3) | 6 (10) | <0.001b |
| HbA1c (%) | 5.6 (4.9–7.2) | – | ||
The data are shown as means ± standard deviation, medians and percentiles (p25th - p75th) or as indicated.
T2D, Type-2 diabetes; BMI, Body mass index; WC, Waist circumference; BP, Blood pressures; HDL-c, High-density lipoprotein cholesterol; LDL-c, Low-density lipoprotein cholesterol; HbA1c, Glycosylated hemoglobin.
t-test.
Chi-square test.
Mann-Whitney test.
Fig. 1.
Relative expression of lncRNAs in serum or isolated serum exosome from T2D diagnosed patients or controls subjects. The median with interquartile range of relative expression for H19 or MALAT1 are showed.
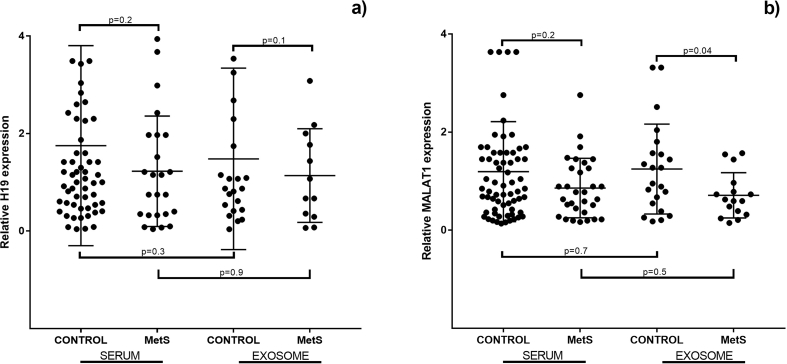
On the other hand, we identified a significant reduction in the relative MALAT1 expression in serum exosome from patients with metabolic syndrome (MetS). The reduction observed in the MetS patients in comparison with the controls was of 48%. A similar pattern was observed for the relative H19 expression from serum or serum exosome and the relative MALAT1 expression from serum. However, these reductions were not significant (Fig. 2). Moreover, we analyzed the changes in relative lncRNAs expression among diabetic patients under good glucose control (<7% HbA1c) compared to diabetics with poor glycemic control (≥7% HbA1c). We identified that the relative expression of serum H19 was over 2-fold higher in patients with poor glycemic control compared to diabetics under good glucose control (Table 2).
Fig. 2.
Relative expression of lncRNAs in serum or isolated serum exosome from metabolic syndrome (MetS) patients or controls subjects. The median with interquartile range of relative expression for H19 or MALAT1 are showed.
Table 2.
Changes in relative expression of lncRNAS in patients with uncontrolled diabetes.
| Source | lncRNA | Controlled <7% HbA1c (n = 43) |
Uncontrolled ≥7% HbA1c (n = 17) |
Pa |
|---|---|---|---|---|
| Serum | H19 | 0.75 (0.34–1.63) | 1.6 (0.98–2.42) | 0.039 |
| MALAT1 | 0.73 (0.28–1.26) | 0.55 (0.36–0.81) | 0.547 | |
| Serum exosomes | H19 | 0.67 (0.29–1.77) | 1.55 (0.41–2.68) | 0.525 |
| MALAT1 | 0.73 (0.39–0.97) | 0.77 (0.26–1.28) | 0.947 |
The data are shown as medians and percentiles (p25th - p75th). HbA1c: glycated hemoglobin.
Mann-Whitney test.
We identified a significant reduction in the expression of MALAT1 from serum (β = −0.62; 95% CI: -0.96, −0.27) and serum exosomes (β = −0.70; 95% CI: -1.25, −0.15) in individuals with T2D. In addition, we found reduced MALAT1 expression in exosomes from subjects with MetS (β = −0.64; 95% CI: -1.19, −0.09). Moreover, low levels of HDL-c were related to a decrease in the relative expression of MALAT1 (β = −0.47; 95% CI: -0.83, −0.12) in serum (Table 3). Through principal component analysis (PCA), we identified that serum-expression levels of MALAT1, total cholesterol and HDL-c levels correlated with the component 2 (factor loads 0.46, 0.44 or 0.45 respectively). Moreover, H19 levels of exosomes and waist circumference were positively correlated with the component 3, with factor loads of 0.43 for each one (Table S1 and Fig. S1). In addition, the second component (serum MALAT1, total cholesterol and HDL-c levels) was negatively associated with diabetic patients (OR = 0.24; 95% CI: 0.11–0.54; p = 0.001) (Fig. 3a). Similar results were observed for component 3 (exosomes levels of H19 and waist circumference), but this result was not significant (OR = 0.69, 95% CI: 0.16–3; p = 0.62) (Fig. 3b).
Table 3.
Relation between the levels of expression of H19 and MALAT1 with type-2 diabetes, metabolic syndrome, and biochemical measurements.
| Serum |
Exosomes |
|||||||
|---|---|---|---|---|---|---|---|---|
| H19 |
MALAT1 |
H19 |
MALAT1 |
|||||
| β (95% CI) | p | β (95% CI) | p | β (95% CI) | p | β (95% CI) | p | |
| T2D | −0.24 (−1.0, 0.56) | 0.555 | −0.62 (−0.96, −0.27) | < 0.001 | −0.43 (−1.5, 0.68) | 0.444 | −0.70 (−1.25, −0.15) | 0.013 |
| MetS | −0.53 (−1.41, 0.35) | 0.240 | −0.33 (−0.71, 0.06) | 0.099 | −0.34 (−1.48, 0.81) | 0.565 | −0.64 (−1.19, −0.09) | 0.023 |
| WC, cm ≥ 102 male/88 female |
−0.22 (−1.02, 0.58) | 0.591 | 0.01 (−0.35, 0.38) | 0.946 | 0.53 (−0.55, 1.62) | 0.336 | −0.50 (−1.07, 0.06) | 0.080 |
| BP, mmHg ≥135/85 |
−0.26 (−1.19, 0.67) | 0.589 | −0.21 (−0.66, 0.25) | 0.374 | 0.80 (−0.48, 2.08) | 0.223 | −0.25 (−0.98, 0.47) | 0.495 |
| TG, mg/dL ≥ 150 | −0.42 (−1.22, 0.37) | 0.298 | −0.03 (−0.40, 0.33) | 0.855 | −0.42 (−1.54, 0.70) | 0.465 | −0.07 (−0.66, 0.51) | 0.804 |
| HDL-c, mg/dL <40 male/50 female |
0.34 (−0.46, 1.13) | 0.406 | −0.47 (−0.83, −0.12) | 0.009 | −0.49 (−1.62, 0.63) | 0.389 | −0.15 (−0.76, 0.44) | 0.618 |
| HbA1c ≥ 7% | 1.07 (−0.05, 2.19) | 0.060 | −0.14 (−0.45, 0.16) | 0.375 | 0.42 (−1.38, 2.23) | 0.645 | −0.03 (−0.80, 0.74) | 0.945 |
Β, Regression coefficient calculated by generalized linear models adjusted for age; CI, Confidence interval; T2D, Type-2 diabetes; MetS, Metabolic syndrome; WC, Waist circumference; BP, Blood pressure; TG, Triglycerides; HDL-c, High-density lipoprotein cholesterol; HbA1c, Glycosylated hemoglobin.
Fig. 3.
Logistic regression of principal component analysis on risk of T2D. The ORs (●) and 95% Confidence Interval (black line) obtained from multiple logistic regression (adjusted by age and sex) are shown. a) PCA estimated to serum MALAT1 (Component 1: glucose, waist circumference, and triglycerides; Component 2: MALAT1, total cholesterol and HDL-c); b) PCA estimated to exosome H19 (Component 1: total cholesterol and triglycerides; Component 2: Glucose and HDL-c; Component 3: H19 and waist circumference).
4. Discussion
An increase in the serum levels of EVs has been reported in individuals with obesity, MetS, insulin resistance and diabetes.19 Additionally, it has been demonstrated that in T2D the secreted exosomes of skeletal muscle, visceral adipose tissue and hepatocytes, can transfer both functional proteins and RNAs that regulate the metabolic function of distant tissues [5,25,26]. We analyzed the relationship between the level of expression of H19 and MALAT1 lncRNAs of T2D patients. This is the first study to analyze the level of lncRNAs expression in serum and in exosomes isolated from serum. We identified a significant reduction in the expression of MALAT1 from serum in diabetic patients compared to individuals without diabetes, as well as a significant increase in H19 expression levels in diabetic subjects with poor glycemic control. Moreover, we identified that T2D, MetS, and decreased levels of HDL-c had a significant association with decreased levels of MALAT1 derived from serum or exosomes.
H19-lncRNA is a highly conserved, imprinted gene expressed only in maternal allele. Both maternal and paternal H19 alleles are expressed at the first stage of embryonal development. However, only maternal chromosomes express H19 after 10 weeks of gestation. H19 has been associated with decreased expression in diabetic patients. Gao et al. recognized that H19 is reduced approximately five times in the skeletal muscle of diabetic subjects and in mice with insulin resistance in comparison to its controls [17]. However, in another study realized in liver biopsies obtained from subjects with T2D and nondiabetic controls, an increased expression of H19 in the diabetic patients’ liver was reported [27]. To test whether increased hepatic H19 expression, seen in T2D patients, other researchers conducted a study in treated mice with a high-fat diet (HFD). Compared with normal chow–fed mice, the HFD-fed mice had an elevated fasting glucose. They also found that HFD-fed mice increased hepatic H19 expression and impaired glucose homeostasis. Results that the same researchers verified when inducing overexpression of H19 in the liver of mice and identified that the H19-overexpressed mice were glucose intolerant and insulin resistant. The researchers suggest that an elevated level of H19 in the liver augments hepatic glucose production and impairs glucose homeostasis [20]. Moreover, H19 overexpression has been reported in patients with coronary artery disease in comparison with the controls, though without statistical significance [28].
On the other hand, the MALAT1 lncRNA can act as a transcriptional regulator of numerous genes, including some genes involved in metastasis and cell migration in cancer, for which it has been implicated in cell cycle regulation [29]. Other studies discovered relationship between the reduction of MALAT1 in diabetic animal models with the reduction of the microvascular dysfunction. In addition, it has been identified that MALAT1 expression levels have been associated with retinopathy and diabetic nephropathy, through the promotion of angiogenesis in these organs [30,31]. The frequency of any microvascular dysfunction is strongly associated with the glycemic control evaluated through HbAc1 [32]. In this context, the 71.7% of patients diagnosed T2D included in this work, showed a good glycemic control (evaluated using the HbAc1 levels); for this reason, the reduction in MALAT1 levels could be explicate by the lower risk of the microvascular dysfunction. However, more studies are necessary for evaluating the plausible association between the MALAT1 levels and the development of microvascular dysfunction.
Moreover, high concentrations of glucose in the individual or cellular environment is linked with the changes on MALAT1 levels. Accordingly, the exposure of human umbilical vein endothelial (HUVEC) to high glucose concentration (25mMol ≈ 450 mg/dL) for 12 h increase the MALAT1 levels. However, this effect was not observed when the HUVEC were exposed to low glucose concentration (5mMol ≈ 90 mg/dL) for 12 h. This observation suggested that MALAT1 is upregulated only under high glucose conditions [19]. Similar results were observed in women diagnosed with gestational diabetes mellitus (GDM). The patients were diagnosed using oral glucose tolerance test; the women that showed a blood glucose levels ≥ 180 mg/dL or ≥153 mg/dL after 1 h or 2 h post ingestion of 75 g of glucose respectively. Controls were selected if they showed fasting blood glucose levels ≤ 79.2 mg/dL. The results showed that MALAT1 was upregulated in patients under hyperglycemia state (GDM) [33].
Another study that analyzed the expression levels of various lncRNAs in peripheral blood mononuclear cells (PBMCs) from T2D patients reported significantly increased levels of HOTAIR, MEG3, LET, MALAT1, MIAT, among others, and a significant decrease in the expression of THRL y SALRNA1 in patients with T2D compared to control subjects. In addition, the T2D diagnosed patients showed an 8.1% of HbAc1 suggesting a poor glycemic control associated with hyperglycemia status [34]. In this regard, in the present study only 28% of the T2D diagnosed patients included showed blood glucose levels ≥ 150 mg/dL and only 18% showed HbAc1 ≥8%, these facts could be associated with the no increase of MALAT1 levels, and suggesting that MALAT1 could be increase in T2D diagnosed patient that shows a poor glycemic control. Clearly, larger-scale longitudinal studies in this area are required to probe the role of hyperglycemic states with the upregulation or downregulation of MALAT1.
The difference between our results and the previous reported results, could be due to the type of sample used to analyze the expression of the lncRNAs, or to the limitations of our study, such as the small sample size, the few years of T2D evolution and the use of serum samples stored at −70 °C. In addition, the good glycemic control was a frequently observed in the T2D diagnosed patients included.
5. Conclusions
In summary, increased serum levels of H19-lncRNA were significantly related to diabetic patients with poor glycemic control. A significant reduction of MALAT-lncRNA levels derived from serum or serum exosomes were associated with type-2 diabetes. In addition, a significant decrease in MALAT1 expression levels of serum exosomes in subjects with metabolic syndrome was found. Future studies with a larger sample size, patients classified in strata for different years of evolution of T2D, and the use of recently obtained serum samples are still necessary to further clarify the effect of T2D on the expression of lncRNAs.
Declaration of competing interest
The authors have no conflicts of interest to declare.
Acknowledgements
We thank Nathaniel Raymond Pullen for their assistance in reviewing the English translation of the manuscript. This research was supported by the FOMIX-GUE-CONACYT (Code: 249719) and the Academic Quality Strengthening Program (2018).
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.ncrna.2020.03.001.
Contributor Information
Vianet Argelia Tello-Flores, Email: vatellof@gmail.com.
Adán Valladares-Salgado, Email: adanval@gmail.com.
Marco Antonio Ramírez-Vargas, Email: marvar@uagro.mx.
Miguel Cruz, Email: mcruzl@yahoo.com.
Oscar del-Moral-Hernández, Email: odelmoralh@gmail.com.
José Ángel Cahua-Pablo, Email: jose9@hotmail.com.
Mónica Ramírez, Email: ruanomoni@hotmail.com.
Daniel Hernández-Sotelo, Email: danhs1mx@yahoo.com.
Adakatia Armenta-Solis, Email: adakatia@gmail.com.
Eugenia Flores-Alfaro, Email: efloresa_2@hotmail.com.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
Fig. S1.
Diagram of principal component analysis for the LncRNAs and biochemicals or anthropometric parameters. Shows the loads factor relation for serum MALAT1 “a” or exosome H19 “b”.
References
- 1.Ogurtsova K., da Rocha Fernandes J.D., Huang Y., Linnenkamp U., Guariguata L., Cho N.H., Cavan D., Shaw J.E., Makaroff L.E. IDF Diabetes Atlas: global estimates for the prevalence of diabetes for 2015 and 2040. Diabetes Res. Clin. Pract. 2017;128:40–50. doi: 10.1016/j.diabres.2017.03.024. [DOI] [PubMed] [Google Scholar]
- 2.Prasad R.B., Groop L. Genetics of type 2 diabetes-pitfalls and possibilities. Genes (Basel) 2015;6:87–123. doi: 10.3390/genes6010087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Todorova D., Simoncini S., Lacroix R., Sabatier F., Dignat-George F. Extracellular vesicles in angiogenesis. Circ. Res. 2017;120:1658–1673. doi: 10.1161/CIRCRESAHA.117.309681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Alipoor S.D., Mortaz E., Garssen J., Movassaghi M., Mirsaeidi M., Adcock I.M. Exosomes and exosomal miRNA in respiratory diseases. Mediat. Inflamm. 2016;2016 doi: 10.1155/2016/5628404. 5628404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Xu W., Yang Z., Lu N. From pathogenesis to clinical application: insights into exosomes as transfer vectors in cancer. J. Exp. Clin. Canc. Res. 2016;35:156. doi: 10.1186/s13046-016-0429-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lee M.J., Park D.H., Kang J.H. Exosomes as the source of biomarkers of metabolic diseases. Ann. Pediatr. Endocrinol. Metab. 2016;21:119–125. doi: 10.6065/apem.2016.21.3.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pardo F., Villalobos-Labra R., Sobrevia B., Toledo F., Sobrevia L. Extracellular vesicles in obesity and diabetes mellitus. Mol. Aspect. Med. 2018;60:81–91. doi: 10.1016/j.mam.2017.11.010. [DOI] [PubMed] [Google Scholar]
- 8.He X., Ou C., Xiao Y., Han Q., Li H., Zhou S. LncRNAs: key players and novel insights into diabetes mellitus. Oncotarget. 2017;8:71325–71341. doi: 10.18632/oncotarget.19921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Malik B., Feng F.Y. Long noncoding RNAs in prostate cancer: overview and clinical implications. Asian J. Androl. 2016;18:568–574. doi: 10.4103/1008-682X.177123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Knoll M., Lodish H.F., Sun L. Long non-coding RNAs as regulators of the endocrine system. Nat. Rev. Endocrinol. 2015;11:151–160. doi: 10.1038/nrendo.2014.229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sun X., Wong D. Long non-coding RNA-mediated regulation of glucose homeostasis and diabetes. Am. J. Cardiovasc. Dis. 2016;6:17–25. [PMC free article] [PubMed] [Google Scholar]
- 12.Hermans-Beijnsberger S., van Bilsen M., Schroen B. Long non-coding RNAs in the failing heart and vasculature. Noncoding RNA Res. 2018;3:118–130. doi: 10.1016/j.ncrna.2018.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Beermann J., Piccoli M.T., Viereck J., Thum T. Non-coding RNAs in development and disease: background, mechanisms, and therapeutic approaches. Physiol. Rev. 2016;96:1297–1325. doi: 10.1152/physrev.00041.2015. [DOI] [PubMed] [Google Scholar]
- 14.Chen X., Yan C.C., Zhang X., You Z.H. Long non-coding RNAs and complex diseases: from experimental results to computational models. Briefings Bioinf. 2017;18:558–576. doi: 10.1093/bib/bbw060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Losko M., Kotlinowski J., Jura J. Long noncoding RNAs in metabolic syndrome related disorders. Mediat. Inflamm. 2016;2016:5365209. doi: 10.1155/2016/5365209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Qi P., Zhou X.Y., Du X. Circulating long non-coding RNAs in cancer: current status and future perspectives. Mol. Canc. 2016;15:39. doi: 10.1186/s12943-016-0524-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gao Y., Wu F., Zhou J., Yan L., Jurczak M.J., Lee H.Y., Yang L., Mueller M., Zhou X.B., Dandolo L., Szendroedi J., Roden M., Flannery C., Taylor H., Carmichael G.G., Shulman G.I., Huang Y. The H19/let-7 double-negative feedback loop contributes to glucose metabolism in muscle cells. Nucleic Acids Res. 2014;42:13799–13811. doi: 10.1093/nar/gku1160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Leti F., DiStefano J.K. Long noncoding RNAs as diagnostic and therapeutic targets in type 2 diabetes and related complications. Genes (Basel) 2017;8 doi: 10.3390/genes8080207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Puthanveetil P., Chen S., Feng B., Gautam A., Chakrabarti S. Long non-coding RNA MALAT1 regulates hyperglycaemia induced inflammatory process in the endothelial cells. J. Cell Mol. Med. 2015;19:1418–1425. doi: 10.1111/jcmm.12576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhang N., Geng T., Wang Z., Zhang R., Cao T., Camporez J.P., Cai S.Y., Liu Y., Dandolo L., Shulman G.I., Carmichael G.G., Taylor H.S., Huang Y. Elevated hepatic expression of H19 long noncoding RNA contributes to diabetic hyperglycemia. JCI Insight. 2018;3 doi: 10.1172/jci.insight.120304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gordon A.D., Biswas S., Feng B., Chakrabart S. MALAT1: a regulator of inflammatory cytokines in diabetic complications. Endocrinol Diabetes Metab. 2018;1 doi: 10.1002/edm2.10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hu M., Wang R., Li X., Fan M., Lin J., Zhen J., Chen L., Lv Z. LncRNA MALAT1 is dysregulated in diabetic nephropathy and involved in high glucose-induced podocyte injury via its interplay with β-catenin. J. Cell Mol. Med. 2017;21:2732–2747. doi: 10.1111/jcmm.13189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Alberti K.G., Zimmet P., Shaw J., Group I.E.T.F.C. The metabolic syndrome--a new worldwide definition. Lancet. 2005;366:1059–1062. doi: 10.1016/S0140-6736(05)67402-8. [DOI] [PubMed] [Google Scholar]
- 24.Livak K.J., Schmittgen T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 25.Lawson C., Vicencio J.M., Yellon D.M., Davidson S.M. Microvesicles and exosomes: new players in metabolic and cardiovascular disease. J. Endocrinol. 2016;228:R57–R71. doi: 10.1530/JOE-15-0201. [DOI] [PubMed] [Google Scholar]
- 26.Freeman D.W., Noren Hooten N., Eitan E., Green J., Mode N.A., Bodogai M., Zhang Y., Lehrmann E., Zonderman A.B., Biragyn A., Egan J., Becker K.G., Mattson M.P., Ejiogu N., Evans M.K. Diabetes; 2018. Altered Extracellular Vesicle Concentration, Cargo and Function in Diabetes Mellitus. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nilsson E., Matte A., Perfilyev A., de Mello V.D., Käkelä P., Pihlajamäki J., Ling C. Epigenetic alterations in human liver from subjects with type 2 diabetes in parallel with reduced folate levels. J. Clin. Endocrinol. Metab. 2015;100:E1491–E1501. doi: 10.1210/jc.2015-3204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bitarafan S., Yari M., Broumand M.A., Ghaderian S.M.H., Rahimi M., Mirfakhraie R., Azizi F., Omrani M.D. Association of increased levels of lncRNA H19 in PBMCs with risk of coronary artery disease. Cell J. 2019;20:564–568. doi: 10.22074/cellj.2019.5544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhou X., Liu S., Cai G., Kong L., Zhang T., Ren Y., Wu Y., Mei M., Zhang L., Wang X. Long non coding RNA MALAT1 promotes tumor growth and metastasis by inducing epithelial-mesenchymal transition in oral squamous cell carcinoma. Sci. Rep. 2015;5:15972. doi: 10.1038/srep15972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Liu J.Y., Yao J., Li X.M., Song Y.C., Wang X.Q., Li Y.J., Yan B., Jiang Q. Pathogenic role of lncRNA-MALAT1 in endothelial cell dysfunction in diabetes mellitus. Cell Death Dis. 2014;5 doi: 10.1038/cddis.2014.466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yan B., Tao Z.F., Li X.M., Zhang H., Yao J., Jiang Q. Aberrant expression of long noncoding RNAs in early diabetic retinopathy. Invest. Ophthalmol. Vis. Sci. 2014;55:941–951. doi: 10.1167/iovs.13-13221. [DOI] [PubMed] [Google Scholar]
- 32.Walraven I., Mast M.R., Hoekstra T., Jansen A.D., van der Heijden A.A., Rauh S.P., Rutters F., van’t Riet E., Elders P.J., Moll A.C. Distinct HbA1c trajectories in a type 2 diabetes cohort. Acta Diabetol. 2015;52:267–275. doi: 10.1007/s00592-014-0633-8. [DOI] [PubMed] [Google Scholar]
- 33.Zhang Y., Wu H., Wang F., Ye M., Zhu H., Bu S. Long non-coding RNA MALAT1 expression in patients with gestational diabetes mellitus. Int. J. Gynecol. Obstet. 2018;140:164–169. doi: 10.1002/ijgo.12384. [DOI] [PubMed] [Google Scholar]
- 34.Sathishkumar C., Prabu P., Mohan V., Balasubramanyam M. Linking a role of lncRNAs (long non-coding RNAs) with insulin resistance, accelerated senescence, and inflammation in patients with type 2 diabetes. Hum. Genom. 2018;12:41. doi: 10.1186/s40246-018-0173-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.