Abstract
Objective
The metabolic influence of gut microbiota plays a pivotal role in the pathogenesis of cardiometabolic diseases. Antibiotics affect intestinal bacterial diversity, and long-term usage has been identified as an independent risk factor for atherosclerosis-driven events. The aim of this study was to explore the interaction between gut dysbiosis by antibiotics and metabolic pathways with the impact on atherosclerosis development.
Methods
We combined oral antibiotics with different diets in an Apolipoprotein E-knockout mouse model linking gut microbiota to atherosclerotic lesion development via an integrative cross-omics approach including serum metabolomics and cecal 16S rRNA targeted metagenomic sequencing. We further investigated patients with carotid atherosclerosis compared to control subjects with comparable cardiovascular risk.
Results
Here, we show that increased atherosclerosis by antibiotics was connected to a loss of intestinal diversity and alterations of microbial metabolic functional capacity with a major impact on the host serum metabolome. Pathways that were modulated by antibiotics and connected to atherosclerosis included diminished tryptophan and disturbed lipid metabolism. These pathways were related to the reduction of certain members of Bacteroidetes and Clostridia by antibiotics in the gut. Patients with atherosclerosis presented a similar metabolic signature as those induced by antibiotics in our mouse model.
Conclusion
Taken together, this work provides insights into the complex interaction between intestinal microbiota and host metabolism. Our data highlight that detrimental effects of antibiotics on the gut flora are connected to a pro-atherogenic metabolic phenotype beyond classical risk factors.
Keywords: Atherosclerosis, Antibiotics, Gut microbiota, Dysbiosis, Metabolic diversity, Cross-omics
Highlights
-
•
Antibiotics exacerbate atherosclerosis independently of diet.
-
•
Gut microbiota and metabolic alpha diversity are reduced by antibiotics.
-
•
Pathways connected to atherogenesis are tryptophan and lipid metabolism.
-
•
Metabolic changes are linked to reduced Clostridia and Bacteroidetes in the gut.
1. Introduction
Cardiovascular disease is the leading cause of mortality and morbidity worldwide and is determined by genetic as well as environmental factors. In recent years, studies have identified the gut microbiome as an emerging contributor to human metabolism, also affecting the cardiovascular system [1]. The gut is a complex co-evolved ecosystem harboring trillions of bacteria. Based on their huge genetic potential, they promote many metabolic processes, thus producing a vast array of bioactive metabolites [2,3]. Particularly, the gut microbiota-dependent metabolite trimethylamine-N-oxide (TMAO) has been a major focus of interest due to its association to cardiovascular disease and mortality in several cohorts [[4], [5], [6], [7]]. While TMAO [8] and the presence of gut microbiota compared to germ-free conditions in mice [9,10] have been linked to augmented arterial thrombosis, other studies in human cohorts and mouse models suggest that loss of bacterial diversity and microbial gene richness negatively affects the cardiovascular disease continuum from risk factors such as blood lipids, hypertension and obesity/insulin resistance to ischemic heart disease and heart failure [1,[11], [12], [13], [14], [15], [16], [17], [18]]. Antibiotic treatment reduces bacterial diversity and has major effects on the host metabolism [19]. Studies have shown that alterations of gut bacteria after antibiotic usage are long-lasting, with a reduction of bacterial richness even after a period of up to 2 years after treatment [20]. In childhood, treatment with antibiotics has been associated with late-onset metabolic diseases such as obesity [21]. Recently, the long-term use of antibiotics in the middle and late adulthood was identified as an independent predictor for future cardiovascular events among women [22]. However, mechanisms linking antibiotics treatment to cardiovascular disease development have to date remained relatively unexplored [23].
Therefore we aimed to investigate the metabolic impact of gut microbiota disruption by antibiotics that may underlie atherosclerosis development. We combined broad oral antibiotics treatment with different diets in Apolipoprotein E knockout (ApoE−/−) mice and analyzed metabolic mechanisms via an integrative cross-omics approach combining serum metabolomics, 16S ribosomal RNA (rRNA) targeted metagenomic sequencing, and phenotype. To support our hypothesis, we performed serum metabolomics and fecal 16S rRNA sequencing in humans with atherosclerotic vascular disease. Here, we show that antibiotics treatment increased atherosclerotic lesion size independently of the diet in ApoE−/− mice. We observed reduced gut microbiota alpha diversity, which was linked to the loss of metabolic diversity in the serum and further associated to enhanced atherosclerosis. Weighted correlation network analysis (WGCNA) combined with phenotype-associated filtering of serum metabolomics allowed us to identify specific pathways modulated by antibiotics that were linked to atherogenesis. In detail, these were diminished tryptophan metabolism and altered lipid metabolism. The metabolomics findings were related to reduced microbial functional capacity with reduced tryptophan and increased fatty acid biosynthesis shown by KEGG gene reconstruction of the gut microbiome as well as the loss of certain members of Bacteroidetes and Clostridia revealed by an inter-omics model. Supplementation of tryptophan in the diet was able to restore antibiotics-induced atherosclerosis, thus strengthening the role of reduced microbial tryptophan biosynthesis in antibiotics-induced atherosclerosis. Analysis of human subjects with carotid atherosclerosis compared to control subjects without detectable atherosclerosis with the same cardiovascular risk profile particularly showed diminished serum tryptophan as well as the loss of the same members of Bacteroidetes and Clostridia. Together, our study provides insights between gut microbiota and host metabolism using a non-targeted cross-omics approach. We suggest that increased atherosclerosis through antibiotics-induced dysbiosis is mediated by specific metabolic changes in the host metabolism. These findings may explain the increased cardiovascular risk by long-term use of antibiotics in later adulthood [22].
2. Methods
2.1. Experimental animals
Male Apolipoprotein E knockout mice (B6.129P2-Apoetm1Unc/J) were purchased from Charles River (Wilmington, MA, USA) at the age of 6 weeks. Mice were housed in groups of 3–4 mice in shoebox-sized filtertop cages in a controlled environment (12h daylight cycle, lights off at 08:00 p.m.). Data were controlled for cage effect with no impact on atherosclerotic lesion size, serum metabolome or gut microbiota composition by 16S rRNA targeted metagenomics sequencing (Supplementary Figure S1). Cages were autoclaved and changed twice weekly, bedding was sterilized. Mice had free access to water and food. Mice received either normal chow diet (10% calories from fat; GLP Mucedola Srl, Settimo Milanese, Italy) or a Western diet (45% calories from fat; D12079B; Research Diets, New Brunswick, NJ, USA). Orally largely non-absorbable antibiotics (ampicillin 1 g/L, metronidazole 1 g/L, neomycin 1 g/L, vancomycin 0.5 g/L (Sigma–Aldrich, St. Louis, MO, USA)) [4] were administered in drinking water of the appropriate experimental groups for 10 weeks starting 6 weeks after initiation of the experiment. Mice were sacrificed after 16 weeks at the age of 22 weeks. In the supplementary mouse experiment, 1% L-Tryptophan (Sigma–Aldrich, St. Louis, MO, USA) was mixed into the diet.
All animal procedures are in accordance with the Guide for the Care and Use of Laboratory Animals published by the NIH (publication no. 85–23, revised 1996), approved by the University Hospital of Tor Vergata Animal Care Facility.
2.2. Human cohort
Human subjects with data on carotid atherosclerosis were recruited as part of the FLOROMIDIA cohort. FLOROMIDIA is an exploratory study to investigate OMICS signatures in subjects with a BMI range of 20–60. Predefined clinical phenotypes and OMICS are available for:
-
•
BMI n = 42
-
•
HOMA-IR n = 42
-
•
Euglycemic Hyperinsulinemic clamp (EHC) n = 25
-
•
OGTT n = 38
-
•
Liver echography (steatosis grade) n = 35
-
•
Carotid atherosclerosis data n = 30
-
•
Serum metabolome profile n = 42
-
•
Stool 16S gut microbiome profile n = 42
-
•
Colon transcriptomics n = 37
Recruitment of patients and processing of samples: All subjects gave written informed consent, validated and approved by the ethical committee of Policlinico Tor Vergata University of Rome (Comitato Etico Indipendente, approval number 28-05-2009). The human subject cohort comprised 42 subjects recruited at the Department of Medicine Policlinico Tor Vergata. Sample size was not determined by statistical methods, given the exploratory nature of the project, but is comparable to other studies in the field [15,[24], [25], [26]].
Inclusion criteria: Pre-established inclusion criteria were as follows: all subjects were of Caucasian origin; the subjects reported a stable body weight 3 months preceding the study, were free of any infections, including the use of antibiotics, one month before visit 1 and had no systemic disease.
Exclusion criteria: Pre-established exclusion criteria were: presence of liver disease, specifically HBV/HCV infection and tumor disease, and subjects with thyroid dysfunction were excluded by biochemical work-up. Alcohol consumption >20 g/day was also an exclusion criterion.
Visit 1: enrollment, colonoscopy and colon biopsy (collected in RNAlater, fragmented and immediately flash-frozen in liquid nitrogen before storage at −80°C). Visit 2: one month after visit 1, patients were subjected to EHC, collection of stool and biofluid samples (stored at −80°C). Visit 3: one week after visit 2, patients were subjected to OGTT, liver and carotid echography.
In this manuscript, we used FLOROMIDIA resources to study the link of serum metabolome profile as well as 16S gut microbiome profile to carotid atherosclerosis. Therefore only patients with carotid atherosclerosis data were included in this study. Patients with atherosclerotic plaques were assigned to atherosclerosis group (n = 10), while all other subjects served as control group (n = 20). Baseline characteristics of subjects are displayed in Table 1. Integrated analysis of gut-microbiome>colon transcriptome>metabolome will be described in another manuscript.
Table 1.
Baseline characteristics of human cohort.
| Parameter | Unit | Ctrl | Ath | P-value |
|---|---|---|---|---|
| Number | 20 | 10 | ||
| Age | years | 59.4 ± 9.1 | 64.0 ± 5.8 | 0.105 |
| Gender | % female | 55 | 50 | 0.999 |
| BMI | kg/m2 | 27.7 ± 4.5 | 25.9 ± 3.4 | 0.253 |
| Waist-to-hip ratio | meter | 0.9 ± 0.1 | 0.9 ± 0.1 | 0.152 |
| GFR (CKD-EPI) | mL/min/1.73m2 | 89.0 ± 16.0 | 84.5 ± 22.1 | 0.570 |
| Mean arterial pressure | mmHg | 90.2 ± 7.2 | 90.4 ± 8.0 | 0.956 |
| C-reactive protein | nmol/L | 173.4 ± 299.4 | 118.9 ± 143.8 | 0.556 |
| Cholesterol, total | mmol/L | 5.1 ± 0.9 | 5.1 ± 1.0 | 0.927 |
| Cholesterol, HDL | mmol/L | 1.5 ± 0.4 | 1.6 ± 0.4 | 0.606 |
| Cholesterol, LDL | mmol/L | 3.0 ± 0.9 | 2.9 ± 1.0 | 0.741 |
| Triglycerides | mmol/L | 1.2 ± 0.7 | 1.2 ± 0.8 | 0.951 |
| Apo A1 | μmol/L | 55.2 ± 9.2 | 55.9 ± 5.8 | 0.809 |
| Apo B | μmol/L | 1.8 ± 0.5 | 1.9 ± 0.3 | 0.454 |
| Fasting blood glucose | mmol/L | 5.5 ± 1.7 | 5.0 ± 0.9 | 0.315 |
| HBA1c | mmol/mol | 37.4 ± 11.5 | 35.0 ± 6.1 | 0.471 |
| Insulin | μU/mL | 11.3 ± 6.2 | 13.4 ± 7.3 | 0.454 |
| HOMA-IR | 2.9 ± 2.4 | 3.0 ± 1.7 | 0.952 | |
| Hypoglycemic drugs | % | 5 | 10 | 0.999 |
| Metformin | % | 5 | 10 | 0.999 |
| Statins | % | 20 | 30 | 0.657 |
| Ezetimibe | % | 5 | 0 | 0.999 |
| Aspirin | % | 10 | 20 | 0.584 |
| Anticoagulants | % | 5 | 0 | 0.999 |
| ACE inhibitors | % | 45 | 30 | 0.694 |
| Diuretics | % | 15 | 30 | 0.372 |
| Calcium channel blockers | % | 10 | 0 | 0.540 |
| Beta blockers | % | 5 | 0 | 0.999 |
| Alpha blockers | % | 10 | 10 | 0.999 |
| Proton-pump inhibitors | % | 5 | 0 | 0.999 |
| Probiotics | % | 5 | 10 | 0.999 |
Characteristics of human subjects with data on carotid atherosclerosis as part of the FLOROMIDIA cohort. Ten patients with carotid atherosclerosis (Ath) and 20 control subjects (Ctrl) were included. Analysis by two-sided Student's t-test or Chi-square test for categorical variables. Glomerular filtration rate (GFR) was calculated based on serum creatinine, age, sex and ethnicity by CKD-EPI (Chronic Kidney Disease Epidemiology Collaboration) formula. ACE: Angiotensin-converting enzyme, HOMA-IR: Homeostatic Model Assessment of Insulin Resistance. Data are the mean ± S.D. or percent.
2.3. Histology (mouse)
To determine extent of atherosclerosis and macrophage infiltration in the outflow tract and valve area, the top half of the heart was removed, washed in PBS and transferred into 4% PFA for 48 h. After dehydration with ethanol, hearts were embedded in paraffin. Serial 5 μm sections where all three valve leaflets were visible were stained with hematoxylin and eosin. The slides were then digitalized and plaque size measured by two independent observers (B.A.K. and L.D.A.) blinded to experimental condition using ImageJ software Version 1.50 (NIH, USA; http://rsb.info.nih.gov/ij/). One sample of normal diet antibiotics group had to be excluded from the analysis due to the lack of all three valve leaflets on the same slide.
2.4. Determination of carotid atherosclerosis (human)
Carotid ultrasonography examination was performed by the same trained physician (M.B.) in 30 subjects. Participants were examined in the supine position using the same ultrasound system (MyLab70 ESAOTE). Linear array probes with a transmission frequency of 5–10 MHz were used. Each common carotid artery, internal carotid artery, external carotid artery, and bulb was examined and recorded for the presence atherosclerotic plaques in the longitudinal and transverse planes. Plaque was defined as a focal structure encroaching into the arterial lumen and having a maximal thickness ≥1.5 mm, as measured from the media-adventitia interface to the intima-lumen [[27], [28], [29]].
2.5. Clinical biochemistry (mouse and human)
Human cohort: Plasma glucose concentrations were measured in duplicate by the glucose oxidase method using a Beckman glucose analyzer II (Beckman Instruments, Brea, CA, USA). Total plasma cholesterol was measured by an enzymatic, colorimetric method through the cholesterol esterase–cholesterol oxidase–peroxidase reaction (Cobas CHOL2, Roche, Basel, Switzerland). HDL (high-density lipoprotein) cholesterol was quantified by a homogeneous enzymatic colorimetric assay through the cholesterol esterase–cholesterol oxidase–peroxidase reaction (Cobas HDLC3, Roche, Basel, Switzerland). Total plasma triglycerides were measured by an enzymatic, colorimetric method with glycerol phosphate oxidase and peroxidase (Cobas TRIGL, Roche, Basel, Switzerland). LDL (low-density lipoprotein) cholesterol was calculated using the Friedewald formula. All other parameters were analyzed via routine laboratory testing. Glomerular filtration rate was calculated based on serum creatinine, age, sex, and ethnicity by CKD-EPI (Chronic Kidney Disease Epidemiology Collaboration) formula [30]. Body surface area was estimated according to Mosteller [31].
Mouse: Clinical biochemistry was measured on an automated analyzer (Keylab System, BPC Biosed s.r.l., Rome, Italy): total cholesterol (CHOLESTEROL F.X., KV1018VET), triglycerides (TRIGLYCERIDES, KV1052VET), ALT (GPT/ALT Liquid, KV1034VET) (all kits provided by BPC Biosed s.r.l., Rome, Italy).
2.6. Triglyceride content liver (mouse)
For analysis of triglyceride content of the liver, a triglyceride assay kit was used according to the manufacture's protocol (Abcam, Cambridge, UK).
2.7. Intraperitoneal glucose tolerance test (mouse)
Mice were fasted overnight (16 h) before the test. Glucose was injected intraperitoneally (2 g/kg body weight). Blood samples were taken from the tail vein before and 30, 60, 90 and 120 min after the injection of glucose. Blood glucose was determined with a OneTouch glucometer (Lifescan, Milpitas, CA, USA).
2.8. Flow cytometry analysis (mouse)
Two hundred μL of blood were collected retro-orbitally and anticoagulated with heparin. Red blood cells were lysed with eBioscienc RBC Lysis Buffer (Thermo Fisher Scientific, Waltham, MA, USA) and then stained with CD115-APC, CD11b-FITC and GR1-PerCP (all Milteny Biotec, Bergisch Gladbach, Germany). Samples were analyzed using a FACScalibur (BD Biosciences, San Jose, CA, USA) running BD Cellquest Pro and analyzed with Flow JO (TreeStar Inc., Ashland, OR, USA). Monocytes were defined as CD115 positive and CD11b positive. We used the mouse marker Gr1 (which binds Ly6C) to mark pro-inflammatory monocytes.
2.9. Metabolomics (mouse and human)
Data generation: After blood draw, the serum was immediately shock-frozen and stored at −80 °C until metabolomics analysis. Prior to the extraction process, samples were thawed and 100 μL of each serum sample was pipetted into randomly assigned wells of 2 mL 96-well plates. Human reference plasma samples were pipetted into a well of each 96-well plate. In addition, human pooled serum (Seralab) or pool of aliquots of mouse serum samples were pipetted into six wells of each 96-well plate for human or mouse serum sample sets, respectively. Those samples were used to assess process variability and to serve as technical replicates. One hundred μL of water were pipetted into six wells of each 96-well plate to serve as process blanks. Protein was precipitated and metabolites were extracted with methanol (475 μL for human serum samples or 500 μL for the mouse serum samples) containing four recovery standard compounds to monitor extraction efficiency. After centrifugation, the supernatant was split into four aliquots (100 μL each for human serum sample set) or five aliquots (80 μL each for mouse serum sample set) onto 96-well microplates. To minimize human error, liquid handling was performed on a Hamilton Microlab STAR robot (Hamilton, Reno, NV, USA). Sample extracts were dried on a TurboVap 96 (Zymark, Hopkinton, MA, USA).
For the human serum samples, two aliquots were used for reverse phase (RP)/Ultra Performance Liquid Chromatography-tandem Mass spectrometry (UPLC-MS/MS) analysis in positive and negative electrospray ionization (ESI) mode, while two aliquots were kept as reserves. Prior to UPLC-MS/MS in positive ESI mode, the samples were reconstituted with 50 μL of 0.1% formic acid (FA) and those analyzed in negative ESI mode with 50 μL of 6.5 mM ammonium bicarbonate, pH 8.0. Reconstitution solvents for both ionization modes further contained a cocktail of QC internal standards to monitor instrument performance and also to serve as retention reference markers. Analyses were performed on a linear ion trap LTQ XL mass spectrometer (Thermo Fisher Scientific GmbH, Dreieich, Germany) coupled with a Waters Acquity UPLC system (Waters GmbH, Eschborn, Germany). Two separate columns (2.1 × 100 mm Waters BEH C18 1.7 μm particle) were used for acidic (solvent A: 0.1% FA in water, solvent B: 0.1% FA in methanol) and for basic (A: 6.5 mM ammonium bicarbonate pH 8.0, B: 6.5 mM ammonium bicarbonate in 95% methanol) mobile phase conditions, optimized for positive and negative ESI modes, respectively. After injection of the sample extracts, the columns were developed in a gradient of 99.5% A to 98% B in 11 min run time at 350 μL/min flow rate. The eluent flow was directly connected to the ESI source of the LTQ XL mass spectrometer. Full scan mass spectra (80–1000 m/z) and data dependent MS/MS scans with dynamic exclusion were recorded in turns.
For the mouse serum samples, two aliquots of the extract supernatant were used for analysis by two separate (RP)/UPLC-MS/MS methods with positive ESI mode, one for analysis by (RP)/UPLC-MS/MS with negative ESI mode, and one for analysis by (HILIC)/UPLC-MS/MS with negative ESI mode, and one sample was reserved for backup. Prior to the UPLC-MS/MS runs the dried extract samples were reconstituted with 80 μL of solvents compatible to each of the four methods. The reconstitution solvents contained an internal cocktail of QC standards to monitor instrument performance and that also served as retention reference markers. Analyses were performed on a Waters Acquity UPLC system (Waters GmbH, Eschborn, Germany) coupled with a Thermo Scientific Q-Exactive high resolution accurate mass spectrometry interfaced with a heated electrospray ionization (HESI-II) source and Orbitrap mass analyzer operated at 35,000 mass resolution (Thermo Fisher Scientific GmbH, Dreieich, Germany). One of the aliquots was analyzed using acidic positive ion conditions, chromatographically optimized for more hydrophilic compounds. In this method, the extract was gradient eluted from a C18 column (Waters UPLC BEH C18-2.1 × 100 mm, 1.7 μm) using water, methanol, 0.05% perfluoropentanoic acid (PFPA), and 0.1% FA. The second aliquot was analyzed using acidic positive ion conditions, chromatographically optimized for more hydrophobic compounds. The extract was gradient eluted from the same aforementioned C18 column using water, methanol, acetonitrile, 0.05% PFPA and 0.01% FA, and was operated at an overall higher organic content. The third aliquot was analyzed using basic negative ion optimized conditions using a separate dedicated C18 column. The basic extracts were gradient eluted from the column using water, methanol, and 6.5 mM ammonium bicarbonate pH 8. The fourth aliquot was analyzed via negative ionization following elution from a HILIC column (Waters UPLC BEH Amide 2.1 × 150 mm, 1.7 μm) using a gradient consisting of water, acetonitrile, and 10 mM ammonium formate pH 10.8. The MS analysis alternated between MS and data-dependent MS scans using dynamic exclusion. The scan range varied slighted between methods but covered 70–1000 m/z.
Raw data were extracted, peak-identified and QC processed using the hardware and software of Metabolon (Durham, NC, USA). Compounds were identified using their retention index (RI), accurate mass (+/− 10 ppm) and MS/MS by comparison to library entries maintained by Metabolon (Durham, NC, USA). Lastly, compounds were manually checked and corrected, if necessary, by data analysts at Metabolon using proprietary visualization and interpretation software to confirm the consistency of peak identification among the various samples. Data were delivered with the area under the curve of the compounds' peak as the value at its original scale. For further analysis, compounds were filtered for deficient groups. A compound was removed if it was missing in more than half of the samples within a group and if it was present above this threshold in just one treatment group. Missing values were filled with half the minimum of each compound. For all following analyses, filtered and imputed metabolome data were log2 transformed.
2.10. 16S targeted metagenomic sequencing (mouse and human)
Data generation: The stool of human subjects was collected at visit 2. The cecal content from each mouse was collected directly postmortem. All samples were immediately stored at −80 °C in Biopure tubes (Eppendorf, Hamburg, Germany). DNA was extracted from the cecal content of mice and human stool using the QIAamp DNAStoolMini (Qiagen, Venlo, Netherlands). The bacterial 16S rDNA gene was sequenced (Vaiomer SAS, Labège, France) as previously described [32]. The quality and quantity of extracted nucleic acids were evaluated by gel electrophoresis (1% [w/w] agarose in Tris/borate/ethylenediaminetetraacetic acid 0.5 x) and NanoDrop 2000 UV spectrophotometer (Thermo Fisher Scientific, Waltham, MA, USA). The V3–V4 hyper-variable regions of the 16S rDNA gene were amplified from the DNA extracts during a first PCR step using universal 16S primers. The joint pair length was set to encompass 476 base pairs amplicon and include specificity for the 16S rDNA gene of 95% of the bacteria in the Ribosomal Database Project. For each sample, a sequencing library was generated by the addition of sequencing adapters and multiplexing indexes during a second PCR step as described previously [32,33]. The pool was denatured, diluted and loaded onto the Illumina MiSeq cartridge according to the manufacturer's instructions using MiSeq Reagent Kit v3 (2 × 300 bp Paired-End Reads; Illumina, San Diego, CA, USA). After demultiplexing of the barcoded Illumina paired reads, single read sequences were cleaned and paired for each sample independently into longer fragments. After quality-filtering and alignment against a 16S reference database, a clustering into operational taxonomic units (OTU) with a 97% identity threshold, and a taxonomic assignment were performed in order to determine community profiles. RDP Mothur formatted RDP training set v9 and SILVA v119 Database [34] were used for alignment and taxonomic assignment of sequences into OTUs. One sample of the mouse Western diet antibiotics group was excluded for further analysis as it was a clear outlier and possibly mislabeled during library generation. The generated OTU table was imported and further processed in R using the phyloseq package [35].
Filtering: OTUs that occurred less than six times per sample in the whole dataset were removed. After filtering, 727 OTUs remained in our mouse model and 529 OTUs in the human subjects. Normalization: OTU counts were normalized using the Variance Stabilization of the DESeq2 R package [35,36]. This gave similar results compared to a log2 transformation. Before the normalization, size factors were estimated using the geometric mean. The normalized values were then used for subsequent analysis.
2.11. Bioinformatics analyses
2.11.1. 16S rRNA-based metagenomic and metabolomic alpha diversity indices (mouse)
For the metabolomic diversity area, values were not imputed, filtered or log transformed. The area represents the integrated curve of ion intensity counts and is used analogous to the OTU counts. This violates a few assumptions of the alpha diversity indices: 1) areas are continuous values not ordinal like OTU counts, 2) a high area does not correspond to a high concentration of the metabolite, as metabolites are ionized at different efficiencies by the ESI in the Mass spectrometer, 3) ions without a library entry were excluded in this analysis, so the whole population might not be covered. Only the Observed index can in theory be accurately applied to these data, where the presence of different metabolites can be counted by checking if their area is above zero and not missing.
Generation of alpha diversities for metabolomic data was done using the same way as the metagenomic alpha diversity indices using the diversity function of the vegan package. The calculated diversity indices were:
Observed:
Is the number of metabolites with area >0 and not missing.
Shannon:
where pi is the proportional abundance of the area and R the number of metabolites.
2.11.2. Metabolite cluster generation via WGCNA (mouse)
As a method to reduce the dimensionality of the metabolomics data weighted correlation network analysis (WGCNA) was done using the R WGCNA package [37]. A soft-thresholding power of 4 was chosen based on the scale-free topology fit index-curve. Using this value the unsigned correlation network adjacency was calculated. The topological overlap matrix dissimilarity (TOM) of the adjacency matrix was then clustered using the ‘ward.D2’ method. The resulting tree was cut using a hybrid tree-cutting algorithm that was implemented in the cutreeDynamic function using a deepSplit of 3 and minClusterSize of 20. This resulted in 15 clusters, with no unassigned compounds. The names of the clusters were chosen arbitrarily as colors. The eigenvalues in each sample of the resulting clusters were used for further analyses.
2.11.3. Pathway enrichment analysis (mouse)
Compounds that were clustered into a WGCNA cluster were subjected to pathway enrichment analysis. The pathway information for each compound was reported by Metabolon (Durham, NC, USA). A one-sided Fisher test was used to determine if a pathway was enriched within a cluster. P-values were then adjusted using Benjamini-Hochberg method and a cut-off of P < 0.05 and q < 0.2 were chosen to determine if a pathway was significantly enriched.
2.11.4. PICRUSt/HUMAnN analysis (mouse)
HMP Unified Metabolic Analysis Network (HUMAnN) [38] is a tool to infer the metabolic pathway information from a microbial community. PICRUSt was used to predict the functional content of the microbiome using 16S rRNA genes [39]. PICRUSt requires annotations from the Green Genes database. OTUs were mapped accroding to Green Genes Database. Following the MiSEQ SOP ([40], visited https://www.mothur.org/wiki/MiSeq_SOP on 01/06/2016) Green Genes (gg_13_8_99) [41] was used as an input to classifiy.seqs of Mothur [40]. Clustering was then done using cluster.split with a cutoff of 0.15 at taxlevel 4 (Order). The sequences were then classified into OTUs via classify.otu at a cutoff of 0.03 and the biom file was generated using make.biom, with gg_13_8_99 as the reference taxonomy. The resulting biom file was then used for PICRUSt. PICRUSt (Version 1.0.0) was run on a Google Cloud Compute Engine instance. The OTUs in the Green genes biom file were normalized by their copy number. The normalized counts were then used to predict the metagenomes with PICRUSt. The resulting biom file from PICRUSt was converted to a tsv file and used for HUMAnN. HUMAnN2 (version 0.7.0) was run on a Google Cloud Compute Engine instance. Each sample was processed with humann2, the results were joined and reported as relative abundances.
2.11.5. Cross-omics correlation network analysis (mouse)
Correlations between aortic lesion size assessed by histology, WGCNA serum metabolite cluster eigenvalues and OTUs were calculated using the spearman correlation coefficient and the cor.test function of the R psych package. OTUs with less than four distinct observations were excluded, so 522 out of 727 OTUs remained. Significance was assessed using Benjamini & Hochberg P-value correction with a cutoff of 0.05. For visualization the 10 WGCNA serum metabolite clusters with the most significant OTU interactions were chosen. In addition to the correlation coefficients, -log10 P-values from the main-effects of the 2-way ANOVA using cluster eigenvalues against the treatment groups were added to show the effect of diet and antibiotics treatment in the clusters. To better visualize interactions only significant correlations between OTU and clusters and between clusters and aortic lesion were kept in the final network. The line thickness is based on the P-value or correlation coefficient, values between atherosclerotic lesion size and the clusters were scaled by the factor 10 to increase readability. The network was constructed and modified via igraph in R and exported to Cytoscape for final adjustment.
2.11.6. Differential analysis of OTU count (human)
OTU counts were transformed for DESeq2 with the phyloseq_to_deseq2 function of the phyloseq package. The DESeq2 package was then used to perform a Wald test to compare differences in OTUs between control versus atherosclerosis subjects [36]. In brief, a generalized linear model is fitted to give the log2 fold change using a negative binomial distribution with estimated sample-specific size factors and gene-specific dispersion parameters. The Wald test (nbiomWaldTest of DESeq2) was then used to test for significance of the log2 fold change.
2.11.7. LEfSe analysis (mouse)
Linear discriminant analysis Effect Size (LEfSe) [42] was used to identify microbial communities that interacted consistently with our treatments. LEfSe was performed using the LEfSe Docker container of the biobakery account (biobakery/LEfSe). The raw counts of the filtered OTU table was used as an input. LEfSE parameters were left at their defaults: alpha for ANOVA and Wilcoxon tests at 0.05, threshold of the logarithmic LDA score at 2.0.
2.11.8. Random Forest of metabolomics (mouse)
Random Forest is a supervised learning algorithm suitable for high dimensional data analysis. It uses an ensemble of classification trees, each of which is grown by random feature selection from a bootstrap sample at each branch. Class prediction is based on the majority vote of the ensemble. Random Forest analysis of murine serum metabolomics was performed using the randomForest R package [43] and visualized by MetaboAnalyst 4.0 [44].
2.11.9. Data availability
The mouse cecal 16S rRNA sequence data can be found under http://www.ncbi.nlm.nih.gov/bioproject/PRJNA595367.
The human stool 16S rRNA sequence data can be found under http://www.ncbi.nlm.nih.gov/bioproject/PRJNA595382.
2.12. Statistical analysis
Statistical approach of bioinformatic analyses is described in the appropriate sections above. Data were analyzed using GraphPad Prism, R and SPSS. The statistical analyses were carried out using unpaired two-tailed Student's t-test, one-way analysis of variance (ANOVA) or ANOVA with repeated measures when appropriate. Data are presented as means ± standard deviation (S.D.). A P-value < 0.05 was considered to be statistically significant.
2.13. Software used
-
-
R version 3.4.4 (2018-03-15)
-
-R packages
-
○Vegan 2.5–5
-
○WGCNA 1.63
-
○phyloseq 1.22.3
-
○DESeq2 1.18.1
-
○randomForest
-
○ggplot2 3.2.1
-
○psych 1.8.12
-
○
-
-
LefSE
-
-
Python 2.7.12
-
-
Google Cloud Compute Engine instances: n1-standard-1 with ubuntu 14.04
-
-
FastQC v0.11.3
-
-
Cytoscape 3.7.0
-
-
Mothur v1.34.4
-
-
FROGS v1.4.0
-
-
MetaboAnalyst 4.0
-
-
GraphPad Prism 7.0a
-
-
IBM SPSS Statistics Version 25
-
-
Image J Version 1.50
3. Results
3.1. Exacerbated atherosclerosis by antibiotics is linked to reduced microbial and metabolic diversity
To examine the effect of long-term antibiotics treatment on atherosclerosis development, mice were subjected to a mix of orally largely non-absorbable antibiotics (ABX) treated either with normal chow diet (ND) or a Western-type diet (WD) (Figure 1A). As expected, we observed that WD increased atherosclerosis development at aortic root level (Figure 1B). Independently of the diet, ABX further augmented atherosclerotic lesion size, while no interaction effect between diet and ABX was noticed (Figure 1B). Metabolic phenotyping including body weight, glucose tolerance, standard fasting lipid profile and liver analysis did not reveal a specific effect explaining antibiotics-induced atherosclerosis progression in our model (Supplementary Figure S2A-F). No impact of antibiotics on circulating M1/M2 macrophages differentiation was observed by flow cytometry (Supplementary Figure S2G,H).
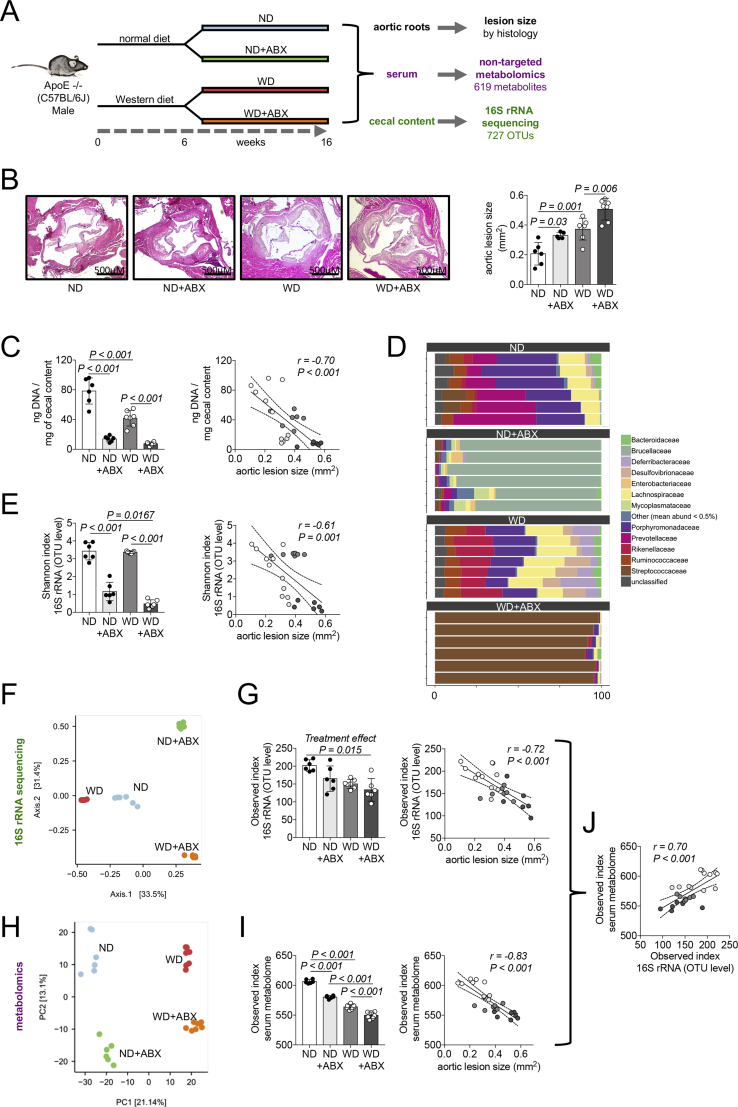
Figure 1.
Exacerbated atherosclerosis by antibiotics is linked to reduced microbial and metabolic diversity. (A) Flowchart showing the cross-omics approach to reveal gut microbiome related pathways underlying atherosclerosis progression after antibiotics treatment ND: normal diet, WD: Western diet, ABX: antibiotics treatment. (B) Micrographs of aortic roots stained with hematoxylin and eosin stain to evaluate extend of atherosclerosis (representative images) and quantification of aortic lesion size (data are the mean ± S.D., n = 5–7 per group). (C) DNA concentration of cecal content as indicator of gut bacteria quantity (data are the mean ± S.D, n = 6–7 per group) and Pearson correlation to aortic lesion size. (D) Relative abundance of cecal bacteria at family level. (E) Alpha diversity of cecal microbiome by Shannon index (data are the mean ± S.D., n = 6–7 per group) and Pearson correlation to aortic lesion size. (F) Principal coordinate analysis plot of 16S rRNA sequencing data of cecal content. (G) Alpha diversity of cecal microbiome by Observed index (data are the mean ± S.D., n = 6–7 per group) and Pearson correlation to aortic lesion size. (H) Principal component analysis of serum metabolomics. (I) Alpha diversity of serum metabolome measured by Observed index (data are the mean ± S.D., n = 6–7 per group) and Pearson correlation to aortic lesion size. (J) Correlation between serum metabolome alpha diversity and cecal microbiome alpha diversity (both measured by Observed index).
Given the major impact of ABX on the bacterial gut flora, we next evaluated the antibiotics effect on gut bacteria. Mice treated by ABX had increased weight of cecal content with a more liquid consistency (Supplementary Figure S3A), which has been described in antibiotics-treated and germ-free mice before [45]. Quantitative analysis of cecal content DNA – as an indicator of bacterial count – exhibited large reductions of DNA by ABX in both diets (Figure 1C). We performed 16S ribosomal rRNA targeted metagenomic sequencing of cecal content for deeper analysis of gut microbiome composition. Clear separation between all four groups was revealed by principal coordinate analysis (Figure 1F) as well as low variance within the different groups identified, for example, by relative abundance on family level (Figure 1D). To obtain a picture of gut bacterial diversity, we estimated alpha diversity by different indices, which revealed loss of diversity by antibiotics treatment (Figure 1E,G). Both the loss of bacterial quantity and microbial alpha diversity in the cecum had a strong negative correlation to atherosclerotic lesion size (Figure 1C,E,G).
Linear discriminant analysis (LDA) Effect Size (LEfSe) algorithm [42,46] (to gain deeper insights into the gut microbiome differences between the groups taking account of the complex taxonomic levels) revealed that ND untreated group was mainly characterized by Porphyromonadaceae and Prevotellaceae of class Bacteroidetes, as well as Ruminococcaceae and Lachnospiraceae, members of class Clostridia. All of the latter but Prevotellaceae were also among the top bacteria in untreated WD mice (Supplementary Figure S3B and Supplementary Table S1). LEfSe analysis of antibiotics-treated groups exposed a highly dysbiotic microbiome including members of Actinobacteria, Bacilli, Tenericutes, Alpha-, Beta- and Gammaproteobacteria as predominant bacterial classes. Ruminococcaceae as well as some members of Bacteroidetes had significant LDA scores in ND-ABX, but not in the WD-ABX group (Supplementary Figure S3B and Supplementary Table S1), therefore suggesting a potential protective effect regarding dysbiosis of normal compared to Western diet in antibiotics-treated animals.
We further aimed to evaluate the metabolic impact of reduced microbial diversity by antibiotics. The blood metabolome has been used to predict gut microbiota diversity before [47]. Therefore we performed non-targeted serum metabolomics in the mice (Supplementary Table S2). Principle component analysis as well as Random Forest classification of serum metabolomics (Figure 1G and Supplementary Figure S4) showed a distinct separation of all groups indicating a strong effect of diet on the metabolome, but also of antibiotic treatment. Next, we investigated the alpha diversity of the metabolome by calculation of Observed index, the only applicable alpha diversity index for metabolomics data. We found a clear reduction of metabolome alpha diversity by diet as well as antibiotics treatment analogous to the decrease of microbial diversity (Figure 1G,I). Gut microbiota and serum metabolome alpha diversity presented a strong correlation to atherosclerotic lesion size (Figure 1E,G,I) as well as to each other (Figure 1J).
3.2. A distinct metabolic signature links gut flora metabolism to atherosclerosis
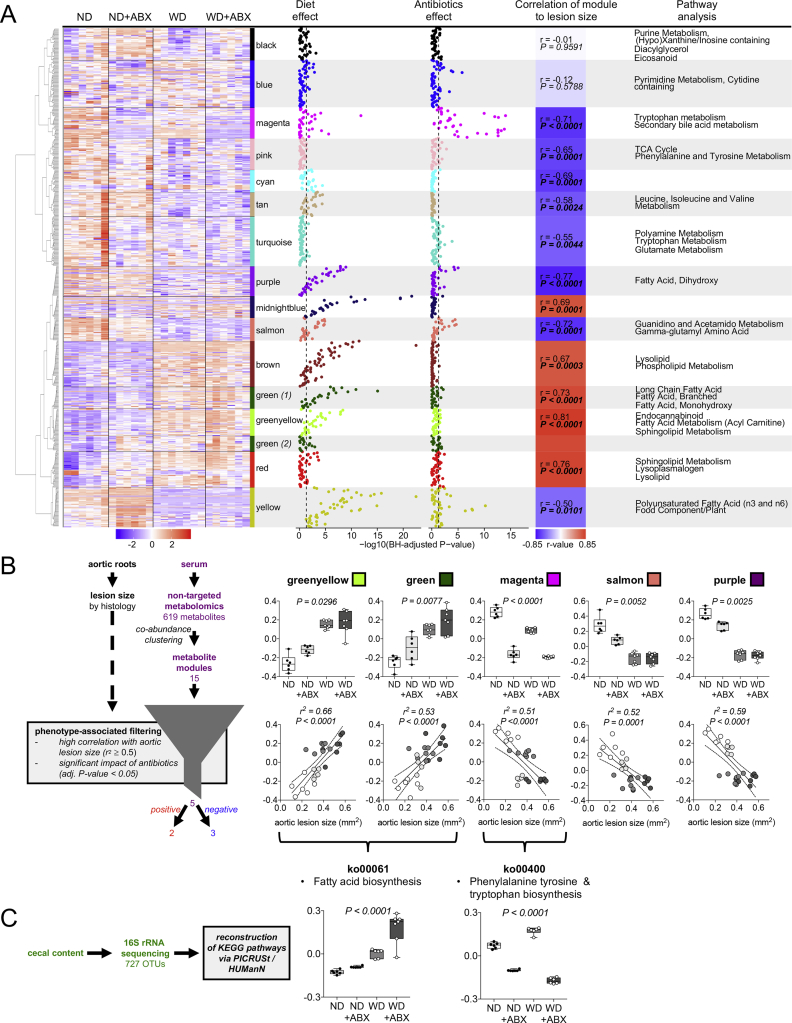
We then asked which particular metabolic changes underlie antibiotics-induced atherosclerosis exacerbation. To identify specific metabolic pathways that explain atherosclerosis progression by antibiotics, co-abundance clustering by weighted correlation network analysis (WGCNA) of metabolites was performed [24,48]. A total of 619 chemically-identified metabolites were clustered and a set of 15 metabolite clusters could be established (Figure 2A). Cluster names were chosen arbitrarily as colors. Significant overrepresented pathways in the clusters were identified (Figure 2A). Manhattan plots of single metabolites of each cluster as well as 2-way ANOVA analysis of eigenvalues of the metabolite clusters allowed us to identify clusters that were impacted by diet, antibiotics or both (Figure 2A and Supplementary Table S3). Diet had a significant influence on all metabolite clusters shown by the 2-way ANOVA analysis with the greatest impact on clusters related to lipid metabolism (Figure 2A and Supplementary Table S3). Analysis of antibiotics treatment effect revealed an impact on 10 of the 15 clusters with the strongest effect on metabolite cluster magenta (tryptophan metabolism and secondary bile acid metabolism), as well as blue (pyrimidine metabolism, cytidine containing) and yellow (polyunsaturated fatty acids and food component/plant) (Figure 2A and Supplementary Table S3). To identify pathways that were modulated by antibiotics and associated to atherosclerosis, we subjected the metabolite clusters to phenotype-associated filtering [48]. To discover strong candidates that may modulate atherosclerosis, only metabolite clusters with a high correlation to lesion size (r2 ≥ 0.5; Benjamini-Hochberg-adjusted P < 0.05) were selected as well as those with a significant impact of antibiotics as shown by 2-way ANOVA (Benjamini-Hochberg-adjusted P < 0.05) (Figure 2B and Supplementary Table S3). The algorithm identified five metabolite clusters matching these criteria: two with positive correlation (greenyellow; green) and three with negative correlation to aortic lesion size (magenta; salmon; purple). Cluster magenta, previously identified as the cluster with the highest impact of antibiotics, was enriched by tryptophan metabolism and secondary bile acid metabolism (Figure 2A). Top metabolites of magenta particularly belonged to gut flora-derived tryptophan derivatives, which was exposed by their chemical indole-structure (Supplementary Figure S5 and Supplementary Table S2) [2]. Tryptophan degradation products by human enzymes, kynurenate and kynurenine, were not among the metabolites in cluster magenta and were not altered by antibiotics treatment (Supplementary Table S2).
Figure 2.
A distinct metabolic signature links gut flora metabolism to atherosclerosis. (A) Clustering of serum metabolites by weighted correlation network analysis (WGCNA) resulting in 15 metabolites clusters (names of the clusters were chosen arbitrarily as colors). Manhattan plots show impact of treatments (diet and antibiotics) on single metabolites of each metabolite cluster. The dashed lines indicate a Benjamini-Hochberg-adjusted P-value <0.05. Eigenvalues of clusters were used for Pearson correlation analysis to atherosclerotic lesion size assessed by histology. Pathway enrichment analysis was performed by a one sided Fisher test based on the pathway annotations. All n = 6–7 per group. Treatment effect of single metabolites by Benjamini-Hochberg-adjusted 2-way-ANOVA. Pearson correlation between WGCNA metabolite eigenvalues to lesion size (P-values Benjamini-Hochberg-adjusted). Pathway enrichment analysis by one sided Fisher test (P-values were adjusted using Benjamini-Hochberg method and a cut-off of q < 0.2 was chosen to determine if a pathway was significantly enriched). ND: normal diet, WD: Western diet, ABX: antibiotics treatment. (B) Phenotype-associated filtering of WGCNA metabolite clusters by significant impact of antibiotics treatment and high correlation to atherosclerotic lesion size revealed 5 metabolite clusters matching these criteria. Upper graphs: eigenvalues of clusters. Benjamini-Hochberg-adjusted P-values indicate impact of ABX by 2-way-ANOVA (Boxplots: Center line: median; box limits: 25-75th percentiles; whiskers: min. to max., n = 6–7 per group). Lower graphs: Pearson correlation of eigenvalues of clusters to aortic lesion size (n = 5–7 per group). All P-values were corrected for multiple testing using the Benjamini–Hochberg criterion. (C) Prediction of functional content of the cecal content microbiome using HMP Unified Metabolic Analysis Network (HUMAnN) and Phylogenetic Investigation of Communities by Reconstruction of Unobserved States (PICRUSt). Two KEGG pathways matching the metabolomics findings are shown. Benjamini-Hochberg-adjusted P-values indicate impact of ABX by 2-way-ANOVA (Boxplots: Center line: median; box limits: 25-75th percentiles; whiskers: min. to max., n = 6 – 7 per group).
Clusters salmon and purple were also influenced by antibiotics, but to a lesser extent. Enriched pathways were guanidino and acetamido metabolism/gamma-glutamyl amino acids (cluster salmon) and dihydroxy fatty acids (cluster purple) (Figure 2A, Supplementary Figure S5 and Supplementary Table S2). Pathways significantly enriched in cluster greenyellow and green were particularly associated to lipid metabolism (Figure 2A, Supplementary Figure S5 and Supplementary Table S2).
To validate that the observed metabolic changes by antibiotics in our model are based on modulation of gut microbiota, we analyzed bacterial KEGG gene pathways in the cecal content. Given the limitation that full sequencing data were not available, we reconstructed the microbial metabolic KEGG pathways of 16S rRNA sequencing data via PICRUSt/HUMAnN [24,46,49]. We found that KEGG pathway ko00061 fatty acid synthesis fit our findings of metabolite cluster green and greenyellow, whereas ko00400 phenylalanine, tyrosine and tryptophan biosynthesis matched our metabolomics findings of cluster magenta, thus providing evidence for a direct link between microbial functional capacity and the serum metabolome (Figure 2C and Supplementary Table S4).
3.3. Atherosclerosis-linked serum metabolome is associated to reduction of certain Bacteroidetes and Clostridia in the gut
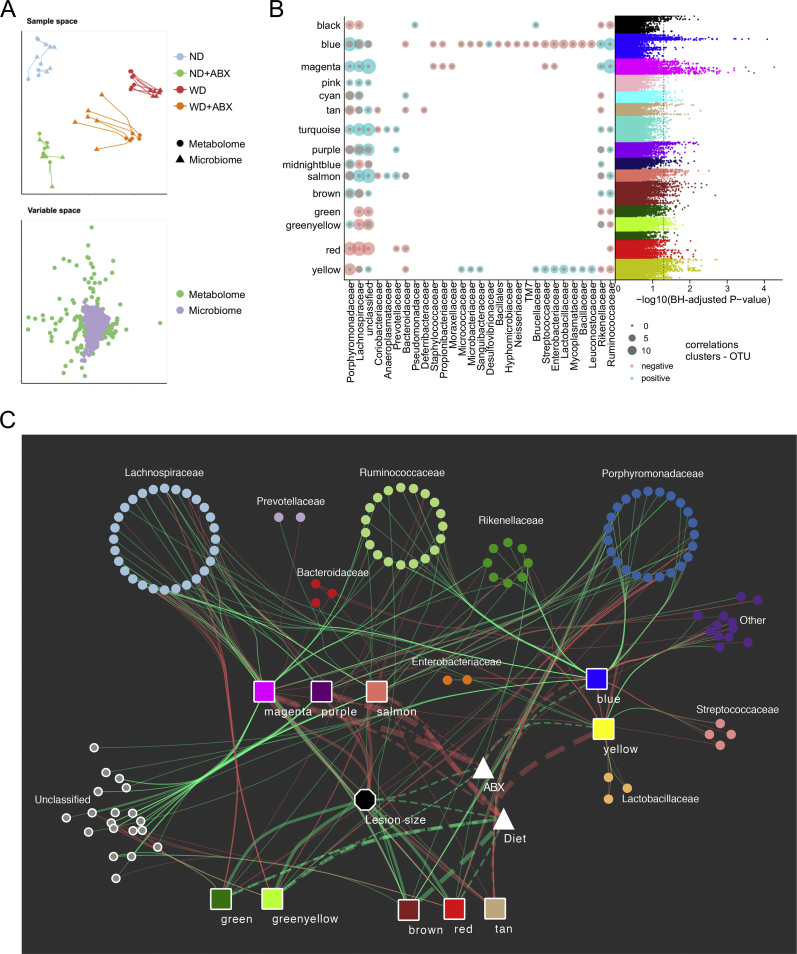
To connect specific changes in the gut microbiome to atherosclerosis via serum metabolome, we created an integrative cross-omics model including serum metabolite clusters, bacterial OTUs and atherosclerotic lesion size (Figure 3A–C). Visual inspection and Coinertia analysis with Monte–Carlo test (P = 0.01) confirmed a general overlap of the datasets (Figure 3A). The visualized integrative cross-omics model included the 10 clusters with the most OTU significant interactions and additionally atherosclerotic lesion size by histology as phenotype and type of treatment (Figure 3C).
Figure 3.
Atherosclerosis-linked serum metabolome is associated to reduction of certain Bacteroidetes and Clostridia in the gut. (A) Sample and variable space between serum metabolomics and cecal 16S rRNA sequencing data sets showing a good overlap of both data sets. (B) Number of significant positive (blue)/negative (red) Spearman correlations (Benjamini-Hochberg adjusted p-value < 0.05) between log transformed metabolite areas and OTU counts, grouped by WGCNA cluster assignment (y-axis) and OTU family (x-axis). To the right: Manhattan plot showing Benjamini-Hochberg adjusted P-values of Spearman correlations between serum metabolites assigned to WGCNA metabolite clusters and cecal OTU counts (n = 6–7 per group). The dashed line indicates an adjusted P-value <0.05. (C) Integrative cross-omics analysis including aortic lesion size by histology as phenotype, OTUs with more than four distinct observations (522/727), aortic lesion size and cluster eigenvalues were analyzed using Spearman correlation. P-values were adjusted using Benjamini-Hochberg and significance was assessed at adjusted P-value < 0.05. The 10 metabolite clusters with the most OTU significant interactions were kept. Only OTU – cluster and cluster – aortic lesion interactions are displayed. -log10 P-values from the main-effect ANOVA using cluster eigenvalues against the treatment groups were added to show the effect of diet and antibiotics in the clusters. The line thickness is based on the -log10 P-value of the ANOVA or the correlation coefficient, values between aortic lesion size and the clusters were scaled by the factor 10 to increase readability. Solid lines: Spearman correlation; dashed lines: -log10 P-value of ANOVA; Triangle: main effect of ANOVA; Octagon: phenotype; squares: WGNCA metabolite clusters; dots: OTUs; red lines: negative correlation/main effect; green lines: positive correlation/main effect.
Metabolite cluster magenta, which was previously identified as the cluster with highest impact of antibiotics treatment, also revealed the highest number of correlations between bacterial OTUs and metabolites of the cluster shown by Manhattan plot (Figure 3B). Our cross-omics analysis discovered that cluster magenta had positive correlations to certain Clostridia (Lachnospiraceae, Ruminococcaceae) and Bacteroidetes (Porphyromonadaceae, Rikenellaceae) (Figure 3B,C). Streptococcacae, Staphylococcacae, Propionibactericaeae and Enterobacteriaceae OTUs showed a weak negative correlation to the cluster. However, magenta was mainly characterized by gut-derived metabolites that have been associated to gut alpha diversity before [47]. Therefore we hypothesized that the loss of certain bacterial species rather than the increase of others by antibiotics is responsible for a reduction of metabolites of the cluster magenta. Blue, another cluster highly impacted by antibiotics, also exhibited a high number of correlations between its metabolites and OTUs (Figure 3B,C). However, no relationship to atherosclerosis was observed.
Analysis of the other metabolic clusters related to lesion size disclosed less diverse correlations to bacterial OTUs (Figure 3B,C). Both metabolite clusters, which were positively associated with atherosclerosis (green; greenyellow) and mainly characterized by lipid metabolism, showed negative correlations, particularly to Lachnospiraceae (Figure 3B,C). Clusters purple and salmon exhibited similar correlations to OTU as magenta, but additionally to Prevotellaceae of class Bacteroidetes.
In summary, our model proposes that mainly three bacterial families contributed to an altered serum metabolome associated to antibiotics-induced atherosclerosis. In detail, these were two members of class Clostridia, Lachnospiraceae and Ruminococcaceae, as well as a member of Bacteroidetes, Porphyromonadaceae.
3.4. Tryptophan supplementation reverses in part antibiotics-induced atherosclerosis
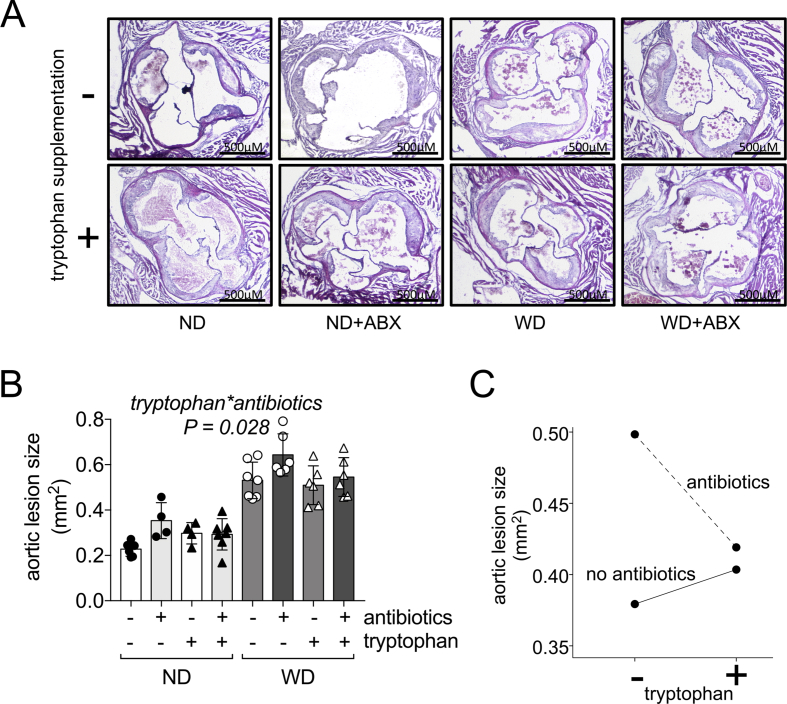
Phenotype-associated filtering revealed five different clusters that may drive antibiotics-induced atherosclerosis progression (Figure 2B). Magenta was identified as the metabolite cluster with the strongest impact of antibiotics and majorly linked to changes in gut bacteria. The top ranked metabolites in this cluster belonged to tryptophan metabolism (Supplementary Figure S5), while we observed that microbial tryptophan biosynthesis was reduced by antibiotics (Figure 2C). Therefore we asked if reduced microbial tryptophan biosynthesis by antibiotics contributes to atherosclerosis and whether supplementation of tryptophan could restore the atherosclerotic phenotype induced by antibiotics. To test this hypothesis, a separate set of ApoE −/− mice was treated by the same protocol as before either on ND or WD receiving the same antibiotics cocktail. Groups were further divided into 1% tryptophan supplementation in their diet and no supplementation during the time of antibiotics treatment. Histology of aortic roots was assessed after 16 weeks of the experiment in analogy with the previous mouse experiment. In a 3-way ANOVA model including antibiotics treatment, with diet and tryptophan supplementation as independent variables and lesion size as a dependent variable, we observed in accordance to our previous experiment that antibiotics treatment (P = 0.005) and diet (P < 0.001) had independent effects on the degree of atherosclerosis (Figure 4A,B). Tryptophan supplementation alone did not have any impact on atherosclerosis (P = 0.231). By contrast, our model revealed a significant interaction effect of antibiotics treatment and tryptophan supplementation on the reduction of aortic lesion size (P = 0.028) (Figure 4A–C). Hence, taking in account that more than a single pathway is involved in atherosclerosis progression by antibiotics, this model provides evidence that tryptophan supplementation is in part able to restore the antibiotics-induced phenotype.
Figure 4.
Tryptophan supplementation reverses in part antibiotics-induced atherosclerosis. (A) Micrographs of aortic roots stained with hematoxylin and eosin stain to evaluate extend of atherosclerosis of mice with or without supplementation of tryptophan (representative images). ND: normal diet, WD: Western diet, ABX: antibiotics treatment. (B) Quantification of aortic lesion size (data are the mean ± S.D., n = 4–7 per group). The displayed P-value represents the interaction between tryptophan and antibiotics using a 3-way ANOVA model including diet, antibiotics and tryptophan supplementation as independent variables and aortic lesion size as dependent variable. (C) Estimated marginal means between antibiotics- and tryptophan-treated mice.
3.5. Antibiotics-linked atherogenic metabolic pathways and fecal bacteria are altered in patients with carotid artery disease
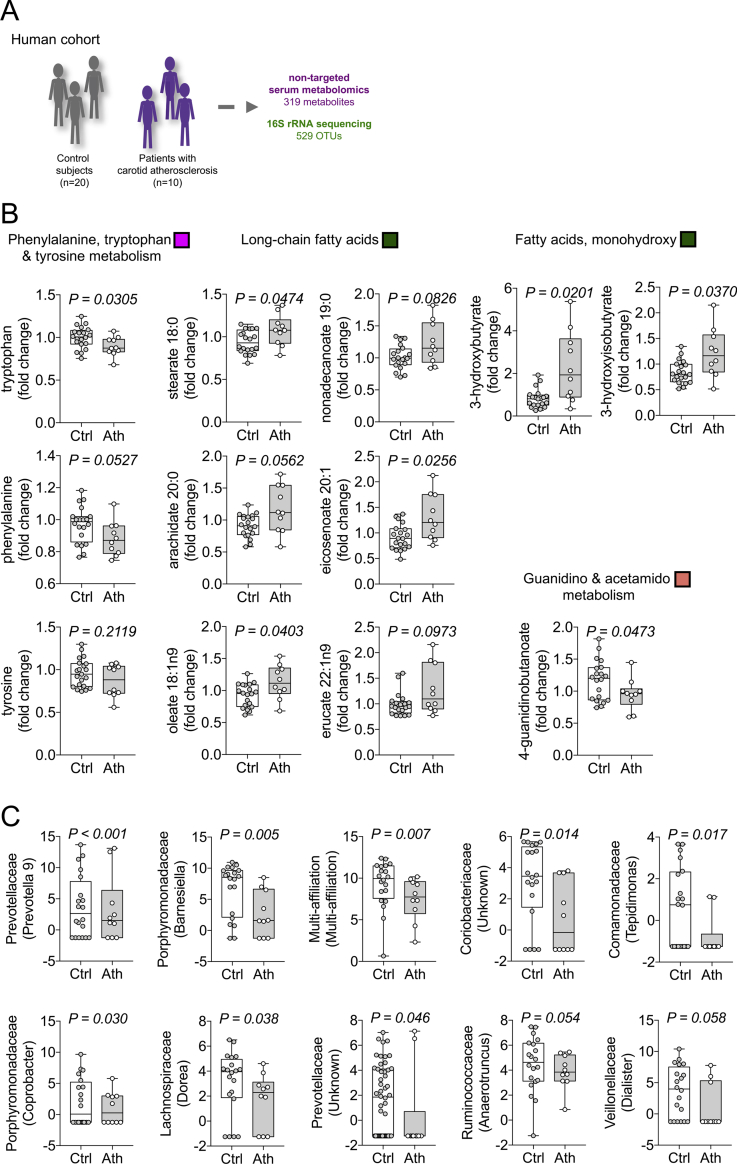
To endorse that pro-atherogenic metabolic pathways modulated by antibiotics in our mouse model are also linked to atherosclerosis in humans, we performed the same metabolomics approach in a cohort of 10 patients with carotid atherosclerosis diagnosed by duplex sonography and 20 control subjects (Figure 5A) with no significant difference in baseline characteristics including age, gender, body mass index and renal function, blood pressure, diabetes-related parameters, medication, and standard serum lipid profile (Table 1). Our metabolomics approach detected 319 chemically identified metabolites (Supplementary Table S5). In patients with carotid atherosclerosis, our analyses revealed a similar metabolic phenotype as the one induced by antibiotics in our mouse model. Particularly, patients with atherosclerosis exhibited lower levels of tryptophan (Figure 5B).
Figure 5.
Antibiotics-linked atherogenic metabolic pathways and fecal bacteria are altered in patients with carotid artery disease. (A) Serum and feces of patients with carotid atherosclerosis (Ath, n = 10) diagnosed by duplex sonography and control subjects (Ctrl, n = 20) were analyzed via metabolomics and 16S rRNA targeted sequencing. (B) 319 chemically-identified metabolites were compared to the previously identified atherosclerosis-linked pathways impacted by antibiotics treatment in the mouse mode (Boxplots: Center line: median; box limits: 25-75th percentiles; whiskers: min. to max.). Colors indicate corresponding WGCNA metabolite cluster in the mouse model. (C) Top 10 operational taxonomic units (OTU) of fecal 16S analysis differing between control and atherosclerosis group revealed by Wald test (Data are variance stabilized OTU counts. Boxplots: Center line: median; box limits: 25-75th percentiles; whiskers: min. to max.)
We also observed metabolic changes in correspondence to other pathways identified in our mouse model. Long-chain fatty acids were increased in the atherosclerosis group (Figure 5B) comparable to the findings in mouse WGCNA metabolite cluster green and bacterial KEGG pathway ko00061 (Figure 2A,C). We found monohydroxy fatty acids also to be modulated in our patients cohort (Figure 5B) as well as mouse WGCNA metabolite cluster green (Figure 2A,C). In addition, 4-guanidinobutanoate, a metabolite of guanidino metabolism, was found to be reduced in patients with carotid atherosclerosis (Figure 5B) and among the top metabolites of WGCNA cluster salmon in our mouse model (Figure 2A and Supplementary Figure S5).
Analysis of fecal 16S sequencing data in our human cohort revealed only a trend towards lower alpha diversity in the diseased group (Supplementary Figure S6). However, among the top 10 bacteria differing between both groups shown by Wald test [50], we found a reduction of the members of four bacterial families, which have been diminished by antibiotics and linked to increased atherosclerosis: Lachnospiraceae, Ruminococcaceae, Porphyromonadaceae and Prevotellaceae (Figure 5C and Supplementary Table S6).
4. Discussion
The relationship between gut microbiota and host metabolism remains largely elusive, but recent breakthrough data suggest a significant role in the progression of cardiovascular disease [1,51]. A major emerging concept is the multi-faceted role of the microbiome playing either a positive or negative role, depending on the circumstances. Gut-derived TMAO has been identified as a biomarker of cardiovascular mortality [6,7] and microbiota-associated pathways have been linked to augmented arterial thrombosis [[8], [9], [10]]. On the other hand, the use of antibiotics has long-term effects on bacterial richness [20] and negatively affects metabolic parameters such as insulin sensitivity and obesity [21,52]. Recently, exposure to antibiotics has been identified as novel risk factor for cardiovascular events [22].
To explore gut microbiota-related metabolic pathways connected to atherosclerosis progression, we integrated data on phenotype, gut microbiome and serum metabolome in a murine model of atherosclerosis challenged with oral antibiotics. Our model points towards exacerbated atherosclerosis by antibiotics-induced dysbiosis, particularly due to a decrease of tryptophan metabolites, but also other pathways, mainly related to lipid metabolism. These metabolic alterations were largely facilitated by loss of few bacterial families only, notably Ruminococcaceae and Lachnospiraceae (both Clostridia) as well as Porphyromonadaceae (Bacteroidetes). Functional reconstruction of targeted 16S rRNA metagenomic sequencing data confirmed reduced tryptophan and increased fatty acid biosynthesis in the gut microbiome, thus indicating a direct impact on the serum pool of these metabolites. In patients with carotid atherosclerosis compared to subjects with a similar cardiovascular risk profile, our serum metabolome analyses revealed a similar metabolic phenotype as the one induced by antibiotics in our mouse model. This included reduced tryptophan and increased long-chain fatty acids. Clustering of serum metabolites by WGCNA and phenotype-associated filtering identified a metabolite cluster, magenta, mainly characterized by tryptophan metabolism, that was highly affected by antibiotics treatment and inversely correlated to atherosclerotic lesion size. Based on these findings and reduced microbial tryptophan biosynthesis by antibiotics treatment, we showed that tryptophan supplementation was in part able to restore the antibiotics-induced atherosclerosis phenotype. Our functional mouse experiment therefore suggests a previously unknown protective effect of tryptophan supplementation in antibiotics-induced atherosclerosis progression.
Our work complements other mouse studies investigating the influence of gut microbiota on atherosclerosis. While our study revealed a diet-independent effect of antibiotics on atherosclerosis development with no effect on the fasting standard lipid profile, Lindskog Jonsson et al. as well as Stepankova et al. revealed increased atherosclerosis accompanied by elevations in serum cholesterol in germ-free compared to conventionally-raised ApoE−/− mice [53,54]. This effect was observed only in a normal chow diet, but not in a Western diet. Similar observations were made in the low-density lipoprotein receptor knockout (LDLR−/−) mouse model [10]. In contrast, others studies using ApoE−/− mice found either no effect of germ-free conditions on atherosclerosis (on Western diet) [55], or even reduced lesion size (on a chow diet) [56]. Neither study detected alterations in serum cholesterol. Studies with antibiotics also revealed conflicting results. Rune et al. observed reduced plaque size and cholesterol levels by ampicillin treatment in Western diet-treated ApoE−/− mice [57], whereas Gosh et al. found reduced lesion and cholesterol by combination of neomycin and polymyxin B in LDLR−/− mice [58]. In contrast, another study found an increase of serum cholesterol by broad-spectrum antibiotics in both knockout models, but did not evaluate atherosclerotic lesion size [59]. Using the same antibiotics protocol as in our study (ampicillin, metronidazole, neomycin and vancomycin), Wang and colleagues demonstrated reduced lesion size by antibiotics in ApoE−/− mice fed with a choline-rich chow diet, presumably by reducing gut microbiota-derived TMAO. However, in mice receiving a chow diet without choline supplementation, antibiotics treatment resulted in a trend towards increased lesion size, which was similar to our study [4]. In contrast to the findings of Wang, Lindskog Jonsson et al. found no effect of choline supplementation on atherosclerotic lesion size in ApoE−/− mice on a normal chow diet, although TMAO levels were significantly elevated by choline treatment in conventionally-raised mice. As expected, we found markedly reduced levels of TMAO by antibiotics (Supplementary Figure S5), thus suggesting no contribution of TMAO to the phenotype in our study.
Various factors may explain these potentially conflicting results. Although most studies have been performed on the same background (C57BL/6), different breeding/housing conditions, age, diet and feeding regime as well as experimental protocol might lead to substantial differences in the gut microbiome. Those experimental difference might impact the microbiota composition of conventionally-raised/non-antibiotics-treated control groups in different studies [45,60]. Furthermore, broad-spectrum antibiotics effectively deplete gut microbiota, but do not establish germ-free conditions. Overgrowth of antibiotic-resistant species rather than loss of commensal bacteria by antibiotics might explain differences between antibiotics-treated and germ-free mice [61]. In our study, antibiotic treatment resulted in a remarkably increase of Brucellaceae relative abundance in normal diet and Streptococcacae in Western diet (Figure 1D). Our functional KEGG gene reconstruction revealed increased capacity of fatty acid biosynthesis (Figure 2C). Thus, bacterial overgrowth of species with increased capacity of fatty acid biosynthesis might contribute to increased plaque size by antibiotics in our study.
However, our data are in line with human studies showing that dysbiosis, such as through antibiotics treatment, is associated with an unfavorable metabolic phenotype [18,52] as well as cardiovascular outcome [22,62]. Lately, the same antibiotics cocktail used in our study induced detrimental effects in a mouse model of myocardial infarction [63].
As expected, antibiotics had a major impact on gut bacteria leading to major dysbiosis. This finding was highly correlated with the extent of atherosclerosis. There is increasing evidence that dysbiotic changes in the gut microbiome are linked to cardiovascular disease [16,64]. In our study, we observed a loss of bacterial diversity and an increase of some Proteobacteria in antibiotics-treated mice. In a cohort of almost 4000 patients, blood measurements of 16S markers for common bacteria phyla correlated negatively and 16S markers for Proteobacteria positively with the onset of cardiovascular events [62]. In another study including 617 middle-aged women, pulse wave velocity (PWV) as a marker for arterial stiffness was negatively correlated with intestinal alpha diversity. The main bacterial family with a negative association to PWV after adjusting for covariates was Ruminococcaceae [14]. Intriguingly, loss of Ruminococcaceae was one of the main drivers of metabolic changes linked to atherosclerosis in our mouse model and among the top 10 bacteria differing between patients with and without atherosclerosis in our cohort.
Antibiotics treatment highly affects the host serum metabolome and has been linked to metabolic perturbations in humans and mice [52,65,66]. Here, however, we show first that the loss of gut microbiota diversity by antibiotics is directly associated with the loss of metabolic diversity in the host. Previous studies found a link between diminished short-chain fatty acids by antibiotics and the cardiometabolic phenotype [63,65]. In accordance, others suggested a protective role of gut-derived short-chain fatty acids, particularly butyrate, on atherosclerosis [[67], [68], [69]]. We observed a trend towards reduction of short-chain fatty acid metabolites by antibiotics in mice on normal diet, but not on Western diet (Supplementary Figure S7). The low fiber content of the Western diet might account for these diet-specific differences. We cannot exclude that diminished short-chain fatty acids by antibiotics contribute to increased atherosclerosis in normal diet-treated animals, but this finding does not explain the diet-independent effect of antibiotics on atherosclerosis in our study.
Our non-targeted cross-omics approach identified particularly tryptophan metabolism, as well as alteration in lipid metabolism including long-chain fatty acids as main, diet-independent, facilitators of antibiotics-induced atherosclerosis progression. Our additional data in humans are limited in power and do not allow a full integrative analysis. Considering this limitation, our study suggests that reduced tryptophan and increased long-chain fatty acid metabolism are respectively linked to atherosclerosis in humans as well. These results are in agreement with previous studies. Low levels of serum tryptophan have been shown to predict cardiovascular mortality in patients with coronary heart disease [70], while diminished tryptophan and microbiota-linked tryptophan derivatives levels have been linked to advanced atherosclerosis in study by Cason et al. [71]. In this study, tryptophan and indolepropionate, one of the top 10 metabolites in the cluster magenta in our study, were further negatively correlated with ankle-brachial index as a surrogate for overall atherosclerotic disease burden [71]. In a combined host-gut bacteria model based on available genome annotation information on human intestinal bacteria, tryptophan metabolites have been predicted as major bioactive microbiota metabolites [2]. Our results highlight a link between atherosclerosis and reduced bacterial tryptophan synthesis accompanied by diminished microbiota tryptophan serum metabolites. Gut-derived tryptophan derivatives have been recognized as essential microbial interspecies as well interkingdom signaling molecules to communicate with the host [72]. In end-stage chronic kidney disease, some tryptophan derivatives such as indoxyl sulfate and indoleacetate reach high concentrations due to a lack of renal clearance and are associated with cardiovascular mortality [73,74]. However, our data contribute to the concept, in which an equilibrium of tryptophan derivatives from microbiota is essential to maintain host integrity. Lately, it has be shown that exacerbation of blood pressure by high salt intake was facilitated by changes in the gut microbiome linked to the reduction of gut-derived tryptophan metabolites affecting Th17 response [75].
Molecular targets of indoles that may affect atherosclerosis remain to be defined. Several microbial tryptophan catabolites have been identified as modulators of aryl hydrocarbon receptor (Ahr) [76,77]. Ahr expression has been linked to atherosclerosis in humans and mice [78,79]. However, the effects of Ahr activation on the vasculature remain ambiguous and depend on the type of agonist, experimental model as well as species [76,77,80]. In an ApoE−/− mouse model, activation of Ahr increased atherosclerosis [81], whereas loss of Ahr was protective on vascular aging [82]. On the other side, activation of Ahr by indoles has been shown to balance intestinal mucosal reactivity via interleukin-22 [76]. In a complementary study, Fatkhullina et al. were able to demonstrate that interleukin-22 response reduced diet-induced atherosclerosis in LDL−/− mice by repressing pro-atherogenic gut bacteria [83]. In summary, these studies indicate a protective effect of certain indoles on atherosclerosis via interleukin-22 upon activation of Ahr.
Similar conflicting results have been revealed for the xenobiotic sensor pregnane X receptor (PXR), which can be activated by gut microbiota-derived indolepropionate [84]. In a mouse model of colitis, a Clostridium strain mediated protective effects via indolepropionate production and consecutive PXR activation [84]. Administration of indolepropionate in a rat model of steatohepatitis was able to restore gut dysbiosis and reduce endotoxin leakage, a mechanism that has been linked to atherosclerosis before [85]. On the contrary, studies with ApoE−/− mice indicate a detrimental effect of PXR activation by a specific agonists, respectively a protective role of PXR knockout [86,87].
Together, these data suggest that xenobiotic receptors such as PXR and Ahr exhibit either positive or negative effects depending on the circumstances. Further, species differences in receptor activation must be taken in account [80,84]. Hence, data in mice should be interpreted with caution and might not be fully translatable to humans.
Our study has limitations. The scope of our study was to give insights in metabolic changes that may underlie atherosclerosis development by antibiotics via a multi-omics model. Although we could show that disturbed tryptophan metabolism is in part responsible for increased atherosclerosis by antibiotics, we were not able to provide a full mechanistic model due to the multifaceted impact of gut microbiota on the host metabolism. We propose a metabolic effect of antibiotics on atherosclerosis, but the impact of the immune system has been studied only to a minor extend. Future studies must integrate the cross-talk between metabolic alterations, immune response, molecular targets and atherosclerosis. In addition, other pathways that were correlated to atherosclerosis in our study should be validated in future works to obtain a complete picture. In fact, secondary bile acid metabolism was another pathway reduced in cluster magenta and negatively correlated to atherosclerotic plaque size. Secondary bile acids are able to modulate pro-inflammatory cytokine response as well as hepatic glucose and lipoprotein metabolism in part through activation of the Takeda G protein-coupled receptor 5 and farnesoid X receptor [88]. A direct link between secondary bile acid metabolism and atherosclerosis remains to be established, but a specific agonist for both bile acid receptors was able to reduce atherosclerotic burden in an ApoE−/− mouse model [89].
Further, our human study was of exploratory nature to investigate metabolic pathways linked to atherosclerosis beyond classical risk factors. These findings need to be validated in a larger cohort and any intervention strategies including antibiotics treatment must also be addressed in future studies.
5. Conclusion
In summary, our data show detrimental effects of antibiotics on atherosclerosis development independently of diet. Taking advantage of the negative impact of antibiotics on atherogenesis, we provide comprehensive data on the complex interaction between gut microbiome and host metabolism. Our study supports the evidence that dysbiotic changes, such as by antibiotics, are linked to metabolic diseases and increased cardiovascular risk. Therefore, restrictive use of antibiotics might not only be important with regards to antimicrobial resistance, but also to maintain gut microbiota integrity to prevent cardiometabolic diseases.
We advise others to interpret our data in the context of other mouse studies with various results. Our study contributes to the concept of a multi-faceted role of microbiota on cardiovascular disease, which depends on different circumstances such as diet, species, and experimental protocol.
Author contributions
Conceptualization, B.A.K. and M.F.; Methodology, B.A.K. and M.F.; Development or design of methodology, B.A.K. and M.F.; Software, M.H.; Formal Analysis, B.A.K. and M.H.; Investigation, B.A.K., L.D.A, M.B., O.A.P., G.Mo., G.Mi., A.A., J.A., R.S., C.G., M.M., R.B. and R.M.; Resources, B.A.K, M.H., and M.F.; Data Curation, B.A.K and M.H.; Writing – Original Draft, B.A.K and M.F.; Writing – Review & Editing, B.A.K., R.S., B.S., N.M. and M.F.; Visualization, B.A.K., M.H., and M.F.; Supervision, N.M., B.S., and M.F.; Project Administration, M.F.; Funding Acquisition, B.A.K, N.M. and M.F.
Acknowledgements
M.F. laboratory was in part funded by EU-FP7 FLORINASH [grant agreement ID: 241913], Ministry of University (MIUR) Progetti di Ricerca di Interesse Nazionale (PRIN) [protocol number 2015MPESJS_004 and 2017FM74HK], Fondazione Roma call for Non-Communicable Diseases NCD 2014, EU-FP7 EURHYTHDIA [grant agreement ID: 278397]. B.A.K. was supported by grants from the Deutsche Herzstiftung (DHS)[S/08/15], Deutsche Stiftung für Herzforschung (DSHF)[F-43-16] and RWTH Aachen University (START) and a PhD fellowship from University of Rome Tor Vergata; N.M. was supported by the German Research Foundation (DFG) [SFB TRR 219 (projects M−03 and M−05)].
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.molmet.2020.100976.
Conflict of interest
None declared.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
References
- 1.Brown J.M., Hazen S.L. Microbial modulation of cardiovascular disease. Nature Reviews Microbiology. 2018;16(3):171–181. doi: 10.1038/nrmicro.2017.149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sridharan G.V., Choi K., Klemashevich C., Wu C., Prabakaran D., Pan L.B. Prediction and quantification of bioactive microbiota metabolites in the mouse gut. Nature Communications. 2014;5:5492. doi: 10.1038/ncomms6492. [DOI] [PubMed] [Google Scholar]
- 3.Nicholson J.K., Holmes E., Kinross J., Burcelin R., Gibson G., Jia W. Host-gut microbiota metabolic interactions. Science. 2012;336(6086):1262–1267. doi: 10.1126/science.1223813. [DOI] [PubMed] [Google Scholar]
- 4.Wang Z., Klipfell E., Bennett B.J., Koeth R., Levison B.S., Dugar B. Gut flora metabolism of phosphatidylcholine promotes cardiovascular disease. Nature. 2011;472(7341):57–63. doi: 10.1038/nature09922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tang W.H.W., Wang Z., Kennedy D.J., Wu Y., Buffa J.A., Agatisa-Boyle B. Gut microbiota-dependent trimethylamine N-oxide (TMAO) pathway contributes to both development of renal insufficiency and mortality risk in chronic kidney disease. Circulation Research. 2015;116(3):448–455. doi: 10.1161/CIRCRESAHA.116.305360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Schuett K., Kleber M.E., Scharnagl H., Lorkowski S., März W., Niessner A. Trimethylamine-N-oxide and heart failure with reduced versus preserved ejection fraction. Journal of the American College of Cardiology. 2017;70(25):3202–3204. doi: 10.1016/j.jacc.2017.10.064. [DOI] [PubMed] [Google Scholar]
- 7.Koeth R.A., Wang Z., Levison B.S., Buffa J.A., Org E., Sheehy B.T. Intestinal microbiota metabolism of L-carnitine, a nutrient in red meat, promotes atherosclerosis. Nature Medicine. 2013;19(5):576–585. doi: 10.1038/nm.3145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhu W., Gregory J.C., Org E., Buffa J.A., Gupta N., Wang Z. Gut microbial metabolite TMAO enhances platelet hyperreactivity and thrombosis risk. Cell. 2016:1–14. doi: 10.1016/j.cell.2016.02.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jäckel S., Kiouptsi K., Lillich M., Hendrikx T., Khandagale A., Kollar B. Gut microbiota regulate hepatic von Willebrand factor synthesis and arterial thrombus formation via Toll-like receptor-2. Blood. 2017;130(4):542–553. doi: 10.1182/blood-2016-11-754416. [DOI] [PubMed] [Google Scholar]
- 10.Kiouptsi K., Jäckel S., Pontarollo G., Grill A., Kuijpers M.J.E., Wilms E. The microbiota promotes arterial thrombosis in low-density lipoprotein receptor-deficient mice. mBio. 2019;10(5) doi: 10.1128/mBio.02298-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kelly T.N., Bazzano L.A., Ajami N.J., He H., Zhao J., Petrosino J.F. Gut microbiome associates with lifetime cardiovascular disease risk profile among bogalusa heart study participants. Circulation Research. 2016;119(8):956–964. doi: 10.1161/CIRCRESAHA.116.309219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rothschild D., Weissbrod O., Barkan E., Kurilshikov A., Korem T., Zeevi D. Environment dominates over host genetics in shaping human gut microbiota. Nature. 2018;555(7695):210–215. doi: 10.1038/nature25973. [DOI] [PubMed] [Google Scholar]
- 13.Fu J., Bonder M.J., Cenit M.C., Tigchelaar E., Maatman A., Dekens J.A.M. The gut microbiome contributes to a substantial proportion of the variation in blood lipids. Circulation Research. 2015;117(9):817–824. doi: 10.1161/CIRCRESAHA.115.306807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Menni C., Lin C., Cecelja M., Mangino M., Matey-Hernandez M.L., Keehn L. Gut microbial diversity is associated with lower arterial stiffness in women. European Heart Journal. 2018;39(25):2390a–2397a. doi: 10.1093/eurheartj/ehy226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hoyles L., Fernández-Real J.M., Federici M., Serino M., Abbott J., Charpentier J. Molecular phenomics and metagenomics of hepatic steatosis in non-diabetic obese women. Nature Medicine. 2018;24(7):1070–1080. doi: 10.1038/s41591-018-0061-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Karlsson F.H., Fåk F., Nookaew I., Tremaroli V., Fagerberg B., Petranovic D. Symptomatic atherosclerosis is associated with an altered gut metagenome. Nature Communications. 2012;3:1245. doi: 10.1038/ncomms2266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tang W.H.W., Li D.Y., Hazen S.L. Dietary metabolism, the gut microbiome, and heart failure. Nature Reviews Cardiology. 2019;16(3):137–154. doi: 10.1038/s41569-018-0108-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Le Chatelier E., Nielsen T., Qin J., Prifti E., Hildebrand F., Falony G. Richness of human gut microbiome correlates with metabolic markers. Nature. 2013;500(7464):541–546. doi: 10.1038/nature12506. [DOI] [PubMed] [Google Scholar]
- 19.Perez-Cobas A.E., Gosalbes M.J., Friedrichs A., Knecht H., Artacho A., Eismann K. Gut microbiota disturbance during antibiotic therapy: a multi-omic approach. Gut. 2012;62(11):1–11. doi: 10.1136/gutjnl-2012-303184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Korpela K., Salonen A., Virta L.J., Kekkonen R.A., Forslund K., Bork P. Intestinal microbiome is related to lifetime antibiotic use in Finnish pre-school children. Nature Communications. 2016;7:1–8. doi: 10.1038/ncomms10410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Saari A., Virta L.J., Sankilampi U., Dunkel L., Saxen H. Antibiotic exposure in infancy and risk of being overweight in the first 24 Months of life. Pediatrics. 2015;135(4):617–626. doi: 10.1542/peds.2014-3407. [DOI] [PubMed] [Google Scholar]
- 22.Heianza Y., Zheng Y., Ma W., Rimm E.B., Albert C.M., Hu F.B. Duration and life-stage of antibiotic use and risk of cardiovascular events in women. European Heart Journal. 2019;0:1–8. doi: 10.1093/eurheartj/ehz231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pothineni N.V.K., Subramany S., Kuriakose K., Shirazi L.F., Romeo F., Shah P.K. Infections, atherosclerosis, and coronary heart disease. European Heart Journal. 2017;38(43):3195–3201. doi: 10.1093/eurheartj/ehx362. [DOI] [PubMed] [Google Scholar]
- 24.McHardy I.H., Goudarzi M., Tong M., Ruegger P.M., Schwager E., Weger J.R. Integrative analysis of the microbiome and metabolome of the human intestinal mucosal surface reveals exquisite inter-relationships. Microbiome. 2013;1(1):17. doi: 10.1186/2049-2618-1-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wu H., Esteve E., Tremaroli V., Khan M.T., Caesar R., Mannerås-Holm L. Metformin alters the gut microbiome of individuals with treatment-naive type 2 diabetes, contributing to the therapeutic effects of the drug. Nature Medicine. 2017;23(7):850–858. doi: 10.1038/nm.4345. [DOI] [PubMed] [Google Scholar]
- 26.Laurans L., Venteclef N., Haddad Y., Chajadine M., Alzaid F., Metghalchi S. Genetic deficiency of indoleamine 2,3-dioxygenase promotes gut microbiota-mediated metabolic health. Nature Medicine. 2018;24(8):1113–1120. doi: 10.1038/s41591-018-0060-4. [DOI] [PubMed] [Google Scholar]
- 27.Nambi V., Chambless L., Folsom A.R., He M., Hu Y., Mosley T. Carotid intima-media thickness and presence or absence of plaque improves prediction of coronary heart disease risk. The ARIC (atherosclerosis risk in communities) study. Journal of the American College of Cardiology. 2010;55(15):1600–1607. doi: 10.1016/j.jacc.2009.11.075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Naqvi T.Z., Lee M.S. Carotid intima-media thickness and plaque in cardiovascular risk assessment. Journal of the American College of Cardiology: Cardiovascular Imaging. 2014;7(10):1025–1038. doi: 10.1016/j.jcmg.2013.11.014. [DOI] [PubMed] [Google Scholar]
- 29.Wolf P.A., Agostino R.B.D., Ph D. Carotid intima–media thickness and cardiovascular events. New England Journal of Medicine. 2011;365(17):1640–1642. doi: 10.1056/NEJMc1109714. [DOI] [PubMed] [Google Scholar]
- 30.Levey A.S., Stevens L.A., Schmid C.H., Zhang Y., Castro A.F., Feldman H.I. A new equation to estimate glomerular filtration rate. Annals of Internal Medicine. 2009;150(9):604–612. doi: 10.7326/0003-4819-150-9-200905050-00006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mosteller R.D. Simplified calculation of body-surface area. The New England Journal of Medicine. 1987;317(17):1098. doi: 10.1056/NEJM198710223171717. [DOI] [PubMed] [Google Scholar]
- 32.Lluch J., Servant F., Païssé S., Valle C., Valière S., Kuchly C. The characterization of novel tissue microbiota using an optimized 16S metagenomic sequencing pipeline. PloS One. 2015;10(11) doi: 10.1371/journal.pone.0142334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Païssé S., Valle C., Servant F., Courtney M., Burcelin R., Amar J. Comprehensive description of blood microbiome from healthy donors assessed by 16S targeted metagenomic sequencing. Transfusion. 2016;56(5):1138–1147. doi: 10.1111/trf.13477. [DOI] [PubMed] [Google Scholar]
- 34.Pruesse E., Quast C., Knittel K., Fuchs B.M., Ludwig W., Peplies J. SILVA: a comprehensive online resource for quality checked and aligned ribosomal RNA sequence data compatible with ARB. Nucleic Acids Research. 2007;35(21):7188–7196. doi: 10.1093/nar/gkm864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.McMurdie P.J., Holmes S. Waste not, want not: why rarefying microbiome data is inadmissible. PLoS Computational Biology. 2014;10(4) doi: 10.1371/journal.pcbi.1003531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Love M.I., Huber W., Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biology. 2014;15(12) doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Langfelder P., Horvath S. WGCNA: an R package for weighted correlation network analysis. BMC Bioinformatics. 2008;9 doi: 10.1186/1471-2105-9-559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Franzosa E.A., McIver L.J., Rahnavard G., Thompson L.R., Schirmer M., Weingart G. Species-level functional profiling of metagenomes and metatranscriptomes. Nature Methods. 2018;15(11):962–968. doi: 10.1038/s41592-018-0176-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Langille M.G.I., Zaneveld J., Caporaso J.G., McDonald D., Knights D., Reyes J.A. Predictive functional profiling of microbial communities using 16S rRNA marker gene sequences. Nature Biotechnology. 2013;31(9):814–821. doi: 10.1038/nbt.2676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kozich J.J., Westcott S.L., Baxter N.T., Highlander S.K., Schloss P.D. Development of a dual-index sequencing strategy and curation pipeline for analyzing amplicon sequence data on the miseq illumina sequencing platform. Applied and Environmental Microbiology. 2013;79(17):5112–5120. doi: 10.1128/AEM.01043-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.DeSantis T.Z., Hugenholtz P., Larsen N., Rojas M., Brodie E.L., Keller K. Greengenes, a chimera-checked 16S rRNA gene database and workbench compatible with ARB. Applied and Environmental Microbiology. 2006;72(7):5069–5072. doi: 10.1128/AEM.03006-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Segata N., Izard J., Waldron L., Gevers D., Miropolsky L., Garrett W.S. Metagenomic biomarker discovery and explanation. Genome Biology. 2011;12(6):R60. doi: 10.1186/gb-2011-12-6-r60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Liaw A., Wiener M. Classification and regression by randomForest. R News. 2002;2/3:18–22. [Google Scholar]
- 44.Chong J., Xia J. MetaboAnalystR: an R package for flexible and reproducible analysis of metabolomics data. Bioinformatics (Oxford, England) 2018;34(24):4313–4314. doi: 10.1093/bioinformatics/bty528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bayer F., Ascher S., Pontarollo G., Reinhardt C. Antibiotic treatment protocols and germ-free mouse models in vascular research. Frontiers in Immunology. 2019;10 doi: 10.3389/fimmu.2019.02174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Grasset E., Puel A., Charpentier J., Collet X., Christensen J.E., Tercé F. A specific gut microbiota dysbiosis of type 2 diabetic mice induces GLP-1 resistance through an enteric NO-dependent and gut-brain Axis mechanism. Cell Metabolism. 2017;25(5):1075–1090. doi: 10.1016/j.cmet.2017.04.013. e5. [DOI] [PubMed] [Google Scholar]
- 47.Wilmanski T., Rappaport N., Earls J.C., Magis A.T., Manor O., Lovejoy J. Blood metabolome predicts gut microbiome α-diversity in humans. Nature Biotechnology. 2019;37(10):1217–1228. doi: 10.1038/s41587-019-0233-9. [DOI] [PubMed] [Google Scholar]
- 48.Pedersen H.K., Gudmundsdottir V., Nielsen H.B., Hyotylainen T., Nielsen T., Jensen B.A.H. Human gut microbes impact host serum metabolome and insulin sensitivity. Nature. 2016;535(7612):376–381. doi: 10.1038/nature18646. [DOI] [PubMed] [Google Scholar]
- 49.Abubucker S., Segata N., Goll J., Schubert A.M., Izard J., Cantarel B.L. Metabolic reconstruction for metagenomic data and its application to the human microbiome. PLoS Computational Biology. 2012;8(6) doi: 10.1371/journal.pcbi.1002358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Chen Z., Liu J., Ng H.K.T., Nadarajah S., Kaufman H.L., Yang J.Y. Statistical methods on detecting differentially expressed genes for RNA-seq data. BMC Systems Biology. 2011;5(SUPPL. 3) doi: 10.1186/1752-0509-5-S3-S1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Jonsson A.L., Bäckhed F. Role of gut microbiota in atherosclerosis. Nature Reviews Cardiology. 2017 doi: 10.1038/nrcardio.2016.183. [DOI] [PubMed] [Google Scholar]
- 52.Vrieze A., Out C., Fuentes S., Jonker L., Reuling I., Kootte R.S. Impact of oral vancomycin on gut microbiota, bile acid metabolism, and insulin sensitivity. Journal of Hepatology. 2014;60(4):824–831. doi: 10.1016/j.jhep.2013.11.034. [DOI] [PubMed] [Google Scholar]
- 53.Stepankova R., Tonar Z., Bartova J., Nedorost L., Rossman P., Poledne R. Absence of microbiota (germ-free conditions) accelerates the atherosclerosis in ApoE-deficient mice fed standard low cholesterol diet. Journal of Atherosclerosis and Thrombosis. 2010;17(8):796–804. doi: 10.5551/jat.3285. [DOI] [PubMed] [Google Scholar]
- 54.Lindskog Jonsson A., Caesar R., Akrami R., Reinhardt C., Fåk Hållenius F., Borén J. Impact of gut microbiota and diet on the development of atherosclerosis in apoe −/−mice. Arteriosclerosis, Thrombosis, and Vascular Biology: ATVBAHA. 2018;118:311233. doi: 10.1161/ATVBAHA.118.311233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wright S.D., Burton C., Hernandez M., Hassing H., Montenegro J., Mundt S. Infectious agents are not necessary for murine atherogenesis. Journal of Experimental Medicine. 2000;191(8):1437–1441. doi: 10.1084/jem.191.8.1437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kasahara K., Tanoue T., Yamashita T., Yodoi K., Matsumoto T., Emoto T. Commensal bacteria at the crossroad between cholesterol homeostasis and chronic inflammation in atherosclerosis. Journal of Lipid Research. 2017;58(3):519–528. doi: 10.1194/jlr.M072165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Rune I., Rolin B., Larsen C., Nielsen D.S., Kanter J.E., Bornfeldt K.E. Modulating the gut microbiota improves glucose tolerance, lipoprotein profile and atherosclerotic plaque development in ApoE-deficient mice. PloS One. 2016;11(1):1–16. doi: 10.1371/journal.pone.0146439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ghosh S.S., Bie J., Wang J., Ghosh S. Oral supplementation with non-absorbable antibiotics or curcumin attenuates western diet-induced atherosclerosis and glucose intolerance in LDLR-/- mice--role of intestinal permeability and macrophage activation. PloS One. 2014;9(9) doi: 10.1371/journal.pone.0108577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Le Roy T., Lécuyer E., Chassaing B., Rhimi M., Lhomme M., Boudebbouze S. The intestinal microbiota regulates host cholesterol homeostasis. BMC Biology. 2019:1–18. doi: 10.1186/s12915-019-0715-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Pontarollo G., Kiouptsi K., Reinhardt C. A holobiont view on thrombosis: unravelling the microbiota's influence on arterial thrombus growth. Microbial Cell. 2020;7(1):28–31. doi: 10.15698/mic2020.01.704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Ubeda C., Taur Y., Jenq R.R., Equinda M.J., Son T., Samstein M. Vancomycin-resistant Enterococcus domination of intestinal microbiota is enabled by antibiotic treatment in mice and precedes bloodstream invasion in humans. Journal of Clinical Investigation. 2010;120(12):4332–4341. doi: 10.1172/JCI43918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Amar J., Lange C., Payros G., Garret C., Chabo C., Lantieri O. Blood microbiota dysbiosis is associated with the onset of cardiovascular events in a large general population: the D.E.S.I.R. study. PloS One. 2013;8(1) doi: 10.1371/journal.pone.0054461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Tang T.W.H., Chen H.C., Chen C.Y., Yen C.Y.T., Lin C.J., Prajnamitra R.P. Loss of gut microbiota alters immune system composition and cripples postinfarction cardiac repair. Circulation. 2019;139(5):647–659. doi: 10.1161/CIRCULATIONAHA.118.035235. [DOI] [PubMed] [Google Scholar]
- 64.Jie Z., Xia H., Zhong S.-L., Feng Q., Li S., Liang S. The gut microbiome in atherosclerotic cardiovascular disease. Nature Communications. 2017;8(1):845. doi: 10.1038/s41467-017-00900-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Zarrinpar A., Chaix A., Xu Z.Z., Chang M.W., Marotz C.A., Saghatelian A. Antibiotic-induced microbiome depletion alters metabolic homeostasis by affecting gut signaling and colonic metabolism. Nature Communications. 2018;9(1):2872. doi: 10.1038/s41467-018-05336-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Reijnders D., Goossens G.H., Hermes G.D.A., Neis E.P.J.G., van der Beek C.M., Most J. Effects of gut microbiota manipulation by antibiotics on host metabolism in obese humans: a randomized double-blind placebo-controlled trial. Cell Metabolism. 2016;24(1):63–74. doi: 10.1016/j.cmet.2016.06.016. [DOI] [PubMed] [Google Scholar]
- 67.Kasahara K., Krautkramer K.A., Org E., Romano K.A., Kerby R.L., Vivas E.I. Interactions between Roseburia intestinalis and diet modulate atherogenesis in a murine model. Nature Microbiology. 2018;3(12):1461–1471. doi: 10.1038/s41564-018-0272-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Aguilar E.C., Leonel A.J., Teixeira L.G., Silva A.R., Silva J.F., Pelaez J.M.N. Butyrate impairs atherogenesis by reducing plaque inflammation and vulnerability and decreasing NFκB activation. Nutrition, Metabolism, and Cardiovascular Diseases: NMCD. 2014;24(6):606–613. doi: 10.1016/j.numecd.2014.01.002. [DOI] [PubMed] [Google Scholar]
- 69.Brandsma E., Kloosterhuis N.J., Koster M., Dekker D.C., Gijbels M.J.J., Van Der Velden S. A proinflammatory gut microbiota increases systemic inflammation and accelerates atherosclerosis. Circulation Research. 2019;124(1):94–100. doi: 10.1161/CIRCRESAHA.118.313234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Murr C., Grammer T.B., Kleber M.E., Meinitzer A., März W., Fuchs D. Low serum tryptophan predicts higher mortality in cardiovascular disease. European Journal of Clinical Investigation. 2015;45(3):247–254. doi: 10.1111/eci.12402. [DOI] [PubMed] [Google Scholar]
- 71.Cason C.A., Dolan K.T., Sharma G., Tao M., Kulkarni R., Helenowski I.B. Plasma microbiome-modulated indole- and phenyl-derived metabolites associate with advanced atherosclerosis and postoperative outcomes. Journal of Vascular Surgery. 2018;68(5):1552–1562. doi: 10.1016/j.jvs.2017.09.029. e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Lee J.-H., Wood T.K., Lee J. Roles of indole as an interspecies and interkingdom signaling molecule. Trends in Microbiology. 2015;23(11):707–718. doi: 10.1016/j.tim.2015.08.001. [DOI] [PubMed] [Google Scholar]
- 73.Barreto F.C., Barreto D.V., Liabeuf S., Meert N., Glorieux G., Temmar M. Serum indoxyl sulfate is associated with vascular disease and mortality in chronic kidney disease patients. Clinical Journal of the American Society of Nephrology: CJASN. 2009;4(10):1551–1558. doi: 10.2215/CJN.03980609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Dou L., Sallée M., Cerini C., Poitevin S., Gondouin B., Jourde-Chiche N. The cardiovascular effect of the uremic solute indole-3 acetic acid. Journal of the American Society of Nephrology: JASN. 2015;26(4):876–887. doi: 10.1681/ASN.2013121283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Wilck N., Matus M.G., Kearney S.M., Olesen S.W., Forslund K., Bartolomaeus H. Salt-responsive gut commensal modulates TH17 axis and disease. Nature. 2017;551(7682):585–589. doi: 10.1038/nature24628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Zelante T., Iannitti R., Cunha C., DeLuca A., Giovannini G., Pieraccini G. Tryptophan catabolites from microbiota engage aryl hydrocarbon receptor and balance mucosal reactivity via interleukin-22. Immunity. 2013;39(2):372–385. doi: 10.1016/j.immuni.2013.08.003. [DOI] [PubMed] [Google Scholar]
- 77.Bock K.W. Human AHR functions in vascular tissue: pro- and anti-inflammatory responses of AHR agonists in atherosclerosis. Biochemical Pharmacology. 2019;159:116–120. doi: 10.1016/j.bcp.2018.11.021. [DOI] [PubMed] [Google Scholar]
- 78.Kerley-Hamilton J.S., Trask H.W., Ridley C.J.A., Dufour E., Lesseur C., Ringelberg C.S. Inherent and benzo[a]pyrene-induced differential aryl hydrocarbon receptor signaling greatly affects life span, atherosclerosis, cardiac gene expression, and body and heart growth in mice. Toxicological Sciences. 2012;126(2):391–404. doi: 10.1093/toxsci/kfs002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Huang S., Shui X., He Y., Xue Y., Li J., Li G. AhR expression and polymorphisms are associated with risk of coronary arterial disease in Chinese population. Scientific Reports. 2015;5:8022. doi: 10.1038/srep08022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Hubbard T.D., Murray I.A., Bisson W.H., Lahoti T.S., Gowda K., Amin S.G. Adaptation of the human aryl hydrocarbon receptor to sense microbiota-derived indoles. Scientific Reports. 2015;5:12689. doi: 10.1038/srep12689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Wu D., Nishimura N., Kuo V., Fiehn O., Shahbaz S., Van Winkle L. Activation of aryl hydrocarbon receptor induces vascular inflammation and promotes atherosclerosis in apolipoprotein E-/- mice. Arteriosclerosis, Thrombosis, and Vascular Biology. 2011;31(6):1260–1267. doi: 10.1161/ATVBAHA.110.220202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Eckers A., Jakob S., Heiss C., Haarmann-Stemmann T., Goy C., Brinkmann V. The aryl hydrocarbon receptor promotes aging phenotypes across species. Scientific Reports. 2016;6 doi: 10.1038/srep19618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Fatkhullina A.R., Peshkova I.O., Dzutsev A., Aghayev T., McCulloch J.A., Thovarai V. An interleukin-23-interleukin-22 Axis regulates intestinal microbial homeostasis to protect from diet-induced atherosclerosis. Immunity. 2018 doi: 10.1016/j.immuni.2018.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Venkatesh M., Mukherjee S., Wang H., Li H., Sun K., Benechet A.P. Symbiotic bacterial metabolites regulate gastrointestinal barrier function via the xenobiotic sensor PXR and Toll-like receptor 4. Immunity. 2014;41(2):296–310. doi: 10.1016/j.immuni.2014.06.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Zhao Z.H., Xin F.Z., Xue Y., Hu Z., Han Y., Ma F. Indole-3-propionic acid inhibits gut dysbiosis and endotoxin leakage to attenuate steatohepatitis in rats. Experimental & Molecular Medicine. 2019;51(9) doi: 10.1038/s12276-019-0304-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Sui Y., Xu J., Rios-Pilier J., Zhou C. Deficiency of PXR decreases atherosclerosis in apoE-deficient mice. Journal of Lipid Research. 2011;52(9):1652–1659. doi: 10.1194/jlr.M017376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Zhou C., King N., Chen K.Y., Breslow J.L. Activation of PXR induces hypercholesterolemia in wild-type and accelerates atherosclerosis in apoE deficient mice. Journal of Lipid Research. 2009;50(10):2004–2013. doi: 10.1194/jlr.M800608-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Chávez-Talavera O., Tailleux A., Lefebvre P., Staels B. Bile acid control of metabolism and inflammation in obesity, type 2 diabetes, dyslipidemia, and nonalcoholic fatty liver disease. Gastroenterology. 2017 doi: 10.1053/j.gastro.2017.01.055. [DOI] [PubMed] [Google Scholar]
- 89.Jadhav K., Xu Y., Xu Y., Li Y., Xu J., Zhu Y. Reversal of metabolic disorders by pharmacological activation of bile acid receptors TGR5 and FXR. Molecular Metabolism. 2018;9(January):131–140. doi: 10.1016/j.molmet.2018.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The mouse cecal 16S rRNA sequence data can be found under http://www.ncbi.nlm.nih.gov/bioproject/PRJNA595367.
The human stool 16S rRNA sequence data can be found under http://www.ncbi.nlm.nih.gov/bioproject/PRJNA595382.