Abstract
Objectives
AmpC-β-lactamase production is an under-recognized antibiotic resistance mechanism that renders Gram-negative bacteria resistant to common β-lactam antibiotics, similar to the well-known ESBLs. For infection control purposes, it is important to be able to discriminate between plasmid-mediated AmpC (pAmpC) production and chromosomal-mediated AmpC (cAmpC) hyperproduction in Gram-negative bacteria as pAmpC requires isolation precautions to minimize the risk of horizontal gene transmission. Detecting pAmpC in Escherichia coli is challenging, as both pAmpC production and cAmpC hyperproduction may lead to third-generation cephalosporin resistance.
Methods
We tested a collection of E. coli strains suspected to produce AmpC. Elaborate susceptibility testing for third-generation cephalosporins, WGS and machine learning were used to develop an algorithm to determine ampC genotypes in E. coli. WGS was applied to detect pampC genes, cAmpC hyperproducers and STs.
Results
In total, 172 E. coli strains (n=75 ST) were divided into a training set and two validation sets. Ninety strains were pampC positive, the predominant gene being blaCMY-2 (86.7%), followed by blaDHA-1 (7.8%), and 59 strains were cAmpC hyperproducers. The algorithm used a cefotaxime MIC value above 6 mg/L to identify pampC-positive E. coli and an MIC value of 0.5 mg/L to discriminate between cAmpC-hyperproducing and non-cAmpC-hyperproducing E. coli strains. Accuracy was 0.88 (95% CI=0.79–0.94) on the training set, 0.79 (95% CI=0.64–0.89) on validation set 1 and 0.85 (95% CI=0.71–0.94) on validation set 2.
Conclusions
This approach resulted in a pragmatic algorithm for differentiating ampC genotypes in E. coli based on phenotypic susceptibility testing.
Introduction
Escherichia coli is an important pathogen in both community and healthcare-associated infections.1,2 ESBL-producing E. coli have spread worldwide, restricting available treatment options. Although to a lesser degree, acquired AmpC β-lactamases in E. coli are also emerging as a potential threat to the activity of broad-spectrum penicillins and third-generation cephalosporins (3GCs). Acquired AmpC β-lactamases are encoded on plasmids and hence transferable between species. The prevalence of plasmid-mediated AmpC (pAmpC) β-lactamases in E. coli clinical isolates reported in the literature varies between 0.06% and 10.1%;3,4 however, variance in prevalence is likely to be influenced by diagnostic strategies used in these studies, and there are also regional differences in prevalence. In the Netherlands, a country with low levels of antimicrobial resistance, a pAmpC prevalence between 0.6% and 1.3% was found in E. coli isolates recovered from faecal samples in the community.5,6 Recently, Harris et al.7 described pAmpC as the second most common group (17.1%) of 3GC-hydrolysing β-lactamases in E. coli bloodstream infections in Australia, New Zealand and Singapore. Different types of plasmid-mediated ampC (pampC) genes have been detected in Enterobacterales, with blaCMY-2 as the most common AmpC-encoding resistance gene. Other, less frequently isolated AmpC β-lactamase genes are other varieties of blaCMY, as well as blaDHA, blaACT, blaACC, blaMIR, blaMOX, blaFOX and blaCFE. Depending on the type of pAmpC β-lactamase, the hydrolysing capability might vary.8,9
E. coli naturally carries a chromosomal-mediated ampC (campC) gene, but unlike in other Enterobacterales this gene is non-inducible.8 In E. coli AmpC production is regulated by promoter and attenuator mechanisms resulting in constitutive low-level ampC expression and hence allows the use of β-lactam antibiotics to treat these E. coli infections in the absence of other resistance mechanisms. Various mutations in the promoter/attenuator region of E. coli may cause constitutive hyperexpression of campC. These E. coli strains may then become resistant to cephamycins, broad-spectrum penicillins or even 3GCs, making it difficult to differentiate these strains phenotypically from pAmpC enzyme production.
In contrast to hyperexpressed campC genes, pampC genes are capable of spreading this resistance mechanism to other bacteria within a hospital setting by horizontal gene transfer.10,11 This poses a greater threat to infection control than pure clonal transfer. Consequently, pAmpC production in E. coli requires active detection and contact precautions for colonized or infected patients, as recommended by different guidelines;12,13 however, this is often ignored due to the more cumbersome identification in the microbiological laboratory.
Current commercial phenotypic AmpC confirmation tests fail to reliably discriminate between pAmpC and constitutive hyperproduction of the chromosomal-mediated AmpC (cAmpC).14 In E. coli, an approach solely based on phenotypic testing has a high sensitivity to detect pAmpC production, but lacks specificity as it detects a high number of isolates that overproduce cAmpC, resulting in unnecessary patient isolation precautions with increased unnecessary healthcare costs. PCR is capable of detecting various pampC genes.15 The recommended method for detection of pAmpC production in Enterobacterales according to the EUCAST guidelines is to screen isolates for cefoxitin MICs >8 mg/L combined with phenotypic resistance to cefotaxime and/or ceftazidime.16 Confirmation is advised in a two-step algorithm using cloxacillin synergy detection and PCR to discriminate pampC from hyperexpressed campC in E. coli. Several studies suggest the screening of isolates in a similar fashion.17,18 However, molecular tests are not always available in laboratories and are relatively expensive and often time-consuming.
The aim of this present study was to evaluate various diagnostic approaches through determining the MICs of specific cephalosporins, two commercial AmpC disc-diffusion confirmation tests and WGS to develop an algorithm to detect pAmpC production in ESBL-negative and cefoxitin-resistant E. coli.
Materials and methods
Overall study design
Three datasets consisting of E. coli cefoxitin-resistant and ESBL-negative strains were identified. Most strains were suspected of having either a pAmpC or a cAmpC resistance mechanism. All strains were subjected to WGS to obtain the genotypes [pampC, campC, promoter mutations (hyperproducer) and absence of both (negative)] and subjected to Etests and two AmpC disc-diffusion confirmation tests. The training set contained a wide variety of phenotypes and was used as input for constructing an algorithm to classify the three genotypes (pampC, hyperproducer and negative). The most accurate algorithm was selected as the final algorithm and validated in two validation sets. Validation set 1 was used to validate the algorithm and represents the epidemiology in a Dutch hospital setting. Due to a low number of pampC-positive strains and restricted geographical background we broadened the representation of suspect AmpC-producing isolates in a second validation set (validation set 2). An extensive description of the selection of samples in the training set, validation set 1 and validation set 2 can be found in the Supplementary Materials and methods (available at JAC Online).
Etests and AmpC disc-diffusion confirmation tests
Deep-frozen samples of the selected strains were recultured on Columbia III agar (BD Diagnostic Systems, Sparks, MD, USA) or blood agar (Media production, Elizabeth-Tweesteden Hospital, Tilburg, the Netherlands) prior to testing. Strains were tested using Etest (bioMérieux, Marcy-l’Étoile, France) to determine the MICs of cefotaxime, ceftazidime and cefoxitin. Etests were placed on Mueller–Hinton (Oxoid Ltd, Altrincham, Cheshire, England) culture plates, which were placed in the oven within 15 min and incubated for 16–20 h under an O2 atmosphere at 36°C. Exact MIC values were noted. The presence of AmpC was phenotypically confirmed using the AmpC Confirm Kit (Rosco Diagnostica A/S, Taastrup, Denmark) according to the manufacturer’s guidelines. A second phenotypical confirmation with the D68C AMPC + ESBL detection set (MAST Group Ltd, Bootle, UK) was performed according to the manufacturer’s guidelines. From both confirmation tests the zone inhibition differences, measured in millimetres, were recorded for further use.
DNA isolation, library preparation and DNA sequencing
For logistical reasons DNA isolation, library preparation and DNA sequencing were performed at two different centres. For training and validation set 2, bacterial DNA was extracted by a CTAB-based method and a paired-end 2 × 150 bp library was sequenced using an Illumina NextSeq500 sequencer (Illumina, San Diego, CA, USA) (see the Supplementary Materials and methods). For validation set 1, bacterial DNA was extracted using the MagNA Pure LC Total Nucleic Acid Kit - High Performance on a MagNA Pure LC instrument (Roche Diagnostics International Ltd, Rotkreuz, Switzerland) according to the manufacturer’s protocol. A 2 × 300 bp paired-end library was sequenced on an Illumina MiSeq sequencer (Illumina, San Diego, CA, USA) (Supplementary Materials and methods).
WGS analyses
Sequence reads were demultiplexed and merged to obtain fastq files for each sample. Reads were quality assessed and adapter trimmed by Trim_galore (version 0.4.1)19 followed by a custom NextSeq read cleaning script to remove reads containing six or more As and Gs introduced by the sequencing chemistry. Read coverage was calculated by dividing the number of sequence bases for each sample by the length of E. coli K-12 strain C3026 (RefSeq: NZ_CP014272.1). Samples not exceeding 30× read coverage were excluded for further analyses and samples containing >120× read coverage were subsampled to 120×. Reads were de novo assembled to create contigs by SPAdes (version 3.11.1)20 using default settings and k-mer sizes 21, 41, 61, 81 and 101. MLST STs were derived from the contigs using mlst (version 2.5 pubMLST, 31 October 2017).21,22
Plasmid-mediated ampC detection
To detect pampC genes, contigs were BLASTed (version 2.2.30+)23 against the ResFinder database (2018-02-16)24 using abricate (version 0.5).25 Genes that had a coverage of ≥90% and a sequence identity >75% were interpreted as present. To circumvent the absence of genes due to wrong assembly, pampC genes were validated using KMA (version 0.14.3)26 with the ResFinder database (2018–02-16), which is a method that uses raw sequences as input. Genes were marked present if KMA matched >90% coverage and >90% identity. Finally, pampC genes were considered present if both methods reported an identical gene and the strain was labelled pampC accordingly.
Detection of ampC hyperproducer genotype
The promoter and attenuator region of campC was extracted from all samples to obtain a similar 271 bp fragment, as described by Peter-Getzlaff et al.27 The sequence of each strain was aligned against the promoter/attenuator region of the campC gene of the E. coli K-12 strain MG1655 (GenBank accession number U00096.3) using AliView (version 1.23).28 Strains were labelled cAmpC hyperproducer when promoter mutations were found, as reported by Caroff et al.29 and Tracz et al.30
Creating an algorithm based on the training set
For the decision tree model, Recursive Partitioning And Regression Trees (RPART), an R package (version 4.1-13), was used; this is an implementation of Classification and Regression Tree (CART), a statistical technique to solve classification problems, developed by Breiman et al.31 RPART was used to create a decision tree model to classify strains based on Etest MICs, AmpC Confirm Kit or D68C test results into a pampC, hyperproducer or negative class. Model optimization and cross-validation were performed within the caret R package (version 6.0-80) in R (version 3.5.1).32 The RPART model was trained to optimize for accuracy and by using seed 825 to be able to reproduce model creation. The cross-validation was performed using a 10-fold three-times-repeated cross-validation using the repeatedcv parameter. Student’s t-test was used to compare model performances (P=0.05). A two-class model was derived from the three-class model by combining the negatives with the hyperproducer class and recalculating the statistics.
Results
Training set
Between January 2014 and March 2018, 267 E. coli strains that had cefoxitin MICs >8 mg/L and were ESBL negative were found in the laboratory information management system at Radboudumc. Out of these strains, 98 were selected for further testing. Eleven of these strains could not be retrieved from the freezer and three strains were identified as not being E. coli by MALDI-TOF MS. This resulted in a training set of 84 E. coli strains. MICs determined using the BD Phoenix System indicated that the training set likely consisted of a wide variety of different resistance phenotypes. A substantial proportion of strains were resistant to both ceftazidime and ceftriaxone (42.9%, n=36) (Table 1), 20 strains (23.8%) were susceptible to 3GCs and 28 strains (33.3%) were intermediate or resistant to at least one of the 3GCs. WGS results revealed that 32 of 84 E. coli strains (38.1%) contained blaCMY-2 and 29.8% (n=25) showed known mutations in the ampC promoter region and were therefore labelled as hyperproducers, 20.2% (n=17) were negative for both pampC genes and mutations in the promoter region of campC and were classified as negative (Figure S1A, available as Supplementary data at JAC Online).
Table 1.
BD Phoenix System susceptibility of 84 E. coli strains in the training set
| Cefoxitin (R>8 mg/L)a | Ceftriaxone (S ≤1 mg/L; R >4 mg/L)b | Ceftazidime (S ≤1 mg/L; R >4 mg/L)b | n | Percentage |
|---|---|---|---|---|
| R | S | S | 20 | 23.81 |
| R | S | I | 13 | 15.48 |
| R | S | R | 8 | 9.52 |
| R | R | S | 1 | 1.19 |
| R | I | I | 1 | 1.19 |
| R | R | I | 1 | 1.19 |
| R | I | R | 4 | 4.76 |
| R | R | R | 36 | 42.86 |
| total=84 | total=100.00 |
R, resistant; S, susceptible; I, intermediate.
MIC cut-off adapted from EUCAST guideline on detection of resistance mechanisms v2.0.
MIC breakpoints according to EUCAST clinical breakpoints for bacteria v.9.0.
Validation sets 1 and 2
Validation set 1 consisted of 47 clinical E. coli strains. WGS results showed that 72.3% (n=34) of the strains were hyperproducers and 12.8% (n=6) were pampC and hyperproduction negative. Two pampC variants were found, 12.8% (n=6) blaCMY-2 and 2.1% (n=1) blaDHA-1 (Figure S1B). To cope with the low number of pampC-positive strains in validation set 1, validation set 2 (n=41) consisted of pampC-positive strains with mainly blaCMY-2 (97.6%) (Figure S1C).
Genomic composition
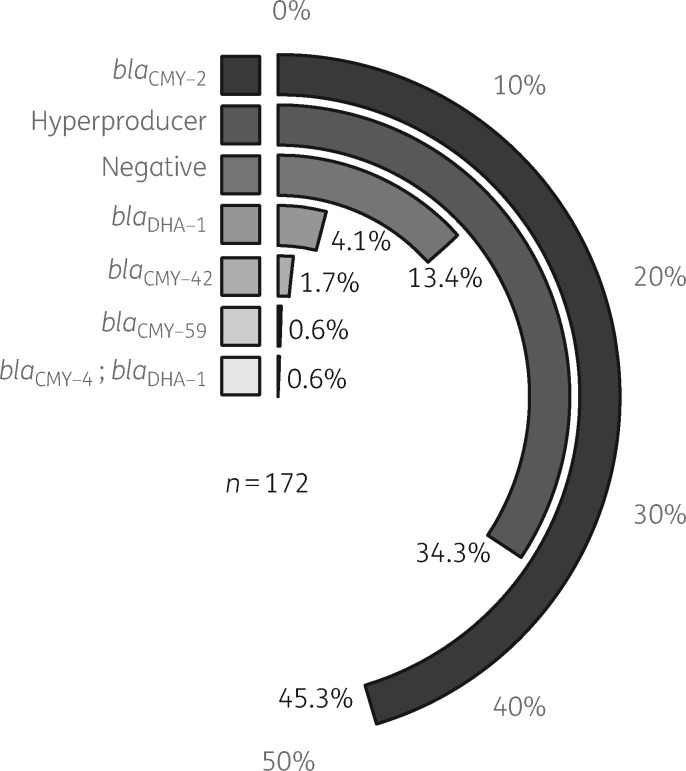
In total, the 172 E. coli strains represented 75 different MLST STs, of which ST131 (8.14%, n=14), ST38 (6.98%, n=12) and ST73 (6.98%, n=12) were the most prevalent. Furthermore, the STs of 13 strains were unknown (see Table S1). For the identification of pampC genes we found that there was 100% concordance between the tools abricate and KMA, which supports the accurate detection of pampC genes from WGS data. Overall, in 172 E. coli strains, blaCMY-2 was the most prevalent (45.3%) resistance mechanism followed by hyperproducers (34.3%) (Figure 1).
Figure 1.
Clockplot showing the distribution of ampC genotypes in all 172 E. coli strains. The key is sorted in decreasing order of occurrence. Half a circle indicates 50%; each genotype fills part of the circle to indicate the percentage of each genotype.
Etests and AmpC disc diffusion confirmation tests
By combining the WGS results with the Etest results, we found higher median MICs of cefoxitin, ceftazidime and cefotaxime for strains that harbour a pampC gene (cefoxitin median=256 mg/L; ceftazidime median=10 mg/L; cefotaxime median=12 mg/L) compared with hyperproducers (cefoxitin median=48 mg/L; ceftazidime median=2 mg/L; cefotaxime median=1.5 mg/L) and negatives (cefoxitin median=32 mg/L; ceftazidime median=0.38 mg/L; cefotaxime median=0.38 mg/L) (Figure S2). Furthermore, zone inhibition differences found with the AmpC Confirm Kit showed higher zone inhibition differences in the pampC strains (ceftazidime + cloxacillin versus ceftazidime median=12 mm; cefotaxime + cloxacillin versus cefotaxime median=8 mm) compared with negative strains (ceftazidime + cloxacillin versus ceftazidime median=3 mm; cefotaxime + cloxacillin versus cefotaxime median=1 mm). However, pampC-positive strains showed more overlap with the hyperproducer group (ceftazidime + cloxacillin versus ceftazidime median=8 mm; cefotaxime + cloxacillin versus cefotaxime median=7 mm) as compared with the AmpC-negative group (Figure S3). The boxplots of the D68C test illustrate that there was no clear separation between hyperproducer (D68C C-A median=15 mm; D68C D-B median=14 mm) and pampC-positive strains (D68C C-A median=15 mm; D68C D-B median=15 mm) based on zone inhibition differences (Figure S4).
MICs in relation to the presence of different ampC genes
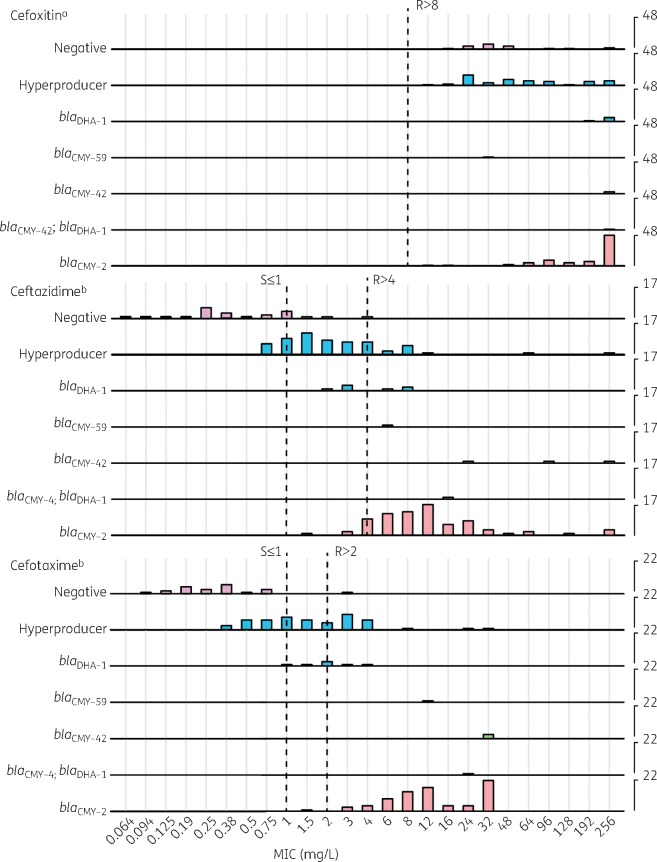
A ridge plot was generated to visualize the Etest MICs for each genotype for all 172 E. coli strains (Figure 2). The plot reveals that negative strains showed MICs of ceftazidime of ≤4 mg/L and of cefotaxime of ≤3 mg/L. For hyperproducers, MICs of ceftazidime were predominantly in the range of 0.75–12 mg/L and cefotaxime MICs were in the range of 0.38–4 mg/L. Isolates that harboured blaCMY showed ceftazidime MICs of 1.5–256 mg/L and cefotaxime MICs of 1.5–32 mg/L. In contrast, blaDHA-1-positive strains showed lower MICs of 3GCs (ceftazidime 2–8 mg/L and cefotaxime 1–4 mg/L), which overlapped with MIC ranges for hyperproducing strains.
Figure 2.
Ridge plot of Etest MICs for 172 E. coli strains grouped by genotype. The x-axis indicates MICs in mg/L. The left-hand y-axis indicates genotypes of strains. The right-hand y-axis indicates number of counts for each MIC; counts are scaled for each Etest to enhance visibility. R, resistant; S, susceptible. aMIC cut-off adapted from EUCAST guideline on detection of resistance mechanisms v2.0. bMIC breakpoints according to EUCAST clinical breakpoints for bacteria v.9.0. This figure appears in colour in the online version of JAC and in black and white in the print version of JAC.
Performance of susceptibility tests to predict ampC type
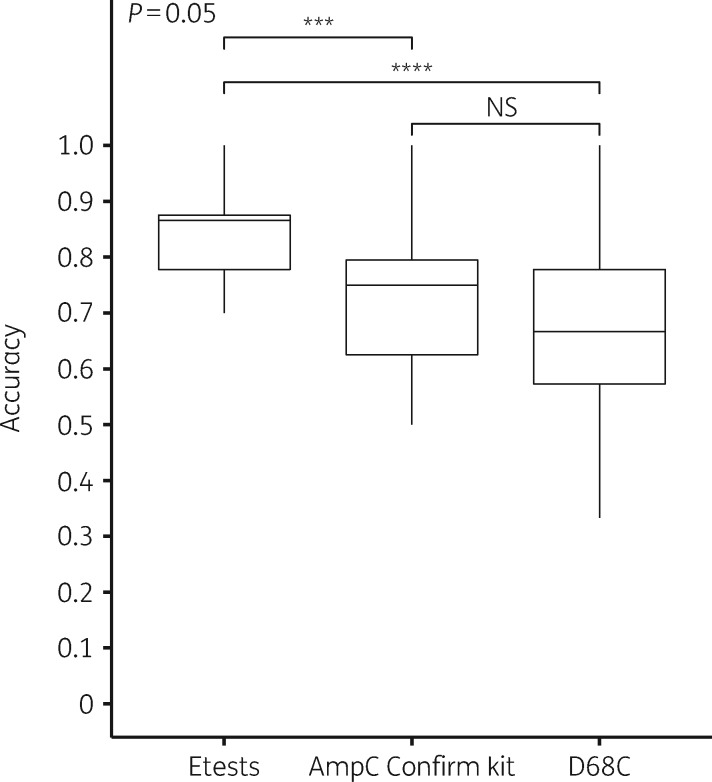
Training of the RPART model and the cross-validation on the training set (n=84) were performed to predict whether strains have a negative, hyperproducer or pampC genotype. The model indicated that training with Etest MICs performed best (Figure 3). It had the highest average accuracy (0.83) and the performance was significantly better than the AmpC Confirm Kit (0.73) and the D68C test (0.67). Furthermore, cross-validation using Etest MICs resulted in the smallest quartile, implying that the model could be extra stable when applied to other datasets. Therefore, we selected the decision tree trained on Etest MICs as the final decision tree model to test performance on training and validation sets.
Figure 3.
Boxplots of model performance. The x-axis indicates the performance using Etests, AmpC Confirm Kit and MAST D68C. The y-axis indicates accuracy based on 10-fold three-times-repeated cross-validation using all 84 E. coli strains of the training set. ***P ≤ 0.001; ****P ≤ 0.0001; NS, not significant.
Model description and performance
The final RPART model contained two decisions and performance was evaluated on the training set (n=84). In the first decision—cefotaxime with an MIC of ≥6 mg/L for all pampC strains (n=42)—34 were correctly classified as pampC positive (n=34/42). In the second decision, samples with cefotaxime MIC <6 mg/L were divided by a cefotaxime MIC breakpoint of 0.50 mg/L. With an MIC breakpoint of cefotaxime <0.50 mg/L, 16 strains were correctly classified as negative (n=16/17). With a cefotaxime MIC ≥0.50 mg/L, all hyperproducer strains except one were correctly classified as hyperproducers (n=24/25). However, nine strains categorized as hyperproducers were either negative (n=1/17) or pampC positive (n=8/42) (Figure 4). This resulted in an overall model accuracy of 0.88 (95% CI=0.79–0.94) (Table 2). From an infection control perspective, it is most important to distinguish pampC from non-pampC. Therefore, we recalculated the performance from a three-class model to a two-class model. The negative and hyperproducer classes were merged to a non-pampC class. The two-class model resulted in an accuracy of 0.90 (95% CI=0.82–0.96) with a sensitivity and specificity of 0.81 (95% CI=0.66–0.91) and 1.00 (95% CI=0.92–1.00), respectively (Table 2 and Table S2).
Figure 4.
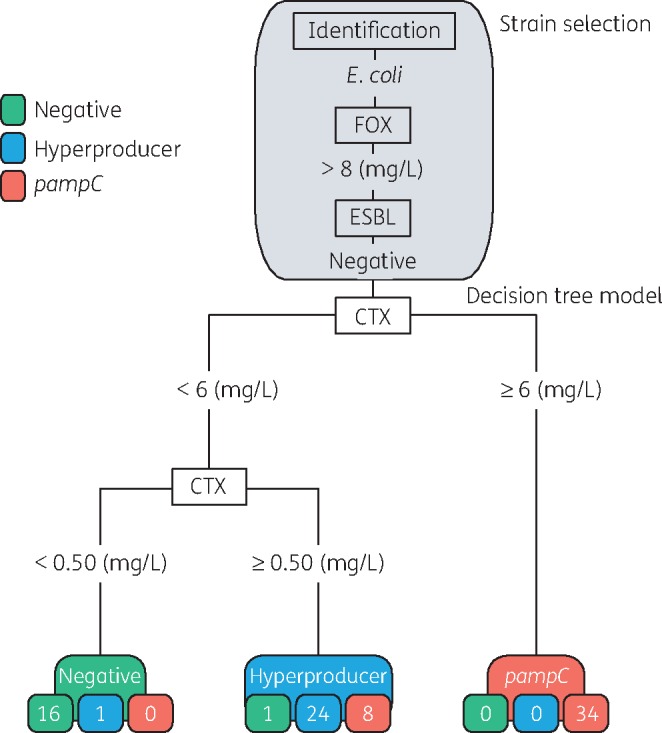
Strain selection and decision tree model based on 84 E. coli strains of the training set. The grey area corresponds to strain selection. The numbers in the coloured boxes indicate the numbers of strains classified according to the decision tree. The decision tree cut-off value for pampC is ≥6 mg/L. For the negative strains the cut-off is <0.50 mg/L. For the hyperproducer strains the cut-off is set to <6 mg/L followed by ≥0.50 mg/L. Cut-offs are based on Etest values. CTX, cefotaxime; FOX, cefoxitin.
Table 2.
Accuracy of final decision tree model trained using the 84 E. coli strains of the training set on all datasets
| Dataset | n | Three-class model, percentage accuracy (95% CI) | Two-class model, percentage accuracy (95% CI) |
|---|---|---|---|
| Training set | 84 | 0.88 (0.79–0.94) | 0.90 (0.82–0.96) |
| Validation set 1 | 47 | 0.79 (0.64–0.89) | 0.91 (0.80–0.98) |
| Validation set 2 | 41 | 0.85 (0.71–0.94) | 0.85 (0.71–0.94) |
Model performance on validation sets
To perform a more extensive evaluation of the decision tree, the model was tested by using two validation sets as input, as described in the Materials and methods section. The performance of the algorithm on validation set 1 (n=47) resulted in an accuracy of 0.79 (95% CI=0.64–0.89) (Table 2). Using validation set 2 (n=41), the decision tree achieved an accuracy of 0.85 (95% CI=0.71–0.94) (Table 2). More details on the performance of the two- and three-class models can be found in the confusion matrix in Table S2 and the confusion matrix in Table S3.
Discussion
To the best of our knowledge, this is the first study that combined susceptibility testing, WGS and simple supervised machine learning to develop a user-friendly algorithm to determine the likelihood of pampC in cefoxitin-resistant and ESBL-negative E. coli strains (Figure 4). The decision tree requires a single cefotaxime Etest as input, is easy to apply in most laboratory settings and results in an accurate detection of pampC-positive strains.
Timely and more accurate identification of pampC isolates improves infection control practices and minimizes unnecessary and costly isolation measures. In the current setting a genotypic confirmation is recommended to differentiate between pAmpC and cAmpC production in cefoxitin-resistant E. coli as phenotypic confirmation is not reliable.16 Our comparison of the AmpC Confirm Kit, D68C test and Etests shows that disc-diffusion zone differences are useful to detect AmpC production in general, but are inadequate to differentiate between pAmpC and cAmpC production (Figures S2–S4). Therefore, rapid and accurate differentiation is needed to further improve infection control policies. Introducing an Etest in combination with the proposed algorithm illustrates that accurate phenotypic detection and identification of pampC harbouring E. coli is feasible.
A relationship between 3GC resistance and the presence of pAmpC has been reported in the literature.17,18,33 Although pAmpC-producing E. coli isolates in this present study showed higher MICs of 3GCs than isolates without pampC genes, the distributions of MIC between pAmpC and hyperproducing cAmpC isolates overlap. This overlap was mainly caused by the E. coli strains that produced DHA-1 enzymes. Edquist et al.18 also concluded that clinical resistance to cefotaxime and/or ceftazidime as a screening criterion for pAmpC might be useful, although discriminatory performance was more prominent when using disc diffusion as compared with MIC testing by Etest strips. In their study, a multiplex PCR was performed to detect pampC genes, but there was no verification for cAmpC hyperproducers in the strain collection used. Our WGS results reliably show that phenotypic hyperproduction of cAmpC β-lactamase can be caused by mutations in the ampC promoter region.18 No conclusions can be drawn about mutations resulting in elevated AmpC production, in addition to previously mentioned mutations. However, there is evidence that alterations of the AmpC β-lactamase34,35 or changes in membrane permeability may lead to differences in cephalosporin resistance.36 Further analysis on the incorrectly classified campC isolates is needed to exclude causes of cephalosporin resistance, other than the known promoter mutations.
ACT-1, DHA-1, DHA-2 and CMY-13 are inducible while other pAmpC β-lactamases (such as CMY-2) are constitutively expressed.8,9,37 Reisbig et al.37 previously reported that absence of the ampD gene in combination with the inducible ACT-1 pampC gene increased MICs of 3GCs. If we assume that the inducible blaDHA group might have a similar mechanism, we would expect higher ceftazidime and cefotaxime MICs in the absence of ampD; our strains with blaDHA showed lower MICs of 3GCs compared with the non-inducible blaCMY-2 genes, so we can infer that ampD might be present; however, further analysis on the influence of ampD on blaDHA expression is needed.
Additionally, Reisbig et al.37 described the contribution of plasmid copy number of ACT-1 and MIR-1 pampC genes to 3GC resistance. They concluded that plasmid copy number probably impacts MIC values for pampC-positive strains; however, this was not substantiated.38 We were able to accurately detect pampC-positive strains even without measuring plasmid copy numbers.
A strength of the present study is that ST information on E. coli in our datasets was provided, which made it possible to exclude clonal origin, in contrast to other studies.17,18,33 Though ST131, ST73 and ST38 predominated, a wide variety of STs was represented in our collection (Table S2). This is in line with other studies that report higher prevalence of ST131 and ST73 in human samples39,40 and ST38 in animal samples.41
bla CMY-2 is the predominant pampC gene in Enterobacterales in the Netherlands, which is consistent with the number of blaCMY-2-positive strains in our datasets.5,42 The CMY group, including blaCMY-2, is the most prevalent pampC gene.8 It should be noted that a higher prevalence of other pampC genes could influence algorithm outcomes. For example, blaACC will be omitted because it has a cefoxitin-susceptible phenotype.9 Moreover, blaDHA-1 was included in our panels and use of the decision tree model resulted in a lower discriminatory value for this pampC variant. So, our decision tree is probably most optimal in settings with relatively high amounts of blaCMY.
Strains from validation set 1 were only sequenced when D68C was positive for AmpC. Analyses of the MICs for D68C-negative samples illustrate that MICs of cefotaxime are <6 mg/L (Figure S5). Moreover, it seems unlikely that these strains would have contained pampC, as previous studies have shown high sensitivity and specificity with the D68C test for the detection of AmpC production.14,43
This present study focused on E. coli, being the most common and well-studied pathogen.1 Nonetheless, there are other species with inducible expression of campC, such as Enterobacter spp., Citrobacter freundii and Pseudomonas aeruginosa.8 Our study outcomes may not be extrapolated to these other species.
In conclusion, the use of a cefotaxime MIC test is an inexpensive and relatively quick method to detect pAmpC-producing E. coli. Therefore, the proposed decision tree could serve as a good alternative to EUCAST guidelines, which include cloxacillin synergy testing in combination with PCR. A comparison between the two algorithms in a clinical setting may be of interest for future studies.
WGS combined with machine learning algorithms is useful to improve laboratory and infection control methods.44,45 We used a simplified version of machine learning, which is directly applicable in current settings. Results show great potential for further optimization of present microbiological methods. Future work may use an extensive amount of data and state-of-the-art machine learning to improve accuracy of β-lactamase detection.
Supplementary Material
Acknowledgements
Many thanks to M. Janssens (Laboratory for Medical Microbiology and Immunology, Elisabeth-Tweesteden Hospital, Tilburg, the Netherlands) and S. van Leest (Laboratory for Microbiology, Microvida, location Bravis Hospital, the Netherlands) for providing samples that were included in validation set 1. Many thanks to K. T. Veldman and D. J. Mevius (Department of Bacteriology and Epidemiology, Wageningen Bioveterinary Research, Lelystad, the Netherlands), E. A. Reuland (Medical Microbiology and Infection Control, Amsterdam UMC location VUmc, Amsterdam, the Netherlands) and W. H. F. Goessens (Erasmus University Medical Center, Rotterdam, Netherlands) for providing samples that were included in validation set 2. Many thanks to R. Bosboom (Rijnstate, Arnhem, the Netherlands) and P. Vos (Check-Points, Wageningen, the Netherlands) for providing additional strains. We thank both H. W. Hoogewerf and L. van der Wiel for their input on and discussions about machine learning.
Funding
This research was partly funded by INTERREG health-i-care consortium 17 (European Union funding programme).
Transparency declarations
None to declare.
Supplementary data
Supplementary Materials and methods, Figures S1 to S5 and Tables S1 to S3 are available as Supplementary data at JAC Online.
References
- 1. Weinstein RA, Gaynes R, Edwards JR.. Overview of nosocomial infections caused by Gram-negative bacilli. Clin Infect Dis 2005; 41: 848–54. [DOI] [PubMed] [Google Scholar]
- 2. Pitout JDD. Extraintestinal pathogenic Escherichia coli: a combination of virulence with antibiotic resistance. Front Microbiol 2012; 3: 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Jørgensen RL, Nielsen JB, Friis-Møller A. et al. Prevalence and molecular characterization of clinical isolates of Escherichia coli expressing an AmpC phenotype. J Antimicrob Chemother 2010; 65: 460–4. [DOI] [PubMed] [Google Scholar]
- 4. Ding H, Yang Y, Lu Q. et al. The prevalence of plasmid-mediated AmpC β-lactamases among clinical isolates of Escherichia coli and Klebsiella pneumoniae from five children’s hospitals in China. Eur J Clin Microbiol Infect Dis 2008; 27: 915–21. [DOI] [PubMed] [Google Scholar]
- 5. Reuland EA, Halaby T, Hays JP. et al. Plasmid-mediated AmpC: prevalence in community-acquired isolates in Amsterdam, the Netherlands, and risk factors for carriage. PLoS One 2015; 10: e0113033.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Van Hoek A, Schouls L, Van Santen MG. et al. Molecular characteristics of extended-spectrum cephalosporin-resistant Enterobacteriaceae from humans in the community. PLoS One 2015; 10: e0129085.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Harris PNA, Ben Zakour NL, Roberts LW. et al. Whole genome analysis of cephalosporin-resistant Escherichia coli from bloodstream infections in Australia, New Zealand and Singapore: high prevalence of CMY-2 producers and ST131 carrying blaCTX-M-15 and blaCTX-M-27. J Antimicrob Chemother 2018; 73: 634–42. [DOI] [PubMed] [Google Scholar]
- 8. Jacoby GA. AmpC β-lactamases. Clin Microbiol Rev 2009; 22: 161–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Philippon A, Arlet G, Jacoby GA.. Plasmid-determined AmpC-type β-lactamases. Antimicrob Agents Chemother 2002; 46: 1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. San Millan A. Evolution of plasmid-mediated antibiotic resistance in the clinical context. Trends Microbiol 2018; 26: 978–85. [DOI] [PubMed] [Google Scholar]
- 11. Rozwandowicz M, Brouwer MSM, Fischer J. et al. Plasmids carrying antimicrobial resistance genes in Enterobacteriaceae. J Antimicrob Chemother 2018; 73: 1121–37. [DOI] [PubMed] [Google Scholar]
- 12. Australian Guidelines on Prevention and Control of Infection in Healthcare 2010. www.nhmrc.gov.au.
- 13.RCPI. Guidelines for the Prevention and Control of Multi-Drug Resistant Organisms (MDRO) Excluding MRSA in the Healthcare Setting 2014. http://www.hpsc.ie.
- 14. Ingram PR, Inglis TJJ, Vanzetti TR. et al. Comparison of methods for AmpC β-lactamase detection in Enterobacteriaceae. J Med Microbiol 2011; 60: 715–21. [DOI] [PubMed] [Google Scholar]
- 15. Javier Pérez-Pérez F, Hanson ND.. Detection of plasmid-mediated AmpC β-lactamase genes in clinical isolates by using multiplex PCR. J Clin Microbiol 2002; 40: 2153–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Martinez L, Simonsen GS.. EUCAST Guidelines for Detection of Resistance Mechanisms and Specific Resistances of Clinical and/or Epidemiological Importance, Version 2.0. 2017. http://www.eucast.org/fileadmin/src/media/PDFs/EUCAST_files/Resistance_mechanisms/EUCAST_detection_of_resistance_mechanisms_170711.pdf. [Google Scholar]
- 17. Polsfuss S, Bloemberg GV, Giger J. et al. Practical approach for reliable detection of AmpC β-lactamase-producing Enterobacteriaceae. J Clin Microbiol 2011; 49: 2798–803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Edquist P, Ringman M, Liljequist BO. et al. Phenotypic detection of plasmid-acquired AmpC in Escherichia coli—evaluation of screening criteria and performance of two commercial methods for the phenotypic confirmation of AmpC production. Eur J Clin Microbiol Infect Dis 2013; 32: 1205–10. [DOI] [PubMed] [Google Scholar]
- 19. Martin M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet J 2011; 17: 10–2. [Google Scholar]
- 20. Bankevich A, Nurk S, Antipov D. et al. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol 2012; 19: 455–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Seemann T. mlst https://github.com/tseemann/mlst.
- 22. Jolley KA, Maiden M.. BIGSdb: scalable analysis of bacterial genome variation at the population level. BMC Bioinformatics 2010; 11: 595.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Altschul SF, Gish W, Miller W. et al. Basic local alignment search tool. J Mol Biol 1990; 215: 403–10. [DOI] [PubMed] [Google Scholar]
- 24. Camacho C, Coulouris G, Avagyan V. et al. BLAST+: architecture and applications. BMC Bioinformatics 2009; 10: 421.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Seemann T. abricate https://github.com/tseemann/abricate.
- 26. Clausen P, Aarestrup FM, Lund O.. Rapid and precise alignment of raw reads against redundant databases with KMA. BMC Bioinformatics 2018; 19: 307.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Peter-Getzlaff S, Polsfuss S, Poledica M. et al. Detection of AmpC β-lactamase in Escherichia coli: comparison of three phenotypic confirmation assays and genetic analysis. J Clin Microbiol 2011; 49: 2924–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Larsson A. AliView: a fast and lightweight alignment viewer and editor for large datasets. Bioinformatics 2014; 30: 3276–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Caroff N, Espaze E, Gautreau D. et al. Analysis of the effects of −42 and −32 ampC promoter mutations in clinical isolates of Escherichia coli hyperproducing AmpC. J Antimicrob Chemother 2000; 45: 783–8. [DOI] [PubMed] [Google Scholar]
- 30. Tracz DM, Boyd DA, Hizon R. et al. ampC gene expression in promoter mutants of cefoxitin-resistant Escherichia coli clinical isolates. FEMS Microbiol Lett 2007; 270: 265–71. [DOI] [PubMed] [Google Scholar]
- 31. Breiman L, Friedman JH, Olshen RA. et al. Classification and Regression Trees. New York: Chapman and Hall, 1984. [Google Scholar]
- 32.R Core Team. R: A Language and Environment for Statistical Computing. Vienna: R Foundation for Statistical Computing, 2018. https://www.R-project.org/. [Google Scholar]
- 33. Aarestrup FM, Hasman H, Veldman K. et al. Evaluation of eight different cephalosporins for detection of cephalosporin resistance in Salmonella enterica and Escherichia coli. Microb Drug Resist 2010; 16: 253–61. [DOI] [PubMed] [Google Scholar]
- 34. Nordmann P, Poirel L, Nordmann P.. Extension of the hydrolysis spectrum of AmpC β-lactamase of Escherichia coli due to amino acid insertion in the H-10 helix. J Antimicrob Chemother 2007; 60: 490–4. [DOI] [PubMed] [Google Scholar]
- 35. Mammeri H, Galleni M, Nordmann P.. Role of the Ser-287-Asn replacement in the hydrolysis spectrum extension of AmpC β-lactamases in Escherichia coli. Antimicrob Agents Chemother 2009; 53: 323–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Martínez-Martínez L. Extended-spectrum β-lactamases and the permeability barrier. Clin Microbiol Infect 2008; 14: 82–9. [DOI] [PubMed] [Google Scholar]
- 37. Reisbig MD, Hossain A, Hanson ND.. Factors influencing gene expression and resistance for Gram-negative organisms expressing plasmid-encoded ampC genes of Enterobacter origin. J Antimicrob Chemother 2003; 51: 1141–51. [DOI] [PubMed] [Google Scholar]
- 38. Hanson ND. AmpC β-lactamases: what do we need to know for the future? J Antimicrob Chemother 2003; 52: 2–4. [DOI] [PubMed] [Google Scholar]
- 39. Doumith M, Day M, Ciesielczuk H. et al. Rapid identification of major Escherichia coli sequence types causing urinary tract and bloodstream infections. J Clin Microbiol 2015; 53: 160–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Miajlovic H, Aogáin M, Mac Collins CJ. et al. Characterization of Escherichia coli bloodstream isolates associated with mortality. J Med Microbiol 2016; 65: 71–9. [DOI] [PubMed] [Google Scholar]
- 41. Pietsch M, Irrgang A, Roschanski N. et al. Whole genome analyses of CMY-2-producing Escherichia coli isolates from humans, animals and food in Germany. BMC Genomics 2018; 19: 601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Den Drijver E, Verweij JJ, Verhulst C. et al. Decline in AmpC β-lactamase-producing Escherichia coli in a Dutch teaching hospital (2013-2016). PLoS One 2018; 13: e0204864.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Nourrisson C, Tan RN, Hennequin C. et al. The MAST® D68C test: an interesting tool for detecting extended-spectrum β-lactamase (ESBL)-producing Enterobacteriaceae. Eur J Clin Microbiol Infect Dis 2015; 34: 975–83. [DOI] [PubMed] [Google Scholar]
- 44. Quainoo S, Coolen JPM, van Hijum S. et al. Whole-genome sequencing of bacterial pathogens: the future of nosocomial outbreak analysis. Clin Microbiol Rev 2017; 30: 1015–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Nguyen M, Wesley Long S, McDermott PF. et al. Using machine learning to predict antimicrobial MICs and associated genomic features for nontyphoidal Salmonella. J Clin Microbiol 2019; 57: e01260-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.