Abstract
Background:
Paratuberculosis and tuberculosis (TB) caused by Mycobacterium avium paratuberculosis (MAP) and Mycobacterium tuberculosis complex (MTC), respectively are economically important, chronic debilitating diseases affecting the dairy herds and are also potential zoonotic threats.
Aims:
Differential diagnosis of paratuberculosis and TB in blood samples of cattle and buffaloes.
Methods:
In this study, an in-house developed multiplex polymerase chain reaction (PCR) targeting MAP, Mycobacterium bovis and Mycobacterium smegmatis was used in blood samples (buffy coat) parallel with IS900 PCR and esxB PCR for diagnosis of paratuberculosis and TB, respectively; in a total of 202 cattle and buffaloes.
Results:
Out of 202 animals, 12 (5.9%) and 17 (8.4%) animals were positive for MAP by multiplex PCR and IS900 PCR, respectively; from which only 8 (4%) animals were positive by both tests; whereas 4 and 9 animals were exclusively positive by multiplex PCR and IS900 PCR, respectively. None of the animals were found to be positive for M. bovis and M. smegmatis by the multiplex PCR. However, the esxB PCR detected 13 (6.4%) animals positive for TB. In fact, 3 (1.5%) animals were found to be co-infected by both paratuberculosis and TB.
Conclusion:
The in-house multiplex PCR detected MAP in buffy coat and there was a fair degree of agreement between the multiplex PCR and IS900 PCR in detection of MAP DNA though the latter detected more number of animals to be positive for MAP. Besides, esxB PCR showed a high diagnostic potential and can be used for diagnosis of TB from blood.
Key Words: Blood samples, Multiplex PCR, Paratuberculosis, Tuberculosis
Introduction
Paratuberculosis or Johne’s disease (JD), caused by Mycobacterium avium paratuberculosis (MAP), is a chronic and progressive enteritis of domestic and wild ruminants, and is endemic in cattle of developing countries (OIE, 2019a). Mycobacterium avium paratuberculosis has impact on food safety and is also associated with Crohn’s disease, which is a chronic inflammatory bowel disease in humans (Hermon-Taylor, 2009 ▶; Singh et al., 2010 ▶; Carvalho et al., 2012 ▶). Polymerase chain reaction (PCR) assays using primers specific for F57, ISMav2, ISMAP02, and ISMAP04 elements have been used for specific detection of MAP DNA. However, the higher number of copies of IS900 element in comparison to other IS elements makes IS900-based detection very sensitive (Singh et al., 2010 ▶; Carvalho et al., 2012 ▶).
Tuberculosis (TB) is also an infectious chronic debilitating disease of zoonotic importance, affecting a wide range of livestock, domestic and wild animals, besides humans (OIE, 2019b). It is caused by the pathogenic mycobacterial species, Mycobacterium tuberculosis complex (MTC) comprised of Mycobacterium bovis, M. tuberculosis, Mycobacterium africanum, Mycobacterium canneti, Mycobacterium microti, Mycobacterium pinnipedii, and Mycobacterium caprae (Olsen et al., 2010 ▶). Tuberculosis in milch animal is mainly caused by M. bovis (OIE, 2009 ▶). Molecular diagnosis of TB using esxB (CFP-10) PCR targeting esxB gene present in the region of difference 1 (RD1) region of the pathogenic mycobacterial species, is a fast diagnostic tool having higher sensitivity and specificity (Dikshit et al., 2012 ▶; Brahma et al., 2017 ▶).
Opportunistic mycobacterial infections due to environmental mycobacteria may also occur in immunocompromised animals. One such example is the Mycobacterium smegmatis, which is a saprophytic, rapid growing, atypical, non-tuberculous mycobacteria (NTM) (Quinn et al., 2011) and has the potential of causing opportunistic infection. Mycobacterium smegmatis has been reported from relapsing polygranulomatous mastitis in cattle (Siqueira et al., 2016 ▶) and was also detected in milk samples among other atypical mycobacteria, which may be a public health concern (Franco et al., 2013 ▶; Bolaños et al., 2017 ▶).
Therefore, compared to the traditional culture and microscopy method of diagnosis, the molecular diagnostic technique using Blood PCR is rapid and has a high potential for diagnosis of JD (Bhide et al., 2006 ▶; Singh et al., 2013 ▶), as well as TB (Zali et al., 2014 ▶), especially during the bacteremic stage in subclinical or clinical stage. Therefore, in this study an in-house developed multiplex-PCR which differentiates mycobacterial infections caused by MAP, M. bovis and M. smegmatis was used in blood samples (buffy coat) of cattle and buffaloes for differential diagnosis of JD, TB and M. smegmatis infection. Further, this multiplex PCR was similarly complemented by conventional IS900 PCR and esxB (CFP-10) PCR for comparison of the diagnostic potential for MAP and MTC, respectively.
Materials and Methods
Collection of blood samples and DNA extraction
A total of 202 animals (42 cattle and 160 buffaloes) aged 2 years and above were selected randomly from an organized dairy farm in Ludhiana. Whole blood samples were collected and buffy coat was obtained from 1 ml each of whole blood samples by centrifugation at 12,000 g for 10 min and then washed with phosphate-buffered saline (PBS) for 2-3 times by re-centrifugation until white pellet is obtained. The buffy coat was subjected to DNA extraction using QIAamp DNA blood mini kit (Qiagen). The DNA sample was stored at -20°C until further use.
In-house developed multiplex PCR
Multiplex PCR primers (Brahma et al., 2017) designed with the help of in-silico PCR targeting three mycobacterial species: MAP, M. bovis, and M. smegmatis (Table 1) were used.
Table 1.
Primer sequences for the in-house designed multiplex PCR
| Target organism and strain | Primer sequence | Location of primer gene sequence | Size of PCR product | |
|---|---|---|---|---|
|
M. avium subsp. paratuberculosis MAP4 CP005928.1 |
Forward | 5´- CGCGCGTACC TGACAAAAC - 3´ |
562055 - 562037 |
187 bp |
| Reverse | 5´- TCACCCTGAC ACTGACAGACA - 3´ |
561869 - 561889 |
||
|
M. bovis strain SP38 CP015773.1 |
Forward | 5´- GATGGTGGAA CACGACCACT - 3´ |
4138314 - 4138333 |
571 bp |
| Reverse | 5´- TTGATCGACC GTTCCGGTTT - 3´ |
4138865 - 4138884 |
||
|
M. smegmatis MC2 155 CP009494.1 |
Forward | 5´- ACCATGTCTAT CTCAGTGTGCT - 3´ |
3877883 - 3877904 |
628 bp |
| Reverse | 5´- ACGCTCGAGGT CCACTACAA - 3´ |
3878510 - 3878491 |
||
Brahma et al. (2017), assessed the sensitivity of the in-house multiplex primers by using ten-fold serial dilution of the standard DNA of M. tuberculosis (IMTECH, Chandigarh), MAP (GENEKAM, Germany) and M. Smegmatis (Microbiologics). Specificity of the primers were cross-tested individually against genomic DNA of M. tuberculosis, MAP, M. smegmatis, and NTM species (Mycobacterium fortuitum and Mycobacterium kansasii) and non-mycobacterial species (Brucella abortus, Pasteurella multocida and Escherichia coli).
The DNA extracted from the buffy coat was amplified by in-house multiplex PCR primers and PCR conditions were followed as per Brahma et al. (2017) ▶.
IS 900 PCR
The DNA extracted from the buffy coat was also amplified by MAP species specific PCR based on the insertion sequence IS900. The sequences are as follows: Forward (IS900/150C): 5´- CCG CTA ATT GAG AGA TGC GAT TGG - 3´ and Reverse (IS900/921): 5´- AAT CAA CTC CAG CAG CAG CGC GGC CTC G -3´ designed to amplify a 229 bp target sequence (Vary et al., 1990 ▶; Singh et al., 2008 ▶). Reaction volume and PCR cycling conditions were followed as per Brahma et al. (2017).
esx B (CFP-10) PCR
The primer sequences for CFP-10 were as follows: Forward: 5´-ATG GCA GAG ATG AAG ACC GAT GCC GCT-3´ and Reverse: 5´-TCA GAA GCC CAT TTG CGA GGA CAG CGC C-3´ (Dikshit et al., 2012 ▶) giving a product size of 302 bp. Sample DNA from buffy coat was amplified by esxB (CFP-10) PCR, for detection of MTC. Reaction volume and PCR cycling conditions were followed as per Brahma et al. (2017) with a little modification, i.e., out of total 25 µL reaction volume, the DNA template was increased from 6 µL to 8 µL and nuclease free water was reduced from 4.5 µL to 2.5 µL, so as to increase the chances of detection, whereas the other reagent compositions were unchanged.
Results
In-house developed multiplex PCR
The sensitivity of M. bovis, MAP, and M. smegmatis primers were as little as 170 fg/μL, 300 fg/μL, and 51 fg/μL of genomic DNA, respectively. None of the organisms other than the specific standard DNA had amplification at the standardised annealing temperature i.e. 65.5-68°C which clearly indicates the specificity of in-house multiplex PCR (Brahma et al., 2017 ▶).
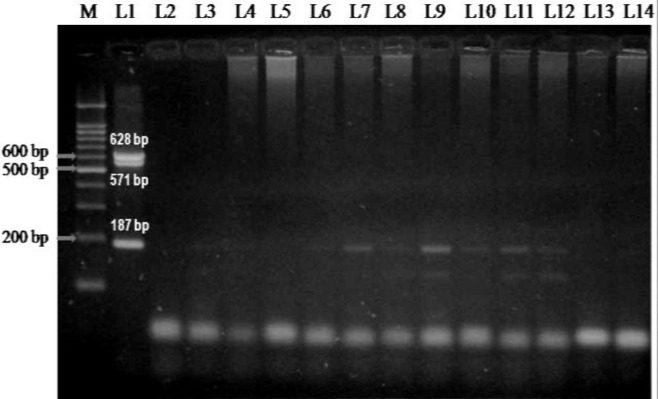
Out of 202 animals, 12 (5.9%) were found to be positive for MAP infection by in-house multiplex PCR (Fig. 1). However, none of the animals were found to be positive for M. bovis and M. smegmatis.
Fig. 1.
Amplification of DNA from the buffy coat using in-house developed multiplex primers. Lane M: 100 bp plus ladder. L1: Positive control (M. avium paratuberculosis, M. tuberculosis, and M. smegmatis), L2: Negative control, and L3-L14: Blood samples
IS 900 PCR
The analytical sensitivity of the IS900 PCR was upto 30 fg/µL of MAP DNA (GENEKAM, Germany) and no other bacteria other than the target species was detected indicating its specificity (Brahma et al., 2017 ▶).
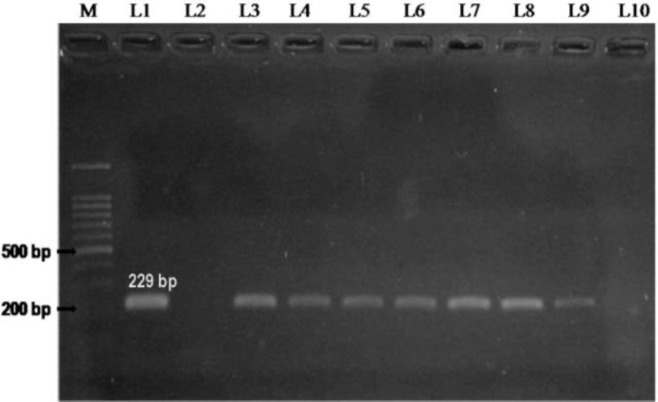
Out of a total 202 animals screened for JD, a total of 17 (8.4%) animals were positive by IS900 PCR (Fig. 2). However, only 8 (4%) out of 17 animals were positive by both the multiplex and IS900 PCR; whereas 4 and 9 animals were exclusively positive by multiplex PCR and IS900 PCR, respectively.
Fig. 2.
Amplification of DNA from the buffy coat using IS900 primers. Lane M: 100 bp plus ladder. L1: Positive control (M. avium paratuberculosis), L2: Negative control, and L3-L10: Blood samples
esx B (CFP-10) PCR
The analytical sensitivity of esxB (CFP-10) primers were upto 800 fg/µL of M. tuberculosis DNA (IMTECH, Chandigarh) and none of the NTM or other bacteria were detected (Brahma et al., 2017 ▶).
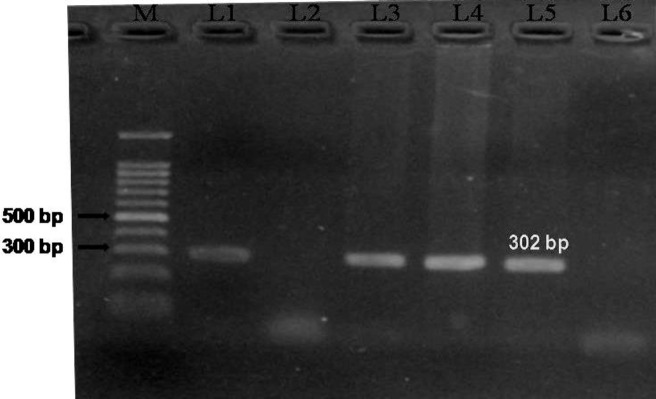
Out of the 202 animals screened for TB, only 13 (6.4%) animals were found to be positive for TB by esxB PCR (Fig. 3). In fact, the combined results of IS900 and esxB PCR revealed incidences of co-infection with both JD and TB in 3 animals which had debilitated health condition.
Fig. 3.
Amplification of DNA from the buffy coat using esxB (CFP-10) primers. Lane M: 100 bp plus ladder. L1-L4: Blood samples, L5: Positive control (M. tuberculosis), and L6: Negative control
Discussion
In this study, the in-house developed multiplex PCR detected MAP in blood samples though it did not detect M. bovis and M. smegmatis. In contrast, Brahma et al. (2017) ▶, using the same multiplex PCR detected both MAP and M. bovis from tissue samples, though the sensitivity for detection of M. bovis was comparatively less than the esxB PCR. Amplification of M. bovis in multiplex PCR may be inhibited in blood samples due to PCR inhibitors or it could be presence of DNA in undetectably low concentration or due to presence of low copy number of the target sequence. In other studies by different workers, the accuracy of multiplex PCR for differential detection of M. bovis and M. tuberculosis was found to be 100% in terms of specificity and could detect as little as 20 pg of genomic DNA (Shah et al., 2002 ▶; Bakshi et al., 2005 ▶).
IS900 PCR-based detection of MAP from white blood cells (WBCs) has also been reported by some workers (Naser et al., 2004 ▶; Stott et al., 2005 ▶). The reason for detection of MAP in more number of samples by IS900 PCR compared to the in-house developed multiplex PCR could be due to the higher copy number of IS900 element as reported by Vary et al. (1990) ▶ and Singh et al. (2010) ▶. However, using tissue samples, Brahma et al. (2017) ▶ detected MAP DNA in 2 out of 2 histopathologically JD positive cases by the in-house multiplex PCR.
In other studies, it has been found that MAP could be readily detected in blood from cattle with disseminated infections, and possibly in the blood of infected young and asymptomatic animals (Juste et al., 2005 ▶). Blood PCR targeting IS900 gene was rapid, highly sensitive, and specific for detecting MAP infection at any stage of infection in any age of goats (Singh et al., 2010 ▶). Mycobacterium avium paratuberculosis DNA was also detected in WBCs from about 11% of 262 Indian cattle of unspecified age or disease status (Bhide et al., 2006 ▶). Singh et al. (2013) ▶ found substantial agreement between “microscopy” and blood PCR detecting 39.3 and 13.1% positive by faecal microscopy and blood PCR, respectively; though Blood PCR detected more cases from heavy MAP shedders in microscopy. It may be assumed that low positivity in blood PCR may be due to presence of MAP in blood stream for a limited period (Singh et al., 2013 ▶). Gwozdz et al. (2000) ▶ contrarily showed poor performance of “blood PCR” to detect subclinically infected sheep. Barrington et al. (2003) ▶ had also recorded lower sensitivity of “blood-PCR” in comparison to PCR applied on milk, liver and faecal samples of advanced subclinically infected cows. The overall sensitivity of blood PCR for JD diagnosis from different studies was 11-66%. However, Youssef et al. (2014) ▶, reported that, out of 150 faecal samples, 31.33%, 29.33%, and 42.66% were found positive whereas MAP DNA was detected in only 4.67%, 3.33%, and 5.33% of milk samples by conventional IS900 PCR, conventional F57 PCR and IS900 real-time (RT) PCR, respectively. Gümüşsoy et al. (2015) ▶, in PCR of milk and stool samples, detected MAP DNA in 20 (13.61%) and 42 (28.57%) samples, respectively. However, MAP is only shed via milk in a small proportion of cows with subclinical JD for a limited period of time (Khol et al., 2013 ▶) and the shedding of MAP in faeces and milk is not synchronized (Gao et al., 2009 ▶).
Several PCR systems have also been developed for the diagnosis of TB, e.g., PCR amplification of esxA and esxB genes targeting ESAT-6 and CFP-10 proteins, respectively present in pathogenic mycobacterial species can be used for detection of M. tuberculosis as well as M. bovis (Pinxteren et al., 2000 ▶; Rogerson et al., 2006 ▶; Dikhsit et al., 2012 ▶). Using esxB PCR on tissue samples Brahma et al. (2017) ▶ detected all 7 out of 7 histopathologically TB positive cases, indicating its efficiency in TB diagnosis. Zali et al. (2014) ▶, using JB21 and JB22 primers specific for M. bovis, detected M. bovis positive in 8% of blood and lymph node samples of cattle. In this study, PCR targeting esxB (CFP-10) gene in blood samples detected 13 (6.4%) TB positive animals which were not detected by the in-house multiplex PCR, thus, indicating the early diagnostic potential of esxB (CFP-10) PCR. After all, 3 of the TB infected animals found to be positive for JD indicates presence of co-infection with both the diseases within the herd. Infection of either JD or TB may make the animals more susceptible to co-infection due to the immuno-compromised condition (Brahma et al., 2017 ▶).
In conclusion, “Blood PCR” using buffy coat was rapid, highly sensitive and specific, and the combination of various PCR techniques increased the chances of successful diagnosis of both JD and TB infection in cattle and buffaloes. The use of multiplex-PCR was able to detect MAP and differentiate from other mycobacterial infections in blood samples, thus providing a rapid diagnosis and its confirmation for the specific disease of concern, JD. However, the probable limitation is that, a large-scale study is required to determine whether this PCR assay is adequate for JD and TB control program.
Acknowledgements
The authors are very grateful to DBT (Department of Biotechnology), Government of India for providing funds for the present work through a Project (BT/PR5776/MED/30/928/2012) and to the Director of Research, Guru Angad Dev Veterinary and Animal Sciences University, Ludhiana for providing the necessary facilities.
References
- Bakshi CS, Shah DH, Verma R, Singh RK, Malik M. Rapid differentiation of Mycobacterium bovis and Mycobacterium tuberculosis based on a 127-kb fragment by a single tube multiplex-PCR. Vet. Microbiol. 2005;109:211–216. doi: 10.1016/j.vetmic.2005.05.015. [DOI] [PubMed] [Google Scholar]
- Barrington GM, Gay JM, Eriks IS, Davis WC, Evermann JF, Emerson C, O’Rourke JL, Hamilton MJ, Bradway DS. Temporal patterns of diagnostic results in serial samples from cattle with advanced paratuberculosis infections. J. Vet. Diagn. Invest. 2003;15:195–200. doi: 10.1177/104063870301500219. [DOI] [PubMed] [Google Scholar]
- Bhide M, Chakurkar E, Tkacikova L, Barbudhe S, Novak M, Mikula I. IS900-PCR-based detection and characterization of Mycobacterium avium subsp paratuberculosis from buffy coat of cattle and sheep. Vet. Microbiol. 2006;112:33–41. doi: 10.1016/j.vetmic.2005.10.004. [DOI] [PubMed] [Google Scholar]
- Bolaños CAD, de Paula CL, Guerra ST, Franco MMJ, Ribeiro MG. Diagnosis of mycobacteria in bovine milk: an overview. Rev. Inst. Med. Trop. São Paulo. 2017;59:e40. doi: 10.1590/S1678-9946201759040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brahma D, Narang D, Chandra M, Gupta K, Singh A, Kaur G. Diagnosis of Mycobacterial infections (Tuberculosis and Paratuberculosis) in tissue samples using molecular (inhouse multiplex PCR, PCR and TaqMan real-time PCR), histopathology and immuno-histochemical techniques. Trop. Biomed. 2017;34:1–17. [PubMed] [Google Scholar]
- Carvalho IA, Vinicius CEB, Souza IM, Zanardo LG, Filho JDR, Gomes MJP, Moreira MAS. Diagnosis of Paratuberculosis in cattle: microbiological culture, serology and PCR. Braz. J. Microbiol. 2012;43:581–585. doi: 10.1590/S1517-83822012000200020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dikshit M, Sharma RJ, Adsool AD, Chaphalkar SR. ESAT-6 and CFP-10 proteins of Mycobacteriumtuberculosis in making diagnostic tool for TB. J. Biotechnol. Lett. 2012;3:28–30. [Google Scholar]
- Franco MM, Paes AC, Ribeiro MG, Pantoja JC, Santos AC, Miyata M, Leite CQ, Motta RG, Listoni FJ. Occurrence of mycobacteria in bovine milk samples from both individual and collective bulk tanks at farms and informal markets in the southeast region of Sao Paulo, Brazil. BMC Vet. Res. 2013;9:85. doi: 10.1186/1746-6148-9-85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao A, Odumeru J, Raymond M, Hendrick S, Duffield T, Mutharia L. Comparison of milk culture, direct and nested polymerase chain reaction (PCR) with fecal culture based on samples from dairy herds infected with Mycobacterium avium subsp paratuberculosis. Can. J. Vet. Res. 2009;73:58–64. [PMC free article] [PubMed] [Google Scholar]
- Gümüşsoy KS, İça T, Abay S, Aydin F, Hizlisoy H. Serological and molecular diagnosis of paratuberculosis in dairy cattle. Turk. J. Vet. Anim. Sci. 2015;39:147–153. [Google Scholar]
- Gwozdz JM, Thompson KG, Manktelow BW, Murray A, West DM. Vaccination against para-tuberculosis of lambs already infected experimentally with Mycobacterium avium subspecies paratuberculosis. Aust. Vet. J. 2000;78:560–566. doi: 10.1111/j.1751-0813.2000.tb11902.x. [DOI] [PubMed] [Google Scholar]
- Hermon-Taylor J. Mycobacterium avium subspecies paratuberculosis, Crohn’s disease and the Doomsday scenario. Gut Pathog. 2009;1:15. doi: 10.1186/1757-4749-1-15. doi: 10.1186/1757-4749-1-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Juste RA, Garrido JM, Geijo MV, Elguezabal N, Aduriz G, Atxaerandio R, Sevilla I. Comparison of blood PCR and ELISA for detection of Mycobacterium avium subsp paratuberculosis infection in sheep and cattle. J. Vet. Diagn. Invest. 2005;17:354–359. doi: 10.1177/104063870501700409. [DOI] [PubMed] [Google Scholar]
- Khol JL, Wassertheurer M, Sodoma E, Revilla-Fernández S, Damoser J, Österreicher E, Dünser M, Kleb U, Baumgartner W. Long-term detection of Mycobacterium avium subspecies para-tuberculosis in individual and bulk tank milk from a dairy herd with a low prevalence of Johne’s disease. J. Dairy Sci. 2013;96:3517–3524. doi: 10.3168/jds.2012-6466. [DOI] [PubMed] [Google Scholar]
- Naser SA, Ghobrial G, Romero C, Valentine JF. Culture of Mycobacterium avium subspecies paratuberculosis from the blood of patients with Crohn’s disease. Lancet. 2004;364:1039–1044. doi: 10.1016/S0140-6736(04)17058-X. [DOI] [PubMed] [Google Scholar]
- OIE. Chap. 2.4.6: Bovine tuberculosis. Terrestrial Manual. Paris, France: World Organization for Animal Health; 2009. [Google Scholar]
- Olsen I, Barletta RG, Thoen CO. Mycobacterium. In: Gyles CL, Prescott JF, editors. Pathogenesis of bacterial infections in animals. 4th Edn. Ames, Iowa: Wiley-Blackwell; 2010. pp. 113–132. [Google Scholar]
- Pinxteren LAHV, Ravn P, Agger EM, Pollock J, Andersen P. Diagnosis of tuberculosis based on the two specific antigens ESAT-6 and CFP-10. Clin. Diagn. Lab. Immunol. 2000;7:155–160. doi: 10.1128/cdli.7.2.155-160.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogerson BJ, Jung YJ, LaCourse R, Ryan L, Enright N, North RJ. Expression levels of Mycobacterium tuberculosis antigen encoding genes versus production levels of antigen specific T cells during stationary level lung infection in mice. Immunology. 2006;118:195–201. doi: 10.1111/j.1365-2567.2006.02355.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shah DH, Verma R, Bakshi CS, Singh RK. A multiplex-PCR for the differentiation of Mycobacterium bovis and Mycobacterium tuberculosis. FEMS Microbiol. Lett. 2002;214:39–43. doi: 10.1111/j.1574-6968.2002.tb11322.x. [DOI] [PubMed] [Google Scholar]
- Singh SV, Singh PK, Gupta S, Chaubey KK, Singh B, Kumar A, Singh AV, Kumar N. Comparison of microscopy and blood-PCR for the diagnosis of clinical Johne’s disease in domestic ruminants. Iran. J. Vet. Res. 2013;14:345–349. [Google Scholar]
- Singh PK, Singh SV, Kumar H, Sohal JS, Singh AV. Diagnostic application of IS900 PCR using blood as a source sample for the detection of Mycobacterium avium subspecies paratuberculosis in early and subclinical cases of caprine paratuberculosis. Vet. Med. Int. 2010 doi: 10.4061/2010/748621. Article ID 748621, 8 pages. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh SV, Singh AV, Singh R, Sharma S, Shukla N, Mishra S, Singh PK, Sohal JS, Kumar H, Patil PK, Misra P, Sandhu KS. Sero-prevalence of bovine Johne’s disease in buffaloes and cattle population of north India using indigenous ELISA kit based on native Mycobacterium avium subsp paratuberculosis ‘Bison type’ genotype of goat origin. Comp. Immunol. Microbiol. Infect. Dis. 2008;31:419–433. doi: 10.1016/j.cimid.2007.06.002. [DOI] [PubMed] [Google Scholar]
- Siqueira FM, Lopes CE, Snell GG, Gomes MJP. Identification of Mycobacterium smegmatis in bovine Mastitis. Acta Sci. Vet. 2016;44(Suppl. 1):166. [Google Scholar]
- Stott AW, Jones GM, Humphry RW, Gunn GJ. Financial incentive to control paratuberculosis (Johne’s disease) on dairy farms in the United Kingdom. Vet. Rec. 2005;156:825–831. doi: 10.1136/vr.156.26.825. [DOI] [PubMed] [Google Scholar]
- Vary PH, Anderson PR, Green E, Taylor JH, McFadden JJ. Use of highly specific DNA probes and the polymerase chain reaction to detect Mycobacterium paratuberculosis in Johne’s disease. J. Clin. Microbiol. 1990;28:933–937. doi: 10.1128/jcm.28.5.933-937.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Youssef DGS, Sallam FA, Darwish SF, Amin AS. Evaluation of conventional and real-time PCR assays for molecular diagnosis of Johne’s disease in dairy cattle. Int. J. Curr. Microbiol. App. Sci. 2014;3:969–981. [Google Scholar]
- Zali MHS, Farajnia F, Yahyapour H, Moslemzadeh T, Hashempour A. Detection of Mycobacterium bovis in cattle suspected to tuberculosis by PCR method in Urmia-Iran. BEPLS. 2014;3:152–157. [Google Scholar]