Abstract
Microtubule nucleation is controlled by the γ-tubulin ring complex (γTuRC) and related γ-tubulin complexes, providing spatial and temporal control over the initiation of microtubule growth. Recent structural work has shed light on the mechanism of γTuRC-based microtubule nucleation, confirming the long-standing hypothesis that it functions as a microtubule template. Crystallographic analysis of the first non-γ-tubulin γTuRC component (GCP4) has resulted in a new appreciation of the relationships among all γTuRC proteins, leading to a refined model of their organization and function. The structures have also suggested an unexpected mechanism for regulating γTuRC activity via conformational modulation of the complex component GCP3. New experiments on γTuRC localization extend these insights, suggesting a direct link between attachment at specific cellular sites and activation.
Introduction
The microtubule cytoskeleton is critically important for both the spatial and temporal organization of eukaryotic cells, playing a central role in functions as diverse as intracellular transport, organelle positioning, motility, signaling, and cell division. The ability to play this variety of roles requires microtubules to be arranged in complex arrays capable of rapid reorganization. Microtubules themselves are highly dynamic polymers that switch between cycles of growth and depolymerization, and cells have evolved a variety of ways to manipulate the basic polymer dynamics to achieve precise control of the organization and reorganization of the microtubule cytoskeleton. While many different mechanisms are used to regulate microtubule dynamics, at a fundamental level the cell achieves control by manipulating the rates of microtubule assembly and microtubule catastrophe, as well as the timing and location of the nucleation events that give rise to new microtubules.
Microtubules are hollow tubes of about 250 Å in diameter that are assembled from α–β-tubulin heterodimers in a GTP-dependent manner (Fig. 1a). The tubulin subunits make two types of filament contacts: longitudinal contacts run the length of the microtubule forming protofilaments, and lateral contacts between protofilaments (generally α-tubulin to α-tubulin and β-tubulin to β-tubulin) form the circumference of the microtubule1, 2. Microtubule geometry is not fixed, however; the more flexible lateral contacts can accommodate between 11 and 16 protofilaments3, yielding microtubules of different diameter when assembled in vitro from purified tubulin4. In vivo, though, almost all microtubules have thirteen protofilaments5–7, suggesting that one level of cellular control involves defining a unique microtubule geometry. Thirteen-fold symmetry is likely preferred because it is the only geometry in which protofilaments run straight along the microtubule length as opposed to twisting around the microtubule, allowing processively tracking motor proteins to always remain on the same face of the structure. An unusual feature of thirteen-protofilament microtubules is that, as a consequence of their helical symmetry, a “seam” is formed from lateral α-tubulin–β-tubulin interactions8, 9, which are generally presumed to be weaker than α–α or β–β tubulin lateral contacts. The mechanism by which cells ensure thirteen protofilament geometry has long been a mystery.
Figure 1. Microtubule assembly.
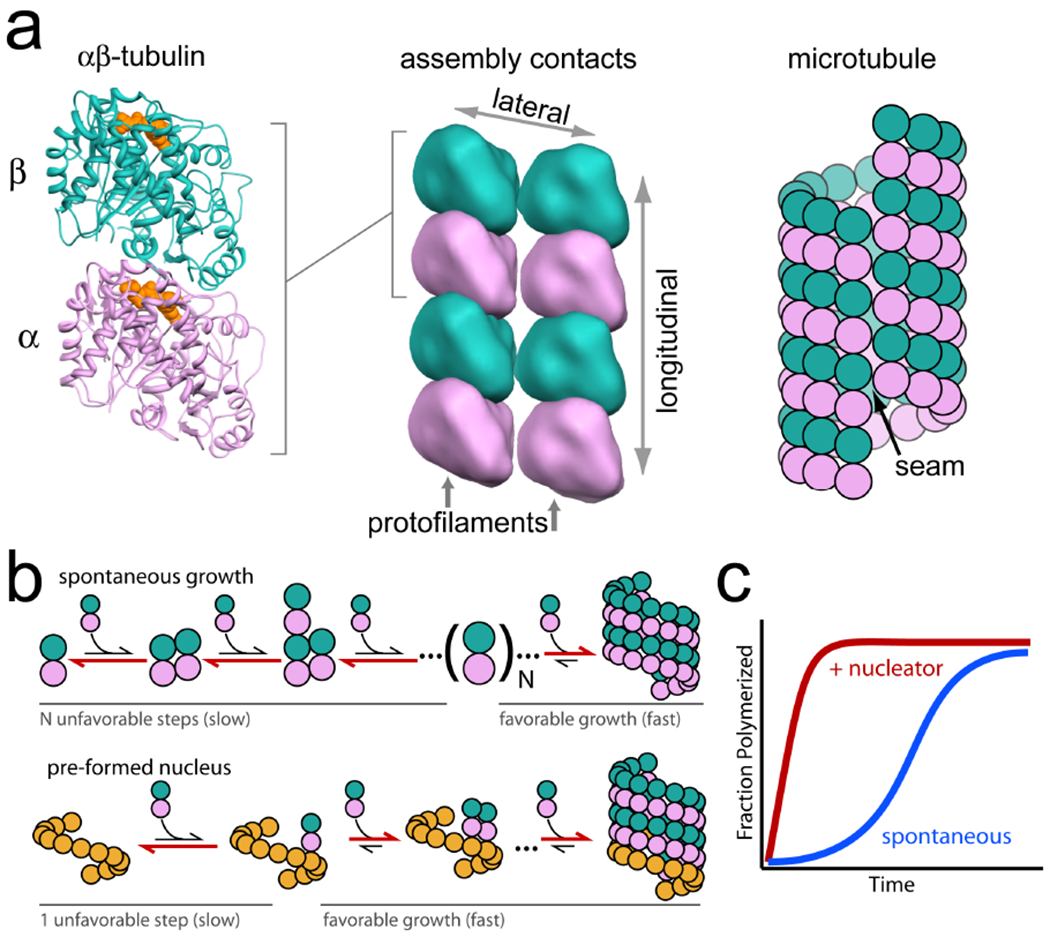
a) The αβ-tubulin heterodimer is the fundamental repeating subunit of microtubules. When bound to GTP (indicated in orange, left panel) heterodimers come together through two types of contacts (indicated by double-headed arrows): GTP-mediated longitudinal contacts between α-tubulin and β-tubulin that form protofilaments, and lateral α-tubulin to α-tubulin and β-tubulin to β-tubulin contacts that form between protofilaments. The addition of tubulin subunits to this lattice yields the hollow microtubule. In 13-protofilament microtubules a ‘seam’ is formed as a result of lateral α-tubulin-β-tubulin interactions. b) Spontaneous microtubule growth in vitro occurs in two stages: a relatively slow phase through unstable early assembly intermediates, and a rapid elongation phase. In early steps the assembly energetics favor disassembly over assembly, but after a sufficiently large oligomer (denoted here by N) is formed, assembly is energetically favored and elongation proceeds rapidly. Pre-formed nuclei allow microtubule growth to bypass the slow phase, providing spatial and temporal control over new microtubule growth (Adapted from Ref. 10). c) In bulk assembly assays, the presence of a nucleator causes rapid polymerization, bypassing the lag phase observed during spontaneous growth.
Another key difference between microtubule assembly in vivo and in vitro is with regard to how new microtubules are initiated. In vitro, microtubule growth must proceed through small, early assembly intermediates, in which disassembly is energetically favored over assembly to result in slow initial growth10. After a sufficiently large oligomer has been achieved, microtubule growth becomes energetically favorable and the addition of tubulin heterodimers proceeds rapidly (Fig. 1b). Significantly, rather than relying on the spontaneous initiation of new microtubules, cells have evolved specialized nucleation sites in vivo that bypass the early, slower growth phase. These nucleation sites are largely found at microtubule organizing centers (MTOCs).
More than a century ago the centrosome was identified as the primary MTOC in animal cells11. The centrosome, organized around a pair of centrioles, serves as the central anchor point for microtubules within the cell, defining a polar microtubule array12. In fungi the functional analog of the centrosome is the spindle pole body, a large multilayered structure embedded in the nuclear envelope that nucleates microtubules on both the cytoplasmic and nuclear faces13. Plants, on the other hand, have no centrosome equivalent, but nevertheless have highly organized acentrosomal microtubule arrays14.
Despite the variation in MTOC morphology, they all rely on γ-tubulin, a homolog of α- tubulin and β-tubulin, for nucleating microtubules. γ-tubulin was first discovered in Aspergillus nidulans genetic screens as a suppressor of a β-tubulin mutation15, and subsequently found localized at all MTOCs16–21. Purification of γ-tubulin from animal and yeast cells showed it to be part of larger complexes, which can directly nucleate microtubule growth in vitro22–26. γ-Tubulin is essential for normal microtubule organization in every organism in which it has been studied, and it is nearly ubiquitous throughout eukaryotes. Moreover, it is also involved in nucleation from non-MTOC sites within cells, such as the chromosome-mediated nucleation pathway27, and in plants28, which lack centrosome-like structures, suggesting that it is critical for the initiation of all new microtubules in vivo.
In this Review we focus on recent advances in our understanding of the mechanism of γ-tubulin based microtubule nucleation. We begin with a brief review of the components of γ-tubulin complexes and previous models for their assembly and mechanism of nucleation. We then describe recent structures of key components that lead us to a new model for the organization of γ-tubulin complexes. We also explore the growing body of work on γ-tubulin complex localization, which increasingly appears to be linked with regulation of nucleating activity.
γTuSC and γTuRC nucleating complexes
Early biochemical characterization of γ-tubulin showed that it was part of larger complexes that did not include α-tubulin or β-tubulin. When γ-tubulin was purified from Drosophila melanogaster embryos or Xenopus laevis eggs it was found to be part of a ~2.2 MDa complex with at least six other proteins, GCP2-6 (GCP: γ-tubulin complex protein) and NEDD1. The complex had a striking ring shape in electron micrographs, leading to the name γ-tubulin ring complex (γTuRC)24 γTuRC dissociates under high salt conditions to yield a stable 300 kDa subcomplex of γ-tubulin associated with two other proteins (GCP2 and GCP3), which is dubbed the γ-tubulin small complex (γTuSC)29 (Box1). Importantly, purified γTuSC has a much lower microtubule nucleating activity than the intact γTuRC29, suggesting that the assembly state of γ-tubulin is important in determining its activity.
Box 1. γ-tubulin complex proteins and prior models for their assembly and action.
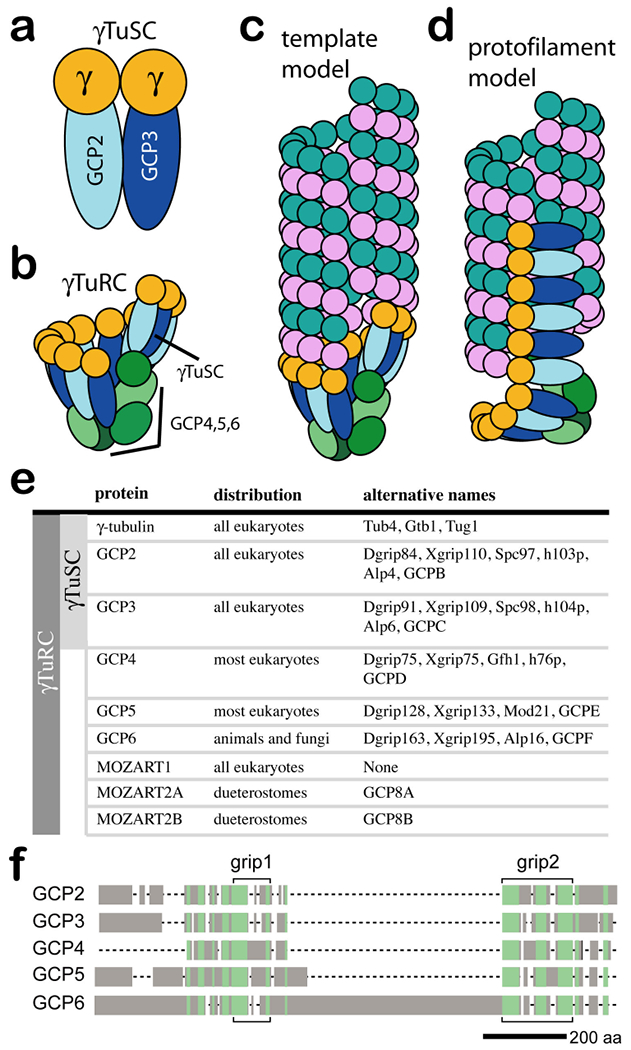
The γ-tubulin small complex (γTuSC) is the conserved, essential core of the microtubule nucleating machinery, and it is found in nearly all eukaryotes. γTuSC has two copies of γ-tubulin and one each of γ-tubulin complex protein 2 (GCP2) and GCP3 (see the figure, part a). In many eukaryotes, multiple γTuSCs assemble with GCP4, GCP5 and GCP6 into the γ-tubulin ring complex (γTuRC) (see the figure, part b). Previous models of γTuRC assembly suggested that GCP4, GCP5 and GCP6 together function as a cap-like scaffold for arranging multiple γTuSCs into a distinctive ring shape. This view depicts a model with six γTuSCs (12 γ-tubulins), which would leave a gap in the template, owing to the fact that microtubules are made up of 13 profilaments. The most widely accepted model for the mechanism of γTuRC-based nucleation, the template model, suggests that γTuRC acts as a template, presenting a ring of γ-tubulins that make longitudinal contacts with α-tubulin–β-tubulin (see the figure, part c). The protofilament model, on the other hand, suggests that the γTuRC unfurls to present a γ-tubulin protofilament, which would nucleate through lateral contacts with αβ-tubulin (see the figure, part d). A complete list of proteins that are thought to be part of γTuSC and γTuRC, including the more recently identified MOZART1 and MOZART2 proteins, are listed in part e of the figure, along with alternative names for each protein. The five GCPs share regions of homology, although with very low levels of sequence identity (as low as 15% identity between different GCP families). Two homologous regions, grip1 and grip2, initially defined the homology82 (see the figure, part f). Regions of more distant homology were later shown to be more widely dispersed in the GCP sequences32, 50 (green shading in part f of the figure).
Saccharomyces cerevisiae and closely related yeast are unusual as they appear to have lost all of the γTuRC-specific components, retaining only γTuSC25, 26, 30. This supports the view that γTuSC is the core of the nucleating machinery, sufficient in itself for proper microtubule organization. The apparent simplicity of the budding yeast γ-tubulin complex has made it an attractive model for elucidating the mechanisms of microtubule nucleation. And yet, an apparent contradiction has remained unresolved: budding yeast have only the weakly-nucleating γTuSC, yet are perfectly capable of nucleating microtubules.
The GCP family of γ-tubulin complex components.
In addition to γ-tubulin itself, microtubule nucleating complexes include a family of five homologous γ-tubulin complex proteins (GCPs)31–33 (Box 1). γTuSC consists of two copies of γ-tubulin and one each of GCP2 and GCP3. γTuRC is composed of multiple copies of γTuSC plus GCP4, GCP5 and GCP6. GCP2 and GCP3 are found in almost all eukaryotes and are essential for proper microtubule organization, suggesting that they form the core of the nucleating machinery. Most eukaryotes also possess GCP4 and GCP5, while GCP6 appears to be a recent addition in the animal and fungal lineages.
Although they constitute a unique family of homologous proteins, the overall sequence identity between GCPs is quite low (less than 15% identity overall in most comparisons between GCP groups). Homology has only been confidently predicted in two short segments, the grip1 and grip2 motifs31, which are unique to the GCPs. Almost nothing has been known about the specific functions of these motifs, although it was speculated that they might participate in conserved protein-protein interactions32. The overall size of GCPs varies more than two-fold (ranging from ~70–210 kDa), with numerous insertions and/or deletions, suggesting different functionality for each family member. Outside of the grip1 and grip2 motifs that define the GCP family, none of the members has any other identifiable motifs conserved with other protein families.
It is important to note that the various γ-tubulin complex components were initially described by different researchers in different organisms, leading to an at times confusing litany of names used for homologous proteins. Here, we have adopted the generic GCP designation33 for GCP2–6 and prefer to limit its use to this family to indicate their common evolutionary origin. Box 1 includes a list of the different names that have been used for each component.
Non-GCP family components of γTuRC.
Recently, two small proteins with no homology to the GCP family — MOZART1 and MOZART2 — were described as integral γTuRC components in human cell lines34, 35. It appears that, due to their small size, these proteins were overlooked in earlier γTuRC pull-down experiments. When either protein is immunoprecipitated from cells it is found in complex with all of the γTuRC components. MOZART1, which is found in most eukaryotes, appears to play a role in γTuRC recruitment to MTOCs. MOZART2A and MOZART2B, found only in the deuterostome lineage (that is, echinoderms, chordates, hemichordates and xenoturbellida), are specifically involved in γTuRC recruitment to interphase centrosomes but do not seem to play a role in γTuRC assembly. NEDD1 also frequently copurifies with γTuRC, but does not appear to be an integral component of the complex. Rather, it is now clear that NEDD1 is a localization factor, important for both centrosomal and non-MTOC localization of γTuRC, for example within the mitotic spindle36–38.
All of the core γTuRC components have been identified through co-purification, but it should be noted that a large number of proteins co-precipitate with γTuRC at lower stoichiometries. Many of these interacting proteins may be factors that help γTuRC attach to the MTOC, or play transient roles in γTuRC regulation. However, given the recent experience with MOZART1 and MOZART2, it would not be surprising to find that our list of γTuRC components is incomplete, with additional integral γTuRC components yet to be discovered.
Stoichiometry of γTuRC components.
The precise stoichiometry of γTuRC components remains unclear. A study in human cells showed that the complex contains multiple copies of the γTuSC components and GCP4, but only a single copy of GCP5 (no determination could be made about the copy number of GCP6)32. A more recent study has quantified the ratio of components in human γTuRC from gels of purified complex, and estimated the stoichiometry of the complex to be 14 γ-tubulins, 12 copies of GCP2 or GCP3, 2-3 copies of GCP4, and a single copy of GPC539. However, this quantification should be viewed as preliminary, as GCP6 was present at less than one copy per γTuRC, raising the possibility of heterogeneity in the sample. Interestingly, the stoichiometry inferred in this study has more γ-tubulins than GCP2 and GCP3, suggesting a small portion of γ-tubulin in γTuRC is not directly incorporated into γTuSCs.
γTuRC assembly and action: old models
It has been assumed that γ-tubulin nucleates by forming oligomers that mimic an early assembly intermediate of αβ-tubulin, with either lateral or longitudinal microtubule-like lattice contacts between γ-tubulins. Nucleation should then proceed through direct interactions of γ-tubulin with αβ-tubulin through lattice-like contacts. Generating models for the arrangement of γ-tubulin within γTuRC, and for the mechanism of γ-tubulin-based microtubule nucleation, are therefore two aspects of the same problem. Lines of evidence from structural and biochemical studies have provided some insight into both problems.
Imaging of γTuRC by electron microscopy — both two-dimensional images24, 29 and a low-resolution three-dimensional structure40 — revealed a unique lock-washer shape with repeating subunits around the circumference and a diameter and helical pitch similar to a microtubule. γTuSCs were proposed to form the repeating wall of the ring. An apparent cap-like feature at the base of γTuRC, seen in the low-resolution structure, was thought to be formed from GCP4–6. Given its position, the asymmetric cap was predicted to act as a scaffold for arranging γTuSCs into a defined ring shape (Box 1).
In vitro, γTuRC was shown to interact specifically with microtubule minus ends where it functions as a cap to prevent microtubule growth in the minus direction41. This was consistent with electron micrograph images showing closed structures at the ends of microtubules, whether nucleated by γTuRCs in vitro40–42or attached to MTOCs in vivo43. Synthesis of these data led to the ‘template model’, which suggests that the γ-tubulins in γTuRC function as a microtubule template, making lateral contacts with each other around the ring and longitudinal contacts with α-tubulin (Figure 2b,c).
Figure 2. The structure of γTuSC.
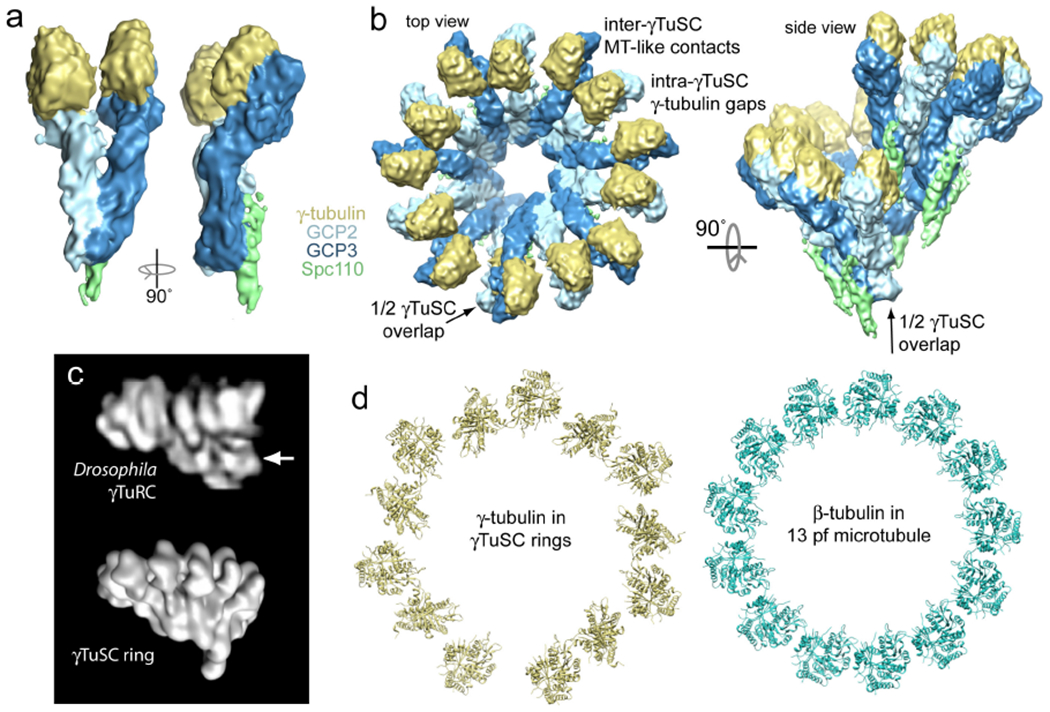
a) The 8 Å cryo-electron microscopy (EM) structure of S. cerevisiae γTuSC bound to the attachment factor Spc110 is shown. This γTuSC is a single subunit of a large γTuSC oligomer (see panel b). In this view, the N-termini of GCP2 and GCP3 are at bottom, with their C-terminal domains near the top interacting with γ-tubulin. In the structure, the two γ-tubulins are held separated from each other in a configuration incompatible with the microtubule lattice, which partially explains the relatively low nucleating capacity of free γTuSC relative to γTuRC. b) Top-down and side views of the γTuSC ring are shown. The ring has six and a half γTuSCs per turn, which arise due to a half γTuSC overlap between the first and seventh subunits in the ring (see side view). This yields thirteen γ-tubulins per turn, matching the in vivo microtubule protofilament number. The conformation of γTuSC is unchanged in the ring structure, such that the intra-γTuSC gap between γ-tubulins remains. However, microtubule-like lateral interactions are observed between γ-tubulins at the inter-γTuSC interface (Ref. 49). c) The low-resolution negative stain EM reconstruction of a single Drosophila melanogaster γTuRC (top, Ref. 40) closely resembles the γTuSC ring shown in panel b, rendered here at lower resolution for comparison (bottom). The region of γTuRC originally interpreted as a GCP4,5,6 cap is indicated with an arrow; this region appears to correspond to the N-terminal regions of GCP2 and GCP3 instead. d) Comparison of γ-tubulin positions in γTuSC rings and αβ-tubulin in the microtubule shows a mismatch in geometry, with alternating contacts and gaps in the γ-tubulin arrangement (Ref. 49).
While the model is compelling in its simplicity, the experimental data were insufficient to define the specific number of γTuSCs in the ring, leading to questions as to how the pairs of γ-tubulins within the γTuSCs could nucleate microtubules with an odd number of protofilaments. Two possibilities were generally offered: six γTuSCs (twelve γ-tubulins) might form an incomplete ring, leaving a gap at the location of the thirteenth protofilament, or seven γTuSCs (fourteen γ-tubulins) could form a ring with one extra γ-tubulin that does not interact with the microtubule.
An alternative hypothesis – the ‘protofilament model’ - was proposed early on, in which γ-tubulins would make longitudinal contacts with each other around the ring44, 45. This seemed reasonable, a priori, as longitudinal contacts are much stronger than lateral contacts, and rings of longitudinally-interacting tubulin or its bacterial homolog FtsZ have been observed. Moreover, electron micrographs of γTuRCs indicated that the structure might be quite flexible, suggesting it could potentially unfurl to present a single protofilament of γ-tubulins that would nucleate through lateral contacts with α-tubulin and β-tubulin. However, the weight of evidence now strongly supports the template model.
Although a template mechanism of nucleation has been the dominant model for γTuRC function for over a decade, it has remained unproven, and several important questions have persisted. What is the mode of interaction (lateral or longitudinal) between γ-tubulin and αβ-tubulin? Why is γ-tubulin nucleating capacity weaker in γTuSC than in γTuRC, and how does S. cerevisiae, which only has γTuSC, efficiently nucleate microtubules? How are 13-protofilament microtubules nucleated when γ-tubulins enter the complex in pairs through γTuSC? And, finally, what are the structural and functional roles of the non γ-tubulin components of γTuRC? Several recent advances have provided insight into these questions, generating a more complete framework for understanding γ-tubulin based microtubule nucleation.
Structural insight into γTuRC function
A thorough, mechanistic understanding of microtubule nucleation by γ-tubulin will require a high-resolution structural model of γTuRC. This is a daunting task. The large size and compositional complexity of γTuRC have made it a challenging target for recombinant expression, and to date only small quantities of heterogeneous material have been purified from native sources (for example, D. melanogaster embryos29, X. laevis eggs24, and human cell lines32). An alternative strategy that has recently borne fruit has been to determine high-resolution structures of individual γTuRC components by crystallography and electron microscopy, and integrate these into a model of γTuRC.
γ-tubulin crystal structure.
The crystal structure of monomeric human γ-tubulin was determined bound to GTP and to GDP10, 46. γ-tubulin is very similar to α-tubulin and β-tubulin in its overall fold, consistent with the expectation that it is capable of making lattice-like contacts with the microtubule. Small differences on the microtubule lattice surfaces may give rise to differences in γ-tubulin interaction affinities at those sites, influencing the strength of γ-tubulin-γ-tubulin assembly interactions or γ-tubulin-microtubule interactions. Importantly, in the two γ-tubulin crystal forms the individual γ-tubulins make lateral contacts with the same contact region used by αβ-tubulin in microtubule lateral interactions, suggesting that this is their preferred mode of interaction. The crystal packing provided support for the template model of microtubule nucleation, which predicts lateral interactions between γ-tubulins and longitudinal interactions between γ-tubulin and αβ-tubulin.
The structure of γTuSC.
The structure of free S. cerevisiae γTuSC was initially determined at 25 Å by negative stain single particle electron microscopy (EM)47 (its V-shaped structure was later confirmed at higher resolution by cryo-EM (see below)), and the subunit arrangement and the orientations of GCP2 and GCP3 in the structure were determined by direct labeling experiments48. The arms of the V-shaped structure are composed of GCP2 and GCP3, which have similar overall shapes and dimerize through their N-terminal domains at the base of the V. The tips of the V contain g-tubulin, which interacts with C-terminal domains of GCP2 and GCP3 (Figure 2a). Surprisingly, the two γ-tubulins in the structure are held separate from each other, not making the anticipated lateral contacts required to match the microtubule lattice. This mismatch provides a partial explanation for the weaker nucleating activity of free γTuSC — each γ-tubulin remains totally independent, rather than forming a microtubule-like assembly intermediate that could facilitate microtubule assembly. Thus, the structure of γTuSC suggests that it is in an ‘off’ state, which raises the possibility of regulation at the level of γTuSC conformation.
γTuSC assembles with microtubule-like symmetry.
Purified S. cerevisiae γTuSC has a weak tendency to spontaneously assemble in vitro into ring-shaped structures that closely resemble γTuRC49. The ring assemblies are only formed under a narrow range of buffer conditions, and their heterogeneity and instability made them an extremely challenging subject for structure determination. However, it was discovered that copurification of γTuSC with the N-terminal domain of the S. cerevisiae attachment factor Spc110 (which links the γTuSC complex to the core of the spindle pole body) dramatically stabilizes γTuSC assembly. So much so that, when associated with Spc110, γTuSC rings continue to grow, yielding extended helical filaments of laterally associated γTuSCs that are very well suited to cryo-EM reconstruction. The 8 Å structure of this γTuSC filament provided a breakthrough in our understanding of γTuSC assembly, with important implications for the mechanism of microtubule nucleation49.
The most striking feature of the γTuSC oligomer structure is that there are 6 Ɖ γTuSCs per helical turn, due to a half-subunit overlap between the first and seventh subunits (Figure 3b). This gives thirteen γ-tubulins per turn, matching the in vivo microtubule protofilament number, with a helical pitch very similar to a microtubule. There is remarkable similarity between a single ring of γTuSC and the low-resolution structure of γTuRC, strongly suggesting that γTuSC assemblies like these constitute the core of γTuRC (Figure 2c). This finding also resolved the paradox of how budding yeast efficiently nucleate microtubules with only γTuSC — they can form γTuRC-like structures from γTuSC alone.
Figure 3. The GCP4 crystal structure defines the core structure of all the GCPs.
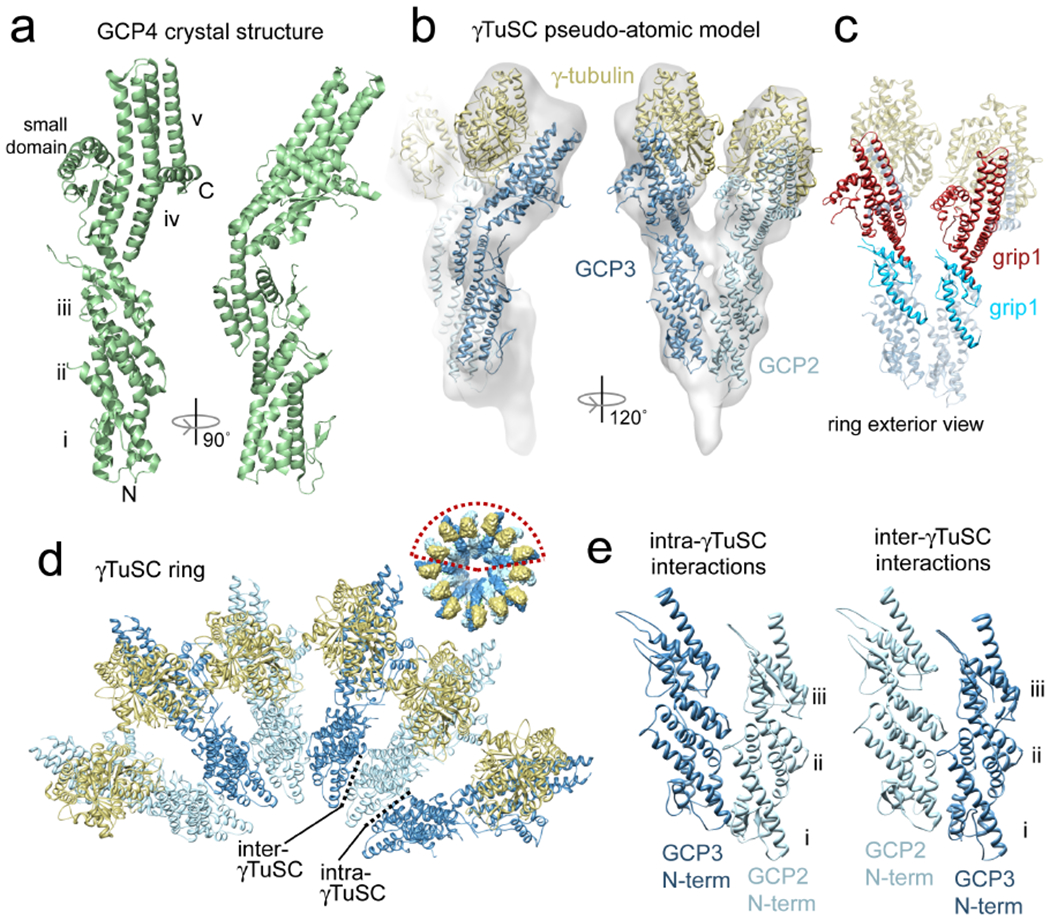
a) The γ-tubulin complex protein 4 (GCP4) crystal structure is shown in two orthogonal views. In the view on the left the five α-helical bundles (i-v), small domain labeled, and N-terminus and C-terminus are labeled. The C-terminal domain, consisting of bundle iv, bundle v and the small domain was shown to directly bind γ-tubulin. b) A pseudo-atomic model of γTuSC. The γ-tubulin crystal structure (gold) and the GCP4 crystal structure (blue) as a stand in for GCP2and GCP3 were fit into the γTuSC cryo-EM reconstruction (semi-transparent surface). The model reveals interaction surfaces between complex components. c) The model also shows the positions of the conserved grip1 and grip2 domains in GCP2 and GCP3 in the context of the full γTuSC. Grip2 is clearly involved in γ-tublin binding. The role of grip1 is more ambiguous; it forms part of the lateral contact surfaces between γTuSCs, as well as part of the faces of GCP2 and GCP3 that are exposed on the outside of the γTuSC ring. d) When the pseudo-atomic model from panel b is fit into the cryo-EM structure of the γTuSC ring (inset), it also reveals the surfaces of GCP2 and GCP3 that are important for oligomerization. γTuSCs interact with each other primarily through the sides of bundles i and ii (Ref. 50). e) The N-terminal domains of GCP2 and GCP3 are shown making intra- and inter-γTuSC contacts, with helical bundles i-iii labeled. Equivalent surfaces of the N-terminal domains of GCP2 and GCP3 are involved in both intra-γTuSC and inter-γTuSC interactions, indicating that a single assembly rule determines the organization of the ring structure. However, the affinities have been modulated such that the stronger intra-γTuSC interactions yield a stable complex, while the weaker inter-γTuSC interactions allow the assembly of γTuSCs into rings to be reversible.
The increased resolution of the γTuSC subunit allowed the precise orientation of each γ-tubulin to be determined. Both γ-tubulin minus ends are buried in the interaction surface with GCP2 and GCP3 and their lateral surfaces are all facing adjacent γ-tubulins. Moreover, each plus end is fully exposed, strongly suggesting that this surface interacts via longitudinal contacts with the minus ends of αβ-tubulin. The combination of the γ-tubulin geometry and its orientation provides the strongest evidence to date that γ-tubulin complexes function as microtubule templates. Indeed, the γTuSC rings likely provide the constraint that ensures thirteen protofilament microtubules in vivo. It is important to note that the thirteen-fold architecture of the oligomer is defined almost entirely by the conformations of, and interactions between, GCP2 and GCP3, with only minor contacts between γ-tubulins within the ring. The problem of how an odd-protofilament geometry can be templated from a complex with an even number of subunits is also now resolved — the half-γTuSC overlap ensures that, at most, thirteen γ-tubulins are exposed for interaction with αβ-tubulin.
While the symmetry of γ-tubulin in γTuSC rings is similar to microtubule symmetry, it is not a perfect match. There are no major conformational changes to the individual γTuSCs upon oligomerization; the two γ-tubulins within each γTuSC are still held apart. However, contacts between γ-tubulins of adjacent γTuSCs in the ring are nearly identical in both their spacing and relative orientation to microtubule lateral interactions, giving rise to an alternating pattern around the ring of contacting γ-tubulin pairs separated by gaps (Figure 2d). It is important to note that the relative orientation of the γ-tubulins in the ring is determined primarily by interactions between GCP2 and GCP3, which have far greater surface areas in contact than the γ-tubulins.
The nucleating activity of the Spc110-stabilized oligomers was only slightly greater than the heterogeneous γTuSC rings assembled in the absence of Spc11049, and both had much lower nucleation levels than have been reported for γTuRC29. However, under conditions in which γTuSC remains monomeric its nucleating activity was completely eliminated, suggesting that assembly of γTuSCs is required even for low levels of nucleation activity49. The imprecise match between the γ-tubulin geometry and microtubule geometry explains the modest levels of microtubule nucleation observed from the γTuSC oligomers, which likely arises just from the pairs of properly spaced γ-tubulins between γTuSCs.
GCP4 crystal structure: a model for the GCP family.
A major advance toward the full understanding of γ-tubulin complexes was achieved recently by the determination of the crystal structure of human GCP450. GCP4 has a unique fold, forming an elongated structure from five α-helical bundles with a pronounced kink between the third and fourth bundle, and a small domain flanking the fourth and fifth bundles (Figure 3a). The crystal structure itself is incomplete, as it is missing several large loops due to their inherent flexibility. Nonetheless, GCP4 fits remarkably well into the γTuSC cryo-EM structure in the positions of GCP2 and GCP3, with only small adjustments necessary in the bend angle between the third and fourth helical bundles. The remarkably good match between GCP4 and GCP2 and GCP3 demonstrates an unexpectedly strong conservation of the overall fold of the GCP family proteins. Previously, sequence homology had only been identified in the short grip1 and grip2 motifs of the GCP family proteins31–33 (Box 1), but the structural similarity of GCP2 and GCP3 to GCP4 prompted a reexamination of sequence similarity. Using the GCP4 crystal structure and predicted secondary structures of the remaining GCPs as guides, a more accurate alignment of the entire family was possible, showing small islands of sequence conservation scattered throughout the proteins. The regions of strongest conservation were predominantly buried in the protein, defining a structural core, with highly variable loop regions allowing for numerous insertions and/or deletions. GCP4 is the shortest of the GCPs, being almost entirely composed of homologous regions. The strong conservation of the overall fold between GCP4 and GCP2 and GCP3, along with the more expansive sequence homology now evident, allows us to use GCP4 as a model for the core of all the other GCPs.
This work also demonstrated a direct interaction with high affinity between GCP4 and γ-tubulin, showing not only structural but functional conservation in the GCP family. The binding activity of GCP4 was localized within its C-terminal domain, which is precisely the region juxtaposed to γ-tubulin when GCP4 and γ-tubulin are fit into the γTuSC cryo-EM structure49. This is also consistent with the direct labeling experiments that showed the C-termini of GCP2 and GCP3 interact with γ-tubulin48. Indeed, the surfaces involved in γ-tubulin binding are among the most conserved in the GCP family, and include the grip2 motif. Earlier work with the D. melanogaster proteins had also suggested that γ-tubulin binds directly to GCP5 and GCP636. The conservation of sequence and structure suggests that all of the GCPs directly bind γ-tubulin; as explored more fully below, this has important implications for understanding γTuRC organization.
A pseudo-atomic model of γTuSC.
Using the GCP4 crystal structure as a template, homology models of GCP2 and GCP3 were generated and fit into the γTuSC cryo-EM structure, along with the crystal structure of γ-tubulin, to create a pseudo-atomic model of γTuSC50 (Figure 3b). The γTuSC model predicts the surfaces involved in γ-tubulin–GCP2 and GCP3 interactions. The model also reveals the positions of the gripl and grip2 motifs, and suggest functions which were previously unknown (Figure 3c). The grip2 motif is clearly involved in the γ-tubulin binding surface, consistent with in vitro binding experiments with GCP4 and γ-tubulin. The role of gripl is less clear; it forms part of the lateral interaction surfaces suggesting it plays a role in γTuSC assembly, but also forms part of the surface of GCP2 and GCP3 exposed on the outer surface of the ring, suggesting it may be a binding site for other proteins that interact with γTuSC.
The pseudo-atomic model of γTuSC also provides insight into the nature of assembly contacts in γTuSC oligomers (Figure 3d). The intra- and inter-γTuSC interactions between GCP2 and GCP3 are very similar— essentially the interactions along the base of a γTuSC ring are the same all the way around, and primarily involve contacts between helical bundles i and ii (Figure 3e). There appears to be a single assembly rule guiding interactions between GCP2 and GCP3, whether within or between γTuSCs. Changes at these interaction surfaces appear to have tuned the affinities to give very strong binding to hold together individual γTuSCs, but weaker interactions driving their reversible assembly into γTuSC rings.
Conformational regulation of γTuSC
The mismatch between the γ-tubulins in γTuSC rings and microtubule geometry was interpreted as an “off” state of γTuSC, in which the nucleating complex is fully assembled but conformationally inactivated49. However, the γ-tubulins were arranged such that small movements could realign them into microtubule-like contacts (Figure 4a) The key to conformational activation may lie in the inherent flexibility of GCP3, observed as a hinge-like motion in negative stain EM reconstructions47 (Figure 4b). GCP4 was predicted by normal mode analysis to have a flex point at the position equivalent to the GCP3 hinge50 (Figure 4c). The GCP4 crystal structure provides a detailed view of the hinge point, allowing for a more precise model of the observed flexibility in GCP3, which appears to rely on rearrangement of hydrophobic interactions between the domains on either side of the hinge. Using the geometry of the thirteen-protofilament microtubule as a guide, we have developed a model for γTuSC activation in which GCP3 straightens at its hinge point. This rearrangement in GCP3 is sufficient to bring the two γ-tubulins in γTuSC into the exact microtubule lattice spacing49 (Figure 4d). In the context of the γTuSC ring, straightening of GCP3 to close the gap between each pair of intra-γTuSC γ-tubulins would create a perfect template for microtubule assembly49 (Figure 4e).
Figure 4. A model for the conformational activation of γTuSC.
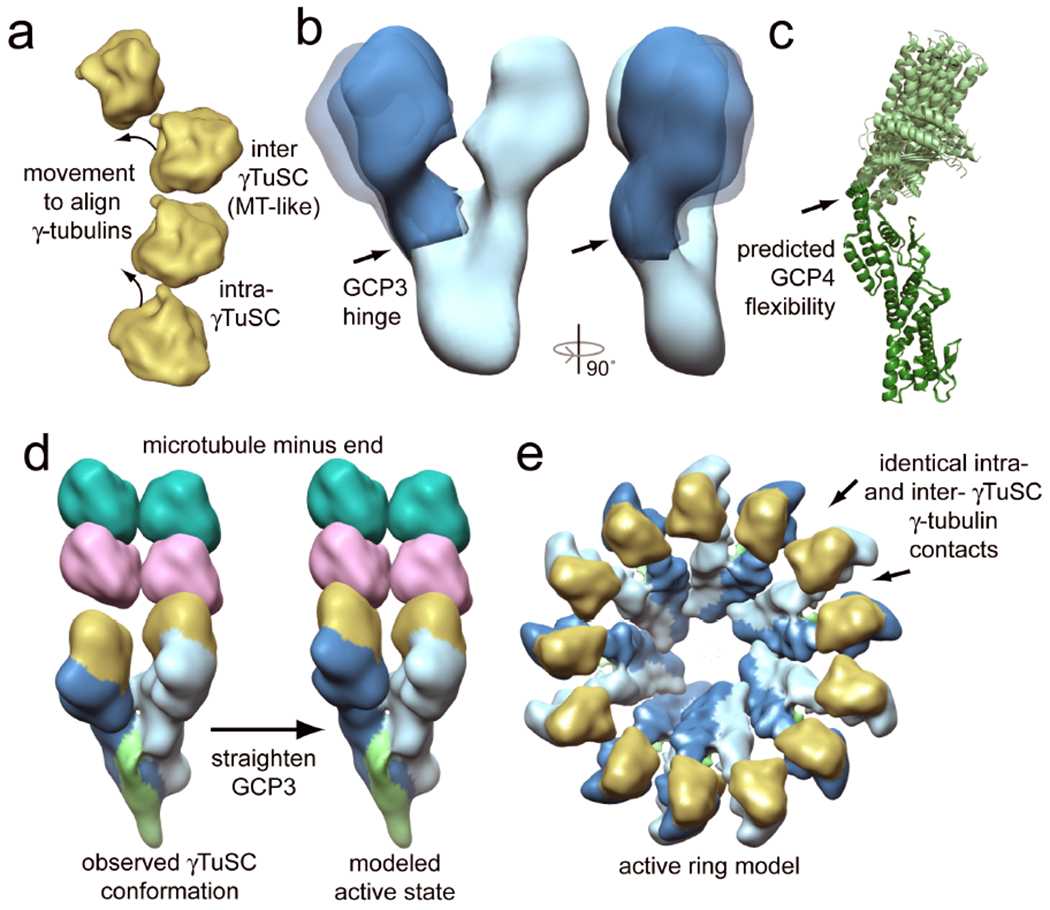
a) The γ-tubulins of two adjacent γTuSCs from the γTuSC ring are shown in a top-down view. The inter-γTuSC contact is the same as a microtubule lateral contact, but the intra-γTuSC arrangement does not match the microtubule lattice. Arrows indicate the approximate motions that would align the intra-γTuSC contacts to match the microtubule lattice. b) The negative stain electron microscopy (EM) reconstruction of free γTuSC revealed flexibility at a hinge point in GCP3, resulting in varying distances between the two γ-tubulins. (Ref. 47) c) Normal mode analysis of the GCP4 crystal structure predicts flexibility at the indicated position, near the equivalent hinge point in GCP3. This suggests conservation of flexibility in the GCPs (Ref. 50). d) A model for the conformational activation of γTuSC through the straightening of GCP3. In the observed conformation the two γ-tubulins are held apart, so that they can’t both be making contacts with the microtubule. However, straightening at the GCP3 hinge point by 23° would close the intra-γTuSC–γ-tubulin gaps, bringing all of the γ-tubulins in the ring to microtubule lattice like spacing (Ref. 49). e) In this modeled state, γ-tubulin in the ring would adopt perfect thirteen-protofilament microtubule geometry, serving as a potent microtubule nucleator.
This model remains to be tested to determine whether such a conformational change in GCP3 is possible, and if so what might mediate the rearrangement. One possible mechanism is post-translational modification of γTuSC components; indeed, all three of the γTuSC components are phosphorylated at different points during the cell cycle by different kinases, including Cdk1 and Mps151–53. Another possibility is that the conformation is changed through allosteric interactions with γTuSC-binding proteins. Although less likely, nucleotide binding and hydrolysis by γ-tubulin may also play a role in regulating the conformation of the complex.
Another possibility is that the predicted conformational change occurs only after microtubule growth has begun. That is, perhaps pairs of protofilaments begin to grow from the properly-spaced γ-tubulins between γTuSCs, and lateral association of the nascent protofilaments drives straightening of GCP3. Regulation might then be achieved by modification of the stiffness of the GCP3 hinge. However, growth in this way would seem to be much less favorable than growth from a properly-formed γ-tubulin nucleus with the correct geometry, and would function more as a minus-end anchor than as a nucleator.
Conformational regulation of nucleating activity is not an entirely new concept. A very similar mechanism is at play in actin nucleation by the Arp2/3 complex. In this case, the nucleating complex is assembled with the actin homologs Arp2 and Arp3 held separated from each other54. The complex is then activated by a structural rearrangement that brings Arp2 and Arp3 together with F-actin like contacts, creating a nucleus for actin filament growth55, 56. It is striking that evolution appears to have converged on similar mechanisms for regulating nucleation activity in these two very different filament systems.
A new model of γTuRC assembly
The recent progress in understanding γ-tubulin complex structures has led us directly to a revised γTuRC model. As described above, previous models of γTuRC assembly posited a repeating ring of γTuSC organized by a scaffolding cap composed of GCP4, GCP5 and GCP6 (Box 1). The roles of GCP4, GCP5 and GCP6 in our model of γTuRC assembly must be revisited in light of several important findings. First, γTuSC spontaneously assembles ring structures with microtubule-like symmetry without GCP4, GCP5 and GCP6 (Fig. 2), negating the necessity of a scaffolding role for these three proteins. Second, the overall structure and ability to bind γ-tubulin is conserved in GCP2, GCP3 and GCP4 (Fig. 3), suggesting that all of the GCPs directly bind γ-tubulin. Third, a single GCP assembly rule appears to define interactions between GCPs (Fig. 4e), suggesting that all of the GCPs assemble into γTuRC through equivalent conserved surfaces.
Structural roles of GCP4, GCP5 and GCP6.
In light of these findings, we propose a new model for γTuRC structure in which GCP4, GCP5 and GCP6 are incorporated directly into the ring structure, each binding directly to γ-tubulin (Figure 5). This model nicely explains why the observed ratio of γ-tubulin to GCP2 and GCP3 is greater than one39. Based on the γTuSC ring structure, the region at the base of the earlier γTuRC structure, which was originally interpreted as a scaffolding cap, appears to consist of the N-terminal regions of the GCPs (Figure 2c). Indeed, the similarity between the γTuRC structures and the γTuSC ring structure is quite striking, suggesting that the entire γTuRC consists of a ring of γTuSC-like structures.
Figure 5. A revised model of γTuRC assembly.
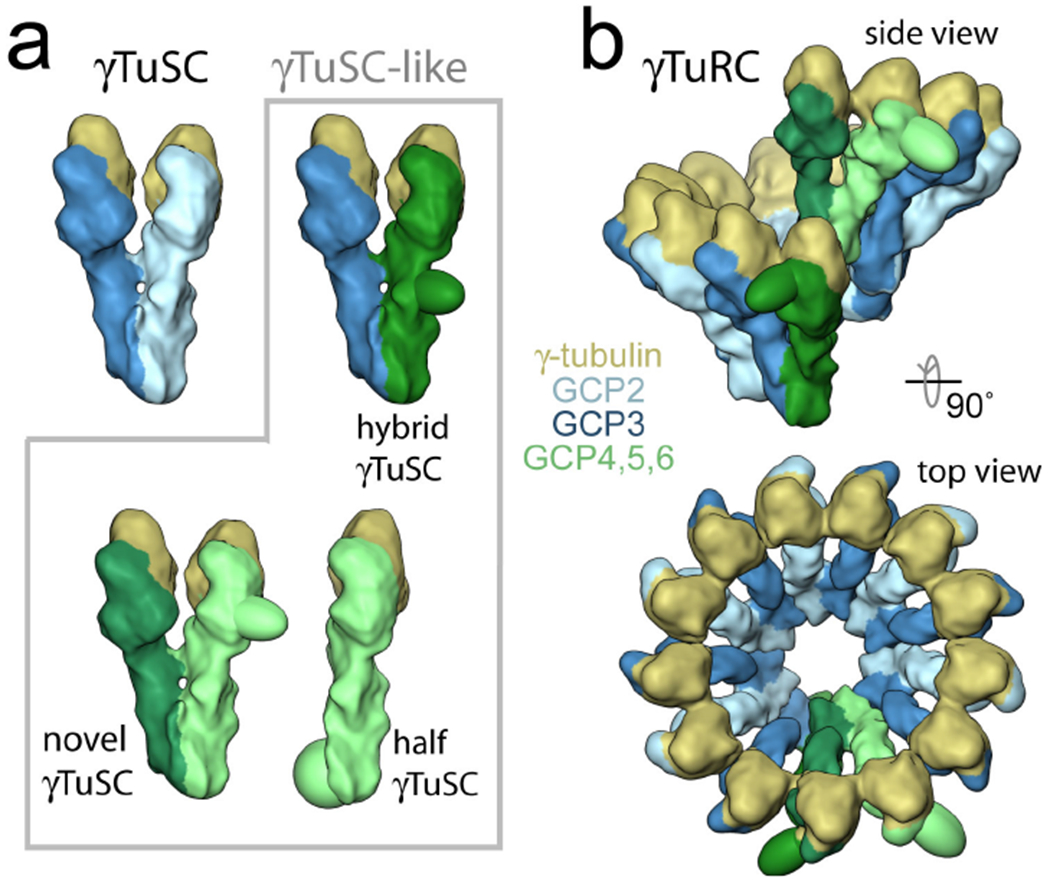
a) The overall structure and ability to bind γ-tubulin are conserved between the γTuRC-specific GCP4 and the γTuSC components GCP2 and GCP3, suggesting that all GCP-family members act as γTuSC-like components. The γTuRC-specific GCPs (GCP4, GCP5 and GCP6, shown in green) may function in one of three γTuSC-like complexes: as γ-tubulin-binding half γTuSCs, as hybrid γTuSCs with GCP2 or GCP3 (shown in blue), or interacting with each other to form novel γTuSC-like complexes. b) Through conserved lateral interactions, GCP4, GCP5 and GCP6 could be directly incorporated into the ring structure, as opposed to forming a cap structure as in previous models (see Box 1). While GCP4, GCP5 and GCP6 might incorporate at any position within the ring, it is most attractive to think of them interacting at the ends, where they might function to initiate or terminate ring formation and to stabilize the ring at the overlap (Ref. 49).
In the model GCP4, GCP5 and GCP6 interact with each other, and with GCP2 and GCP3, via the lateral GCP assembly rule. One can imagine GCP4, GCP5 and GCP6 acting as γTuSC-like complexes in one of three modes: as half γTuSCs with a single GCP binding one γ-tubulin; as hybrid γTuSCs, where a γTuRC-specific GCP replaces GCP2 or GCP3 in the γTuSC; or as completely novel γTuSCs composed of two γTuRC-specific GCPs (Figure 5a). Different GCPs may assemble through different modes. High-resolution homology modeling of the other GCPs based on the GCP4 crystal structure may prove useful in determining which GCPs directly interact with each other, as well as the potential limitations on assembly interactions at some surfaces (that is, inserts at some positions near lateral interaction surfaces might be predicted to interfere with further assembly in that direction). γ-Tubulin bound GCP4, GCP5 and GCP6 could then substitute for γTuSC GCPs within the ring by the GCP assembly rule (Figure 5b).
The positions of GCP4, GCP5 and GCP6 within the ring are unclear. While they could potentially insert at any position in the ring, some indirect evidence suggests that the three interact directly with each other. Loss of any one of GCP4, GCP5 or GCP6 destabilizes γTuRCs57–61, suggesting that these GCPs function as a unit to stabilize a well-defined ring. Studies in Aspergillus nidulans59 and Schizosaccharomyces pombe62 have also demonstrated a hierarchical localization dependence for GCP4, GCP5 and GCP6, suggesting that they directly interact with each other in γTuRC. In our view, the best place to position GCP4, GCP5 and GCP6 would be at the ends of the ring where the half-γTuSC overlap occurs. In this location they could efficiently initiate or terminate γTuSC assembly and could stabilize the ring by interacting with each other across the overlap. By interacting with each other at the ends of the ring, GCP4, GCP5 and GCP6 would also be able to define a single ring structure, as opposed to the elongated helical filaments that can be formed from γTuSC alone.
The structure of γTuSC oligomers did not reveal how many γTuSCs are required to form a functional microtubule nucleation site – it was consistent with both previous models, with either twelve γ-tubulins and a gap or fourteen γ-tubulins and an overlap. A consequence of our model, with GCP4, GCP5 and GCP6 at opposite ends of the ring but interacting with each other, is the prediction that γTuRC will have an overlap, allowing GCP4, GCP5 and GCP6 to be close enough to interact while also ensuring a well-defined ring.
In the model, GCP4, GCP5 and GCP6 define the position of the microtubule seam, where αβ-tubulin lateral interactions occur; at this position, a single lateral interaction would be formed between γ-tubulin and α-tubulin. Direct stabilization of the weaker α-tubulin to β-tubulin lateral contacts at the seam could potentially play a role in the nucleation mechanism of γTuRC. It should also be noted that the γTuRC model is only consistent with nucleation of a B-lattice configuration (α-tubulin–α-tubulin and β-tubulin-β-tubulin lateral interactions, with the exception of the seam, as depicted in Figure 1a) and not with an A-lattice configuration (α-tubulin-β-tubulin lateral interactions at each site in the microtubule).
While the overall structure and γ-tubulin binding function of the GCP family proteins are conserved, there remains a great deal of variation within the family, largely in the form of multiple insertions/deletions within the sequences (Box 1). These regions are likely responsible for unique functionality of the GCPs, and could serve to alter assembly interactions to ensure incorporation at unique sites within the ring, and to act as unique attachment sites to confer γTuRC-specific localization.
Roles of GCPs in localization.
A clear distinction exists between the γTuRC components that are required for its centrosomal and spindle localization. Depletion of either GCP2 or GCP3 from D. melanogaster S2 cells eliminates the localization of γ-tubulin at centrosomes and spindles and results in gross abnormalities in microtubule organization. However, depletion of GCP4, GCP5 and GCP6 — either singly or all three simultaneously — eliminates the spindle, but not centrosomal, localization of γ-tubulin in S2 cells as well as in the yeast A. nidulans57, 59, 63. Surprisingly, the GCP4, GCP5 and GCP6 depleted cells are still able to nucleate microtubules from the centrosome and to assemble mitotic spindles. This is perhaps less puzzling in light of the ability of γTuSC to assemble ring structures without GCP4, GCP5 and GCP649. These rings, while less stable without GCP4, GCP5 and GCP6, would then be bound to the centrosome through γTuSC-specific attachment, where they could nucleate microtubules.
γTuRC attachment and activation
In animal cells the majority of γTuRC (80%) is soluble in the cytoplasm64. However, its nucleating activity seems to be limited to specific locations in the cell, such as the centrosome or spindle pole body, or within the mitotic spindle. While a considerable number of proteins are known to bind to cytoplasmic γTuRC in both interphase and mitosis, including NEDD1, MOZART1, MOZART2A, MOZART2B, and NME35–39, 65, none of them appear to be sufficient to stimulate nucleation. This raises the possibility that binding of γ-tubulin complexes by attachment factors directly induces their nucleating activity. As discussed above, one level of activation likely involves a conformational change in GCP3 to reorganize the γ-tubulin geometry; direct binding of attachment factors may allosterically induce the predicted conformational change in GCP3.
Attachment factors can be roughly categorized in two groups: centrosomal (or spindle pole body) and non-centrosomal, and are discussed below.
Centrosomal attachment factors.
The primary mode of centrosomal attachment appears to be through interaction with γTuSC components, as γTuSC localization is unaffected by the absence of other γTuRC components. This is demonstrated in budding yeast which lack GCP4, GCP5 and GCP6, and also by the knock-down of these GCPs either singly or altogether in animal cells. This suggests a conserved mechanism for direct γTuSC attachment to MTOCs, analogous to the way in which the attachment factor Spc110 links γTuSC to the spindle pole body in budding yeast (Figure 6a). When fully-assembled γTuRCs are present, there may also be redundant mechanisms for centrosomal attachment that function through the γTuRC-specific proteins (Figure 6b).
Figure 6. Modes of γTuSC- and γTuRC-specific attachment.
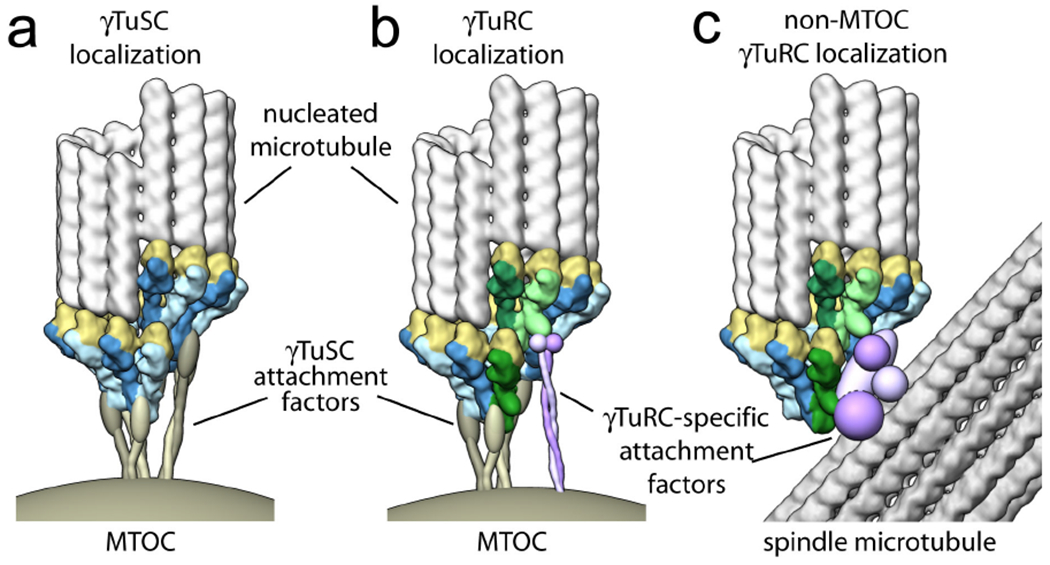
a) A conserved mechanism exists for direct γTuSC attachment to the microtubule organizing center (MTOC). In budding yeast, the γTuSC is attached to the nuclear face of the MTOC by Spc110, which not only serves to localize γTuSC but also to promote its assembly into rings. b) In organisms with complete γTuRCs an analogous means of γTuSC-mediated attachment must exist, as γTuSC localizes at the MTOC even when all of the γTuRC-specific components (GCP4, GCP5 and GCP6, green) are depleted. Redundant γTuRC-specific attachment factors may also exist at the MTOC (purple). c) Localization of nucleating complexes at non-MTOC sites within the cell is largely dependent on the presence all three γTuRC-specific proteins. For example, γTuRC localization to existing microtubules within the mitotic spindle by augmin (purple) requires GCP4, GCP5 and GCP6 (Ref. 49).
In the case of budding yeast, direct binding to the attachment factor Spc110 is not sufficient to fully activate γTuSC in vitro, although this may be due to the use of a truncated form of Spc11049. In animal cells, several centrosomal proteins have been described to bind or activate γ-tubulin complexes, including pericentrin, CG-Nap/AKAP450, ninein, and Cep19266–70. These are all large structural proteins forming coiled-coil interactions, and all are putative scaffolding components of a fibrous pericentriolar matrix, such as seen in reconstructions of the pericentriolar material in which γTuRCs are embedded71. For some of these proteins, an interaction with GCP2 and GCP3 has been proposed, but it is unclear whether this interaction is direct or indirect66,67
Non-centrosomal attachment factors.
In contrast to γTuSC-mediated localization at MTOCs, attachment of γ-tubulin complexes at other sites appears to depend largely on the γTuRC-specific GCPs (GCP4, GCP5 and GCP6). The recently discovered eight-subunit augmin complex is a non-centrosomal γTuRC attachment factor, important for γTuRC localization within the mitotic spindle63, 72–76. Depletion of augmin components leads to loss of γTuRC localization within the spindle, but does not affect centrosomal localization63, 72, 73, 77 Depletion of GCP4, GCP5, GCP6 or NEDD-1 also results in loss of γ-tubulin localization within the spindle37, 57, suggesting that augmin may interact with γTuRC through one or all of these components78.
Based on these data, it has been proposed that augmin links γTuRCs to the surface of spindle microtubules, where they function as secondary nucleation sites for additional spindle microtubules72. A similar function has been suggested for Mto1, a γTuRC attachment factor that binds along microtubules in fission yeast cytoplasmic arrays62. The regular arrangement of microtubule arrays that result from Mto1 or augmin sites in fission yeast, D. melanogaster, and human cells, suggests γTuRC is bound to the microtubules in a defined geometry which dictates the orientation of freshly nucleated microtubules. This would be consistent with observations in the acentrosomal micotubule arrays of plants, where γTuRC is recruited to the surface of existing microtubules and nucleates new microtubules with a well-defined branch angle28, 79.
A clear link between the localization of γTuRC and the activation of nucleation was demonstrated in S. pombe – when the cytoplasmic attachment factor Mtol is deleted cytoplasmic microtubule nucleation is completely abolished62. Other studies suggest a similar activation ability for a class of proteins that includes Mto1, centrosomin in D. melanogaster, and Cdk5rap2 and myomegalin in vertebrates80. In contrast to Mto1 which is a specific cytoplasmic attachment factor, centrosomin, Cdk5rap2, and myomegalin are found both at the centrosome and in the cytoplasm, and may therefore participate in both centrosomal and cytoplasmic recruitment of γTuRCs. All these proteins are related by the presence of an ~60 amino acid motif that has been dubbed the γTuRC-mediated nucleation activator (γTuNA) motif39. Overexpression of protein fragments containing γTuNA strongly induces cytoplasmic microtubule nucleation in a γ-tubulin-dependent manner in both human and D. melanogaster cells39, 81. Moreover, γTuNA itself directly binds γTuRC and greatly enhances its ability to nucleate microtubules in vitro, providing a direct functional link between the localization and activation of γTuRC. It remains unclear how, and via which γTuRC components, the γTuNA induces microtubule activation, as binding seems to occur only if the intact γTuRC is present39.
Conclusions
The recent structural studies described above have enhanced our understanding of γ-tubulin based microtubule nucleation. γ-Tubulin complexes have been shown to form microtubule templates that almost certainly nucleate microtubules through longitudinal contacts with α-tubulin and β-tubulin. This activity appears to be regulated, at least in part, through the conformation of GCP3. Which actors modulate γTuRC activity, and by what mechanism, remain pressing questions in understanding γTuRC regulation. Increasingly, it appears that attachment of γTuRC, both centrosomal and non-centrosomal, is correlated with an increase in its nucleating activity; the observation that the small γTuNA motif enhances γTuRC nucleation activity provides another tool for understanding the mechanism of attachment-factor based enhancement, and whether this correlates directly with the predicted change in GCP3.
Another major question in understanding γ-tubulin complex function is the role of nucleotide binding and hydrolysis in nucleation. γ-Tubulin and β-tubulin have similar affinities and basal hydrolysis rates for GTP. However, it remains an open question whether formation of longitudinal contacts with α-tubulin stimulates hydrolysis of the GTP bound by γ-tubulin (as it does for GTP bound β-tubulin), and whether hydrolysis weakens the α-tubulin-γ-tubulin interaction (as it does the α-tubulin-β-tubulin interaction). For example, complete hydrolysis of GTP on γTuRC could facilitate release of bound microtubules.
Our revised model for γTuRC assembly, with GCP4, GCP5, and GCP6 interacting with γTuSC as part of the ring itself, provides a new framework for future studies aimed at elucidating the mechanistic basis of γTuRC function, regulation and localization. In particular, it will now be important to determine the individual functions of GCP4, GCP5, and GCP6, the specific interactions they make with each other and with γTuSC, and their positions within γTuRC. To this end, structural work and modeling of individual components, as well as a higher resolution structure of γTuRC itself, will be necessary to provide an accurate pseudo-atomic model of the entire γTuRC. This model will doubtless prove invaluable in generating specific, testable hypotheses about γTuRC function and regulation.
Online Summary.
γTuSC alone can assemble into ring complexes with microtubule-like symmetry.
The structure of γ-tubulin complexes suggests they serve as microtubule templates.
The γ-tubulin complex proteins (GCPs) are conserved in sequence, overall structure, and ability to bind γ-tubulin.
The conformation of γTuSC may play a role in regulating nucleating activity.
A revised model of γTuRC assembly with all GCPs incorporated into the ring.
Attachment and activation of γTuRC are linked.
Glossary
- Microtubule catastrophe
The rapid depolymerization of microtubules that occurs when GTP has been hydrolyzed in all subunit up to the growing tip
- Microtubule organizing centers (MTOCs)
Primary sites of microtubule nucleation in the cell, including centrosomes in animal cells and the spindle pole body in yeast
- Chromosome-mediated nucleation
The pathway by which new microtubules are nucleated around chromosomes in response to a Ran gradient
- Deuterostome lineage
One of the two superphyla of more complex animals, including
- single particle electron microscopy (EM)
A method for combining two-dimensional images of molecules into a three-dimensional structure
- normal mode analysis
A computational method for predicting the flexibility of a protein structure based on its shape
- acentrosomal microtubule arrays
Ordered arrays of microtubules formed in the absence of a microtubule organizing center
References
- 1.Nogales E, Whittaker M, Milligan RA & Downing KH High-resolution model of the microtubule. Cell 96, 79–88 (1999). [DOI] [PubMed] [Google Scholar]
- 2.Nogales E, Wolf SG & Downing KH Structure of the alpha beta tubulin dimer by electron crystallography. Nature 391, 199–203 (1998). [DOI] [PubMed] [Google Scholar]
- 3.Chretien D & Wade RH New data on the microtubule surface lattice. Biol Cell 71, 161–74 (1991). [DOI] [PubMed] [Google Scholar]
- 4.Sui H & Downing KH Structural basis of interprotofilament interaction and lateral deformation of microtubules. Structure 18, 1022–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ledbetter MC & Porter KR Morphology of Microtubules of Plant Cell. Science 144, 872–4 (1964). [DOI] [PubMed] [Google Scholar]
- 6.Tilney LG et al. Microtubules: evidence for 13 protofilaments. J Cell Biol 59, 267–75 (1973). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Evans L, Mitchison T & Kirschner M Influence of the centrosome on the structure of nucleated microtubules. J Cell Biol 100, 1185–91 (1985). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mandelkow EM, Schultheiss R, Rapp R, Muller M & Mandelkow E On the surface lattice of microtubules: helix starts, protofilament number, seam, and handedness. J Cell Biol 102, 1067–73 (1986). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.McEwen B & Edelstein SJ Evidence for a mixed lattice in microtubules reassembled in vitro. J Mol Biol 139, 123–45 (1980). [DOI] [PubMed] [Google Scholar]
- 10.Rice LM, Montabana EA & Agard DA The lattice as allosteric effector: structural studies of alphabeta- and gamma-tubulin clarify the role of GTP in microtubule assembly. Proc Natl Acad Sci U S A 105, 5378–83 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wilson EB The Cell in Development and Heredity (Macmillan, New York, 1928). [Google Scholar]
- 12.Azimzadeh J & Bornens M Structure and duplication of the centrosome. J Cell Sci 120, 2139–42 (2007). [DOI] [PubMed] [Google Scholar]
- 13.Jaspersen SL & Winey M The budding yeast spindle pole body: structure, duplication, and function. Annu Rev Cell Dev Biol 20, 1–28 (2004). [DOI] [PubMed] [Google Scholar]
- 14.Wasteneys GO & Ambrose JC Spatial organization of plant cortical microtubules: close encounters of the 2D kind. Trends Cell Biol 19, 62–71 (2009). [DOI] [PubMed] [Google Scholar]
- 15.Oakley CE & Oakley BR Identification of gamma-tubulin, a new member of the tubulin superfamily encoded by mipA gene of Aspergillus nidulans. Nature 338, 662–4 (1989). [DOI] [PubMed] [Google Scholar]
- 16.Oakley BR, Oakley CE, Yoon Y & Jung MK Gamma-tubulin is a component of the spindle pole body that is essential for microtubule function in Aspergillus nidulans. Cell 61, 1289–301 (1990). [DOI] [PubMed] [Google Scholar]
- 17.Zheng Y, Jung MK & Oakley BR Gamma-tubulin is present in Drosophila melanogaster and Homo sapiens and is associated with the centrosome. Cell 65, 817–23 (1991). [DOI] [PubMed] [Google Scholar]
- 18.Stearns T, Evans L & Kirschner M Gamma-tubulin is a highly conserved component of the centrosome. Cell 65, 825–36 (1991). [DOI] [PubMed] [Google Scholar]
- 19.Sobel SG & Snyder M A highly divergent gamma-tubulin gene is essential for cell growth and proper microtubule organization in Saccharomyces cerevisiae. J Cell Biol 131, 1775–88 (1995). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Horio T et al. The fission yeast gamma-tubulin is essential for mitosis and is localized at microtubule organizing centers. J Cell Sci 99 ( Pt 4), 693–700 (1991). [DOI] [PubMed] [Google Scholar]
- 21.Spang A, Geissler S, Grein K & Schiebel E gamma-tubulin-like Tub4p of Saccharomyces cerevisiae is associated with the spindle pole body substructures that organize microtubules and is required for mitotic spindle formation. J Cell Biol 134, 429–41 (1996). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Raff JW, Kellogg DR & Alberts BM Drosophila gamma-tubulin is part of a complex containing two previously identified centrosomal MAPs. J Cell Biol 121, 823–35 (1993). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Stearns T & Kirschner M In vitro reconstitution of centrosome assembly and function: the central role of gamma-tubulin. Cell 76, 623–37 (1994). [DOI] [PubMed] [Google Scholar]
- 24.Zheng Y, Wong ML, Alberts B & Mitchison T Nucleation of microtubule assembly by a gamma-tubulin-containing ring complex. Nature 378, 578–83 (1995). [DOI] [PubMed] [Google Scholar]
- 25.Knop M, Pereira G, Geissler S, Grein K & Schiebel E The spindle pole body component Spc97p interacts with the gamma-tubulin of Saccharomyces cerevisiae and functions in microtubule organization and spindle pole body duplication. Embo J 16, 1550–64 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Geissler S et al. The spindle pole body component Spc98p interacts with the gamma-tubulin-like Tub4p of Saccharomyces cerevisiae at the sites of microtubule attachment. Embo J 15, 3899–911 (1996). [PMC free article] [PubMed] [Google Scholar]
- 27.Mishra RK, Chakraborty P, Arnaoutov A, Fontoura BM & Dasso M The Nup107–160 complex and gamma-TuRC regulate microtubule polymerization at kinetochores. Nat Cell Biol 12, 164–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Murata T et al. Microtubule-dependent microtubule nucleation based on recruitment of gamma-tubulin in higher plants. Nat Cell Biol 7, 961–8 (2005). [DOI] [PubMed] [Google Scholar]
- 29.Oegema K et al. Characterization of two related Drosophila gamma-tubulin complexes that differ in their ability to nucleate microtubules. J Cell Biol 144, 721–33 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Vinh DB, Kern JW, Hancock WO, Howard J & Davis TN Reconstitution and characterization of budding yeast gamma-tubulin complex. Mol Biol Cell 13, 1144–57 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gunawardane RN et al. Characterization and reconstitution of Drosophila gamma-tubulin ring complex subunits. J Cell Biol 151, 1513–24 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Murphy SM et al. GCP5 and GCP6: two new members of the human gamma-tubulin complex. Mol Biol Cell 12, 3340–52 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Murphy SM, Urbani L & Stearns T The mammalian gamma-tubulin complex contains homologues of the yeast spindle pole body components spc97p and spc98p. J Cell Biol 141, 663–74 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Teixido-Travesa N et al. The gammaTuRC revisited: a comparative analysis of interphase and mitotic human gammaTuRC redefines the set of core components and identifies the novel subunit GCP8. Mol Biol Cell 21, 3963–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hutchins JR et al. Systematic analysis of human protein complexes identifies chromosome segregation proteins. Science 328, 593–9. [DOI] [PMC free article] [PubMed] [Google Scholar]; Identifies two new γTuRC components, MOZART1 and MOZART2, and shows that MOZART1 depletion results in abnormal spindles and reduced γ-tubulin recruitment to mitotic spindle poles, suggesting a role for MOZART1 in γTuRC localization.
- 36.Gunawardane RN, Martin OC & Zheng Y Characterization of a new gammaTuRC subunit with WD repeats. Mol Biol Cell 14, 1017–26 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Luders J, Patel UK & Stearns T GCP-WD is a gamma-tubulin targeting factor required for centrosomal and chromatin-mediated microtubule nucleation. Nature cell biology 8, 137–47 (2006). [DOI] [PubMed] [Google Scholar]
- 38.Haren L et al. NEDD1-dependent recruitment of the gamma-tubulin ring complex to the centrosome is necessary for centriole duplication and spindle assembly. The Journal of cell biology 172, 505–15 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Choi YK, Liu P, Sze SK, Dai C & Qi RZ CDK5RAP2 stimulates microtubule nucleation by the gamma-tubulin ring complex. J Cell Biol 191, 1089–95 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]; Shows that a class of γTuRC attachment factors that includes Cdk5rap2, centrosomin, Mto1, and myomegalin also stimulated γTuRC activity both in vivo and in vitro, and demonstrates that a short sequence motif, γTuNA, directly binds γTuSC and is sufficient stimulate microtubule nucleation.
- 40.Moritz M, Braunfeld MB, Guenebaut V, Heuser J & Agard DA Structure of the gamma-tubulin ring complex: a template for microtubule nucleation. Nat Cell Biol 2, 365–70 (2000). [DOI] [PubMed] [Google Scholar]
- 41.Wiese C & Zheng Y A new function for the gamma-tubulin ring complex as a microtubule minus-end cap. Nat Cell Biol 2, 358–64 (2000). [DOI] [PubMed] [Google Scholar]
- 42.Keating TJ & Borisy GG Immunostructural evidence for the template mechanism of microtubule nucleation. Nat Cell Biol 2, 352–7 (2000). [DOI] [PubMed] [Google Scholar]
- 43.Byers B, Shriver K & Goetsch L The role of spindle pole bodies and modified microtubule ends in the initiation of microtubule assembly in Saccharomyces cerevisiae. J Cell Sci 30, 331–52 (1978). [DOI] [PubMed] [Google Scholar]
- 44.Erickson HP & Stoffler D Protofilaments and rings, two conformations of the tubulin family conserved from bacterial FtsZ to alpha/beta and gamma tubulin. J Cell Biol 135, 5–8 (1996). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Erickson HP Gamma-tubulin nucleation: template or protofilament? Nat Cell Biol 2, E93–6 (2000). [DOI] [PubMed] [Google Scholar]
- 46.Aldaz H, Rice LM, Stearns T & Agard DA Insights into microtubule nucleation from the crystal structure of human gamma-tubulin. Nature 435, 523–7 (2005). [DOI] [PubMed] [Google Scholar]
- 47.Kollman JM et al. The structure of the gamma-tubulin small complex: implications of its architecture and flexibility for microtubule nucleation. Mol Biol Cell 19, 207–15 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Choy RM, Kollman JM, Zelter A, Davis TN & Agard DA Localization and orientation of the gamma-tubulin small complex components using protein tags as labels for single particle EM. J Struct Biol 168, 571–4 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kollman JM, Polka JK, Zelter A, Davis TN & Agard DA Microtubule nucleating gamma-TuSC assembles structures with 13-fold microtubule-like symmetry. Nature 466, 879–82 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]; Demonstrates the ability of γTuSC to spontaneously assemble into rings, presents the structure of γTuSC rings which have thirteen-fold symmetry but are in an off state due to the conformation of γTuSC components.
- 50.Guillet V et al. Crystal structure of gamma-tubulin complex protein GCP4 provides insight into microtubule nucleation. Nat Struct Mol Biol 18, 915–919 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]; Presents the first atomic structure of a GCP, GCP4, which is very similar to GCP2 and GCP3 in γTuSC, providing the basis for a pseudo-atomic model of γTuSC and a new model for γTuRC organization.
- 51.Keck JM et al. A cell cycle phosphoproteome of the yeast centrosome. Science 332, 1557–1561 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Vogel J et al. Phosphorylation of gamma-tubulin regulates microtubule organization in budding yeast. Dev Cell 1, 621–31 (2001). [DOI] [PubMed] [Google Scholar]
- 53.Lin TC et al. Phosphorylation of the Yeast gamma-tubulin Tub4 Regulates Microtubule Function. PLoS One 6, e19700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Robinson RC et al. Crystal structure of Arp2/3 complex. Science 294, 1679–84 (2001). [DOI] [PubMed] [Google Scholar]
- 55.Volkmann N et al. Structure of Arp2/3 complex in its activated state and in actin filament branch junctions. Science 293, 2456–9 (2001). [DOI] [PubMed] [Google Scholar]
- 56.Rodal AA et al. Conformational changes in the Arp2/3 complex leading to actin nucleation. Nat Struct Mol Biol 12, 26–31 (2005). [DOI] [PubMed] [Google Scholar]
- 57.Verollet C et al. Drosophila melanogaster gamma-TuRC is dispensable for targeting gamma-tubulin to the centrosome and microtubule nucleation. J Cell Biol 172, 517–28 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Vogt N, Koch I, Schwarz H, Schnorrer F & Nusslein-Volhard C The gammaTuRC components Grip75 and Grip128 have an essential microtubule-anchoring function in the Drosophila germline. Development 133, 3963–72 (2006). [DOI] [PubMed] [Google Scholar]
- 59.Xiong Y & Oakley BR In vivo analysis of the functions of gamma-tubulin-complex proteins. J Cell Sci 122, 4218–27 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]; Demonstrates a hierarchical organization of γTuRC-specific GCPs in Aspergillus nidulans.
- 60.Zhang L, Keating TJ, Wilde A, Borisy GG & Zheng Y The role of Xgrip210 in gamma-tubulin ring complex assembly and centrosome recruitment. J Cell Biol 151, 1525–36 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Izumi N, Fumoto K, Izumi S & Kikuchi A GSK-3beta regulates proper mitotic spindle formation in cooperation with a component of the gamma-tubulin ring complex, GCP5. J Biol Chem 283, 12981–91 (2008). [DOI] [PubMed] [Google Scholar]
- 62.Anders A, Lourenco PC & Sawin KE Noncore components of the fission yeast gamma-tubulin complex. Mol Biol Cell 17, 5075–93 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Goshima G et al. Genes required for mitotic spindle assembly in Drosophila S2 cells. Science 316, 417–21 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Moudjou M, Bordes N, Paintrand M & Bornens M gamma-tubulin in mammalian cells: the centrosomal and the cytosolic forms. Journal of cell science 109 ( Pt 4), 875–87 (1996). [DOI] [PubMed] [Google Scholar]
- 65.Teixido-Travesa N et al. The gammaTuRC revisited: a comparative analysis of interphase and mitotic human gammaTuRC redefines the set of core components and identifies the novel subunit GCP8. Molecular biology of the cell 21, 3963–72 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]; Describes a new core γTuRC component, GCP8 (MOZART2), and shows that it is necessary for interphase localization of γTuRC to the centrosome but not necessary for γTuRC assembly.
- 66.Takahashi M, Yamagiwa A, Nishimura T, Mukai H & Ono Y Centrosomal proteins CG-NAP and kendrin provide microtubule nucleation sites by anchoring gamma-tubulin ring complex. Mol Biol Cell 13, 3235–45 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Zimmerman WC, Sillibourne J, Rosa J & Doxsey SJ Mitosis-specific anchoring of gamma tubulin complexes by pericentrin controls spindle organization and mitotic entry. Mol Biol Cell 15, 3642–57 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Delgehyr N, Sillibourne J & Bornens M Microtubule nucleation and anchoring at the centrosome are independent processes linked by ninein function. J Cell Sci 118, 1565–75 (2005). [DOI] [PubMed] [Google Scholar]
- 69.Gomez-Ferreria MA et al. Human Cep192 is required for mitotic centrosome and spindle assembly. Curr Biol 17, 1960–6 (2007). [DOI] [PubMed] [Google Scholar]
- 70.Zhu F et al. The mammalian SPD-2 ortholog Cep192 regulates centrosome biogenesis. Curr Biol 18, 136–41 (2008). [DOI] [PubMed] [Google Scholar]
- 71.Moritz M, Braunfeld MB, Sedat JW, Alberts B & Agard DA Microtubule nucleation by gamma-tubulin-containing rings in the centrosome. Nature 378, 638–40 (1995). [DOI] [PubMed] [Google Scholar]
- 72.Goshima G, Mayer M, Zhang N, Stuurman N & Vale RD Augmin: a protein complex required for centrosome-independent microtubule generation within the spindle. J Cell Biol 181, 421–9 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]; Describes the augmin complex, showing that it is critical for γTuRC-specific localization within the mitotic spindle which leads to amplification of microtubules within the spindle.
- 73.Lawo S et al. HAUS, the 8-subunit human Augmin complex, regulates centrosome and spindle integrity. Curr Biol 19, 816–26 (2009). [DOI] [PubMed] [Google Scholar]
- 74.Uehara R et al. The augmin complex plays a critical role in spindle microtubule generation for mitotic progression and cytokinesis in human cells. Proc Natl Acad Sci U S A 106, 6998–7003 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Meireles AM, Fisher KH, Colombie N, Wakefield JG & Ohkura H Wac: a new Augmin subunit required for chromosome alignment but not for acentrosomal microtubule assembly in female meiosis. J Cell Biol 184, 777–84 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Wainman A et al. A new Augmin subunit, Msd1, demonstrates the importance of mitotic spindle-templated microtubule nucleation in the absence of functioning centrosomes. Genes Dev 23, 1876–81 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Bucciarelli E et al. Drosophila Dgt6 interacts with Ndc80, Msps/XMAP215, and gamma-tubulin to promote kinetochore-driven MT formation. Curr Biol 19, 1839–45 (2009). [DOI] [PubMed] [Google Scholar]
- 78.Zhu H, Coppinger JA, Jang CY, Yates JR 3rd & Fang G FAM29A promotes microtubule amplification via recruitment of the NEDD1-gamma-tubulin complex to the mitotic spindle. J Cell Biol 183, 835–48 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Nakamura M, Ehrhardt DW & Hashimoto T Microtubule and katanin-dependent dynamics of microtubule nucleation complexes in the acentrosomal Arabidopsis cortical array. Nat Cell Biol 12, 1064–70. [DOI] [PubMed] [Google Scholar]
- 80.Sawin KE, Lourenco PC & Snaith HA Microtubule nucleation at non-spindle pole body microtubule-organizing centers requires fission yeast centrosomin-related protein mod20p. Curr Biol 14, 763–75 (2004). [DOI] [PubMed] [Google Scholar]
- 81.Terada Y, Uetake Y & Kuriyama R Interaction of Aurora-A and centrosomin at the microtubule-nucleating site in Drosophila and mammalian cells. J Cell Biol 162, 757–63 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Wiese C & Zheng Y Microtubule nucleation: gamma-tubulin and beyond. J Cell Sci 119, 4143–53 (2006). [DOI] [PubMed] [Google Scholar]


