Abstract
PLK4 has emerged as a prime target for cancer therapeutics and its overexpression is frequently observed in various types of human cancer. Recent studies have further revealed an unexpected oncogenic activity of PLK4 in regulating cancer cell migration and invasion. However, the molecular basis behind PLK4’s role in these processes still remains only partly understood. Our previous work demonstrated that an intact CEP85-STIL binding interface is necessary for robust PLK4 activation and centriole duplication. Here we show that CEP85 and STIL are also required for directional cancer cell migration. Mutational and functional analyses reveal that the interactions between CEP85, STIL and PLK4 are essential for effective directional cell motility. Mechanistically, we show that PLK4 can drive the recruitment of CEP85 and STIL at the leading edge of cells to promote protrusive activity, and that downregulation of CEP85 and STIL leads to a reduction in ARP2 phosphorylation and reorganization of the actin cytoskeleton, which in turn impairs cell migration. Collectively, our studies provide molecular insight into the important role of the CEP85-STIL complex in modulating PLK4 driven cancer cell migration.
Keywords: Centriole, centrosomes, cell motility and actin
Introduction
Cell migration is a fundamental cellular process essential for animal. In cancer, unscheduled increase in the motile property of cancer cells can promote intervasation and metastasis, this step-wise process relying on the spatiotemporal control of microtubule and actin dynamics (De Pascalis and Etienne-Manneville, 2017, Mayor and Etienne-Manneville, 2016, Paul et al., 2016). As the primary microtubule-organizing center, the centrosome plays a crucial role in the control of cell motility. In directional migration, centrosomes are repositioned towards the leading edge of cells to nucleate microtubules and to direct intracellular trafficking frontward (Ananthakrishnan and Ehrlicher, 2007, Rørth, 2007). Centrosomes also act as actin-organizing centers through PCM-dependent recruitment of WASH and ARP2/3 complex, providing an alternate mechanism for centrosome-based control of cell migration (Farina et al., 2016).
The centrosome consists of a pair of centrioles surrounded by pericentriolar material (Azimzadeh and Marshall, 2010, Gönczy, 2012). Centrioles must duplicate once per cell cycle to ensure that two centrosomes are present in mitosis to organize the spindle poles, failure to do so leading to a variety of diseases like cancer, microcephaly and ciliopathies (Strnad and Gönczy, 2008, Nigg and Raff, 2009, Arquint et al., 2014). PLK4 is the most upstream regulator of centriole duplication, its kinase activity essential to initiate this process. Increased PLK4 activity leads to centriole amplification, while its inhibition prevents centriole formation (Habedanck et al., 2005, Kleylein-Sohn et al., 2007). PLK4 activity is tightly controlled through trans-autophosphorylation which targets itself for degradation (Cunha-Ferreira et al., 2009, Guderian et al., 2010, Holland et al., 2010, Rogers et al., 2009). Additionally, direct interaction of STIL with PLK4 has been shown to further stimulate its kinase activity, possibly by promoting PLK4 autophosphorylation (Arquint and Nigg, 2016, Moyer et al., 2015, Ohta et al., 2014). More recently, we have reported that binding of CEP85 to STIL is critical for robust PLK4 activation and efficient centriole assembly (Liu et al., 2018).
PLK4-directed centrosome amplification is sufficient to induce aneuploidy and promote tumor initiation in flies and in mice, when the P53 pathway is partially inhibited (Levine et al., 2017, Basto et al., 2008). Moreover, excess PLK4 expression has also been linked to a distinct cellular-invasion mechanism through increased centrosomal-based microtubule nucleation and increase of RAC1 activity at the cortex (Godinho et al., 2014). A more recent study also revealed unexpected oncogenic activity for PLK4 in regulating ARP2/3-mediated actin rearrangement to promote cancer cell migration and invasion in a RAC1 and CDC42 independent manner (Kazazian et al., 2017). It still remains elusive, however, how PLK4 mediates contextual control of actin remodelling and cell motility. In addition, CEP192, an upstream regulator of PLK4, has been shown to participate in the control of directional migration (O’Rourke et al., 2014, Fung et al., 2018).
Here we show that the CEP85-STIL protein module participates in directed cell motility. Using structure-guided information, we find that disrupting the CEP85-STIL binding interface negatively affects directional cell migration. Mechanistically, CEP85 and STIL can be recruited to the cell cortex in PLK4 kinase-dependent manner. Downregulation of CEP85 or STIL significantly reduces phosphorylation of ARP2, thus impairing actin re-organization and cell motility.
Results
CEP85 regulates directional cell migration
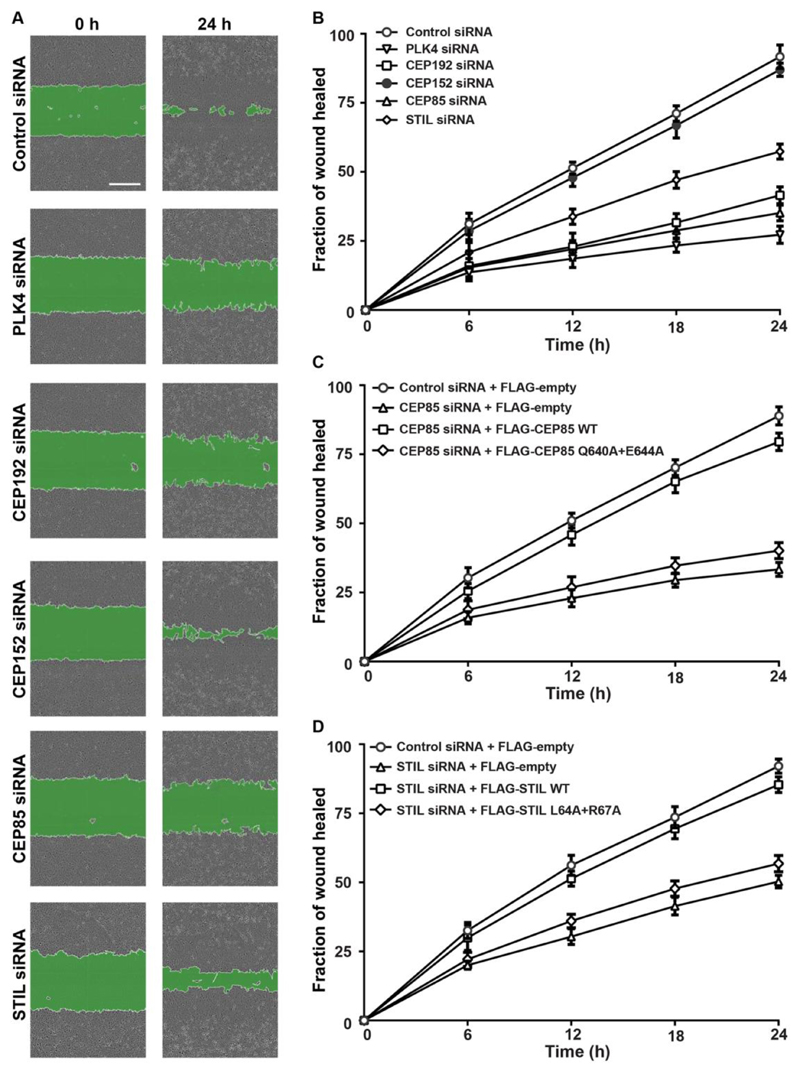
To identify additional components involved in PLK4-driven cell migration, we sought to examine the role of PLK4-associated centriole duplication factors in this process. Towards this, we used siRNAs to deplete CEP192, CEP152, CEP85 and STIL in U-2 OS cells and measured the effect of their depletion on directional cell migration using conventional wound healing assays. Western blot analyses confirmed efficient depletion of each of these proteins in U-2 OS cells (sFigure 1A). Our results show that the downregulation of CEP85 in U-2 OS cells significantly supressed wound healing to levels comparable to those observed upon PLK4 or CEP192 depletion (Figure 1A-B). In agreement with these observations, we found that the reduction of PLK4, CEP192 or CEP85 levels exhibited a marked decrease in transwell cell migration (sFigure 1D-F). STIL depletion also prevented efficient wound closure and led to reduced transwell cell migration, albeit in a less pronounced manner (Figure 1 A-B and sFigure 11D-F). Interestingly, although CEP152 is known to act as a scaffold for PLK4 activity (Sonnen et al., 2013, Park et al., 2014, Dzhindzhev et al., 2010), no noticeable defects in would healing was observed upon its depletion, indicating it does not act in this pathway. We next determined that the wound healing defects observed were not caused indirectly through impaired cell cycle progression, centrosome loss or positioning of the Golgi apparatus. Indeed, short-term treatment with centrinone B, a PLK4 inhibitor (Wong et al., 2015), significantly impaired wound healing (sFigure 1B-C), ruling out the possibility that centrosome loss was responsible for the observed defect. Consistently with this, depletion of PLK4, CEP192, CEP152, CEP85 and STIL, under the low-serum conditions used in the migration assays, led to centrosome loss in ~10% of cells (sFigure 2A-B), with no obvious defects in Golgi repositioning in the direction of the wound (sFigure 2A, C), and cell cycle progression (sFigure 2D-K). These phenotypes are unlikely to be large enough to account for the cell migration defects observed upon CEP85 depletion.
Figure 1. CEP85 and STIL are required for directional cell migration.
(A). Representative images from scratch-wound healing assays of U-2 OS cells transfected with different siRNAs. Scale bar, 400 μm. (B-D). Quantification of wound closure efficiency shown in A as measured by the percentage of wound area recovered (n = 3/experiment, three independent experiments).
Previous work established that CEP85 directly interacts with STIL to enable robust PLK4 kinase activation (Liu et al., 2018). To determine whether the CEP85-STIL interaction is also functionally important for PLK4-dependent cell migration (Rosario et al., 2014, Kazazian et al., 2017), we first depleted endogenous CEP85 or STIL in U-2 OS cells and examined the ability of RNAi-resistant wild-type (WT) CEP85 or STIL or the CEP85 Q640A+E644A or STIL L64A+R67A mutants (that compromise the CEP85-STIL interaction) to rescue cell migration (Liu et al., 2018). We observed that compared to wild-type CEP85 or STIL, the expression of their mutants could not fully rescue the wound healing defect in CEP85 or STIL depleted cells (Figure 1C-D). These data suggest that a functional CEP85-STIL binding interface is required for efficient directional cell migration.
The binding of CEP85 to PLK4 but not CEP192 is required for cell migration
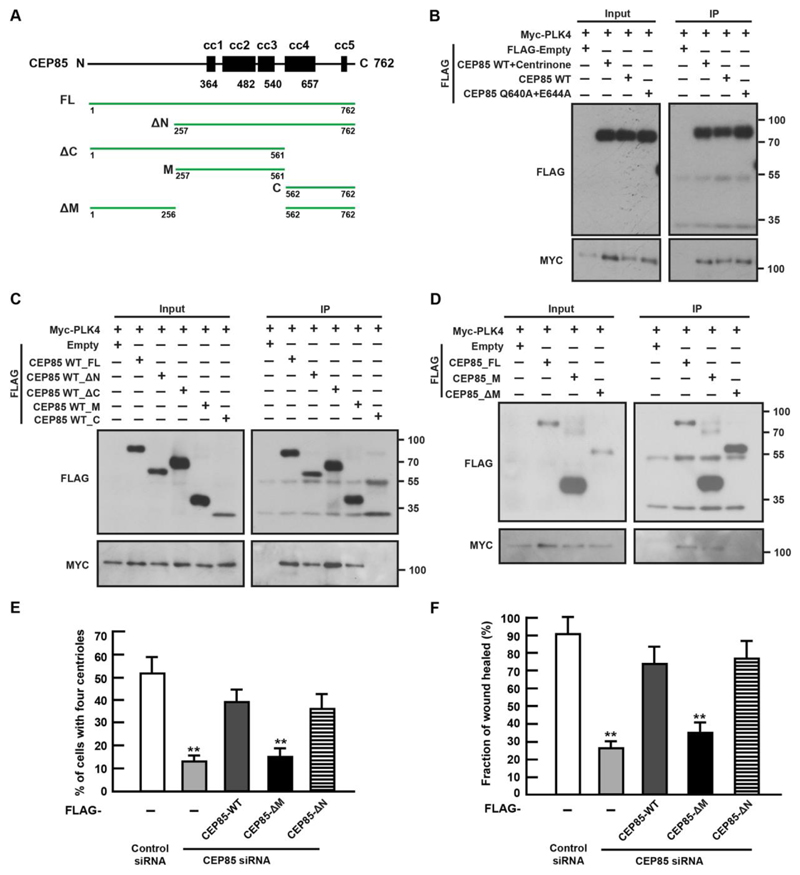
Using BioID, we and others have previously identified PLK4 and CEP192 as putative proximity interaction partners of CEP85 (Firat-Karalar et al., 2014, Liu et al., 2018). To build on these results, we conducted co-IP analysis and found that both wild-type CEP85 and the Q640A+E644A mutant could interact with PLK4, and that treatment with centrinone B, had no effect on this association (Figure 2A-B). These data suggest that the binding of CEP85 to PLK4 is independent of the CEP85-STIL interaction and PLK4 kinase activity. To determine which region in CEP85 is responsible for this interaction, we transiently expressed a set of CEP85 truncation mutants in U-2 OS cells for co-IP analysis. The results indicate that the middle region of CEP85 is responsible for its interaction with PLK4 (Figure 2C-D). Intriguingly, the CEP85 ΔM mutant that was defective for PLK4 binding displayed reduced centriole localization, implying a potential role for this interaction in centriolar targeting of CEP85 (sFigure 3B). To further explore the putative CEP85-CEP192 interaction (Liu et al., 2018, Firat-Karalar et al., 2014), we used the yeast-two-hybrid assay and found that the N-terminal region of CEP85 can indeed associate with CEP192 (sFigure 3A). This putative interaction is not required for centrosome localization of CEP85 (sFigure 3B). Next, we set out to investigate the ability of CEP85 ΔN and ΔM mutants to rescue centriole duplication and cell migration in U-2 OS cells with CEP85 depletion. Our results indicate that the expression of CEP85 ΔM mutant could not efficiently rescue these phenotypes compared to wild-type CEP85 and CEP85 ΔN mutant, indicative of a potential role for the CEP85-PLK4 interaction in cell migration and centriole assembly (Figure 2E-F). To test this hypothesis, we sought to shorten their binding region and generated a set of truncation mutants in the middle region of CEP85 for co-IP analysis (sFigure 4A). We found that both M1 and M2 can interact with PLK4 (albeit M2 appears to interact better with PLK4 than M1), implying that the entire M domain is important for robust PLK4 association (sFigure 4B). To functionally assess the effect of deleting the M2 region, we expressed an RNAi-resistant ΔM2 mutant in CEP85-depleted cells and found that it could not fully rescue the centriole duplication and cell motility phenotypes (sFigure 4C-D). Together these results suggest that reducing the ability of PLK4 to interact with CEP85 negatively affects its ability to drive cell migration and centriole duplication. Furthermore, these findings could confirm the physiological relevance of the PLK4-CEP85 interaction.
Figure 2. CEP85 interacts functionally with PLK4.
(A). Domain overview of human CEP85. cc coiled-coil. Vectors that were used in this work are shown by green lines. (B-D). Detection of expressed FLAG-CEP85 WT, Q640A+E644A mutant or fragments coimmunoprecipitating with Myc- PLK4. (E-F). U-2 OS cells expressing FLAG or the siRNA-resistant FLAG-CEP85 transgene were transfected with control or CEP85 siRNA for 72 h. (E). The graph indicates the percentage of cells with four centrioles at normal serum conditions (n = 100, three independent experiments). (F). The graph indicates the percentage of the percentage of wound area recovered at low serum conditions (n = 3/experiment, three independent experiments). Two-tailed t-test was performed for all p-values (**p < 0.01, *p <0.05).
During directional cell migration, CEP192 has previously been shown to affect microtubule dynamics (O’Rourke et al., 2014). Consistent with this observation, Western blot analysis indicated that CEP192 depletion led to a marked increase in the levels of less dynamic acetylated microtubules, while depletion of CEP85, STIL or PLK4 had no noticeable effect (sFigure 3C). These results suggest that the PLK4 interaction might be required for CEP85’s ability to drive centriole duplication and cell migration, independently of CEP192.
CEP85 and STIL localize at the leading edge
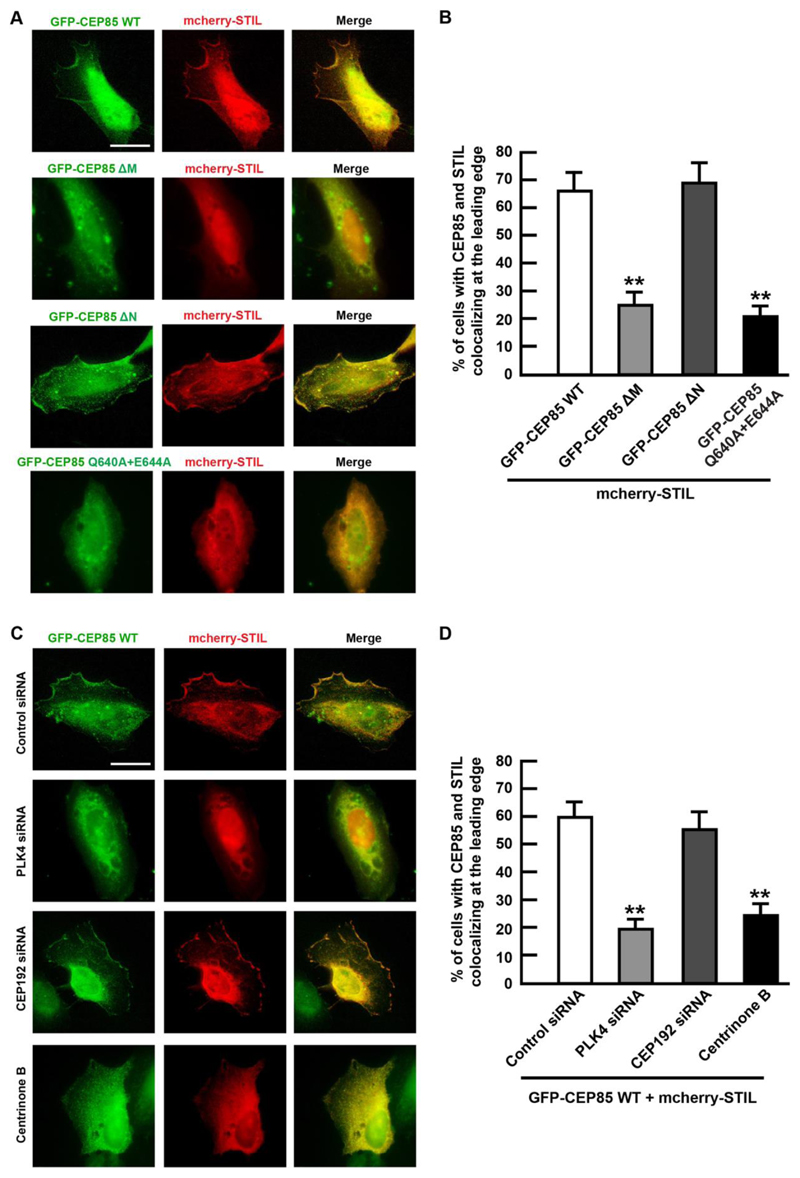
We next sought to determine if CEP85 and STIL can localize to the cell cortex. To test it, we transiently over-expressed mCherry-STIL in U-2 OS cells harbouring tetracycline (Tet)-inducible GFP-tagged CEP85. Our results revealed that over-expressed wild-type CEP85 was able to colocalize with STIL at the cell cortex (Figure 3A-B and sFigure 3D). In contrast, expression of the CEP85 ΔM and Q640A+E644A mutants failed to localize CEP85 and STIL at the cortex, suggesting that CEP85 binding to PLK4 and STIL is essential for their cortical localization (Figure 3A-B and sFigure 3D).
Figure 3. CEP85 and STIL localization at the cell cortex.
(A). Representative images of U-2 OS cells expressing GFP-CEP85 WT, ΔM, ΔN and Q640A+E644A mutants, transfected with mCherry-STIL at low serum conditions. (B). The graph shows the percentage of cells with the described localization pattern (n = 100/experiment, three independent experiments). (C). U-2 OS cells expressing GFP-CEP85 WT and mCherry-STIL were either treated with centrinone B (1 μM) for 24 h or transfected with different siRNAs for 72 h at low serum conditions. (D). The graph shows the percentage of cells with the described localization pattern (n = 100/experiment, three independent experiments). Two-tailed t-test was performed for all p-values (**p < 0.01, *p <0.05). All error bars represent SD and scale bars are 20 μm.
To test this hypothesis, we depleted endogenous STIL in U-2 OS cells using siRNA and induced the expression of RNAi resistant mcherry-STIL WT and mutant to levels comparable to endogenous STIL (sFigure 3H). The results suggest that wild-type STIL still localizes to the cortex, whereas the L64A+R67A mutations perturbs its cortical localization (sFigure 3F-G), suggesting that CEP85 binding is critical for STILs cortical localization. Next, we examined the effect of knocking down PLK4 and CEP192 on the cortical localization of CEP85 and STIL in U-2 OS cells. Our results indicate that the depletion of PLK4, or treatment with centrinone B, but not CEP192 depletion, perturbed the localization of CEP85 and STIL at the cortex (Figure 3C-D and sFigure 3E). Together, our results suggest that PLK4 is an upstream regulator of CEP85 and STIL during cell migration.
CEP85 and STIL regulate the actin cytoskeleton
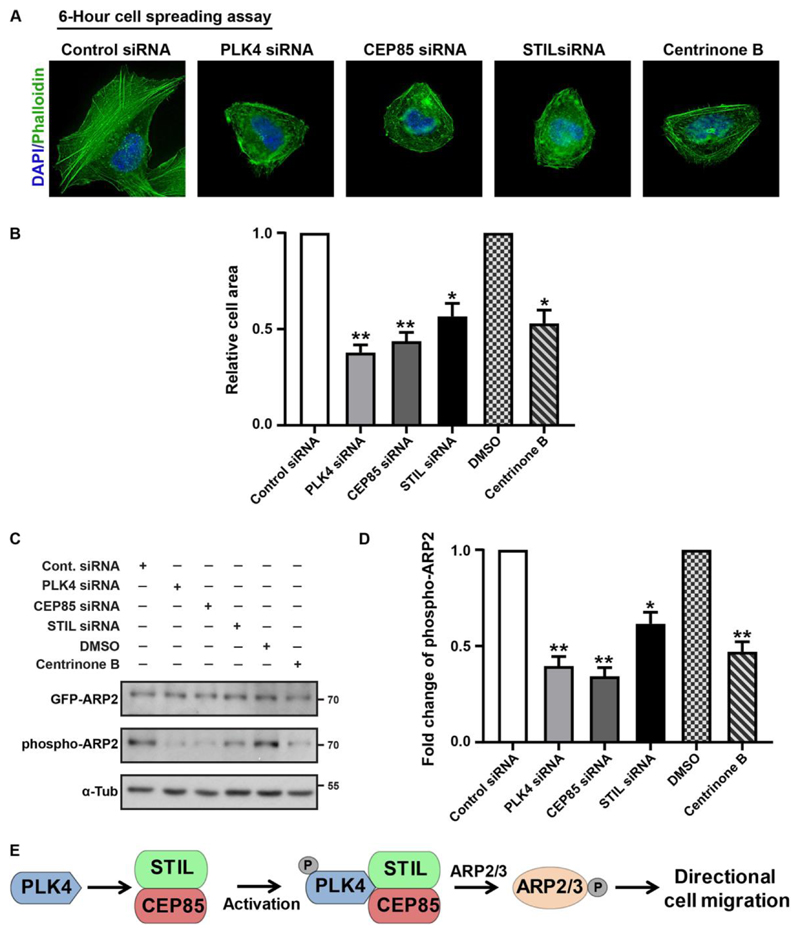
It has been shown that PLK4 phosphorylates ARP2 to mediate actin cytoskeletal rearrangement (Kazazian et al., 2017). Hence, we sought to determine whether CEP85 and STIL, that act to facilitate PLK4 activation (Liu et al., 2018), also contribute to this regulation. In this regard, we first use the canonical cell spreading assay in response to the stimulus of replating. Our results indicate that depletion of CEP85 and STIL adversely affected the cell spreading along with significant cell size reduction, an effect that was comparable to PLK4 depletion or centrinone B treatment (Figure 4A-B). We also confirm that the majority of those siRNA transfected cells maintained their centrosomes in the spreading assay (sFigure 4F), suggesting that the phenotypes observed are independent of the centrosome. To further explore the molecular mechanism, we aim to determine whether CEP85 and STIL impact PLK4-dependent phosphorylation of ARP2 (Kazazian et al., 2017). Using an antibody specific for phosphorylated ARP2, we observed that ARP2 phosphorylation at T237/T238 was significantly reduced in cells treated with centrinone B or depleted of PLK4, CEP85 and STIL (Figure 4C-D). Together, these results indicate that the PLK4-CEP85-STIL module is required for ARP2/3 activation during directional cell migration.
Figure 4. CEP85 and STIL regulate ARP2/3 mediated actin reorganization.
(A). Representative images of spreading assays in control, PLK4, CEP85 or STIL siRNA transfected or centrinone B treated U-2 OS cells. Cells were stained with Alexa Fluor 488 Phalloidin. Scale bar 20 μm. (B). The graph shows the quantification of relative cell area (n = 100/experiment, three independent experiments). (C). Western blot showing the levels of phospho- and total GFP-ARP2 in different conditions. (D). Quantification of fold change in phospho-ARP2 relative to total GFP-ARP2 (n = 2/experiment, six independent experiments). (E). A model for how PLK4-CEP85-STIL operates in the control of directional cell migration. CEP85 and STIL act downstream of PLK4 and their interaction facilitates PLK4 activation and subsequent ARP2 phosphorylation, which further regulates ARP2/3 mediated actin assembly and directed cell migration. Two-tailed t-test was performed for all p-values (**p < 0.01, *p <0.05). All error bars represent SD.
The work described here identifies an unexpected function of the centrosomal PLK4-CEP85-STIL module in directional cell migration. We find that PLK4 is required for the recruitment of CEP85 and STIL to the cell cortex, which is dependent on PLK4 kinase activity and its interaction with CEP85. This is consistent with our previous work indicating that the interaction between CEP85 and STIL is required for robust PLK4 kinase activation (Liu et al., 2018). This synergy promotes ARP2 phosphorylation in order to drive ARP2/3 dependent actin re-organization and directional cell migration. Overall, our data suggest that PLK4-CEP85-STIL module plays a pivotal role in the regulation of both centriole duplication and cell motility.
It has emerged that centrosomes can act as actin-organizing centers through ARP2/3-based actin nucleation (Farina et al., 2016). The ARP2/3 complex consists of two actin related proteins (ARP2 and ARP3) and five accessory proteins, serving as unique nucleation sites for new actin filaments (Goley and Welch, 2006). Phosphorylation on residues T237, T238, and Y202 of ARP2 is required for stabilizing the ARP2/3 complex in an active conformation, which promotes efficient assembly of new actin (LeClaire et al., 2008). In the model proposed by Kazazian and colleagues, PLK4 is required for phosphorylation of ARP2 at the T237/T238 residues and ARP2 activity is critical for PLK4-driven cell migration (Kazazian et al., 2017). Our studies reveal the molecular basis of this regulation and characterize the CEP85-STIL complex as a novel modulator of PLK4-ARP2/3 mediated actin filament assembly and cell motility. Given the traditional view that these proteins functionally associate at the centrosomes and regulate centrosome-based actin nucleation, it must now also be considered that the PLK4-CEP85-STIL module can also control actin reorganization at the leading edge.
PLK4 is also implicated in the control of non-directional cell motility (Luo et al., 2019). In this model, CEP192 interacts with both PLK4 and AURKB. In response to exosome-WNT signalling stimulation, DVL2 initiates the recruitment of CEP192, PLK4 and AURKB to cell protrusions where PLK4 and AURKB act redundantly to drive formin-dependent actin reorganization (Luo et al., 2019). In contrast, as presented here, in the case of directional migration, PLK4 activity alone is required to drive the process. These observations raise the tantalizing possibility that other PLK4- and CEP192-associated components may be differentially regulated to control different types of cancer cell motility mediated by diverse actin nucleators, allowing for intricate contextual control of many forms of cell motility. This isgenetic amplification of PLK4 and its overexpression at the mRNA and protein level are frequently observed in such as breast, lung, pancreatic, rectum, and stomach (sFigure 4E).
It will be important to dissect mechanistically, how the CEP85-STIL complex coordinates with PLK4 to activate ARP2/3-dependent actin polymerization and how this complex balances its centrosomal and non-centrosomal functions. Overall, our findings illuminate previously unappreciated regulatory mechanisms that participate in the control of directional cell migration.
Materials and Methods
Cell culture
U-2 OS cells were grown under standard conditions, and were purchased from ATCC. U-2 OS T-REx cells were cultured in McCoy 5A medium with 10% FBS, 2mM GlutaMAX, zeocin (100 μg/ml) and blasticidin (3 μg/ml). U-2 OS T-REx cells with Tet-inducible Myc-tagged PLK4 were a kind gift from E. Nigg, and were maintained in 10% FBS, 2mM GlutaMAX and G418 (0.5 mg/mL). HCT116 cells were maintained in McCoy’s 5A medium with L-glutamine (Life Technologies) supplemented with 10% FBS (Life Technologies) and 1% Penicillin/Streptomycin (Life Technologies). All human cell lines were cultured in a humidified 5% CO2 atmosphere at 37°C. All cell lines were tested without mycoplasma contamination.
RNA interference
Luciferase duplex GL2 (CGUACGCGGAAUACTTCG) from Dharmacon was used as a control. The siRNAs against human PLK4 (M-005036-02-0005), CEP192 (L-032250-01-0005) and CEP152 (M-022241-01) were purchased from Dharmacon. CEP85 and STIL were depleted using the following siRNA oligonucleotide sequences: human CEP85 siRNA, 5'-CCUAGAGCAGGAAGUGGCUCAAGAA-3′ and human STIL siRNA, 5'-GCUCCAAAC AGUUUCUGCUGGAAU-3 ’ (Liu et al., 2018). Transfections were performed using Lipofectamine RNAiMax (Invitrogen) according to the manufacturer’s protocol.
Cloning and stable cell lines
GFP or FLAG tagged CEP85 fragments were recombined into pcDNA5/FRT/TO vector backbone (Life Technologies) using a tetracycline-inducible CMV promoter. U-2 OST-REx cells carrying full length CEP85 and STIL or CEP85 Q640A+E644A and STIL L64A/R67A mutants were generated in our previous study (Liu et al., 2018). U-2 OS cells expressing Tet-inducible FLAG-tagged siRNA resistant CEP85 ΔM and ΔN transgenes were generated as previously described (Liu et al., 2018).
Wound healing assay
U-2 OS cells were seeded into 96-well plates and transfected with the indicated siRNAs and cultured at low serum concentrations (0.5% FBS). At 48 h post transfection, a single scratch wound was generated using IncuCyte™ Wound Maker (Essen BioScience). Then cells were washed three times with PBS and cultured in serum-free medium for wounding healing assays. IncuCyte™ live-cell imaging systems (Essen BioScience) were utilized to conduct the 96-well scratch wound cell migration assay according to the manufacturer’s protocol. Imaging was taken at 1 h intervals up to 24 h using a 10X objective lens. Image analysis was performed using ImageJ to measure the area of healed wound at t=6 h, 12 h, 18 h and 24 h.
For measuring the axis of nucleus-centrosome-Golgi apparatus, cells were fixed 0 or 6 h after generating wounds and stained with DAPI and antibodies specific to Pericentrin and GM130. Axis of nucleus-centrosome-Golgi apparatus was determined from the middle of the nucleus to the middle point between centrosome and Golgi apparatus. Angles between leading edge and axis of nucleus-centrosome-Golgi apparatus were measured with Image J. Radar graphs were drawn in RStsudio (Ver 1.2.1335) with ggplot2 library.
Transwell assay
1.0 × 106 U-2 OS cells were seeded on 6-well plates and then indicated siRNAs were transfected next day. One day after siRNA transfection, cells were grown in media containing 0.5% FBS for 24 h. For migration assay, 5.0 × 104 cells were spread into the transwell (Millipore Sigma, CLS3422) with 0.1 ml of media containing 0.5% FBS and then the transwells were inserted into 24 wells containing 0.6 ml of media supplemented with 10% FBS. After 24 h cells were fixed with 4% PFA for 2 minutes and cold methanol for 10 minutes. After removing cells inside the transwell, cells were stained with 0.5% crystal violet for 20 minutes. Cells were imaged on a Deltavision Elite DV imaging system (GE Healthcare) equipped with a sCMOS 2048x2048 pixels2 camera (GE Healthcare).
Spreading assays
U-2 OS cells were transfected with the indicated siRNAs and cultured at low serum conditions. At 72 h post transfection, cells were trypsinized and replated onto 12-well plate with glass coverslips for another 6 h. Next, cells were fixed by 4% PFA and immunostained with Alexa Fluor 488 Phalloidin for labeling F-actin. Imaging were acquired on a Deltavision Elite DV imaging system equipped with a 60×/1.4 NA objective and a sCMOS 2048x2048 pixels2 camera (GE Healthcare). Z stacks (0.2 μm apart) were used, and images were deconvolved and projected using softWoRx (v6.0, Applied Precision). Quantification of cell area based on F-actin signal was performed using ImageJ.
Immunofluorescence microscopy
Centrin and PCNT staining were performed following standard protocols as previously described (Liu et al., 2018). For the Phalloidin staining, cells were fixed by 4% PFA at room temperature for 20 minutes and treated with permeabilization buffer (0.5 % Triton X-100 in PBS) for another 10 minutes. Cells were then blocked in 1% BSA in PBS for one hour and incubated with Alexa Fluor 488 Phalloidin (Invitrogen A12379) and DAPI in blocking solution for another one hour. After a final wash three times with PBS for 5 minutes each, cells were inverted and mounted on glass slides with standard mounting solution (ProLong Gold antifade, Molecular Probes). Cells were imaged on a Deltavision Elite DV imaging system (GE Healthcare) equipped with a sCMOS 2048x2048 pixels2 camera (GE Healthcare). Z stacks (0.2 μm apart) were collected, and images were deconvolved and projected using softWoRx (v6.0, Applied Precision).
Co-Immunoprecipitation
The respective 293T or U-2 OS stable lines were seeded into 10-cm2 dishes and 24 h later transfected with 4 ¼g of plasmid DNA and incubated with tetracycline (2μg/ml). At 48 h post-transfection, transfected cells were washed with 1x PBS, then harvested and lysed immediately in lysis buffer (50mM HEPES pH8; 100mM KCl; 2mM EDTA; 10% Glycerol; 0.1% NP-40; 1mM DTT; protease inhibitors (Roche)) for 30 minutes on ice. Lysates were frozen in dry ice for 5 minutes, then thawed and centrifuged for 20 minutes at 16,000xg at 4ºC. Cleared supernatant were incubated with anti-FLAG M2 Affinity Gel (Sigma-Aldrich) for 3 hours at 4ºC. A fraction of the protein extracts (Inputs) were saved before the incubation with the beads. After the incubation, the beads were pelleted and washed three times in lysis buffer. Samples (Inputs and IPs) were prepared by addition of Laemmli buffer and boiling at 95ºC for 5 minutes. Immunopurified proteins were analyzed by immunoblotting with the indicated antibodies.
Western Blots
Cells were lysed in Laemmli sample supplemented with a phosphatase inhibitor cocktail and benzonase nuclease. For phospho-ARP2 T237+T238 detection, cells were treated with 5.0 mmol/L pervanadate for 10 minutes before lysis. Proteins were loaded to 8% SDS-PAGE gel for electrophoresis and then electroblotted onto PVDF membranes (Immobilon-P, Millipore). Membranes were incubated with primary antibodies in TBST (TBS, 0.1% Tween-20) in 5% skim milk powder or 5% BSA for the anti-ARP2 (phospho T237 + T238) antibody overnight at 4°C. Membranes were washed 3x10 minutes in TBST, and then incubated with secondary antibodies conjugated to HRP for 1 h at room temperature. Western blots were developed using SuperSignal reagents (Thermo Scientific). Mouse polyclonal antibody used in this study is anti-CEP85 (H00064793-B01P, dilution 1:500; Abnova). Mouse monoclonal antibody used in this study are anti-α-tubulin (Clone DM1A, T6199, dilution 1:15000; Sigma-Aldrich), anti-GFP (11814460001, dilution 1:2000; Roche), anti-FLAG (F3165, dilution 1:1000; Sigma), and anti-acetylated tubulin (T6793, dilution 1:2000; Sigma). Rabbit polyclonal antibodies used in this study are anti-CEP192 (A302-324A-1, dilution 1:1000; Bethyl Laboratories), anti-ARP2 phospho T237 + T238 (ab119766, dilution 1:500; Abcam) and anti-STIL (A302-441A, dilution 1:500; Bethyl Laboratories). Goat polyclonal antibody used in this study is anti-C-Myc (ab19234, dilution 1:500; Abcam).
Flow cytometry
U-2 OS cells were cultured at low serum conditions and harvested 72 h post-siRNA transfection from 10 cm plates. Fixation was performed using 4% paraformaldehyde for 10 min at RT followed by permeabilization with 0.1% Triton-X for 15min at RT. Cells were washed and resuspended in wash buffer (PBS+4% FBS). DAPI was added at 1:100 final dilutions and cells were incubated in the dark at RT for 15min. Flow cytometric analysis was performed on Fortessa X-20 (BD) using the UV laser for excitation and DAPI fluorescence was measured at 461nm. 50,000 events were recorded for each sample. Doublet cell population was gated out and cell cycle distribution analysis was modeled using ModFit analysis and cells in G1, S and G2/M phases were quantified.
Statistical methods
Two-tailed unpaired student t-tests were performed for all p-values. Individual p-values, experiment sample numbers and the number of replicates for statistical testing were indicated in corresponding figure legends. Unless otherwise mentioned, all error bars are SD, and the asterisk placeholders for p-values are **p<0.01 and *p<0.05.
Supplementary Material
Acknowledgments
U-2 OS Tet-inducible PLK4 cells were from E. Nigg. Y.L. was funded by a CIHR Doctoral Award. This work was supported by a CIHR Foundation and Project Grants (#167279, MOP#130507 and 142192), the Krembil Foundation and the Ontario Ministry for Research and Innovation.
Footnotes
Data availability
The authors declare that all data supporting the findings of this study can be found within the paper and its supplementary information files or from the corresponding author upon reasonable request.
Competing interests
The authors declare no competing financial interests.
Author contributions
Y.L. performed the cell biological experiments, J.K. the transwell cell migration and Golgi repositioning assays, V.S. and Y.L. the flow cytometry, R.P. the PLK4 expression analysis in cancers. M.C. drew the model for transwell migration assay and M.V.B performed the yeast two-hybrid experiments. The paper was written by Y.L. and L.P.
References
- Ananthakrishnan R, Ehrlicher A. The Forces Behind Cell Movement. International Journal of Biological Sciences. 2007;3:303–317. doi: 10.7150/ijbs.3.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arquint C, Gabryjonczyk AM, Nigg EA. Centrosomes as signalling centres. Philos Trans R Soc Lond B Biol Sci. 2014;369 doi: 10.1098/rstb.2013.0464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arquint C, Nigg EA. The PLK4-STIL-SAS-6 module at the core of centriole duplication. Biochem Soc Trans. 2016;44:1253–1263. doi: 10.1042/BST20160116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Azimzadeh J, Marshall WF. Building the centriole. Current Biology. 2010;20:R816–25. doi: 10.1016/j.cub.2010.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basto R, Brunk K, Vinadogrova T, Peel N, Franz A, Khodjakov A, Raff JW. Centrosome amplification can initiate tumorigenesis in flies. Cell. 2008;133:1032–1042. doi: 10.1016/j.cell.2008.05.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bauer M, Cubizolles F, Schmidt A, Nigg EA. Quantitative analysis of human centrosome architecture by targeted proteomics and fluorescence imaging. The EMBO journal. 2016;35:2152–2166. doi: 10.15252/embj.201694462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cunha-ferreira I, Rodrigues-martins A, Bento I, Riparbelli M, Zhang W, Laue E, Callaini G, Glover DM, Bettencourt-dias M. The SCF/Slimb ubiquitin ligase limits centrosome amplification through degradation of SAK/PLK4. Current Biology. 2009;19:43–49. doi: 10.1016/j.cub.2008.11.037. [DOI] [PubMed] [Google Scholar]
- De pascalis C, Etienne-manneville S. Single and collective cell migration: the mechanics of adhesions. Molecular biology of the cell. 2017;28:1833–1846. doi: 10.1091/mbc.E17-03-0134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dzhindzhev NS, YU QD, Weiskopf K, Tzolovsky G, Cunha-ferreira I, Riparbelli M, Rodrigues-martins A, Bettencourt-dias M, Callaini G, Glover DM. Asterless is a scaffold for the onset of centriole assembly. Nature. 2010;467:714. doi: 10.1038/nature09445. [DOI] [PubMed] [Google Scholar]
- Farina F, Gaillard J, GuÉrin C, CoutÉ Y, Sillibourne J, Blanchoin L, ThÉry M. The centrosome is an actin-organizing center. Nature cell biology. 2016;18:65–75. doi: 10.1038/ncb3285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Firat-karalar ELIFN, Rauniyar N, Yates John R, iii, Stearns T. Proximity interactions among centrosome components identify regulators of centriole duplication. Current Biology. 2014;24:664–670. doi: 10.1016/j.cub.2014.01.067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fung E, Richter C, Yang H-B, SchÄffer I, Fischer R, Kessler BM, Bassermann F, D'angiolella V. FBXL13 directs the proteolysis of CEP192 to regulate centrosome homeostasis and cell migration. EMBO reports. 2018;19:e44799. doi: 10.15252/embr.201744799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Godinho SA, Picone R, Burute M, Dagher R, Su Y, Leung CT, Polyak K, Brugge JS, Thery M, Pellman D. Oncogene-like induction of cellular invasion from centrosome amplification. Nature. 2014;510:167–171. doi: 10.1038/nature13277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- GOLEY ED, WELCH MD. The ARP2/3 complex: an actin nucleator comes of age. Nature Reviews Molecular Cell Biology. 2006;7:713. doi: 10.1038/nrm2026. [DOI] [PubMed] [Google Scholar]
- Gönczy P. Towards a molecular architecture of centriole assembly. Nat Rev Mol Cell Biol. 2012;13:425–435. doi: 10.1038/nrm3373. [DOI] [PubMed] [Google Scholar]
- Guderian G, Westendorf J, Uldschmid A, Nigg EA. Plk4 transautophosphorylation regulates centriole number by controlling βTrCP-mediated degradation. Journal of Cell Science. 2010;123:2163–2169. doi: 10.1242/jcs.068502. [DOI] [PubMed] [Google Scholar]
- Habedanck R, Stierhof Y-D, Wilkinson CJ, Nigg EA. The Polo kinase Plk4 functions in centriole duplication. Nat Cell Biol. 2005;7:1140–1146. doi: 10.1038/ncb1320. [DOI] [PubMed] [Google Scholar]
- Holland AJ, Lan W, Niessen S, Hoover H, Cleveland DW. Polo-like kinase 4 kinase activity limits centrosome overduplication by autoregulating its own stability. The Journal of Cell Biology. 2010;188:191–198. doi: 10.1083/jcb.200911102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kazazian K, Go C, Wu H, Brashavitskaya O, Xu R, Dennis JW, Gingras A-C, Swallow CJ. Plk4 Promotes Cancer Invasion and Metastasis through Arp2/3 Complex Regulation of the Actin Cytoskeleton. Cancer Research. 2017;77:434–447. doi: 10.1158/0008-5472.CAN-16-2060. [DOI] [PubMed] [Google Scholar]
- Kleylein-sohn J, Westendorf J, Le clech M, Habedanck R, Stierhof Y-D, Nigg EA. Plk4-induced centriole biogenesis in human cells. Developmental Cell. 2007;13:190–202. doi: 10.1016/j.devcel.2007.07.002. [DOI] [PubMed] [Google Scholar]
- Leclaire LL, Baumgartner M, Iwasa JH, Mullins RD, Barber DL. Phosphorylation of the Arp2/3 complex is necessary to nucleate actin filaments. The Journal of Cell Biology. 2008;182:647–654. doi: 10.1083/jcb.200802145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levine MS, Bakker B, Boeckx B, Moyett J, Lu J, Vitre B, Spierings DC, Lansdorp PM, Cleveland DW, Lambrechts D, Foijer F, et al. Centrosome Amplification Is Sufficient to Promote Spontaneous Tumorigenesis in Mammals. Developmental Cell. 2017;40:313–322.e5. doi: 10.1016/j.devcel.2016.12.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, Gupta GD, Barnabas DD, Agircan FG, Mehmood S, Wu D, Coyaud E, Johnson CM, Mclaughlin SH, Andreeva A, Freund SMV, et al. Direct binding of CEP85 to STIL ensures robust PLK4 activation and efficient centriole assembly. Nature Communications. 2018;9 doi: 10.1038/s41467-018-04122-x. 1731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo Y, Barrios-rodiles M, Gupta GD, Zhang YY, Ogunjimi AA, Bashkurov M, Tkach JM, Underhill AQ, Zhang L, Bourmoum M, Wrana JL, et al. Atypical function of a centrosomal module in WNT signalling drives contextual cancer cell motility. Nature Communications. 2019;10 doi: 10.1038/s41467-019-10241-w. 2356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maniswami RR, Prashanth S, Karanth AV, Koushik S, Govindaraj H, Mullangi R, Rajagopal S, Jegatheesan SK. PLK4: a link between centriole biogenesis and cancer. Expert Opinion on Therapeutic Targets. 2018;22:59–73. doi: 10.1080/14728222.2018.1410140. [DOI] [PubMed] [Google Scholar]
- Mayor R, Etienne-Manneville S. The front and rear of collective cell migration. Nature Reviews Molecular Cell Biology. 2016;17:97. doi: 10.1038/nrm.2015.14. [DOI] [PubMed] [Google Scholar]
- Moyer TC, Clutario KM, Lambrus BG, Daggubati V, Holland AJ. Binding of STIL to Plk4 activates kinase activity to promote centriole assembly. The Journal of Cell Biology. 2015;209:863–878. doi: 10.1083/jcb.201502088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nigg EA, Raff JW. Centrioles, centrosomes, and cilia in health and disease. Cell. 2009;139:663–78. doi: 10.1016/j.cell.2009.10.036. [DOI] [PubMed] [Google Scholar]
- O’rourke BP, Gomez-Ferreria MA, Berk RH, Hackl AMU, Nicholas MP, O’rourke SC, Pelletier L, Sharp DJ. Cep192 Controls the Balance of Centrosome and Non-Centrosomal Microtubules during Interphase. PLOS ONE. 2014;9:e101001. doi: 10.1371/journal.pone.0101001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohta M, Ashikawa T, Nozaki Y, Kozuka-hata H, Goto H, Inagaki M, Oyama M, Kitagawa D. Direct interaction of Plk4 with STIL ensures formation of a single procentriole per parental centriole. Nature Communications. 2014;5 doi: 10.1038/ncomms6267. 5267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park S-Y, Park J-E, Kim T-S, Kim JH, Kwak M-J, Ku B, Tian L, Murugan RN, Ahn M, Komiya S, Hojo H, et al. Molecular basis for unidirectional scaffold switching of human Plk4 in centriole biogenesis. Nat Struct Mol Biol. 2014;21:696–703. doi: 10.1038/nsmb.2846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paul CD, Mistriotis P, Konstantopoulos K. Cancer cell motility: lessons from migration in confined spaces. Nature Reviews Cancer. 2016;17:131. doi: 10.1038/nrc.2016.123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogers GC, Rusan NM, Roberts DM, Peifer M, Rogers SL. The SCF(Slimb) ubiquitin ligase regulates Plk4/Sak levels to block centriole reduplication. The Journal of Cell Biology. 2009;184:225–239. doi: 10.1083/jcb.200808049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rørth P. Collective guidance of collective cell migration. Trends in Cell Biology. 2007;17:575–579. doi: 10.1016/j.tcb.2007.09.007. [DOI] [PubMed] [Google Scholar]
- Rosario CO, Kazazian K, Zih FSW, Brashavitskaya O, Haffani Y, Xu RSZ, George A, Dennis JW, Swallow CJ. A novel role for Plk4 in regulating cell spreading and motility. Oncogene. 2014;34:3441. doi: 10.1038/onc.2014.275. [DOI] [PubMed] [Google Scholar]
- Sonnen KF, Gabryjonczyk A-M, Anselm E, Stierhof Y-D, Nigg EA. Human Cep192 and Cep152 cooperate in Plk4 recruitment and centriole duplication. Journal of Cell Science. 2013;126:3223–3233. doi: 10.1242/jcs.129502. [DOI] [PubMed] [Google Scholar]
- Strnad P, GÖnczy P. Mechanisms of procentriole formation. Trends in Cell Biology. 2008;18:389–396. doi: 10.1016/j.tcb.2008.06.004. [DOI] [PubMed] [Google Scholar]
- Wong YL, Anzola JV, Davis RL, Yoon M, Motamedi A, Kroll A, Seo CP, Hsia JE, Kim SK, Mitchell JW, Mitchell BJ, et al. Reversible centriole depletion with an inhibitor of Polo-like kinase 4. Science. 2015;348:1155–1160. doi: 10.1126/science.aaa5111. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.