Abstract
Objective
The aim of this study was to identify susceptibility genes associated with hereditary predisposition to uveal melanoma (UM) in patients with no detectable germline BAP1 alterations.
Design
Retrospective case series from academic referral centers.
Participants
Cohort of 154 UM patients with high risk of hereditary cancer defined as patients with one or more of the following: 1) familial UM; 2) young age (<35 years) at diagnosis; 3) personal history of other primary cancers; 4) family history of ≥2 primary cancers with no detectable mutation or deletion in BAP1 gene.
Methods
Whole exome sequencing and/or cancer gene panel were carried out. Probands included 27 with familial UM, one with bilateral UM, one with congenital UM, and 125 UM patients with strong personal and/or family histories of cancer. Functional validation of variants was carried out by immunohistochemistry, RT-PCR and genotyping.
Main outcome and measures
Clinical characterization of UM patients with germline alterations in known cancer genes.
Results
We identified actionable pathogenic variants in eight known hereditary cancer predisposition genes (PALB2, MLH1, MSH6, CHEK2, SMARCE1, ATM, BRCA1 and CTNNA1) in nine patients, including 3/27 (11%) with familial UM and 6/127 (4.7%) with high-risk for cancer. Two patients had pathogenic variants in CHEK2 and PALB2; while variants in the other genes each occurred in one patient. Biallelic inactivation of PALB2 and MLH1 was observed in tumors from the respective patients. The frequencies of pathogenic variants in PALB2, MLH1 and SMARCE1 in UM patients were significantly higher than the observed frequencies in non-cancer controls [p=0.02, OR: 8.9 (1.5–30.6); p=0.04, OR: 25.4 (1.2–143); p= 0.001, OR: 2047 (52–4.5e15), respectively].
Conclusions
The study provides moderate evidence of gene/disease association of germline mutations in PALB2 and MLH1 with hereditary predisposition to UM. It also identifies several other candidate susceptibility genes. The results suggest locus heterogeneity in predisposition to UM. Genetic testing for hereditary predisposition to cancer is warranted in UM patients with strong personal and/or family history of cancers.
Precis
Genes associated with predisposition to cancer contribute to the etiology of uveal melanoma. Genetic testing for cancer genes is warranted in uveal melanoma patients with strong personal and/or family history of cancers.
Familial aggregation of uveal melanoma (familial UM) is rare 1,2 However, UM clusters in families with other cancers in up to 12% of cases 3,4 This, together with the rarity of the disease and lack of strong environmental causes suggests that genetic factors play an important role in UM etiology. Currently, BRCA-associated-protein 1 (BAP1) is the only gene with definitive evidence of association with hereditary predisposition to UM. Germline pathogenic variants in BAP1 have been reported in at least 80 UM patients5. The frequency of BAP1 pathogenic variants is 3–4% in UM with strong personal and family history of cancer 6–9 and about 20% in familial UM10. Germline pathogenic variants in BRCA2 have been reported in a total of six UM patients 11–13, MBD4 in four14, 15, while only case reports of UM associated with several other genes, including BRCA111, MLH116, CDKN2A17, FLCN18, 19 and MSH620 are documented. This suggests the existence of other high penetrance drivers of UM which have yet to be identified. In this study we assessed coding genetic alterations in UM patients with strong personal and/or family history of cancer and no detectable genetic alteration in BAP1.
MATERIALS AND METHODS
Samples
Patients were accrued from The Ohio State University Medical Center, Cleveland Clinic Foundation or referral to our program. Patients were included if they met one of the following criteria: 1) familial UM defined as a UM with 1st, 2nd or 3rd degree family member with UM;; 2) young age (<35 years) at diagnosis; 3) personal history of other primary cancers; 4) family history of ≥2 primary cancers with no detectable mutation or deletion in BAP1 gene. The age cutoff of less than 35 years was chosen because it is 15–25 years younger than the median age of diagnosis in UM general population. Similar age cut off points have been used in clinical cancer genetic practice for germline genetic testing in other cancers22–24. Approval for this project was obtained from the Institutional Review Board at The Ohio State University (2006C0045) and Cole Eye Institute. Informed consents were obtained prior to testing.
DNA and RNA Extraction
Germline DNA was extracted from peripheral blood mononuclear cells (PBMCs) according to a published protocol 25 RNA was extracted using Trizol (Lifetechnology) from PBMCs and from tumor and non-tumor tissues in patients who underwent enucleation. DNA from tissue was extracted using Qiagen DNeasy or All prep RNA/DNA/miRNA kits (Qiagen, Valencia, CA).
Whole Exome Sequencing (WES) and Cancer Gene Panel
WES was carried out for six UM probands, two relatives and one UM tumor at the Genetic Resources Core Facility, Johns Hopkins Institute of Genetic Medicine, Baltimore, MD, according to their published protocol26 utilizing Human All Exon V5 (Agilent) and HiSeq 2000 sequencer (Illumina) at a depth of 20X. The remaining 23 probands, seven relatives and three tumors were sequenced to an average depth of 104X at Nationwide Children’s Hospital (Columbus, OH) utilizing the SureSelect Human All Exon V6 plus COSMIC (Agilent) and the HiSeq 4000 sequencer (Illumina) to produce paired-end 150bp reads. One sample was sequenced on the two platform for validation. The analysis of the sequencing data was conducted using the Churchill pipeline 27, in which the data was aligned to GRCh37 using Burrows-Wheeler Aligner-MEM28, deduplicated using SAMBLASTER, and variants were jointly called across all samples using GATK’s HaplotypeCaller. Variants with a maximum frequency >0.005 as observed in the ExAC (Exome Aggregation Consortium) without TCGA (The Cancer Genome Atlas), the 1000 Genome Project and the EVS (Exome Variant Server) were excluded. Furthermore, any variants that did not segregate in other affected family members were removed.
We focused our primary analysis on pathogenic or likely pathogenic alterations defined as null variants (stop gain, start loss, frame shift, canonical splice site ≤2 base pairs from exons) and missense variants reported as pathogenic in ClinVar29.
For missense variants of uncertain significance we prioritized those with multiple lines of computational evidence supporting pathogenicity (SIFT 30: ≤0.05, a Genomic Evolutionary Rate Profiling (GERP) score of >2 and Polyphen 231: ≥0.453) in genes with established associations with hereditary cancer from the COSMIC cancer genes census32, as well as genes involved in DNA repair33, 34
The BROCA multi-gene panel testing of 30 DNA repair/hereditary predisposition genes assay sequences all exons and flanking intronic sequences of tested genes35, 36, Supplement Table 1 Large deletions and duplications were detected using methods described by Nord et al. 201137. Confirmation of the variants and assessment of segregation in families were carried out by direct (Sanger) sequencing.
Quantitative RT-PCR
TaqMan 5’ nuclease quantitative (real-time) RT-PCR assays were carried out using pre-developed assays from Applied Biosystems according to the manufacturer’s protocol. Reactions were carried out in triplicate. The endogenous controls GUSB and/or PP1A were tested in separate reactions. Relative expression levels were assessed by the comparative threshold cycle (CT) method.
Microsatellite Instability Assay
A custom panel of ten microsatellite markers, labeled with either FAM or HEX dyes, was used (BAT25, BAT26, BAT40, NR27, NR21, NR22, D2S123, D3S1260, D17S250 and D5S346). This included the five NCI panel38.
Immunohistochemistry for Assessment of Candidate Gene Product Expression
Immunostaining for MLH1 and MSH6 was carried out at the Department of Pathology (OSU) Clinical Laboratory according to standard practices. Immunostaining for other candidate gene products was carried out in the laboratory of M. Abdel-Rahman using published protocols39–41 Supplement Table 2..
Statistical Analysis
GnomAD Loss-of-Function Intolerance Metric was obtained from gnomAD v2.1.1 at https://gnomad.broadinstitute.org on June 28, 2019. The metric is ratio of the observed number of loss-of-function variants in the gene in gnomAD to what would be expected if such variants were selectively neutral. The conditional maximum likelihood estimate of the odds ratio and Fisher’s exact test two-sided mid-p value for the null hypothesis that the odds ratio equals 1 were obtained using the exact2×2 package in R 42, 43 The two-sided mid-p value was calculated by doubling the smaller of the one-sided p-values. The 95% confidence interval (CI) was obtained by inverting the test based on the two-sided mid-p value using this same package 42, 43
RESULTS
Patient Population
163 unrelated UM patients were accrued. WES was carried out on germline DNA of 29 unrelated UM patients, nine of their relatives and available UM tissues of four patients. The probands for WES included 25 with familial UM, one with bilateral UM, one with congenital UM,21 and two patients with strong family history of cancers, (Table 1). The relatives included six with UM, one with ovarian cancer and the unaffected parents of the congenital UM. Two UM relatives were first degree, while the remaining four were third degree. The cancer gene panel was assessed on 125 additional UM patients with personal and/or family histories suggestive of high-risk hereditary predisposition to cancer, (Table 1).
Table 1.
Patient Demographics and Cancer Phenotype
| Patients studied by WES | Pathogenic variants1 | Patients studied by multi-gene panel | Pathogenic variants1 | |
|---|---|---|---|---|
| Total | 29 | 4 | 125 | 5 |
| Male/Female | 12/17 | 0/4 | 54/71 | 4/1 |
| Age at UM diagnosis (median, range) | 55 (0.25–79) | 49 (46–67) | 58 (15–87) | 58 (50–89) |
| Patients diagnosed UM at young age (<35) | 2 | 0 | 12 | 0 |
| - Age of patients (years) | 0.25, 25 | 15, 19, 23, 27 (x4), 28 (x2), 29, 31, 33 | ||
| Treatment | ||||
| - Enucleation | 5 | 2 | 37 | 2 |
| TNM Stage | ||||
| - pT1 | 4 | 0 | 13 | 1 |
| - pT2 | 5 | 2 | 26 | 0 |
| - pT3 | 5 | 0 | 43 | 1 |
| - pT4 | 1 | 0 | 8 | 0 |
| - Unknown | 14 | 2 | 37 | 3 |
| Clinical outcome | ||||
| - Average Follow Up (months) | 78 | 68 | 102 | 90.8 |
| - Alive | 22 | 3 | 89 | 4 |
| - Deceased | 6 | 1 | 36 | 1 |
| - Unknown | 1 | 0 | 0 | 0 |
| - Metastatic UM | 5 | 1 | 29 | 2 |
| Bilateral or multiple UM | 1 | 0 | 4 | 0 |
| Personal /family history of cancer*: | 29 | 4 | 125 | 6 |
| - BAP1-TPDS | 18 | 2 | 31 | 0 |
| - Lynch Syndrome | 0 | 0 | 2 | 1 |
| - Breast Ovarian | 3 | 1 | 14 | 2 |
| - Familial Melanoma | 0 | 0 | 3 | 0 |
| - Nonspecific | 10 | 1 | 83 | 3 |
| Family History of UM + | 25 | 3 | 2 | 0 |
| - BAP1-TPDS | 15 | 2 | 1 | 0 |
| - Lynch Syndrome | 0 | 0 | 0 | 0 |
| - Breast Ovarian | 1 | 0 | 0 | 0 |
| - Nonspecific | 9 | 1 | 1 | 0 |
Pathogenic variants (null or confirmed pathogenic missense) in established hereditary cancer predisposition gene
WES: whole exome sequencing
BAP1-TPDS: BAP1 tumor predisposition syndrome
Cancer syndrome classification is based on clinical presentation. Numbers are greater than total because more than one individual may be at risk for more than one syndrome.
Pathogenic Alterations in Established Cancer Genes
We identified actionable pathogenic variants in eight known hereditary cancer predisposition genes (PALB2, MLH1, MSH6, CHEK2, SMARCE1, ATM, BRCA1 and CTNNA1) accross nine patients, including 3/27 (11.1%) patients with familial UM and 6/127 (4.7%) UM patients with strong personal and/or family history of cancer, (Table 2 and 3).
Table 2.
Pathogenic and Likely Pathogenic Variants Detected in Known Cancer Predisposition Genes
| Gene | Transcript | Variant (dbSNP) | Effect | Proband cancer | Cancer in Family | Heterozygous Cancer Risk1 |
|---|---|---|---|---|---|---|
| A- Whole Exome Sequencing | ||||||
| CHEK2 | NM_007194.3 | c.1100delC:p.Thr367Metfs (rs555607708) | Frameshift | UM, RCC, basal cell carcinoma | CM, breast | Breast |
| PALB2 | NM_024675.3 | c.3201+1G>C (rs587776423) | Splice site | UM, RCC | UM | Breast, Pancreatic |
| SMARCE1 | NM_003079.4 | c.373G>T, p.Glu125* (not reported) | Stopgain | UM, uterine | UM, Skin, | Meningioma |
| MLH1 | NM_001258271.1 | c.200G>A, p.Gly67Glu (rs63749939) | Non-synonymous | UM, breast (sebaceous adenocarcinoma) | UM, colon, skin, bladder, prostate, Hodgkins | Colon, ovarian, endometrial |
| B- BROCA Multi-Gene Panel | ||||||
| MSH6 | NM_000179.2 | c.C2731T: p.Arg911* (rs63751017) | Non-synonymous | UM, endometrial, BCC, SCC | Lynch syndrome in family | Colon, endometrial |
| BRCA1 | NM_007300.3 | c.C2603G: pSer868* (rs80356925) | Non-synonymous | UM, prostate | Breast × 3, stomach, mycosis fungoides, lymphoma, cervical, Skin, Liver | Breast, ovarian |
| ATM | NM_000051.3 | c.2251(−10)T>G (rs730881346) | Splice site | UM, lung | None | Breast, pancreatic |
| CHEK2 | NM_007194.3 | c.T470C:p.Ile157Thr (rs17879961) | Non-synonymous | UM | Pancreatic, breast × 2, colon | Breast |
| PALB2 | NM_024675.3 | c.49–1G>A (not reported) | Splice site | Breast, pancreatic | ||
| CTNNA1 | NM_001903 | c.1072_1075del AAAG: Arg360Valfs*8 (not reported) | Frameshift | UM | None | Gastric |
Pathogenic and likely pathogenic is based on ClinVar
Total of 156 unrelated proband with UM
Only the most commonly associated cancer types are listed.
BCC: Basel Cell Carcinoma; CM: Cutaneous Melanoma; RCC: Renal Cell Carcinoma; UM: Uveal Melanoma
Table 3.
Frequencies of null variants in the cancer genes in UM cohort and controls from public database
| Gene | gnomAD Loss-of-Function Intolerance Metric (90% CI)* | UM Cohort** | gnomAD Control Cohort*** | Odds Ratio (95% CI)**** | P-value**** |
|---|---|---|---|---|---|
| # Pathogenic Variant Carriers/# Individuals (%) | |||||
| BAP1 | 0.12 (0.06 – 0.28) | 7/172 (4.1%) | 2/59095 (0.0034%) | 1255, 95% CI: 277 – 8192 | <2.2e-16 |
| BRCA1 | 0.73 (0.59 – 0.92) | 1/156 (0.64%) | 153/59095 (0.26%) | 2.5 (0.1 – 12.6) | 0.40 |
| PALB2 | 0.76 (0.58 – 1.01) | 2/156 (1.3%) | 86/59095 (0.15%) | 8.9 (1.5 – 30.6) | 0.02 |
| MLH1 | 0.37 (0.25 – 0.57) | 1/156 (0.64%) | 15/59095 (0.025%) | 25.4 (1.2 – 143.2) | 0.04 |
| MSH6 | 0.34 (0.23 – 0.50) | 1/156 (0.64%) | 65/59095 (0.11%) | 5.9 (0.3 – 30.1) | 0.17 |
| CHEK2 | 1.15 (0.87 – 1.53) | 2/156 (1.3%) | 409/59095 (0.69%) | 1.9 (0.3 – 6.3) | 0.39 |
| CTNNA1 | 0.17 (0.10 – 0.31) | 1/156 (0.64%) | 17/59095 (0.029%) | 22.4 (1.1 – 124.8) | 0.05 |
| ATM | 0.60 (0.51 – 0.71) | 1/156 (0.64%) | 198/59095 (0.34%) | 1.9 (0.1 – 9.7) | 0.51 |
| RECQL4 | 0.96 (0.77 – 1.21) | 1/29 (3.4%) | 232/59095 (0.39%) | 9.1 (0.4 – 48.2) | 0.11 |
| SMARCE1 | 0.04 (0.01 – 0.19) | 1/29 (3.4%) | 1/59095 (0.0017%) | 2047 (52 – 4.5e15) | 0.001 |
| TP53AIP1 | 1.73 (0.82 – 1.95) | 1/29 (3.4%) | 987/59095 (1.7%) | 2.1 (0.1 – 11.1) | 0.47 |
| DLEC1 | 0.74 (0.61 – 0.91) | 1/29 (3.4%) | 844/59095 (1.4%) | 2.4 (0.1 – 13.0) | 0.41 |
| MMS19 | 0.27 (0.18 – 0.42) | 1/29 (3.4%) | 33/59095 (0.056%) | 63.8 (3.0 – 354.2) | 0.02 |
| POLI | 0.61 (0.41 – 0.93) | 1/29 (3.4%) | 60/59095 (0.10%) | 35.1 (1.7 – 191.2) | 0.03 |
This metric is the ratio of the observed number of loss-of-function variants in the gene in gnomAD to what would be expected if such variants were selectively neutral. Values closer to zero indicate loss-of-function intolerance (fewer than expected). Obtained from gnomAD v2.1.1 at https://gnomad.broadinstitute.org on June 28, 2019.
One UM patient with large germline deletion of BAP1 was not counted in order to make a valid comparison with gnomAD.
The gnomAD non-Finnish European non-cancer cohort comprised 59,095 unrelated individuals with data (51,377 with exomes and 7,718 with whole genomes). The number of carriers of pathogenic variants meeting the same criteria used in the UM cohort was estimated for each gene by the number of variant alleles at missense variants reported as pathogenic or likely pathogenic in ClinVar and null (stop, large deletion, frameshift, canonical splicing) variants not reported as benign and/or variants of uncertain significance in ClinVar. This approximation assumes that multiple pathogenic variant alleles do not appear on the same haplotype and that there are no individuals who are compound heterozygotes or homozygotes for pathogenic variants. To the extent that these assumptions are not met, the frequency of carriers in the control cohort would be overestimated, which would bias associations toward the null. No pathogenic or likely pathogenic missense variants were reported in ClinVar for for DLEC1, MMS19, POLI, and TP53AIP1.
Carrying pathogenic variants in genes highlighted in bold was associated with higher odds of UM (P ≤ 0.05).
Prioritized Variants in other Potential Cancer Genes
Given the rarity of UM, for the 29 UM patients studied by WES we focused our analysis on rare variants (≤ 0.005 minor allele frequency (MAF) in the general population) with strong evidence of pathogenesis (stop gain, start loss, frame shift, canonical splice site ≤2 base pairs from exons) and missense variants reported as pathogenic in ClinVar29. Of the 527 variants identified, 493 were null and 34 missense. In addition to pathogenic variants in four known cancer predisposition genes, CHEK2, MLH1, PALB2 and SMARCE1 (Table 2), pathogenic variants in five other genes were detected, Table 4. This included three (RECQL4, MMS19 and POLI) genes associated with DNA damage repair, one (DLEC1) reported as a tumor suppressor in UNIPROT (http://www.uniprot.org/keywords/KW-0043), and TP53AIP1, which has been suggested as a cutaneous melanoma (CM) susceptibility gene 44
Table 4.
Null Variants in Genes Associated with Cancer
| Gene | Transcript | Variant (dbSNP) | Effect | Proband Cancer | Cancer in Family (1st and 2nd degree) | Heterozygous Cancer Risk1 |
|---|---|---|---|---|---|---|
| DLEC1 | NM_007335.2 | c.5068_5071dupAACA:p.Ser1691fs (rs577236284) | frameshift | UM | UM, CM, SCC, BCC, thyroid | - |
| MMS19 | NM_022362.4 | c.733_734delCT:p.Leu245fs | frameshift | UM, CM | UM, skin | - |
| POLI | NM_007195.2 | c.729_732delAACA:p.Thr244fs (rs747005866) | frameshift | UM | UM, CASU, Leukemia, Hodgkins, Lung, colon, stomach, prostate | - |
| TP53AIP1 | NM_022112.2 | c.63dupG:p.Gln22fs (rs141395772) | frameshift | UM | UM, CM, breast, ovarian | Cutaneous melanoma |
| RECQL4 | NM_004260.3 | c.1573delT, p.Cys525fs (rs386833845) | frameshift | UM, RCC | UM, colon, cervical, lung | Osteosarcoma |
Total of 29 unrelated proband with UM assessed by whole exome sequencing
Only the most commonly associated cancer types are listed.
BCC: Basel Cell Carcinoma; CASU: Cancer Site Unknown; CM: Cutaneous Melanoma; RCC: Renal Cell Carcinoma; SCC: Squamous Cell Carcinoma; UM: Uveal Melanoma
In addition, a total of 1969 unique missense variants predicted as deleterious by multiple computational tools were identified by WES. Out of these, 47 were in cancer associated genes in 21 patients including: RET, MSH3, MSH6, FANCD2, FANCM, RB1 and WT1, with established associations with hereditary cancer predisposition. The remaining genes are associated with DNA damage repair and/or are proposed as tumor suppressors, (Supplemental Table 3). In six patients, no variants in suspected cancer genes were detected, while in 16 patients, more than one variant (range 2–6) was detected (Supplementary Table 3). Variants in DAPK1 and MSH3 were each seen in two patients, variants in LZSTS1 in three patients, while variants in SRRM2 were seen in four patients, including one with two different variants. Variants in all the other genes were seen once, Supplement Table 3.
Validation of Potential Candidate Genes
i-. PALB2
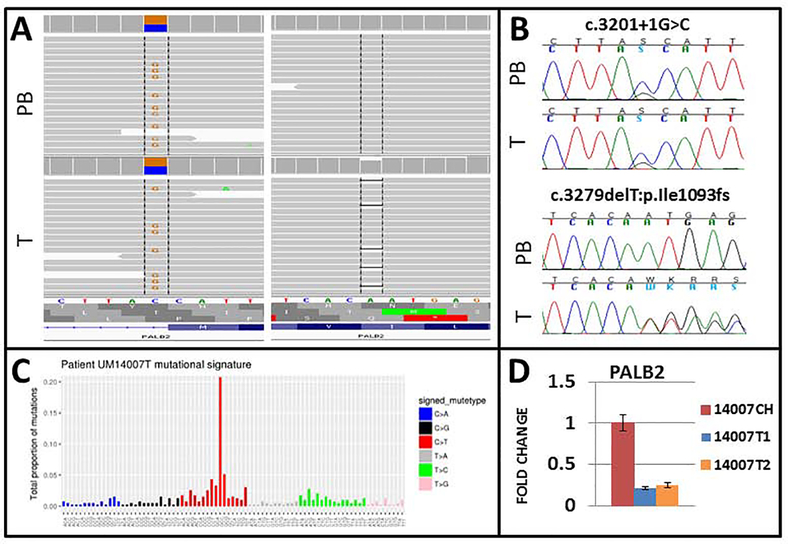
A canonical splice site variant, c.3201+1G>C, in PALB2 was detected in a patient with a personal history of UM and RCC, both diagnosed at age 67 years. Family history was positive for a maternal relative with UM. WES of the tumor identified biallelic inactivation of PALB2, with a somatic truncating mutation, c.3279delT:p.Ile1093fs. A 75–80% decrease in the expression of PALB2 was detected in the UM compared to the non-tumor choroid of the same patient, confirming the biallelic inactivation of PALB2 (Figure 1).
Figure 1. UM patient with germline PALB2 mutation with evidence of biallelic inactivation of PALB2 in the tumor.
A) Whole exome sequencing of germline DNA (PB) and tumor tissue (T) identified a germline canonical splice site variant c.3201+1G>C with a somatic frameshift mutationc.3279delT:p.Ile1093fs in PALB2. B) Variants were confirmed by direct sequencing. C), Mutation signatures from the patient’s tumor were significant for SBS39 and SBS1 which are commonly reported in sporadic UM. D) The expression of PALB2 mRNA was significantly lower in the tumor tissue from the patient compared to the matching non-tumor choroid.
A different canonical splice site variant, c.49–1G>A, was identified in another patient in the replication cohort. The 63year-old male presented with UM at age 51 years with no personal history of other cancers but a family history of breast and pancreatic cancers in his mother, breast cancer in his maternal aunt and a maternal first cousin, and colon cancer in a maternal aunt. In addition to PALB2, this patient had a likely pathogenic non-synonymous variant, c.T470C:p.Ile157Thr (rs17879961) in CHEK2. No tumor tissue was available for the assessment of biallelic inactivation.
One additional patient, a female presenting with UM at age 65 years, breast cancer at age 67 years and multiple non-melanoma skin cancers (ages 62–67 years), had a missense variant of uncertain significance (VUS), c.3418T>G :p.Trp1140Gly (rs62625283) in PALB2. Family history was positive for non-melanoma skin cancers, leukemia and prostate cancer. Recent, biochemical studies of this variant suggests that it has a deleterious effect on the DNA damage repair function of PALB2 and its interaction with BRCA245. The patient was treated by brachytherapy and tumor tissue was not available for evaluation of biallelic inactivation.
We further surveyed a database of patients with germline mutations in PALB2 for cases of UM. We identified one additional proband, a 65 year-old female who was diagnosed with bilateral breast cancer at 36 and 56 years. She was subsequently diagnosed with a UM at 62 years, which was managed by enucleation. The family history was notable for a male breast cancer in her brother at 67 years. Genetic testing identified a pathogenic germline PALB2 c.3113G>A variant, which results in the generation of a premature stop codon and has been previously reported on multiple occasions46. To check for somatic pathogenic variants elsewhere in PALB2 and in other genes previously associated with cancer, DNA was extracted from tumor tissue and analyzed using the Illumina TruSight Cancer panel, but no additional pathogenic variant in PALB2 was detected and there was no evidence of loss of heterozygosity.
ii. MLHI
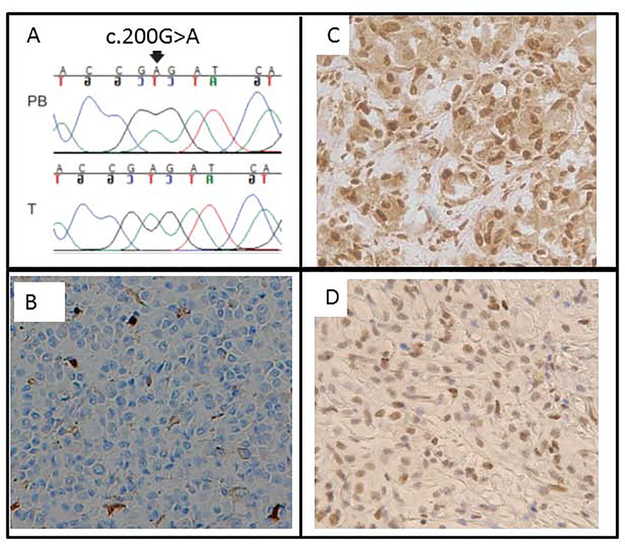
A missense pathogenic variant, c.200G>A, p.Gly67Glu (rs63749939), in MLH1 was detected in a female diagnosed with UM at age 49 years, breast cancer at age 39 and a sebaceous adenocarcinoma. Her family cancer history was significant, with a brother diagnosed with UM at age 45 and Hodgkin’s lymphoma at age 32, the mother diagnosed with colon cancer at age 39, the father diagnosed with prostate, urinary bladder and skin cancers, and a maternal and paternal grandfathers with colon cancers. All family members with cancers were deceased, with no tissue samples available. Archival tumor tissue was available from the UM for the proband. Sequencing of the tumor showed a relative decrease in the allele frequency of the wild type allele. Genotyping with markers in the vicinity of MLH1 (D3S3135) showed loss of heterozygosity, and immunostaining showed loss of MLH1 protein expression in the tumor (Figure 2) but no microsatellite instability was observed. Taken together, these results confirm somatic biallelic inactivation of MLH1 in the UM.
Figure 2: Assessment of biallelic inactivation in tumors from patients with germline variants in MLH1, SMARCE1 and MSH6.
A) Direct sequencing confirmedthe germline variant c.200G>A, p.Gly67Glu and showed allelic imbalance in the tumor tissue. The variant is reported as pathogenic in ClinVar (rs63749939). B) Immunohistochemistry of tumor and tumor with adjacent non-tumor tissue show loss of nuclear expression of MLH1 protein in all tumor cells with preserved expression in the nuclei in non-tumor tissue. C) and D) A patient showed a null variant in SMARCE1 with variant of uncertain significance in MSH6. Immunohistochemistry showed preserved nuclear expression of SMARCE1 (C) and MSH6 (D) in the tumor.
Two additional germline variants in MLH1 were identified in our cohort. One, c.1039–8T>A (rs193922367) was reported benign in ClinVar, while the other one, c.277A>G:p.Ser93Gly (rs41295282) is rare in the general population (MAF 0.00003 in ExAC and 0.00008 in GO-ESP)but recently reported as likely benign by multiple clinical laboratories. Tumor tissue was not available from that patient for assessment of biallelic inactivation. One additional VUS in MLH1 was identified in one of the sporadic UM patients47.
iii. SMARCE1
A novel truncating pathogenic variant, c.373G>T:p.Glu125*, was detected in a female patient who was diagnosed with UM at age 46 and endometrial carcinoma at age 51. The patient had a family history of multiple non-melanoma skin cancer in mother and a maternal aunt and UM in a maternal aunt. An unreported missense VUS in MSH6, c.2589C>G:p.Cys863Trp, was also identified in the proband. Immunohistochemistry for SMARCE1 showed strong nuclear expression in the endometrial cancer and UM tumor cells. MSH6 loss of expression was observed in the endometrial cancer and only weak expression was observed in the UM tumor cells. The tumor showed areas of necrosis and there was insufficient material to study somatic mutation and copy number alteration.
DISCUSSION
Using WES and a cancer gene panel we identified actionable pathogenic variants in eight known hereditary cancer predisposition genes: PALB2, MLH1, MSH6, CHEK2, SMARCE1, ATM, BRCA1 and CTNNA1 in nine UM patients, including three (11%) with familial UM and six with strong personal and/or family history of cancer (4.7%). Combined with our previous study of germline mutation in BAP1 in the same cohort48, pathogenic variants in established hereditary predisposition genes were detected in 9/33 (27.3%) familial UM patients and 8/129 (6.1%) of UM patients with strong personal and/or family history of cancer48. This suggests the importance of referring these patients and their family members for cancer genetic counseling and genetic testing for proper management of risk of other cancers.
One of the major challenges in the field of cancer genetics is to establish evidence of gene-disease associations. Among the eight cancer predisposition genes with pathogenic variants observed in our cohort only two (PALB2 and MLH1) showed moderate evidence of an association with hereditary predisposition to UM49. The evidence included the higher frequency of pathogenic variants in the UM cohort compared to the general population, the biallelic inactivation of the gene product in tumors and the observation of pathogenic variants in more than one unrelated UM patient. For MLH1, evidence included a previously reported UM case with a truncating mutation16 in addition to the case identified in this study. For PALB2 three unrelated cases were identified with pathogenic variants, two identified by screening a UM cohort and the third by screening a PALB2 cohort. One additional case had a VUS which may be pathogenic based on recent experimental evidence 45. The personal and family history of the patient with a pathogenic variant in MLH1 was consistent with Lynch syndrome50 and the personal and/or family histories of the three patients with PALB2 were consistent with the reported cancer phenotype for the gene51. It is not clear whether UM hereditary risk is unique to MLH1 or could be associated with other mismatch repair genes especially that germline pathogenic variant in MSH6 was observed in one UM patient in our cohort and has been reported in another patient20. Validation studies in larger cohorts will be needed. One potential explanation of the rarity of UM in subjects with germline pathogenic variant in MLH1 and PALB2 is the 4–6/million general population prevalence of UM so even a 100-fold increase in the odds ratio would still be observed as a rare phenotype.
SMARCE1 is another potential candidate identified in our cohort. Our data provide only limited evidence for its potential association with UM predisposition based on the significantly higher frequency of truncating mutation observed in UM patients compared to general population. So far seven probands/families have been reported with germline mutations in SMARCE1: six with meningiomas52, 53 and one with Coffin-Siris Syndrome and anaplastic astrocytoma54. It is noteworthy that evidence of biallelic inactivation of SMARCE1 was observed in some but not all of these patients. Further studies of the potential role of SMARCE1 in UM tumorigenesis is warranted.
A germline truncating variant in TP53AIP1 was identified in one of our patients. This variant was the same variant detected in a report suggesting TP53AIP1 as a predisposition gene to CM44. Although a significant difference was observed between cases and controls in that study, the high frequency (2%), of TP53AIP1 truncating variants in the non-cancer general population largely preclude it as a predisposition gene to UM or CM.
It is worth noting that VUS in two genes with suggested role in hereditary predisposition to cancer, DAPK155 and SRRM2 56, were observed in more than one proband in our cohort; however, we did not identify additional supporting evidence of their role in predisposition to UM.
Pathogenic and likely pathogenic variants in more than one cancer associated gene were observed in several patients most notably PALB2/CHEK2 in one and SMARCE1/MSH6 in another, supplement Table 1. Also VUS in established and/or potential cancer genes were observed in several of UM probands suggesting multi-gene etiology in a subset of UM patients.
Combined with previous reports of germline pathogenic variants of several known cancer genes such as BRCA2 11–13, BRCA111, MLH116 CDKN2A17, FLCN18, 19 and MSH620, our study suggests that in UM patients with strong personal and/or family history of cancer a panel testing of at least the known-cancer genes is warranted. WES can be used as a research tool. Given the high frequency of somatic BAP1 pathogenic alterations in UM, IHC screening of the tumors for BAP1 has very limited role in predicting patients with germline mutation.
We selected 35 year-old as cut off point for patients with early age of onset of their tumors which represents 15–25 years younger than the reported median age of onset of UM57. Other investigators have used younger than 18 or 21 year-old to define pediatric and young UM, respectively58, 59 but no assessment of germline pathogenic variants were carried out. The median age of onset of patients with pathogenic variants in this study, 51 years (range 39–89), was slightly lower than the median age of patients with no pathogenic variants, median 58 years (range 3 months-87 years), but the difference was not statistically significant. Younger age of onset has been observed in patients with germline BAP1 pathogenic variants 5 and the frequency of pathogenic variants is much higher (19%) in subjects with young age of onset ≤ 30 of their tumors compared to general UM population (1–2%). We recommend screening UM patients developing their tumors at young age for at least germline pathogenic variants in BAP1.
In conclusion, our results suggest locus heterogeneity in predisposition to UM. Actionable pathogenic variants in established cancer genes, other than BAP1, were identified in 3/27 (11%) familial UM and 6/129 (4.7%) of UM with high-risk for hereditary cancer. In addition, possibly deleterious variants were detected in several other genes in the DNA-damage pathway. Our results provide moderate evidence of association of PALB2 and MLH1 with predisposition to UM. Genetic testing for hereditary predisposition to cancer is warranted in UM patients with strong personal and/or family history of cancers. Future studies to explore multigenic factors, gene rearrangements and noncoding elements using a whole genome approach are warranted.
Supplementary Material
Acknowledgments
Financial Support: This work was supported by the Patti Blow Research Fund in Ophthalmology, and funds from the Ohio Lions Eye Research Foundation, the R21CA191943 and R21CA219884 grants from the National Cancer Institute (PI: Abdel-Rahman, MH), R35 CA197458 (PI. King M.C.), the National Eye Institute grant K08EY022672 (PI: Cebulla), a cancer center core grant 2P30CA016058–40 and award number 8UL1TR000090–05 from the National Center For Advancing Translational Sciences. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health. MT was supported by funding from the European Union Seventh Framework Program (2007Y2013)/ European Research Council (Grant No. 310018). The funding organizations had no role in the design or conduct of this research.
Footnotes
Conflict of Interest: Dr. Walsh reports personal fees from Color Genomics, outside the submitted work. Dr. Stacey reports personal fees from Immunocore, outside the submitted work. No conflicting relationship exists for the other authors.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- 1.Abdel-Rahman MH, Pilarski R, Cebulla CM, et al. Germline BAP1 mutation predisposes to uveal melanoma, lung adenocarcinoma, meningioma, and other cancers. J Med Genet 2011;48(12):856–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Abdel-Rahman MH, Pilarski R, Ezzat S, et al. Cancer family history characterization in an unselected cohort of 121 patients with uveal melanoma. Fam Cancer 2010;9(3):431–8. [DOI] [PubMed] [Google Scholar]
- 3.Nagy R, Sweet K, Eng C. Highly penetrant hereditary cancer syndromes. Oncogene 2004;23(38):6445–70. [DOI] [PubMed] [Google Scholar]
- 4.Lu KH, Wood ME, Daniels M, et al. American Society of Clinical Oncology Expert Statement: Collection and Use of a Cancer Family History for Oncology Providers. Journal of Clinical Oncology 2014;32(8):833–+. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Walpole S, Pritchard AL, Cebulla CM, et al. Comprehensive Study of the Clinical Phenotype of Germline BAP1 Variant-Carrying Families Worldwide. J Natl Cancer Inst 2018;110(12): 1328–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Turunen JA, Markkinen S, Wilska R, et al. BAP1 Germline Mutations in Finnish Patients with Uveal Melanoma. Ophthalmology 2016. [DOI] [PubMed] [Google Scholar]
- 7.Gupta MP, Lane AM, DeAngelis MM, et al. Clinical Characteristics of Uveal Melanoma in Patients With Germline BAP1 Mutations. JAMA Ophthalmol 2015;133(8):881–7. [DOI] [PubMed] [Google Scholar]
- 8.Popova T, Hebert L, Jacquemin V, et al. Germline BAP1 mutations predispose to renal cell carcinomas. Am J Hum Genet 2013;92(6):974–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Aoude LG, Vajdic CM, Kricker A, et al. Prevalence of germline BAP1 mutation in a population-based sample of uveal melanoma cases. Pigment Cell Melanoma Res 2013;26(2):278–9. [DOI] [PubMed] [Google Scholar]
- 10.Rai K, Pilarski R, Boru G, et al. Germline BAP1 alterations in familial uveal melanoma. Genes Chromosomes Cancer 2017;56(2):168–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cruz C, Teule A, Caminal Jm, et al. Uveal melanoma and BRCA1/BRCA2 genes: a relationship that needs further investigation. J Clin Oncol 2011;29(34):e827–9. [DOI] [PubMed] [Google Scholar]
- 12.Iscovich J, Abdulrazik M, Cour C, et al. Prevalence of the BRCA2 6174 del T mutation in Israeli uveal melanoma patients. Int J Cancer 2002;98(1):42–4. [DOI] [PubMed] [Google Scholar]
- 13.Sinilnikova OM, Egan KM, Quinn JL, et al. Germline brca2 sequence variants in patients with ocular melanoma. Int J Cancer 1999;82(3):325–8. [DOI] [PubMed] [Google Scholar]
- 14.Johansson PA, Stark A, Palmer JM, et al. Prolonged stable disease in a uveal melanoma patient with germline MBD4 nonsense mutation treated with pembrolizumab and ipilimumab. Immunogenetics 2019. [DOI] [PubMed] [Google Scholar]
- 15.Rodrigues M, Mobuchon L, Houy A, et al. Outlier response to anti-PD1 in uveal melanoma reveals germline MBD4 mutations in hypermutated tumors. Nat Commun 2018;9(1):1866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lobo J, Pinto C, Freitas M, et al. Ovarian metastasis from uveal melanoma with MLH1/PMS2 protein loss in a patient with germline MLH1 mutated Lynch syndrome: consequence or coincidence? Virchows Arch 2017;470(3):347–52. [DOI] [PubMed] [Google Scholar]
- 17.Kannengiesser C, Avril MF, Spatz A, et al. CDKN2A as a uveal and cutaneous melanoma susceptibility gene. Genes Chromosomes Cancer 2003;38(3):265–8. [DOI] [PubMed] [Google Scholar]
- 18.Marous CL, Marous MR, Welch RJ, et al. Choroidal Melanoma, Sector Melanocytosis, and Retinal Pigment Epithelial Microdetachments in Birt-Hogg-Dube Syndrome. Retin Cases Brief Rep 2017. [DOI] [PubMed] [Google Scholar]
- 19.Fontcuberta IC, Salomao DR, Quiram PA, Pulido JS. Choroidal melanoma and lid fibrofoliculomas in Birt-Hogg-Dube syndrome. Ophthalmic Genet 2011;32(3):143–6. [DOI] [PubMed] [Google Scholar]
- 20.Baglietto L, Lindor NM, Dowty JG, et al. Risks of Lynch syndrome cancers for MSH6 mutation carriers. J Natl Cancer Inst 2010;102(3):193–201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Singh AD, Schoenfield LA, Bastian BC, et al. Congenital uveal melanoma? Surv Ophthalmol 2016;61(1):59–64. [DOI] [PubMed] [Google Scholar]
- 22.Riley BD, Culver JO, Skrzynia C, et al. Essential elements of genetic cancer risk assessment, counseling, and testing: updated recommendations of the National Society of Genetic Counselors. J Genet Couns 2012;21 (2):151–61. [DOI] [PubMed] [Google Scholar]
- 23.Weitzel JN, Blazer KR, MacDonald DJ, et al. Genetics, genomics, and cancer risk assessment: State of the Art and Future Directions in the Era of Personalized Medicine. CA Cancer J Clin 2011;61(5):327–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hampel H, Sweet K, Westman JA, et al. Referral for cancer genetics consultation: a review and compilation of risk assessment criteria. J Med Genet 2004;41(2):81–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Miller SA, Dykes DD, Polesky HFrn. A simple salting out procedure for extracting DNA from human nucleated cells. Nucleic Acids Res 1988;16(3):1215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Fitzgerald LM, Kumar A, Boyle EA, et al. Germline Missense Variants in the BTNL2 Gene Are Associated with Prostate Cancer Susceptibility. Cancer Epidemiol Biomarkers Prev 2013;22(9):1520–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kelly BJ, Fitch JR, Hu Y, et al. Churchill: an ultra-fast, deterministic, highly scalable and balanced parallelization strategy for the discovery of human genetic variation in clinical and population-scale genomics. Genome Biol 2015;16:6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Li H, Durbin R. Fast and accurate long-read alignment with Burrows-Wheeler transform. Bioinformatics 2010;26(5):589–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Landrum MJ, Lee JM, Benson M, et al. ClinVar: improving access to variant interpretations and supporting evidence. Nucleic Acids Res 2018;46(D1):D1062–D7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kumar P, Henikoff S, Ng PC. Predicting the effects of coding non-synonymous variants on protein function using the SIFT algorithm. Nat Protoc 2009;4(7):1073–81. [DOI] [PubMed] [Google Scholar]
- 31.Adzhubei IA, Schmidt S, Peshkin L, et al. A method and server for predicting damaging missense mutations. Nat Methods 2010;7(4):248–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rahman N Realizing the promise of cancer predisposition genes. Nature 2014;505(7483):302–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Milanowska K, Krwawicz J, Papaj G, et al. REPAIRtoire--a database of DNA repair pathways. Nucleic Acids Res 2011;39(Database issue):D788–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wood RD, Mitchell M, Lindahl T. Human DNA repair genes, 2005. Mutat Res 2005;577(1–2):275–83. [DOI] [PubMed] [Google Scholar]
- 35.Walsh T, Casadei S, Lee MK, et al. Mutations in 12 genes for inherited ovarian, fallopian tube, and peritoneal carcinoma identified by massively parallel sequencing. Proc Natl Acad Sci U S A 2011; 108(44):18032–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Walsh T, Lee MK, Casadei S, et al. Detection of inherited mutations for breast and ovarian cancer using genomic capture and massively parallel sequencing. Proc Natl Acad Sci U S A 2010; 107(28):12629–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Nord AS, Lee M, King MC, Walsh T. Accurate and exact CNV identification from targeted high-throughput sequence data. BMC Genomics 2011;12:184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Boland CR, Thibodeau SN, Hamilton SR, et al. A National Cancer Institute Workshop on Microsatellite Instability for cancer detection and familial predisposition: development of international criteria for the determination of microsatellite instability in colorectal cancer. Cancer Res 1998;58(22):5248–57. [PubMed] [Google Scholar]
- 39.Abdel-Rahman MH, Boru G, Massengill J, et al. MET oncogene inhibition as a potential target of therapy for uveal melanomas. Invest Ophthalmol Vis Sci 2010;51(7):3333–9. [DOI] [PubMed] [Google Scholar]
- 40.Abdel-Rahman MH, Yang Y, Zhou XP, et al. High frequency of submicroscopic hemizygous deletion is a major mechanism of loss of expression of PTEN in uveal melanoma. J Clin Oncol 2006;24(2):288–95. [DOI] [PubMed] [Google Scholar]
- 41.Abdel-Rahman MH, Craig EL, Davidorf FH, Eng C. Expression of vascular endothelial growth factor in uveal melanoma is independent of 6p21-region copy number. Clin Cancer Res 2005;11(1):73–8. [PubMed] [Google Scholar]
- 42.Fay MP. Confidence intervals that match Fisher’s exact or Blaker’s exact tests. Biostatistics 2010;11(2):373–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Fay MP. Two-sided Exact Tests and Matching Confidence Intervals for Discrete Data. R Journal 2010;2(1):53–8. [Google Scholar]
- 44.Benfodda M, Gazal S, Descamps V, et al. Truncating mutations of TP53AIP1 gene predispose to cutaneous melanoma. Genes Chromosomes Cancer 2018;57(6):294–303. [DOI] [PubMed] [Google Scholar]
- 45.Rodrigue A, Margaillan G, Torres Gomes T, et al. A global functional analysis of missense mutations reveals two major hotspots in the PALB2 tumor suppressor. Nucleic Acids Res 2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Southey MC, Teo ZL, Dowty JG, et al. A PALB2 mutation associated with high risk of breast cancer. Breast Cancer Res 2010;12(6):R109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Huang KL, Mashl RJ, Wu Y, et al. Pathogenic Germline Variants in 10,389 Adult Cancers. Cell 2018;173(2):355–70 e14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Boru G, Grosel TW, Pilarski R, et al. Germline large deletion of BAP1 and decreased expression in non-tumor choroid in uveal melanoma patients with high risk for inherited cancer. Genes Chromosomes Cancer 2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Strande NT, Riggs ER, Buchanan AH, et al. Evaluating the Clinical Validity of Gene-Disease Associations: An Evidence-Based Framework Developed by the Clinical Genome Resource. Am J Hum Genet 2017;100(6):895–906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lynch HT, de la Chapelle A. Hereditary colorectal cancer. N Engl J Med 2003;348(10):919–32. [DOI] [PubMed] [Google Scholar]
- 51.Tischkowitz M, Xia B. PALB2/FANCN: recombining cancer and Fanconi anemia. Cancer Res 2010;70(19):7353–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Raffalli-Ebezant H, Rutherford SA, Stivaros S, et al. Pediatric intracranial clear cell meningioma associated with a germline mutation of SMARCE1: a novel case. Childs Nerv Syst 2015;31(3):441–7. [DOI] [PubMed] [Google Scholar]
- 53.Evans LT, Van Hoff J, Hickey WF, et al. SMARCE1 mutations in pediatric clear cell meningioma: case report. J Neurosurg Pediatr 2015;16(3):296–300. [DOI] [PubMed] [Google Scholar]
- 54.Lin B, Kesserwan C, Quinn EA, et al. Anaplastic Astrocytoma in a Child With Coffin-Siris Syndrome and a Germline SMARCE1 Mutation: A Case Report. J Pediatr Hematol Oncol 2018. [DOI] [PubMed] [Google Scholar]
- 55.Wei QX, Claus R, Hielscher T, et al. Germline allele-specific expression of DAPK1 in chronic lymphocytic leukemia. PLoS One 2013;8(1):e55261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Tomsic J, He H, Akagi K, et al. A germline mutation in SRRM2, a splicing factor gene, is implicated in papillary thyroid carcinoma predisposition. Sci Rep 2015;5:10566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kaliki S, Shields Cl. Uveal melanoma: relatively rare but deadly cancer. Eye (Lond) 2017;31(2):241–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Fry MV, Augsburger JJ, Correa ZM. Clinical Features, Metastasis, and Survival in Patients Younger Than 21 Years With Posterior Uveal Melanoma. Jama Ophthalmology 2019;137(1):75–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Al-Jamal RT, Cassoux N, Desjardins L, et al. The Pediatric Choroidal and Ciliary Body Melanoma Study: A Survey by the European Ophthalmic Oncology Group. Ophthalmology 2016; 123(4):898–907. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.