Abstract
Background
Musk deer can produce musk which has high medicinal value and is closely related to human health. Viruses in forest musk deer both threaten the health of forest musk deer and human beings.
Methods
Using viral metagenomics we investigated the virome in 85 faeces samples collected from forest musk deer.
Results
In this article, eight novel CRESS-DNA viruses were characterized, whole genomes were 2148 nt–3852 nt in length. Phylogenetic analysis indicated that some viral genomes were part of four different groups of CRESS-DNA virus belonging in the unclassified CRESS-DNA virus, Smacoviridae, pCPa-like virus and pPAPh2-like virus. UJSL001 (MN621482), UJSL003 (MN621469) and UJSL017 (MN621476) fall into the branch of unclassified CRESS-DNA virus (CRESSV1–2), UJSL002 (MN621468), UJSL004 (MN621481) and UJSL007 (MN621470) belong to the cluster of Smacoviridae, UJSL005 (MN604398) showing close relationship with pCPa-like (pCRESS4–8) clusters and UJSL006 (MN621480) clustered into the branch of pPAPh2-like (pCRESS9) virus, respectively.
Conclusion
The virome in faeces samples of forest musk deer from Chengdu, Sichuan province, China was revealed, which further characterized the diversity of viruses in forest musk deer intestinal tract.
Keywords: Forest musk deer, Faeces, Viral metagenomics, CRESS-DNA virus, Complete genome
Introduction
Forest musk deer is a national protected animal, mainly distributed in Sichuan province, Guangxi province and other places, China [1, 2]. The death of forest musk deer occurs mainly in the young musk deer. Diseases were the most important factor in causing fawn death [3]. There have been studies on the diagnosis and prevention of some known diseases [4–7], but there is a lack of research on the unknown etiology.
Viruses with small circular rep-encoding ssDNA (CRESS-DNA) genomes encode a replication associated protein (Rep), mainly includes Circoviridae [8], Genomoviridae.
[9], Smacoviridae [10], Geminiviridae, Nanoviridae and Bacilladnaviridae [11]. These are widely found in various environments [12, 13], plant samples [14–17], dragonflies and damselflies [18–21], mosquitoes [22], rats [23], bats [24], duck [25], cattle [26], pigs [27, 28], dogs [29], human [30–33], and turkey [34]. CRESS-DNA genomes typically encode a replication initiator protein (Rep) and a capsid protein (Cap) [35].
In this study, the virus community in the intestinal tract of forest musk deer was analyzed by virus metagenomics. The results of this study put forward for the first time on CRESS-DNA viruses propagating among forest musk deer.
Materials and methods
Samples
In 2016, 85 forest musk deer faeces samples were collected from Chengdu, Sichuan province, China. Samples were collected by disposable materials and transported to the laboratory on dry-ice and store in the − 80 °C refrigerator. Samples were put into 1.5 ml tubes containing phosphate buffered saline (PBS). The supernatants of fecal samples were collected after vigorous eddy current for 5 min and centrifugation for 10 min (15,000 g) [36, 37].
Viral metagenomic analysis
500 μl of supernatant was filtered through a 0.45 μm filter (Millipore) to remove eukaryotic and bacterial cell sized particles. The viral particle enrichment filtrate was then treated with uncleases to digest nonparticle protected nucleic acid at 37 °C for 90 min [38]. Remaining total nucleic acid, protected from digestion with in viral capsids, were then extracted using the QiaAmp Mini Viral RNA kit (Qianen) according to manufacturer’s protocol [37, 39, 40]. Eight separate pools of nucleic acids from 85 faecal specimens were generated randomly, of which six contained ten faecal apecimens, the other one contained 12 faecal specimens and another one contained 13 faecal specimens. These eight viral nucleic acid pools, containing both DNA and RNA viral sequences, were then subjected to RT reactions with SuperScript III reverse transcriptase (Invitrogen) and 100 pmol of a random hexamer primer, followed by a single round of DNA synthesis using Klenow fragment polymerase [37, 41]. Eight libraries were constructed using Nextera XT DNA Sample Preparation Kit (Illumina) and sequenced using the MiSeq Illumina platform with 250 bases paired ends with dual barcoding for each library. The data is processed using an internal analysis pipeline running on a 32-nodes Linux cluster. Clonal reads were removed, and low quality sequence tails were trimmed with Phred quality score ten as the threshold. The adapter is trimmed using of VecScreen’s default parameters, NCBI BLASTn, with specialized parameters designed for adapter removal [42]. After deleting repeated reads and reads less than 50 in length followed by de novo assembly [43]. The contigs and singlets were matched against an internal viral proteome database using BLASTx with an E-value cutoff of < 10–5. BLASTx were used to identify viral sequences in order to annotated viral proteins available in GenBank’s viral RefSeq database [44].
Genome acquisition and PCR screening
Putative open reading frames (ORFs) in the circular genomes were predicted by Geneious software version 2019.0.3 [45], and the stem-loop in the circular genomes were located through the The Mfold [24] (Table 1 and Fig. 1b). If the whole genome sequence of the virus was not obtained through sequence reads analysis, inverse PCR was needed. Two whole genomes of UJSL004 and UJSL005 were acquired by screen PCR and inverse PCR. Primers are shown in an additional file (see Additional file 1). The PCR conditions in screen PCR were: 95 °C for 5 min, 31 cycles 95 °C for 30 s, 50 °C (for the first round) or 57 °C (for the second round) for 30 s and 72 °C for 40 s, a final extension at 72 °C for 5 min, resulting in an expected amplicon of 300 bp–500 bp. The PCR conditions in inverse PCR of UJSL004 were: 95 °C for 5 min, 35 cycles 95 °C for 30 s, 50 °C (for the first round) or 55 °C (for the second round) for 30 s and 72 °C for 1.5 min, a final extension at 72 °C for 5 min, resulting in an expected amplicon of 1000 bp. The PCR conditions in inverse PCR of UJSL005 were: 95 °C for 5 min, 35 cycles 95 °C for 30 s, 50 °C (for the first round) or 51 °C (for the second round) for 30 s and 72 °C for 1.5 min, a final extension at 72 °C for 5 min, resulting in an expected amplicon of 1000 bp.
Table 1.
Loop sequences of these CRESS-DNA virus CRESS-DNA, small circular rep-encoding ssDNA
| strain name | Loop sequence | sequence length |
|---|---|---|
| UJSL001 | ATTCTTCTACGCTT | 14 |
| UJSL002 | GCCACCCTCGAC | 12 |
| UJSL003 | AGTATGAGGT | 10 |
| UJSL004 | AGGCTCATCATAT | 13 |
| UJSL005 | CCAACCCCCCAAG | 13 |
| UJSL006 | GCTTAATATTACC | 13 |
| UJSL007 | ATAGTTCACT | 10 |
| UJSL017 | ACCTGAATATT | 11 |
Fig. 1.
The whole genome sequence structure and the stem-loop structures of the eight CRESS-DNA viruses isolated from forest musk deer. a. The genomic organization of the eight CRESS-DNA viruses identified in the forest musk deer faeces samples. b. The stem-loop structures of the eight CRESS-DNA viruses isolated from forest musk deer
Phylogenetic analysis
The Rep protein sequences of these novel virus were homology alignment with the reference sequences in GenBank using the ClustalW program in MEGA7.0. Phylogenetic analyses were constructed using full-length rep protein of novel virus and other genetically close relatives [22, 46]. Save the aligned sequence as a Nexus form file, which was used to construct the phylogenetic tree using Bayes’ theorem in Mrbayes3.2.7 program. Using mixed models and Markov chain Monte Carlo (MCMC) methods. In phylogenetic analyses, tree samples are typically most divergent, so we introduced the average standard deviation of split frequencies (ASDSF) in MrBayes to allow quantitative evaluation of similarity among these samples. MrBayes allow users to set cut-off frequency (default value 0.10, [47–49]). We used the “sump” and “sumt” commands to get more detailed diagnostic information after the run has completed.
Results
The 85 faeces samples of the eight libraries generated a total of 6, 153, 736 unique sequence reads using illumine Miseq sequencing runs with 250 base pair terminals. The Ensemble program was used to read the de novo assembly sequence [43] and BLASTx was used to compare it with Genbank’s non-redundant protein database. The results indicated that CRESS-DNA virus accounted for the main part of the total mammalian virus readings, with 4, 775 reads showing sequence similarity to the CRESS-DNA virus, 462 reads related to viruses from Smacoviridae and 473 reads sequence similar to the virus of Circoviridae. Table 2 list the detailed information.
Table 2.
Characterization of the viral sequence reads in forest musk deer faeces samples. nt nucleotides, aa amino acids
| Library ID (Total unique reads) | Family | Genus | Strain name | Accession number | No. of nt in the genome | No. of aa in protein | GC-content(%) | GenBank no. of the matches | Aa identities with the match(s) | Max_Contig | Total reads | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Rep (Direction) | Cap (Direction) | |||||||||||
| L1(1136670) | Circoviridae | Circovirus | UJSL017 | MN621476 | 3025 | 355(−) | 380(+) | 42.6 | KU043411.1 | 62.54% | 2497 | 1123 |
| L2(991928) | None | None | UJSL001 | MN621482 | 3518 | 399(−) | 190(+) | 41 | KY487934.1 | 48.76% | 3448 | 583 |
| L3(307679) | Circoviridae | None | UJSL002 | MN621468 | 2665 | 283(−) | 372(+) | 50.8 | NC_030125.1 | 69.09% | 3919 | 205 |
| None | None | UJSL003 | MN621469 | 3212 | 281(−) | 173(+) | 46.5 | MH617688.1 | 44.00% | 3330 | 206 | |
| L5(777594) | Circoviridae | None | UJSL004 | MN621481 | 2866 | 390(−) | 204(+) | 40.6 | NC_039054.1 | 75.39% | 2053 | 41 |
| L6(530538) | Circoviridae | None | UJSL005 | MN604398 | 3852 | 428(−) | 692(+) | 42.4 | NC_026635.1 | 32.63% | 3155 | 473 |
| L7(1237606) | None | None | UJSL006 | MN621480 | 2148 | 327(+) | 203(+) | 32.4 | MK858258.1 | 58.86% | 2253 | 2863 |
| L8(802429) | ||||||||||||
| L9(369292) | None | None | UJSL007 | MN621470 | 2526 | 280(−) | 337(+) | 45.6 | MH500284.1/MH500317.1 | 95.54% | 2672 | 216 |
| Total(6153736) | ||||||||||||
CRESS-DNA genomes
Four complete CRESS-DNA genomes showing the highest sequence identity to CRESS-DNA virus. Genomes were 3518 nt (UJSL001, from library 2), 3212 nt (UJSL003, from library 3), 2148 nt (UJSL006, from library 7) and 3025 nt (UJSL017, from library 1) in length. As shown in Fig. 1a, the genomes of UJSL001, UJSL003 and UJSL017 contained two bidirectional ORFs while UJSL006 is in the same direction, encoding the putative Rep and Cap proteins. BLASTp search in GenBank based on the protein sequence of Rep showed UJSL001 shared the highest identity of 48.76% to unclassified circular virus (KY487934.1), UJSL003 shared the highest sequence identity of 44.00% to unclassified ssDNA viruses (MH617688.1), UJSL006 shared the highest sequence identity of 58.86% to an unclassified circular DNA viruses (MK858258.1) and UJSL017 shared the highest identity of 62.54% to unclassified ssDNA viruses (KU043411.1) (Table 2).
Three complete CRESS-DNA genomes showing the highest identity to Smacovirus. Genomes were 2665 nt (UJSL002, from library 3), 2866 nt (UJSL004, from library 5) were obtained through inverse PCR, and 2526 nt (UJSL007, from library 9) in length, respectively. Figure 1a manifested the genomic organization of UJSL002, UJSL004 and UJSL007, where the predicted Rep and Cap of the three viruses are differently arranged. BLASTp search in GenBank based on the protein sequence of Rep showed UJSL002 shared the highest sequence identity of 69.09% to a Bovine faeces associated smacovirus5 (NC_030125.1), UJSL004 shared the highest sequence identity of 75.39% to a Bovismacovirus (NC_039054.1) and UJSL007 shared the highest sequence identity of 95.54% to two Porprismacovirus (MH500284.1 and MH500317.1) (Table 2).
A complete CRESS-DNA genome showing the highest sequence identity to Circoviridae. Genome was 3852 nt (UJSL005, from library 6) in length. UJSL005 genome was acquired through inverse PCR based on a large contigs from library 6 and Sanger sequencing. Figure 1a indicated the genomic organization of UJSL005, where the predicted Rep and Cap of the UJSL005 in the opposite direction. BLASTp search in GenBank based on the protein sequence of Rep showed UJSL005 shared the highest sequence identity of 32.63% to unclassified Circoviridae (NC_026635.1) (Table 2).
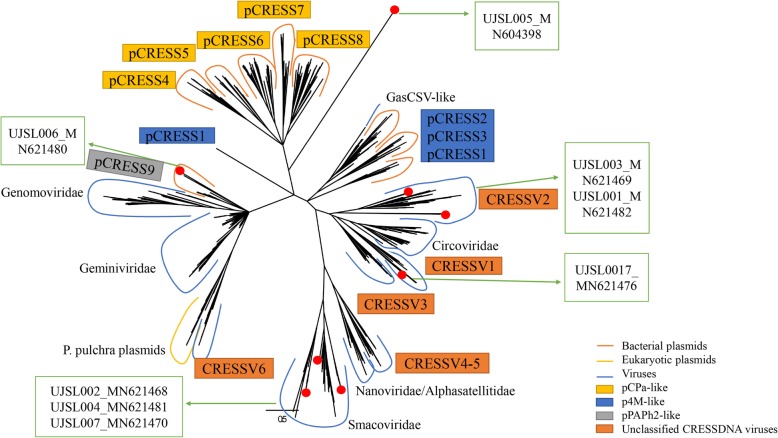
Based on the alignment of the Rep amino acid sequences herein detected with the best matches of BLASTp search in GenBank and those of representative CRESS-DNA genomes including 6 groups of unclassified CRESS-DNA virus (CRESSV1–6), two GasCSV-like viruses, Bacterial plasmids (pCRESS1–9) and a small group of Eukaryotic plasmids (P. pulchra plasmids) from GenBank, a phylogenetic tree was constructed [50–52]. For phylogenetic analyses, we used a dataset with 672 sequences of the Rep amino acid (Fig. 2) (Additional file 2).
Fig. 2.
Phylogenetic analysis and genomic organization of the novel CRESS-DNA virus identified in forest musk deer. Phylogenetic analysis was performed based on the amino acid sequence of Rep protein. A total of 672 alignment sequences were included 8 CRESS-DNA virus identified here, their best BLASTp matches in GenBank based on the Rep proteins, and the representative of all classified families of CRESS-DNA virus as well as 6 groups of unclassified CRESS-DNA virus (CRESSV1–6), two GasCSV-like viruses, Bacterial plasmids (pCRESS1–9) and a small group of Eukaryotic plasmids (P. pulchra plasmids). All clads are shown with curves and the names are shown beside the corresponding clades. Viruses identified in this study were labeled with red colored dots and the virus names and sequence accession numbers are shown in green arrow and text box beside their corresponding
UJSL001, UJSL003 and UJSL017 fall into the branch of unclassified CRESS-DNA virus (CRESSV1–2), UJSL001 and UJSL003 belong to the cluster of CRESSV2, UJSL001 showing close relationship with CRESS_AUM21936, UJSL003 showing close relationship with CRESS_AXH77830 (Fig. 3a) (see Additional file 3) and UJSL017 belong to the cluster of CRESSV1, showing close relationship with CRESSV1_KJ206566 and CRESSV1_KU043411 (Fig. 3e) (see Additional file 7). UJSL002, UJSL004 and UJSL007 belong to the cluster of Smacoviridae (Fig. 3b) (see Additional file 4), UJSL005 fall into the branch showing close relationship with pCPa-like (pCRESS4–8) clusters (Fig. 3c) (see Additional file 5) and UJSL006 fall into pPAPh2-like (pCRESS9) clusters, showing close relationship with pCRESS9_KXT29032 (Fig. 3d) (see Additional file 6).
Fig. 3.
Phylogenetic analysis of novel virus detected from forest musk deer in different clads. The accession clusters name is shown on the right side of the tree and the novel virus name are marked with red color. a. Phylogenetic analysis were based on the Rep protein amino acid sequence of UJSL001, UJSL003 and 12 similar viral amino acid sequences in the clads of unclassified CRESS-DNA virus (CRESSV2). b. Phylogenetic trees were constructed with the Rep protein amino acid sequence of UJSL002, UJSL004, UJSL007 and 15 similar viral amino acid sequences in the clads of Smacoviridae. c. Phylogenetic trees were constructed used the Rep protein amino acid sequence of UJSL005 and 15 the similar viral amino acid sequences into pCPa-like (Pcress4–8) clusters. d. Phylogenetic trees were constructed with UJSL006 and 11 similar viral amino acid sequences into pPAPh2-like (Pcress9) clusters. e. Phylogenetic were constructed by Rep protein amino acid sequence of UJSL017 and 12 viruses amino acid sequences that are relatively close together in the clads of unclassified CRESS-DNA virus (CRESSV1)
Nucleotide sequence accession numbers
The viral genomes described in detail here were deposited in GenBank under the following accession numbers: MN604398, MN621468- MN621470, MN621480- MN621482 and MN621476.
Discussion
Our report describes viral nucleic acids enriched in forest musk deer faeces, shows that CRESS-DNA virus sequences are present in all libraries and have the most reads compared to other viruses. This suggests that these viruses are likely to replicate in forest musk deer host cells, but there is no evidence for this. Based on phylogenetic analysis, four different groups of CRESS-DNA genomes in forest musk deer faeces were detected, which belonged to CRESS-DNA virus, Smacoviridae, pCPa-like virus (pCRESS4–8) and pPAPh2-like (pCRESS9). For the first time, CRESS-DNA virus in the faeces of forest musk deer was mentioned, which was beneficial to further understanding of the genetic and evolutionary diversity of these viruses.
CRESS-DNA viruses with small, circular replication-associated protein (Rep)-encoding single stranded (CRESS) DNA genomes, are largely identified based on conserved rolling circle replication proteins [11]. It consists of a large group of highly specific viruses that can infect many types of host [53]. These virus included: Circoviridae [39], which can infect vertebrates, Geminiviridae [14] and Nanoviridae [54] which can infect plants. The genomes of Circoviridae range in size from 1.7 to 2.1 kb and contain two major ORFs, which encode Rep and Cap proteins. According to the International Committee on Taxonomy of Viruses (ICTV), the ssDNA has genomes between 1.7-6 kb. Eight CRESS-DNA virus extracted in this study, the genomes range in size from 2.1 kb to 3.5 kb. Previous research on the stem-loop structure of diverse circovirus and cycloviruses, a highly conserved stem-loop structure is also found [31, 52, 55], because they study multiple viruses of the same genus. Eight viruses in our study located in different genera based on rep protein phylogenetic analysis, so the stem-loop structure sequences are different from each other.
In the recent years, a large number of CRESS-DNA genomes have been determined in human and any other mammals, birds, insects, plants, fungi, and environment samples which bringing to light a high level of genetic diversity among these virus [25, 26, 31, 33, 52, 56]. Although use metagenomics to identify these viruses from forest musk deer faeces, we cannot rule out that they may also represent food contaminants and environmental pollution [57]. These viruses exploit host polymerases for DNA synthesis and code for proteins that modulate the host’s cell cycle favourably for virus multiplication [58]. There are reports that the virus is associated with disease, but it has not been proven to cause the disease directly [59, 60]. And the effects and disease correlation of these viruses on the health of forest musk deer need further study.
In conclusion, this study is the first to discover a variety of new CRESS-DNA viruses in the intestinal tract of forest musk deer and analyze their genomic characteristics, which is of great significance for the study of forest musk deer virus and the genetic and evolutionary diversity of CRESS-DNA virus. At the same time, the host adaptability and pathogenicity of the new CRESS-DNA virus need further study.
Conclusions
The virome in faeces samples of forest musk deer from Chengdu included the viruses showing sequence similarity to CRESS-DNA viruses, where eight divergent genomes of CRESS-DNA viruses were identified in detail. The contents include genome protein structure, stem-loop structure and rep protein phylogenetic analysis. Although CRESS-DNA virus is prevalent in forest musk deer, its pathogenicity has not been known. This study increased the knowledge of the diversity of viruses in forest musk deer faeces.
Supplementary information
Acknowledgements
Not applicable.
Abbreviations
- BLAST
Basic local alignment search tool
- CRESS
Circular rep-encoding single stranded
- GasCSV
Gastropod-associated circular ssDNA virus
- NCBI
National Center for Biotechnology Information
- ORF
Open reading frame
- pCPa
Circular plasmids from plant-pathogenic
- PCR
Polymerase chain reaction
- pPAPh2
Plasmids Plant Pathogens of Phytoplasma
- P. pulchra plasmids
Pyropia pulchra plasmids
Authors’ contributions
WZ and GQ conceived the study. QL performed most of the experiments. QL and WZ wrote the paper. All authors participated the experiments and read and approved the final manuscript.
Funding
This work was partly supported by National Key Research and Development Programs of China No. 2017YFC1200201, Science and Technology Department of Sichuan Province No. 2018SZDX0037, Jiangsu Provincial Key Research and Development Projects No. BE2017693, National Natural Science Foundation of China No. 81741062 and Chengdu technology hui min technology research and development project No. 2015-HM01–00234-SF.
Availability of data and materials
The viral genomes described in detail here were deposited in GenBank under the following accession numbers: MN604398, MN621468- MN621470, MN621480- MN621482 and MN621476.
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Qi Liu, Email: liuqi_11807@163.com.
Hao Wang, Email: 863251487@qq.com.
Yu Ling, Email: 13505522945@163.com.
Shi-Xing Yang, Email: johnsonyang1979@163.com.
Xiao-Chun Wang, Email: chun_xiao@163.com.
Rui Zhou, Email: 18326905093@163.com.
Yu-Qing Xiao, Email: 379387268@qq.com.
Xu Chen, Email: chenxu710810145@qq.com.
Jie Yang, Email: 1647517479@qq.com.
Wei-Guo Fu, Email: fuweiguo@ujs.edu.cn.
Wen Zhang, Email: z0216wen@yahoo.com.
Gui-Lan Qi, Email: 497006351@qq.com.
Supplementary information
Supplementary information accompanies this paper at 10.1186/s12985-020-01332-y.
References
- 1.Yang QS, Meng XX, Xia L, Feng ZJ. Conservation status and causes of decline of musk deer (Moschus spp.) in China. Biol Conserv. 2003;109:333–342. doi: 10.1016/S0006-3207(02)00159-3. [DOI] [Google Scholar]
- 2.Zou FD, Yue BS, Xu L, Zhang Y. Isolation and characterization of microsatellite loci from forest musk deer (Moschus berezovskii) Zool Sci. 2005;22:593–598. doi: 10.2108/zsj.22.593. [DOI] [PubMed] [Google Scholar]
- 3.Xu ZQ, Xu HF. Population characteristics and fawn survival in musk deer (Moshus moschiferus) Acta Theriologica Sinica. 2003;1:17–21. [Google Scholar]
- 4.Fu WL, Fu CM, Wang JM, Cai YH. Diagnosis and treatment of deep part abscess on the rearward of forest musk deer throat. J Econ Anim. 2010;4:222–224. [Google Scholar]
- 5.Li L, Wang BB, Ge YF, Wan QH. Major histocompatibility complex class II polymorphisms in forest musk deer (Moschus berezovskii) and their probable association with purulent disease. Int J Immunogenet. 2014;41:401–412. doi: 10.1111/iji.12135. [DOI] [PubMed] [Google Scholar]
- 6.Sun X, Cai R, Jin X, Shafer ABA, Hu X, Yang S, Li Y, Qi L, Liu S, Hu D. Blood transcriptomics of captive forest musk deer (Moschus berezovskii) and possible associations with the immune response to abscesses. Sci Rep. 2018;8(1):599. doi: 10.1038/s41598-017-18534-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhao K, Tian Y, Yue B, Wang H, Zhang X. Virulence determinants and biofilm production among Trueperella pyogenes recovered from abscesses of captive forest musk deer. Arch Microbiol. 2013;195(3):203–209. doi: 10.1007/s00203-013-0869-7. [DOI] [PubMed] [Google Scholar]
- 8.Breitbart M, Delwart E, Rosario K, Segalés J, Varsani A, ICTV report consortium ICTV virus taxonomy profile:Circoviridae. J Gen Virol. 2017;98(8):1997–1998. doi: 10.1099/jgv.0.000871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Krupovic M, Ghabrial SA, Jiang D, Varsani A. Genomoviridae: a new family of widespread single-stranded DNA viruses. Arch Virol. 2016;161(9):2633–2643. doi: 10.1007/s00705-016-2943-3. [DOI] [PubMed] [Google Scholar]
- 10.Varsani A, Krupovic M. Correction to: Smacoviridae : a new family of animal-associated single-stranded DNA viruses. Arch Virol. 2018;163(11):3213–3214. doi: 10.1007/s00705-018-3960-1. [DOI] [PubMed] [Google Scholar]
- 11.Guo Z, He Q, Tang C, Zhang B, Yue H. Identification and genomic characterization of a novel CRESS DNA virus from a calf with severe hemorrhagic enteritis in China. Virus Res. 2018;255:141–146. doi: 10.1016/j.virusres.2018.07.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bistolas K, Jackson EW, Watkins JM, Rudstam LG, Hewson I. Distribution of circular single-stranded DNA viruses associated with benthic amphipods of genus Diporeia in the Laurentian Great Lakes. Freshw Biol. 2017;62(7):1220–1231. doi: 10.1111/fwb.12938. [DOI] [Google Scholar]
- 13.Reavy B, Swanson MM, Cock PJ, Dawson L, Freitag TE, Singh BK, Torrance L, Mushegian AR, Taliansky M. Distinct circular single-stranded DNA viruses exist in different soil types. Appl Environ Microbiol. 2015;81(12):3934. doi: 10.1128/AEM.03878-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Brown JK, Merritt R, Nelson Characterisation of watermelon curly mottle virus, a geminivirus distinct from squash leaf curl virus. Ann Appl Biol. 1989;115(2):243–252. doi: 10.1111/j.1744-7348.1989.tb03383.x. [DOI] [Google Scholar]
- 15.Bryan D, Harrison HB, Bock KR, Guthrie EJ, Gina Meredith M, ATKINSON Plant viruses with circular single-stranded DNA. Nature. 1977;270(5639):760–762. doi: 10.1038/270760a0. [DOI] [Google Scholar]
- 16.Howarth AJ, Goodman RM. Plant viruses with genomes of single-stranded DNA. Trends Biochem Sci. 1982;7(5):180–182. doi: 10.1016/0968-0004(82)90133-5. [DOI] [Google Scholar]
- 17.Ikegami M, Yazaki K, Honda Y, Iwaki M, Fujii H, Morinaga T, Miura K. Single-stranded DNA in Mung bean yellow mosaic virus. Microbiol Immunol. 2013;29(8):783–789. doi: 10.1111/j.1348-0421.1985.tb00881.x. [DOI] [PubMed] [Google Scholar]
- 18.Dayaram A, Potter KA, Pailes R, Marinov M, Rosenstein DD, Varsani A. Identification of diverse circular single-stranded DNA viruses in adult dragonflies and damselflies (Insecta: Odonata) of Arizona and Oklahoma, USA. Infect Genet Evol. 2015;30:278–287. doi: 10.1016/j.meegid.2014.12.037. [DOI] [PubMed] [Google Scholar]
- 19.Islam SU, Lin W, Wu R, Lin C, Islam W, Arif M, Du Z, Wu Z. Complete genome sequences of three novel cycloviruses identified in a dragonfly (Odonata: Anisoptera) from China. Arch Virol. 2018;163(9):1–5. doi: 10.1007/s00705-018-3876-9. [DOI] [PubMed] [Google Scholar]
- 20.Rosario K, Dayaram A, Marinov M, Ware J, Kraberger S, Stainton D, Breitbart M, Varsani A. Diverse circular ssDNA viruses discovered in dragonflies (Odonata: Epiprocta) J Gen Virol. 2012;93(Pt 12):2668. doi: 10.1099/vir.0.045948-0. [DOI] [PubMed] [Google Scholar]
- 21.Rosario K, Marinov M, Stainton D, Kraberger S, Wiltshire EJ, Collings DA, Walters M, Martin DP, Breitbart M, Varsani A. Dragonfly cyclovirus, a novel single-stranded DNA virus discovered in dragonflies (Odonata: Anisoptera) J Gen Virol. 2011;92(Pt 6):1302. doi: 10.1099/vir.0.030338-0. [DOI] [PubMed] [Google Scholar]
- 22.Garigliany MM, Börstler J, Jöst H, Badusche M, Desmecht D, Schmidt-Chanasit J, Cadar D. Characterization of a novel circo-like virus in Aedes vexans mosquitoes from Germany: evidence for a new genus within the Circoviridae family. J Gen Virol. 2015;96(Pt 4):915–920. doi: 10.1099/vir.0.000036. [DOI] [PubMed] [Google Scholar]
- 23.Hansen TA, Fridholm H, Frøslev TG, Kjartansdóttir KR, Willerslev E, Nielsen LP, Hansen AJ. New type of papillomavirus and novel circular single stranded DNA virus discovered in urban Rattus norvegicus using circular DNA enrichment and Metagenomics. PLoS One. 2015;10(11):e0141952. doi: 10.1371/journal.pone.0141952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ge X, Li J, Peng C, Wu L, Yang X, Wu Y, Zhang Y, Shi Z. Genetic diversity of novel circular ssDNA viruses in bats in China. J Gen Virol. 2011;92(Pt 11):2646–2653. doi: 10.1099/vir.0.034108-0. [DOI] [PubMed] [Google Scholar]
- 25.Fehér E, Kaszab E, Forró B, Bali K, Marton S, Lengyel G, Bányai K. Genome sequence of a mallard duck origin cyclovirus, DuACyV-1. Arch Virol. 2017;162(12):1–5. doi: 10.1007/s00705-017-3566-z. [DOI] [PubMed] [Google Scholar]
- 26.Wang H, Li S, Mahmood A, Yang S, Wang X, Shen Q, Shan T, Deng X, Li J, Hua X, Cui L, Delwart E, Zhang W. Plasma virome of cattle from forest region revealed diverse small circular ssDNA viral genomes. Virol J. 2018;15(1):11. doi: 10.1186/s12985-018-0923-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cheung AK, Ng TF, Lager KM, Bayles DO, Alt DP, Delwart EL, Pogranichniy RM, Kehrli ME., Jr A divergent clade of circular single-stranded DNA viruses from pig feces. Arch Virol. 2013;158(10):2157–2162. doi: 10.1007/s00705-013-1701-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sikorski A, Argüello-Astorga GR, Dayaram A, Dobson RC, Varsani A. Discovery of a novel circular single-stranded DNA virus from porcine faeces. Arch Virol. 2013;158(1):283–289. doi: 10.1007/s00705-012-1470-0. [DOI] [PubMed] [Google Scholar]
- 29.Li L, McGraw S, Zhu K, Leutenegger CM, Marks SL, Kubiski S, Gaffney P, Dela Cruz FN, Jr, Wang C, Delwart E, Pesavento PA. Circovirus in tissues of dogs with vasculitis and hemorrhage. Emerg Infect Dis. 2013;19(4):534–541. doi: 10.3201/eid1904.121390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tan le V, van Doorn HR, Nghia HD, Chau TT, Tu le TP, de Vries M, Canuti M, Deijs M, Jebbink MF, Baker S, Bryant JE, Tham NT, BKrong NT, Boni MF, Loi TQ, Phuong le T, Verhoeven JT, Crusat M, Jeeninga RE, Schultsz C, Chau NV, Hien TT, van der Hoek L, Farrar J, de Jong MD. Identification of a new cyclovirus in cerebrospinal fluid of patients with acute central nervous system infections. mBio. 2013;4(3):00231–00213. doi: 10.3391/mbi.2013.4.3.05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Li L, Kapoor A, Slikas B, Bamidele OS, Wang C, Shaukat S, Masroor MA, Wilson ML, Ndjango JB, Peeters M, Gross-Camp ND, Muller MN, Hahn BH, Wolfe ND, Triki H, Bartkus J, Zaidi SZ, Delwart E. Multiple diverse circoviruses infect farm animals and are commonly found in human and chimpanzee feces. J Virol. 2010;84(4):1674–1682. doi: 10.1128/JVI.02109-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Phan TG, Mori D, Deng X, Rajindrajith S, Ranawaka U, Fan Ng TF, Bucardo-Rivera F, Orlandi P, Ahmed K, Delwart E. Small circular single stranded DNA viral genomes in unexplained cases of human encephalitis, diarrhea, and in untreated sewage. Virology. 2015;482(12):98–104. doi: 10.1016/j.virol.2015.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Altan E, Del Valle MJ, Deng X, Phan TG, Sadeghi M, Delwart EL. Small circular rep-encoding single-stranded DNA genomes in Peruvian diarrhea Virome. Genome Announcements. 2017;5(38):e00822–e00817. doi: 10.1128/genomeA.00822-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Reuter G, Boros Á, Delwart E, Pankovics P. Novel circular single-stranded DNA virus from turkey faeces. Arch Virol. 2014;159(8):2161–4. [DOI] [PubMed]
- 35.Fontenele RS, Lacorte C, Lamas NS, Schmidlin K, Varsani A, Ribeiro SG. Single Stranded DNA Viruses Associated with Capybara Faeces Sampled in Brazil. Viruses. 2019;11(8):710. [DOI] [PMC free article] [PubMed]
- 36.Qi D, Shan T, Liu Z, Deng X, Zhang Z, Bi W, Owens JR, Feng F, Zheng L, Huang F, Delwart E, Hou R, Zhang W. A novel polyomavirus from the nasal cavity of a giant panda (Ailuropoda melanoleuca) Virol J. 2017;14(1):207. doi: 10.1186/s12985-017-0867-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhang W, Li L, Deng X, Kapusinszky B, Pesavento PA, Delwart E. Faecal virome of cats in an animal shelter. J Gen Virol. 2014;95(11):2553–2564. doi: 10.1099/vir.0.069674-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kapoor A, Victoria J, Simmonds P, Slikas E, Chieochansin T, Naeem A, Shaukat S, Sharif S, Alam MM, Angez M, Wang C, Shafer RW, Zaidi S, Delwart E. A highly prevalent and genetically diversified Picornaviridae genus in south Asian children. Proc Natl Acad Sci U S A. 2008;105(51):20482–20487. doi: 10.1073/pnas.0807979105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhang W, Li L, Deng X, Blümel J, Nübling CM, Hunfeld A, Baylis SA, Delwart E. Viral nucleic acids in human plasma pools. Transfusion. 2016;56(9):2248–2255. doi: 10.1111/trf.13692. [DOI] [PubMed] [Google Scholar]
- 40.Victoria JG, Kapoor A, Li L, Blinkova O, Slikas B, Wang C, Naeem A, Zaidi S, Delwart E. Metagenomic Analyses of Viruses in Stool Samples from Children with Acute Flaccid Paralysis. J Virol. 2009;83(9):4642–4651. doi: 10.1128/JVI.02301-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Liu Z, Yang S, Wang Y, Shen Q, Yang Y, Deng X, Zhang W, Delwart E. Identification of a novel human papillomavirus by metagenomic analysis of vaginal swab samples from pregnant women. Virol J. 2016;13(1):122. doi: 10.1186/s12985-016-0583-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhang W, Yang S, Shan T, Hou R, Liu Z, Li W, Guo L, Wang Y, Chen P, Wang X, Feng F, Wang H, Chen C, Shen Q, Zhou C, Hua X, Cui L, Deng X, Zhang Z, Qi D, Delwart E. Virome comparisons in wild-diseased and healthy captive giant pandas. Microbiome. 2017;5(1):90. doi: 10.1186/s40168-017-0308-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Deng X, Naccache SN, Ng T, Federman S, Li L, Chiu CY, Delwart EL. An ensemble strategy that significantly improves de novo assembly of microbial genomes from metagenomic next-generation sequencing data. Nucleic Acids Res. 2015;43(7):e46. doi: 10.1093/nar/gkv002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Robert A, Edwards FR. Viral metagenomics. Nat Rev Microbiol. 2005;3(6):504–510. doi: 10.1038/nrmicro1163. [DOI] [PubMed] [Google Scholar]
- 45.Kearse M, Moir R, Wilson A, Stones-Havas S, Cheung M, Sturrock S, Buxton S, Cooper A, Markowitz S, Duran C, Thierer T, Ashton B, Meintjes P, Drummond A. Geneious basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics. 2012;28(12):1647–1649. doi: 10.1093/bioinformatics/bts199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Castrignano SB, Nagasse-Sugahara TK, Garrafa P, Monezi TA, Barrella KM, Mehnert DU. Identification of circo-like virus-Brazil genomic sequences in raw sewage from the metropolitan area of São Paulo: evidence of circulation two and three years after the first detection. Mem Inst Oswaldo Cruz. 2017;112(3):175–181. doi: 10.1590/0074-02760160312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Huelsenbeck JP, Ronquist F. MRBAYES: Bayesian inference of phylogenetic trees. Bioinformatics. 2001;17(8):754–755. doi: 10.1093/bioinformatics/17.8.754. [DOI] [PubMed] [Google Scholar]
- 48.Ronquist F, Huelsenbeck JP. MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics. 2003;19(12):1572–1574. doi: 10.1093/bioinformatics/btg180. [DOI] [PubMed] [Google Scholar]
- 49.Ronquist F, Teslenko M, van der Mark P, Ayres DL, Darling A, Höhna S, Larget B, Liu L, Suchard MA, Huelsenbeck JP. MrBayes 3.2: efficient Bayesian phylogenetic inference and model choice across a large model space. Syst Biol. 2012;61(3):539–542. doi: 10.1093/sysbio/sys029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Dayaram A, Goldstien S, Zawar-Reza P, Gomez C, Harding JS, Varsani A. Novel ssDNA virus recovered from estuarine Mollusc (Amphibola crenata) whose replication associated protein (rep) shares similarities with rep-like sequences of bacterial origin. J Gen Virol. 2013;94(5):1104–1110. doi: 10.1099/vir.0.050088-0. [DOI] [PubMed] [Google Scholar]
- 51.Kazlauskas D, Varsani A, Koonin EV, Krupovic M. Multiple origins of prokaryotic and eukaryotic single-stranded DNA viruses from bacterial and archaeal plasmids. Nat Commun. 2019;10(1):3425. doi: 10.1038/s41467-019-11433-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Rosario K, Breitbart M, Harrach B, Segalés J, Delwart E, Biagini P, Varsani A. Revisiting the taxonomy of the family Circoviridae: establishment of the genus Cyclovirus and removal of the genus Gyrovirus. Arch Virol. 2017;162(5):1447–1463. doi: 10.1007/s00705-017-3247-y. [DOI] [PubMed] [Google Scholar]
- 53.Rosario K, Duffy S, Breitbart M. A field guide to eukaryotic circular single-stranded DNA viruses: insights gained from metagenomics. Arch Virol. 2012;157(10):1851–1871. doi: 10.1007/s00705-012-1391-y. [DOI] [PubMed] [Google Scholar]
- 54.Hassan-Sheikhi P, Heydarnejad J, Massumi H, Kraberger S, Varsani A. Novel nanovirus and associated alphasatellites identified in milk vetch plants with chlorotic dwarf disease in Iran. Virus Res. 2020;276:197830. doi: 10.1016/j.virusres.2019.197830. [DOI] [PubMed] [Google Scholar]
- 55.Mankertz A, Caliskan R, Hattermann K, Hillenbrand B, Kurzendoerfer P, Mueller B, Schmitt C, Steinfeldt T, Finsterbusch T. Molecular biology of porcine circovirus: analyses of gene expression and viral replication. Vet Microbiol. 2003;98(2):81–88. doi: 10.1016/j.vetmic.2003.10.014. [DOI] [PubMed] [Google Scholar]
- 56.Belák S, Karlsson OE, Blomström AL, Berg M, Granberg F. New viruses in veterinary medicine, detected by metagenomic approaches. Vet Microbiol. 2013;165(1–2):95–101. doi: 10.1016/j.vetmic.2013.01.022. [DOI] [PubMed] [Google Scholar]
- 57.Kraberger S, Argüello-Astorga GR, Greenfield LG, Galilee C, Law D, Martin DP, Varsani A. Characterisation of a diverse range of circular replication-associated protein encoding DNA viruses recovered from a sewage treatment oxidation pond. Infect Genet Evol. 2015;31:73–86. doi: 10.1016/j.meegid.2015.01.001. [DOI] [PubMed] [Google Scholar]
- 58.Gronenborn B. Nanoviruses: genome organisation and protein function. Vet Microbiol. 2004;98(2):103–109. doi: 10.1016/j.vetmic.2003.10.015. [DOI] [PubMed] [Google Scholar]
- 59.O'Dea MA, Kabay MJ, Carr J, Wilcox GE, Richards RB. Porcine circovirus-associated disease in weaner pigs in Western Australia. Aust Vet J. 2011;89(4):122–130. doi: 10.1111/j.1751-0813.2010.00680.x. [DOI] [PubMed] [Google Scholar]
- 60.Biagini P. Human circoviruses. Vet Microbiol. 2004;98(2):95–101. doi: 10.1016/j.vetmic.2003.10.004. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The viral genomes described in detail here were deposited in GenBank under the following accession numbers: MN604398, MN621468- MN621470, MN621480- MN621482 and MN621476.