Abstract
Acinetobacter baumannii causes nosocomial infections due to its multidrug resistance and high environmental adaptability. Colistin is a polypeptide antibacterial agent that targets lipopolysaccharide (LPS) and is currently used to control serious multidrug-resistant Gram-negative bacterial infections, including those caused by A. baumannii. However, A. baumannii may acquire colistin resistance by losing their LPS. In mouse models, LPS-deficient A. baumannii have attenuated virulence. Nevertheless, the mechanism through which the pathogen is cleared by host immune cells is unknown. Here, we established colistin-resistant A. baumannii strains and analyzed possible mechanisms through which they are cleared by neutrophils. Colistin-resistant, LPS-deficient strains harbor mutations or insertion sequence (IS) in lpx genes, and introduction of intact lpx genes restored LPS deficiency. Analysis of interactions between these strains and neutrophils revealed that compared with wild type, LPS-deficient A. baumannii only weakly stimulated neutrophils, with consequent reduced levels of reactive oxygen species (ROS) and inflammatory cytokine production. Nonetheless, neutrophils preferentially killed LPS-deficient A. baumannii compared to wild-type strains. Moreover, LPS-deficient A. baumannii strains presented with increased sensitivities to antibacterial lysozyme and lactoferrin. We revealed that neutrophil-secreted lysozyme was the antimicrobial factor during clearance of LPS-deficient A. baumannii strains. These findings may inform the development of targeted therapeutics aimed to treat multidrug-resistant infections in immunocompromised patients who are unable to mount an appropriate cell-mediated immune response.
Keywords: Acinetobacter baumannii, colistin, LPS, lysozyme, neutrophil
Introduction
Acinetobacter baumannii is an aerobic Gram-negative bacillus that is widely distributed in nature. A. baumannii tolerates both damp and dry environments and proliferates easily in hospitals and on medical devices, causing nosocomial infections (Dijkshoorn et al., 2007; Maragakis and Perl, 2008; Munoz-Price and Weinstein, 2008). On the healthy human skin, A. baumannii is usually harmless; however, it may cause opportunistic infections in immunocompromised patients (Chu et al., 1999; Dijkshoorn et al., 2007; Maragakis and Perl, 2008; Munoz-Price and Weinstein, 2008). A. baumannii infection is characterized by pneumonia, sepsis, urinary tract infection, meningitis, and other conditions. The bacteria have been implicated as the source of ventilator-associated pneumonia (VAP) and urinary tract and blood vessel infections from contaminated catheters and surgical wound infections (Cisneros and Rodríguez-Baño, 2002; Garnacho-Montero et al., 2005; Dijkshoorn et al., 2007; Maragakis and Perl, 2008; Munoz-Price and Weinstein, 2008). A. baumannii is sometimes responsible for outbreaks among ICU (intensive care unit) patients. This pathogen requires constant surveillance in health care institutions (Garnacho-Montero et al., 2015; Leao et al., 2016).
The pathogenicity of A. baumannii may be explained by its bacterial capsule, biofilm formation, secretory systems, high adhesion capacity, strong affinity for iron, and other factors not yet identified (Mcconnell et al., 2013; Weber et al., 2016; Garcia-Patino et al., 2017; Wong et al., 2017). Previously, we investigated the interaction of A. baumannii with neutrophils and found that it inhibits neutrophil extracellular traps (NETs), a neutrophilic biological defense mechanism that traps and inactivates bacteria (Kamoshida et al., 2018). Moreover, we found that A. baumannii spreads throughout the body by flagging and hijacking neutrophils in a process known as “bacterial immunity taxi” (Kamoshida et al., 2015, 2016). Thus, it is necessary to study the interaction between A. baumannii and host immune cells for a better understanding of this bacterial pathogen.
Acinetobacter baumannii rapidly and readily acquires drug resistance. It harbors a gene encoding β-lactamase, has poor outer membrane permeability to antibacterial agents, acquires resistance through foreign genes harbored by plasmids, and readily undergoes biofilm formation (Perez et al., 2007; Peleg et al., 2008). Nosocomial infections caused by A. baumannii have become severe due to the high rate of drug resistance in the organism. Multidrug-resistant A. baumannii (MDRA) has proliferated and dispersed globally. Despite the close attention of medical professionals, MDRA is still difficult to manage and remains a critical issue. The number of fatalities ascribed to A. baumannii infections continues to rise worldwide (Dijkshoorn et al., 2007; Peleg et al., 2008).
Colistin is a polypeptide antibacterial drug that targets lipopolysaccharide (LPS) on the surfaces of Gram-negative bacteria and is the last resort against serious infections caused by multidrug-resistant Gram-negative bacteria such as MDRA (Levin et al., 1999; Falagas and Kasiakou, 2005; Maragakis and Perl, 2008). A. baumannii acquires colistin resistance at a rate of 10–8–10–9 (Moffatt et al., 2010; Cai et al., 2012). In the event that colistin proves ineffective against MDRA, there may be no other drugs available to treat the infection. Colistin cannot be used efficaciously against MDRA without first elucidating its resistance mechanism. A. baumannii acquires resistance to colistin by completely losing its LPS (Moffatt et al., 2010). LPS deficiency in A. baumannii causes various changes in the organism, including its relative sensitivity to antibacterial drugs (Moffatt et al., 2010; Carretero-Ledesma et al., 2018). Compared to wild type, the virulence of LPS-deficient A. baumannii is low, as found in mouse models (Moffatt et al., 2013; Beceiro et al., 2014; Carretero-Ledesma et al., 2018). However, if and how immune cells contribute to bacterial clearance remains ill-defined.
Neutrophils are immune cells that play a pivotal role during the initial immune response to various bacterial infections. Neutrophils migrate toward infection sites and act against bacteria through phagocytosis and by producing toxic factors such as reactive oxygen species (ROS) (Nathan, 2006; Nauseef and Borregaard, 2014). Moreover, neutrophil granules have molecules, such as lysozyme, lactoferrin, LL-37, and elastase with antimicrobial activity that are important for immune defense against bacterial infections (Ellison and Giehl, 1991; Borregaard et al., 2007). Thus, understanding neutrophil–bacterial interaction is very important in controlling infection. In the present study, we established colistin-resistant strains to investigate effects of neutrophil-secreted lysozyme on LPS-deficient A. baumannii and examined clearance mechanism of this bacteria by neutrophils. Colistin-resistant A. baumannii (CRAb) are often isolated in clinical settings, particularly in ICU patients with immunodeficiency conditions such as neutropenia (Qureshi et al., 2015). This study may help to treat multidrug-resistant infections concluding colistin-resistant bacteria in immunocompromised patients who are unable to mount an appropriate cell-mediated immune response such as neutropenia.
Materials and Methods
Bacterial Strains, Plasmids, and Neutrophils
Wild-type A. baumannii ATCC 19606 was used as the reference strain (Kamoshida et al., 2015, 2016, 2018). Bacteria were grown in LB (Luria-Bertani) broth (BD Biosciences, San Diego, CA, United States) for 16 h at 37°C shaking. Colistin-resistant variants of ATCC 19606 (CRAb) were isolated by directly plating the parent strain onto LB agar containing 10 μg/ml colistin sulfate (Sigma-Aldrich Corp., St. Louis, MO, United States).
The pMU125 plasmid (Escherichia coli–Acinetobacter shuttle vector pWH1266 with gfp) was kindly provided by Dr. Luis Actis of Miami University, Miami, FL, United States (Dorsey et al., 2002). To construct the plasmid, pKAMO was prepared by In-Fusion cloning (Takara Bio Inc., Shiga, Japan), ampicillin resistance gene in pMU125 was replaced with kanamycin resistance gene from pHSG298 and gfp was replaced with a multiple cloning site sequence (5′-CTCGAGGAGCTCAGTACTGCGGCCGCAGGTACCGCA TGC-3′: XhoI–SacI–ScaI–NotI–KpnI–SphI). lpxC DNA fragment (primer pairs: 5′-ACAGAACAGAATTCTATGGTG AAACAGCGTACTCTCA-3′ and 5′-TCTGAGATGGTCGACT TATGTCACACTCACGTATGGA-3′) from ATCC 19606 was ligated to pKAMO to construct the lpxC expression plasmid pKAMO-lpxC. CRAb-5 was transformed by electroporation with pKAMO-lpxC and pKAMO (control vector) to construct CRAb-LPS and CRAb-mock, respectively. The bacteria were selected on LB agar containing 50 μg/ml kanamycin.
Neutrophils were isolated from the peripheral blood of healthy human volunteers (n = 10) as previously described (Kamoshida et al., 2017). The heparinized blood was mixed with 1% (w/v) dextran (MW 200,000) in saline to sediment erythrocytes. The supernatant was subjected to density gradient centrifugation in a Lymphosepar I (Wako Pure Chemical Industries Ltd., Osaka, Japan). The neutrophils were purified from the cell pellets under hypotonic conditions to lyse any remaining erythrocytes. Neutrophil purity was >95%. Samples from independent donors were used for each set of experiments. Written informed consent was obtained from all study participants in accordance with the Declaration of Helsinki. The study was approved by the Ethical Review Committee of the School of Medicine of Teikyo University, Tokyo, Japan.
Neutrophil and Bacterial Co-cultures
The bacteria were cultured on LB broth containing 50 μg/ml kanamycin at 37°C for 16 h, rinsed with phosphate-buffered saline (PBS), and resuspended in fresh RPMI 1640 (Sigma-Aldrich). Bacteria [MOI (multiplicity of infection) 50, 5 × 107 CFU/ml] and neutrophils (106 cells/ml) were co-cultured in RPMI 1640 containing 2% human serum and maintained in standing still suspension at 37°C under a 5% CO2 atmosphere for the indicated amount of time. When required by experiments, 5 μg/ml cytochalasin B (simultaneous addition) was used to inhibit neutrophil phagocytosis in suspension co-cultures.
PCR-Sanger Sequencing
Genome regions containing lpx genes were amplified by colony PCR using the following primers: lpxA: 5′-TGGTAA TGCAGAAGCGCGGTATCTACAA-3′ and 5′-ATCCTCTAGA GTCGACCAATATTCAAAGTCTGAAGAAGCA-3′; lpxC: 5′- TCAGCAACGTAAGTAATTTAGCGTACA-3′ and 5′-GCCA AGCTTTACTACGTTTGGCAAGCAA-3′; lpxD: 5′-AAGCTT GCATGCGTTAAGCAAGCTGCTGAGCAATTACGAA-3′ and 5′-CCAATAAGAATGGGTAACGATGCGGCAA-3′.
The PCR products were treated with EXOSAP-IT (Thermo Fisher Scientific, Waltham, MA, United States) and followed by cycle-sequencing reaction with Big Dye Terminator v3.1 (Applied Biosystems, Foster City, CA, United States) using the primers listed above. Direct sequencing process was performed in 3730xl Genetic Analyzer (Applied Biosystems).
Limulus Amebocyte Lysate Assay
Limulus amebocyte lysate assay was performed using a Toxicolor LS-50M set (Seikagaku Corporation, Tokyo, Japan), according to the manufacturer’s instructions. Overnight A. baumannii cultures (after adjusted OD600 = 0.1) were diluted in sterile, pyrogen-free water (Otsuka Pharmaceutical Co., Ltd., Tokyo, Japan).
Growth Curve
Bacterial culture density was adjusted to OD600 = 0.005 from overnight cultures. The bacteria were grown in LB broth for 8 h at 37°C, shaking 135 rpm using BioShaker BR-23FP (Taitec, Saitama, Japan). Culture OD600 was measured every hour using BioPhotometer plus (Eppendorf, Hamburg, Germany).
Lipopolysaccharide Analysis by Polyacrylamide Gel Electrophoresis (PAGE)
Lipopolysaccharide was purified from 16-h cultures (adjusted OD600 = 0.1) of A. baumannii using LPS Extraction kit (iNtRON Biotechnology, Seoul, South Korea), according to the manufacturer’s instructions. LPS preparations were separated on 15% (w/v) SDS-PAGE as previously described (Kamoshida et al., 2012). To visualize LPS, the gel was stained with 2D-Silver Stain Reagent II (Cosmo Bio, Tokyo, Japan).
Determination of Minimum Inhibitory Concentrations
Minimum inhibitory concentrations for antimicrobial drugs and egg white lysozyme (Sigma-Aldrich), human lactoferrin (Sigma-Aldrich), LL-37 (Peptide Institute, Osaka, Japan), human serum, and H2O2 were determined by microdilution in LB broth according to the Clinical and Laboratory Standards Institute protocol (M07 Methods for Dilution Antimicrobial Susceptibility Tests for Bacteria That Grow Aerobically, 11th Edition). Bacterial culture density was adjusted to OD600 = 0.001 from overnight cultures and cultured for 16 h at 37°C. A twofold dilution series was used for each molecule to set up the MIC studies. The concentration range was between 256 and 0.007 μg/ml for antimicrobial drugs, 1000 and 0.002 μg/ml for lysozyme, 4000 and 4 μg/ml for lactoferrin, 5000 and 5 nM for LL-37, 40 and 0.02% for serum, and 10 and 0.01% for H2O2. Survival rates were calculated as percentages of the uninhibited pure bacterial culture.
Detection of ROS and Superoxide
ROS and superoxide production levels in neutrophil and bacterial co-cultures were measured by flow cytometry (FACSCanto II, BD Biosciences) and a total ROS/superoxide detection kit (Enzo Life Sciences, Farmingdale, NY, United States). Neutrophils induced with 100 nM phorbol-12-myristate-13-acetate (PMA) served as positive controls.
Measurements of Cytokine Levels
Cytokines were evaluated by BD cytometric bead array (CBA) system (BD Biosciences) [human inflammation kit with interleukin (IL)-8, IL-1β, IL-6, IL-10, tumor necrosis factor (TNF), and IL-12p70)]. Supernatants were obtained from suspension co-culture of neutrophils and bacteria. The supernatants of neutrophils stimulated with PMA (100 nM) served as positive controls.
Lysozyme Measurement in Neutrophils
Lysozyme secreted by neutrophils into supernatants was measured by enzyme-linked immunosorbent assay (ELISA) (Assaypro, St. Charles, MO, United States). Supernatants were obtained from neutrophils co-cultured with bacteria (MOI 50) in RPMI 1640 containing 2% human serum at 37°C for 4 h in 5% CO2. Lysozyme was removed by mixing 1.0 ml of the neutrophil supernatant with 5 μg/ml anti-lysozyme antibody (Abcam, Cambridge, United Kingdom) conjugated with protein G–Sepharose [0.05 ml; 50% (w/v) suspension] and incubating at 4°C for 16 h. The supernatant was recovered by low-speed centrifugation.
Bacterial Clearance by Neutrophils
Neutrophil and bacteria (MOI 50) were co-cultured for 4 h. Appropriate dilutions of co-culture suspension were spread onto LB agar plates and incubated overnight at 37°C to count bacteria colonies and evaluate cell viability. Bacterial clearance capacity of co-culture supernatants of neutrophils and CRAb-5 was assessed by using CRAb-LPS or CRAb-mock strains. Bacteria (5 × 107 CFU/ml) were incubated for 4 h. Cell suspension was then inoculated onto LB agar plates containing kanamycin to count viable cells. To test bacterial clearance in supernatants without lysozymes produced by neutrophils, co-culture supernatants were cleared of lysozyme as described above, prior to use in clearance capacity experiments.
Statistical Analysis
Data are expressed as the mean ± standard deviation (SD) and were compared by unpaired Student’s t-test and two-way ANOVA. Differences with P < 0.05 were considered statistically significant.
All methods were carried out in accordance with relevant guidelines and regulations.
Results
CRAb Strains Lack LPS and Exhibit Attenuated Growth
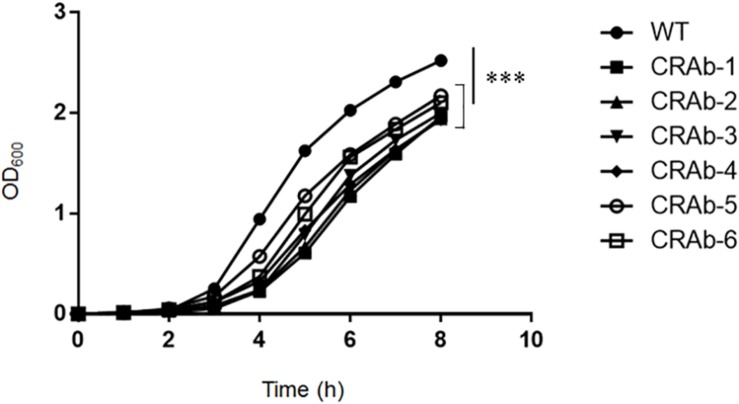
Acinetobacter baumannii acquires colistin resistance at a frequency in the order of 10–8–10–9. We used colistin to select for and establish six CRAb strains and analyzed lpxA, lpxC, and lpxD genes, previously reported to participate in colistin resistance (Moffatt et al., 2010). We confirmed the presence of non-synonymous mutation and/or gene rearrangement in some or all of lpx genes in all six CRAb strains (Table 1). CRAb-1, CRAb-2, and CRAb-5 presented with non-synonymous point mutations or depletions of intact genes. In CRAb-6, the ISAba11 sequence was inserted into lpxC, resulting in LpxC truncation at C-terminal amino acid 160 (Moffatt et al., 2011). In CRAb-3 and CRAb-4, lpxC or lpxA was not amplified, suggesting deletion of these regions or insertion of large insertion sequences. We measured bacterial endotoxin using the LAL assay. Endotoxin levels were below the detection limit in most of the CRAb strains (Table 1). Endotoxin production was detected only for CRAb-5 but at a considerably lower level than that of the wild type (ATCC 19606). Therefore, disruption of lpxA, lpxC, and lpxD genes in A. baumannii may have accounted for the observed LPS deficiency leading to colistin resistance. All CRAb strains grew slower than the wild type (Figure 1), indicating the importance of LPS synthetic pathway in normal growth.
TABLE 1.
Lpx mutation analysis and limulus amebocyte lysate (LAL) assay of colistin-resistant Acinetobacter baumannii (CRAb) strains.
| Strain | IpxA | IpxC | IpxD | AA change | EU/ml |
| ATCC19606 | WT | WT | WT | 69 × 103 ± 27 × 103 | |
| CRAb-1 | 382-384GTA→- | WT | WT | LpxA V128- (deletion of 128 valine) | 0.2> |
| CRAb-2 | WT | 312T→A | 934G→T | LpxD E312* (truncation of C-terminal 45 aa)T LpxC D104E | 0.2> |
| CRAb-3 | WT | no band | WT | Not detected | 0.2> |
| CRAb-4 | No band | WT | WT | Not detected | 0.2> |
| CRAb-5 | WT | 573T→G | WT | LpxC F191L | 1.5 ± 1.2 |
| CRAb-6 | WT | 420T::X | WT | LpxC D141* (truncation of C-terminal 160 aa) | 0.2> |
AA: amino acid EU: endotoxin units.
FIGURE 1.
Growth curve of colistin-resistant Acinetobacter baumannii (CRAb) strains. ATCC 19606 (parent) and CRAb strains preadjusted to OD600 = 0.005 were grown in LB broth at 37°C for 8 h. OD600 was measured hourly. Data are means ± SD; n = 3 per group. ∗∗∗P < 0.001.
Establishment of LPS Complementation Strain of CRAb
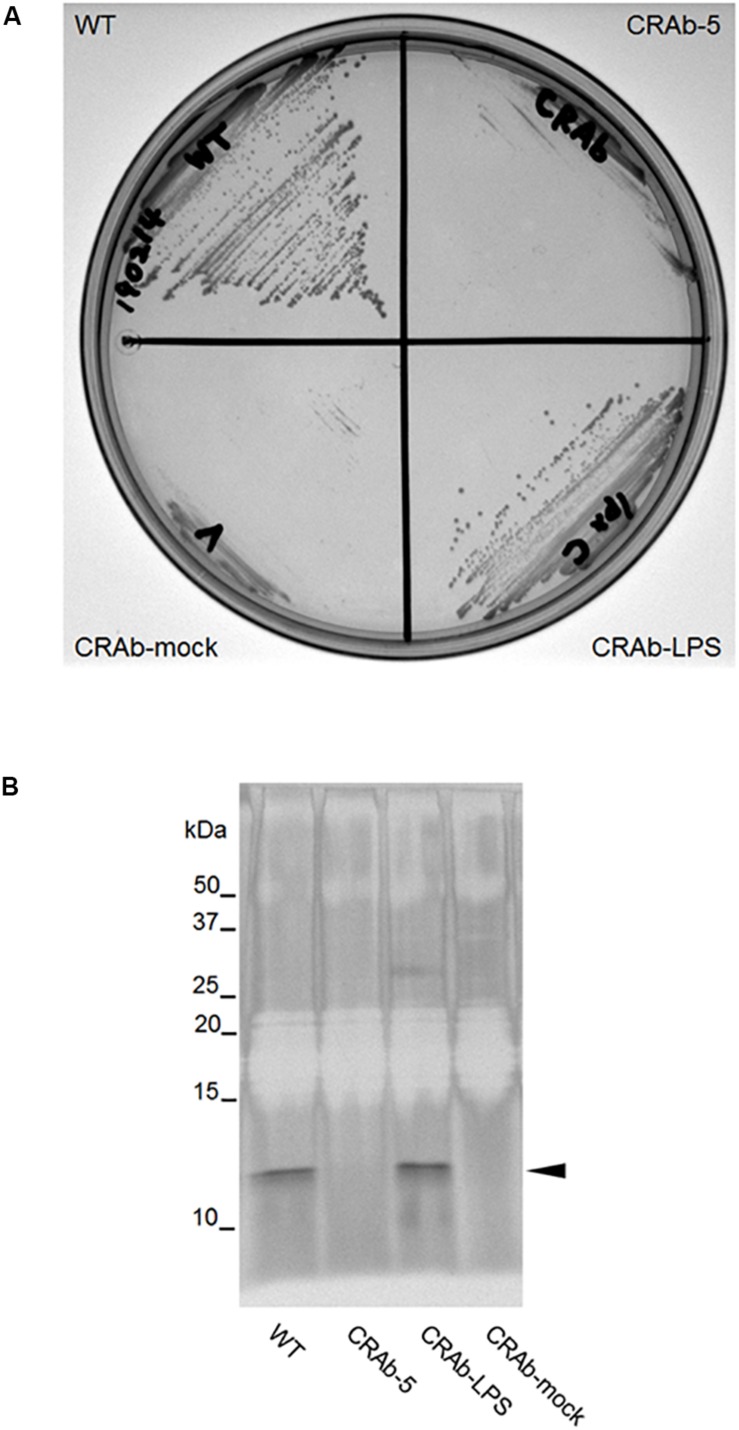
It was predicted that CRAb-5 is deficient in intact lpxC and lost most of its LPS. To examine the differences between LPS-positive and LPS-negative A. baumannii, we constructed a complementation strain by re-introducing intact lpxC into CRAb-5. The complemented lpxC mutant (CRAb-LPS) recovered its displayed same colony size as the wild type (Figure 2A). The growth of CRAb-mock, a strain harboring the control vector, was similar to the parental CRAb-5 strain. To assess the capacity of the wild type, CRAb-5, CRAb-LPS, and CRAb-mock to produce LPS, we purified and analyzed LPS from their cultures. Wild-type and CRAb-LPS produced migrating lipid A in the molecular mass range of 10–15 kDa (Moffatt et al., 2010). CRAb-5 and CRAb-mock produced no detectable lipid A (Figure 2B). Thus, CRAb-LPS, the lpxC complementation strain, recovered LPS production and clarified the difference between LPS-positive and LPS-negative A. baumannii.
FIGURE 2.
Confirmation of lipopolysaccharide (LPS) complementation strain of colistin-resistant Acinetobacter baumannii (CRAb). (A) LPS complementation strain colony formation. ATCC 19606 (WT), CRAb-5, CRAb-LPS, and CRAb-mock were streaked on LB agar plates, cultured at 37°C for 18 h to confirm colony formation. LPS was purified from colonies. LPS preparations were separated by electrophoresis and visualized by silver stain (B). Arrow: lipid A.
MICs of Various Antimicrobial Drugs Against LPS-Deficient A. baumannii Strains
First, we evaluated the MICs of polymixins (colistin and polymyxin B). LPS-negative CRAb-5 and CRAb-mock were highly resistant to both drugs. In contrast, wild-type A. baumannii (ATCC 19606) and the LPS-restoring CRAb-LPS were sensitive to both drugs (Table 2). Moreover, susceptibility to β-lactams, such as meropenem, imipenem, and cefozopran, drugs that target peptidoglycan biosynthesis, was higher in CRAb-5 and CRAb-mock than the LPS-positive strains. The LPS-negative strains were also more sensitive to aminoglycosides such as amikacin. However, CRAb-5 and CRAb-mock were not more sensitive than their LPS-positive counterparts to tetracyclines such as minocycline and tigecycline, sulbactam, or the quinolone ciprofloxacin (Table 2). Thus, LPS deficiency in A. baumannii not only causes the organism to acquire colistin and polymyxin B resistance but also increases its sensitivity to β-lactams and several other antibiotics.
TABLE 2.
Minimum inhibitory concentrations (MICs) of various antimicrobial drugs against lipopolysaccharide-deficient Acinetobacter baumannii strains.
| WT | CRAb-5 | CRAb-LPS | CRAb-mock | |
| ATCC19606 | LPS-deficient | +intact IpxC | +control vector | |
| Colistin | 0.5 | >256 | 0.5 | >256 |
| Polymyxin B | 0.25 | 128 | 0.5 | 64 |
| Minocycline | 0.25 | 0.125 | 0.25 | 0.06 |
| Tigecycline | 0.5 | 0.25 | 0.5 | 0.125 |
| Sulbactam | 0.5 | 0.5 | 0.5 | 0.5 |
| Meropenem | 0.5 | 0.007> | 0.5 | 0.007> |
| Imipenem | 8 | 1 | 8 | 1 |
| Cefozopran | 8 | 2 | 8 | 1 |
| Amikacin | 16 | 4 | 16 | 2 |
| Ciprofloxacin | 0.06 | 0.06 | 0.06 | 0.03 |
Drug concentrations are in μg/ml.
LPS-Defective A. baumannii Stimulates Lower Levels of ROS and Inflammatory Cytokines in Neutrophils
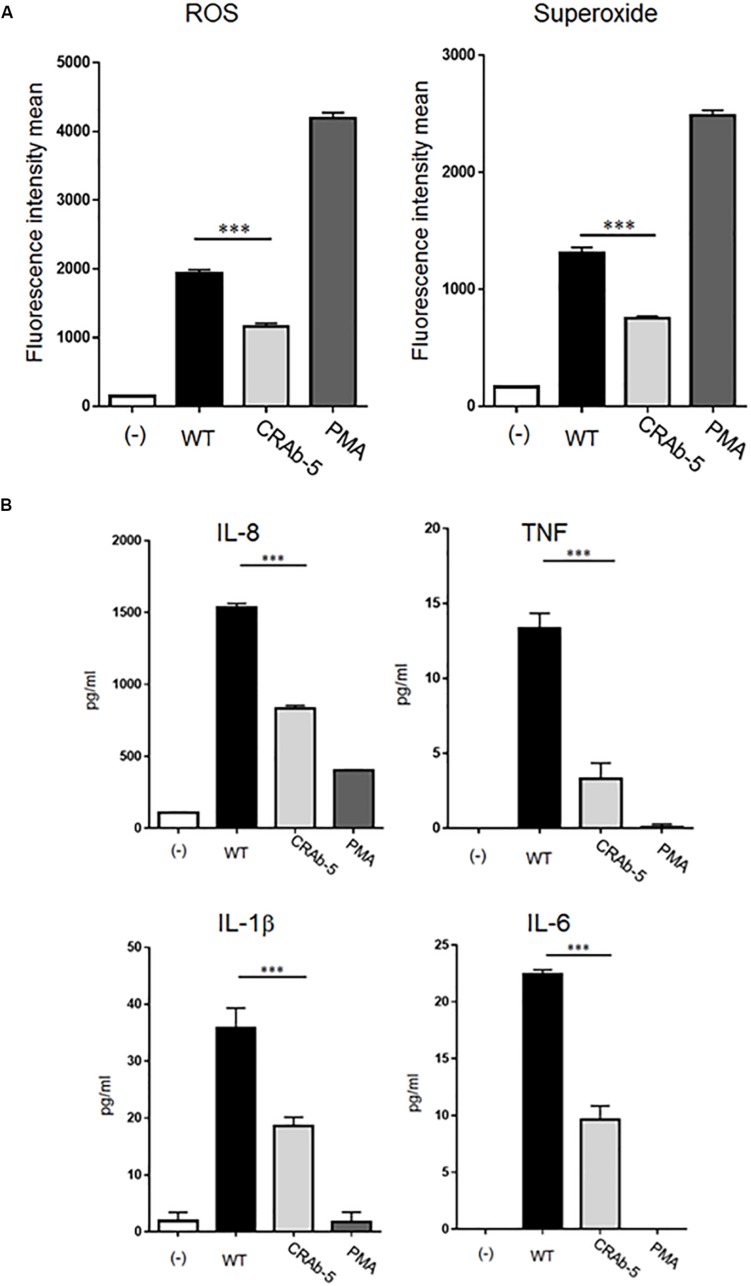
Studies in mouse models demonstrated that LPS-deficient A. baumannii is less virulent than wild-type strains (Moffatt et al., 2013; Beceiro et al., 2014; Carretero-Ledesma et al., 2018). However, the mechanism of immune clearance of LPS-deficient bacterial pathogens like A. baumannii is not known. Based on the important roles of neutrophils in initial host immune responses, we examined interactions between neutrophils and LPS-deficient A. baumannii. First, we evaluated ROS and superoxide production in neutrophils exposed to A. baumannii and found lower levels in neutrophils induced by CRAb-5 than in those challenged by wild-type A. baumannii (Figure 3A). Similar results were obtained with CRAb-3 and CRAb-6 (Supplementary Figure S1). Furthermore, ROS levels were lower in neutrophils induced by CRAb-mock (empty vector control strain) than those by the LPS-restoring strain, CRAb- LPS (Supplementary Figure S2). These observations remained the same in experiments performed with formalin-killed bacteria (Supplementary Figure S3).
FIGURE 3.
Measurements of inflammatory response molecules secreted by neutrophils. ROS and superoxide expression levels in neutrophils challenged by bacteria (ATCC 19606: WT, colistin-resistant A. baumannii: CRAb-5) were measured by flow cytometry and a total ROS/superoxide detection kit (A). PMA (100 nM)-stimulated neutrophils served as positive controls. Data are means ± SD; n = 5 per group. ∗∗∗P < 0.001. (B) Inflammatory cytokines such as interleukin (IL)-8, tumor necrosis factor (TNF), IL-1β, and IL-6 produced by neutrophils in response to bacterial stimulation were measured by the cytometric bead array system. Supernatants were obtained by co-culturing neutrophils and bacteria for 4 h. The supernatants of neutrophils alone stimulated with PMA (100 nM) served as positive controls. Data are means ± SD; n = 4 per group. ∗∗∗P < 0.001.
We next assessed inflammatory cytokines production by A. baumannii-stimulated neutrophils. Interleukin (IL)-8, tumor necrosis factor (TNF), IL-1β, and IL-6 levels were lower in neutrophils challenged with CRAb-5 than in those exposed to the wild type (Figure 3B). IL-10 and IL-12p70 were below detection limits. Collectively, these results suggest that LPS- deficient CRAb-5 strain is less effective than the parent strain at stimulating neutrophilic ROS, superoxide, and inflammatory cytokine production.
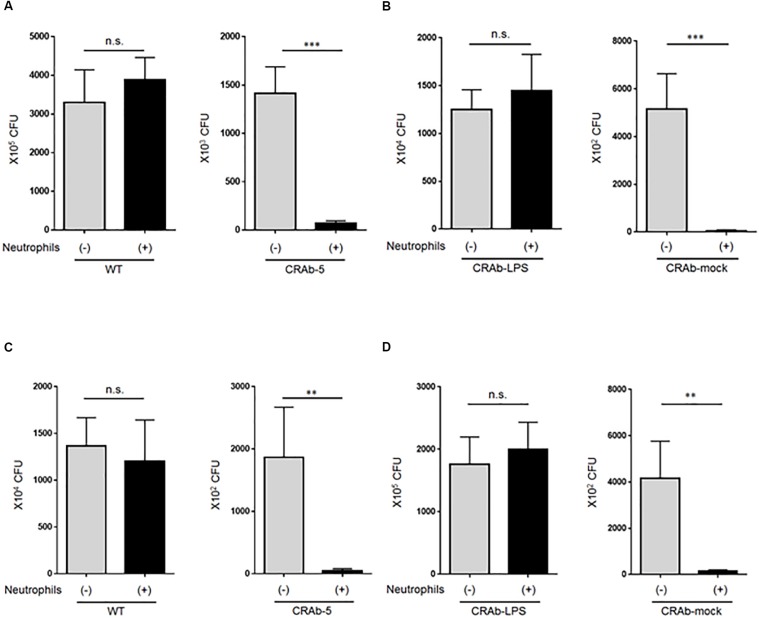
LPS-Deficient A. baumannii Strains Are More Prone to Neutrophil Clearance
To explore the impact of neutrophils on A. baumannii clearance, we first asked whether LPS-deficient CRAb-5 could still be killed by neutrophils. Wild-type A. baumannii or CRAb-5 and neutrophils were co-cultured for 4 h and the surviving bacteria were counted. We found that CRAb-5 was significantly killed by neutrophils, whereas the wild type was not (Figure 4A). CRAb-3 and CRAb-6 strains were also significantly killed by neutrophils (Supplementary Figure S4). Notably, CRAb-mock were killed by neutrophils but not CRAb-LPS (Figure 4B). We then repeated bacterial clearance experiments in the presence of neutrophil phagocytosis inhibition or with using neutrophil-CRAb-5 co-cultivation supernatants. The suppression of neutrophil phagocytosis by cytochalasin B did not prevent killing of CRAb-5 killing (Figure 4C). The observation of neutrophil-CRAb-5 co-cultivation resulted in nearly no phagocytosis of the pathogen (Supplementary Figure S5). Further, bacterial culture with co-culture supernatant killed LPS-deficient CRAb-mock but not LPS-restoring CRAb-LPS (Figure 4D). These results suggest that neutrophils killed LPS-deficient A. baumannii strains but not the wild type, with released factors present in neutrophils supernatant.
FIGURE 4.
Assay of lipopolysaccharide-deficient Acinetobacter baumannii strain clearance by neutrophils. (A) ATCC 19606 (WT), colistin-resistant A. baumannii (CRAb-5), (B) CRAb-LPS, or CRAb-mock and neutrophils were co-cultured for 4 h. Surviving bacteria were then counted. In panel (C), cytochalasin B was used to inhibit neutrophil phagocytosis in co-cultures. Clearance capacity of 4-h neutrophil/CRAb-5 co-culture supernatant was evaluated by adding supernatant to CRAb-LPS or CRAb-mock and incubating for 4 h. Viable cells were then counted by plating on kanamycin agar (D). Data are means ± SD; n = 6 per group. n.s., not significant, ∗∗∗P < 0.001, ∗∗P < 0.01. Means are representative of three experiments.
LPS-Deficient A. baumannii Strains Differentially Respond to Various Neutrophilic Bactericidal Molecules
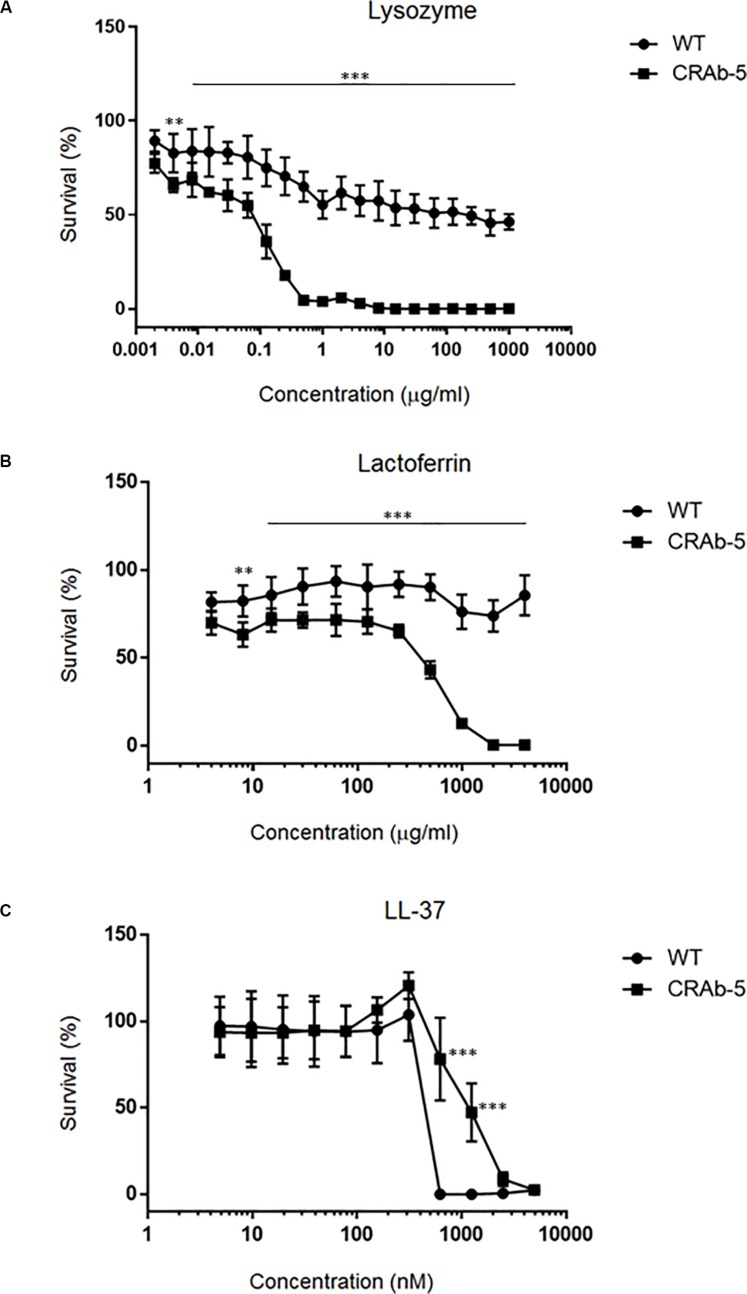
Since neutrophils evidently secreted extracellular substances lethal to LPS-negative A. baumannii strains (Figure 4D), we examined bactericidal effects of known neutrophilic factors on LPS-deficient A. baumannii. The bactericidal activities of lysozyme and lactoferrin were higher against LPS-deficient A. baumannii (CRAb-5, CRAb-mock) than they were against LPS-positive wild type and CRAb-LPS. LPS-deficient CRAb-5 was killed by only 0.6 μg/ml lysozyme, whereas the wild type survived 1000 μg/ml lysozyme (Figure 5A). Lactoferrin killed CRAb-5 at 1000 μg/ml while the wild type survived 4000 μg/ml (Figure 5B). In contrast, the antibacterial activity of the cathelicidin peptide LL-37 was weaker against LPS-deficient than it was against LPS-positive A. baumannii strains. LL-37 killed the wild type at 625 nM but CRAb-5 survived 2500 nM LL-37 (Figure 5C). The resistance to serum was slightly lower in the LPS-negative A. baumannii strains. The bactericidal efficacies of H2O2 were nearly the same against both LPS-positive and LPS-negative A. baumannii (Table 3).
FIGURE 5.
Dose–response curves of bactericidal ability of various biomolecules against lipopolysaccharide-deficient Acinetobacter baumannii strains. A twofold dilution series of (A) lysozyme (maximum concentration 1000 μg/ml), (B) lactoferrin (maximum concentration 4000 μg/ml), or (C) LL-37 (maximum concentration 5000 nM) was prepared and the viabilities of the ATCC 19606 (wild type) or colistin-resistant A. baumannii strain (CRAb-5) were assessed. Survival rates were calculated as percentages of the uninhibited, pure bacterial culture. Data are means ± SD; n = 3 per group. ∗∗∗P < 0.001, ∗∗P < 0.01.
TABLE 3.
Bactericidal activity of various biomolecules against lipopolysaccharide-deficient Acinetobacter baumannii strains.
| WT | CRAb-5 | CRAb-LPS | CRAb-mock | |
| Lysozyme | >1000 μg/ml | 0.6 μg/ml | >1000 μg/ml | 0.6 μg/ml |
| Lactoferrin | >1000 μg/ml | 1000 μg/ml | >1000 μg/ml | 1000 μg/ml |
| LL-37 | 625 nM | 2500 nM | 625 nM | 1250 nM |
| Human-Serum | 40% | 20% | 20% | 10% |
| H202 | 0.02% | 0.02% | 0.02% | 0.02% |
Neutrophil-Secreted Lysozyme Kills LPS-Deficient A. baumannii Strains
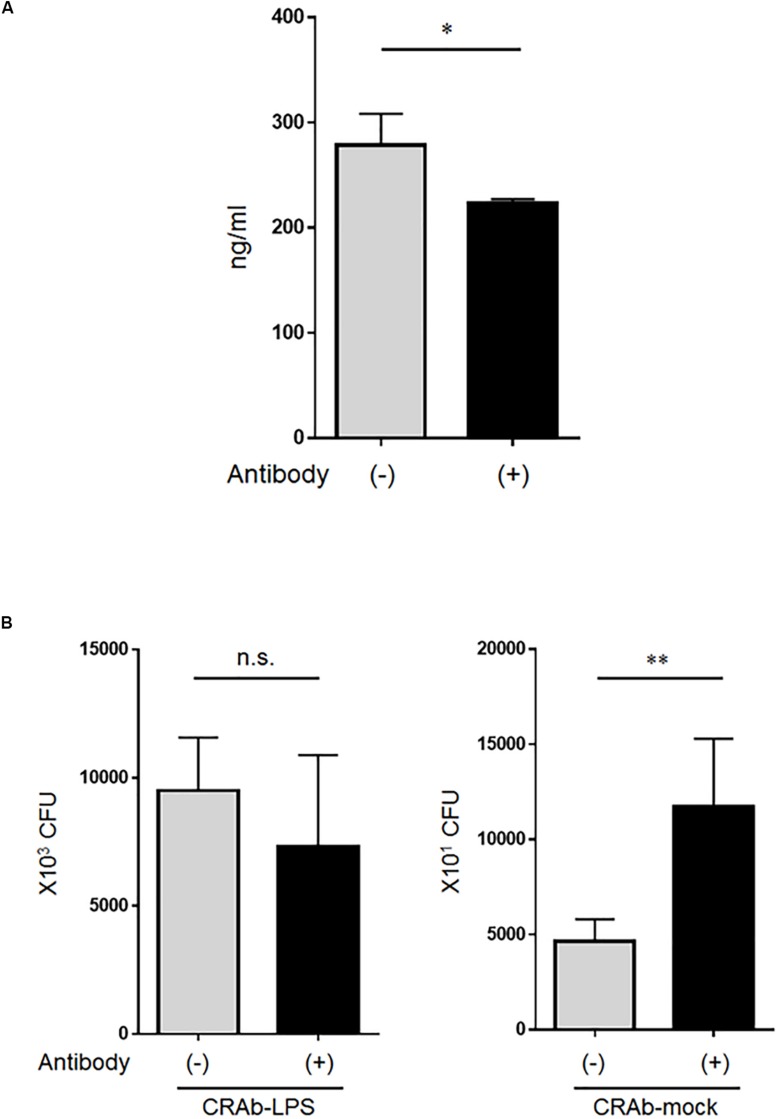
Whereas low lysozyme concentrations killed LPS-deficient A. baumannii strains, even high lysozyme concentrations had no apparent adverse effect on the LPS-positive strains. Therefore, we speculated that the bactericidal factor in the neutrophil supernatant may have been lysozyme. Lysozyme at ∼280 ng/ml was detected in the co-cultured bacteria (CRAb-5)/neutrophil supernatant and reacting the supernatant with anti-lysozyme antibody reduced its amount (Figure 6A). Bacterial clearance experiments were conducted on these supernatants to determine the effects of lysozyme on LPS-deficient A. baumannii. Treatment of co-culture supernatant with anti-lysozyme decreased the killing effects on LPS-deficient CRAb-mock but had no apparent deleterious effects on LPS-restoring CRAb-LPS (Figure 6B). This observation suggests that neutrophilic lysozyme sufficiently kills LPS-deficient A. baumannii.
FIGURE 6.
Effect of lysozyme released by neutrophils on lipopolysaccharide-deficient Acinetobacter baumannii strains. (A) Neutrophilic lysozyme secreted into culture supernatants was measured by ELISA. Supernatants were obtained from neutrophils co-cultured with colistin-resistant A. baumannii for 4 h. Lysozymes were neutralized in other supernatants with anti-lysozyme antibody. Data are means ± SD; n = 4 per group. (B) Bacterial clearance experiments were performed using these supernatants. The supernatants were added to colistin-resistant A. baumannii (CRAb)-LPS or CRAb-mock and viable bacteria were counted by plating on kanamycin agar. Data are means ± SD; n = 6 per group. n.s., not significant, ∗∗P < 0.01, ∗P < 0.05. Means are representative of three experiments.
Discussion
In the present study, we established CRAb strains and analyzed the mechanism through which neutrophils clear them. Previous studies reported that relative to the wild type, the virulence of LPS-deficient A. baumannii is low in mouse models (Moffatt et al., 2013; Beceiro et al., 2014; Carretero-Ledesma et al., 2018). This was based on the comparatively reduced inflammatory cytokine (TNF-α and IL-6) levels in the blood of mice subjected to the LPS-deficient A. baumannii (Carretero-Ledesma et al., 2018). Moreover, murine macrophage-like cell lines lacking receptors indicated that LPS-deficient A. baumannii received only weak NF-κB signals from TLR4 (Moffatt et al., 2013). It is known that neutrophils play an important role in the control of bacterial infection. However, the mechanism involved has not yet been elucidated. With a focus on the interaction between neutrophils and LPS-deficient A. baumannii, we found that neutrophils significantly killed LPS-deficient A. baumannii better than wild-type strains. Furthermore, LPS-deficient A. baumannii strains have increased susceptibility to antibacterial proteins and β-lactam antibacterial drugs and might be less able to survive in vivo due to reduced proliferation ability. Inflammatory cytokine production by neutrophils was relatively low in response to challenge induced by LPS-deficient strains, suggesting that these strains are lesser activators of neutrophils than wild-type A. baumannii. In LPS-deficient A. baumannii strains, it is conceivable that a decrease in the induction of inflammatory cytokines would minimize subsequent immune responses. This minor attenuation of immune response by LPS-deficient bacteria may not be a problem in healthy people; however, in patients with immune diseases such as neutropenia, infection may be worsened due to lesser neutrophil activation and reduced inflammatory cytokine production.
Lysozyme occurs in the cytoplasmic granules of macrophages and neutrophils (Leffell and Spitznagel, 1972). It catalyzes the hydrolysis of peptidoglycan, which is a major component of bacterial cell walls. Bacterial cell walls subjected to lysozyme lose their integrity and the cell undergoes lysis (Kirby, 2001). The LPS in the cell walls of Gram-negative bacteria inhibits lysozyme binding and prevents lysis (Ohno and Morrison, 1989; Brandenburg et al., 1998). Furthermore, LPS, the outer membrane of Gram-negative bacteria, serves as a selectively permeable barrier that keeps large molecules, such as lysozyme, from accessing periplasmic peptidoglycan (Nikaido, 2003). Loss of LPS likely leads, which in turn leads to increased access of lysozyme to periplasmic peptidoglycan. Wild-type A. baumannii and the complementary strains derived from it were less susceptible to lysozyme because of their LPS content. In contrast, CRAb-5 and CRAb-mock lacked LPS and were easier targets for lysozyme. Lysozyme has enhanced activity against colistin-resistant strains lacking LPS (Garcia-Quintanilla et al., 2014). Using complementary strains and neutrophil-derived lysozyme, we demonstrated that bacterial clearance by the enzyme was influenced by bacterial LPS status. Further, high concentrations of the antibacterial biomolecule lactoferrin killed only LPS-deficient A. baumannii strains. The primary function of lactoferrin is to sequester free iron, which is an essential substrate for bacterial growth (Masson et al., 1969). Its secondary role is to bind bacterial wall LPS. This reaction alters membrane permeability, especially at high lactoferrin concentrations (Farnaud and Evans, 2003; Drago-Serrano et al., 2012). LL-37 is a cationic antimicrobial present in macrophage and neutrophil lysosomes (Turner et al., 1998; Durr et al., 2006). Two conflicting reports on CRAb strains lacking LPS demonstrated increased or decreased resistance to LL-37 (Moffatt et al., 2013; Garcia-Quintanilla et al., 2014). We found that LPS-deficient A. baumannii strains were resistant to LL-37 (Table 3 and Figure 5C). LL-37 is positively charged. Whereas wild-type A. baumannii strains has anionic LPS, LPS-deficient strains lack this negatively charged molecule, making it difficult for cationic LL-37 to bind to them. Reportedly, LPS-deficient A. baumannii strains change the compositions of the outer membrane and increase the amount of molecules such as lipoproteins (Henry et al., 2012; Boll et al., 2016). Recently, secondary compensatory mutations for LPS deficiency have been reported in A. baumannii (Powers and Trent, 2018). The observed differential responses of A. baumannii strains in our study to biomolecules might be due to subtle differences in the outer membrane. A more detailed analysis of the outer membrane composition is thus warranted.
Previous studies reported that A. baumannii often acquires colistin resistance. Here, mutations were found in lpxA, lpxC, and lpxD of the CRAb strains we established. These genes are involved in the early stages of LPS biosynthesis (Raetz and Whitfield, 2002). To the best of our knowledge, only A. baumannii and A. nosocomialis acquire resistance to colistin through complete LPS deficiency (Moffatt et al., 2010; Vila-Farres et al., 2015). CRAb-6 was created by inserting the ISAba11 sequence into lpxC (data not shown). This transformation was reported earlier by Moffatt et al. (2011). Thus, the acquisition of colistin resistance by LPS deficiency seems to be a frequently occurring resistance mechanism in A. baumannii. A mutation introduced into the two-component pmrAB system and LPS modification with phosphoethanolamine both induced colistin resistance in A. baumannii (Adams et al., 2009; Beceiro et al., 2011). The pmrAB mutation resembled the wild type in terms of growth rate, biofilm formation, and mouse model pathogenicity (Beceiro et al., 2014). Recently, plasmid-mediated colistin resistance gene mcr (mobilized colistin resistance) was identified and investigated (Liu et al., 2016). This gene encodes phosphatidylethanolamine transferase, which modifies LPS, and therefore, causes colistin resistance (Hinchliffe et al., 2017). Since neutrophils seemed to exclusively target LPS-deficient A. baumannii in the current study, future studies should examine the interactions between neutrophils and LPS modification and colistin resistance in A. baumannii.
The LPS-deficient strains are reported to have less fitness (Beceiro et al., 2014; Mu et al., 2016), and so had our CRAb strains (Figures 1, 2A). The LPS-deficient strains might have problems growing on agar. The LPS-deficient strain had fewer colonies on agar even when the OD600, indicating the number of planktonic cells in broth, was almost the same value as the parent strain. By establishment of the LPS complement strain of CRAb, we could show that LPS was involved in bacterial fitness (Figure 2A). Furthermore, sensitivities to β-lactam drugs and other antimicrobials were increased in LPS-deficient A. baumannii strains compared with those of LPS-positive strains possibly because of the differences in membrane composition of the strains. Our findings suggested that β-lactam drugs may be more effective in patients with neutropenia comorbid with CRAb infection. Therefore, co-use of the colistin and β-lactams may be useful for treatment to A. baumannii infection in patients with neutropenia. Moreover, LPS-deficient A. baumannii are comparatively more susceptible to disinfectants such as chlorhexidine, sodium dodecyl sulfate, and ethanol than LPS-positive strains (Carretero-Ledesma et al., 2018). A few studies have provided direct evidence that LPS-deficient A. baumannii demonstrate lower proliferation, mobility, and biofilm formation capability than their LPS-positive counterparts (Moffatt et al., 2010; Beceiro et al., 2014; Dafopoulou et al., 2015; Carretero-Ledesma et al., 2018; Farshadzadeh et al., 2018).
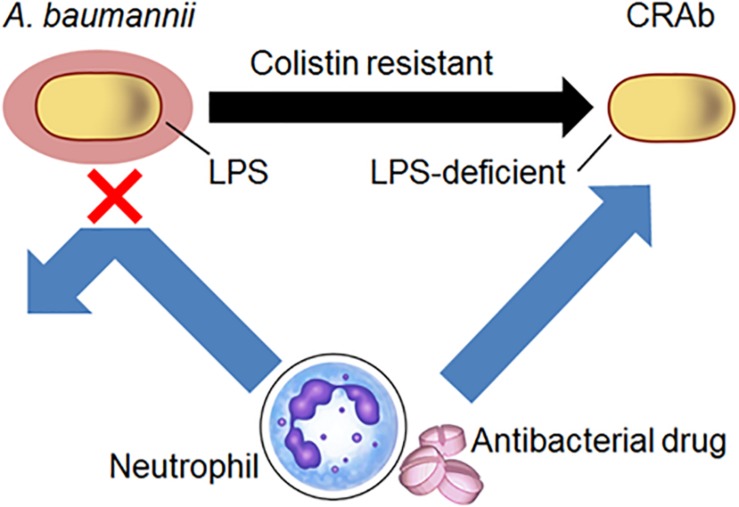
Although A. baumannii frequently acquires colistin resistance, LPS-deficient strains are nonetheless susceptible to host immunity, antibacterial drugs, and disinfectants. Clearly, our studies indicate that A. baumannii lacking LPS are resistant to colistin. We demonstrated a predilection for neutrophil-derived lysozyme in killing LPS-deficient A. baumannii strains and showed that these strains have increased susceptibility to antibacterial proteins from neutrophils and antibacterial drugs such as β-lactam (Figure 7). These findings are substantial in the current combat against colistin-resistant bacteria and control of A. baumannii infections in hospitals especially in immunocompromised patients with neutropenia.
FIGURE 7.
Schematic diagram of the study. Acinetobacter baumannii acquire resistance to colistin by LPS deficiency. Neutrophil-derived lysozyme and antibacterial drugs such as β-lactam were killed by LPS-deficient A. baumannii strains.
Data Availability Statement
All datasets generated for this study are included in the article/Supplementary Material.
Ethics Statement
The studies involving human participants were reviewed and approved by the Ethical Review Committee of the School of Medicine of Teikyo University, Tokyo, Japan. The patients/participants provided their written informed consent to participate in this study.
Author Contributions
GK designed and performed the experiments, analyzed and discussed the data, and wrote the manuscript. TA performed the experiments. NT performed the DNA analysis experiments and discussed the data. YSu, YSa, DK, TH, DY, TK-U, SN, YU, ST-N, TU, and TM-A performed a portion of experiments and discussed the data. MO and YO supervised the study and co-wrote the manuscript.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
The authors thank Dr. Naoki Hayashi (Kyoto Pharmaceutical University, Kyoto, Japan), Dr. Teruo Akuta, Mr. Keitaro Imaizumi (Kyokuto Pharmaceutical Industrial Co. Ltd., Tokyo, Japan), and Dr. Hirotoshi Kikuchi (Teikyo University School of Medicine, Tokyo, Japan) for their helpful comments and input on this study. The authors thank Ms. Chizuru Miyazaki (Teikyo University School of Medicine, Tokyo, Japan) for her technical assistance and Mr. Yuichi Nakamura for preparing the illustration.
Abbreviations
- IL
interleukin
- LPS
lipopolysaccharide
- MDRA
multidrug-resistant Acinetobacter baumannii
- MOI
multiplicity of infection
- PBS
phosphate-buffered saline
- PMA
phorbol-12-myristate-13-acetate
- ROS
reactive oxygen species
- TNF
tumor necrosis factor.
Footnotes
Funding. This work was supported in part by the Ministry of Education, Culture, Sports, Science, and Technology of Japan (JSPS KAKENHI; No. 17K16230 to GK and No. 17K10032 to YO), the Private University Research Branding Project, and the Science Research Promotion Fund of the Promotion and Mutual Aid Corporation for Private Schools of Japan.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2020.00573/full#supplementary-material
References
- Adams M. D., Nickel G. C., Bajaksouzian S., Lavender H., Murthy A. R., Jacobs M. R., et al. (2009). Resistance to colistin in Acinetobacter baumannii associated with mutations in the PmrAB two-component system. Antimicrob. Agents Chemother. 53 3628–3634. 10.1128/aac.00284-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beceiro A., Llobet E., Aranda J., Bengoechea J. A., Doumith M., Hornsey M., et al. (2011). Phosphoethanolamine modification of lipid A in colistin-resistant variants of Acinetobacter baumannii mediated by the pmrAB two-component regulatory system. Antimicrob. Agents Chemother. 55 3370–3379. 10.1128/aac.00079-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beceiro A., Moreno A., Fernandez N., Vallejo J. A., Aranda J., Adler B., et al. (2014). Biological cost of different mechanisms of colistin resistance and their impact on virulence in Acinetobacter baumannii. Antimicrob. Agents Chemother. 58 518–526. 10.1128/aac.01597-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boll J. M., Crofts A. A., Peters K., Cattoir V., Vollmer W., Davies B. W., et al. (2016). A penicillin-binding protein inhibits selection of colistin-resistant, lipooligosaccharide-deficient Acinetobacter baumannii. Proc. Natl. Acad. Sci. U.S.A. 113 E6228–E6237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borregaard N., Sorensen O. E., Theilgaard-Monch K. (2007). Neutrophil granules: a library of innate immunity proteins. Trends Immunol. 28 340–345. 10.1016/j.it.2007.06.002 [DOI] [PubMed] [Google Scholar]
- Brandenburg K., Koch M. H., Seydel U. (1998). Biophysical characterisation of lysozyme binding to LPS Re and lipid A. Eur. J. Biochem. 258 686–695. 10.1046/j.1432-1327.1998.2580686.x [DOI] [PubMed] [Google Scholar]
- Cai Y., Chai D., Wang R., Liang B., Bai N. (2012). Colistin resistance of Acinetobacter baumannii: clinical reports, mechanisms and antimicrobial strategies. J. Antimicrob. Chemother. 67 1607–1615. 10.1093/jac/dks084 [DOI] [PubMed] [Google Scholar]
- Carretero-Ledesma M., García-Quintanilla M., Martín-Peña R., Pulido M. R., Pachón J., Mcconnell M. J. (2018). Phenotypic changes associated with Colistin resistance due to Lipopolysaccharide loss in Acinetobacter baumannii. Virulence 9 930–942. 10.1080/21505594.2018.1460187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu Y. W., Leung C. M., Houang E. T., Ng K. C., Leung C. B., Leung H. Y., et al. (1999). Skin carriage of acinetobacters in Hong Kong. J. Clin. Microbiol. 37 2962–2967. 10.1128/jcm.37.9.2962-2967.1999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cisneros J. M., Rodríguez-Baño J. (2002). Nosocomial bacteremia due to Acinetobacter baumannii: epidemiology, clinical features and treatment. Clin. Microbiol. Infect. 8 687–693. 10.1046/j.1469-0691.2002.00487.x [DOI] [PubMed] [Google Scholar]
- Dafopoulou K., Xavier B. B., Hotterbeekx A., Janssens L., Lammens C., De E., et al. (2015). Colistin-Resistant Acinetobacter baumannii clinical strains with deficient biofilm formation. Antimicrob. Agents Chemother. 60 1892–1895. 10.1128/aac.02518-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dijkshoorn L., Nemec A., Seifert H. (2007). An increasing threat in hospitals: multidrug-resistant Acinetobacter baumannii. Nat. Rev. Microbiol. 5 939–951. 10.1038/nrmicro1789 [DOI] [PubMed] [Google Scholar]
- Dorsey C. W., Tomaras A. P., Actis L. A. (2002). Genetic and phenotypic analysis of Acinetobacter baumannii insertion derivatives generated with a transposome system. Appl. Environ. Microbiol. 68 6353–6360. 10.1128/aem.68.12.6353-6360.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drago-Serrano M. E., De La Garza-Amaya M., Luna J. S., Campos-Rodriguez R. (2012). Lactoferrin-lipopolysaccharide (LPS) binding as key to antibacterial and antiendotoxic effects. Int. Immunopharmacol. 12 1–9. 10.1016/j.intimp.2011.11.002 [DOI] [PubMed] [Google Scholar]
- Durr U. H., Sudheendra U. S., Ramamoorthy A. (2006). LL-37, the only human member of the cathelicidin family of antimicrobial peptides. Biochim. Biophys. Acta 1758 1408–1425. 10.1016/j.bbamem.2006.03.030 [DOI] [PubMed] [Google Scholar]
- Ellison R. T., III, Giehl T. J. (1991). Killing of gram-negative bacteria by lactoferrin and lysozyme. J. Clin. Invest. 88 1080–1091. 10.1172/jci115407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falagas M. E., Kasiakou S. K. (2005). Colistin: the revival of polymyxins for the management of multidrug-resistant gram-negative bacterial infections. Clin. Infect. Dis. 40 1333–1341. 10.1086/429323 [DOI] [PubMed] [Google Scholar]
- Farnaud S., Evans R. W. (2003). Lactoferrin—a multifunctional protein with antimicrobial properties. Mol. Immunol. 40 395–405. 10.1016/s0161-5890(03)00152-4 [DOI] [PubMed] [Google Scholar]
- Farshadzadeh Z., Taheri B., Rahimi S., Shoja S., Pourhajibagher M., Haghighi M. A., et al. (2018). Growth rate and biofilm formation ability of clinical and laboratory-evolved colistin-resistant strains of Acinetobacter baumannii. Front. Microbiol. 9:153. 10.3389/fmicb.2018.00153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Patino M. G., Garcia-Contreras R., Licona-Limon P. (2017). The immune response against Acinetobacter baumannii, an emerging pathogen in nosocomial infections. Front. Immunol. 8:441. 10.3389/fimmu.2017.00441 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Quintanilla M., Pulido M. R., Moreno-Martinez P., Martin-Pena R., Lopez-Rojas R., Pachon J., et al. (2014). Activity of host antimicrobials against multidrug-resistant Acinetobacter baumannii acquiring colistin resistance through loss of lipopolysaccharide. Antimicrob. Agents Chemother. 58 2972–2975. 10.1128/aac.02642-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garnacho-Montero J., Dimopoulos G., Poulakou G., Akova M., Cisneros J. M., De Waele J., et al. (2015). Task force on management and prevention of Acinetobacter baumannii infections in the ICU. Intensive Care Med. 41 2057–2075. 10.1007/s00134-015-4079-4 [DOI] [PubMed] [Google Scholar]
- Garnacho-Montero J., Ortiz-Leyba C., Fernandez-Hinojosa E., Aldabo-Pallas T., Cayuela A., Marquez-Vacaro J. A., et al. (2005). Acinetobacter baumannii ventilator-associated pneumonia: epidemiological and clinical findings. Intensive Care Med. 31 649–655. 10.1007/s00134-005-2598-0 [DOI] [PubMed] [Google Scholar]
- Henry R., Vithanage N., Harrison P., Seemann T., Coutts S., Moffatt J. H., et al. (2012). Colistin-resistant, lipopolysaccharide-deficient Acinetobacter baumannii responds to lipopolysaccharide loss through increased expression of genes involved in the synthesis and transport of lipoproteins, phospholipids, and poly-beta-1,6-N-acetylglucosamine. Antimicrob. Agents Chemother. 56 59–69. 10.1128/aac.05191-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hinchliffe P., Yang Q. E., Portal E., Young T., Li H., Tooke C. L., et al. (2017). Insights into the mechanistic basis of plasmid-mediated colistin resistance from crystal structures of the catalytic domain of MCR-1. Sci. Rep. 7:39392. 10.1038/srep39392 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamoshida G., Kikuchi-Ueda T., Nishida S., Tansho-Nagakawa S., Kikuchi H., Ubagai T., et al. (2017). Spontaneous formation of neutrophil extracellular traps in serum-free culture conditions. FEBS Open Biol. 7 877–886. 10.1002/2211-5463.12222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamoshida G., Kikuchi-Ueda T., Nishida S., Tansho-Nagakawa S., Ubagai T., Ono Y. (2018). Pathogenic bacterium Acinetobacter baumannii Inhibits the formation of neutrophil extracellular traps by suppressing neutrophil adhesion. Front. Immunol. 9:178. 10.3389/fimmu.2018.00178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamoshida G., Kikuchi-Ueda T., Tansho-Nagakawa S., Nakano R., Nakano A., Kikuchi H., et al. (2015). Acinetobacter baumannii escape from neutrophil extracellular traps (NETs). J. Infect. Chemother. 21 43–49. 10.1016/j.jiac.2014.08.032 [DOI] [PubMed] [Google Scholar]
- Kamoshida G., Matsuda A., Katabami K., Kato T., Mizuno H., Sekine W., et al. (2012). Involvement of transcription factor Ets-1 in the expression of the alpha3 integrin subunit gene. FEBS J. 279 4535–4546. 10.1111/febs.12040 [DOI] [PubMed] [Google Scholar]
- Kamoshida G., Tansho-Nagakawa S., Kikuchi-Ueda T., Nakano R., Hikosaka K., Nishida S., et al. (2016). A novel bacterial transport mechanism of Acinetobacter baumannii via activated human neutrophils through interleukin-8. J. Leukoc. Biol. 100 1405–1412. 10.1189/jlb.4ab0116-023rr [DOI] [PubMed] [Google Scholar]
- Kirby A. J. (2001). The lysozyme mechanism sorted – after 50 years. Nat. Struct. Biol. 8 737–739. [DOI] [PubMed] [Google Scholar]
- Leao A. C., Menezes P. R., Oliveira M. S., Levin A. S. (2016). Acinetobacter spp. are associated with a higher mortality in intensive care patients with bacteremia: a survival analysis. BMC Infect. Dis. 16:386. 10.1186/s12879-016-1695-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leffell M. S., Spitznagel J. K. (1972). Association of lactoferrin with lysozyme in granules of human polymorphonuclear leukocytes. Infect. Immun. 6 761–765. 10.1128/iai.6.5.761-765.1972 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levin A. S., Barone A. A., Penco J., Santos M. V., Marinho I. S., Arruda E. A., et al. (1999). Intravenous colistin as therapy for nosocomial infections caused by multidrug-resistant Pseudomonas aeruginosa and Acinetobacter baumannii. Clin. Infect. Dis. 28 1008–1011. 10.1086/514732 [DOI] [PubMed] [Google Scholar]
- Liu Y. Y., Wang Y., Walsh T. R., Yi L. X., Zhang R., Spencer J., et al. (2016). Emergence of plasmid-mediated colistin resistance mechanism MCR-1 in animals and human beings in China: a microbiological and molecular biological study. Lancet Infect. Dis. 16 161–168. 10.1016/s1473-3099(15)00424-7 [DOI] [PubMed] [Google Scholar]
- Maragakis L. L., Perl T. M. (2008). Acinetobacter baumannii: epidemiology, antimicrobial resistance, and treatment options. Clin. Infect. Dis. 46 1254–1263. 10.1086/529198 [DOI] [PubMed] [Google Scholar]
- Masson P. L., Heremans J. F., Schonne E. (1969). Lactoferrin, an iron-binding protein in neutrophilic leukocytes. J. Exp. Med. 130 643–658. 10.1084/jem.130.3.643 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mcconnell M. J., Actis L., Pachon J. (2013). Acinetobacter baumannii: human infections, factors contributing to pathogenesis and animal models. FEMS Microbiol. Rev. 37 130–155. 10.1111/j.1574-6976.2012.00344.x [DOI] [PubMed] [Google Scholar]
- Moffatt J. H., Harper M., Adler B., Nation R. L., Li J., Boyce J. D. (2011). Insertion sequence ISAba11 is involved in colistin resistance and loss of lipopolysaccharide in Acinetobacter baumannii. Antimicrob. Agents Chemother. 55 3022–3024. 10.1128/aac.01732-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moffatt J. H., Harper M., Harrison P., Hale J. D., Vinogradov E., Seemann T., et al. (2010). Colistin resistance in Acinetobacter baumannii is mediated by complete loss of lipopolysaccharide production. Antimicrob. Agents Chemother. 54 4971–4977. 10.1128/aac.00834-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moffatt J. H., Harper M., Mansell A., Crane B., Fitzsimons T. C., Nation R. L., et al. (2013). Lipopolysaccharide-deficient Acinetobacter baumannii shows altered signaling through host Toll-like receptors and increased susceptibility to the host antimicrobial peptide LL-37. Infect. Immun. 81 684–689. 10.1128/iai.01362-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mu X., Wang N., Li X., Shi K., Zhou Z., Yu Y., et al. (2016). The effect of colistin resistance-associated mutations on the fitness of Acinetobacter baumannii. Front. Microbiol. 7:1715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munoz-Price L. S., Weinstein R. A. (2008). Acinetobacter infection. N. Engl. J. Med. 358 1271–1281. [DOI] [PubMed] [Google Scholar]
- Nathan C. (2006). Neutrophils and immunity: challenges and opportunities. Nat. Rev. Immunol. 6 173–182. 10.1038/nri1785 [DOI] [PubMed] [Google Scholar]
- Nauseef W. M., Borregaard N. (2014). Neutrophils at work. Nat. Immunol. 15 602–611. 10.1038/ni.2921 [DOI] [PubMed] [Google Scholar]
- Nikaido H. (2003). Molecular basis of bacterial outer membrane permeability revisited. Microbiol. Mol. Biol. Rev. 67 593–656. 10.1128/mmbr.67.4.593-656.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohno N., Morrison D. C. (1989). Lipopolysaccharide interaction with lysozyme. Binding of lipopolysaccharide to lysozyme and inhibition of lysozyme enzymatic activity. J. Biol. Chem. 264 4434–4441. [PubMed] [Google Scholar]
- Peleg A. Y., Seifert H., Paterson D. L. (2008). Acinetobacter baumannii: emergence of a successful pathogen. Clin. Microbiol. Rev. 21 538–582. 10.1128/cmr.00058-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perez F., Hujer A. M., Hujer K. M., Decker B. K., Rather P. N., Bonomo R. A. (2007). Global challenge of multidrug-resistant Acinetobacter baumannii. Antimicrob. Agents Chemother. 51 3471–3484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Powers M. J., Trent M. S. (2018). Phospholipid retention in the absence of asymmetry strengthens the outer membrane permeability barrier to last-resort antibiotics. Proc. Natl. Acad. Sci. U.S.A. 115 E8518–E8527. 10.1073/pnas.1806714115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qureshi Z. A., Hittle L. E., O’hara J. A., Rivera J. I., Syed A., Shields R. K., et al. (2015). Colistin-resistant Acinetobacter baumannii: beyond carbapenem resistance. Clin. Infect. Dis. 60 1295–1303. 10.1093/cid/civ048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raetz C. R., Whitfield C. (2002). Lipopolysaccharide endotoxins. Annu. Rev. Biochem. 71 635–700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner J., Cho Y., Dinh N. N., Waring A. J., Lehrer R. I. (1998). Activities of LL-37, a cathelin-associated antimicrobial peptide of human neutrophils. Antimicrob. Agents Chemother. 42 2206–2214. 10.1128/aac.42.9.2206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vila-Farres X., Ferrer-Navarro M., Callarisa A. E., Marti S., Espinal P., Gupta S., et al. (2015). Loss of LPS is involved in the virulence and resistance to colistin of colistin-resistant Acinetobacter nosocomialis mutants selected in vitro. J. Antimicrob. Chemother. 70 2981–2986. 10.1093/jac/dkv244 [DOI] [PubMed] [Google Scholar]
- Weber B. S., Hennon S. W., Wright M. S., Scott N. E., De Berardinis V., Foster L. J., et al. (2016). Genetic dissection of the type vi secretion system in acinetobacter and identification of a novel peptidoglycan hydrolase, tagx, required for its biogenesis. mBio 7:e01253-16. 10.1128/mBio.01253-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong D., Nielsen T. B., Bonomo R. A., Pantapalangkoor P., Luna B., Spellberg B. (2017). Clinical and Pathophysiological Overview of Acinetobacter Infections: a Century of Challenges. Clin. Microbiol. Rev 30 409–447. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All datasets generated for this study are included in the article/Supplementary Material.