Abstract
Objective
To investigate the molecular characteristics of carbapenem-resistant Enterobacteriaceae (CRE) from county hospitals in China.
Materials and Methods
Forty-three sequential non-duplicate CRE strains (including 33 Klebsiella pneumoniae isolates, 4 Enterobacter cloacae isolates, 3 Escherichia coli isolates, 1 Serratia marcescens, 1 Morganella morganii and 1 Citrobacter freundii) were collected from 4 county hospitals and 2 municipal hospitals. Antimicrobial susceptibility testing was conducted by broth microdilution method, using 3-aminophenylboronic acid and EDTA and the modified carbapenem inactivation method (mCIM) to screen phenotype of carbapenemase. β-Lactamases were characterized by polymerase chain reaction (PCR) and DNA sequencing. The transferability of blaNDM-5 was investigated by transformation experiment. Clonal relatedness was evaluated by pulsed-field gel electrophoresis and multilocus sequence typing .
Results
The results of antimicrobial susceptibility testing indicated that 43 CRE strains were resistant to most of the antimicrobial agents, except tigecycline and colistin. Overall, 93%, 93%, and 97.7% of these strains were resistant to imipenem, meropenem, and ertapenem, respectively. PCR and DNA sequencing indicated that 67.4% (29/43) were blaKPC-2 positive isolates, in which 3.4% (1/29) was coproduced with blaNDM-1. In addition, 7.0% (3/43), 4.7% (2/43), 4.7% (2/43), 2.3% (1/43), 2.3% (1/43) were blaNDM-1, blaNDM-16, blaNDM-4, blaNDM-5, blaIMP-4 positive isolates, respectively. The 29 blaKPC-2-positive isolates belonged to 12 different PFGE type and designated as ST11 (n=20) and ST15, ST39, ST116, ST667, ST2245, ST2338. The plasmid bearing blaNDM-5 could be transferred into recipient E. coli J53 through transformation.
Conclusion
Our study indicated the dissemination of CRE between the tertiary hospitals and secondary hospitals.
Keywords: Enterobacteriaceae, blaKPC-2, blaNDM-1
Introduction
Carbapenems are often used as a last resort to treat Gram-negative bacteria infections. The emergence and rapid spread of carbapenemases in Enterobacteriaceae clinical isolates which were the most frequent pathogens of causing nosocomial infections is becoming a great public health problem worldwide.1,2 According to the CHINET surveillance network reported (www.chinets.com), the rates of Klebsiella pneumoniae resistance to imipenem and meropenem increased markedly from 2005 to 2014, from 1.3% to14.6% and from 0% to 15.2%, respectively.3 Currently, three main classes of carbapenemases have been identified including class A, B, and D type carbapenemases (these groups do not solely contain carbapenemases). Klebsiella pneumoniae carbapenemases (KPC) was first reported in the United States in the late 1990s and since then worldwide, with a marked endemicity in the United States, Greece, and Italy.4,5
Because genes encoding carbapenemase are often located in plasmids, and these plasmids often carry mobile genetic elements and a variety of resistance genes, causing resistance to spread between different bacteria, different cities and different countries. Currently, the study about carbapenemases mainly focused on the Enterobacteriaceae clinical isolates collected from large cities and hospitals, rarely involving county hospitals. Our study aimed to investigate the prevalence of carbapenem-resistant Enterobacteriaceae (CRE) clinical isolates in county hospital and to provide basis for prevention and control of nosocomial infections.
Materials and Methods
Bacterial Isolates
A sequential non-duplicated collection of 43 CRE (33 Klebsiella pneumonia isolates, 4 Enterobacter cloacae isolates, 3 Escherichia coli isolates, 1 Serratia marcescens, 1 Morganella morganii and 1 Citrobacter freundii) were routine obtained in clinical microbiology laboratory from January to June in 2017. From 4 county hospitals (usually called secondary hospital, the numbers range of beds is 200–480) and 2 municipal hospitals (usually called municipal hospital, the numbers of beds is 1300 and 1600, respectively) in Zhejiang province in China. Of 43 isolates, 58.1% (25/43) were isolated from sputum, followed by urine (20.9%, 9/43), blood (11.6%, 5/43), secretion sample (7.0%, 3/43) and abdominal fluid (2.3%, 1/43). All clinical isolates were resistant to at least one of carbapenem (ertapenem, imipenem or meropenem) and were identified using the VITEK 2 Compact system. E. coli ATCC 25922, Salmonella ser. Braenderup H9812, and E. coli J53 were used as the quality control for antimicrobial susceptibility testing, reference marker for PFGE, and recipient strain for transformation, respectively.
Antimicrobial Susceptibility Testing and β-Lactamase Characterization
Antimicrobial susceptibility testing was performed using broth microdilution method and results were interpreted following the criteria of the Clinical and Laboratory Standards Institute.6 The breakpoints proposed by European Committee on Antimicrobial Susceptibilities Testing (EUCAST) were used for colistin and tigecycline.7 The carbapenem inactivation method (CIM) were used for phenotypic detection of carbapenemase.8 The presence of genes encoding β-lactamase, including CTX-M-type extended-spectrumβ-lactamases (ESBLs), plasmid-born AmpC β-lactamases, and carbapenemases, were investigated by PCR using primers previously described.9–11 Polymerase chain reaction (PCR) amplicons were sequenced and the DNA sequences obtained were compared with those available in the NCBI GenBank database using BLAST searches (https://blast.ncbi.nlm.nih.gov/Blast.cgi).
Transfer of Carbapenemase Resistance
Transformation experiment was carried out with E. coli J53 as the recipient to determine the transferability of the carbapenemase gene, as described previously.12
Bacterial Genotyping
Whole-cell DNA of clinical strains embedded in agarose gel plugs, was separated by PFGE for 33 K. pneumoniae isolates and 3 E. coli isolates. Clonal relationships were analyzed using PFGE of XbaⅠ-digested genomic DNA as previously described,13 and the results were analyzed according to the criteria proposed by Tenover et al.14 MLST for these isolates was performed as described previously.15
Results
Antimicrobial Susceptibility Testing
The results of antimicrobial susceptibility testing indicated the resistant rate of 43 CRE isolates to imipenem, meropenem, and ertapenem was 93%, 93%, and 97.7% with the MIC50/MIC90 of >16/>16 mg/L, >16/>16mg/L and >32/>32 mg/L, respectively. 46.5%, 90.3% and 93.0% were susceptible to amikacin, tigecycline and colistin, respectively (Table 1). For three colistin-resistant strains, one was K. pneumoniae, and the other two strains were Serratia marcescens and Morganella morganii which was intrinsically resistant to colistin.
Table 1.
Resistance and Susceptibility of All the Strains to Antimicrobial Agents
| Antimicrobial Agent | %R | %S | MIC50 | MIC90 | MIC Range |
|---|---|---|---|---|---|
| Piperacillin | 97.7 | 2.3 | >256 | >256 | ≤2–>256 |
| Cefoperazone-sulbactam | 93 | 4.7 | >128 | >128 | ≤1–>128 |
| Piperacillin-tazobactam | 95.3 | 2.3 | >256 | >256 | ≤2–>256 |
| Cefazolin | 100 | 0 | >32 | >32 | >32–>32 |
| Cefuroxime | 100 | 0 | >64 | >64 | 32–>64 |
| Ceftazidime | 93 | 4.7 | >32 | >32 | ≤0.25–>32 |
| Cefepime | 97.7 | 2.3 | >32 | >32 | ≤0.25–>32 |
| Cefotaxime | 95.3 | 2.3 | >32 | >32 | ≤0.25–>32 |
| Cefmetazole | 79.1 | 18.6 | >64 | >64 | 4–>64 |
| Moxalactam | 76.7 | 9.3 | >64 | >64 | ≤0.5–>64 |
| Aztreonam | 93 | 7 | >128 | >128 | ≤1–>128 |
| Ertapenem | 97.7 | 2.3 | >32 | >32 | ≤0.25–>32 |
| Imipenem | 93 | 0 | >16 | >16 | 2–>16 |
| Meropenem | 93 | 7 | >16 | >16 | ≤0.125–>16 |
| Amikacin | 51.2 | 46.5 | >128 | >128 | 2–>128 |
| Gentamicin | 67.4 | 27.9 | >128 | >128 | ≤1–>128 |
| Ciprofloxacin | 81.4 | 18.6 | >8 | >8 | ≤0.06–>8 |
| Levofloxacin | 67.4 | 23.3 | 16 | >16 | ≤0.125–>16 |
| Trimethoprim-sulfamethoxazole | 53.5 | 46.5 | >32 | >32 | ≤0.25–>32 |
| Colistin | 7.0 | 93 | 0.25 | 0.5 | ≤0.125–>16 |
| Nitrofurantoin | 67.4 | 18.6 | 128 | 256 | 8–>128 |
| Tigecycline | 9.7 | 90.3 | 0.5 | 1 | 0.25–2 |
Abbreviations: R, resistance; S, susceptible.
β-Lactamase Characterization
The result of CIM indicated that 97.7% (42/43) of these CRE isolates were carbapenemase-producing. PCR and DNA sequencing indicated that 67.4% (29/43) were blaKPC-2 positive isolates, in which 3.4% (1/29) was coproduced with blaNDM-1. In addition, 7.0% (3/43), 4.7% (2/43), 4.7% (2/43), 2.3% (1/43), 2.3% (1/43) were blaNDM-1, blaNDM-16, blaNDM-4, blaNDM-5, blaIMP-4 positive isolates, respectively (Table 2). PCR was used to detect genes encoding OXA, and VIM type carbapenemases but did not result in amplicons with any of the clinical isolates. No carbapenemase gene was detected for four isolates in this study and only ESBL or plasmid-mediated AmpC were positive including blaCTX-M-15 (K. pneumoniae), blaDHA-1 (M. morgannii), blaCMY-2 (E. coli) and blaSHV-12 (E. coli). Two isolates were negative for all of β-lactamase gene detected in this study.
Table 2.
Distribution of Gene Type of 43 Strains of CRE
| Gene Type | Strains (Number) | MIC Range (mg/L) | PFGE | MLST | |||
|---|---|---|---|---|---|---|---|
| Imipenem | Meropenem | Colistin | Tigecycline | ||||
| KPC-2+CTX-M+14+SHV-11 | K. pneumoniae (8) | 8–>16 | 8–>16 | <=0.125–1 | 0.25–1 | A1,A2,B,D,G3,H,J1(2) | ST11(8) |
| KPC-2+CTX-M-14+SHV-12 | K. pneumoniae (6) | >16 | >16 | 0.25–0.5 | 0.25–2 | A3(2),A4,B,G2,M | ST11(6) |
| KPC-2+CTX-M-15+CTX-M-14+SHV-11 | K. pneumoniae (3) | 8–>16 | 16–>16 | 0.25–0.5 | 0.25–1 | A1,D,J2 | ST11(3) |
| KPC-2 | K. pneumoniae (3) | >16 | 8->16 | <=0.125–>16 | 0.25-2 | O(3) | ST116(1) |
| KPC-2+SHV-11 | K. pneumoniae (2) | 16–>16 | >16 | 0.25 | 0.5–1 | C,I | ST11(2) |
| NDM-4 | E. cloacae (2) | 4-4 | 8-8 | 0.25 | 0.25–0.5 | - | - |
| CTX-M-55 | K. pneumoniae (1) | 32 | >16 | 0.25 | 0.25 | K | ST2536 |
| DHA-1 | M. morganii (1) | 4 | <=0.125 | >16 | 1 | - | - |
| CMY-2 | E. coli (1) | 2 | 1 | 0.25 | 0.5 | S | ST58 |
| KPC-2+CTX-M-55 | K. pneumoniae (1) | >16 | >16 | 0.25 | 0.5 | K | ST2338 |
| KPC-2+SHV-2a | K. pneumoniae (1) | >16 | >16 | 0.25 | 2 | G1 | ST2245 |
| KPC-2+CTX-M-15+SHV-LEN | K. pneumoniae (1) | >16 | >16 | 0.25 | 0.25 | F | ST39 |
| KPC-2+CTX-M-14+SHV-31 | K. pneumoniae (1) | >16 | >16 | 0.25 | 0.5 | H | ST11 |
| KPC-2+CTX-M-15+SHV-28 | K. pneumoniae (1) | 8 | 8 | 0.25 | 0.25 | E | ST15 |
| KPC-2+CTX-M-15+CTX-M-14+SHV-12 | K. pneumoniae (1) | >16 | >16 | 0.25 | 0.25 | G1 | ST667 |
| KPC-2+NDM-1+SHV-12 | C. freundii (1) | 8 | 8 | 0.5 | 1 | - | - |
| SHV-12 | E. coli (1) | 4 | 1 | 0.5 | 0.25 | Q | ST506 |
| NDM-1 | E. cloacae (1) | 4 | 16 | 0.25 | 0.25 | - | - |
| NDM-1+SHV-12 | E. cloacae (1) | 2 | 4 | <0.125 | 0.25 | - | - |
| NDM-5+CTX-M-55+SHV-12 | E. coli (1) | >16 | >16 | 0.5 | 0.5 | R | ST2 |
| NDM-16+SHV-12 | K. pneumoniae (1) | 8 | 16 | 0.25 | 0.5 | N | ST705 |
| NDM-16+CTX-M-15+SHV-LEN | K. pneumoniae (1) | 8 | 16 | 0.25 | 0.5 | L | ST39 |
| IMP-4+CTX-M-15+SHV-11 | K. pneumoniae (1) | 2 | 4 | 0.25 | 0.25 | L | ST1110 |
| Negative | K. pneumoniae (1), S. marcescen (1) | >16 | 16->16 | 0.5->16 | 0.25–2 | - | - |
Transfer of Carbapenemase Resistance
One E. coli bearing blaNDM-5 coupled with blaCTX-M-55 and blaSHV-12 were random selected for transformation experiment with E. coli J53 as the recipient. The plasmid bearingblaNDM-5 was successfully transferred from donor to recipient E. coli J53, and the transformation exhibited high resistant to carbapenems, consistent with the detection of blaNDM-5. The MICs of imipenem, meropenem, and ertapenem for the transformant were >16, >16, 32 mg/L. The MICs of meropenem increased more than 1000 times compared with the recipient.
Bacterial Genotyping
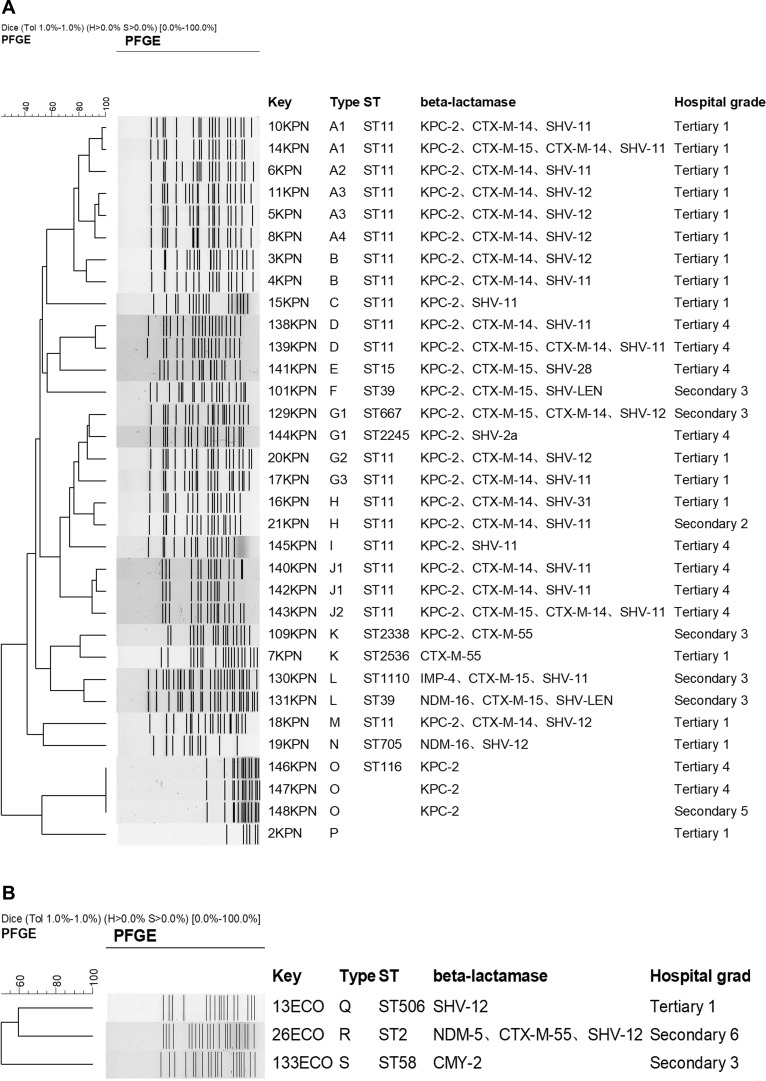
PFGE shows 12 different types for 33 K. pneumoniae isolates, and ST11 (60.6%, 20/33) was the predominant clone. The other ST type were ST2, ST15, ST39, ST58, ST116, ST506, ST667, ST705, ST1110, ST2245, ST2338, ST2536 (Figure 1A). PFGE shows 3 different types for 3 E. coli isolates (Figure 1B). For K. pneumoniae isolates, most of isolates were dissemination in the same hospital. However, PFGE type G1 (isolated from hospital secondary 3 and tertiary 4), H (isolated from hospital secondary 2 and tertiary 1), K (isolated from hospital secondary 3 and tertiary 1) and O (isolated from hospital secondary 5 and tertiary 4) isolates were dissemination among different hospitals (Figure 1A).
Figure 1.
DNA fingerprints and β-lactamases distribution of K. pneumoniae isolates (A) and E. coli isolates (B).
Discussion
Although carbapenems are the most widely antimicrobial spectrum of antibacterial drugs for Enterobacteriaceae clinical isolates, infections caused by CRE associated with significant morbidity and mortality are increasing year by year.16 In this study, 43 CRE isolates were highly resistant to most common antimicrobial agents except tigecycline, colistin, trimethoprim-sulfamethoxazole and amikacin with the sensitive rate for 90.3%, 93%, 46.5%and 46.5%, respectively. Although more than 90% of isolates were susceptible to tigecycline and colistin, tigecycline is not recommended for bloodstream infection because of the low concentration, and currently colistin is also not available for patients in China. The limitation of available therapeutic regimens for the infection caused by CRE is the great challenge. For the purpose to limit the spread of CRE, early monitoring of CRE infection or colonization on admission may play a more important role for timely control of the spread of CRKP.17 As Gorrie reported, the carriage frequencies of K. pneumoniae were about 6% among ICU patients admitted directly from the community, and 19% among those with recent healthcare contact. Gut colonization on admission was significantly associated with subsequent infection, 49% of K. pneumoniae infections were caused by the patients’ own unique strain, and 48% of screened patients with infections were positive for prior colonization.18
Studies had shown that the resistance mechanism of carbapenem-resistant Enterobacteriaceae clinical strains isolated from children patients, adult patients or from the different geographical distribution were the difference.19 In China, for the CRE strains isolated from children patients, the main type of carbapenemases mediated resistance to carbapenems was metallo-β-lactamases including blaNDM-1 and blaIMP, while blaKPC was predominant among adult patients. However, the diversity of carbapenemases among CRE isolated from county hospitals and municipal hospitals was not unequivocal. Our study indicated 67.4% (29/43) were blaKPC-2 positive isolates, in which 3.4% (1/29) was coproduced with blaNDM-1. 20.9% (9/43) were producing metallo-β-lactamases including blaNDM-1, blaNDM-16, blaNDM-4, blaNDM-5, and blaIMP-4. For 26 K. pneumoniae clinical strains collected from municipal hospitals, 3.8% (1/26) was blaNDM-16 positive. However, 28.6% (2/7) was blaNDM-16 positive or blaIPM-4 positive isolates among 7 K. pneumoniae clinical strains collected from county hospitals. The diversification of carbapenemases among CRE from different patients may strengthen the difficulty for infection control to limit the spread of CRE in clinical facilities.20–22 In this study, we also studied the relation for the CRE strains collected from tertiary hospitals and secondary hospitals. PFGE shows 12 different types for 33 K. pneumoniae isolates, and ST11 (60.6%, 20/33) was the predominant clone which was the main ST type for CRE. Except for one strain was isolated from secondary hospital, the other ST11 CRE were all from tertiary hospitals.
As reported,23–25 ST11 K pneumoniae clone is the dominant clone of KPC-producing K. pneumoniae in the world. ST11 clone would be good colonizers to capture plasmids and these isolates with ST11 clone will be easily transmitted between patients. For controlling the dissemination of CRE especially for ST11 KPC-producing K. pneumoniae among health-care facilities, rapid and accuracy detection of carbapenemases is critical for preventing and controlling outbreaks of CRE, however, there is no perfect method is suitable for detecting all types of carbapenemases because some carbapenemase producers demonstrate low resistance level to carbapenems.26,27 Currently, mCIM method was recommended by CLSI to detect the suspected carbapenemases among Enterobacteriaceae clinical isolates. Although mCIM has high sensitivity and specificity for confirmation of carbapenemase-producing strains, it is time-consuming for incubating 18–20h to read the final result and the false negative for some carbapenemase including OXA-type carbapenemase. Currently, several molecular technologies for Rapid detection of CRE, particularly carbapenemase-producing CRE have been developed for the automated detection of target genes including class A, class B and class D type carbapenemases directly from the clinical samples with high sensitivity and specificity,28 however, it only can detect the known gene, nor for the novel carbapenemase gene. In the future, we should investigate the novel carbapenemase gene for 6 strains which were carbapenemase-negative isolates determined by PCR including one was negative by mCIM.
Acknowledgments
This work was supported by the National Natural Science Foundation of China (grant no. 81871690, 81902101), self-financing program of science and technology plan project of Lishui city, Zhejiang province (2016ZCGYX15), Chinese Hospital Infection Fund (ZHYY2015-0015) and CHINET Antimicrobial Surveillance Network (grant WI207259). The funders had no role in study design, data collection and analysis, decision to publish or preparation of the manuscript.
Disclosure
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
References
- 1.Patel G, Bonomo RA. “Stormy waters ahead”: global emergence of carbapenemases. Front Microbiol. 2013;4(48):1–17. doi: 10.3389/fmicb.2013.00048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Skov RL, Monnet DL. Plasmid-mediated colistin resistance (mcr-1 gene): three months later, the story unfolds. Euro Surveill. 2016;21(9):30155. doi: 10.2807/1560-7917.ES.2016.21.9.30155 [DOI] [PubMed] [Google Scholar]
- 3.Hu FP, Guo Y, Zhu DM, et al. Resistance trends among clinical isolates in China reported from CHINET surveillance of bacterial resistance, 2005–2014. Clin Microbiol Infect. 2016;22(Suppl1):S9–S14. doi: 10.1016/j.cmi.2016.01.001 [DOI] [PubMed] [Google Scholar]
- 4.Nordmann P. Carbapenemase-producing Enterobacteriaceae: overview of a major public health challenge. Med Mal Infect. 2014;44:51–56. doi: 10.1016/j.medmal.2013.11.007 [DOI] [PubMed] [Google Scholar]
- 5.Bradford PA, Bratu S, Urban C, et al. Emergence of carbapenem-resistant Klebsiella species possessing the class A carbapenem-hydrolyzing KPC-2 and inhibitor-resistant TEM-30 beta-lactamases in New York City. Clin Infect Dis. 2004;39:55–60. doi: 10.1086/421495 [DOI] [PubMed] [Google Scholar]
- 6.Clinical and Laboratory Standards Institute. Performance Standards for Antimicrobial Susceptibility Testing; Twenty-Seventh Informational Supplement. CLSI Document M100-S27. Wayne, PA: Clinical and Laboratory Standards Institute; January, 2017. [Google Scholar]
- 7.European Committee on Antimicrobial Susceptibilities Testing (EUCAST) Breakpoint tables for interpretation of MICs and zone diameters. EUCAST 2014. Available from: http://www.eucast.org/fileadmin/src/media/PDFs/EUCAST_files/Breakpoint_tables/Breakpoint_table_v_4.0.pdf. Accessed September9, 2014.
- 8.van der Zwaluw K, de Haan A, Pluister GN, Bootsma HJ, de Neeling AJ, Schouls LM. The carbapenem inactivation method (CIM), a simple and low-cost alternative for the Carba NP test to assess phenotypic carbapenemase activity ingram-negative rods. PLoS One. 2015;10:e0123690. doi: 10.1371/journal.pone.0123690 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Woodford N, Fagan EJ, Ellington MJ. Multiplex PCR for rapid detection of genes encoding CTX-M extended-spectrum (beta) - lactamases. J Antimicrob Chemother. 2006;57:154–155. doi: 10.1093/jac/dki412 [DOI] [PubMed] [Google Scholar]
- 10.Perez-Perez FJ, Hanson ND. Detection of plasmid-mediated AmpC beta-lactamase genes in clinical isolates by using multiplex PCR. J Clin Microbiol. 2002;40:2153–2162. doi: 10.1128/JCM.40.6.2153-2162.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Poirela L, Walsh TR, Cuvillier V, et al. Multiplex PCR for detection of acquired carbapenemase genes. Diagn Microbiol Infect Dis. 2011;70:119–123. doi: 10.1016/j.diagmicrobio.2010.12.002 [DOI] [PubMed] [Google Scholar]
- 12.Borgia S, Lastovetska O, Richardson D, et al. Outbreak of carbapenem-resistant Enterobacteriaceae containing blaNDM-1, Ontario, Canada. Clin Infect Dis. 2012;55:e109–e117. doi: 10.1093/cid/cis737 [DOI] [PubMed] [Google Scholar]
- 13.Schlesinger J, Navon-Venezia S, Chmelnitsky I, et al. Extended-spectrum beta-lactamases among Enterobacter isolates obtained in Tel Aviv, Israel. Antimicrob Agents Chemother. 2005;49:1150–1156. doi: 10.1128/AAC.49.3.1150-1156.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tenover FC, Arbeit RD, Goering RV, et al. Interpreting chromosomal DNA restriction patterns produced by pulsed-field gel electrophoresis: criteria for bacterial strain typing. J Clin Microbiol. 1995;33:2233–2239. doi: 10.1128/JCM.33.9.2233-2239.1995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Diancourt L, Passet V, Verhoef J, Grimont PA, Brisse S. Multilocus sequence typing of Klebsiella pneumoniae nosocomial isolates. J Clin Microbiol. 2005;43:4178–4182. doi: 10.1128/JCM.43.8.4178-4182.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Manenzhe RI, Zar HJ, Nicol MP, et al. The spread of carbapenemase-producing bacteria in Africa: a systematic review. J Antimicrob Chemother. 2015;70:23–40. doi: 10.1093/jac/dku356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pannaraj PS, Bard JD, Cerini C, Weissman SJ. Pediatric carbapenem-resistant Enterobacteriaceae in Los Angeles, California, a high-prevalence region in the United States. Pediatr Infect Dis J. 2015;34:11–16. doi: 10.1097/INF.0000000000000471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gorrie CL, Mirčeta M, Wick RR. Gastrointestinal Carriage Is a Major Reservoir of Klebsiella pneumoniae Infection in Intensive Care Patients. Clin Infect Dis. 2017;65:208–215. doi: 10.1093/cid/cix270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhang R, Liu L, Zhou H, et al. Nationwide Surveillance of Clinical Carbapenem-resistant Enterobacteriaceae (CRE) Strains in China. EBioMedicine. 2017;19:98–106. doi: 10.1016/j.ebiom.2017.04.032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rafailidis PI, Falagas ME. Options for treating carbapenem-resistant Enterobacteriaceae. Curr Opin Infect Dis. 2014;27:479–483. doi: 10.1097/QCO.0000000000000109 [DOI] [PubMed] [Google Scholar]
- 21.Zhuochao JF. Risk factors and etiological analysis of multi drug resistant bacteria infection in elderly patients with stroke associated pneumonia. Chin J Anti Infect Agents. 2012;37:795–800. [Google Scholar]
- 22.Munoz MA, Welcome FL, Schukken YH, et al. Molecular epidemiology of two Klebsiella pneumoniae mastiffs outbreaks on a dairy farm in New York State. J Clin Microbiol. 2007;45:3964–3971. doi: 10.1128/JCM.00795-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Qi Y, Wei ZQ, Ji SJ, Du XX, Shen P, Yu YS. ST11, the dominant clone of KPC-producing Klebsiella pneumoniae in China. J Antimicrob Chemother. 2011;66:307–312. doi: 10.1093/jac/dkq431 [DOI] [PubMed] [Google Scholar]
- 24.Yang QW, Jia XM, Zhou ML, et al. Emergence of ST11-K47 and ST11-K64 hypervirulent carbapenem-resistant Klebsiella pneumoniae in bacterial liver abscesses from China: a molecular, biological, and epidemiological study. Emerg Microbes Infect. 2020;9(1):320–331. doi: 10.1080/22221751.2020.1721334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Oteo J, Pérez-Vázquez M, Bautista V, et al. The spread of KPC-producing Enterobacteriaceae in Spain: WGS analysis of the emerging high-risk clones of Klebsiella pneumoniae ST11/KPC-2, ST101/KPC-2 and ST512/KPC-3. J Antimicrob Chemother. 2016;71(12):3392–3399. doi: 10.1093/jac/dkw321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Carre RA, Fortineau N, Nordmann P. Use of ChromID extended-spectrum β-lactamase medium for detecting carbapenemase-producing Enterobacteriaceae. J Clin Microbiol. 2010;48:1913–1914. doi: 10.1128/JCM.02277-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nordmann P, Naas T, Poirel L. Global spread of Carbapenemase producing Enterobacteriaceae. Emerg Infect Dis. 2011;17:1791–1798. doi: 10.3201/eid1710.110655 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Moore NM, Cantón R, Carretto E, Peterson LR, Sautter RL, Traczewski MM; Carba-R Study Team. Rapid identification of five classes of carbapenem resistance genes directly from rectal swabs by use of the xpert carba-R assay. J Clin Microbiol. 2017;55:2268–2275. doi: 10.1128/JCM.00137-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Citations
- European Committee on Antimicrobial Susceptibilities Testing (EUCAST) Breakpoint tables for interpretation of MICs and zone diameters. EUCAST 2014. Available from: http://www.eucast.org/fileadmin/src/media/PDFs/EUCAST_files/Breakpoint_tables/Breakpoint_table_v_4.0.pdf. Accessed September9, 2014.