Abstract
Mechanical cues regulate the function of a broad range of stem cells in culture and in tissue. For example, soft substrates promote the neuronal differentiation of neural stem cells (NSCs) by suppressing cytoskeletal contractility. However, the mechanisms that link cytoskeletal signaling to the transcriptional regulatory processes that ultimately govern stiffness-dependent NSC fate commitment are not fully understood. Here, we show that Angiomotin (AMOT), which can bind both F-actin and the neurosuppressive transcriptional coactivator Yes-associated protein (YAP), is critical for mechanotransduction in NSCs. On soft substrates, loss of AMOT substantially reduces neurogenesis, whereas on stiff substrates, loss of AMOT negates the rescue of neurogenesis normally induced by pharmacologic inhibition of myosin activity. Furthermore, overexpression of a phospho-mimetic S175E AMOT mutant, which has been established to enhance AMOT–YAP binding, increases β-catenin activity and rescues neurogenesis on stiff substrates. Together, our data identify AMOT as an important intermediate signal transducer that allows NSCs to sense and respond to extracellular stiffness cues.
INTRODUCTION
Neural stem cells (NSCs) have been implicated as key cellular effectors of adult neuroplasticity and have been explored as potential cell-replacement sources in therapies for neurodegenerative disease (Lledo et al., 2006; Yasuhara et al., 2006; Kim et al., 2013). In adult mammals, NSCs are confined to two specific regions of the central nervous system, the subventricular zone of the lateral ventricles, and the subgranular zone (SGZ) of the hippocampal dentate gyrus (Gage, 2000). NSCs within the SGZ are of particular interest as they play critical roles in learning, memory formation, behavioral and mood regulation, and disease pathology (Shors et al, 2001; Kempermann et al., 2004; Winner et al., 2008; David et al., 2009; Trouche et al., 2009; Boldrini et al., 2018). Accordingly, there has been an interest in precisely elucidating the extracellular signaling inputs and intracellular molecular pathways that regulate NSC behavior, with the goal of enhancing our basic understanding of NSC physiology and uncovering novel means to reliably control NSC function for regenerative medicine applications.
Within their endogenous neurogenic niches, NSCs receive an array of microenvironmental biochemical and biophysical cues that tightly control their behavior. A number of potent soluble and cell–cell biochemical signals that regulate NSCs have been identified, including: Wnt3, which promotes hippocampal neurogenesis; Notch, which helps maintain NSCs; and astrocyte-presented ephrins, which inhibit NSC growth and promote neurogenesis (Lie et al., 2005; Jiao et al., 2008; Imayoshi et al., 2010; Ashton et al., 2012; Yao et al., 2016). However, mechanical inputs such as substrate stiffness are being increasingly recognized as important regulators of NSC self-renewal and fate commitment as well. For example, we have previously reported that substrate stiffness strongly biases neuronal versus astrocytic differentiation (Saha et al., 2008). However, while it is clear that these inputs impact NSCs, the intracellular mechanisms that link stiffness inputs to gene regulation remain incompletely understood.
In past work, we showed that cellular contractile forces mediated by the Rho family GTPases RhoA and Cdc42 strongly modulate mechanosensitive fate commitment in hippocampal NSCs (Keung et al., 2011). We later demonstrated that stiff matrices increase levels of Yes-associated protein (YAP), which acts within a temporally restricted 12-to 36-h window to inhibit the proneurogenic transcription factor and Wnt pathway effector β-catenin (Rammensee et al., 2016). However, the key signaling events that link cytoskeletal mechanics to changes in the activity of transcriptional regulators such as YAP have remained unclear.
Angiomotin (AMOT) binds both actin and YAP and is thus a candidate protein to bridge this signaling gap (Ernkvist et al., 2006; Zhao et al., 2011; Mana-Capelli et al., 2014; Wang et al., 2015). An exciting recent study has shown that AMOT inhibits YAP during neural differentiation in an engineered H9 human ESC cell line, confirming the significance of this signaling axis during neurogenesis (Zaltsman et al., 2019). However, the importance of AMOT’s regulation of YAP is still unknown in the context of stiffness-sensitive adult neurogenesis, which is a related but distinct biological process.
Importantly, AMOT’s actin binding activity is known to be inhibited by large tumor suppressor kinase (LATS) phosphorylation, which could thereby regulate AMOT’s interactions with YAP (Chan et al., 2013; Dai et al., 2013). Indeed, recent studies have confirmed that phospho-mimetic or non–actin-binding mutants of AMOT display enhanced binding to and inhibition of YAP, demonstrating that the AMOT–YAP interaction could be mechanistically linked with the actin cytoskeleton and that AMOT phosphorylation is a potentially functionally important consequence of the Hippo pathway kinase cascade (Adler et al., 2013; Hirate et al., 2013; Mana-Capelli et al, 2014; Moleirinho et al., 2017). Interestingly, while most initial studies indicated that AMOT inhibits YAP, several recent reports have shown that AMOT can also promote YAP activity, particularly in the nucleus (Yi et al., 2013; Lv et al., 2016; Ragni et al., 2017; Liu et al., 2018). Further, AMOT is found at the plasma membrane in some cell types and can colocalize with Merlin, ZO1, and other components of adherens or tight junctions while also shuttling YAP to the membrane to inhibit its activity (Wells et al., 2006; Yi et al., 2011; Zhao et al, 2011). Therefore, while the AMOT–YAP interaction seems largely conserved, the nature and consequence of that interaction, the subcellular component or structure in which it occurs, and its regulation by stiffness cues or cell mechanics vary with tissue and cellular context. Intriguingly, AMOT and YAP were recently found to play critical roles in dendritic morphogenesis in cultured hippocampal neurons, motivating further study into how these proteins can affect other functions within the brain (Rojek et al., 2019).
Here, we have sought to explore whether AMOT is an important regulator of mechanosensitive neuronal differentiation of hippocampal NSCs by studying the nature of its interaction with YAP and the impact of its phosphorylation state and subcellular localization on NSC behavior. Overall, we find that AMOT promotes neurogenesis in a mechanosensitive manner. On soft substrates, increased AMOT phosphorylation promotes cytoplasmic AMOT localization, which is consistent with previous reports (Moleirinho et al., 2017). Overexpressed phospho-mimetic AMOT displays the same cytoplasmic localization and antagonizes YAP activation while promoting β-catenin activity and neurogenesis. On stiff substrates, AMOT preferentially localizes to the nucleus and is hypo-phosphorylated compared with soft substrates, allowing YAP to inhibit β-catenin and suppress neurogenesis. These studies advance our understanding of the intracellular events that govern mechanosensitive NSC fate commitment, a major obstacle toward precise and reliable usage of NSCs in tissue engineering and regenerative medicine applications.
RESULTS
Loss of AMOT reduces neurogenesis
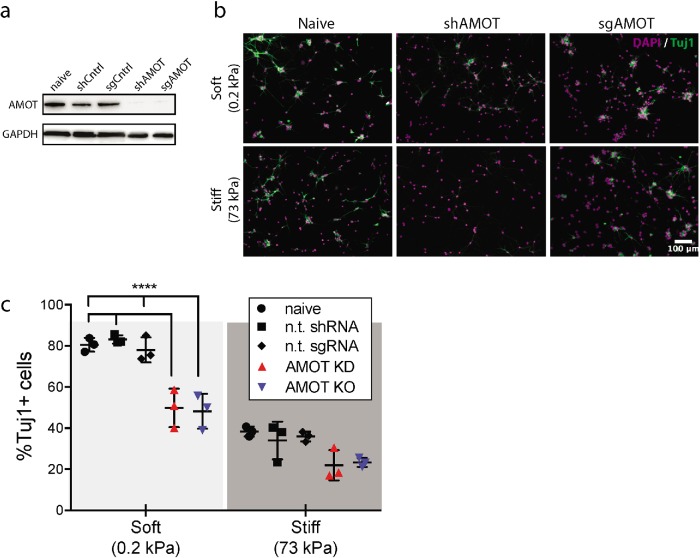
To investigate the potential importance of AMOT in stiffness-sensitive NSC differentiation, we eliminated functional expression of the AMOT gene with targeted short hairpin RNA (shRNA) or single guide RNA (sgRNA) + Cas9-mediated silencing (Figure 1a). Both approaches utilized lentiviral delivery of constructs encoding the requisite silencing machinery. Following transduction, cells were differentiated on either soft (0.2 kPa) or stiff (72 kPa) polyacrylamide gels under media conditions that support both neuronal and astrocytic differentiation (1 µM retinoic acid + 1% fetal bovine serum [FBS]) and immunostained for the neuronal marker Tuj1, which demonstrated that shAMOT and sgAMOT cells exhibited ∼40% reduced neurogenesis compared with their controls (Figure 1, b and c). In particular, loss of AMOT reduced neurogenesis on soft gels to levels similar to what we observed for naive and control cells on stiff substrates (Figure 1c). Importantly, differentiation on the two stiffnesses was not associated with changes in cell density, consistent with a mechanism in which stiffness cues and AMOT instruct fate commitment decisions rather than selecting for specific lineages (Supplemental Figure S1). These results implicate AMOT as an important effector of proper neurogenic differentiation of hippocampal NSCs and a potential mediator of robust neurogenesis under mechanical conditions that more closely mimic the mechanical properties of the in vivo niche.
FIGURE 1:
AMOT promotes neurogenesis on soft substrates. (a) Western blot showing protein depletion and gene KO of AMOT in NSCs using shRNA and CRISPR/Cas9. (b) Representative immunofluorescence images of naive, AMOT KD (shAMOT), and AMOT KO (sgAMOT) NSCs after culture in mixed differentiation conditions (1 µM retinoic acid + 1% FBS) on soft (0.2 kPa) or stiff (73 kPa) substrates. Cells were fixed and stained for DAPI (magenta) and Tuj1 (green), a neuronal marker. Bar = 100 µm. (c) Quantification of neurogenesis was measured by the percentage of Tuj1+ cells after 6 d of differentiation. Error bars represent SD (n = 3 gels). ****p < 0.0001 by two-way analysis of variance (ANOVA) followed by Tukey’s post-hoc test, n.s. = not significant (p > 0.05).
Phosphorylation of AMOT regulates its proneurogenic effect
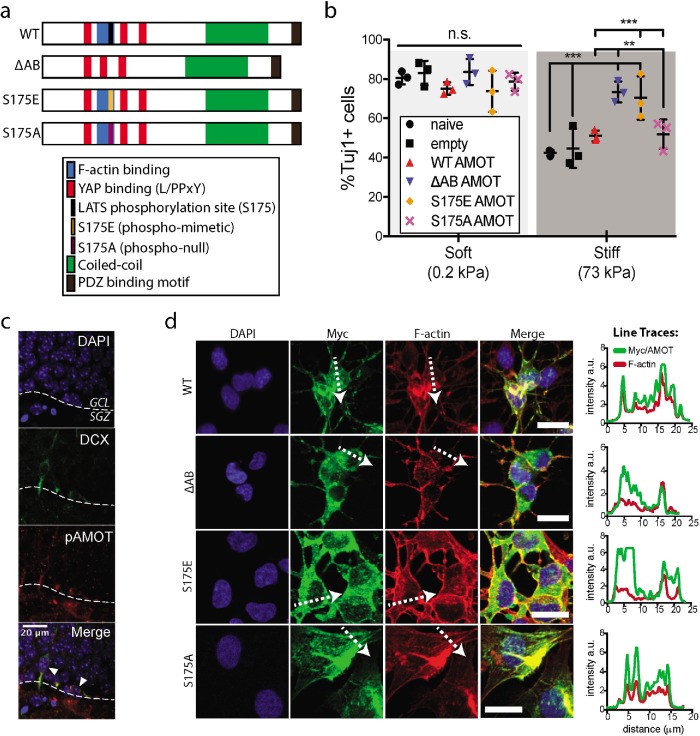
To further examine whether AMOT is a proneurogenic factor in NSCs, we next overexpressed AMOT variants and conducted differentiation experiments analogous to those with the shAMOT and sgAMOT cells. We hypothesized that F-actin binding may critically regulate AMOT function, as previous studies have described a competitive binding mechanism between AMOT binding to F-actin versus YAP (Mana-Capelli et al., 2014; Nakajima et al., 2017). Although the degree of F-actin polymerization and cytoskeletal dynamics in general could influence this balance, previous studies have shown that phosphorylation by LATS at Ser-175 (S175) within the F-actin binding region potently inhibits AMOT’s ability to bind F-actin and promotes the AMOT–YAP interaction (Adler et al., 2013). To examine the importance of the phosphorylation state and F-actin binding on AMOT’s impact on NSC behavior, we generated cell lines overexpressing phospho-null (S175A) or phospho-mimetic (S175E) AMOT, as well as another AMOT variant with a deletion of 10 AA necessary for functional F-actin binding (ΔAB) that includes S175 (Figure 2a). The cDNA were variants of AMOT’s full-length p130 isoform, which contains the functional F-actin binding domain and S175 residue, which are not present in the truncated p80 isoform.
FIGURE 2:
AMOT phosphorylation and actin-binding regulate its neurogenic effect. (a) Schematic drawings depicting protein sequence of AMOT variants that were cloned into retroviral plasmids before being packaged and used to generate stable AMOT overexpression NSC cell lines. (b) Quantification of neurogenesis as the percentage of Tuj1+ cells after 6 d of differentiation. Error bars represent SD (n = 3 gels). (c) Fluorescent immunohistochemical staining of a mouse hippocampal section. Tissue section was fixed and stained for DAPI (blue), DCX (green), an immature neuronal marker, and pAMOT (red). White arrowheads indicate DCX+/pAMOT+ cells. (d) Left: representative immunofluorescence images of WT, ΔAB, S175E, and S175A AMOT overexpression NSCs after 24 h of differentiation on stiff substrates. Myc antibody detects a C-terminal epitope tag only present on exogenous AMOT. Right: plotted intensity line traces from the Myc/AMOT and F-actin channels correlating to white dotted arrows shown on the left. Bars = 20 µm. **p < 0.005, ***p < 0.001 by two-way ANOVA followed by Tukey’s post-hoc test, n.s. = not significant (p > 0.05).
Strikingly, while overexpression of wild-type (WT) or S175A AMOT did not rescue neurogenesis on stiff substrates, ΔAB and S175E AMOT increased the fraction of Tuj1+ cells on stiff gels by more than 1.5-fold compared with Venus YFP controls (Figure 2b). By comparison, there was no significant difference between the various cell lines on soft substrates, indicating that the levels of neurogenesis seen in the controls apparently approach the maximum NSC neurogenic capacity in the conditions used. Importantly, the engineered ablation (Figure 1) or overexpression of various genetic variants of AMOT (Figure 2, a and b) did not impact the proliferative capacity of NSCs as measured by EdU incorporation assays (Supplemental Figure S2), supporting the idea that these perturbations instruct neurogenic fate commitment rather than select for or against committed progeny. This result is consistent with our past finding that stiffness is an instructive differentiation cue that does not significantly impact NSC proliferation or apoptosis (Keung et al., 2011). We sought to further explore the importance of AMOT phosphorylation in in vivo neurogenesis by examining immature DCX+ neurons within the hippocampus. Strikingly, we observed instances of clear pAMOT and DCX colocalization within early committed neurons of the granular cell layer (Figure 2c). Together, these findings suggest that AMOT phosphorylation promotes neurogenesis in hippocampal NSCs.
AMOT phosphorylation regulates its subcellular localization
Consistent with AMOT’s reported F-actin binding capabilities and the N-terminal region functionally associated with that interaction, exogenous S175A and WT AMOT displayed colocalization with actin-based structures. Conversely, ΔAB and S175E AMOT were not found associated with F-actin, validating both AMOT’s F-actin binding capacity and the effect of S175 phosphorylation on AMOT-F-actin binding in NSCs (Figure 2d).
Interestingly, further examination of the localization of the overexpressed AMOT variants revealed that ΔAB and S175E AMOT were significantly enriched in the cytoplasm, consistent with a recent report showing that phosphorylated AMOT is preferentially localized to cytoplasm and plasma membrane, whereas unphosphorylated AMOT localizes to the nucleus (Moleirinho et al., 2017). Together, these results strongly indicate that AMOT is not only proneurogenic in NSCs but also that this effect is significantly regulated by its S175 phosphorylation state and localization. Since LATS itself has been shown to be negatively regulated by actin assembly (Wada et al., 2011; Yu et al., 2012b; Zhao et al., 2012), these results are consistent with a signaling axis in which increased cytoskeletal signaling in response to biophysical inputs have an inhibitory effect on AMOT’s activity by inhibiting LATS and increasing unphosphorylated AMOT.
Substrate stiffness influences endogenous AMOT protein phosphorylation and localization
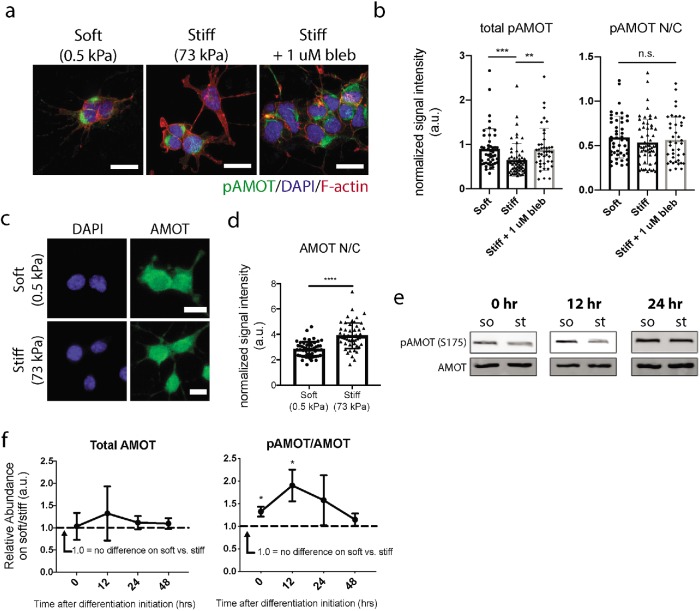
To examine whether stiffness could influence the localization of endogenous pAMOT in a manner consistent with observations of overexpressed S175E AMOT, we immunostained pAMOT in NSCs that had been differentiated on soft and stiff substrates. We also included a condition with cells cultured on stiff substrates in the presence of 1 µM blebbistatin to test whether stiffness-dependent cytoskeletal tension is an essential regulator of pAMOT localization. Importantly, the cells were fixed after 24 h of differentiation, which is within the 12- to 36-h time frame in which NSC fate commitment is sensitive to extracellular stiffness input that we have previously described (Rammensee et al., 2016). We observed that endogenous pAMOT is clearly localized within the cytoplasm in differentiating NSCs, and that inhibition of myosin contractility with blebbistatin apparently increased cytoplasmic pAMOT on stiff substrates (Figure 3a). Further quantification revealed that pAMOT is preferentially localized to the cytoplasm relative to the nucleus, as indicated by an N/C ratio <1 (Figure 3b). Interestingly, this localization pattern was consistent throughout all three conditions, but the cells differentiated on soft substrates or treated with blebbistatin on stiff substrates displayed higher overall pAMOT levels than cells differentiated on stiff substrates (Figure 3b).
FIGURE 3:
Stiffness regulates AMOT phosphorylation and localization. (a) Representative immunofluorescence images of DAPI (blue), F-actin (red), and endogenous pAMOT (green) in NSCs after 24 h of differentiation on soft, stiff, or stiff + 1 µM blebbistatin culture conditions. (b) Quantification of total pAMOT intensity and nuclear/cytoplasmic localization ratio after 24 h of differentiation in various conditions. **p < 0.005, ***p < 0.001 by one-way ANOVA followed by Tukey’s post-hoc test, n.s. = not significant (p > 0.05). (c) Representative immunofluorescence images of AMOT (green) in NSCs after 24 h of differentiation on soft (0.5 kPa) or stiff (73 kPa) substrates (blue = DAPI). (d) Quantification of AMOT nuclear/cytoplasmic localization measured after 24 h of differentiation. Error bars represent SD (n = 54 cells for soft, n = 53 cells for stiff). ****p < 0.0001 by unpaired t test. (e) Representative Western blots and quantification of pAMOT/AMOT protein levels in cells differentiated for 0, 12, and 24 h of differentiation on soft vs. stiff substrates. (f) pAMOT and AMOT Western blot band intensities were normalized to GAPDH and β-actin, and the pAMOT/AMOT ratio of the normalized values on soft vs. stiff was calculated for each trial (n = 3 gels per timepoint). *p < 0.05 by one-sample t test against hypothetical value of 1.0.
Because we found that AMOT’s phosphorylation state is a key regulator of its impact on neurogenesis (Figure 2b) and that pAMOT showed preferential localization to the cytoplasm, we next tested whether we could detect differences in total AMOT localization on soft versus stiff substrates. Cells on soft substrates displayed increased cytoplasmic AMOT compared with cells on stiff substrates (Figure 3, c and d). This supports a mechanism where soft substrates promote AMOT phosphorylation, which correlates with increased overall AMOT cytoplasmic localization.
To further verify the importance of AMOT phosphorylation in mechanosensitive fate commitment, we conducted Western blots to measure pAMOT within the critical early time window. We observed that while total AMOT levels did not vary with stiffness, the fraction of pAMOT was higher at 12 and 36 h of differentiation on soft substrates (Figure 3, e and f). Together, these data show that AMOT phosphorylation and localization are significantly altered during NSC fate commitment on various stiffnesses and may be a critical mechanism of mechanosensitive neurogenesis.
AMOT is regulated by Rho GTPase signaling
Rho GTPases have been reported to mediate many cellular processes including cytoskeletal remodeling, proliferation, migration, and differentiation through their modulation of the actin cytoskeleton (Takano et al., 1998; Yang et al., 2007; Leone et al., 2010; Keung et al., 2011; Zegers and Friedl, 2014; Hoon et al., 2016). In NSCs, we have shown that the Rho GTPases RhoA and Cdc42 are activated in response to increased substrate stiffness, and that constitutive activation of these proteins reduces neurogenesis in vitro and in vivo (Keung et al., 2011). Therefore, we tested whether AMOT is regulated by Rho GTPase signaling and potentially responsible for the influence of Rho GTPase activity on stiffness-sensitive NSC fate commitment.
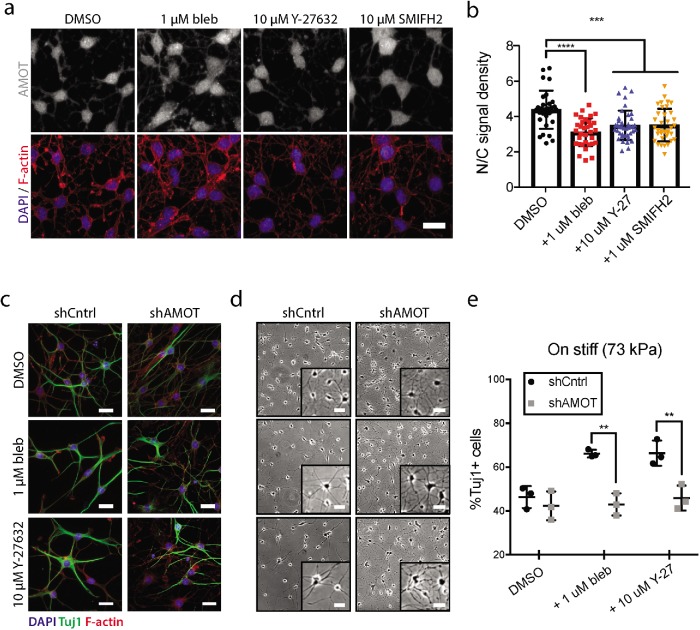
To investigate functional contributions of Rho GTPase signaling, we used pharmacological inhibitors for ROCK (Y-27632), formins (SMIFH2), and myosin II (blebbistatin), which all lie downstream of Rho activation. As shown above, NSCs undergoing differentiation on stiff substrates display enrichment of AMOT in the nucleus (Figure 4, a and b). However, treatment with any of the inhibitors for the first 24 h of differentiation reduced the degree of nuclear AMOT localization and resulted in a subcellular distribution similar to that of cells cultured on soft substrates (Figures 4b and 3c). Therefore, stiffness-induced myosin contractility mediated by Rho GTPase influences AMOT subcellular localization during NSC fate commitment.
FIGURE 4:
Rho GTPase pathway influences stiffness-sensitive neurogenesis by regulating AMOT. (a) Representative immunofluorescence images of AMOT (gray), DAPI (blue), and F-actin (red) differentiated for 24 h on stiff (73 kPa) substrates in the presence of inhibitors for myosin II (blebbistatin), ROCK (Y-27632), formins (SMIFH2), or DMSO control. (b) Quantification of AMOT nuclear/cytoplasmic localization measured from immunofluorescence images taken after 24 h of differentiation in the presence of inhibitors or DMSO. Error bars represent SD (n = 30 for DMSO, n = 33 for bleb, n = 40 for Y-27632, n = 36 for SMIFH2). ***p < 0.001, ****p < 0.0001 by one-way ANOVA followed by Tukey’s post-hoc test. (c) Representative immunofluorescence images of nontargeting control cells (shCntrol) and AMOT KD (shAMOT) cells stained after 6 d of differentiation on stiff substrates in the presence of bleb, Y-27632, or DMSO. Cells were fixed and stained for DAPI (blue), Tuj1 (green), and F-actin (red). (d) Representative bright-field images of shCntrl and shAMOT cells. (e) Quantification of neurogenesis was measured by the percentage of Tuj1+ cells after 6 d of differentiation on stiff substrates. Error bars represent SD (n = 3 gels). Bars = 20 µm. **p < 0.005 by two-way ANOVA followed by Tukey’s post-hoc test, n.s. = not significant (p > 0.05).
Inhibition of ROCK, formins, and myosin are all expected to convert cells into low-tension states that are reminiscent of cells cultured on soft substrates, and application of these inhibitors often phenocopies effects of soft or confined substrates on stem cell differentiation (McBeath et al., 2004; Engler et al., 2006). Accordingly, we have previously shown that treatment with blebbistatin and Y-27632 rescues neurogenesis on stiff substrates (Keung et al., 2011). To test whether AMOT was responsible for this effect, we differentiated AMOT KD (shAMOT) or nontargeting scramble cells on stiff substrates for 6 d before fixing and staining for neuronal markers (Figure 4, c and e). Indeed, we found that while blebbistatin and Y-27632 treatment both rescued neurogenesis in scramble control cells, shAMOT cells were unresponsive to inhibitor treatment (Figure 4e). In addition, consistent with previous reports, we observed that blebbistatin and Y-27632 treatment influenced cellular morphology in scramble cells, which displayed increased neurite length and decreased neurite branching when compared with dimethyl sulfoxide (DMSO) treatment controls (Yu et al., 2012a; Chen et al., 2013; Figure 4d). Interestingly, AMOT KD desensitized neurite length and branching to these inhibitors (Figure 4d), suggesting that AMOT may not only be influenced by cytoskeletal dynamics but may also be an active participant in Rho/ROCK-dependent cytoskeletal remodeling.
AMOT regulates neurogenesis through YAP and β-catenin
Multiple reports have shown that YAP is a crucial molecular rheostat within cells and can direct stem cell self-renewal and differentiation in a stiffness-sensitive manner (Lian et al., 2010; Dupont et al., 2011, 2016). We have previously shown that while YAP is increased in differentiating NSCs on stiff substrates, its neurosuppressive effect is mediated by its interaction with β-catenin rather than its canonical downstream effectors, the TEAD transcription factors (Rammensee et al., 2016). Previous studies in other cell systems have indicated that AMOT can inhibit YAP via direct binding, and that this inhibition is enhanced when AMOT is phosphorylated (Adler et al., 2013; Mana-Capelli et al., 2014). However, no studies to date have experimentally investigated whether the AMOT–YAP interaction is important in mechanotransductive contexts, though it has been hypothesized (Low et al., 2014). Therefore, we examined whether AMOT’s proneurogenic effect is regulated by substrate stiffness through its phosphorylation and ability to impact YAP and/or β-catenin activity.
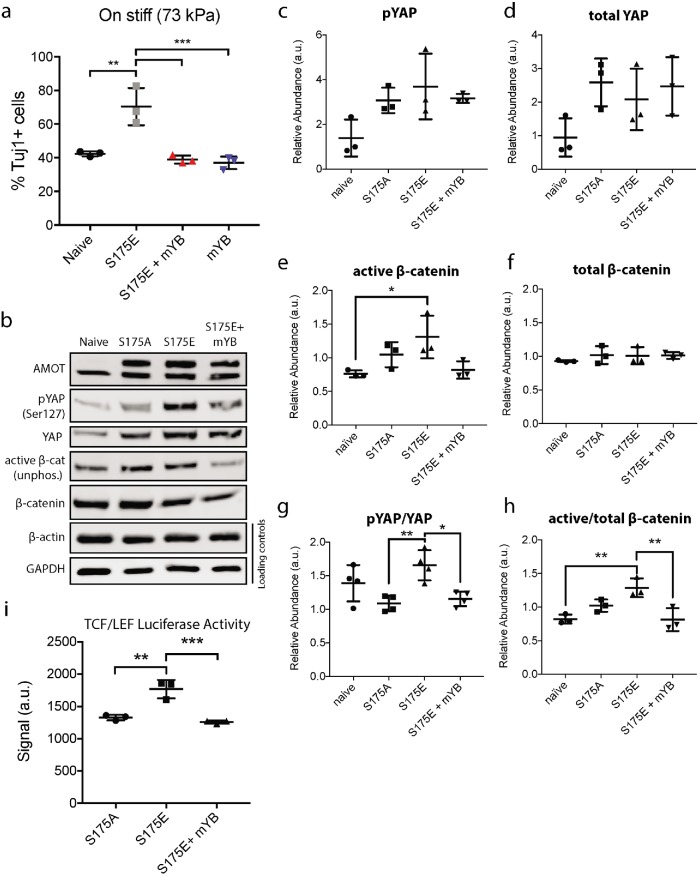
AMOT is known to bind to YAP’s WW domains through three L/PPxY motifs near AMOT’s N-terminus. Therefore, we generated an AMOT variant that has the phospho-mimetic S175E mutation as well as mutations at its three L/PPxY motifs to determine whether an interaction with YAP is essential for S175E AMOT’s proneurogenic function (S175E + mYB). Strikingly, unlike the S175E mutant, overexpressed S175E + mYB AMOT completely lacked the capacity to rescue neurogenesis on a stiff substrate (Figure 5a). While this result indicated that S175E AMOT must bind YAP to promote neurogenesis, we sought to further test whether YAP binding capacity impacts downstream YAP and β-catenin activity. Importantly, we found that S175E cells displayed increased YAP phosphorylation, a marker for YAP inactivation, and active (phosphorylated) β-catenin by Western blotting compared with S175E + mYB cells (Figure 5, b–h). This is consistent with a recent study that described how AMOT can directly contribute to Hippo-mediated phosphorylation of YAP by acting as a scaffold (Mana-Capelli and McCollum, 2018). To further test whether AMOT functionally impacts β-catenin-mediated transcription, we generated cells carrying a 7xTFP luciferase reporter for β-catenin/TCF/LEF transcriptional activity and overexpressed the various AMOT mutant variants. After 24 h of differentiation on stiff substrates, we collected and analyzed lysates for β-catenin/TCF/LEF-driven luciferase expression. β-Catenin/TCF activity was higher in S175E cells than in S175A or S175E + mYB cells (Figure 5i). Taken together, these experiments indicate that AMOT phosphorylation promotes neurogenesis by inhibiting YAP and promoting β-catenin activity (Figure 6).
FIGURE 5:
Phosphorylated AMOT promotes neurogenesis by inhibiting YAP and promoting β-catenin activity. (a) Quantification of neurogenesis from NSCs overexpressing phospho-mimetic (S175E), YAP nonbinding (mYB), or S175E + mYB AMOT on stiff substrates (73 kPa). Error bars represent SD (n = 3 gels). (b) Representative Western blots of lysates collected from phosho-null (S175A), S175E, and S175E + mYB cells after 24 h of differentiation on stiff substrates to confirm AMOT overexpression and measure pYAP, total YAP, active β-catenin, total β-catenin, and loading controls (GAPDH and β-actin). (c–h) Quantification of Western blot bands normalized to both GAPDH and β-actin (n = 3). (i) Quantification of luminescence signal from cells overexpressing S175A, S175E, or S175E + mYB AMOT in addition to a 7xTFP TCF/LEF luciferase reporter construct. Cells were differentiated for 24 h on stiff substrates before lysate collection. (n = 3). *p < 0.05, **p < 0.005, ***p < 0.001 by one-way ANOVA followed by Tukey’s post-hoc test.
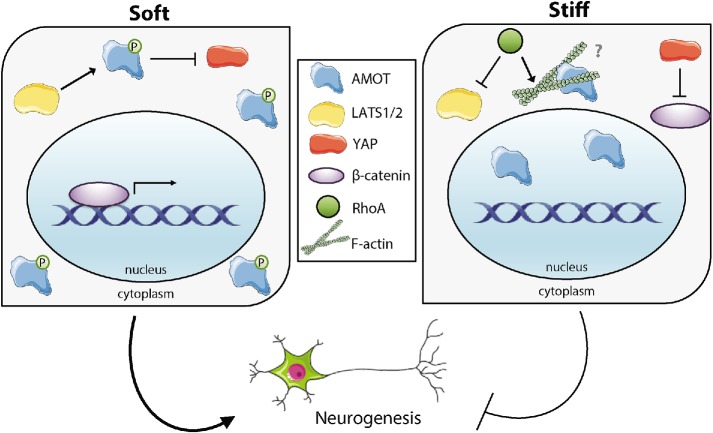
FIGURE 6:
AMOT coordinates stiffness-sensitive neurogenesis through its impact on YAP, which is regulated by LATS phosphorylation.
DISCUSSION
Recently, it has become clear that extracellular stiffness and other biophysical cues are key inputs for stem cell regulation, but the intracellular mechanisms that transduce these signals remain incompletely elucidated. Our recent findings that YAP influences mechanosensitive differentiation in NSCs through a noncanonical interaction with β-catenin (Rammensee et al., 2016) motivated us to more closely examine mechanisms regulating YAP. To address these questions, we investigated AMOT, which has been shown to link F-actin and YAP in prior studies that did not study stem cells or cellular mechanics (Adler et al., 2013; Mana-Capelli et al., 2014). Specifically, we interrogated the importance of AMOT in stiffness-sensitive neurogenesis, the effect of AMOT phosphorylation and direct AMOT–YAP binding, and how Rho-mediated signaling influences AMOT activity. The results presented here describe a critical role for AMOT, whose phosphorylation leads to interaction with and inhibition of YAP in the cytoplasm and consequently enables β-catenin-mediated neurogenesis. Furthermore, Rho pathway activation suppresses neurogenesis on stiff substrates by regulating AMOT and influencing its localization.
Previous studies have shown that stem cells can display “mechanical memory” where earlier presentation of mechanical cues can influence long-term cell behavior (Keung et al., 2011; Yang et al., 2014; Rammensee et al., 2016; Peng et al., 2017). Further exploring this concept, we discovered that NSCs are sensitive to stiffness inputs only within an early, relatively narrow time window of differentiation in culture (12–36 h). Therefore, intracellular signaling events that ultimately influence fate commitment should also occur within a similar time frame. For example, YAP levels in NSCs differentiating on stiff substrates are higher compared with NSCs on soft substrates, and this difference is maximized between 24 and 48 h (Rammensee et al., 2016). Strikingly consistent with this timing, we found that AMOT phosphorylation, but not total AMOT, was most increased 12 and 36 h after differentiation initiation on soft substrates versus stiff substrates. This is distinct from signaling in pluripotent ESCs, where total AMOT was up-regulated during neurogenesis to inhibit YAP (Zaltsman et al., 2019). Therefore, the approaches we employed in this work and its findings further delineate the intricate mechanisms by which stiffness input can direct NSC fate commitment through a Rho-AMOT-YAP-β-catenin signaling axis, providing crucial insights into the continuous cascade of mechanotransductive events within adult NCSs.
Most studies to date have reported that AMOT inhibits YAP through direct binding, which apparently sequesters YAP in the cytoplasm to prevent YAP’s transcriptional impact in the nucleus. However, one study has reported that in hepatic epithelial cells, AMOT promotes YAP nuclear translocation by sterically blocking its interaction with LATS in the cytoplasm and then forms a complex with YAP-TEAD within the nucleus to coregulate target gene transcription (Yi et al., 2013). These seemingly conflicting reports highlight that while AMOT clearly interacts with YAP, the nature of this interaction is particularly important and highly context dependent. Through the use of phospho-mimetic and YAP nonbinding mutants of AMOT, we found that in NSCs, AMOT inhibits YAP in the cytoplasm by promoting YAP phosphorylation and that this interaction is enhanced by AMOT’s own phosphorylation. The importance of AMOT phosphorylation on its binding with YAP is consistent with previous reports (Adler et al., 2013), and our findings are consistent with a model where AMOT may act as a scaffolding protein to promote YAP phosphorylation by LATS while AMOT is itself phosphorylated. Overall, the role of the canonical Hippo pathway in stiffness-sensitive YAP regulation remains unclear. In our study, likely AMOT phosphorylation by LATS at S175 enhanced AMOT’s inhibition of YAP and promotion of neurogenesis, suggesting that AMOT’s effects are Hippo dependent. Interestingly, a recent study (Meng et al., 2018) showed that the Ras family GTPase RAP2 inhibits YAP/TAZ in a stiffness-sensitive manner by promoting LATS activity, which could potentially be upstream of AMOT in NSCs.
In addition, we observed that a significant fraction of endogenous AMOT is localized in the nucleus and that this fraction is enriched even further on stiff substrates. Given previous reports that AMOT can promote YAP activity in the nucleus (Yi et al., 2013; Lv et al., 2016; Ragni et al., 2017; Liu et al., 2018), this raises the interesting possibility that AMOT has opposite effects on YAP and, ultimately, neurogenesis, in the cytoplasm versus the nucleus in NSCs and that AMOT phosphorylation is a master regulator of this process. Phosphorylation also regulates AMOT’s F-actin binding, which may influence AMOT’s ability to bind YAP. However, it is yet unclear whether AMOT endogenously binds to F-actin in NSCs or whether this interaction is promoted by increased F-actin polymerization, which would be consistent with increased YAP activity and decreased neurogenesis on stiff substrates. When we overexpressed WT or S175A AMOT, we observed thick F-actin fibers that colocalized with AMOT (Figure 2c), which is likely due to AMOT’s known actin-bundling activity. These actin bundles were not observed in naive or S175E AMOT overexpressing NSCs. Therefore, while AMOT phosphorylation is influenced by stiffness and promotes neurogenesis by inhibiting YAP, the importance of AMOT binding to F-actin is still unclear and is open for further investigation in NSC fate commitment.
NSCs and other stem cells have strong potential for tissue engineering and the development of novel cell-based treatments for injury and disease. One of the major obstacles that these exciting applications face is insufficient understanding of the mechanisms that regulate cell behavior, and the capacity to control NSC behavior by modulating how cells sense and respond to biophysical cues thus offers additional targets for therapeutic intervention. Toward addressing this need, our study identifies AMOT as a key molecular messenger and sheds light on how pathways controlling cytoskeletal remodeling directly impact transcriptional events that ultimately govern stem cell self-renewal and differentiation in vitro. In future studies, it will be valuable to directly manipulate AMOT expression and phosphorylation in vivo and determine their impact on endogenous neurogenesis. With increased understanding of the components of mechanotransductive pathways and the nature of their interactions, we could further delineate how stem cells respond to proper and aberrant niche signals in vivo as well as more effectively engineer microenvironments to reliably control stem cell behavior in novel tissue engineering approaches.
MATERIALS AND METHODS
Constructs and antibodies
shAMOT, shCntrl, sgAMOT, and sgAMOT oligonucleotide inserts were obtained from Elim Biopharmaceuticals. shRNA inserts were designed with AgeI- and EcoRI-based overhangs and cloned into the pLKO.1 vector, a gift from David Root (Broad Institute; Addgene plasmid #10878; Moffat et al., 2006). sgRNA inserts were designed with vector-specific BsmBI-based overhangs and cloned into the lentiCRISPR v2 vector, a gift from Feng Zhang (Broad Institute; Addgene plasmid # 52961; Sanjana et al., 2014). pcDNA4 plasmids with cDNA encoding WT, ΔAB, S175E, and mYB AMOT were a generous gift from D. McCollum (University of Massachusetts Medical School, Worcester, MA). The AMOT cDNA sequences were PCR-amplified, digested with SfiI and PmeI, and cloned into the pCLPIT vector. S175A and S175E + mYB AMOT cDNA sequences were generated using the QuikChange Site-Directed Mutagenesis system (Agilent Technologies) before also being digested and cloned into the pCLPIT vector.
Primary antibodies and dilutions used were as follows: Tuj1 (1:750; BioLegend 801201), AMOT (1:1000 for Western blotting, 1:250 for immunocytochemistry; Bethyl Laboratories A303-303A), c-Myc (1:250; Santa Cruz Biotechnology A-14), pAMOT (1:1000 for Western blotting, 1:250 for immunocytochemistry; EMD Millipore ABS1045), YAP (1:1000; Cell Signaling Technologies 4912S), pYAP (1:1000; Cell Signaling Technologies 4911S), β-catenin (1:1000; Cell Signaling Technologies 2698S), active β-catenin (1:1000; Cell Signaling Technologies 8814S), β-actin (1:100,000; Sigma-Aldrich A1978), and GAPDH (1:1000; Sigma-Aldrich G8795).
siRNA and sgRNA sequences
siRNA targeting rat amot mRNA (gagaaagccatgaggaaca) and a scramble control (gatgcatgttgatagacgtaa) were designed using the online tool Dharmacon siDesign. sgRNA targeting the rat amot genomic locus (gatggatgctacgagaagg) and a scramble control (gcactaccagagctaactca) were designed using the online tool E-CRISP (Heigwer et al., 2014).
Viral packaging and transduction
CLPIT vectors encoding the various human AMOT cDNA were packaged as retroviruses via calcium phosphate–based transfection into HEK 293T cells (10 µg transfer vector + 6 µg pCMV gag-pol + 4 µg pcDNA3 IVS VSV-G per 10-cm-diameter plate). Supernatants were collected 48 and 72 h posttransfection and pooled before filtration and ultracentrifugation to purify the viral particles. Vectors encoding shRNA or sgRNA were similarly packaged into lentiviruses (10 µg transfer vector + 7.5 µg psPAX2 + 2.5 µg pMD2.G per 10-cm-diameter plate) before viral purification. Cells were transduced in all cases at an MOI of 1–2 as calculated by puromycin resistance titer and selected with 0.6 µg/ml puromycin for at least 4 d (Peltier and Schaffer, 2010).
Polyacrylamide gel synthesis and functionalization
Polyacrylamide-bis precursor solutions were made for each stiffness by mixing various concentrations of acrylamide monomer and bis-acrylamide cross-linker (Bio-Rad). Solution compositions to achieve various final polymerized stiffnesses were as follows: 0.2 kPa = 3% acrylamide + 0.04% bis, 0.5 kPa = 3% acrylamide + 0.1% bis, 72 kPa = 10% acrylamide + 0.3% bis (Keung et al., 2011). Polyacrylamide gels were synthesized on 19- or 25-mm glass coverslips with 0.1% TEMED + 1% ammonium persulfate. Polyacrylamide gels were functionalized with laminin conjugation via sulfo-SANPAH (Thermo-Fisher).
Cell culture and differentiation
Adult rat hippocampal NSCs were isolated from adult female Fischer 344 rats (Charles River, Wilmington, MA) as described in Palmer et al. (1999) and were cultured in DMEM/F12 with N2 supplement (Life Technologies) on tissue-culture polystyrene plates that had been coated with poly-ornithine and laminin. Growth conditions for NSCs included 20 ng/ml FGF-2, whereas mixed differentiation conditions included 1% FBS + 1 µM retinoic acid. For full differentiation experiments on polyacrylamide substrates, NSCs were first seeded onto the gels in growth media for 16–18 h before the coverslips were then transferred into new wells with mixed differentiation media. The NSCs were allowed to differentiate for 6 d with 50% media changes every 2 d before fixation.
Cell fixation and immunocytochemistry
Cells were fixed in 4% paraformaldehyde (Alfa Aesar) for 10 min before washing in phosphate-buffered saline (PBS) and permeabilization in 5% goat serum + 0.5% Triton X-100 (Sigma-Aldrich) for 10 min. Permeabilized cells were blocked in 5% goat serum for 1 h at room temperature before immunostaining. Primary and secondary antibody solutions were also made in 5% goat serum.
Brightfield and immunofluorescence image acquisition
Brightfield and epifluorescence images were taken using a Nikon Eclipse Ti Microscope, Hamamatsu Photonics K.K. C10600-10B-H camera, 10× objective lens, and native NIS-Elements AR 5.02.00 software. Samples were submerged in PBS during image acquisition. Nuclei were labeled with a 4′,6-diamidino-2-phenylindole (DAPI) stain (Sigma-Aldrich) and GFAP or Tuj1 were labeled with either a 488 or a 633 dye-conjugated secondary antibody (Thermo-Fisher). Confocal images were taken using a Prairie Technologies 2-photon and confocal microscope, QuantEM 512SC camera, 60X objective lens, and native Prairie View software. Samples were submerged in PBS during image acquisition. Nuclei were labeled with a DAPI stain, F-actin was labeled with an Alexa Fluor 546 Phalloidin (Thermo-Fisher), and other targets were labeled with either a 488 or 633 dye–conjugated secondary antibody. All image processing and analysis was carried out in the free ImageJ software.
Nuclear to cytoplasmic ratio quantification
To detect N/C staining, z-stack images of NSCs were acquired in 1-µm increments using the Prairie Technologies confocal microscope described above. Analysis was conducted in ImageJ, where the z-stack images were projected into a 2D image using the sum of intensities method. Cell bodies were traced manually using the phalloidin channel and the intensity of the channel of interest for each cell was measured. The DAPI channel was used to detect the nuclei and ROIs were created for each individual nucleus. These nuclear ROIs were then used to measure the intensity of the channel of interest within each nucleus. The nuclear intensities were subtracted from the whole-cell intensity of each cell before the nuclear/cytoplasmic value was calculated for each cell.
Luciferase assay
AMOT S175A, S175E, and S175E + mYB cells expressing ectopic AMOT to similar levels were further transduced with a lentiviral construct encoding a 7xTFP TFC/LEF luciferase reporter, which is responsive to β-catenin-TCF/LEF–based transcription. Lysates were collected after 24 h of differentiation in mixed conditions on stiff (73 kPa) substrates using the provided lysis buffer from the Luciferase Assay System kit (Promega). The Luciferase Assay was carried out according to the manufacturer’s protocol in a 96-well plate format and measurements were taken with a plate-reading luminometer.
Supplementary Material
Acknowledgments
We kindly thank Dannel McCollum (University of Massachusetts Medical School, Worcester, MA) for sharing his lab’s pCDNA constructs encoding WT, S175E, S175A, and mYB AMOT. We also thank Kira Mosher (University of California, Berkeley, Berkeley, CA) for sharing her immunohistochemical staining expertise and performing some of the staining and imaging, as well as Qianhe Zhang (University of California, Berkeley, Berkeley, CA) for her experimental assistance. This project was supported by the National Institutes of Health (R01NS074831 to S.K. and D.V.S.) and a Siebel Scholarship to P.H.K.
Abbreviations used:
- AMOT
Angiomotin
- FBS
fetal bovine serum
- LATS
large tumor suppressor kinase
- NCS
neural stem cell
- PBS
phosphate-buffered saline
- sgRNA
single guide RNA
- SGZ
subgranular zone
- shRNA
short hairpin RNA
- WT
wild type
- YAP
Yes-associated protein.
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E19-11-0602) on January 15, 2020.
REFERENCES
- Adler JJ, Johnson DE, Heller BL, Bringman LR, Ranahan WP, Conwell MD, Sun Y, Hudmon A, Wells CD. (2013). Serum deprivation inhibits the transcriptional co-activator YAP and cell growth via phosphorylation of the 130-kDa isoform of Angiomotin by the LATS1/2 protein kinases. Proc Natl Acad Sci USA , 17368–17373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashton RS, Conway A, Pangarkar C, Bergen J, Lim KIl, Shah P, Bissell M, Schaffer DV. (2012). Astrocytes regulate adult hippocampal neurogenesis through ephrin-B signaling. Nat Neurosci , 1399–1406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boldrini M, Fulmore CA, Tartt AN, Simeon LR, Pavlova I, Poposka V, Rosoklija GB, Stankov A, Arango V, Dwork AJ, et al (2018). Human hippocampal neurogenesis persists throughout aging. Cell Stem Cell , 589–599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan SW, Lim CJ, Guo F, Tan I, Leung T, Hong W. (2013). Actin-binding and cell proliferation activities of angiomotin family members are regulated by Hippo pathway-mediated phosphorylation. J Biol Chem , 37296–37307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen K, Zhang W, Chen J, Li S, Guo G. (2013). Rho-associated protein kinase modulates neurite extension by regulating microtubule remodeling and vinculin distribution. Neural Regen Res , 3027–3035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai X, She P, Chi F, Feng Y, Liu H, Jin D, Zhao Y, Guo X, Jiang D, Guan KL, et al (2013). Phosphorylation of angiomotin by Lats1/2 kinases inhibits F-actin binding, cell migration, and angiogenesis. J Biol Chem , 34041–34051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- David DJ, Samuels BA, Rainer Q, Wang JW, Marsteller D, Mendez I, Drew M, Craig DA, Guiard BP, Guilloux JP, et al (2009). Neurogenesis-dependent and -independent effects of fluoxetine in an animal model of anxiety/depression. Neuron , 12–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dupont S. (2016). Role of YAP/TAZ in cell-matrix adhesion-mediated signalling and mechanotransduction. Exp Cell Res , 42–53. [DOI] [PubMed] [Google Scholar]
- Dupont S, Morsut L, Aragona M, Enzo E, Giulitti S, Cordenonsi M, Zanconato F, Le Digabel J, Forcato M, Bicciato S, et al (2011). Role of YAP/TAZ in mechanotransduction. Nature , 179–184. [DOI] [PubMed] [Google Scholar]
- Engler AJ, Sen S, Sweeney HL, Discher DE. (2006). Matrix elasticity directs stem cell lineage specification. Cell , 677–689. [DOI] [PubMed] [Google Scholar]
- Ernkvist M, Aase K, Ukomadu C, Wohlschlegel J, Blackman R, Veitonmäki N, Bratt A, Dutta A, Holmgren L. (2006). p130-Angiomotin associates to actin and controls endothelial cell shape. FEBS J , 2000–2011. [DOI] [PubMed] [Google Scholar]
- Gage FH. (2000). Mammalian neural stem cells. Science , 1433–1438. [DOI] [PubMed] [Google Scholar]
- Heigwer F, Kerr G, Boutros M. (2014). E-CRISP: Fast CRISPR target site identification. Nat Methods , 122–123. [DOI] [PubMed] [Google Scholar]
- Hirate Y, Hirahara S, Inoue KI, Suzuki A, Alarcon VB, Akimoto K, Hirai T, Hara T, Adachi M, Chida K, et al (2013). Polarity-dependent distribution of angiomotin localizes hippo signaling in preimplantation embryos. Curr Biol , 1181–1194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoon JL, Tan MH, Koh C-G. (2016). The regulation of cellular responses to mechanical cues by Rho GTPases. Cells , 17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imayoshi I, Sakamoto M, Yamaguchi M, Mori K, Kageyama R. (2010). Essential roles of notch signaling in maintenance of neural stem cells in developing and adult brains. J Neurosci , 3489–3498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiao J-w, Feldheim DA, Chen DF. (2008). Ephrins as negative regulators of adult neurogenesis in diverse regions of the central nervous system. Proc Natl Acad Sci USA , 8778–8783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kempermann G, Wiskott L, Gage FH. (2004). Functional significance of adult neurogenesis. Curr Opin Neurobiol , 186–191. [DOI] [PubMed] [Google Scholar]
- Keung AJ, De Juan-Pardo EM, Schaffer DV, Kumar S. (2011). Rho GTPases mediate the mechanosensitive lineage commitment of neural stem cells. Stem Cells , 1886–1897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim SU, Lee HJ, Kim YB. (2013). Neural stem cell-based treatment for neurodegenerative diseases. Neuropathology , 491–504. [DOI] [PubMed] [Google Scholar]
- Leone DP, Srinivasan K, Brakebusch C, McConnell SK. (2010). The Rho GTPase Rac1 is required for proliferation and survival of progenitors in the developing forebrain. Dev Neurobiol , 659–678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lian I, Kim J, Okazawa H, Zhao J, Zhao B, Yu J, Chinnaiyan A, Israel MA, Goldstein LSB, Abujarour R, et al (2010). The role of YAP transcription coactivator in regulating stem cell self-renewal and differentiation. Genes Dev , 1106–1118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lie D-C, Colamarino SA, Song H-J, Désiré L, Mira H, Consiglio A, Lein ES, Jessberger S, Lansford H, Dearie AR, et al (2005). Wnt signalling regulates adult hippocampal neurogenesis. Nature , 1370–1375. [DOI] [PubMed] [Google Scholar]
- Liu Y, Lu Z, Shi Y, Sun F. (2018). AMOT is required for YAP function in high glucose induced liver malignancy. Biochem Biophys Res Commun , 1555–1561. [DOI] [PubMed] [Google Scholar]
- Lledo P-M, Alonso M, Grubb MS. (2006). Adult neurogenesis and functional plasticity in neuronal circuits. Nat Rev Neurosci , 179–193. [DOI] [PubMed] [Google Scholar]
- Low BC, Pan CQ, Shivashankar GV, Bershadsky A, Sudol M, Sheetz M. (2014). YAP/TAZ as mechanosensors and mechanotransducers in regulating organ size and tumor growth. FEBS Lett , 2663–2670. [DOI] [PubMed] [Google Scholar]
- Lv M, Li S, Luo C, Zhang X, Shen Y, Sui Y, Wang F, Wang X, Yang J, Liu P, et al (2016). Angiomotin promotes renal epithelial and carcinoma cell proliferation by retaining the nuclear YAP. Oncotarget , 12393–12403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mana-Capelli S, McCollum D. (2018). Angiomotins stimulate LATS kinase autophosphorylation and act as scaffolds that promote Hippo signaling. J Biol Chem , 18230–18241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mana-Capelli S, Paramasivam M, Dutta S, McCollum D. (2014). Angiomotins link F-actin architecture to Hippo pathway signaling. Mol Biol Cell , 1676–1685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McBeath R, Pirone DM, Nelson CM, Bhadriraju K, Chen CS. (2004). Cell shape, cytoskeletal tension, and RhoA regulate stem cell lineage commitment. Dev Cell , 483–495. [DOI] [PubMed] [Google Scholar]
- Meng Z, Qiu Y, Lin KC, Kumar A, Placone JK, Fang C, Wang KC, Lu S, Pan M, Hong AW, et al (2018). RAP2 mediates mechanoresponses of the Hippo pathway. Nature , 655–660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moffat J, Grueneberg DA, Yang X, Kim SY, Kloepfer AM, Hinkle G, Piqani B, Eisenhaure TM, Luo B, Grenier JK, et al (2006). A lentiviral RNAi library for human and mouse genes applied to an arrayed viral high-content screen. Cell , 1283–1298. [DOI] [PubMed] [Google Scholar]
- Moleirinho S, Hoxha S, Mandati V, Curtale G, Troutman S, Ehmer U, Kissil JL. (2017). Regulation of localization and function of the transcriptional co-activator YAP by angiomotin. Elife , e23966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakajima H, Yamamoto K, Agarwala S, Terai K, Fukui H, Fukuhara S, Ando K, Miyazaki T, Yokota Y, Schmelzer E, et al (2017). Flow-dependent endothelial YAP regulation contributes to vessel maintenance. Dev Cell , 523–536.e6. [DOI] [PubMed] [Google Scholar]
- Palmer TD, Markakis EA, Willhoite AR, Safar F, Gage FH. (1999). Fibroblast growth factor-2 activates a latent neurogenic program in neural stem cells from diverse regions of the adult CNS. J Neurosci , 8487–8497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peltier J, Schaffer DV. (2010). Viral packaging and transduction of adult hippocampal neural progenitors. Methods Mol Biol , 103–116. [DOI] [PubMed] [Google Scholar]
- Peng T, Liu L, MacLean AL, Wong CW, Zhao W, Nie Q. (2017). A mathematical model of mechanotransduction reveals how mechanical memory regulates mesenchymal stem cell fate decisions. BMC Syst Biol , 55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ragni CV, Diguet N, Le Garrec JF, Novotova M, Resende TP, Pop S, Charon N, Guillemot L, Kitasato L, Badouel C, et al (2017). Amotl1 mediates sequestration of the Hippo effector Yap1 downstream of Fat4 to restrict heart growth. Nat Commun , 14582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rammensee S, Kang MS, Georgiou K, Kumar S, Schaffer DV. (2016). Dynamics of mechanosensitive neural stem cell differentiation. Stem Cells , 497–506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rojek KO, Krzemien´ J, Dolez˙yczek H, Boguszewski PM, Kaczmarek L, Konopka W, Rylski M, Jaworski J, Holmgren L, Prószyn´ski TJ. (2019). Amot and Yap1 regulate neuronal dendritic tree complexity and locomotor coordination in mice. PLoS Biol , e3000253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saha K, Keung AJ, Irwin EF, Li Y, Little L, Schaffer DV, Healy KE. (2008). Substrate modulus directs neural stem cell behavior. Biophys J , 4426–4438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanjana NE, Shalem O, Zhang F. (2014). Improved vectors and genome-wide libraries for CRISPR screening. Nat Methods , 783–784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shors TJ, Miesegaes G, Beylin A, Zhao M, Rydel T, Gould E. (2001). Neurogenesis in the adult is involved in the formation of trace memories. Nature , 372–376. [DOI] [PubMed] [Google Scholar]
- Takano H, Komuro I, Oka T, Shiojima I, Hiroi Y, Mizuno T, Yazaki Y. (1998). The Rho family G proteins play a critical role in muscle differentiation. Mol Cell Biol , 1580–1589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trouche S, Bontempi B, Roullet P, Rampon C. (2009). Recruitment of adult-generated neurons into functional hippocampal networks contributes to updating and strengthening of spatial memory. Proc Natl Acad Sci USA , 5919–5924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wada K-I, Itoga K, Okano T, Yonemura S, Sasaki H. (2011). Hippo pathway regulation by cell morphology and stress fibers. Development , 3907–3914. [DOI] [PubMed] [Google Scholar]
- Wang W, Li N, Li X, Tran MK, Han X, Chen J. (2015). Tankyrase inhibitors target YAP by stabilizing angiomotin family proteins. Cell Rep , 524–532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wells CD, Fawcett JP, Traweger A, Yamanaka Y, Goudreault M, Elder K, Kulkarni S, Gish G, Virag C, Lim C, et al (2006). A Rich1/Amot complex regulates the Cdc42 GTPase and apical-polarity proteins in epithelial cells. Cell , 535–548. [DOI] [PubMed] [Google Scholar]
- Winner B, Rockenstein E, Lie DC, Aigner R, Mante M, Bogdahn U, Couillard-Despres S, Masliah E, Winkler J. (2008). Mutant α-synuclein exacerbates age-related decrease of neurogenesis. Neurobiol Aging , 913–925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang C, Tibbitt MW, Basta L, Anseth KS. (2014). Mechanical memory and dosing influence stem cell fate. Nat Mater , 645–652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang L, Wang L, Geiger H, Cancelas JA, Mo J, Zheng Y. (2007). Rho GTPase Cdc42 coordinates hematopoietic stem cell quiescence and niche interaction in the bone marrow. Proc Natl Acad Sci USA , 5091–5096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao PJ, Petralia RS, Mattson MP. (2016). Sonic hedgehog signaling and hippocampal neuroplasticity. Trends Neurosci , 840–850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yasuhara T, Matsukawa N, Hara K, Yu G, Xu L, Maki M, Kim SU, Borlongan CV. (2006). Transplantation of human neural stem cells exerts neuroprotection in a rat model of Parkinson’s Disease. J Neurosci , 12497–12511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yi C, Shen Z, Stemmer-Rachamimov A, Dawany N, Troutman S, Showe LC, Liu Q, Shimono A, Sudol M, Holmgren L, et al (2013). The p130 isoform of angiomotin is required for Yap-mediated hepatic epithelial cell proliferation and tumorigenesis. Sci Signal , ra77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yi C, Troutman S, Fera D, Stemmer-Rachamimov A, Avila JL, Christian N, Persson NL, Shimono A, Speicher DW, Marmorstein R, et al (2011). A tight junction-associated merlin-angiomotin complex mediates merlin’s regulation of mitogenic signaling and tumor suppressive functions. Cancer Cell , 527–540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu P, Santiago LY, Katagiri Y, Geller HM. (2012a). Myosin II activity regulates neurite outgrowth and guidance in response to chondroitin sulfate proteoglycans. J Neurochem , 1117–1128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu FX, Zhao B, Panupinthu N, Jewell JL, Lian I, Wang LH, Zhao J, Yuan H, Tumaneng K, Li H, et al (2012b). Regulation of the Hippo-YAP pathway by G-protein-coupled receptor signaling. Cell , 780–791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaltsman Y, Masuko S, Bensen JJ, Kiessling LL. (2019). Angiomotin regulates YAP localization during neural differentiation of human pluripotent stem cells. Stem Cell Rep , 869–877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zegers MM, Friedl P. (2014). Rho GTPases in collective cell migration. Small GTPases , e28997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao B, Li L, Lu Q, Wang LH, Liu CY, Lei Q, Guan KL. (2011). Angiomotin is a novel Hippo pathway component that inhibits YAP oncoprotein. Genes Dev , 51–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao B, Li L, Wang L, Wang CY, Yu J, Guan KL. (2012). Cell detachment activates the Hippo pathway via cytoskeleton reorganization to induce anoikis. Genes Dev , 54–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.