Abstract
Chest X-ray radiography and computed tomography, the two mainstay modalities in thoracic radiology, are under active investigation with deep learning technology, which has shown promising performance in various tasks, including detection, classification, segmentation, and image synthesis, outperforming conventional methods and suggesting its potential for clinical implementation. However, the implementation of deep learning in daily clinical practice is in its infancy and facing several challenges, such as its limited ability to explain the output results, uncertain benefits regarding patient outcomes, and incomplete integration in daily workflow. In this review article, we will introduce the potential clinical applications of deep learning technology in thoracic radiology and discuss several challenges for its implementation in daily clinical practice.
Keywords: Artificial intelligence, Deep learning, Chest radiograph, Chest X-ray, Computed tomography
INTRODUCTION
In recent years, artificial intelligence (AI) and deep learning (DL) have become highlighted technologies across society, including in the field of medicine. The concept of DL is not brand-new (1,2), but the recent rapid growth of computing power and digital data have enabled its success in various fields of application, such as speech recognition (3), natural language processing (4), self-driving vehicles (5), and medicine (6,7).
One of the most successful areas of DL is computer vision. A specific type of DL algorithm called the convolutional neural network (CNN) has played a central role in this success. In 2012, a DL algorithm called “AlexNet,” using a CNN architecture, won the annual ImageNet Large Scale Visual Recognition Challenge, which is the biggest competition in the image recognition field (8), exhibiting a much lower error rate than the winning algorithm from the previous year (16% vs. 26%) (9). In 2015, the winning algorithm of the competition called “ResNet,” based on a CNN, exhibited an error rate of 3.6%, surpassing human-level performance (10).
For medical image analyses, CNN-based DL models showed expert or beyond-expert level performances in various tasks, including the diagnosis of skin cancer from skin photographs (11), diagnosis of diabetic retinopathy from fundus photographs (12,13), and detection of breast cancer metastasis from pathologic slides (14). These initial successes raised expectations that DL-based medical image analysis tools would soon be implemented in daily practice. Recently, it has been asserted that radiologists could and should be educated consumers by understanding the value of AI tools in clinical practices and evaluating their performance before their clinical implementation (15).
Chest X-ray (CXR) ragiographic images and computed tomography (CT), which are the two pillars of thoracic radiology, have been the most actively investigated imaging modalities for various computer-aided image analyses. Some investigations have shown promising results using conventional computer-aided image analyses (16,17,18,19,20,21,22,23), but few of them have been implemented in actual clinical practice because of their suboptimal performances (24,25). It is now anticipated that DL technology will overcome the limitations in performance shown by conventional computer-aided image analyses and be implemented in the daily practice of thoracic radiology (17,26). Indeed, there have been several early investigations reporting the surprisingly high performance of DL technologies in thoracic radiology, particularly CXRs (27,28,29,30,31,32).
The aim of this review article is to introduce potential applications of DL technology in the field of thoracic radiology (Table 1) and possible scenarios of implementation in the clinical workflow. In addition, we aim to discuss challenges in the application of DL in routine clinical practice.
Table 1. Task-Based Classification of Potential Applications of Deep Learning Technology in Field of Thoracic Radiology.
| Detection of abnormalities |
| Detection of lung nodule on CXR (30,43) or chest CT (69) |
| Image classification |
| Classification of lung nodules according to morphology (71) |
| Classification of lung nodules according to likelihood of malignancy (72,73,74) |
| Diagnosis of specific diseases (active tuberculosis (28,29,44), lung cancer (75,77), COPD (85), pulmonary fibrosis (81,84)) |
| Prediction of patient prognosis or treatment response (76,85,86) |
| Image segmentation |
| Organ segmentation (lung (95,96), pulmonary lobes (97), airway (98)) |
| Lung nodule segmentation (99,100) |
| Image generation |
| Image neutralization (108,109,110) |
| Image quality improvement (image noise reduction) (114,115,116) |
COPD = chronic obstructive pulmonary disease, CT = computed tomography, CXR = chest X-ray
Lung Nodule Detection on CXR
Lung nodule detection on CXR is important because lung nodules may represent lung cancer. However, this can often be challenging and lung nodules are not uncommonly missed by radiologists (33,34). Therefore, a computer-aided detection (CAD) system for lung nodules is by far the most investigated task of CAD with respect to CXRs (25,35,36,37,38). Investigations in this field began in the 1960s (39). Regarding conventional CAD systems, multiple studies reported a potential to enhance radiologists' performance (38,40,41,42); however, their standalone performance was suboptimal for a clinical implementation, resulting in many false-positive nodules (1.7–3.3 false positive results per image) (25).
Recent investigations based on DL suggest a potential to overcome the limitations of conventional CAD systems (30,43). In a study by Nam et al. (30), a DL algorithm exhibited a per-nodule sensitivity of 70–82% with 0.02–0.34 false-positives per image for the detection of malignant lung nodules on CXRs. In another study by Cha et al. (43), a DL algorithm showed a 76.8% per-nodule sensitivity at 0.3 false-positives per image. In both studies, the performance of the DL algorithms was better than that of the radiologists. Although it is difficult to directly compare the performances in the two studies because of the differences in the test dataset, the above-radiologist performances of DL suggest the potential of its implementation in daily practice (Fig. 1).
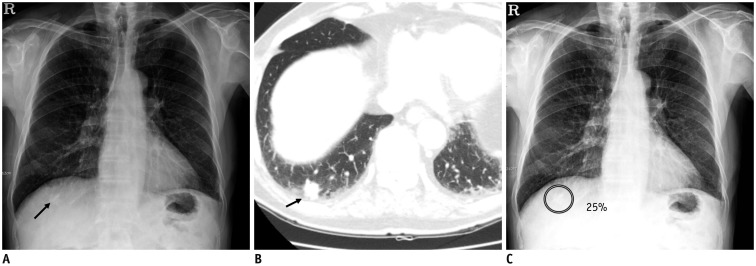
Fig. 1. Detection of lung nodules on chest X-ray.
A. Chest X-ray image shows nodular opacity at juxta-diaphragmatic right basal lung (arrow). B. Corresponding CT image shows 1.5-cm solid nodule at right lower lobe of lung (arrow). C. DL algorithm successfully detected nodule with output probability score of 25% (Courtesy of authors, DL algorithm is same as that in study by Nam et al. (30)). CT = computed tomography, DL = deep learning
Detection of Multiple Abnormalities on CXR
In addition to lung nodules, the DL-based algorithm has shown good performance in various thoracic diseases, such as pulmonary tuberculosis (area under receiver operating characteristic curve [AUC], 0.83–0.99) (28,29,44,45), pneumonia (maximum AUC in internal validation, 0.93) (46), and pneumothorax (AUC, 0.82–0.91) (32,47), and in the evaluation of medical devices on CXRs (48,49,50). However, algorithms specific to a single disease or abnormality may have limited value in real clinical practice, as the interpretation of CXR requires the assessment of various diseases and abnormalities in the thorax.
In 2017, Wang et al. (51) reported a large open-source dataset including 112120 CXRs from 30805 patients, which were labeled for 14 thoracic abnormalities, provided by the U.S. National Institute of Health (50). The authors reported benchmark performances of DL algorithm for the identification of various abnormalities; they showed AUCs ranging from 0.66 (for identification of pneumonia) to 0.87 (for identification of hernia) with an average AUC value of 0.75 (51). Several subsequent studies reported better performances in detecting specific abnormalities using the same dataset (average AUC values, 0.76–0.81) (31,52,53). More recently, additional large-scale open-source CXR datasets labeled for various abnormalities have been released (e.g., CheXpert dataset (54): 224316 CXRs labeled for 14 findings from Stanford Hospital, US; MIMIC-CXR dataset (55): 371920 CXRs labeled for 14 findings from Beth Israel Deaconess Medical Center, US: PADChest dataset (56): 160868 CXRs labeled for 174 findings from Hospital San Juan, Spain) (Table 2). Investigations of DL-based CAD in the detection of multiple abnormalities on CXRs must continue for the time being.
Table 2. Major Large-Scale Open-Source Datasets of CXR.
| Name of Dataset | Distributor | Data Source | No. of Data | Labels | Location |
|---|---|---|---|---|---|
| ChestX-ray14 (51) | US National Institute of Health | National Institute of Health Clinical Center (US) | 112120 CXRs from 30805 patients | 14 radiological findings | https://nihcc.app.box.com/v/ChestXray-NIHCC |
| CheXpert (54) | Stanford University | Stanford Hospital (US) | 224316 CXRs from 65240 patients | 14 radiological findings* | https://stanfordmlgroup.github.io/competitions/chexpert/chexpert/ |
| MIMIC-CXR (55) | Massachusetts Institute of Technology | Beth Israel Deaconess Medical Center (US) | 371920 CXRs from 65383 patients | 14 radiological findings* | https://physionet.org/content/mimic-cxr/2.0.0/ |
| PADChest (56) | Medical Imaging Databank of Valencia Region | Hospital San Juan (Spain) | 160868 CXRs from 67625 patients | 174 radiologic findings; 19 differential diagnoses; 104 anatomic locations | http://bimcv.cipf.es/bimcv-projects/padchest/ |
*Identical labels.
Differentiation of various abnormalities on CXR using DL algorithms can be a challenging task, as various thoracic diseases have overlapping radiologic findings (Fig. 2). In a study by Hwang et al. (27), a DL algorithm could accurately differentiate pneumothorax (accuracy: 95%) from parenchymal diseases (lung cancer, tuberculosis, pneumonia) while exhibiting much lower performance for the differentiation of three parenchymal diseases (accuracy: 21–84%). Despite the limited performance in differential diagnosis, overlapping radiologic findings of various diseases may help the detection of rare, non-targeted diseases using the DL algorithm, considering that training a DL algorithm to cover all diseases that can be found on CXRs is virtually impossible. In our recent study, a DL-based algorithm that had been trained for four diseases (lung cancer, tuberculosis, pneumonia, and pneumothorax) could identify clinically referable CXRs, including those with not only target diseases of the algorithm (sensitivity: 87.9–93.6%), but also non-target diseases of the algorithm (sensitivity: 73.9–82.6%) with an AUC of 0.95, in the emergency department (57). The algorithm also exhibited higher sensitivities compared to those of on-call radiology residents (sensitivity, 65.6%).
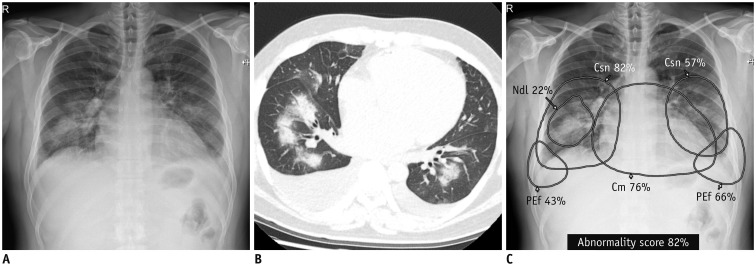
Fig. 2. Detection and differentiation of different abnormalities on chest X-ray.
Chest X-ray (A) and CT (B) obtained on same day from patient with pulmonary edema shows consolidation in both lung fields, bilateral pleural effusion, and mild cardiomegaly. DL algorithm classified chest X-ray image as abnormal, with 82% probability score. C. Algorithm identified Csns, PEf, and Cm on chest X-ray and localized each abnormality separately. Notably, algorithm recognized focal area of dense consolidation in right lower lung field as nodule (Courtesy of authors, DL algorithm is same as that in study by Kim et al. (132)). Cm = cardiomegaly, Csn = consolidation, Ndl = nodule, PEf = pleural effusion
Screening for Tuberculosis on CXR
Detection of pulmonary tuberculosis is another important task of CAD with a high potential for clinical application. The World Health Organization (WHO) recommends systemic screening for active tuberculosis in high-risk populations to reduce the global burden of tuberculosis (58). Although the choice of screening algorithm depends on the population and availability of diagnostic modalities, CXR plays a key role in the suggested screening algorithms (58). However, although CXR has good diagnostic ability for tuberculosis (sensitivity of 87% and specificity of 89% for tuberculosis-related abnormalities) (58,59,60), the number of expert radiologists able to interpret them are limited, especially in high-prevalence countries. In this regard, a commercialized CAD system for tuberculosis (CAD4TB, Delft imaging systems, 's-Hertogenbosch, The Netherlands) has been tested in various screening settings, exhibiting AUCs ranging from 0.71 to 0.84 (58). However, as of 2016, the WHO recommendation is that CAD should be used only in research because of the limited evidence regarding its diagnostic accuracy (61).
DL may boost the performance of CAD systems for tuberculosis. Recent studies using DL algorithms have shown promising performances (AUC, 0.83–0.99) (Fig. 3) (28,29,44,45,62), suggesting a potential for implementation. Furthermore, in a study by Hwang et al. (28), the DL algorithm (AUC, 0.99) outperformed human readers (AUC, 0.75–0.97), including thoracic radiologists and the performance of human readers improved after reviewing the algorithm results (AUC, 0.75–0.97 to 0.85–0.98). Thus far, DL algorithms, however, have been tested on datasets collected for case-control studies (45). Further investigations on the diagnostic performance of DL-based algorithms are required in actual screening or triage situations to prove their applicability in real-world practice.
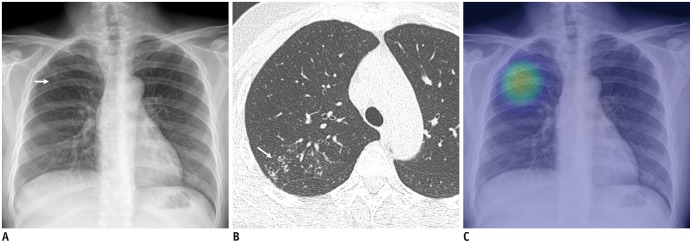
Fig. 3. Identification of chest X-ray with active pulmonary tuberculosis.
A. Chest X-ray of patient with active pulmonary tuberculosis shows subtle nodular infiltration in right upper lung field (arrow). B. Corresponding CT image shows clustered centrilobular nodules and mild bronchiectasis in right upper lobe of lung (arrow). C. DL algorithm successfully detected lesion, with heat map overlaid on chest X-ray (Courtesy of authors, DL algorithm is same as that in study by Hwang et al. (28)).
Lung Cancer Screening with Low-Dose CT
With cumulative evidence of reduced lung cancer mortality following screening using low-dose chest CTs (63,64,65), nationwide systemic lung cancer screening programs have been implemented or are expected to be implemented in the near future (66,67). The workload of radiologists is expected to increase with the implementation of lung cancer screening (68). DL may help radiologists in various aspects of the interpretation of low-dose CTs for lung cancer screening.
Lung nodule detection is a classic task for CAD in the field of chest CTs (19). In 2016, a challenge for lung nodule detection called LUng Nodule Analysis (LUNA) 16 was held (69) and best-performing DL algorithms exhibited over 90% per-nodule sensitivity, ranging from 0.125 to eight false positives per examination (70).
Classification or categorization of lung nodules is another key task in lung cancer screening. The DL algorithm may categorize lung nodules based on their size, location, number, calcification, internal consistency, or existing criteria, such as the Lung CT screening Reporting and Data System (Lung-RADS), to reduce inter-reader variability among radiologists (71) or directly classify each lung nodule based on its likelihood of malignancy (72,73,74).
Per-examination level classification, that is, to classify CT examinations directly into those with and without lung cancer, is another potential strategy. In 2017, Kaggle, a representative online data science competition community, held a competition called “Data Science Bowl 2017” with a task of predicting lung cancer diagnosis within one year of a single CT examination (75). With a similar strategy, the Google AI team published a remarkable study (77). In the study, a DL algorithm evaluated a full set of low-dose chest CT images, with or without a prior CT examination for comparison. The algorithm revealed the likelihood of the subject to be diagnosed with lung cancer. Compared to the Lung-RADS categorization by thoracic radiologists, the algorithm exhibited better performance using a single CT volume (sensitivity, 79.5–95.2% vs. 62.5–90.0%; specificity, 81.3–96.5% vs. 69.7–95.3%; varied by threshold for positive results) and in-par performance with radiologists using two CT volumes (previous and current CTs; sensitivity, 72.5–87.5% vs. 70.0–86.7%; specificity, 84.2–96.5% vs. 83.7–96.3%; varied by threshold for positive results). Although the ability to explain the output and possibility of integration with the current workflow is questionable, the high performance of DL algorithms in the diagnosis of lung cancer, without any intervention by radiologists, is impressive.
Classification of Diffuse Lung Diseases on CT
DL can also be utilized in the interpretation of CTs of patients with diffuse lung diseases. The classification of radiologic findings of interstitial lung disease (ILD) is prone to high intra- and inter-reader variabilities, and DL technologies may help reduce this variability.
Several studies reported that DL algorithms can classify CT findings of ILD (e.g., honeycombing, reticulation, ground-glass opacity, and consolidation) (78,79,80). Furthermore, in a recent study by Walsh et al. (81), DL could classify CTs with fibrotic lung disease according to existing guidelines (82,83), exhibiting overall accuracies of 73.3% and 76.4% for different test datasets. The algorithm exhibited better accuracy than 66% of radiologists. More recently, Christe et al. (84) reported an end-to-end DL algorithm that could segment the lung and airway, classify and segment different findings of lung parenchyma, and finally, classify the examination-level diagnosis based on the current criteria for idiopathic pulmonary fibrosis (83). The algorithm exhibited similar performance to two radiologists (overall accuracy, 56%).
DL can also be utilized to evaluate chronic obstructive pulmonary disease (COPD). In a study by González et al. (85), a DL algorithm could identify CTs with COPD with a C-statistic of 0.856 in a cohort from the COPDGene study. The DL algorithm classified CTs of patients with different stages of COPD, exhibiting accuracies of 51.1% and 29.4% in different cohorts.
Beyond Detection and Diagnosis: DL for Novel Imaging Biomarkers
Most investigations of DL in the field of radiology to date have focused on detection of radiologic abnormalities or identifying diseases. However, the prediction of patient prognosis or therapeutic response may be another potential application of DL. Recently, Lu et al. (86) reported that a DL-based risk score obtained from CXR images could predict long-term all-cause mortality. The authors validated the DL-based risk score in cohorts from the Prostate, Lung, Colorectal, and Ovarian Cancer Screening trial and National Lung Screening Trial and found a graded association of the mortality rate and risk score, independent of age, sex, and the radiologists' interpretation (86). In a study by González et al. (85), the DL algorithm could predict the occurrence of acute respiratory diseases (C-statistic, 0.55–0.64) and death (C-statistic, 0.60–0.72) from chest CT images. In a study by Hosny et al. (76), a DL algorithm could predict 2-year overall survival after radiotherapy (AUC, 0.70) or surgery (AUC, 0.71) for non-small cell lung cancer, outperforming conventional machine learning techniques. DL can also be utilized in radiogenomics research. In a study by Wang et al. (87), a DL algorithm could predict the mutation of epidermal growth factor receptor from CT images, with an AUC of 0.81 in an independent cohort, outperforming the conventional method of using hand-crafted CT features.
Although early investigations have shown the promising performance of novel DL-based imaging biomarkers, outperforming conventional techniques, thorough validation might be warranted for those novel DL-based imaging biomarkers, as the prediction of patient outcomes is a less-intuitive task than lesion detection or image classification.
Applications of AI in Quantitative Imaging Analyses
Segmentation
There have been continuous efforts to extract quantitative biomarkers from chest CT images to evaluate various diseases (88,89,90,91). More recently, radiomics, extracting high-throughput quantitative features from images to predict diagnosis or prognosis, has emerged as an important field of radiologic research (92). The accurate segmentation of specific anatomic structures or pathologic findings of interest might be a gateway for those quantitative image analyses. However, manual segmentation by radiologists is extremely time-consuming and practically impossible in daily practice. DL has shown excellent performance in image segmentation and image classification (93,94). The excellent performance of DL algorithms has been reported in the segmentation of various anatomic structures in chest CTs, including the lung parenchyma (95,96), pulmonary lobes (97), airways (98), and lung nodules (99,100).
Image Neutralization
Another barrier in quantitative image analyses is the variation in reconstructed images caused by variations in scanners, and scanning and reconstruction protocols (101,102,103). For radiomics, those variations have been indicated as the major source of variability in radiomic features, limiting the reproducibility of results and generalizability of radiomics (101,104,105,106). DL can help overcome this barrier by neutralizing images with various image styles. DL can generate a new image with different image textures while maintaining the image content using a specific type of DL algorithm called generative adversarial network (107). Recently, Lee et al. (108) reported that a DL algorithm could convert CT images into those of different reconstruction kernels and reduce the variability in the quantification of emphysema using converted CT images. In subsequent studies, the group reported reduced variability in radiomic features by utilizing DL-based CT image reconstruction kernel conversion (Fig. 4) (109) and slice thickness reduction techniques (110) using DL algorithms.
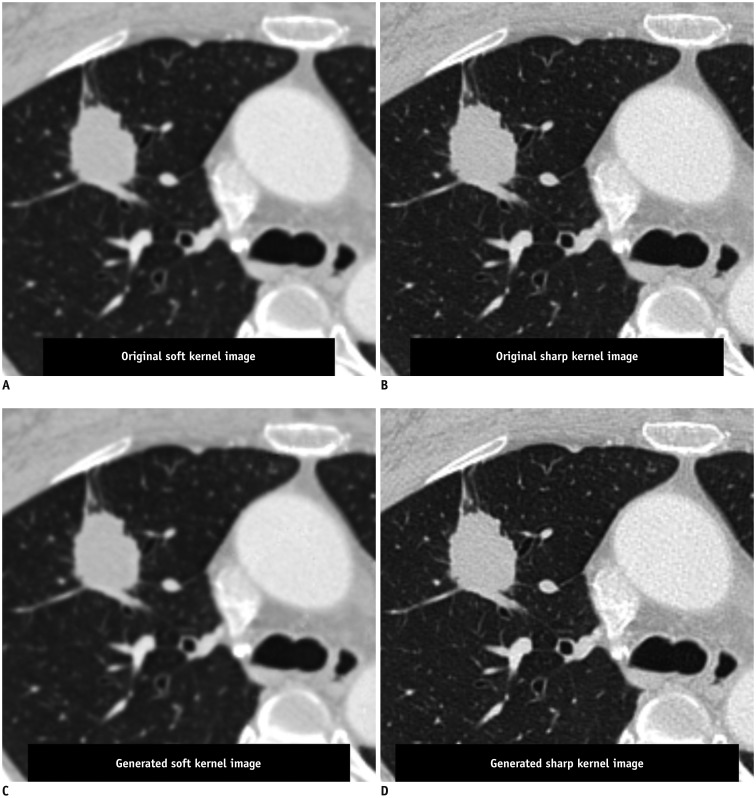
Fig. 4. Conversion of reconstruction kernel on chest CT.
CT images reconstructed with soft kernel (A) and sharp kernel (B) from single scan of patient with lung nodule showing different image textures, which may cause variability in radiomic features of lung nodule. DL algorithm could generate CT image with similar texture to that of soft kernel image from sharp kernel image (C), and vice versa (D). Utilizing generated images with similar textures, variability in radiomic features can be reduced compared to that when using images with different textures (Courtesy of Sang Min Lee, University of Ulsan College of Medicine, Asan Medical Center, DL algorithm is same as that in study by Choe et al. (109)).
Image Quality Improvement
Optimization of the image quality while minimizing the radiation dose are major issues in clinical practice. In the previous decade, the iterative reconstruction (IR) technique achieved remarkable advancements and contributed to image noise reduction on CT images (111,112,113). However, there are several limitations to IR: 1) vendor-specific technologies requiring sinogram data from the CT scanner; 2) over-smoothening of images resulting in the loss of anatomic structures, such as the interlobar fissures; and 3) production of unfamiliar image textures (112). These limitations can potentially be overcome by the utilization of DL-based image generation by providing images with lesser noise but more familiar image style to radiologists, similar to images from a filtered back projection. Some vendors have already demonstrated DL-based noise reduction algorithms (114,115). Furthermore, DL-based noise reduction can be applied independent of vendors or scanners, as the DL algorithm can generate new images from reconstructed CT images, without sinogram data.
The DL may also contribute to the improvement of CXR image quality. In a study by Ahn et al. (116), DL-based software could generate CXR images simulating those obtained with grids from those obtained without the utilization of grids. The subjective image quality and radiologists' preference improved in those generated grid-like images, without the need for an additional radiation dose because of the utilization of grids.
Service Delivery Scenarios of DL Systems in the Clinical Workflow
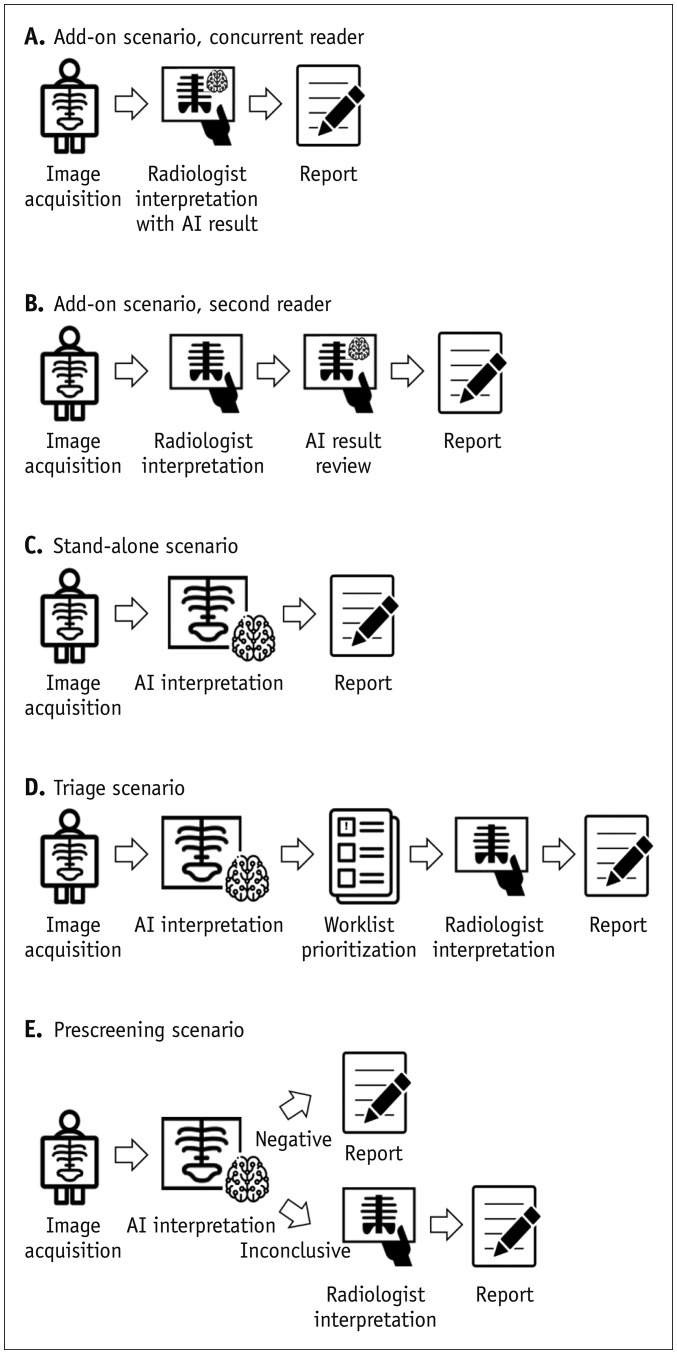
The two classic scenarios of integrating CAD into the clinical workflow are add-on and stand-alone scenarios (Fig. 5) (117,118). In the add-on scenario, radiologists check the results from CAD during (concurrent reader) or after (second reader) image interpretation. In previous studies, the performance of radiologists improved after reviewing the output of DL algorithms when identifying CXRs with malignant nodules (30), active tuberculosis (28), or major thoracic disease (27). In the stand-alone scenario, CAD may automatically classify CXRs without intervention from radiologists. In this scenario, the CAD system may require a more thorough validation of its diagnostic performance and reliability, and should be utilized only in selected clinical situations with narrow tasks (e.g., screening for specific diseases in the healthy population), particularly when the availability of radiologists is limited.
Fig. 5. Delivery scenarios for DL-based CAD systems.
DL-based CAD system can be utilized as assistance tool to enhance diagnostic accuracy of radiologists as concurrent (A) or second reader (B). C. In select situations in which radiologists' interpretations are unavailable, DL-based CAD system may interpret images alone to identify patients requiring referral. D. In triage scenario, CAD system may analyze images before radiologists' interpretations to triage examinations based on presence of findings requiring immediate diagnosis and management and prioritize radiologists' worklists to improve turnaround time for examinations with critical findings. E. Finally, in prescreening scenario, CAD system may analyze large volumes of examinations before radiologists' interpretation to identify clearly negative cases, and radiologists may then only interpret remaining uncertain examinations. AI = artificial intelligence, CAD = computer-aided detection
The third scenario of CAD integration into the clinical workflow can be in the triage of examinations. In this scenario, CAD makes a provisional analysis of each image before radiologists' interpretation and can prioritize the work list in terms of the criticality of the disease or abnormalities. Consequently, when there is a large volume of examinations with limited radiologist availability, such prioritization may help reduce the turnaround time for examinations with critical findings and prevent a delay in treatment. This concept of prioritization has been mainly investigated in the field of neuroradiology, in which the timely diagnosis and management of acute neurologic diseases are critical (119). DL algorithms can automatically identify critical brain CT findings and perform prioritization to minimize delayed diagnosis (120,121). For CXRs, Annarumma et al. (122) utilized a DL algorithm to identify CXRs with critical or urgent findings to prioritize examination. In a simulation study, the median delay for CXRs with critical findings was reduced from 7.2 hours to 43 minutes with the application of DL-based automated prioritization (122).
The last scenario for implementing CAD is a prescreening of negative examinations (123). This scenario could be utilized in selected clinical situations. Very low disease prevalence settings, such as screening of an asymptomatic population (e.g., screening tuberculosis with CXR or screening lung cancer with low-dose CT), with limited availability of experts to interpret images is a typical indication for this scenario. In this scenario, CAD may analyze a large volume of examinations before the interpretation of radiologists to identify clearly negative examinations, and radiologists would interpret the remaining examinations that were positive or inconclusive in the CAD analysis. Such prescreening schemes may help radiologists to reduce the time burden of interpreting large volumes of negative examinations and to focus on more clinically relevant cases. To be utilized as a prescreening tool, the high sensitivity of CAD should be ensured.
Challenges in the Clinical Application of DL
Ability to Explain the DL Algorithm
In order for a DL algorithm to receive credit or the acceptance of radiologists, it should appropriately explain the logical background for the output (124,125,126). For example, let us consider a DL algorithm that can predict lung cancer from screening low-dose CT. In addition to the final output of the algorithm (i.e., the likelihood of lung cancer), radiologists or clinicians may want to know why the algorithm provided the outcome based on their existing knowledge. Was there any pulmonary nodule? What was the size, internal consistency, and location of the nodule? Were there specific features of the nodule or background lung that raised the suspicion of lung cancer? To solve this explainability issue (or “black-box” issue), the most common method is to utilize a saliency map (126,127). By overlaying a saliency map on the input image (Fig. 3), one can visualize the specific areas of the image that contributed to the final output of the DL algorithm. Saliency maps would be good solutions in detection tasks (e.g., detection of pulmonary nodules) or classification algorithms with intuitive tasks (e.g., identification of cardiomegaly). However, it may be insufficient for non-intuitive tasks, such as the diagnosis of specific diseases or prognostication. Radiologic AI should provide interpretability, transparency, reproducibility, and high performance to receive credibility from radiologists and be implemented in clinical practice.
Validation in Actual Clinical Practice
Although previous studies have reported the excellent performance of various DL algorithms in various tasks, most were validated in the algorithms' development setting with retrospectively and conveniently collected datasets (128). Such conveniently collected datasets may have enriched disease prevalence and a narrow disease spectrum. In contrast, the population in the real-word situation may have a much lower disease prevalence and a much broader spectrum of diseases, some of which may not be covered during the development of the DL algorithm (129). Therefore, the excellent performance of the DL algorithm in the algorithms' development setting may not guarantee performance in real-world settings; thus, DL algorithms should be further validated in actual clinical situations before their clinical application. Indeed, in our recent study (57), a DL algorithm to identify CXRs with major thoracic disease showed a decreased performance in a diagnostic cohort comprising patients from the emergency department (AUC, 0.95), compared to that for the test datasets collected for the case-control study (AUC, 0.97–1.00).
The ultimate goal of applying the DL algorithm is improving patient outcomes in clinical practice. However, validation of the DL algorithm with respect to patient outcomes might be a much more challenging task than the validation of diagnostic performance. Improved diagnostic performance does not necessarily mean improved patient outcomes as there are multiple stages between diagnosis and patient outcomes, including patient referral, therapeutic decision making, and patient management (129). To date, little evidence exists regarding the influence of DL algorithms on patient outcomes or on their cost-effectiveness.
Integration into the Daily Clinical Workflow
Even before the rise of DL, a number of software programs for computer-assisted image analysis had demonstrated acceptable performance and passed regulatory approval (17,18,19,21,25). However, most failed to survive in the daily clinical workflow (130,131). Seamless integration with the existing daily clinical workflow is a prerequisite for the utilization of DL in daily clinical practice. Currently, most radiologists perform their daily practice using the picture archiving and communication system (PACS). Therefore, PACS should serve as a platform for the clinical implementation of various DL-based software programs in order for radiologists to utilize these software programs in their routine practice without limitation. In addition, integration with existing PACS or electronic health record systems is essential to enhance the clinical value of DL in improving workflow efficiency in the prioritization of examinations or critical value reporting.
CONCLUSION
In the history of radiology, the introduction of new technologies, including CT, magnetic resonance imaging, and PACS, has dramatically changed the clinical practice of clinicians and radiologists. AI and DL seem to be the “next big thing” in the field of radiology. Although DL is still in its infancy, cumulative evidence suggests that DL has the potential to survive and change clinical practice. CXRs and chest CTs, the two main pillars of thoracic radiology, are now the first target modalities for clinical implementation of DL beyond research.
In this paper, we reviewed the applications of DL in thoracic radiology and discussed different scenarios regarding their implementation in clinical practice. We believe that finding the best clinical scenario where the DL can work well is a substantial task for radiologists prior to its clinical implementation and can provide the initiative to radiologists to determine the evolution of radiology. Therefore, understanding and becoming familiar with the potential of clinical applications and remaining challenges would be essential for radiologists in the era of DL.
Footnotes
The present study was supported by the Seoul National University Hospital research fund (grant number: 03-2019-0190).
Conflicts of Interest: The authors have no potential conflicts of interest to disclose.
References
- 1.LeCun Y, Bengio Y, Hinton G. Deep learning. Nature. 2015;521:436–444. doi: 10.1038/nature14539. [DOI] [PubMed] [Google Scholar]
- 2.LeCun Y, Bottou L, Bengio Y, Haffner P. Gradient-based learning applied to document recognition. Proceedings of the IEEE. 1998;86:2278–2324. [Google Scholar]
- 3.Hinton G, Deng L, Yu D, Dahl GE, Mohamed A, Jaitly N, et al. Deep neural networks for acoustic modeling in speech recognition: the shared views of four research groups. IEEE Signal Process Mag. 2012;29:82–97. [Google Scholar]
- 4.Hirschberg J, Manning CD. Advances in natural language processing. Science. 2015;349:261–266. doi: 10.1126/science.aaa8685. [DOI] [PubMed] [Google Scholar]
- 5.Pendleton S, Andersen H, Du X, Shen X, Meghjani M, Eng Y, et al. Perception, planning, control, and coordination for autonomous vehicles. Machines. 2017;5:6. [Google Scholar]
- 6.Topol EJ. High-performance medicine: the convergence of human and artificial intelligence. Nat Med. 2019;25:44–56. doi: 10.1038/s41591-018-0300-7. [DOI] [PubMed] [Google Scholar]
- 7.Hinton G. Deep learning—a technology with the potential to transform health care. JAMA. 2018;320:1101–1102. doi: 10.1001/jama.2018.11100. [DOI] [PubMed] [Google Scholar]
- 8.Russakovsky O, Deng J, Su H, Krause J, Satheesh S, Ma S, et al. Imagenet large scale visual recognition challenge. Int J Computer Vision. 2015;115:211–252. [Google Scholar]
- 9.Krizhevsky A, Sutskever I, Hinton GE. ImageNet classification with deep convolutional neural networks; 26th conference on neural informatoin processing systems; 2012 December 03–06; Lake Tahoe, USA. [Google Scholar]
- 10.He K, Zhang X, Ren S, Sun J. Deep residual learning for image recognition; The IEEE conference on computer vision and pattern recognition; 2016 June 27–30; Las Vegas, USA. [Google Scholar]
- 11.Esteva A, Kuprel B, Novoa RA, Ko J, Swetter SM, Blau HM, et a. Dermatologist-level classification of skin cancer with deep neural networks. Nature. 2017;542:115–118. doi: 10.1038/nature21056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gulshan V, Peng L, Coram M, Stumpe MC, Wu D, Narayanaswamy A, et al. Development and validation of a deep learning algorithm for detection of diabetic retinopathy in retinal fundus photographs. JAMA. 2016;316:2402–2410. doi: 10.1001/jama.2016.17216. [DOI] [PubMed] [Google Scholar]
- 13.Ting DSW, Cheung CY, Lim G, Tan GSW, Quang ND, Gan A, et al. Development and validation of a deep learning system for diabetic retinopathy and related eye diseases using retinal images from multiethnic populations with diabetes. JAMA. 2017;318:2211–2223. doi: 10.1001/jama.2017.18152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ehteshami Bejnordi B, Veta M, Johannes van Diest P, van Ginneken B, Karssemeijer N, Litjens G, et al. Diagnostic assessment of deep learning algorithms for detection of lymph node metastases in women with breast cancer. JAMA. 2017;318:2199–2210. doi: 10.1001/jama.2017.14585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rubin DL. Artificial intelligence in imaging: the radiologist's role. J Am Coll Radiol. 2019;16:1309–1317. doi: 10.1016/j.jacr.2019.05.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sluimer IC, van Waes PF, Viergever MA, van Ginneken B. Computer-aided diagnosis in high resolution CT of the lungs. Med Phys. 2003;30:3081–3090. doi: 10.1118/1.1624771. [DOI] [PubMed] [Google Scholar]
- 17.van Ginneken B. Fifty years of computer analysis in chest imaging: rule-based, machine learning, deep learning. Radiol Phys Technol. 2017;10:23–32. doi: 10.1007/s12194-017-0394-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.van Ginneken B, ter Haar Romeny BM, Viergever MA. Computer-aided diagnosis in chest radiography: a survey. IEEE Trans Med Imaging. 2001;20:1228–1241. doi: 10.1109/42.974918. [DOI] [PubMed] [Google Scholar]
- 19.Goo JM. A computer-aided diagnosis for evaluating lung nodules on chest CT: the current status and perspective. Korean J Radiol. 2011;12:145–155. doi: 10.3348/kjr.2011.12.2.145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kiraly AP, Odry BL, Godoy MC, Geiger B, Novak CL, Naidich DP. Computer-aided diagnosis of the airways: beyond nodule detection. J Thorac Imaging. 2008;23:105–113. doi: 10.1097/RTI.0b013e318174e8f5. [DOI] [PubMed] [Google Scholar]
- 21.van Ginneken B, Hogeweg L, Prokop M. Computer-aided diagnosis in chest radiography: beyond nodules. Eur J Radiol. 2009;72:226–230. doi: 10.1016/j.ejrad.2009.05.061. [DOI] [PubMed] [Google Scholar]
- 22.Sluimer I, Schilham A, Prokop M, van Ginneken B. Computer analysis of computed tomography scans of the lung: a survey. IEEE Trans Med Imaging. 2006;25:385–405. doi: 10.1109/TMI.2005.862753. [DOI] [PubMed] [Google Scholar]
- 23.van Rikxoort EM, van Ginneken B. Automated segmentation of pulmonary structures in thoracic computed tomography scans: a review. Phys Med Biol. 2013;58:R187–R220. doi: 10.1088/0031-9155/58/17/R187. [DOI] [PubMed] [Google Scholar]
- 24.Pande T, Cohen C, Pai M, Ahmad Khan F. Computer-aided detection of pulmonary tuberculosis on digital chest radiographs: a systematic review. Int J Tuberc Lung Dis. 2016;20:1226–1230. doi: 10.5588/ijtld.15.0926. [DOI] [PubMed] [Google Scholar]
- 25.Schalekamp S, van Ginneken B, Karssemeijer N, Schaefer-Prokop CM. Chest radiography: new technological developments and their applications. Semin Respir Crit Care Med. 2014;35:3–16. doi: 10.1055/s-0033-1363447. [DOI] [PubMed] [Google Scholar]
- 26.Lee SM, Seo JB, Yun J, Cho YH, Vogel-Claussen J, Schiebler ML, et al. Deep learning applications in chest radiography and computed tomography: current state of the art. J Thorac Imaging. 2019;34:75–85. doi: 10.1097/RTI.0000000000000387. [DOI] [PubMed] [Google Scholar]
- 27.Hwang EJ, Park S, Jin KN, Kim JI, Choi SY, Lee JH, et al. Development and validation of a deep learning-based automated detection algorithm for major thoracic diseases on chest radiographs. JAMA Netw Open. 2019;2:e191095. doi: 10.1001/jamanetworkopen.2019.1095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hwang EJ, Park S, Jin KN, Kim JI, Choi SY, Lee JH, et al. Development and validation of a deep learning-based automatic detection algorithm for active pulmonary tuberculosis on chest radiographs. Clin Infect Dis. 2019;69:739–747. doi: 10.1093/cid/ciy967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lakhani P, Sundaram B. Deep learning at chest radiography: automated classification of pulmonary tuberculosis by using convolutional neural networks. Radiology. 2017;284:574–582. doi: 10.1148/radiol.2017162326. [DOI] [PubMed] [Google Scholar]
- 30.Nam JG, Park S, Hwang EJ, Lee JH, Jin KN, Lim KY, et al. Development and validation of deep learning-based automatic detection algorithm for malignant pulmonary nodules on chest radiographs. Radiology. 2019;290:218–228. doi: 10.1148/radiol.2018180237. [DOI] [PubMed] [Google Scholar]
- 31.Rajpurkar P, Irvin J, Ball RL, Zhu K, Yang B, Mehta H, et al. Deep learning for chest radiograph diagnosis: a retrospective comparison of the CheXNeXt algorithm to practicing radiologists. PLoS Med. 2018;15:e1002686. doi: 10.1371/journal.pmed.1002686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Taylor AG, Mielke C, Mongan J. Automated detection of moderate and large pneumothorax on frontal chest X-rays using deep convolutional neural networks: a retrospective study. PLoS Med. 2018;15:e1002697. doi: 10.1371/journal.pmed.1002697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Austin JH, Romney BM, Goldsmith LS. Missed bronchogenic carcinoma: radiographic findings in 27 patients with a potentially resectable lesion evident in retrospect. Radiology. 1992;182:115–122. doi: 10.1148/radiology.182.1.1727272. [DOI] [PubMed] [Google Scholar]
- 34.Gavelli G, Giampalma E. Sensitivity and specificity of chest X-ray screening for lung cancer: review article. Cancer. 2000;89:2453–2456. doi: 10.1002/1097-0142(20001201)89:11+<2453::aid-cncr21>3.3.co;2-d. [DOI] [PubMed] [Google Scholar]
- 35.Lee KH, Goo JM, Park CM, Lee HJ, Jin KN. Computer-aided detection of malignant lung nodules on chest radiographs: effect on observers' performance. Korean J Radiol. 2012;13:564–571. doi: 10.3348/kjr.2012.13.5.564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.De Boo DW, Uffmann M, Weber M, Bipat S, Boorsma EF, Scheerder MJ, et al. Computer-aided detection of small pulmonary nodules in chest radiographs: an observer study. Acad Radiol. 2011;18:1507–1514. doi: 10.1016/j.acra.2011.08.008. [DOI] [PubMed] [Google Scholar]
- 37.de Hoop B, De Boo DW, Gietema HA, van Hoorn F, Mearadji B, Schijf L, et al. Computer-aided detection of lung cancer on chest radiographs: effect on observer performance. Radiology. 2010;257:532–540. doi: 10.1148/radiol.10092437. [DOI] [PubMed] [Google Scholar]
- 38.Kligerman S, Cai L, White CS. The effect of computer-aided detection on radiologist performance in the detection of lung cancers previously missed on a chest radiograph. J Thorac Imaging. 2013;28:244–252. doi: 10.1097/RTI.0b013e31826c29ec. [DOI] [PubMed] [Google Scholar]
- 39.Lodwick GS, Keats TE, Dorst JP. The coding of Roentgen images for computer analysis as applied to lung cancer. Radiology. 1963;81:185–200. doi: 10.1148/81.2.185. [DOI] [PubMed] [Google Scholar]
- 40.Kakeda S, Moriya J, Sato H, Aoki T, Watanabe H, Nakata H, et al. Improved detection of lung nodules on chest radiographs using a commercial computer-aided diagnosis system. AJR Am J Roentgenol. 2004;182:505–510. doi: 10.2214/ajr.182.2.1820505. [DOI] [PubMed] [Google Scholar]
- 41.Shiraishi J, Abe H, Li F, Engelmann R, MacMahon H, Doi K. Computer-aided diagnosis for the detection and classification of lung cancers on chest radiographs ROC analysis of radiologists' performance. Acad Radiol. 2006;13:995–1003. doi: 10.1016/j.acra.2006.04.007. [DOI] [PubMed] [Google Scholar]
- 42.van Beek EJ, Mullan B, Thompson B. Evaluation of a real-time interactive pulmonary nodule analysis system on chest digital radiographic images: a prospective study. Acad Radiol. 2008;15:571–575. doi: 10.1016/j.acra.2008.01.018. [DOI] [PubMed] [Google Scholar]
- 43.Cha MJ, Chung MJ, Lee JH, Lee KS. Performance of deep learning model in detecting operable lung cancer with chest radiographs. J Thorac Imaging. 2019;34:86–91. doi: 10.1097/RTI.0000000000000388. [DOI] [PubMed] [Google Scholar]
- 44.Heo SJ, Kim Y, Yun S, Lim SS, Kim J, Nam CM, et al. Deep learning algorithms with demographic information help to detect tuberculosis in chest radiographs in annual workers' health examination data. Int J Environ Res Public Health. 2019;16:E250. doi: 10.3390/ijerph16020250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Harris M, Qi A, Jeagal L, Torabi N, Menzies D, Korobitsyn A, et al. A systematic review of the diagnostic accuracy of artificial intelligence-based computer programs to analyze chest X-rays for pulmonary tuberculosis. PLoS One. 2019;14:e0221339. doi: 10.1371/journal.pone.0221339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zech JR, Badgeley MA, Liu M, Costa AB, Titano JJ, Oermann EK. Variable generalization performance of a deep learning model to detect pneumonia in chest radiographs: a cross-sectional study. PLoS Med. 2018;15:e1002683. doi: 10.1371/journal.pmed.1002683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Park S, Lee SM, Kim N, Choe J, Cho Y, Do KH, et al. Application of deep learning-based computer-aided detection system: detecting pneumothorax on chest radiograph after biopsy. Eur Radiol. 2019;29:5341–5348. doi: 10.1007/s00330-019-06130-x. [DOI] [PubMed] [Google Scholar]
- 48.Lee H, Mansouri M, Tajmir S, Lev MH, Do S. A deep-learning system for fully-automated Peripherally Inserted Central Catheter (PICC) tip detection. J Digit Imaging. 2018;31:393–402. doi: 10.1007/s10278-017-0025-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lakhani P. Deep convolutional neural networks for endotracheal tube position and X-ray image classification: challenges and opportunities. J Digit Imaging. 2017;30:460–468. doi: 10.1007/s10278-017-9980-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Singh V, Danda V, Gorniak R, Flanders A, Lakhani P. Assessment of critical feeding tube malpositions on radiographs using deep learning. J Digit Imaging. 2019;32:651–655. doi: 10.1007/s10278-019-00229-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wang X, Peng Y, Lu L, Lu Z, Bagheri M, Summers RM. ChestX-ray8: hospital-scale chest X-ray database and benchmarks on weakly-supervised classification and localization of common thorax diseases; The IEEE conference on computer vision and pattern recognition; 2017 July 21–26; Honolulu, USA. [Google Scholar]
- 52.Yao L, Poblenz E, Dagunts D, Covington B, Bernard D, Lyman K. Learning to diagnose from scratch by exploiting dependencies among labels. Cornell University; 2017. [updated Feb 2018]. [Accessed January 8, 2020]. Available at: https://arxiv.org/abs/1710.10501. [Google Scholar]
- 53.Baltruschat IM, Nickisch H, Grass M, Knopp T, Saalbach A. Comparison of deep learning approaches for multi-label chest X-ray classification. Sci Rep. 2019;9:6381. doi: 10.1038/s41598-019-42294-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Irvin J, Rajpurkar P, Ko M, Yu Y, Ciurea-Ilcus S, Chute C, et al. CheXpert: a large chest radiograph dataset with uncertainty labels and expert comparison; The thirty-third AAAI conference on artificial intelligence (AAAI-19); 2019 January 27–February 1; Honolulu, USA. [Google Scholar]
- 55.Johnson AEW, Pollard TJ, Greenbaum NR, Lungren MP, Deng C, Peng Y, et al. MIMIC-CXR-JPG: a large publicly available database of labeled chest radiographs. Cornell University; 2019. [updated Nov 2019]. [Accessed January 8, 2020]. Available at: https://arxiv.org/abs/1901.07042. [Google Scholar]
- 56.Bustos A, Pertusa A, Salinas JM, de la Iglesia-Vayá M. PadChest: a large chest X-ray image dataset with multi-label annotated reports. Cornell University; 2019. [updated Feb 2019]. [Accessed January 8, 2020]. Available at: https://arxiv.org/abs/1901.07441. [DOI] [PubMed] [Google Scholar]
- 57.Hwang EJ, Nam GJ, Lim WH, Park SJ, Jeong YS, Kang JH, et al. Deep learning for chest radiograph diagnosis in the emergency department. Radiology. 2019;293:573–580. doi: 10.1148/radiol.2019191225. [DOI] [PubMed] [Google Scholar]
- 58.World Health Organization. Systematic screening for active tuberculosis: principles and recommendations. Geneva: WHO Document Production Services; 2013. [PubMed] [Google Scholar]
- 59.den Boon S, White NW, van Lill SW, Borgdorff MW, Verver S, Lombard CJ, et al. An evaluation of symptom and chest radiographic screening in tuberculosis prevalence surveys. Int J Tuberc Lung Dis. 2006;10:876–882. [PubMed] [Google Scholar]
- 60.van't Hoog AH, Meme HK, Laserson KF, Agaya JA, Muchiri BG, Githui WA, et al. Screening strategies for tuberculosis prevalence surveys: the value of chest radiography and symptoms. PLoS One. 2012;7:e38691. doi: 10.1371/journal.pone.0038691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.World Health Organization. Chest radiography in tuberculosis detection: summary of current WHO recommendations and guidance on programmatic approaches. Geneva: World Health Organization; 2016. [Google Scholar]
- 62.Santosh KC, Antani S. Automated chest X-ray screening: can lung region symmetry help detect pulmonary abnormalities? IEEE Trans Med Imaging. 2018;37:1168–1177. doi: 10.1109/TMI.2017.2775636. [DOI] [PubMed] [Google Scholar]
- 63.Black WC, Gareen IF, Soneji SS, Sicks JD, Keeler EB, Aberle DR, et al. Cost-effectiveness of CT screening in the National Lung Screening Trial. N Engl J Med. 2014;371:1793–1802. doi: 10.1056/NEJMoa1312547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.National Lung Screening Trial Research Team. Aberle DR, Adams AM, Berg CD, Black WC, Clapp JD, Fagerstrom RM, et al. Reduced lung-cancer mortality with low-dose computed tomographic screening. N Engl J Med. 2011;365:395–409. doi: 10.1056/NEJMoa1102873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Tanoue LT, Tanner NT, Gould MK, Silvestri GA. Lung cancer screening. Am J Respir Crit Care Med. 2015;191:19–33. doi: 10.1164/rccm.201410-1777CI. [DOI] [PubMed] [Google Scholar]
- 66.Moyer VA U.S. Preventive Services Task Force. Screening for lung cancer: U.S. Preventive Services Task Force recommendation statement. Ann Intern Med. 2014;160:330–338. doi: 10.7326/M13-2771. [DOI] [PubMed] [Google Scholar]
- 67.Fintelmann FJ, Bernheim A, Digumarthy SR, Lennes IT, Kalra MK, Gilman MD, et al. The 10 pillars of lung cancer screening: rationale and logistics of a lung cancer screening program. Radiographics. 2015;35:1893–1908. doi: 10.1148/rg.2015150079. [DOI] [PubMed] [Google Scholar]
- 68.Smieliauskas F, MacMahon H, Salgia R, Shih YC. Geographic variation in radiologist capacity and widespread implementation of lung cancer CT screening. J Med Screen. 2014;21:207–215. doi: 10.1177/0969141314548055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Setio AAA, Traverso A, de Bel T, Berens MSN, Bogaard CVD, Cerello P, et al. Validation, comparison, and combination of algorithms for automatic detection of pulmonary nodules in computed tomography images: the LUNA16 challenge. Med Image Anal. 2017;42:1–13. doi: 10.1016/j.media.2017.06.015. [DOI] [PubMed] [Google Scholar]
- 70.LUng nodule analysis 2016. LUNA16 Web site. [Accessed October 10, 2019]. https://luna16.grand-challenge.org/. Published 2016.
- 71.Ciompi F, Chung K, van Riel SJ, Setio AAA, Gerke PK, Jacobs C, et al. Towards automatic pulmonary nodule management in lung cancer screening with deep learning. Sci Rep. 2017;7:46479. doi: 10.1038/srep46479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Shen W, Zhou M, Yang F, Yu D, Dong D, Yang C, et al. Multi-crop convolutional neural networks for lung nodule malignancy suspiciousness classification. Pattern Recognit. 2017;61:663–673. [Google Scholar]
- 73.Causey JL, Zhang J, Ma S, Jiang B, Qualls JA, Politte DG, et al. Highly accurate model for prediction of lung nodule malignancy with CT scans. Sci Rep. 2018;8:9286. doi: 10.1038/s41598-018-27569-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Nishio M, Sugiyama O, Yakami M, Ueno S, Kubo T, Kuroda T, et al. Computer-aided diagnosis of lung nodule classification between benign nodule, primary lung cancer, and metastatic lung cancer at different image size using deep convolutional neural network with transfer learning. PLoS One. 2018;13:e0200721. doi: 10.1371/journal.pone.0200721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Data science bowl 2017. Kaggle Web site. [Accessed October 10, 2019]. https://www.kaggle.com/c/data-science-bowl-2017. Published May, 2017.
- 76.Hosny A, Parmar C, Coroller TP, Grossmann P, Zeleznik R, Kumar A, et al. Deep learning for lung cancer prognostication: a retrospective multi-cohort radiomics study. PLoS Med. 2018;15:e1002711. doi: 10.1371/journal.pmed.1002711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Ardila D, Kiraly AP, Bharadwaj S, Choi B, Reicher JJ, Peng L, et al. End-to-end lung cancer screening with three-dimensional deep learning on low-dose chest computed tomography. Nat Med. 2019;25:954–961. doi: 10.1038/s41591-019-0447-x. [DOI] [PubMed] [Google Scholar]
- 78.Anthimopoulos M, Christodoulidis S, Ebner L, Christe A, Mougiakakou S. Lung pattern classification for interstitial lung diseases using a deep convolutional neural network. IEEE Trans Med Imaging. 2016;35:1207–1216. doi: 10.1109/TMI.2016.2535865. [DOI] [PubMed] [Google Scholar]
- 79.Kim GB, Jung KH, Lee Y, Kim HJ, Kim N, Jun S, et al. Comparison of shallow and deep learning methods on classifying the regional pattern of diffuse lung disease. J Digit Imaging. 2018;31:415–424. doi: 10.1007/s10278-017-0028-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Gao M, Bagci U, Lu L, Wu A, Buty M, Shin HC, et al. Holistic classification of CT attenuation patterns for interstitial lung diseases via deep convolutional neural networks. Comput Methods Biomech Biomed Eng Imaging Vis. 2018;6:1–6. doi: 10.1080/21681163.2015.1124249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Walsh SLF, Calandriello L, Silva M, Sverzellati N. Deep learning for classifying fibrotic lung disease on high-resolution computed tomography: a case-cohort study. Lancet Respir Med. 2018;6:837–845. doi: 10.1016/S2213-2600(18)30286-8. [DOI] [PubMed] [Google Scholar]
- 82.Raghu G, Collard HR, Egan JJ, Martinez FJ, Behr J, Brown KK, et al. An official ATS/ERS/JRS/ALAT statement: idiopathic pulmonary fibrosis: evidence-based guidelines for diagnosis and management. Am J Respir Crit Care Med. 2011;183:788–824. doi: 10.1164/rccm.2009-040GL. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Lynch DA, Sverzellati N, Travis WD, Brown KK, Colby TV, Galvin JR, et al. Diagnostic criteria for idiopathic pulmonary fibrosis: a Fleischner Society white paper. Lancet Respir Med. 2018;6:138–153. doi: 10.1016/S2213-2600(17)30433-2. [DOI] [PubMed] [Google Scholar]
- 84.Christe A, Peters AA, Drakopoulos D, Heverhagen JT, Geiser T, Stathopoulou T, et al. Computer-aided diagnosis of pulmonary fibrosis using deep learning and CT images. Invest Radiol. 2019;54:627–632. doi: 10.1097/RLI.0000000000000574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.González G, Ash SY, Vegas-Sánchez-Ferrero G, Onieva Onieva J, Rahaghi FN, Ross JC, et al. Disease staging and prognosis in smokers using deep learning in chest computed tomography. Am J Respir Crit Care Med. 2018;197:193–203. doi: 10.1164/rccm.201705-0860OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Lu MT, Ivanov A, Mayrhofer T, Hosny A, Aerts HJWL, Hoffmann U. Deep learning to assess long-term mortality from chest radiographs. JAMA Netw Open. 2019;2:e197416. doi: 10.1001/jamanetworkopen.2019.7416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Wang S, Shi J, Ye Z, Dong D, Yu D, Zhou M, et al. Predicting EGFR mutation status in lung adenocarcinoma on computed tomography image using deep learning. Eur Respir J. 2019;53:1800986. doi: 10.1183/13993003.00986-2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Kim SS, Jin GY, Li YZ, Lee JE, Shin HS. CT quantification of lungs and airways in normal Korean subjects. Korean J Radiol. 2017;18:739–748. doi: 10.3348/kjr.2017.18.4.739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Lynch DA, Al-Qaisi MA. Quantitative computed tomography in chronic obstructive pulmonary disease. J Thorac Imaging. 2013;28:284–290. doi: 10.1097/RTI.0b013e318298733c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Nambu A, Zach J, Schroeder J, Jin G, Kim SS, Kim YI, et al. Quantitative computed tomography measurements to evaluate airway disease in chronic obstructive pulmonary disease: relationship to physiological measurements, clinical index and visual assessment of airway disease. Eur J Radiol. 2016;85:2144–2151. doi: 10.1016/j.ejrad.2016.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Bartholmai BJ, Raghunath S, Karwoski RA, Moua T, Rajagopalan S, Maldonado F, et al. Quantitative computed tomography imaging of interstitial lung diseases. J Thorac Imaging. 2013;28:298–307. doi: 10.1097/RTI.0b013e3182a21969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Aerts HJ, Velazquez ER, Leijenaar RT, Parmar C, Grossmann P, Carvalho S, et al. Decoding tumour phenotype by noninvasive imaging using a quantitative radiomics approach. Nat Commun. 2014;5:4006. doi: 10.1038/ncomms5006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Long J, Shelhamer E, Darrell T. Fully convolutional networks for semantic segmentation; The IEEE conference on computer vision and pattern recognition (CVPR); 2015 June 7–12; Boston, USA. [DOI] [PubMed] [Google Scholar]
- 94.Ronneberger O, Fischer P, Brox T. U-net: convolutional networks for biomedical image segmentation. In: Navab N, Hornegger J, Wells W, Frangi A, editors. Medical image computing and computer-assisted intervention–MICCAI 2015. 3rd ed. Basel: Springer Nature; 2015. pp. 234–241. [Google Scholar]
- 95.Dong X, Lei Y, Wang T, Thomas M, Tang L, Curran WJ, et al. Automatic multiorgan segmentation in thorax CT images using U-net-GAN. Med Phys. 2019;46:2157–2168. doi: 10.1002/mp.13458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Park B, Park H, Lee SM, Seo JB, Kim N. Lung segmentation on HRCT and volumetric CT for diffuse interstitial lung disease using deep convolutional neural networks. J Digit Imaging. 2019;32:1019–1026. doi: 10.1007/s10278-019-00254-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Park J, Yun J, Kim N, Park B, Cho Y, Park HJ, et al. Fully automated lung lobe segmentation in volumetric chest CT with 3D U-net: validation with intra- and extra-datasets. J Digit Imaging. 2020;33:221–230. doi: 10.1007/s10278-019-00223-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Yun J, Park J, Yu D, Yi J, Lee M, Park HJ, et al. Improvement of fully automated airway segmentation on volumetric computed tomographic images using a 2.5 dimensional convolutional neural net. Med Image Anal. 2019;51:13–20. doi: 10.1016/j.media.2018.10.006. [DOI] [PubMed] [Google Scholar]
- 99.Aresta G, Jacobs C, Araújo T, Cunha A, Ramos I, van Ginneken B, et al. iW-Net: an automatic and minimalistic interactive lung nodule segmentation deep network. Sci Rep. 2019;9:11591. doi: 10.1038/s41598-019-48004-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Wang S, Zhou M, Liu Z, Liu Z, Gu D, Zang Y, et al. Central focused convolutional neural networks: developing a data-driven model for lung nodule segmentation. Med Image Anal. 2017;40:172–183. doi: 10.1016/j.media.2017.06.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Kim H, Park CM, Lee M, Park SJ, Song YS, Lee JH, et al. Impact of reconstruction algorithms on CT radiomic features of pulmonary tumors: analysis of intra- and inter-reader variability and inter-reconstruction algorithm variability. PLoS One. 2016;11:e0164924. doi: 10.1371/journal.pone.0164924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Kim H, Goo JM, Ohno Y, Kauczor HU, Hoffman EA, Gee JC, et al. Effect of reconstruction parameters on the quantitative analysis of chest computed tomography. J Thorac Imaging. 2019;34:92–102. doi: 10.1097/RTI.0000000000000389. [DOI] [PubMed] [Google Scholar]
- 103.Chen B, Barnhart H, Richard S, Colsher J, Amurao M, Samei E. Quantitative CT: technique dependence of volume estimation on pulmonary nodules. Phys Med Biol. 2012;57:1335–1348. doi: 10.1088/0031-9155/57/5/1335. [DOI] [PubMed] [Google Scholar]
- 104.Berenguer R, Pastor-Juan MDR, Canales-Vázquez J, Castro-García M, Villas MV, Mansilla Legorburo F, et al. Radiomics of CT features may be nonreproducible and redundant: influence of CT acquisition parameters. Radiology. 2018;288:407–415. doi: 10.1148/radiol.2018172361. [DOI] [PubMed] [Google Scholar]
- 105.Meyer M, Ronald J, Vernuccio F, Nelson RC, Ramirez-Giraldo JC, Solomon J, et al. Reproducibility of CT radiomic features within the same patient: influence of radiation dose and CT reconstruction settings. Radiology. 2019;293:583–591. doi: 10.1148/radiol.2019190928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Yip SS, Aerts HJ. Applications and limitations of radiomics. Phys Med Biol. 2016;61:R150–R166. doi: 10.1088/0031-9155/61/13/R150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Goodfellow IJ, Pouget-Abadie J, Mirza M, Xu B, Warde-Farley D, Ozair S, et al. Generative adversarial nets; Advances in neural information processing systems 27 (NIPS 2014); 2014 December 8–13; Montreal, Canada. [Google Scholar]
- 108.Lee SM, Lee JG, Lee G, Choe J, Do KH, Kim N, et al. CT image conversion among different reconstruction kernels without a sinogram by using a convolutional neural network. Korean J Radiol. 2019;20:295–303. doi: 10.3348/kjr.2018.0249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Choe J, Lee SM, Do KH, Lee G, Lee JG, Lee SM, et al. Deep learning-based image conversion of CT reconstruction kernels improves radiomics reproducibility for pulmonary nodules or masses. Radiology. 2019;292:365–373. doi: 10.1148/radiol.2019181960. [DOI] [PubMed] [Google Scholar]
- 110.Park S, Lee SM, Do KH, Lee JG, Bae W, Park H, et al. Deep learning algorithm for reducing CT slice thickness: effect on reproducibility of radiomic features in lung cancer. Korean J Radiol. 2019;20:1431–1440. doi: 10.3348/kjr.2019.0212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Singh S, Khawaja RD, Pourjabbar S, Padole A, Lira D, Kalra MK. Iterative image reconstruction and its role in cardiothoracic computed tomography. J Thorac Imaging. 2013;28:355–367. doi: 10.1097/RTI.0000000000000054. [DOI] [PubMed] [Google Scholar]
- 112.Padole A, Ali Khawaja RD, Kalra MK, Singh S. CT radiation dose and iterative reconstruction techniques. AJR Am J Roentgenol. 2015;204:W384–W392. doi: 10.2214/AJR.14.13241. [DOI] [PubMed] [Google Scholar]
- 113.Geyer LL, Schoepf UJ, Meinel FG, Nance JW, Jr, Bastarrika G, Leipsic JA, et al. State of the art: iterative CT reconstruction techniques. Radiology. 2015;276:339–357. doi: 10.1148/radiol.2015132766. [DOI] [PubMed] [Google Scholar]
- 114.GE gets FDA nod for deep-learning CT reconstruction. AuntMinnie.com Web site. [Accessed October 22, 2019]. https://www.auntminnie.com/index.aspx?sec=sup&sub=cto&pag=dis&ItemID=125242/. Published April 22, 2019.
- 115.Canon gets FDA nod for AI-powered CT reconstruction. AuntMinnie.com Web site. [Accessed October 22, 2019]. https://www.auntminnie.com/index.aspx?sec=sup&sub=cto&pag=dis&ItemID=125783/. Published June 18, 2019.
- 116.Ahn SY, Chae KJ, Goo JM. The potential role of grid-like software in bedside chest radiography in improving image quality and dose reduction: an observer preference study. Korean J Radiol. 2018;19:526–533. doi: 10.3348/kjr.2018.19.3.526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Bossuyt PM, Irwig L, Craig J, Glasziou P. Comparative accuracy: assessing new tests against existing diagnostic pathways. BMJ. 2006;332:1089–1092. doi: 10.1136/bmj.332.7549.1089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Tang A, Tam R, Cadrin-Chênevert A, Guest W, Chong J, Barfett J, et al. Canadian association of radiologists white paper on artificial intelligence in radiology. Can Assoc Radiol J. 2018;69:120–135. doi: 10.1016/j.carj.2018.02.002. [DOI] [PubMed] [Google Scholar]
- 119.Zaharchuk G, Gong E, Wintermark M, Rubin D, Langlotz CP. Deep learning in neuroradiology. AJNR Am J Neuroradiol. 2018;39:1776–1784. doi: 10.3174/ajnr.A5543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Titano JJ, Badgeley M, Schefflein J, Pain M, Su A, Cai M, et al. Automated deep-neural-network surveillance of cranial images for acute neurologic events. Nat Med. 2018;24:1337–1341. doi: 10.1038/s41591-018-0147-y. [DOI] [PubMed] [Google Scholar]
- 121.Prevedello LM, Erdal BS, Ryu JL, Little KJ, Demirer M, Qian S, et al. Automated critical test findings identification and online notification system using artificial intelligence in imaging. Radiology. 2017;285:923–931. doi: 10.1148/radiol.2017162664. [DOI] [PubMed] [Google Scholar]
- 122.Annarumma M, Withey SJ, Bakewell RJ, Pesce E, Goh V, Montana G. Automated triaging of adult chest radiographs with deep artificial neural networks. Radiology. 2019;291:196–202. doi: 10.1148/radiol.2018180921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Obuchowski NA, Bullen JA. Statistical considerations for testing an AI algorithm used for prescreening lung CT images. Contemp Clin Trials Commun. 2019;16:100434. doi: 10.1016/j.conctc.2019.100434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Lee JG, Jun S, Cho YW, Lee H, Kim GB, Seo JB, et al. Deep learning in medical imaging: general overview. Korean J Radiol. 2017;18:570–584. doi: 10.3348/kjr.2017.18.4.570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Hosny A, Parmar C, Quackenbush J, Schwartz LH, Aerts HJWL. Artificial intelligence in radiology. Nat Rev Cancer. 2018;18:500–510. doi: 10.1038/s41568-018-0016-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Choo J, Liu S. Visual analytics for explainable deep learning. IEEE Comput Graph Appl. 2018;38:84–92. doi: 10.1109/MCG.2018.042731661. [DOI] [PubMed] [Google Scholar]
- 127.Zhou B, Khosla A, Lapedriza A, Oliva A, Torralba A. Learning deep features for discriminative localization. Cornell University; 2015. [Accessed January 8, 2020]. Available at: https://arxiv.org/abs/1512.04150. [Google Scholar]
- 128.Kim DW, Jang HY, Kim KW, Shin Y, Park SH. Design characteristics of studies reporting the performance of artificial intelligence algorithms for diagnostic analysis of medical images: results from recently published papers. Korean J Radiol. 2019;20:405–410. doi: 10.3348/kjr.2019.0025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Park SH, Han K. Methodologic guide for evaluating clinical performance and effect of artificial intelligence technology for medical diagnosis and prediction. Radiology. 2018;286:800–809. doi: 10.1148/radiol.2017171920. [DOI] [PubMed] [Google Scholar]
- 130.Doi K. Computer-aided diagnosis in medical imaging: historical review, current status and future potential. Comput Med Imaging Graph. 2007;31:198–211. doi: 10.1016/j.compmedimag.2007.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Le AH, Liu B, Huang HK. Integration of computer-aided diagnosis/detection (CAD) results in a PACS environment using CAD–PACS toolkit and DICOM SR. Int J Comput Assist Radiol Surg. 2009;4:317–329. doi: 10.1007/s11548-009-0297-y. [DOI] [PubMed] [Google Scholar]
- 132.Kim M, Park J, Kim KH, Hwang EJ, Park CM, Park S. Development of a deep learning-based algorithm for independent detection of chest abnormalities on chest radiographs; 105th scientific assembly and annual meeting, Radiological Society of North America; 2019 December 01–06; Chicago, USA. [Google Scholar]