Abstract
Cutaneous inflammation is recurrent in systemic lupus erythematosus (SLE), yet mechanisms that drive cutaneous inflammation in SLE are not well-defined. Type I IFNs are elevated in non-lesional SLE skin and promote inflammatory responses. Staphylococcus aureus, known to induce IFN production, could play a role in cutaneous inflammation in SLE. We show here that active cutaneous lupus erythematosus (CLE) lesions are highly colonized (~50%) by S. aureus. To define the impact of IFNs on S. aureus colonization, we examined the effects of type I and type II IFNs on S. aureus adherence and invasion. An increase in adherent S. aureus was observed after exposure to both IFNα and γ whereas IFNγ appeared to inhibit invasion of S. aureus. CLE lesional skin microarray data and RNA-seq data from SLE keratinocytes identified repression of barrier gene expression, such as filaggrin and loricrin, and SLE keratinocytes exhibited increased S. aureus-binding integrins. These SLE-associated changes could be replicated by IFN treatment of keratinocytes. Further, SLE keratinocytes exhibited increased binding to S. aureus. Together, these data suggest that chronic exposure to IFNs induces barrier disruption that allows for higher S. aureus colonization in SLE skin.
INTRODUCTION
Cutaneous inflammation is a frequent and recurrent manifestation of systemic lupus erythematosus (SLE), a devastating autoimmune disease. Triggers for skin flares include exposure to ultraviolet light (UVB) (Chasset and Arnaud 2018; Muskardin and Niewold 2018; Stannard and Kahlenberg 2016), which raises type I interferons (IFNs) in the skin. Keratinocytes are an important source of IFNs, and SLE patients exhibit elevated production of type I IFNs, both at baseline and after UVB exposure, that promotes infiltration of inflammatory cells in affected skin (Meller et al. 2005; Sarkar et al. 2018; Stannard et al. 2017).
One underexplored cause of cutaneous IFN production is microbial dysbiosis. The skin is home to many commensals that provide a living obstruction to colonization by harmful organisms (Grice and Segre 2018; Kong 2011). Staphylococcus aureus, unlike other members of the genus Staphylococcus, is a relatively minor colonizer of the skin and is involved in the pathogenesis of skin-associated diseases such as atopic dermatitis (AD) (Kong 2011; Nakatsuji et al. 2016; Williams and Gallo 2017). S. aureus colonization precedes clinical onset of AD and contributes to the severity of the disease (Meylan et al. 2017; Nakatsuji et al. 2016; Williams and Gallo 2017). We have demonstrated that production of IFNs follows colonization of mice by S. aureus and that S. aureus peptidoglycan induces production of IFNκ in keratinocytes (Stannard et al. 2017; Syed et al. 2015). In addition, disruption of the epithelial barrier, which is important in promotion of S. aureus colonization (Wanke et al. 2013), is able to drive lupus disease activity (Clark et al. 2015). Moreover, S. aureus is the leading cause of bacteremia in lupus patients and its carriage may be associated with disease flares and development of lupus nephritis (Chen et al. 2008; Conti et al. 2016; Hajialilo et al. 2015). Reflecting the importance of S. aureus-driven immune activation, repeated injections of S. aureus superantigen in wild type mice results in the development of a disease that mimics lupus (Chowdhary et al. 2012). Overall, investigations into S. aureus colonization in SLE patients have been limited, and whether IFNs impact colonization by S. aureus is unknown.
In this paper we investigate the role of IFNs in regulation of the colonization by S. aureus. We show that SLE patients are frequently colonized by S. aureus on their rashes. We also demonstrate that exposure to type I IFNs increases S. aureus adherence and that SLE keratinocytes exhibit greater barrier disruption and S. aureus adhesion when compared to matched healthy controls (HC). This suggests that dysregulation of type I IFNs in SLE could lead to a feed-forward loop resulting in greater S. aureus colonization that in turn leads to inflammation and additional production of IFNs.
RESULTS
SLE-associated skin lesions are colonized with S. aureus at a high rate
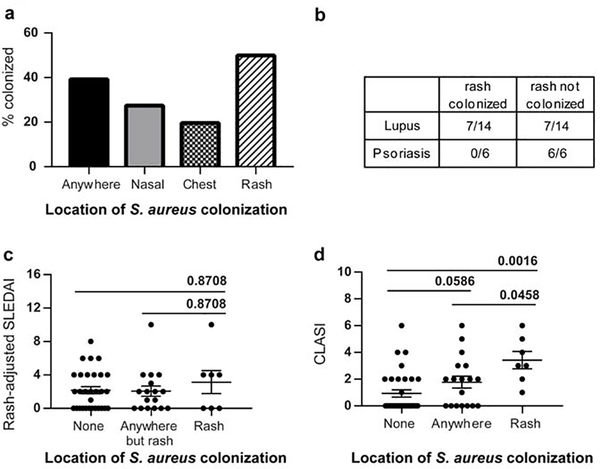
We wished to investigate the colonization frequency of members of the lupus cohort at the University of Michigan (see Table S1 for demographics). Patients (n=54) were tested for S. aureus in their nares, on their chest and on any lupus-related skin lesions. SLE patients were colonized by S. aureus at a rate higher (~40%) than that reported in healthy adults (~30%) and the rate increased further when active skin lesions were sampled (50%) (Fig. 1A). In comparison, psoriatic plaques, which are characterized by a mixed Th1/Th17 signature and low IFN levels (Baliwag et al. 2015), did not exhibit colonization on lesional skin. The colonization rates of SLE lesions and psoriasis thus differed significantly (Fisher’s Exact test; p<0.0001) (Fig. 1B). We next investigated the influence of S. aureus colonization on disease activity. No association between rash-adjusted SLE Disease Activity Index (SLEDAI) scores and colonization by S. aureus were noted (Fig. 1C). CLASI (Cutaneous Lupus Disease Area and Severity Index) is a validated score for severity of active cutaneous involvement in lupus (Robinson and Werth 2015). CLASI activity scores trended higher among colonized individuals (q=0.0586). In addition, significant increases in the CLASI scores were observed when uncolonized and rash colonized patients were compared (q=0.0016). Importantly, CLASI activity scores were also higher when patients with colonized skin lesions were compared to SLE patients colonized in other locations besides the rashes (q=0.0458) (Fig. 1D). These data suggest that either the presence of S. aureus on the rash contributes to higher cutaneous disease activity or the disease activity contributes to an environment conducive to colonization by S. aureus.
Fig.1. S. aureus colonization in SLE patients.
A. Graph represents percent of SLE patients (n=54) with positive colonization by S. aureus at indicated locations. Positive colonization was determined by mauve colonies on Chromagar after 24 hrs incubation followed by confirmation by PCR as outlined in Methods. B. S. aureus colonization rate in SLE is significantly different from the same in psoriasis. Lesions in SLE and psoriasis were swabbed and cultured and checked for S. aureus colonization. Number of samples (rashes) identified as S. aureus positive are presented as a fraction of the total number of samples that were tested. C. Colonization with S. aureus on rashes with respect to systemic disease activity. SLEDAI scores were obtained from all patients who were sampled for S. aureus colonization. Data represents the rash-adjusted SLEDAI scores (see methods) plotted against the colonization status of patients ± SEM. D. Colonization with S. aureus is associated with higher cutaneous disease activity. CLASI scores of patients were obtained for all the patients sampled for S. aureus colonization. Data represents CLASI scores graphed based on their colonization status ± SEM. Data were analyzed by ANOVA on ranks and post-hoc testing using Kruskal-Wallis test was performed using Prism and the computed q values are presented.
SLE lesional skin demonstrates lower barrier gene expression
Loss of function mutations and misregulation of filaggrin (FLG) result in impaired barrier integrity and are important in AD pathogenesis (Nakatsuji et al. 2016). However, barrier gene expression in CLE is not well understood. Thus, we compared normalized data from microarray analysis of SLE lesional skin vs. HC skin (Gene Expression Omnibus (GEO); accession number GSE81071) (Berthier et al. 2019). Expectedly, IFN stimulated genes and IFNs including IFNκ, a regulator in keratinocytes for type I responses, were significantly elevated in cutaneous lupus lesions (1.53-fold change; q=0.0006). Importantly, barrier genes such as loricrin (LOR) and claudin (CLDN1) and transglutaminase (TGM5) were downregulated (Table 1). This suggests that the high IFN environment may downregulate the epidermal barrier, which could promote S. aureus colonization.
Table 1.
Genes misregulated in CLE lesional skin obtained from microarray data analysis.
| Function | Symbol | Description | FC | q-value |
|---|---|---|---|---|
| Barrier genes | LOR | loricrin | 0.79 | 0.0401 |
| CLDN1 | claudin 1 | 0.57 | 0.0007 | |
| CLDN11 | claudin 11 | 0.78 | 0.0401 | |
| TGM5 | Transglutaminase 5 | 0.50 | 0.0000 | |
| KLK1 | kallikrein 1 | 0.66 | 0.0011 | |
| KLK11 | kallikrein 11 | 0.55 | 0.0000 | |
| FLG | filaggrin | 0.95 | 0.3028 | |
| FLG2 | filaggrin family member 2 | 0.77 | 0.2067 | |
| IVL | involucrin | 2.09 | 0.0121 | |
| ITGA5 | integrin subunit alpha 5 | 1.02 | 0.5746 | |
| ITGB1 | integrin subunit beta 1 | 1.04 | 0.5433 | |
| Interferon response genes | IFI44 | interferon induced protein 44 | 17.23 | 0.0000 |
| IFIT1 | interferon induced protein with tetratricopeptide repeats 1 | 5.9 | 0.0000 | |
| Interferon genes | MX1 | MX dynamin like GTPase 1 | 8.67 | 0.0000 |
| OASL | 2’–5’-oligoadenylate synthetase like | 2.35 | 0.0000 | |
| IFNA10 | interferon alpha 10 | 1.7 | 0.0149 | |
| IFNG | interferon gamma | 1.36 | 0.0488 | |
| IFNK | interferon kappa | 1.53 | 0.0006 | |
Red - Upregulation; Blue - Downregulation; q value <0.05 considered significant
Exposure to IFNs results in diminished barrier gene expression
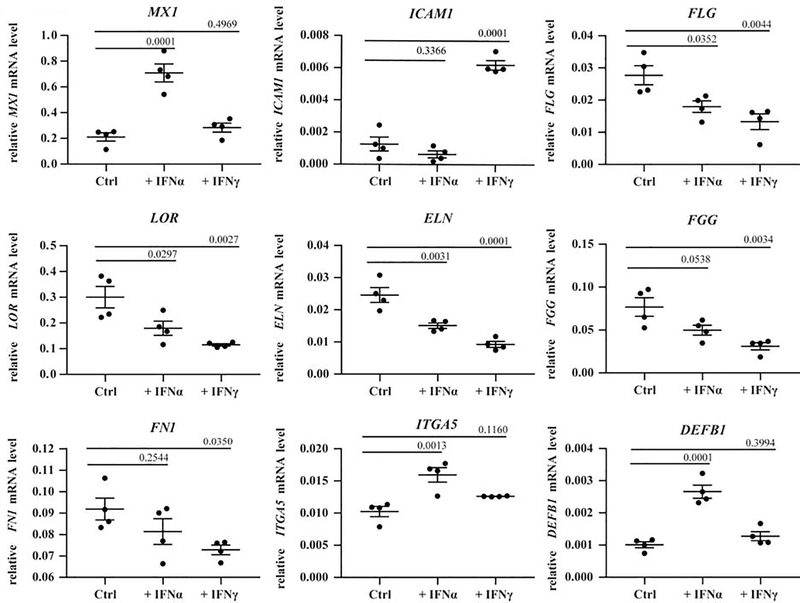
We thus next chose to determine whether the expression of barrier proteins was modulated by IFN exposure in N/TERTs, a human keratinocyte cell line. Following exposure to IFNα or IFNγ for 6 hours, RNA was isolated and gene expression was quantified via real-time PCR. As expected, IFNα treatment resulted in high expression of the transcriptional regulator MX1, and IFNγ treated cells showed high expression of the intercellular adhesion molecule (ICAM1) demonstrating that IFNs utilized for treatment were functional (Fig. 2). Numerous genes involved in the formation of the cornified envelope including FLG, loricrin (LOR), and elastin (ELN), were found to be significantly repressed while tight junction proteins such as desmoglein (DSG1) and FLG2 trended towards decreased expression following IFN exposure (Fig. 2; Fig. S3). Other extracellular matrix molecules such as fibrinogen (FGG) and fibronectin (FN1), which are known to interact with S. aureus, demonstrated reduced expression particularly in the presence of IFNγ. However, α-integrin (ITGA5) was upregulated in IFNα treated (but not IFNγ treated) N/TERTs. Keratinocytes respond to microbial surface molecules (MAMPs) by the production of antimicrobial peptides including β-defensins (DEFB1). IFNα treatment, but not IFNγ, increased DEFB1 expression (Fig. 2). These data suggest that IFN treatment of keratinocytes represses production of proteins that contribute to the integrity of the cornified layer, thus compromising the epithelial barrier while leaving the defensive competences intact.
Fig. 2. Interferon exposure leads to inhibition of several barrier-related genes.
N/TERTs were treated with 1000 U/ml of IFNα and IFNγ for 6 hours followed by expression analysis via qRT-PCR. Graphs represent relative expression to β-actin. Results are from 4 individual experiments ± SEM. Data were analyzed by ANOVA in Prism and the corresponding p values are reported.
IFN exposure increased S. aureus adherence in keratinocytes
Given that IFNs downregulate barrier proteins and upregulate integrins (involved in bacterial adherence) (Clarke and Foster 2006), we next investigated whether IFNs modulated adherence of S. aureus to the N/TERT keratinocyte cell line. Confluent N/TERTs were exposed to washed log phase S. aureus for increasing periods of time to determine the kinetics of S. aureus adhesion with N/TERTs. S. aureus adherence occurred rapidly with ~1.3 X 104 S. aureus CFUs recovered at 30 min. Time of exposure demonstrated a linear relationship with S. aureus adherence with ~1.3 X 105 CFUs recovered after 90 min (Fig. S1A). We also performed assays with gentamicin exposure to determine invasion kinetics since S. aureus is reported to invade keratinocytes and reside within for varying periods of time (Edwards et al. 2011; Garzoni and Kelley 2009; Löffler et al. 2014). The results indicated the same trend observed in the adhesion assays although the number of recovered CFUs were, expectedly, at least two orders of magnitude lower than that observed during adhesion assays (Fig. S1B). These trends were also observed with stationary phase S. aureus (data not shown).
We then determined whether exposure to IFNα or IFNγ influenced S. aureus-keratinocyte interactions. Confluent N/TERTs were treated with 1000 U/ml of either IFNα or IFNγ for 24 hours and then exposed to S. aureus. Consistent with a role for type I IFNs in promoting S. aureus colonization, greater numbers of CFUs were recovered from N/TERTs exposed to IFNα as observed in Fig. 3A (p=0.0397). IFNγ treatment did not promote a significant increase in adherence in comparison to the untreated N/TERTs. Secondly, paired analysis performed on data from multiple experiments also revealed a significant increase in adhesion to N/TERTs post exposure to IFNα (p=0.0056). These trends were replicated when stationary phase S. aureus was utilized (Fig. S2).
Fig 3. IFNα increases adhesion of S. aureus to WT N/TERTs.
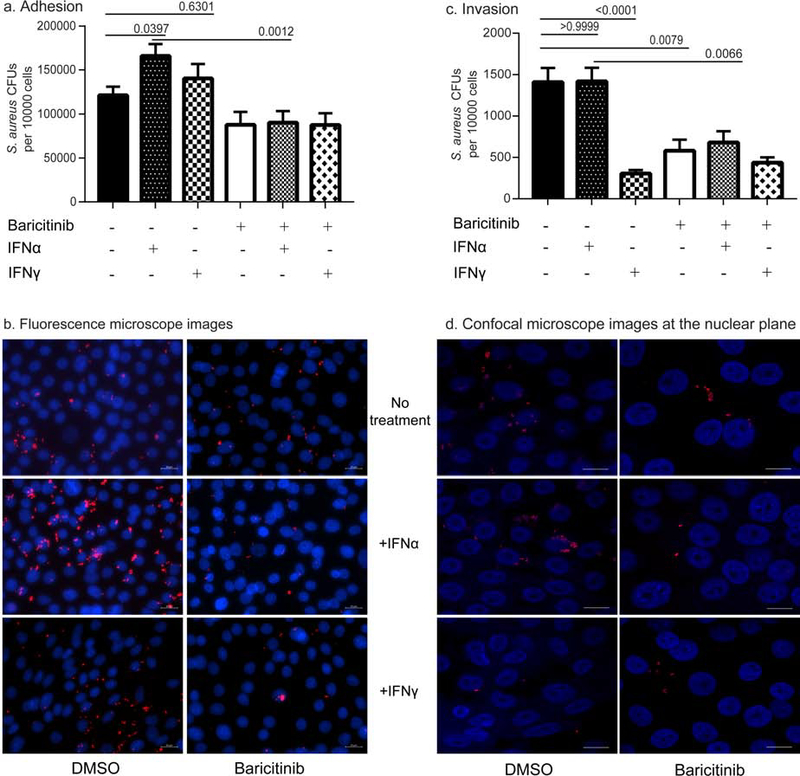
A. N/TERTs were grown to confluence with or without 1,000 U/ml of indicated IFNs. In some instances, N/TERTs were exposed to 10 μM Baricitinib for 60 min before the addition of IFNs. S. aureus adhesion assays were performed as described in methods, and results are presented as S. aureus CFUs recovered per 10,000 N/TERTs. Data presented are from at least three independent experiments ±SEM. Data were analyzed by ANOVA in Prism and significance was reported as computed p values. B. Fluorescence microscopy demonstrating higher S. aureus adhesion among IFN treated N/TERTs. Confluent N/TERTs ± Baricitinib and ± IFNs were treated with S. aureus USA300 expressing dsRed. Hoescht stain was utilized for staining nuclei and representative images from at least three individual experiments are presented. Scale bar-20 μm. C. Exposure to IFNs does not lead to an increase in invasive S. aureus CFUs. N/TERTs were grown to confluence with or without 1,000 U/ml of indicated IFNs. In some instances, N/TERTs were exposed to 10 μM Baricitinib for 60 min before the addition of IFNs. S. aureus invasion assays were performed as described in methods, and results are presented as S. aureus CFUs recovered per 10,000 N/TERTs. Data presented are from at least three independent experiments ±SEM. Data were analyzed by ANOVA in Prism and significance was reported as computed p values. D. Confocal microscopy displaying invasion of S. aureus within N/TERTs. Confluent N/TERTs ± Baricitinib and ± IFNs were treated with S. aureus USA300 expressing dsRed. Hoescht stain was utilized for staining nuclei and representative images obtained in the nuclear plane (0.5 μm sections) from at least three individual experiments are presented. Scale bar-60 μm.
To confirm that the effects on adhesion was due to IFNs, we used baricitinib, a small molecule inhibitor of Janus kinase that blocks signaling of type I and type II IFN receptors (Howell et al. 2018; Kontzias et al. 2012). Preincubation of N/TERTs with baricitinib (10μM) did not change basal rates of S. aureus adherence but blocked increased adhesion induced by IFN exposure (Fig. 3A). To confirm our data from counts, N/TERTs ± IFNs and ± baricitinib exposed to fluorescently labeled S. aureus were evaluated by fluorescence microscopy. As shown in Figure 3B, an increase in S. aureus adhesion was noted with IFNα treatment and this was blocked by the addition of baricitinib. Together, these data indicate that IFNs, and type I IFNs in particular, promote adhesion of S. aureus to keratinocytes.
S. aureus invasion is not affected by IFNα and is actively inhibited by IFNγ
Keratinocytes are non-professional phagocytes that allow S. aureus entry by a fibronectin, integrin and cytoskeleton rearrangement mediated mechanism (Edwards et al. 2011; Löffler et al. 2014). We next examined whether IFN exposure also resulted in higher invasion into N/TERT monolayers. Confluent N/TERTs were exposed to IFNs and invasion assays were performed. IFNα treatment did not result in higher recovered CFUs. In stark contrast, IFNγ strongly repressed S. aureus invasion (p<0.0001) indicating a protective function for type II IFNs in N/TERTs (Fig. 3C). Surprisingly, treatment with baricitinib alone also resulted in a significant decrease in S. aureus invasion. To confirm the invasion data, confocal microscopy was performed on N/TERTs exposed to fluorescent S. aureus. Representative images at the z-planes (0.5 μm sections) of the DAPI labeled nuclei of N/TERT demonstrate identifiable organisms within the cell as evidenced by the presence of fluorescent bacteria in close proximity to nuclei (Fig. 3D) and by counts which reflect the data from the invasion assays.
Keratinocytes from SLE patients demonstrate low expression of barrier-related genes and increased adherence to S. aureus
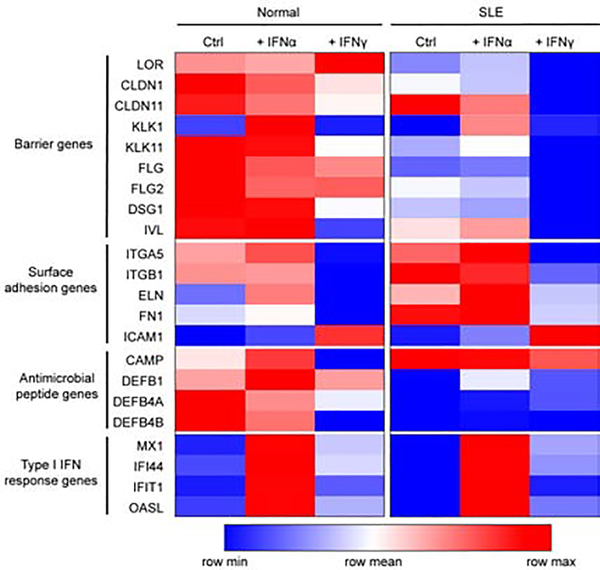
Given that IFNs downregulate barrier genes and non-lesional keratinocytes from SLE patients express elevated type I IFNs (IFNκ) at baseline (Sarkar et al. 2018), we next explored whether barrier genes were differentially expressed in non-lesional SLE vs. control keratinocytes (HC). To this end, keratinocytes were obtained from non-lesional skin and subjected to RNA-seq analysis as reported previously (Tsoi et al. 2019). Barrier genes such as FLG, LOR, IVL, CLDN1 as well as antimicrobial peptides (DEFB) were repressed in SLE keratinocytes in comparison to HC keratinocytes (Fig. 4). In contrast, genes involved in adhesion such as FN1, ITGA5 and ITGB1 were elevated in SLE keratinocytes and even more-so after IFNα treatment. Thus, these data indicate that SLE keratinocytes exhibit impaired barrier formation at baseline and may be more prone to bacterial adhesion.
Fig 4. SLE keratinocytes exhibit lower barrier gene expression.
Heatmap of selected genes from primary control and SLE keratinocytes treated for 6 hours with indicated IFNs. Gene expression was measured via RNAseq. The largest values are displayed as the reddest, the smallest values are displayed as the bluest, and intermediate values are a lighter color of either blue or red.
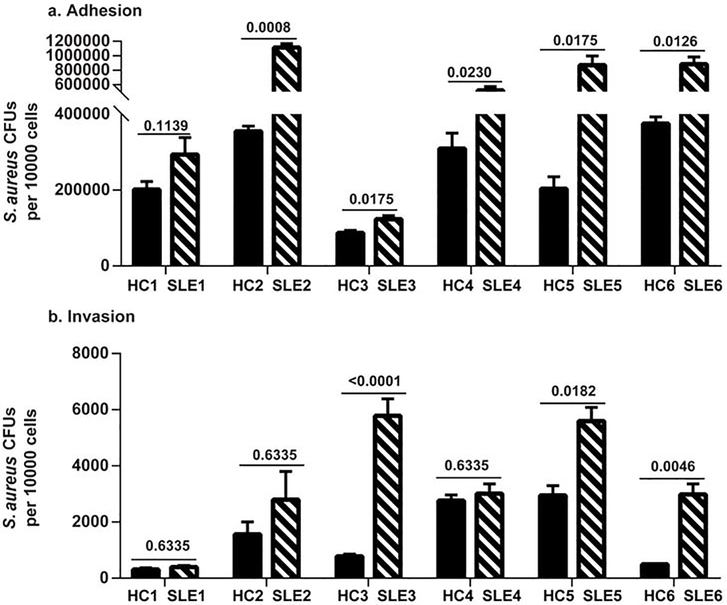
We then utilized non-lesional primary keratinocyte cultures to determine if S. aureus adhesion was increased in SLE patients. 6 sets of age and gender-matched combinations of SLE and HC keratinocytes were used for adhesion and invasion assays. As demonstrated in Fig. 5A, S. aureus demonstrated significantly greater adherence to the majority (5/6) of SLE keratinocytes in comparison to HC keratinocytes. In addition, higher adhered S. aureus CFUs were recovered from both SLE and HC keratinocytes exposed to IFNα (Fig. S4). Some, but not all, SLE keratinocytes demonstrated increased invasion (Fig. 5B). Together, these data suggest that higher colonization of SLE patients is likely due in part to diminished barrier functions and chronic type I IFNs that drive increased adherence.
Fig 5. S. aureus demonstrated greater adherence to SLE keratinocytes compared to HC keratinocytes.
Keratinocytes from non-lesional skin from SLE patients and matched healthy controls (HC) were grown to confluence and exposed for 90 min to washed log phase S. aureus AH1263. S. aureus adhesion (A) and invasion (B) assays were performed as described in methods, and results are presented as S. aureus CFUs recovered per 10,000 keratinocytes. Results are presented as means from experiments performed at least in triplicate from each patient and paired control ± SEM (SLE n=6, HC n=6). Student’s t test was performed on each matched pair and the p values are denoted.
DISCUSSION
Factors that contribute to a propensity for skin inflammation in SLE patients are not well-defined. Here, we demonstrate that cutaneous lesions of SLE patients are characterized by depressed barrier proteins and increased colonization by S. aureus. We also show that type I IFNs can repress barrier gene expression and increase adherence of S. aureus, which serves as a mechanism for our observation that SLE keratinocytes have increased adherence to S. aureus when compared to HCs.
Our study identified colonization rates among SLE patients similar to previous reports by Conti et al. (~21 %) and Hajialilo et al. (~48%) (Conti et al. 2016; Hajialilo et al. 2015). When their strategies for S. aureus identification were examined, it was noted that the latter group utilized mannitol salt agar for growth and did not employ secondary confirmatory strategies. This could have yielded false positive results from mannitol fermentation positive staphylococci (Sirobhushanam et al. 2019). In our study, we utilized Chromagar to identify potentially positive colonies followed by both mannitol salt and PCR confirmation of strains. Despite our rigorous methodology, we determined that SLE lesional skin samples were highly colonized (50%) and demonstrated association with disease activity as evidenced by the higher CLASI scores in colonized patients. These results are in line with AD results, which demonstrate higher colonization on affected skin (Gong et al. 2006). We did not find similar high rates of colonization on psoriasis rashes, which suggests that specific skin lesions may attract and interact with S. aureus. One caveat to consider here is that SLE patients were typically on immunosuppressive and antimalarial medications, which potentially may influence colonization. Further studies will help to delineate this.
Analysis of adhesion and invasion kinetics revealed a linear relationship with time although we did not observe threshold values for adherent S. aureus as had been reported by other groups. This could be due to differences in properties unique to cell lines that are currently unknown (HaCat Vs N/TERTs) (Edwards et al. 2011). Our inquiry into the impact of IFNs on S. aureus colonization showed that N/TERTs exposed to IFNα exhibited increased S. aureus adhesion. Higher expression of ITGA5, a component of the integrin α5β1 known to be involved in S. aureus adhesion and invasion, could be a likely mechanism for promotion of adherence by type I IFNs (Foster et al. 2013). Defense capabilities of N/TERTs (DEFB1 expression) were not compromised by exposure to IFNα suggesting that colonization does not necessarily imply infection.
The effects of IFNs on barrier proteins are likely multi-faceted. Components of the cornified envelope (FLG, FLG2, LOR, IVL) and tight junction components (DSG1) showed lower expression following IFN exposure, indicating a negative impact to the epithelial barrier. Also, barrier genes were repressed both at baseline and in the presence of IFNs in SLE keratinocytes. LOR, IVL, DSG1 and FLG2 are also necessary in the formation of corneocytes and stacked layers of lipid lamellae, critical to the formation of an intact barrier. Formation of an impaired barrier is known to result in penetration of S. aureus into the deeper layers of the skin in AD (Nakatsuji et al. 2016). Furthermore, protective function of commensal microbiota is reversed upon damage to the epidermal barrier (Burian et al. 2017). Loss of filaggrin may have several impacts: natural moisturizing factor is formed via breakdown of filaggrin and lowers skin pH; S. aureus can proliferate rapidly with small increases in pH due to low filaggrin (Cabanillas and Novak 2016; Miajlovic et al. 2010). Also, filaggrin loss has been reported to result in higher photosensitivity (Mildner et al. 2010). It could thus be hypothesized that in addition to promoting S. aureus colonization, barrier compromise due to prolonged IFN exposure leads to inflammatory responses to commensals or other triggers such as UV light.
Treatment with IFNγ also resulted in downregulation of barrier components. Similar results were obtained when neonatal foreskin keratinocytes were exposed to IFNγ leading to significant damage to the epidermal barrier (Banno et al. 2003). Lower FLG expression upon exposure to IFNγ has been reported previously while Noh et al also report the increase in filaggrin protein content upon such exposure (Hvid et al. 2011; Noh et al. 2010). IFNγ exposure leading to low invasion of S. aureus is likely due to the significantly lower expression of surface molecules thus reducing interaction with S. aureus. Also, higher expression of ICAM1 upon exposure to IFNγ likely leads to infiltration into the epidermis by neutrophils contributing to damage (Dustin et al. 1988). Further study of the protective effects of IFNγ are warranted.
SLE keratinocytes, when compared to HC keratinocytes, show higher S. aureus adhesion, supporting our colonization data. Our studies on S. aureus invasion into keratinocytes demonstrated significance in a subset of data, but this was not a defining feature of type I IFN exposure. IFNγ exposure protected against invasion; given that IFNγ did not result in upregulation of ITGA5, these data potentially identify an important target for prevention of invasion. Colonization and disease activity data gathered from longitudinal studies could yield a clearer picture of the role of the skin microbiome in disease progression, including whether there are links to atopy presenting in patients with SLE. We hypothesize that S. aureus colonization could be a part of a feed forward loop where chronic IFN dysregulation in SLE promotes colonization by S. aureus which then induces the production of inflammatory cytokines by keratinocytes that further increase colonization. This could lead not only to long term colonization but may also impact systemic disease development. Addressing skin inflammation by further investigations into barrier restoration and its role in S. aureus colonization and skin could illuminate novel treatment strategies.
In summary, we demonstrate increased colonization in CLE lesions in SLE patients with S. aureus and an increased propensity for SLE keratinocytes to adhere to S. aureus. This is promoted by chronic type I (and possibly type II) IFN exposure through dysregulation of barrier proteins and upregulation of adhesion molecules.
MATERIALS AND METHODS
Human subjects
All patients and healthy control subjects gave written informed consent according to the guidelines of the Declaration of Helsinki. The study protocol was approved by the Institutional Review Board of the University of Michigan Medical School. Patients with SLE fulfilled the criteria for diagnosis (American College of Rheumatology) (Marc C. Hochberg 1997) and were recruited from the University of Michigan Lupus Cohort. Skin biopsies for keratinocyte isolation were obtained from non-lesional skin of the upper thigh. SLEDAI and CLASI scores of the subjects were also recorded for each subject. In order to avoid skewing of the SLEDAI in patients with active rashes (which would overestimate the significance of rash colonization), we adjusted the SLEDAI scores to remove the rash related scores in all subjects.
Cell culture
Primary keratinocytes were isolated and cultured from biopsies as previously described (Aasen and Belmonte 2010; Stannard et al. 2017). In brief, keratinocytes were grown in serum free growth media (Epilife, Cascade Biologics) supplemented with 1% pen-strep (100 U/mL penicillin and 100 μg/mL of streptomycin; Gibco, MD) and 0.25 μg/mL of amphotericin B (Fungizone; Gibco, MD) and human keratinocyte grown serum containing BPE, bovine insulin, hydrocortisone, bovine transferrin, human epidermal growth factor, and 0.1 mM Ca2+. For N/TERTs, see supplementary information.
S. aureus colonization analysis
Patients were swabbed in their nares and on their chest and any lupus-related lesions present with sterile FLOQSwabs (Copan Diagnostics, Murrieta, CA) moistened with sterile PBS. Demographic data as well as associated serological information on the subjects are summarized in Table. S1. 100 μl of vortexed sample was plated on ChromAgar (BD-BBL, Franklin Lakes, NJ) and incubated at 35 ± 2°C for 24 hours. Mauve colored colonies, indicating growth of S. aureus, were picked for further analysis. See supplementary information for S. aureus confirmatory strategies.
Adhesion and invasion assays
Cell adhesion and invasion assays were performed as outlined previously (Edwards et al. 2011). Briefly, N/TERTs were seeded into 24- well plates and grown to confluence. N/TERTs were treated with IFNs at 1000U/mL for 24 hours +/− Baricitinib (10μM) or DMSO as control in DMEM supplemented with 2% FBS (to promote keratinocyte differentiation) (Kontzias et al. 2012). 1:100 dilutions of overnight cultures of S. aureus were grown to mid log phase in tryptic soy broth (~2.5 hours) and washed prior to use. N/TERTs were washed in sterile PBS before adding S. aureus (~20 −30 MOI) suspended in KGM for 90 min. Nonspecifically adhered S. aureus were removed by washing and N/TERTs liberated with 0.25% trypsin. N/TERTs were then quantified in each well via hemocytometer, lysed with 1% Triton X100, serially diluted, and plated on tryptic soy agar (TSA) to determine CFUs. For invasion assays, keratinocytes were treated with KGM containing 200 μg/ml Gentamicin for 60 min in order to kill surface associated S. aureus followed by washing with PBS and CFUs quantification.
See supplementary information for other methods.
Data Availability
Datasets related to the article can be found at GEO (https://www.ncbi.nlm.nih.gov/geo/). RNA-seq data is available in GEO accession number GSE124939; Microarray data from CLE lesional skin is available in GEO accession number GSE81071.
Supplementary Material
Fig S1. A. S. aureus adhesion to WT N/TERTs increases with time of exposure. N/TERTs were grown to confluence and treated for 24 hours with DMEM +2% FBS. S. aureus adhesion assays were performed as described in methods and results were presented as S. aureus CFUs recovered per 10,000 N/TERTs. Data presented are from at least 4 replicates from three independent experiments ±SEM. B. S. aureus invasion to WT N/TERTs increases with time of exposure. N/TERTs were grown to confluence and treated for 24 hours with DMEM +2% FBS. S. aureus invasion assays were performed as described in methods and results were presented as S. aureus CFUs recovered per 10,000 N/TERTs. Data presented are from at least 4 replicates from three independent experiments ±SEM.
Fig. S2. N/TERTs exposed to IFNα allow greater adherence of stationary phase S. aureus. S. aureus was grown overnight and washed cells were allowed to adhere to washed N/TERTs grown to confluence. Data were analyzed by ANOVA using Prism and p values were reported as obtained from posthoc testing.
Fig. S3. Interferon exposure leads to inhibition of several barrier-related genes. N/TERTs were treated with 1000 U/ml of IFNα and IFNγ for 6 hours followed by expression analysis via qRT-PCR. Graphs represent relative expression to β-actin. Results are from 4 individual experiments ± SEM. Data were analyzed by ANOVA in Prism and the corresponding p values are reported.
Fig. S4. HC and SLE keratinocytes demonstrate greater adherence to S. aureus when exposed to IFNα. Keratinocytes were grown to confluence with or without 1,000 U/ml of indicated IFNs. S. aureus adhesion assays were performed as described in methods, and results are presented as S. aureus CFUs recovered per 10,000 keratinocytes. Blue represents HC and green represents SLE keratinocytes. Graphs represent means of three independent experiments for four SLE and three HC keratinocyte cultures.
ACKNOWLEDGEMENTS
This work was supported in part by the Physician Scientist Development award from the Doris Duke Foundation (PI: JMK), R01 AR071384 and R03 AR-066337-01-A1 from the National Institutes of Health National Institute of Arthritis and Musculoskeletal and Skin disease (PI: JMK). This work utilized core services of the University of Michigan Skin Biology and Diseases Resources-based Center (1P30AR075043) and the Applied Systems Biology Core in the O’Brien Renal Center (P30 DK081943). ARH was supported by NIH grant AI133089 and a Merit Award (BX002711) from the Department of Veteran Affairs. NP was partially supported by the U.S. Department of Veterans Affairs. We thank the patients from the Michigan Lupus Cohort for generously sharing their samples for our work. We also thank the University of Michigan Sequencing Core facility and University of Michigan Microscopy Core facility for their equipment and support which enabled us to complete this study.
CONFLICT OF INTEREST
J.E.G. received research funding from AbbVie, SunPharma, Celgene, and Genentech and serves on advisory boards for Novartis, AbbVie, and MiRagen and J.M.K. received research funding from Celgene and serves on advisory boards for AstraZeneca, Boehringer Ingelheim, Bristol Myers Squibb, and Eli Lilly. The other authors have no financial conflicts of interest.
ABBREVIATIONS USED
- IFN
Interferon
- SLE
Systemic Lupus Erythematosus
- CLE
Cutaneous Lupus Erythematosus
- HC
Healthy Control
Footnotes
CRediT STATEMENT
Conceptualization: SS, NP, JEG, JMK
Data Curation: SS, CCB, LCT
Formal Analysis: SS, CCB, LCT, JMK
Funding Acquisition: JMK
Investigation: SS, NP, TJR, GAH, JB
Methodology: SS, NP, MKS, ARH, JEG, JMK
Project Administration: JMK
Resources: MKS, GAH, CD, ARH, JEG
Supervision: JEG, JMK
Writing-original draft: SS, JMK
Writing-review and editing: SS, NP, TJR, CCB, MKS, GAH, LCT, JB, CD, ARH, JEG, JMK
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- Aasen T, Belmonte JCI. Isolation and cultivation of human keratinocytes from skin or plucked hair for the generation of induced pluripotent stem cells. Nat. Protoc 2010;5(2):371–82 [DOI] [PubMed] [Google Scholar]
- Baliwag J, Barnes DH, Johnston A. Cytokines in psoriasis. Cytokine. Elsevier Ltd; 2015;73(2):342–50 Available from: 10.1016/j.cyto.2014.12.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banno T, Adachi M, Mukkamala L, Blumenberg M. Unique keratinocyte-specific effects of interferon-γ that protect skin from viruses, identified using transcriptional profiling. Antivir. Ther 2003;8(6):541–54 [PubMed] [Google Scholar]
- Berthier CC, Tsoi LC, Reed TJ, Stannard JN, Myers EM, Namas R, et al. Molecular Profiling of Cutaneous Lupus Lesions Identifies Subgroups Distinct from Clinical Phenotypes. J. Clin. Med 2019;8(1244):1–15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burian M, Bitschar K, Dylus B, Peschel A, Schittek B. The Protective Effect of Microbiota on S. aureus Skin Colonization Depends on the Integrity of the Epithelial Barrier. J. Invest. Dermatol 2017;137(4):976–9 [DOI] [PubMed] [Google Scholar]
- Cabanillas B, Novak N. Atopic dermatitis and filaggrin. Curr. Opin. Immunol Elsevier Ltd; 2016;42(Figure 1):1–8 Available from: 10.1016/j.coi.2016.05.002 [DOI] [PubMed] [Google Scholar]
- Chasset F, Arnaud L. Targeting interferons and their pathways in systemic lupus erythematosus. Autoimmun. Rev 2018;17(1):44–52 [DOI] [PubMed] [Google Scholar]
- Chen MJ, Tseng HM, Huang YL, Hsu WN, Yeh KW, Wu TL, et al. Long-term outcome and short-term survival of patients with systemic lupus erythematosus after bacteraemia episodes: 6-yr follow-up. Rheumatology. 2008;47(9):1352–7 [DOI] [PubMed] [Google Scholar]
- Chowdhary VR, Tilahun AY, Clark CR, Grande JP, Rajagopalan G. Chronic Exposure to Staphylococcal Superantigen Elicits a Systemic Inflammatory Disease Mimicking Lupus. J. Immunol 2012;189(4):2054–62 Available from: http://www.jimmunol.org/cgi/doi/10.4049/jimmunol.1201097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark KL, Reed TJ, Wolf SJ, Lowe L, Hodgin JB, Kahlenberg JM. Epidermal injury promotes nephritis flare in lupus-prone mice. J. Autoimmun Elsevier Ltd; 2015;65:38–48 Available from: 10.1016/j.jaut.2015.08.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clarke SR, Foster SJ. Surface Adhesins of Staphylococcus aureus. Adv. Microb. Physiol 2006. [DOI] [PubMed] [Google Scholar]
- Conti F, Ceccarelli F, Iaiani G, Perricone C, Giordano A, Amori L, et al. Association between Staphylococcus aureus nasal carriage and disease phenotype in patients affected by systemic lupus erythematosus. Arthritis Res. Ther. Arthritis Research & Therapy; 2016;18(1):177 Available from: http://arthritis-research.biomedcentral.com/articles/10.1186/s13075-016-1079-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dustin ML, Singer KH, Tuck DT, Springer TA. Adhesion of T lymphoblasts to epidermal keratinocytes is regulated by interferon γ and is mediated by intercellular adhesion molecule 1 (ICAM1). J. Exp. Med 1988;167(April):1323–40 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edwards AM, Potter U, Meenan NAG, Potts JR, Massey RC. Staphylococcus aureus keratinocyte invasion is dependent upon multiple high-affinity fibronectin-binding repeats within FnBPA. PLoS One. 2011;6(4):e18899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foster TJ, Geoghegan JA, Ganesh VK, Höök M. Adhesion, invasion and evasion: the many functions of the surface proteins of Staphylococcus aureus. Nat. Rev. Microbiol Nature Publishing Group; 2013;12(1):49–62 Available from: http://www.nature.com/doifinder/10.1038/nrmicro3161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garzoni C, Kelley WL. Staphylococcus aureus: new evidence for intracellular persistence. Trends Microbiol. 2009;17(2):59–65 [DOI] [PubMed] [Google Scholar]
- Gong JQ, Lin L, Lin T, Hao F, Zeng FQ, Bi ZG, et al. Skin colonization by Staphylococcus aureus in patients with eczema and atopic dermatitis and relevant combined topical therapy: a double-blind multicentre randomized controlled trial. Br. J. Dermatol 2006;155(4):680–7 Available from: http://www.ncbi.nlm.nih.gov/pubmed/16965415%5Cnhttp://doi.wiley.com/10.1111/j.1365-2133.2006.07410.x [DOI] [PubMed] [Google Scholar]
- Grice EA, Segre JA. The human skin microbiome. Nat. Rev. Microbiol 2018;16(3):143–55 [DOI] [PubMed] [Google Scholar]
- Hajialilo M, Ghorbanihaghjo A, Khabbazi A, Valizadeh H, Raeisi S, Hasani A, et al. Nasal carriage rate of Staphylococcus aureus among patients with systemic lupus erythematosus and its correlation with disease relapse. Egypt. Rheumatol. Egyptian Society for Joint Diseases and Arthritis; 2015;37(2):81–4 Available from: 10.1016/j.ejr.2014.06.006 [DOI] [Google Scholar]
- Howell MD, Fitzsimons C, Smith P. JAK/STAT inhibitors and other small molecule cytokine antagonists for the treatment of allergic disease. Ann. Allergy. Asthma Immunol Elsevier Inc.; 2018; Available from: http://linkinghub.elsevier.com/retrieve/pii/S1081120618301212%0Ahttp://www.ncbi.nlm.nih.gov/pubmed/29454096 [DOI] [PubMed]
- Hvid M, Johansen C, Deleuran B, Kemp K, Deleuran M, Vestergaard C. Regulation of caspase 14 expression in keratinocytes by inflammatory cytokines - a possible link between reduced skin barrier function and inflammation? Exp. Dermatol 2011;20(8):633–6 [DOI] [PubMed] [Google Scholar]
- Kong HH. Skin microbiome: Genomics-based insights into the diversity and role of skin microbes. Trends Mol. Med Elsevier Ltd; 2011;17(6):320–8 Available from: 10.1016/j.molmed.2011.01.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kontzias A, Kotlyar A, Laurence A, Changelian P, O’Shea JJ. Jakinibs: A new class of kinase inhibitors in cancer and autoimmune disease. Curr. Opin. Pharmacol Elsevier Ltd; 2012;12(4):464–70 Available from: 10.1016/j.coph.2012.06.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Löffler B, Tuchscherr L, Niemann S, Peters G. Staphylococcus aureus persistence in non-professional phagocytes. Int. J. Med. Microbiol Elsevier GmbH.; 2014;304(2):170–6 Available from: 10.1016/j.ijmm.2013.11.011 [DOI] [PubMed] [Google Scholar]
- Hochberg Marc C.. Updating the American College of Rheumatology Revised Criteria for the Classification of Systemic Lupus Erythematosus. Arthritis Rheum. 1997;40(9):1725–34 [DOI] [PubMed] [Google Scholar]
- Meller S, Winterberg F, Gilliet M, Müller A, Lauceviciute I, Rieker J, et al. Ultraviolet radiation-induced injury, chemokines, and leukocyte recruitment: An amplification cycle triggering cutaneous lupus erythematosus. Arthritis Rheum. 2005;52(5):1504–16 [DOI] [PubMed] [Google Scholar]
- Meylan P, Lang C, Mermoud S, Johannsen A, Norrenberg S, Hohl D, et al. Skin Colonization by Staphylococcus aureus Precedes the Clinical Diagnosis of Atopic Dermatitis in Infancy. J. Invest. Dermatol 2017;(October):2497–504 Available from: http://linkinghub.elsevier.com/retrieve/pii/S0022202X1732794X [DOI] [PubMed] [Google Scholar]
- Miajlovic H, Fallon PG, Irvine AD, Foster TJ. Effect of filaggrin breakdown products on growth of and protein expression by Staphylococcus aureus. J. Allergy Clin. Immunol Elsevier Ltd; 2010;126(6):1184–1190.e3 Available from: 10.1016/j.jaci.2010.09.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mildner M, Jin J, Eckhart L, Kezic S, Gruber F, Barresi C, et al. Knockdown of filaggrin impairs diffusion barrier function and increases UV sensitivity in a human skin model. J. Invest. Dermatol 2010;130(9):2286–94 [DOI] [PubMed] [Google Scholar]
- Muskardin TLW, Niewold TB. Type I interferon in rheumatic diseases. Nat. Rev. Rheumatol Nature Publishing Group; 2018;14(4):214–28 Available from: 10.1038/nrrheum.2018.31 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakatsuji T, Chen TH, Two AM, Chun KA, Narala S, Geha RS, et al. Staphylococcus aureus Exploits Epidermal Barrier Defects in Atopic Dermatitis to Trigger Cytokine Expression. J. Invest. Dermatol The Authors; 2016;136(11):2192–200 Available from: 10.1016/j.jid.2016.05.127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noh M, Yeo H, Ko J, Kim HK, Lee CH. MAP17 is associated with the T-helper cell cytokine-induced down-regulation of filaggrin transcription in human keratinocytes. Exp. Dermatol 2010;19(4):355–62 [DOI] [PubMed] [Google Scholar]
- Robinson ES, Werth VP. The role of cytokines in the pathogenesis of cutaneous lupus erythematosus. Cytokine. Elsevier Ltd; 2015;73(2):326–34 Available from: 10.1016/j.cyto.2015.01.031 [DOI] [PubMed] [Google Scholar]
- Sarkar MK, Hile GA, Tsoi LC, Xing X, Liu J, Liang Y, et al. Photosensitivity and type I IFN responses in cutaneous lupus are driven by epidermal-derived interferon kappa. Ann. Rheum. Dis 2018;1–12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sirobhushanam S, Parsa N, Reed TJ, Kahlenberg JM. Chromagar™ requires secondary confirmation strategies to minimize false positive/negative results for detection of Staphylococcus aureus. J. Microbiol. Methods Elsevier; 2019;161(April):71–3 Available from: 10.1016/j.mimet.2019.04.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stannard JN, Kahlenberg JM. Cutaneous lupus erythematosus: updates on pathogenesis and associations with systemic lupus. Curr. Opin. Rheumatol 2016;28(5):453–9 Available from: http://content.wkhealth.com/linkback/openurl?sid=WKPTLP:landingpage&an=00002281-201609000-00002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stannard JN, Reed TJ, Myers E, Lowe L, Sarkar MK, Xing X, et al. Lupus Skin Is Primed for IL-6 Inflammatory Responses through a Keratinocyte-Mediated Autocrine Type I Interferon Loop. J. Invest. Dermatol The Authors; 2017;137(1):115–22 Available from: 10.1016/j.jid.2016.09.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Syed AK, Reed TJ, Clark KL, Boles BR, Kahlenberg JM. Staphlyococcus aureus phenol-soluble modulins stimulate the release of proinflammatory cytokines from keratinocytes and are required for induction of skin inflammation. Infect. Immun 2015;83(9):3428–37 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsoi LC, Hile GA, Berthier CC, Sarkar MK, Reed TJ, Liu J, et al. Hypersensitive IFN Responses in Lupus Keratinocytes Reveal Key Mechanistic Determinants in Cutaneous Lupus. J. Immunol 2019;ji1800650 Available from: http://www.jimmunol.org/lookup/doi/10.4049/jimmunol.1800650 [DOI] [PMC free article] [PubMed]
- Wanke I, Skabytska Y, Kraft B, Peschel A, Biedermann T, Schittek B. Staphylococcus aureus skin colonization is promoted by barrier disruption and leads to local inflammation. Exp. Dermatol 2013;22(2):153–5 [DOI] [PubMed] [Google Scholar]
- Williams MR, Gallo RL. Evidence that Human Skin Microbiome Dysbiosis Promotes Atopic Dermatitis. J. Invest. Dermatol The Authors; 2017;137(12):2460–1 Available from: http://linkinghub.elsevier.com/retrieve/pii/S0022202X17329603 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig S1. A. S. aureus adhesion to WT N/TERTs increases with time of exposure. N/TERTs were grown to confluence and treated for 24 hours with DMEM +2% FBS. S. aureus adhesion assays were performed as described in methods and results were presented as S. aureus CFUs recovered per 10,000 N/TERTs. Data presented are from at least 4 replicates from three independent experiments ±SEM. B. S. aureus invasion to WT N/TERTs increases with time of exposure. N/TERTs were grown to confluence and treated for 24 hours with DMEM +2% FBS. S. aureus invasion assays were performed as described in methods and results were presented as S. aureus CFUs recovered per 10,000 N/TERTs. Data presented are from at least 4 replicates from three independent experiments ±SEM.
Fig. S2. N/TERTs exposed to IFNα allow greater adherence of stationary phase S. aureus. S. aureus was grown overnight and washed cells were allowed to adhere to washed N/TERTs grown to confluence. Data were analyzed by ANOVA using Prism and p values were reported as obtained from posthoc testing.
Fig. S3. Interferon exposure leads to inhibition of several barrier-related genes. N/TERTs were treated with 1000 U/ml of IFNα and IFNγ for 6 hours followed by expression analysis via qRT-PCR. Graphs represent relative expression to β-actin. Results are from 4 individual experiments ± SEM. Data were analyzed by ANOVA in Prism and the corresponding p values are reported.
Fig. S4. HC and SLE keratinocytes demonstrate greater adherence to S. aureus when exposed to IFNα. Keratinocytes were grown to confluence with or without 1,000 U/ml of indicated IFNs. S. aureus adhesion assays were performed as described in methods, and results are presented as S. aureus CFUs recovered per 10,000 keratinocytes. Blue represents HC and green represents SLE keratinocytes. Graphs represent means of three independent experiments for four SLE and three HC keratinocyte cultures.
Data Availability Statement
Datasets related to the article can be found at GEO (https://www.ncbi.nlm.nih.gov/geo/). RNA-seq data is available in GEO accession number GSE124939; Microarray data from CLE lesional skin is available in GEO accession number GSE81071.