Abstract
The world is currently face to face with a pandemic which is spreading rapidly across the globe caused by SARS-CoV-2, a strain of Coronaviruses (CoVs) belonging to subgenus Sarbecovirus of genus Betacoronavirus. World Health Organisation (WHO) on 11 Feb 20 named this disease caused by SARS-CoV-2 as Covid-19. This pandemic is spreading rapidly and more than 20,00,000 cases have occurred globally. The human Coronaviruses discovered in 1960s were considered potentially harmless endemic viruses with seasonal distribution before late 2002. The CoVs are found in a large number of domestic and wild animals and birds. The first pandemic caused by Coronavirus caused by SARS-CoV was recognized in the late 2002 in Guangdong Province and resulted in widespread morbidity and mortality. This was followed by MERS-CoV which began in 2012 in the Arabian peninsula with multiple outbreaks related to it in various parts of the globe. Various studies have suggested how these viruses made their entry from their natural reservoir bats via intermediate host like civets and camels in case of SARS-CoV and MERS-CoV respectively. The intermediate host of the SARS-CoV-2 still needs to be established. The SARS-CoV-2 has 96.2% similarity to the bat Severe Acute Respiratory Syndrome related-Coronavirus (SARSr-CoV RaTG13). SARS-CoV-2 has been found to be more distant in relation to SARS-CoV (79%) and MERS-CoV (50%). At the whole genome sequence level pangolin CoV and SARSr-CoV RaTG13 show 91.02% and 96.2% similarity with SARS-CoV-2 but the S1 subunit of spike protein of pangolin CoV is more closely related to SARS-CoV-2 than SARSr-CoV RaTG13. The genetic analysis of the currently circulating strains of the pandemic have shown 99.98-100% similarity in their genomes implying a recent shift to humans. The animal source of SARS-CoV-2 needs to be identified to implement control measures in the present pandemic. Also, how the virus moves interspecies will help predict and prevent future pandemics.
Keywords: Human coronavirus, COVID-19, SARS CoV-2, SARS-CoV, MERS-CoV
Coronaviruses: origin and evolution
Introduction
A pandemic of acute respiratory illness has shocked the world. Initially reported from Wuhan province, China in Dec 2019, currently, the viral illness is rapidly spreading across the globe. The virus spreads by droplet transmission, contact with infected case or contact with contaminated fomites. The disease was first recognized after a cluster of pneumonia outbreak was reported in late December 2019 from Wuhan, China. A new, human coronavirus (HCoV) was isolated from these cases and identified as a betacoronavirus and provisionally named 2019 novel corona virus (2019-nCoV) using next-generation sequencing technology.1,2 On 11 Feb 2020, the International Committee on taxonomy of Viruses named the virus as “severe acute respiratory syndrome coronavirus 2” (SARS-CoV2) and World Health Organisation announced COVID-19, the name of the new disease caused by it. Many authors have studied the genome sequences of the circulating virus to understand the viral dynamics and the way this new strain has made its way into the human population and lead to the current pandemic. Speculations considering it a laboratory constructed or bioengineered virus have also emerged. Studies to suggest that it naturally evolved from its existing ancestors in zoonotic reservoirs have also been published.3 However the research to establish the real origin of SARS-CoV2, is still underway.
Discovery
Coronavirus (CoV) was first isolated in 1965 by Tyrrell et al. from the nasal washings of a male child.4 Since their discovery in 1965, number of circulating strains of coronaviruses were identified, which were considered harmless pathogens, causing common cold and mild upper respiratory illness.5
Structure
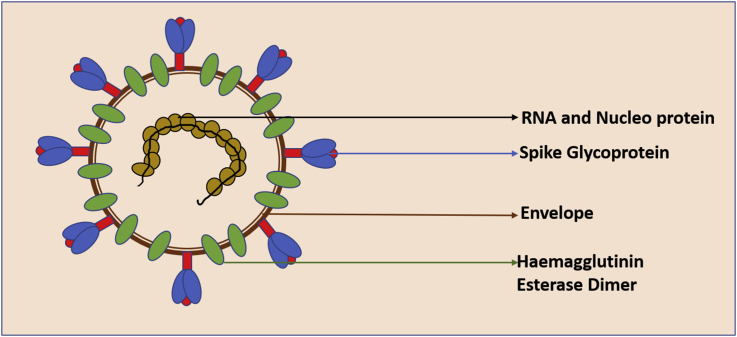
Coronaviruses (CoVs) have large linear positive stranded RNA genomes approximately 30 kb in size (26–32 kb) and they are about 125 nm in diameter,6,7 and comprise four genera (alpha-, beta-, gamma-, and delta-coronavirus).6, 7, 8 The spherical or pleomorphic virions are enveloped and contain a helical nucleocapsid of nucleoproteins (N) associated with the RNA genome. Embedded in the envelope are 20 nm trimer of spike glycoprotein (S), also called peplomers which have a club shaped morphology and facilitate attachment to cells. Envelope also contains integral membrane (M) and envelope (E) proteins. CoVs belonging to the Beta coronavirus lineage have 5–7 nm spikes of an additional membrane glycoprotein hemagglutinin esterase (Fig. 1).7
Fig. 1.
Structure of SARS-CoV2 virus.
Human coronaviruses
Till now Six human CoVs (HCoVs) have been confirmed: HCoV-NL63 and HCoV-229E, which belong to the alpha-coronavirus genus; and HCoV-OC43, HCoV-HKU1, SARS-CoV, and MERS-CoV, belong to the beta-coronavirus genus. SARS-CoV and MERS-CoV are the two major causes of severe pneumonia in humans.5,9, 10, 11 SARS-CoV 2 is the seventh CoV known to infect humans.
SARS-CoV and MERS-CoV
SARS was the first known pandemic caused by a CoV. The disease got recognized in the late 2002 with the outbreak of acute atypical community acquired pneumonia noticed first at Guangdong Province and 29 countries got affected by the spread.5,9,10
The 2003 SARS-CoV pandemic resulted in widespread morbidity and mortality and the same ended in Jun 2003.9,10 This was followed by a novel CoV, which was isolated from a Saudi Arabian patient with severe acute respiratory syndrome in June 2012 and the virus was later named Middle East Respiratory Syndrome - Coronavirus (MERS-CoV). Since then, multiple outbreaks have been reported in or been epidemiologically linked to the Arabian Peninsula.5,11
Besides SARS-CoV and MERS-CoV, the other human coronaviruses are global in their distribution in a seasonal endemic way and are responsible for less than 0.6–2.5% of adult community acquired pneumonia patients.12
Origin
CoVs have been found in large number of domestic and wild mammals and birds. There are studies to suggest that birds and bats are the natural reservoirs of the virus.2,13,14 Coronaviruses also have a potential for interspecies transmission which can also cause zoonotic outbreaks.11 Studies have suggested a bat origin of the HCoV-229E and HCoV-NL63. HCoV-229E have originated from bats with camels acting as intermediate hosts.13, 14, 15, 16 Molecular evolutionary analysis of HCoV-OC43 isolates suggests Bovine CoV (BCoV) is their genetically closest counterpart compared with other CoV species17,18 A high similarity was observed between BCoV, canine respiratory coronavirus (CRCoV) and human coronavirus OC43 (HCoV-OC43).12 The evolution of HCoV-OC43 has been shown to be by recombinant events.12 However the origin of CoV HKU1 is currently unknown.18 Phylogenetic analysis has revealed that 2019-nCoV fell within the subgenus Sarbecovirus of the genus Betacoronavirus. The homology modelling by the authors derived that 2019-nCoV had a similar receptor-binding domain (RBD) structure to that of SARS-CoV, despite amino acid variation at some key residues and the ability of the virus to bind to the Angiotensin converting enzyme 2 (ACE2) in humans.19
The genomic characterization of the novel Coronavirus from Wuhan cluster by various study groups was based on next generation sequencing of samples from bronchoalveolar lavage fluid and cultured isolates. Lu et al. studied the CoV isolated from nine inpatients, eight of whom had visited the Huanan seafood market in Wuhan. Their study showed that 2019-nCoV was related (with 88% identity) to two bat-derived severe acute respiratory syndrome (SARS)-like coronaviruses, bat-SL-CoVZC45 (GenBank accession number MG 772933) and bat-SL-CoVZXC21 (MG772934) and more distant from SARS-CoV (about 79%) and MERS-CoV (about 50%).20 However their study also revealed that S gene of 2019-nCoV had the lowest sequence identity with bat-SL-CoVZC45 and bat-SL-CoVZXC21, at only around 75%.20 Zhou et al. demonstrated that the novel virus has 96.2% similarity to a bat SARS-related Coronavirus (SARSr-CoV; RaTG13 (MN996532.1)).21 Zhang et al. showed that some pangolin CoV genes show higher amino acid sequence identity to SARS-CoV-2 than to RaTG13 genes which included Open Reading Frame 1b (ORF1b), spike protein (97.5% nucleotide identity), Open Reading Frame 7a (ORF7a) and Open Reading Frame10 (ORF10).22 The S1 protein which contains the RBD, is phylogenetically closer to pangolin-CoV than RaTG13 and this RBD region within the S1 was found to be conserved between Pangolin CoV and SARS-CoV2. The CoV spike (S) protein consisting of 2 subunits (S1 and S2), mediates infection of receptor-expressing host cells and the similarity between S1 protein of pangolin CoV to SARS-CoV2 points potential similarity in their pathogenic properties.22 Though the origin of the SARS-CoV 2 is still a debatable topic but the recognition of the intermediate animal host is the crucial step in preventing further dissemination, future outbreaks and blocking the interspecies transmission.
A naturally originated virus
Outbreak of fatal respiratory illnesses in Wuhan, China lead to speculations that SARS-CoV2 could be laboratory manipulated virus, however a study published in Nature Medicine on 17 Mar 20 by Andersen et al. concluded that the SARS-CoV2 is not a laboratory constructed or manipulated virus based on the RBD on the SARS-CoV2.3 SARS-CoV-2 has RBD that has high affinity to ACE2 from humans, ferrets, cats and other species with high receptor homology.21 The receptor binding domain in SARS-CoV2 is different from that of SARS-CoV and the binding of SARS-CoV2 is not optimal based on computational analysis leading to the understanding that there is another mechanism of binding which has arisen out of natural selection of the virus in the human or human like ACE2.3 The other salient finding noted of that study is the presence of a polybasic cleavage site at the junction of S1 and S2, though the role of this is not well established, may allow better cell to cell fusion without affecting viral entry.3, 23, 24
Genetic analysis of circulating strains
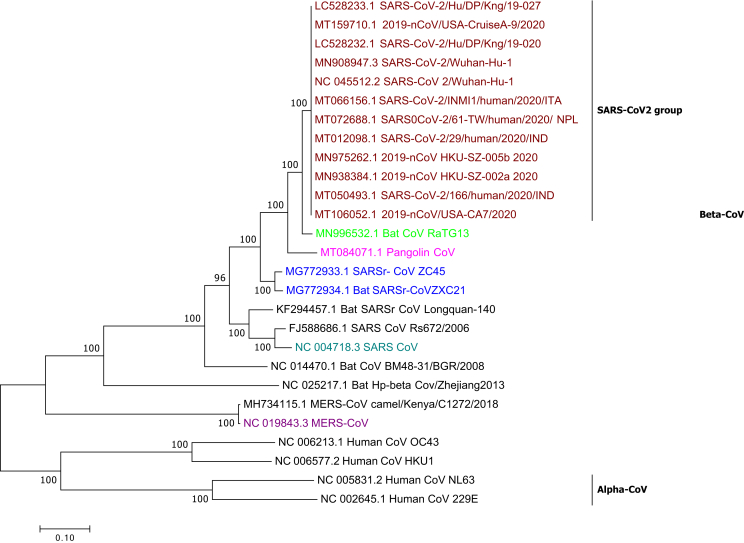
We performed phylogenetic analysis of circulating coronaviruses strains using maximum likelihood method in MEGA software. The near full length sequences of currently circulating strains were randomly selected and downloaded from GeneBank. Recently isolated SARS-CoV2 from India were also included in the study (Accession no. MT050493.1 and MT012098.1). All sequences showed ∼99.98–100% similarity in the nucleotide sequences establishing a relationship between the currently circulating viruses and implying a recent shift to humans (Fig. 2).
Fig. 2.
Phylogenentic Relationship of CoVs Based on the Whole Genome. Maroon text denotes SARS-CoV2. Pink text denotes Pangolin CoV. Green text denotes RaTG13. Blue text denotes Bat SARSr- CoV ZC45 and Bat SARSr-CoVZXC21. Light blue denotes SARS CoV. Purple denotes MERS-CoV.
The L & S type of the circulating SARS-CoV2
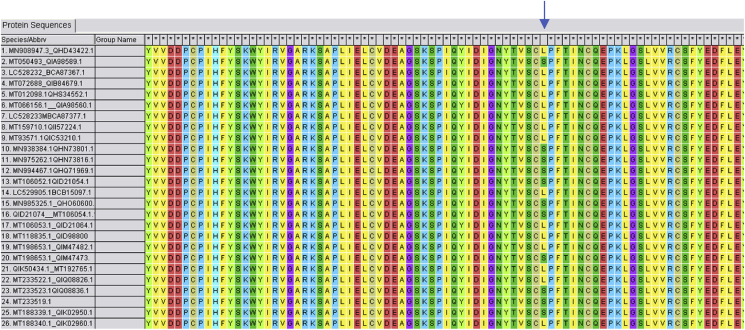
Tang et al. have carried out extensive study on the circulating SARS-CoV2 strains and divided the virus into two major types of SARS-CoV2 define by two single nucleotide poly morphisms (SNPs) that show complete linkage.25 The author analysed 103 SARS-CoV-2 virus strains and found that 101 showed complete linkage between the two SNPs: 72 strains exhibited a “CT” haplotype (defined as “L” type because T28,144 is in the codon of Leucine) and 29 strains exhibited a “TC” haplotype (defined as “S” type because C28,144 is in the codon of Serine) at these two sites. Thus the author proceeded to characterise and categorize the SARS-CoV-2 viruses into two major types, with L being the major type (∼70%) and S being the minor type (∼30%). Multiple sequence alignment in MEGA 7 of amino acid sequences of ORF 8 protein gene of 26 circulating strains from various locations around the globe to look for the frequency of the CT haplotype against the TC haplotype at position 84 was carried out. Our findings show that 11 out of 26 randomly selected strains showed Serine and 15 showed Leucine at position 84 of ORF 8 (57.69%) (Fig. 3).
Fig. 3.
Multiple sequence alignment of protein sequences of 26 circulating strains ORF 8 showing the leucine at position 84 in 15 out of 26 strains.
Why SARS CoV2 pandemic is different from previous CoV outbreak
Although both SARS CoV2 and prior SARS-CoV utilize ACE2 receptors to invade respiratory epithelium, the magnitude of infections caused by SARS-CoV2 is enormous. We are still unable to pinpoint the original reservoir for SARS CoV2. SARS-CoV genome was found to be 99.8% similar to that from civet cats. The similarity of whole genome sequence of SARS CoV2 to pangolins is only 92% and 96.2% to a bat SARS related coronavirus (SARSr-CoV; RaTG13) which is insufficient to prove beyond doubt that these are the sources of the virus. SARS was a relatively rare disease and at the end of the epidemic more than 8000 cases had occurred from 01Nov 2002 to 31 Jul 2003 whereas the ongoing Covid 19 caused by SARS- CoV2 has already caused more than 20, 00, 000 cases across the globe in a span of approximately five months.26,27 Though the mode of spread of both the viruses is almost same, SARS COV2 is far more infectious than SARS-CoV and the reason for the same is yet to be established.
Conclusion
The world today is facing a crisis due to the pandemic caused by SAR-CoV2. Based on the available literature and ongoing research, it is interesting to see that the more virulent forms (SARS-CoV,MERS-CoV and SARS-CoV2) get adapted to humans at some point in time of their evolution and this moment is crucial to the transmission of these viruses to humans. The SARS-CoV 2 appears to have originated in bats and the intermediate host could be pangolins, however it is difficult at this point in time to confidently determine the zoonotic source. Once they enter the human population, it is only a matter of time after which the morbidity and mortality caused by these viruses reach pandemic levels. The whole genome sequence analysis of randomly selected currently circulating human strains of SARS-CoV2 available show ∼99.98–100% similarity suggesting a recent introduction of the virus into humans.
However, within these two types of SARS-CoV-2, L type (∼70%) and S type (∼30%) have been observed by Tang et al. The strains in L type is derived from S type. The L type as per the author's conclusion appears to be more virulent and contagious.25 The human-animal interphase created by either encroachment of natural habitat of wild animals, maintaining domestic animals which get infected or consumption of animals which may be harbouring the viruses is the tipping point as per a large number of studies. It is important to understand viral dynamics so that future outbreaks can be avoided by active surveillance for these viruses. Regulations also need to be formulated to restrict the domestication as well as consumption of wild animals.
Disclosure of competing interest
The authors have none to declare.
References
- 1.Tan W., Zhao X., Ma X. A novel coronavirus genome identified in a cluster of pneumonia cases—Wuhan, China 2019–2020. China CDC Weekly. 2020;2:61–62. [PMC free article] [PubMed] [Google Scholar]
- 2.Zhu N., Zhang D., Wang W. A novel coronavirus from patients with pneumonia in China, 2019. N Engl J Med. 2020 doi: 10.1056/NEJMoa2001017. (published online Jan 24.) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Andersen K.G., Rambaut A., Lipkin W.L., Holmes E.C., Garry R.F. ARTIC Network; 17 February 2020. The Proximal Origin of SARS-CoV-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tyrrell D.A.J., Bynoe M.L. Cultivation of a novel type of common cold virus in organ cultures. Brit. med. J. 1965;1:1467–1470. doi: 10.1136/bmj.1.5448.1467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yin Y., Wunderink R.G. MERS, SARS and other coronaviruses as causes of pneumonia. Respirology. 2018;23:130–137. doi: 10.1111/resp.13196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fehr A.R., Perlman S. Coronaviruses: an overview of their replication and pathogenesis. Methods Mol Biol. 2015;1282:1–23. doi: 10.1007/978-1-4939-2438-7_1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gadsby N.J., Templeton K.E. coronaviruses. In: Caroll Karen C., Pfaller M.A., landry M.L., McAdam A.J., Patel R., editors. 12th ed. vol. 2. ASM Press; Washington DC: 2019. pp. 1606–1624. (Manual of Clinical Microbiology). [Chapter 92] [Google Scholar]
- 8.de Groot R.J., Cowley J.A., Enjuanes L. Order nidovirales. In: King A.M.Q., Adams M.J., Carstens E.B., Lefkowitz E.J., editors. Virus Taxonomy. Ninth Report of the International Committee on Taxonomy of Viruses. Elsevier Academic Press; Amsterdam: 2012. pp. 785–795. [Google Scholar]
- 9.Drosten C., Gunther S., Preiser W. Identification of a novel coronavirus in patients with severe acute respiratory syndrome. N Engl J Med. 2003;348:1967–1976. doi: 10.1056/NEJMoa030747. [DOI] [PubMed] [Google Scholar]
- 10.Ksiazek T.G., Erdman D., Goldsmith C.S. A novel coronavirus associated with severe acute respiratory syndrome. N Engl J Med. 2003;348:1953–1966. doi: 10.1056/NEJMoa030781. [DOI] [PubMed] [Google Scholar]
- 11.Wang Y., Sun J., Zhu A., Zhao J., Zhao J. Current understanding of middle east respiratory syndrome coronavirus infection in human and animal models. J Thorac Dis. 2018;10:S2260–S2271. doi: 10.21037/jtd.2018.03.80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Oong X.Y., Ng K.T., Takebe Y. Identification and evolutionary dynamics of two novel human coronavirus OC43 genotypes associated with acute respiratory infections: phylogenetic, spatiotemporal and transmission network analyses. Emerg Microb Infect. 2017 Jan 4;6(1):e3. doi: 10.1038/emi.2016.132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Li W., Shi Z., Yu M. Bats are natural reservoirs of SARS-like coronaviruses. Science. 2005;310(5748):676–679. doi: 10.1126/science.1118391. [DOI] [PubMed] [Google Scholar]
- 14.Corman V.M., Baldwin H.J., Tateno A.F. Evidence for an ancestral association of human coronavirus 229E with bats. J Virol. 2015;89(23):11858–11870. doi: 10.1128/JVI.01755-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dijkman R., van der Hoek L. Human coronaviruses 229E and NL63: close yet still so far. J Formos Med Assoc. 2009 Apr;108(4):270–279. doi: 10.1016/S0929-6646(09)60066-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Corman V.M., Eckerle I., Memish Z.A. Link of a ubiquitous human coronavirus to dromedary camels. Proc Natl Acad Sci Unit States Am. 2016:201604472. doi: 10.1073/pnas.1604472113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bidokhti M.R.M., Tråvén M., Krishna N.K. Evolutionary dynamics of bovine coronaviruses: natural selection pattern of the spike gene implies adaptive evolution of the strains. J Gen Virol. 2013;94:2036–2349. doi: 10.1099/vir.0.054940-0. [DOI] [PubMed] [Google Scholar]
- 18.Vijgen L., Keyaerts E., Lemey P. Evolutionary history of the closely related group 2 coronaviruses: porcine hemagglutinating encephalomyelitis virus, bovine coronavirus, and human coronavirus OC43. J Virol. 2006;80:7270–7274. doi: 10.1128/JVI.02675-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Woo P.C., Lau S.K., Lam C.S. Discovery of seven novel mammalian and avian coronaviruses in the genus deltacoronavirus supports bat coronaviruses as the gene source of alphacoronavirus and betacoronavirus and avian coronaviruses as the gene source of gammacoronavirus and deltacoronavirus. J Virol. 2012;86:3995–4008. doi: 10.1128/JVI.06540-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lu R., Zhao X., Li J. Genomic characterisation and epidemiology of 2019 novel coronavirus: implications for virus origins and receptor binding. Lancet. 2020 doi: 10.1016/S0140-6736(20)30251-8. Epub 2020/02/03. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhou P., Yang X., Wang X. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature. 2020 doi: 10.1038/s41586-020-2012-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhang T., Wu Q., Zhang Z. Probable pangolin origin of SARS-CoV-2 associated with the COVID-19 outbreak. Curr Biol. March 13 2020 doi: 10.1016/j.cub.2020.03.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Walls A.C., Park Y.J., Tortorici M.A., Wall A., McGuire A.T., Veesler D. Structure, function, and antigenicity of the SARS-CoV-2 spike glycoprotein. Cell. 2020;6(20):30262. doi: 10.1016/j.cell.2020.11.032. S0092–8674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nao N., Yamagishi J., Miyamoto H. Genetic predisposition to acquire a polybasic cleavage site for highly pathogenic avian influenza virus hemagglutinin. mBio. 2017;8 doi: 10.1128/mBio.02298-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tang X., Wu C., Li X. On the origin and continuing evolution of SARS-CoV-2. Natl Sci Rev. 2020 doi: 10.1093/nsr/nwaa036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Summary of Probable SARS Cases with Onset of Illness from 1 November 2002 to 31 July 2003. World Health Organization; 21 April 2004. [Google Scholar]
- 27.Coronavirus COVID-19 Global Cases by the Center for Systems Science and Engineering (CSSE) at Johns Hopkins University (JHU). ArcGIS. Johns Hopkins CSSE. Retrieved 15 April 2020.