Abstract
The worldwide occurring common liver fluke Fasciola hepatica can infect humans and animals and leads to considerable illness and economic loss annually. The aim of this study was to determine the genetic diversity of F. hepatica in Austria. In total, 31 adult flukes isolated from cattle from various regions in Austria were investigated for their cytochrome oxidase subunit 1 (cox1) and nicotinamide dehydrogenase subunit 1 (nad1) gene sequences. It was shown that Austrian isolates of F. hepatica reveal extensive genetic diversity. To the best of our knowledge, these are the first data on the diversity of F. hepatica in Austria.
Keywords: Fasciola hepatica, Digenea, Trematoda, Mitochondrial DNA, Haplotype, Genetic diversity, Austria
Introduction
The parasitic flatworm Fasciola hepatica (Trematoda: Fasciolidae), described already by Linnaeus in 1758, has a worldwide distribution (Mas-Coma et al. 2009). It is transmitted by the oral uptake of plants with attached infective metacercariae and can infest a wide range of mammals including humans. Sheep and cattle are the main final hosts, with several hundreds of million infested, causing substantial production losses (Robinson and Dalton 2009). In Europe, infestation rates in sheep and cattle are highly variable, even within countries. In isolated settings, as e.g., on alpine upland farms prevalences can reach over 90% (Ducheyne et al. 2015; Rinaldi et al. 2015). Galba truncatula is the main intermediate host in Europe and plays an important role for the distribution of F. hepatica (Mas-Coma et al. 2009). Human fasciolosis, caused by F. hepatica and F. gigantica, is considered a re-emerging neglected disease, with 17 million humans assumed to be infested and 180 million at risk. The disease affects mainly children in poor rural areas, particularly high prevalences of human fasciolosis have been reported for Bolivia, Peru, Ecuador, Egypt, Iran, Vietnam, and China (Mas-Coma et al. 2009). In Austria, the main endemic areas of F. hepatica are in the Western federal states (Supperer 1957; Kutzer and Hinaidy 1969; Auer and Aspöck 2014), with domestic as well as game animals being affected. At our institution, we see around 10 human cases of fascioliasis per year, of which, surely some are imported. However, also several autochthonous cases have been recorded, again, mainly from the Western parts of Austria (Auer and Aspöck 2014). Patterns of genetic diversity and population structure may give an insight into the dynamics of dispersal and distribution. The aim of this study was to investigate the diversity of F. hepatica in Austria by comparative sequence analyses of the conserved mitochondrial genes cytochrome oxidase subunit 1 (cox1) and nicotinamide dehydrogenase subunit 1 (nad1).
Material and methods
In total, 31 adult individuals of F. hepatica (Fh) isolated from cattle from various regions in Austria were investigated (Table 1). Eight of these samples (Fh1-Fh6 and Fh30-Fh31) were from the collection of the Natural History Museum Vienna and the Franz Berger GmbH & Co KG, respectively. Thirteen samples (Fh7-Fh19) were freshly collected at the abattoir Alpenrind GmbH, Salzburg, and 10 samples (Fh20-Fh29) were provided by the Austrian Agency for Health and Food Safety (AGES).
Table 1.
Samples of F. hepatica with their year of isolation, host and origin and their respective cox1 and nad1 subtypes
| No. | Year | Host | Origin | Altitude (m a.s.l.) |
cox1 type | nad1 type |
|---|---|---|---|---|---|---|
| FhAT1 | 2004 | Cattle | Mooslandl, Styria | 531 | I | I |
| FhAT2 | 2013 | Cattle | Oberhall, Styria | 574 | I | I |
| FhAT3 | 2013 | Cattle | Weissenbach/Enns, Styria | 430 | VII | I |
| FhAT4 | 2013 | Cattle | Admont, Styria | 640 | II | II |
| FhAT5 | 2013 | Cattle | Johnsbach, Styria | 853 | II | II |
| FhAT6 | 2013 | Cattle | Weißenbach/Enns, Styria | 430 | IX | I |
| FhAT7 | 2015 | Cattle | Hopfgarten im Brixental, Tyrol | 622 | II | II |
| FhAT8 | 2015 | Cattle | Itter, Tyrol | 703 | II | II |
| FhAT9 | 2015 | Cattle | Westendorf, Tyrol | 783 | IV | II |
| FhAT10 | 2015 | Cattle | Eugendorf, Salzburg | 560 | III | II |
| FhAT11* | 2015 | Cattle | Seekirchen/Wallersee, Salzburg | 512 | VIII | I |
| FhAT12* | 2015 | Cattle | Seekirchen/Wallersee, Salzburg | 512 | I | I |
| FhAT13 | 2015 | Cattle | Eugendorf, Salzburg | 560 | II | II |
| FhAT14 | 2015 | Cattle | Zell am Ziller, Tyrol | 575 | I | I |
| FhAT15 | 2015 | Cattle | Feldkirchen, Carinthia | 554 | I | I |
| FhAT16 | 2015 | Cattle | Ottenschlag, Lower Austria | 849 | V | I |
| FhAT17 | 2015 | Cattle | Ebbs, Tyrol | 475 | II | II |
| FhAT18 | 2015 | Cattle | Söll, Tyrol | 703 | I | IV |
| FhAT19 | 2015 | Cattle | Maishofen, Salzburg | 768 | I | I |
| FhAT20 | 2015 | Cattle | Kitzbühel, Tyrol | 762 | I | I |
| FhAT21 | 2015 | Cattle | Leutasch, Tyrol | 1136 | VI | I |
| FhAT22 | 2015 | Cattle | Kössen, Tyrol | 588 | I | I |
| FhAT23 | 2015 | Cattle | Angerberg, Tyrol | 650 | II | V |
| FhAT24 | 2015 | Cattle | Angerberg, Tyrol | 650 | III | III |
| FhAT25 | 2015 | Cattle | Angerberg, Tyrol | 650 | I | I |
| FhAT26 | 2015 | Cattle | Mühlbachl, Tyrol | 995 | I | I |
| FhAT27 | 2015 | Cattle | Neustift, Tyrol | 994 | I | I |
| FhAT28 | 2015 | Cattle | Ellbögen, Tyrol | 1070 | I | I |
| FhAT29 | 2015 | Cattle | Innsbruck, Tyrol | 574 | I | I |
| FhAT30 | 2015 | Cattle | St. Aegyd, Lower Austria | 588 | X | I |
| FhAT31 | 2015 | Cattle | Schwarzenbach, Lower Austria | 510 | I | I |
*flukes deriving from the same cow
PCR
For molecular analysis, 20–25 mg tissue were cut into small pieces with a scalpel. To avoid inclusion of foreign sperm, the tissue was taken from the apical zone of the flukes (Moazeni et al. 2012). Whole-cell DNA was isolated using the QIAamp DNA Mini Kit (QIAGEN, Vienna). The DNA yield was measured with a NanoDrop ND-1000 spectrophotometer (Peqlab, Erlangen, Germany).
Fragments of the mitochondrial DNA (mtDNA) genes cox1 and nad1 were amplified using the forward primer for cox1 (5´-TTGGTTTTTTGGGCATCCT-3′) from Itagaki and Tsutsumi (1998) and the reverse primer for cox1 (5´-AGGCCACCACCAAATAAAAGA-3′) and the forward (5´-TATGTTTTGTACGGGATGAG-3′) and reverse primer (5′- AACAACCCCAACCAACACTTA-3′) for nad1 from Semyenova et al. (2006). The mitochondrial genome of F. hepatica has a length of 14,461 bp, the cox1 gene is located between bp 6,871 and 8,402, and the nad1 gene is located between bp 5,176 and 6,078. The expected size of the cox1 amplicon is ~ 500 bp and the one of the nad1 amplicon ~ 420 bp. The primers were ordered from Microsynth AG (Balgach, Switzerland).
PCR and sequencing were performed as described previously (Husch et al. 2017). In brief, PCR included 15 min of initial denaturation with 95 °C and 30 cycles with 95 °C for 1 min, 56 °C for 2 min, 72 °C for 3 min, and a final extension at 72 °C for 7 min. Bands were visualized under UV light in 2% agarose gels and extracted from the gels using the QIAquick® Gel Extraction Kit (QIAGEN, Vienna). Sequencing PCRs were run with an initial denaturation at 96 °C for 30 s followed by 40 cycles of 96 °C for 10 s, 50 °C for 5 s, and 60 °C for 4 min. Sequences were obtained from both strands in two independent set-ups by direct sequencing with an automated ABI PRISM 310 Sequencer (PE Applied Biosystems, Langen, Germany) and assembled to consensus sequences using GeneDoc (Nicholas et al. 1997).
Sequence analyses
All consensus sequences were blasted against the reference sequences of F. hepatica available in GenBank by using BLAST (Altschul et al. 1990). For subtyping, multiple alignments of all isolates were performed with ClustalX (Thompson et al. 1997). Alignments were manually edited with GeneDoc (Nicholas et al. 1997) to exclude primer regions and to calculate identity scores. All subtypes were compared to the reference sequences of F. hepatica available in GenBank. Identities were evaluated separately for cox1 and nad1. Haplotype analyses were performed using PopART 1.7 (Leigh and Bryant 2015). For haplotype analyses, alignments were trimmed to the lengths of the available reference sequences in GenBank, i.e., to 310 bp for cox1 (AF216697, AP017707, GQ121276, GQ231549, GQ231550, GQ231551, GU112454, JF824670, JF824674, KJ200621, KU555842, KX470584, KX856338, MH561925, MH681796, MK212142, MN006838, X15613) and to 387 bp for nad1 (AF216697, AP017707, KR422393, KR422396, KT893736, KU946972, LC076257, MF287675, X15613), and TCS networks were obtained.
Voucher specimens of all samples were deposited in the Natural History Museum of Vienna, Austria. All sequence data were submitted to GenBank and are available under the following accession numbers: MN507437-MN507467 (cox1) and MN507406- MN507436 (nad1).
Results and discussion
This is the first study on the diversity of F. hepatica in Austria. Cattle positive for F. hepatica were from 475 to 1136 m above sea level and typically in their older age, between 3 and 8 years old. All flukes isolated were adult individuals; the sizes ranged from 2.5 to 3.2 cm. The cox1 fragments had a length of 496 bp in all isolates, and the nad1 fragments had a length of 416–417 bp, depending on the isolate. Altogether, 10 haplotypes were found for cox1 and 5 haplotypes for nad1 (Table 1); however, the nad1 fragment investigated was also shorter. Nevertheless, the diversity level was higher in nad1 compared to cox1, as has been found by others (Semyenova et al. 2006). The differences between the cox1 haplotypes were between 1 and 5 bp (Table 2) and between the nad1 haplotypes were between 1 and 6 bp (Table 3).
Table 2.
Base pair differences between the cox1 subtypes
| I | II | III | IV | V | VI | VII | VIII | IX | X | |
|---|---|---|---|---|---|---|---|---|---|---|
| I | 0 | |||||||||
| II | 4 | 0 | ||||||||
| III | 3 | 1 | 0 | |||||||
| IV | 3 | 1 | 2 | 0 | ||||||
| V | 1 | 5 | 4 | 4 | 0 | |||||
| VI | 1 | 5 | 4 | 4 | 2 | 0 | ||||
| VII | 1 | 3 | 4 | 2 | 2 | 2 | 0 | |||
| VIII | 1 | 3 | 2 | 2 | 2 | 2 | 2 | 0 | ||
| IX | 2 | 2 | 1 | 1 | 3 | 3 | 3 | 1 | 0 | |
| X | 2 | 4 | 5 | 3 | 3 | 3 | 1 | 3 | 4 | 0 |
Table 3.
Base pair differences between the nad1 subtypes
| I | II | III | IV | V | |
|---|---|---|---|---|---|
| I | 0 | ||||
| II | 4 | 0 | |||
| III | 3 | 1 | 0 | ||
| IV | 1 | 5 | 4 | 0 | |
| V | 2 | 2 | 4 | 6 | 0 |
The investigated 31 individuals of F. hepatica clustered into three major groups, termed subtypes I–III, and several more subtypes are only represented by one isolate each. Overall, the typing was rather consistent between cox1 and nad1 (Table 1). The most common subtype was subtype I, represented by 15 isolates for cox1 and by 20 isolates for nad1, and being also the only subtype found in all federal states of Austria investigated. This subtype I (Cox1-I/Nad1-I) reveals a 100% identity (cox1 496/496 bp, nad1 417/417) to one of the reference strains for the mitochondrial genome (AP017707) and 1 bp difference in each gene (fragment) to another mitochondrial genome strain (X15613; cox1 495/496 bp, nad1 416/417), both isolates from the USA. Moreover, it also shows 100% identities with strains from various countries all over the world, of which however, only shorter fragments or only one fragment is available, e.g., from Turkey (GQ121276), South Africa (KT182303), Algeria (MK212144), and Niger (FJ469984). Subtype II (Cox1-II/Nad1-II) shows a 100% identity to strain Geelong isolated in Australia (AF216697; cox1 496/496, nad1 417/417) and also to partly shorter fragments from strains from, e.g., Tunisia (GQ231550), South Africa (KT182261), Italy (JF824674), Denmark (MH561925), Poland (KR422395), Egypt (LC076257), and Iran (GQ175362). Subtype III (Cox1-III/Nad1-III) has 1–3 bp differences per gene, depending on the gene, to the three genome reference strains mentioned above (e.g., X15613; cox1 494/496; nad1 415/417) and again 100% identities to various shorter fragments from strains from all over the world. Haplotype relationships are given in Fig. 1. Interestingly, the flukes Fh11 and Fh12, which derived from the same cow, had the same nad1 haplotype but a different cox1 haplotype. Samples Fh10 and Fh13, deriving from two different individuals but from the same farm, also had the same nad1 haplotype but a different cox1 haplotype. It has been shown previously that a host can hold up to 10 different mitochondrial haplotypes (Elliott et al. 2014; Walker et al. 2007).
Fig. 1.
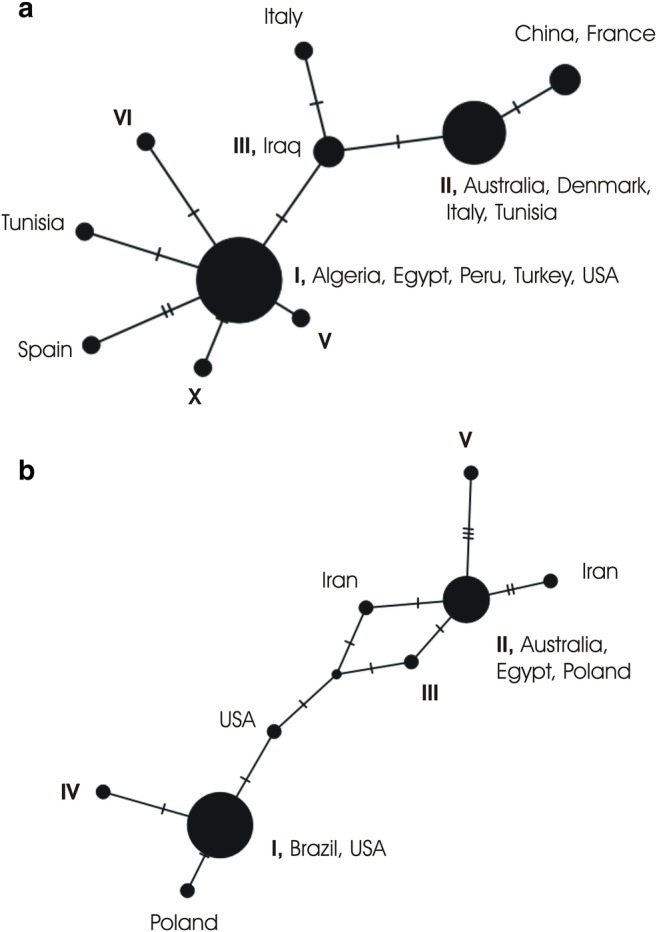
Haplotype networks. a based on the cox1 gene (310 bp). b based on the nad1 gene (387 bp); including all samples from Austria together with the respective reference sequences of F. hepatica from GenBank. Note: As sequences had to be trimmed to the lengths of the respective available reference sequences, not all haplotypes detected among the Austrian samples are represented by individual nodes
Alasaad et al. (2007) and Semyenova et al. (2006) examined the diversity of F. hepatica for several European and non-European countries and also demonstrated high similarities between strains from different continents, which they assume to be mainly due to livestock trafficking. However, depending on the regional setting, the genetic diversity differs dramatically between countries, determined by geography and landscape, on the one hand, but, of course, also on by the predominant type of farming. For example, on the Italian Island of Sardinia, Farjallah et al. (2013) only found three cox1 haplotypes and 5 nad1 haplotypes in 66 isolates from sheep and cattle. Walker et al. (2007) investigated 221 flukes from seven different locations in Ireland and found 18 composite haplotypes for cox3/nad4 and 11 composite haplotypes for cox1/rrna. In a study from the Netherlands, 92 cox3 haplotypes were detected among 422 flukes isolated from 20 cattle from only two farms (Walker et al. 2011). Finally, also the utilization of anthelmintic drugs might have a significant impact on the genetic diversity of flukes (Walker et al. 2007, 2011; Elliott et al. 2014).
In the current study, the highest diversity was found in the Tyrol, with 5 of the 10 cox1 subtypes and all of the 5 nad1 subtypes. However, the Tyrol was also represented by the most isolates. It is considered the main endemic region for F. hepatica in Austria (Auer and Aspöck 2014). This might also be attributed to the prevailing husbandry conditions. The Western Austrian states, particularly the Tyrol, Vorarlberg, and Salzburg, are alpine regions characterized by small farms, while the Eastern Austrian states, particularly Lower Austria, are characterized by flat land with large agricultural holdings. Also, precipitation rates are much higher in Western Austria. In this study, all flukes investigated were from cattle; however, sequences from reference strains with highest identities to our isolates were from different host species (cattle, sheep, or humans) corroborating the known low host specificity of F. hepatica. Today, F. hepatica is the most widely distributed vector-borne parasitic disease (Mas-Coma et al. 2009). A recent study from Armenia suggests that the species might have evolved in temperate Eurasia, where particularly high genetic diversity is found (Aghayan et al. 2019).
In conclusion, F. hepatica was isolated from cattle from up to 1136 m above sea level and typically from older aged animals (>3 years). Altogether, 10 haplotypes for cox1 and 5 for nad1 were detected, most of them with a 100% identity to isolates from all over the world. Most positive cattle were from the Tyrol, where we also saw the highest genetic diversity.
Acknowledgements
The authors wish to particularly thank the abattoir Alpenrind GmbH (and the veterinarians in charge) for enabling the sampling. Moreover, we would like to thank Joseph Ursprung, Hans Weissensteiner, Christian Stuefer, Franz Plank, the Austrian Agency for Health and Food Safety (AGES) and the Franz Berger GmbH & Co KG for providing further specimens. For critical discussions we want to thank Horst Aspöck, Herbert Auer, Verena Mündler, and Nadine Hohensee.
Author contributions
HS, HP and JW: Conceptualization; CH, HS, HP and JW: sampling; CH and IH: lab work; CH and JW: data analyses; CH, HS and JW: Writing.
Funding Information
Open access funding provided by Medical University of Vienna.
Compliance with ethical standards
Conflict of interest
The authors declare that they have no competing interests.
Ethics
Parasites used for this study were obtained from the livers of cattle. All procedures followed the latest legal determinations of veterinarian rules and the animal protection law.
Footnotes
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- Aghayan S, Gevorgian H, Ebi D, Atoyan HA, Addy F, Mackenstedt U, Romig T, Wassermann M. Fasciola spp. in Armenia: genetic diversity in a global context. Vet Parasitol. 2019;268:21–31. doi: 10.1016/j.vetpar.2019.02.009. [DOI] [PubMed] [Google Scholar]
- Alasaad S, Huang CQ, Li QY, Granados JE, García-Romero C, Pérez JM, Zhu XQ. Characterization of Fasciola samples from different host species and geographical localities in Spain by sequences of internal transcribed spacers of rDNA. Parasitol Res. 2007;101:1245–1250. doi: 10.1007/s00436-007-0628-2. [DOI] [PubMed] [Google Scholar]
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215(3):403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- Auer H, Aspöck H. Helminths and helminthoses in Central Europe: general overview and diseases caused by trematodes (flukes) Wien Med Wochenschr. 2014;164:405–413. doi: 10.1007/s10354-014-0316-7. [DOI] [PubMed] [Google Scholar]
- Ducheyne E, Charlier J, Vercruysse J, Rinaldi L, Biggeri A, Demeler J, Brandt C, De Waal T, Selemetas N, Höglund J, Kaba J, Kowalczyk SJ, Hendrickx G. Modelling the spatial distribution of Fasciola hepatica in dairy cattle in Europe. Geospat Health. 2015;9(2):261–270. doi: 10.4081/gh.2015.348. [DOI] [PubMed] [Google Scholar]
- Elliott T, Muller A, Brockwell Y, Murphy N, Grillo V, Toet HM, Anderson G, Sangster N, Spithill TW. Evidence for high genetic diversity of NAD1 and COX1 mitochondrial haplotypes among triclabendazole resistant and susceptible populations and field isolates of Fasciola hepatica (liver fluke) in Australia. Vet Parasitol. 2014;200:90–96. doi: 10.1016/j.vetpar.2013.11.019. [DOI] [PubMed] [Google Scholar]
- Farjallah S, Slimane BB, Piras CM, Amor N, Garippa G, Merella P. Molecular characterization of Fasciola hepatica from Sardinia based on sequence analysis of genomic and mitochondrial gene markers. Exp Parasitol. 2013;135(3):471–478. doi: 10.1016/j.exppara.2013.08.006. [DOI] [PubMed] [Google Scholar]
- Husch C, Sattmann H, Hörweg C, Ursprung J, Walochnik J. Genetic homogeneity of Fascioloides magna in Austria. Vet Parasitol. 2017;243:75–78. doi: 10.1016/j.vetpar.2017.06.007. [DOI] [PubMed] [Google Scholar]
- Itagaki T, Tsutsumi K. Triploid form of Fasciola in Japan: genetic relationships between Fasciola hepatica and Fasciola gigantica determined by ITS-2 sequence of nuclear rDNA. Int J Parasitol. 1998;28(5):777–781. doi: 10.1016/S0020-7519(98)00037-X. [DOI] [PubMed] [Google Scholar]
- Kutzer E, Hinaidy HK. Die parasiten wildebender wiederkäuer Österreichs. Z Parasitenkd. 1969;32:354–368. doi: 10.1007/BF00259648. [DOI] [PubMed] [Google Scholar]
- Leigh JW, Bryant D. PopART: full-feature software for haplotype network construction. Methods Ecol Evol. 2015;6(9):1110–1116. doi: 10.1111/2041-210X.12410. [DOI] [Google Scholar]
- Linnaeus C (1758) Systema Naturae per regna tria naturae secundum classes, ordines, genera, species, cum charactenbus, differentiis, synonymis, locis Editio Decima, reformata. Tom I, pars II — L Salvii, Holmiae: 1–823
- Mas-Coma S, Valero MA, Bargues MD. Fasciola, lymnaeids and human Fascioliasis, with a global overview on disease transmission, epidemiology, evolutionary genetics, molecular epidemiology and control. Adv Parasitol. 2009;69(2):41–146. doi: 10.1016/S0065-308X(09)69002-3. [DOI] [PubMed] [Google Scholar]
- Moazeni M, Sharifiyazdi H, Izadpanah A. Characterization of Fasciola hepatica genotypes from cattle and sheep in Iran using cytochrome C oxidase gene (CO1) Parasitol Res. 2012;110:2379–2384. doi: 10.1007/s00436-011-2774-9. [DOI] [PubMed] [Google Scholar]
- Nicholas KB, Nicholas HB, Jr, Deerfield DW., II GeneDoc: analysis and visualization of genetic variation. EMBNEW News. 1997;4:14. [Google Scholar]
- Rinaldi L, Biggeri A, Musella V, De Waal T, Hertzberg H, Mavrot F, Torgerson PR, Selemetas N, Coll T, Bosco A, Grisotto L, Cringoli G, Catelan D. Sheep and Fasciola hepatica in Europe: the GLOWORM experience. Geospat Health. 2015;9(2):309–317. doi: 10.4081/gh.2015.353. [DOI] [PubMed] [Google Scholar]
- Robinson MW, Dalton JP. Zoonotic helminth infections with particular emphasis on fasciolosis and other trematodiases. Phil Trans R Soc B. 2009;364:2763–2776. doi: 10.1098/rstb.2009.0089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Semyenova SK, Morozova EV, Chrisanfova GG, Gorokhov VV, Arkhipov IA, Moskvin AS, Movsessyan SO, Ryskov AP. Genetic differentiation in eastern Europe and western Asian populations of the liver fluke, Fasciola hepatica, as revealed by mitochondrial NAD1 and COX1 genes. J Parasitol. 2006;92(3):525–530. doi: 10.1645/GE-673R.1. [DOI] [PubMed] [Google Scholar]
- Supperer R. Über die Ursachen der schweren Leberegel-Erkrankungen im Jahre 1956. Wien Tierärztl Mschr. 1957;44:107–109. [Google Scholar]
- Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG. The CLUSTAL X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 1997;25:4876–4882. doi: 10.1093/nar/25.24.4876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker SM, Prodohl PA, Fletcher HL, Hanna RE, Kantzoura V, Hoey EM, Trudgett A. Evidence for multiple mitochondrial lineages of Fasciola hepatica (liver fluke) within infrapopulations from cattle and sheep. Parasitol Res. 2007;101:117–125. doi: 10.1007/s00436-006-0440-4. [DOI] [PubMed] [Google Scholar]
- Walker SM, Johnston C, Hoey EM, Fairweather I, Borgsteede F, Gaasenbeek C, Prodohl PA, Trudgett A. Population dynamics of the liver fluke, Fasciola hepatica: the effect of time and spatial separation on the genetic diversity of fluke populations in the Netherlands. Parasitology. 2011;138:215–223. doi: 10.1017/S0031182010001149. [DOI] [PubMed] [Google Scholar]


