Abstract
Base editing is a form of genome editing that can directly convert a single base (C or A) to another base (T or G), which is of great potential in biomedical applications. The broad application of base editing is limited by its low activity and specificity, which still needs to be resolved. To address this, a simple and quick method for the determination of its activity/specificity is highly desired. Here, we developed a novel system, which could be harnessed for quick detection of editing activity and specificity of base editors (BEs) in human cells. Specifically, multiple cloning sites (MCS) were inserted into the human genome via lentivirus, and base editing targeting the MCS was performed with BEs. The base editing activities were assessed by specific restriction enzymes. The whole process only includes nucleotide-based targeting the MCS, editing, PCR, and digestion, thus, we named it NOTEPAD. This straightforward approach could be easily accessed by molecular biology laboratories. With this method, we could easily determine the BEs editing efficiency and pattern. The results revealed that BEs triggered more off-target effects in the genome than on plasmids including genomic indels (insertions and deletions). We found that ABEs (adenine base editors) had better fidelity than CBEs (cytosine base editors). Our system could be harnessed as a base editing assessment platform, which would pave the way for the development of next-generation BEs.
Keywords: Adenine base editor, Cytosine base editor, Detection, Genome editing, Method
Introduction
CRISPR-Cas9-mediated targeted genome engineering technologies have enabled a broad range of research and medical applications.1,2 CRISPR-Cas9 introduces double-stranded DNA breaks (DSBs) at the target DNA locus, which generate stochastic insertions and deletions (indels) or translocations at the cleavage site.3, 4, 5 Homology-directed repair (HDR) could be harnessed to replace the genomic DNA at the cleavage site with the presence of donor DNA template, in order to achieve precise genome modification.6,7 However, the application of HDR-dependent genome editing is limited by low efficiency and off-target effects caused by continuous, uncontrolled Cas9 and sgRNA (single guide RNA) expression, even after the target locus has been successfully edited.8, 9, 10, 11, 12
A variety of base editors (BEs) including cytosine (CBEs) or adenine base editors (ABEs) as novel genome editing tools have been developed to induce targeted C-to-T or A-to-G base conversions.13, 14, 15 It could proceed more efficiently with single base manipulation.13,15, 16, 17, 18 Specifically, CBEs could efficiently enable the direct, programmable, targeted conversion of a C:G base pair to a T:A base pair and consist of a specific cytidine deaminase domain fused with CRISPR-Cas9.2,13,15 Studies have demonstrated the ability of CBEs and related methods to promote Cas9-directed C to T transformation in mammalian cells15,19,20 or plants,21,22 and expanded CBEs functions have been introduced by the development of the basic editor with a narrowing editing window,17 different protospacer-adjacent motif (PAM) compatibilities,17 small-molecule dependence,23 and enhanced DNA specificity.18 While, the CBEs were unable to induce other forms of base conversion beyond the C to T mutation or with low efficiency conversion to other bases (C to non-T), which limit its application. ABEs, as a base editing tool, could enable the efficient A to G conversion at the target site in mammalian cells, which consists of an adenosine deaminase domain fused to a Cas9 variant domain.24,25 The ABEs could produce editing events in animal and plant genomes, and studies have shown that ABEs were more accurate than CBEs in the editing of animal genomes.26, 27, 28, 29, 30, 31 ABEs greatly broaden the scope of gene editing and together with CBEs could enable programmable conversion of all four bases in genomic DNA. Recent studies show that off-target sites are generated with BEs in a sgRNA-independent manner, which increased the safety concerns of basic editing methods in therapeutic applications.32,33 Furthermore, base conversion between purine and pyrimidine nucleotides cannot be achieved; the efficient editing window was approximately limited to 4–7 or 4–9 (we set and labeled the PAM as positions as 21 to 23). These observations highlight the necessity and urgency of developing more precise BEs. However, how to assess base editing activity in human cells with a simple and quick approach is still a challenge. To address this problem, here we developed a novel method to detect the editing events by BEs in human cells.
Results
Development of a Base Editing Detection Tool
In order to develop a simple and quick method to assess base editing in human cells, we sought to test whether the most commonly used restriction enzyme sites could be harnessed to distinguish single nucleotide changes. Initially, we selected an I-SceI restriction enzyme recognition site as a target site for base editing because it is 18 bases in length, and the enzyme is commercially available. We generated a plasmid harboring an EF1α promoter, I-SceI recognition site, and enhanced green fluorescence protein (EGFP) coding sequence (Figure S1A). Theoretically, any base change of the I-SceI recognition site may render it resistant to I-SceI digestion. However, we found that I-SceI still efficiently cleaves after introduction of point mutations with all the tested mutations except one (Figures S1B and S1C). Thus, the I-SceI enzyme recognition site may be not suitable for the evaluation of base editing.
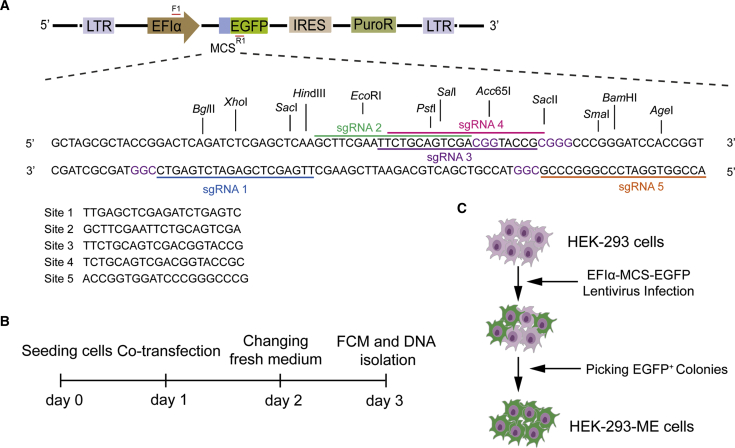
Therefore, we sought to determine whether shorter sequences for restriction enzymes may be better. To address this, we inserted a region of multiple cloning sites (MCS) harboring different restriction enzyme recognition sites in the upstream coding sequence of EGFP for the expression of fusion EGFP to construct plasmids (plasmid of MCS-EGFP, PME plasmids). EGFP has been used to detect the efficiency of genome editing,34,35 to assist in the detection of edits. As we expected, the insertion of a MCS did not affect the expression of EGFP (Figure 1A; Figure S2A). Because the restriction enzyme sites in their MCS may distinguish single nucleotide differences and CBEs-mediated transition of CAG/CAA/CGG into TAG/TAA/TGG (stop codon) may lead to the inactivation of EGFP, this may be applied to the evaluation of base editing. The whole process may only include nucleotide-based targeting the MCS, editing, PCR, and digestion, and thus, we named it NOTEPAD.
Figure 1.
Schematic of NOTEPAD System and Reporter Cell Line
(A) Schematic of the NOTEPAD system. The MCS sequence contains 20 restriction sites, and we designed 5 target sites. Site 1 and site 5 were on the (–) strands (purple base is PAM). (B) Schematic of the transfection experiment. We transfected the plasmids to HEK293 cells (250 ng templates, 250 ng BE expression plasmids, and 125 ng sgRNAs expression plasmids) or HEK293-ME cells (250 ng BE expression plasmids, and 125 ng sgRNAs expression plasmids). The percentage of EGFP (or EGFP disruption) was analyzed by flow cytometry (FCM) or the genomic DNA was isolated for further analysis. (C) Schematic of the HEK293-ME cell line generation. A lentivirus containing EFIα-MCS-EGFP-Puro cassettes was packaged for infecting HEK293 cells. After puromycin selection, colonies of HEK293-ME were picked under a fluorescence microscope.
Detection of Base Editing with the Plasmid-Based NOTEPAD System (Episomal)
We first selected five target sites in the MCS sequence to study whether the editing events of the BEs on the PME plasmid (episomal) could be detected (Figures 1A and 1B). The BE3 used in this study harbors a human cytomegalovirus (CMV) immediate early promoter, rat cytidine deaminase APOBEC1 (apolipoprotein B mRNA editing enzyme, catalytic polypeptide-like 1), a Cas9 variant (Cas9-D10A nickase [nCas9]), and uracil glycosylase inhibitor (UGI).34 Two different ABEs (xCas9-ABE7.10 and ABE7.9) were used in this study. xCas9-ABE7.10 has improved editing targeting scope, efficiency, and DNA specificity.25 The editing window of ABE7.9 (base 4–9) is larger than xCas9-ABE7.10 (base 4–7), counting the PAM as positions 21–23.24
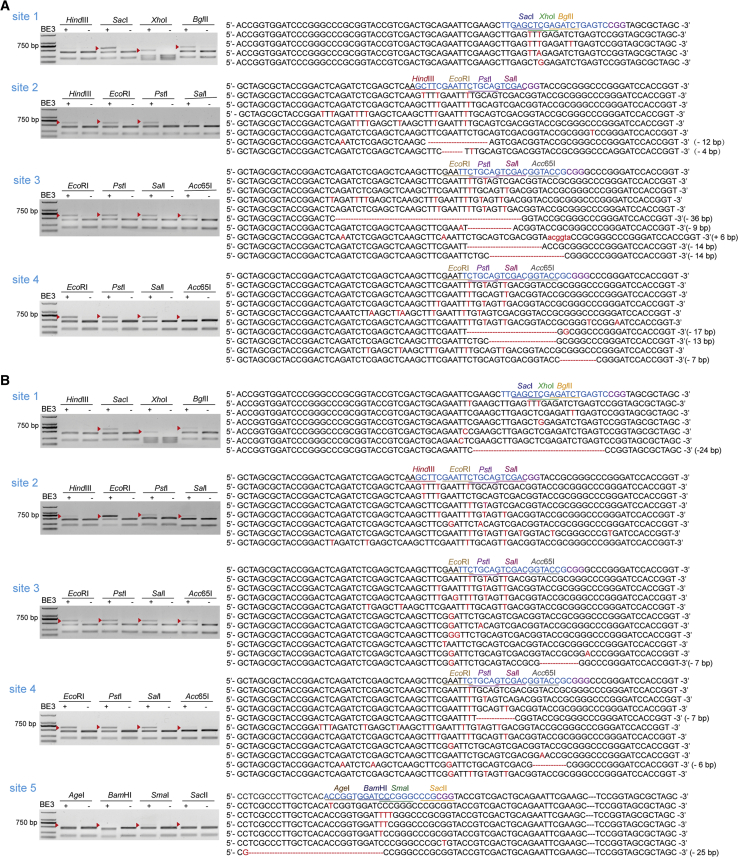
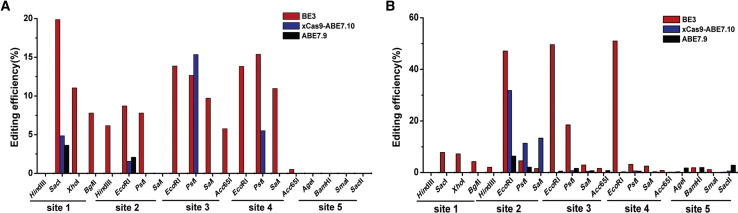
Not surprisingly, with the NOTEPAD method, we clearly observed that BE3 has editing activity at these sites with the exception of site 5 (Figures 2A; Figure S4A). The highest editing efficiency of each site was 19.86% at site 1, 8.71% at site 2, 13.88% at site 3, and 15.37% at site 4 (Figure 4A). We suspected that the lack of activity of site 5 may be due to its GC-rich context. The cytosine of CpG is frequently methylated in mammalian cells, and cytosine methylation strongly inhibits the cytidine deaminase catalysis of certain APOBEC and AID deaminases.36,37 To gain insight into the base editing process, we obtained the resistant-cleavage band sequence information via Sanger sequencing. This showed that BE3 can perform efficient C to T editing in four target sites, but we also found the indel events of site 2 to site 4 (Figure 2A, 28.75% at site 2, 50% at site 3, 33.33% at site 4, the percentage of indels represents the ratio of the indels signal to the total). These results are not consistent with previously reported nCas9-PBE, which conversed cytosine to thymine from position 3 to 9 within the protospacer in rice, wheat, and maize (counting the PAM as positions 21–23).34 This lack of concordance may be partly due to species differences and target sequences. The sequencing results also showed that BE3 could also cause non-C to T base conversion (Figure 2A, 25% at site 1, 5.3% at site 2, 16.7% at site 3, and 11.11% at site 4; the percentage represents the ratio of non-C to T base conversion to the total base conversion) and editing events outside the editing window (Figure 2A, 51.4% at site 2, 44.44% at site 3, and 33.33% at site 4; the percentage represents the signal of outside the editing window to the total), which is consistent with previous reports.2,20,38
Figure 2.
The Editing Events of BE3 Detected with NOTEPAD System
(A) The plasmids expressing PME templates, BE3, and sgRNAs were transfected into HEK293 cells. Genomic DNA was harvested and PCR was performed with primers (F1 and R1) to amplify the DNA sequence flanking of MCS. PCR products were treated with restriction enzymes and then separated with electrophoresis. The resistant-cleavage band sequence information was obtained via Sanger sequencing. (B) The plasmids expressing BE3 and sgRNAs were transfected into HEK293-ME cells. The same protocol was adopted as (A). Blue sequence is target site, purple base is PAM, and red base and ellipsis represent edited events.
Figure 4.
The Editing Efficiency of BEs
(A) The BEs mediated editing efficiency of sites on the plasmid (episomal). (B) The BEs mediated editing efficiency of sites in the genome.
We could only detect editing activity of ABE7.9 at two sites (3.64% at site 1, 2.07% at site 2), while xCas9-ABE7.10 at four sites (4.85% at site 1, 1.56% at site 2, 15.35% at site 3, and 5.51% at site 4) has relative higher activity (Figures S3, S4B, and S4C). This result is consistent with the literature, which showed that xCas9-ABE7.10 has a higher editing activity.25 Notably, CpG-rich regions (site 5) also inhibited the deamination activity of ABE (Figures S4B and S4C). Collectively, the above results illustrated that, at the plasmid level, NOTEPAD could be harnessed as a novel system for the evaluation of base editing efficiency.
Detection of Genomic DNA Base Editing by NOTEPAD System
Because BEs would be applied to the manipulation of the nuclear genome in mammalian or plant cells, we sought to determine the performance of NOTEPAD to the nuclear genome. To address this, a lentivirus containing EF1α-MCS-EGFP-Puro cassettes was packaged and then used to infect HEK293 cells. To make a homogeneous genetic population, we picked single colony (named with HEK293-ME) for further study (Figure 1C; Figure S2).
With HEK293-ME cell line, we investigated the base editing events triggered with BE3 at five target sites (Figure 2B). Similar to the plasmid approach, the editing efficiency of BE3 was lower on site 5 (Figure 2B). Sanger sequencing results revealed that the editing efficiency of BE3 in the MCS region in the genome was similar to that of the plasmids. Off-target effects of indels (14.3% at site 1, 10% at site 3, 20% at site 4, and 14.3% at site 5), non-T base conversion (14.3% at site 1, 9.52% at site 2, 36% at site 3, 23.10% at site 4, and 11.11% at site 5), and editing events outside the editing window (42.9% at site 1, 14.3% at site 2, 90% at site 3, 60% site 4, and 33.33% at site 5) were also observed (Figure 2B). Statistical analysis showed that the genomic editing efficiency of BE3 of each site on the genome was almost higher than that on plasmid templates (Figure 4). The highest editing efficiency on the episomal plasmid at sites 1–4 was 19.86%, 8.71%, 13.88%, and 15.37%, respectively, and the highest editing efficiency on the genomic DNA at sites 1–5 was 7.77%, 47.10%, 49.57%, 50.10%, and 1.85%, respectively. We suspected that this may due to the template levels in the HEK293-ME cells being much lower than that of HEK293 cells, with the same amount of BE, which therefore leads to more editing events at the target sequence in HEK293-ME cells.
We also found that the highest editing efficiency of xCas9-ABE7.10 was almost higher than that of ABE7.9 except site 3 and site 5 (Figure 4B, xCas9-ABE7.10, 31.80% at site 2, 0.79% at site 3, 0.70% at site 4, 0.62% at site 5; ABE 7.9, 6.34% at site 2, 1.63% at site 3, 0.54% at site 4, and 2.84% at site 5) and the latter produced more non-A to G base conversions and indels (Figure 3, indels, xCas9-ABE7.10, 33.33% at site 2; ABE 7.9, 83.33% at site 3, 50% at site 4, and 33.33% at site 5), which is consistent with the literature.25 Notably, the two ABEs rarely trigger A to G editing at site 5, with more indels and non-A to G conversions. We suspected that editing at this site is affected by the CpG. Surprisingly, no editing activity was observed with these two ABEs at site 1, while we could clearly detect its activity with the template of plasmids, indicating that additional factors modulate ABE performance.39, 40, 41 We also observed that ABEs possesses higher activity on the genomic DNA than that on the plasmid, which is consistent with the results of BE3 (Figure 4). Notably, more indel products with the genome-editing group have been detected, compared with that of the episomal group.
Figure 3.
The Editing Results of ABEs Detected with NOTEPAD System
(A) The plasmids expressing xCas9-ABE7.10 and sgRNAs were transfected into HEK293-ME cells, genomic DNA was harvested, and PCR was performed with primers (F1/R1) to amplify the DNA sequence flanking of MCS. PCR products were treated with restriction enzymes and then separated with electrophoresis. The resistant-cleavage band sequence information was obtained via Sanger sequencing. (B) The plasmids expressing ABE7.9 and sgRNAs were transfected into HEK293-ME cells. The same protocol was adopted as (A). Blue sequence is target site, purple base is PAM, and red base and ellipsis are edited events.
Discussion
Base editing is a newly developed genome engineering tool that enables gene editing through irreversible base conversion, which may be developed as a novel gene therapy approach.13,24,28 However, its ubiquitous off-target issues will be a key challenge for both basic research and clinical therapeutic applications.32,33 Therefore, there exists an acute need to develop next-generation BEs with minimized off-target effects, in order to enable robust and safe clinical applications of BE-based gene therapies and researches. To optimize base editing, the development of a simple and quick method to access base editing in human cells is a key step toward it.
So far, deep sequencing is commonly harnessed to assess base editing. This is both time-consuming and costly. Here we developed a method termed NOTEPAD, which is based on the fact that a mutated base triggered with BEs can no longer be cut with a restriction enzyme. Through the digestion with the corresponding restriction enzyme, the non-edited sequence would be totally digested and the edited sequence would be resistant to cleavage. This allows for analysis of the ratio of the edited sequence to the total, thus allowing assessment of editing efficiency. The editing efficiency calculated from the intensity of the bands of PCR/restriction enzyme is generally consistent with that obtained from Next Generation Sequencing (NGS; Table S3). NGS results also confirmed that xCas9-ABE7.10 possesses higher editing activity than ABE7.9. The advantage of the present method includes no requirement of special equipment, simplicity, speed, and low cost.
Initially, we chose the I-SceI restriction enzyme because it recognizes an 18 bp target sequence. However, the fidelity of I-SceI did not meet the detection requirement, with cleavage still observed even in the presence of point mutations (Figure S1). We found that the shorter sequence from MCS could resolve this issue. With NOTEPAD, we could access base editing in human cells on the plasmid (episomal) and on the genomic level. Furthermore, we observed that BE3 could cause indels, off-target effects, and non-T products. Surprisingly, we found CpG-rich regions (site 5) on the plasmid, but not in the genome, inhibited the deamination of BE3, and BE3 triggered non-C to T conversions and editing events outside the editing window on the genomic DNA to a greater extent than on the plasmid. Recently, BE3s have been modified to produce purity without affecting the editing efficiency of BEs,14,18 expanding their targeting scope17,25 and enriching the genome editing toolbox.19,42 Furthermore, hA3A-BE3 has improved efficiency of deamination of methylated cytosine43 and BE-PACE has improved BE3 editing efficiency and target sequence compatibility.44 In order to achieve BE3 applications, it is still essential to reduce off-target effects and non-T conversion products of BE3 to realize precise editing, and expand editing scope.
The results showed that xCas9-ABE7.10 has a higher editing activity than ABE7.9 both on the MCS-containing genomic or plasmid DNA. In particular, the former could trigger the editing of 4 sites, while the latter only triggered editing of 2 sites on the plasmid level. These two ABEs triggered only A to G base conversion on the plasmid, while indels and non-A to G conversions on the genomic DNA have been observed, which highlights that it may be due to certain chromosomal structures and or epigenetic modifications preventing ABE entry.39, 40, 41 Here we mapped the exact integration site of prolentivirus (Figures S5 and S6). ABE adenosine deaminase deamination and nCas9 or xCas9 caused a small amount of indels.25,45 These results may have important implications for mitochondrial DNA base editing for human gene therapy and DNA virus or bacterial-based biological control via base editing. Interestingly, we don’t know whether the reason for the absence of ABE activity at site 1 on genomic DNA is due to the target on the (–) strand (Figure S7). In addition, CpG-rich regions (site 5) on the plasmid but not in the genome inhibited the deamination of ABEs, which was similar to BE3, but such editing results were undesired. Compared with the off-target effects of BE3, we rarely detected editing efficiency of these two ABEs outside the editing window, which is consistent with the literature.32,33
Theoretically, the insertion of EGFP following the MCS could be utilized for the tracking of base editing events via flow cytometry; however, we found this is not the case. Regarding the EGFP negative population, the edited group is almost the same as that of control with the target of plasmid (episomal) (Figure S8A). The reason for that is that HEK293 cells contain multiple editing templates, which cannot all be edited into stop codon simultaneously. Thus, the inactivation of EGFP in the cells can’t still be detected. As to the integrated MCS-EGFP, low levels of inactivation of EGFP were observed, which may due to introducing stop codon or indels causing frameshift (Figure S8B). As to the EGFP-related base editing detection methods,31,34 its limitation includes as follows: it requires the appropriate PAM, the readout is highly depend on single specific site editing, i.e., BFP to GFP or inactive GFP (with stop codon) to wild-type GFP, and the editing window can’t be easily dissected, which could largely resolved by our NOTEPAD system.
So far, with BEs, we could perform a purine to a purine or a pyrimidine to a pyrimidine base conversion, with the new BEs inducing transversions (C to A/G or A to C/T), which would be highly desired to expand the applications of base editing. As to the specificity, new BEs with only targeting editing single nucleotide would be used to realize precise editing. The NOTEPAD system could assist in detecting the base editing events quickly, which would facilitate the engineering of BEs and identification of new BEs. We reason that NOTEPAD system can’t be utilized for detection of all the off-target effects of BEs, especially for the long-distance region of the target sequence, which could be realized with additional whole genome off-target analysis methods, including EndoV-seq and GOTI (genome-wide off-target analysis by two-cell embryo injection).32,46
Materials and Methods
Plasmid Information
The vector plasmid pSin-EFIα-MCS-EGFP-puro (PME) was generated based on the parental vector pSin-EFIα-EGFP-puro with MCS (multiple cloning sites) (Figure 1A). MCS sequence was amplified step by step by primer pairs MCS-F1/R, MCS-F2/R, and MCS-F3/R (Table S2). Plasmid for CBEs was originally from Professor Caixia Gao (Chinese Academy of Sciences; Addgene plasmids #98164). We replaced the maize Ubiquitin-1 (Ubi-1) gene promoter with human CMV immediate early promoter using MfeI and BamHI restriction enzyme, and we named it with BE3. Plasmids for the expression of adenosine deaminase pCMV-ABE7.9 and xCas9(3.7)-ABE(7.10) were directly obtained from Addgene (Addgene plasmids #102918 and #108382, respectively), and we named them with ABE7.9 and xCas9-ABE7.10, respectively. sgRNA oligos were annealed and cloned into vector pJET-U6 with a backbone of pJET1.2 (CloneJET PCR Cloning Kit, Thermo Fisher Scientific), which contains U6 promoter and sgRNA scaffold using a standard protocol (Table S1). Plasmid DNA was isolated by standard techniques. DNA sequencing confirmed the desired specific sequences in the constructs.
Cells and Cell Culture
HEK293 cells were obtained from ATCC (CAT#CRL-1573) and were cultured as previously described.47 HEK293 cells expressing MCS gene were generated by lentiviral transduction. To generate HEK293-ME cell colonies containing MCS and EGFP expression cassettes, we seeded HEK293 cells on day 0 at 3.6 × 105 cells in 6-well plates, and on day 1, PME and lentivirus package plasmids were transfected by TurboFect Transfection Reagent (Thermo Fisher Scientific, MA). The supernatant of cell culture was harvested for the infection, and 200 μL of lentivirus supernatant was added in 6-well plates. After the infection, individual colony with the expression of EGFP was picked under the microscope at day 10∼15. PCR and Sanger sequencing were used for the confirmation of the gene knockin, using specific primers (Table S2; F1, R1, and EF1α-F). To maintain EGFP expression, the HEK293-ME cell culture medium contained puromycin (1 μg/mL).
Flow Cytometry Analysis
The flow cytometry (FCM) protocol was described previously.48 On day 0, 0.9 × 105 HEK293-ME cells were seeded in 24-well plates. On day 1, the cells were co-transfected with BE plasmids (250 ng) and sgRNA plasmids (125 ng) with TurboFect Transfection Reagent (1.5 μL). On day 2, fresh medium was added. On day 3, cells were harvested for genomic DNAs isolation or flow cytometry. The EGFP negative percent was examined via flow cytometry (BD Biosciences, NY) and CellQuest software was used to analyze data. As to the editing test at plasmid level, plasmids harboring the target sequence (250 ng) were co-transfected into HEK293 cells with ABEs or BE3 encoding plasmids.
Editing Efficiency Analysis
Genomic DNA was purified using the standard phenol/chloroform extraction protocols. Amplified products harboring the MCS (PCR primers were F1R1; Table S2) were treated with restriction enzyme. 250 ng total purified PCR products were digested by the corresponding restriction enzymes, and then analyzed on agarose gel. Quantification was based on relative band intensities. Editing efficiency was determined by the formula 100 × (1 – sqrt[(b + c)/(a + b + c)]), where a is the integrated intensity of the undigested PCR product and b and c are the integrated intensities of the cleavage product. Digestion-resistant fragments were also inserted into the vector pJET1.2 (CloneJET PCR Cloning Kit, Thermo Fisher Scientific) and further sequenced on an ABI PRISM 3730 DNA Sequencer (sequencing primers are shown in Table S2).
Identification of Lentiviral Integration Site
To identify the lentiviral integration site, we digested 2 μg genomic DNA from HEK293-ME cell with AciI, ApoI, and TaqI, respectively, and then ligated the digested products with T4 DNA ligase. The amplicons harboring adjacent sequences of prolentivirus were obtained with inverse PCR and nest PCR with primers (inverse F1, inverse R1 for inverse PCR; inverse F2, inverse R2 for nest PCR). The integration site was further confirmed with gene-specific PCR (F2R2, F3R3; Table S2).
NGS and Data Analysis
The NGS protocol and data analysis were described previously.49 The deep sequencing data are available at the NCBI Sequence Read Archive (SRA) under BioProject PRJNA601886 (SRA: SRR10912475; sample accession numbers, SAMN13876266).
Author Contributions
F.G. and J.Z. designed the experiments. X.L., K.Q., T.T., X.H., Y.P., and J.Y. performed experiments. X.L., J.F., R.D., Y.W., J.W., J.Z., and C.L. analyzed the results. X.L. and F.G. wrote the paper.
Conflicts of Interest
The authors declare no competing interests.
Acknowledgments
We thank Bang Wang, Xianglian Ge, Haihua Xie, Fanfan Li, Chenchen Zhou, Xiexie Liu, and Yeqing Liu for technical assistance. This work was supported by grants from the Natural Science Foundation of China (81201181 to F.G.), the Science Technology Project of Zhejiang Province (2017C37176 to F.G.), the Zhejiang Provincial & Ministry of Health Research Fund for Medical Sciences (WKJ-ZJ-1828 to J.Z.), Wenzhou City Grants (Y20160410 to C.L. and C20170007 to J.Z.), and Lin He’s Academician Workstation of New Medicine and Clinical Translation (17331209 to C.L. and 18331105 to J.Z.).
Footnotes
Supplemental Information can be found online at https://doi.org/10.1016/j.omtn.2020.03.004.
Contributor Information
Junzhao Zhao, Email: z.joyce08@163.com.
Feng Gu, Email: gufenguw@gmail.com.
Supplemental Information
References
- 1.Yin H., Kauffman K.J., Anderson D.G. Delivery technologies for genome editing. Nat. Rev. Drug Discov. 2017;16:387–399. doi: 10.1038/nrd.2016.280. [DOI] [PubMed] [Google Scholar]
- 2.Komor A.C., Badran A.H., Liu D.R. CRISPR-Based technologies for the manipulation of eukaryotic genomes. Cell. 2017;168:20–36. doi: 10.1016/j.cell.2016.10.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Davis A.J., Chen D.J. DNA double strand break repair via non-homologous end-joining. Transl. Cancer Res. 2013;2:130–143. doi: 10.3978/j.issn.2218-676X.2013.04.02. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Vilenchik M.M., Knudson A.G. Endogenous DNA double-strand breaks: production, fidelity of repair, and induction of cancer. Proc. Natl. Acad. Sci. USA. 2003;100:12871–12876. doi: 10.1073/pnas.2135498100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hilton I.B., Gersbach C.A. Enabling functional genomics with genome engineering. Genome Res. 2015;25:1442–1455. doi: 10.1101/gr.190124.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wyman C., Kanaar R. DNA double-strand break repair: all’s well that ends well. Annu. Rev. Genet. 2006;40:363–383. doi: 10.1146/annurev.genet.40.110405.090451. [DOI] [PubMed] [Google Scholar]
- 7.Cong L., Ran F.A., Cox D., Lin S., Barretto R., Habib N., Hsu P.D., Wu X., Jiang W., Marraffini L.A., Zhang F. Multiplex genome engineering using CRISPR/Cas systems. Science. 2013;339:819–823. doi: 10.1126/science.1231143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zu Y., Tong X., Wang Z., Liu D., Pan R., Li Z., Hu Y., Luo Z., Huang P., Wu Q. TALEN-mediated precise genome modification by homologous recombination in zebrafish. Nat. Methods. 2013;10:329–331. doi: 10.1038/nmeth.2374. [DOI] [PubMed] [Google Scholar]
- 9.Bodles-Brakhop A.M., Heller R., Draghia-Akli R. Electroporation for the delivery of DNA-based vaccines and immunotherapeutics: current clinical developments. Mol. Ther. 2009;17:585–592. doi: 10.1038/mt.2009.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kay M.A., Glorioso J.C., Naldini L. Viral vectors for gene therapy: the art of turning infectious agents into vehicles of therapeutics. Nat. Med. 2001;7:33–40. doi: 10.1038/83324. [DOI] [PubMed] [Google Scholar]
- 11.Midoux P., Pichon C., Yaouanc J.-J., Jaffrès P.-A. Chemical vectors for gene delivery: a current review on polymers, peptides and lipids containing histidine or imidazole as nucleic acids carriers. Br. J. Pharmacol. 2009;157:166–178. doi: 10.1111/j.1476-5381.2009.00288.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yin H., Xue W., Chen S., Bogorad R.L., Benedetti E., Grompe M., Koteliansky V., Sharp P.A., Jacks T., Anderson D.G. Genome editing with Cas9 in adult mice corrects a disease mutation and phenotype. Nat. Biotechnol. 2014;32:551–553. doi: 10.1038/nbt.2884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Komor A.C., Kim Y.B., Packer M.S., Zuris J.A., Liu D.R. Programmable editing of a target base in genomic DNA without double-stranded DNA cleavage. Nature. 2016;533:420–424. doi: 10.1038/nature17946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Komor A.C., Zhao K.T., Packer M.S., Gaudelli N.M., Waterbury A.L., Koblan L.W., Kim Y.B., Badran A.H., Liu D.R. Improved base excision repair inhibition and bacteriophage Mu Gam protein yields C:G-to-T:A base editors with higher efficiency and product purity. Sci. Adv. 2017;3:o4774. doi: 10.1126/sciadv.aao4774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nishida K., Arazoe T., Yachie N., Banno S., Kakimoto M., Tabata M., Mochizuki M., Miyabe A., Araki M., Hara K.Y. Targeted nucleotide editing using hybrid prokaryotic and vertebrate adaptive immune systems. Science. 2016;353:aaf8729. doi: 10.1126/science.aaf8729. [DOI] [PubMed] [Google Scholar]
- 16.Komor A.C., Badran A.H., Liu D.R. Editing the genome without double-stranded DNA Breaks. ACS Chem. Biol. 2018;13:383–388. doi: 10.1021/acschembio.7b00710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kim Y.B., Komor A.C., Levy J.M., Packer M.S., Zhao K.T., Liu D.R. Increasing the genome-targeting scope and precision of base editing with engineered Cas9-cytidine deaminase fusions. Nat. Biotechnol. 2017;35:371–376. doi: 10.1038/nbt.3803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rees H.A., Komor A.C., Yeh W.-H., Caetano-Lopes J., Warman M., Edge A.S.B., Liu D.R. Improving the DNA specificity and applicability of base editing through protein engineering and protein delivery. Nat. Commun. 2017;8 doi: 10.1038/ncomms15790. 15790–15790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hess G.T., Frésard L., Han K., Lee C.H., Li A., Cimprich K.A., Montgomery S.B., Bassik M.C. Directed evolution using dCas9-targeted somatic hypermutation in mammalian cells. Nat. Methods. 2016;13:1036–1042. doi: 10.1038/nmeth.4038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ma Y., Zhang J., Yin W., Zhang Z., Song Y., Chang X. Targeted AID-mediated mutagenesis (TAM) enables efficient genomic diversification in mammalian cells. Nat. Methods. 2016;13:1029–1035. doi: 10.1038/nmeth.4027. [DOI] [PubMed] [Google Scholar]
- 21.Li J., Sun Y., Du J., Zhao Y., Xia L. Generation of targeted point mutations in rice by a modified CRISPR/Cas9 system. Mol. Plant. 2017;10:526–529. doi: 10.1016/j.molp.2016.12.001. [DOI] [PubMed] [Google Scholar]
- 22.Lu Y., Zhu J.-K. Precise Editing of a target base in the rice genome using a modified CRISPR/Cas9 system. Mol. Plant. 2017;10:523–525. doi: 10.1016/j.molp.2016.11.013. [DOI] [PubMed] [Google Scholar]
- 23.Tang W., Hu J.H., Liu D.R. Aptazyme-embedded guide RNAs enable ligand-responsive genome editing and transcriptional activation. Nat. Commun. 2017;8:15939. doi: 10.1038/ncomms15939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gaudelli N.M., Komor A.C., Rees H.A., Packer M.S., Badran A.H., Bryson D.I., Liu D.R. Programmable base editing of A•T to G•C in genomic DNA without DNA cleavage. Nature. 2017;551:464–471. doi: 10.1038/nature24644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hu J.H., Miller S.M., Geurts M.H., Tang W., Chen L., Sun N., Zeina C.M., Gao X., Rees H.A., Lin Z., Liu D.R. Evolved Cas9 variants with broad PAM compatibility and high DNA specificity. Nature. 2018;556:57–63. doi: 10.1038/nature26155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Liang P., Sun H., Zhang X., Xie X., Zhang J., Bai Y., Ouyang X., Zhi S., Xiong Y., Ma W. Effective and precise adenine base editing in mouse zygotes. Protein Cell. 2018;9:808–813. doi: 10.1007/s13238-018-0566-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yang L., Zhang X., Wang L., Yin S., Zhu B., Xie L., Duan Q., Hu H., Zheng R., Wei Y. Increasing targeting scope of adenosine base editors in mouse and rat embryos through fusion of TadA deaminase with Cas9 variants. Protein Cell. 2018;9:814–819. doi: 10.1007/s13238-018-0568-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Liu Z., Lu Z., Yang G., Huang S., Li G., Feng S., Liu Y., Li J., Yu W., Zhang Y. Efficient generation of mouse models of human diseases via ABE- and BE-mediated base editing. Nat. Commun. 2018;9:2338. doi: 10.1038/s41467-018-04768-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ryu S.-M., Koo T., Kim K., Lim K., Baek G., Kim S.-T., Kim H.S., Kim D.E., Lee H., Chung E., Kim J.S. Adenine base editing in mouse embryos and an adult mouse model of Duchenne muscular dystrophy. Nat. Biotechnol. 2018;36:536–539. doi: 10.1038/nbt.4148. [DOI] [PubMed] [Google Scholar]
- 30.Yan F., Kuang Y., Ren B., Wang J., Zhang D., Lin H., Yang B., Zhou X., Zhou H. Highly efficient A.T to G.C base editing by Cas9n-guided tRNA adenosine deaminase in rice. Mol. Plant. 2018;11:631–634. doi: 10.1016/j.molp.2018.02.008. [DOI] [PubMed] [Google Scholar]
- 31.Li C., Zong Y., Wang Y., Jin S., Zhang D., Song Q., Zhang R., Gao C. Expanded base editing in rice and wheat using a Cas9-adenosine deaminase fusion. Genome Biol. 2018;19:59. doi: 10.1186/s13059-018-1443-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zuo E., Sun Y., Wei W., Yuan T., Ying W., Sun H., Yuan L., Steinmetz L.M., Li Y., Yang H. Cytosine base editor generates substantial off-target single-nucleotide variants in mouse embryos. Science. 2019;364:289–292. doi: 10.1126/science.aav9973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jin S., Zong Y., Gao Q., Zhu Z., Wang Y., Qin P., Liang C., Wang D., Qiu J.-L., Zhang F., Gao C. Cytosine, but not adenine, base editors induce genome-wide off-target mutations in rice. Science. 2019;364:292–295. doi: 10.1126/science.aaw7166. [DOI] [PubMed] [Google Scholar]
- 34.Zong Y., Wang Y., Li C., Zhang R., Chen K., Ran Y., Qiu J.-L., Wang D., Gao C. Precise base editing in rice, wheat and maize with a Cas9-cytidine deaminase fusion. Nat. Biotechnol. 2017;35:438–440. doi: 10.1038/nbt.3811. [DOI] [PubMed] [Google Scholar]
- 35.Bahal R., Ali McNeer N., Quijano E., Liu Y., Sulkowski P., Turchick A., Lu Y.-C., Bhunia D.C., Manna A., Greiner D.L. In vivo correction of anaemia in β-thalassemic mice by γPNA-mediated gene editing with nanoparticle delivery. Nat. Commun. 2016;7:13304. doi: 10.1038/ncomms13304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nabel C.S., Jia H., Ye Y., Shen L., Goldschmidt H.L., Stivers J.T., Zhang Y., Kohli R.M. AID/APOBEC deaminases disfavor modified cytosines implicated in DNA demethylation. Nat. Chem. Biol. 2012;8:751–758. doi: 10.1038/nchembio.1042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mezei P.D., Csonka G.I. Features of the interactions between the methyl-CpG motif and the arginine residues on the surface of MBD proteins. Struct. Chem. 2016;27:1317–1326. [Google Scholar]
- 38.Liang P., Ding C., Sun H., Xie X., Xu Y., Zhang X., Sun Y., Xiong Y., Ma W., Liu Y. Correction of β-thalassemia mutant by base editor in human embryos. Protein Cell. 2017;8:811–822. doi: 10.1007/s13238-017-0475-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Jensen K.T., Fløe L., Petersen T.S., Huang J., Xu F., Bolund L., Luo Y., Lin L. Chromatin accessibility and guide sequence secondary structure affect CRISPR-Cas9 gene editing efficiency. FEBS Lett. 2017;591:1892–1901. doi: 10.1002/1873-3468.12707. [DOI] [PubMed] [Google Scholar]
- 40.Daer R.M., Cutts J.P., Brafman D.A., Haynes K.A. The Impact of chromatin dynamics on Cas9-mediated genome editing in human cells. ACS Synth. Biol. 2017;6:428–438. doi: 10.1021/acssynbio.5b00299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yarrington R.M., Verma S., Schwartz S., Trautman J.K., Carroll D. Nucleosomes inhibit target cleavage by CRISPR-Cas9 in vivo. Proc. Natl. Acad. Sci. USA. 2018;115:9351–9358. doi: 10.1073/pnas.1810062115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yuan J., Ma Y., Huang T., Chen Y., Peng Y., Li B., Li J., Zhang Y., Song B., Sun X. Genetic modulation of RNA splicing with a CRISPR-guided cytidine deaminase. Mol. Cell. 2018;72:380–394.e7. doi: 10.1016/j.molcel.2018.09.002. [DOI] [PubMed] [Google Scholar]
- 43.Wang X., Li J., Wang Y., Yang B., Wei J., Wu J., Wang R., Huang X., Chen J., Yang L. Efficient base editing in methylated regions with a human APOBEC3A-Cas9 fusion. Nat. Biotechnol. 2018;36:946–949. doi: 10.1038/nbt.4198. [DOI] [PubMed] [Google Scholar]
- 44.Thuronyi B.W., Koblan L.W., Levy J.M., Yeh W.-H., Zheng C., Newby G.A., Wilson C., Bhaumik M., Shubina-Oleinik O., Holt J.R. Continuous evolution of base editors with expanded target compatibility and improved activity. Nat. Biotechnol. 2019;37:1070–1079. doi: 10.1038/s41587-019-0193-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Mali P., Aach J., Stranges P.B., Esvelt K.M., Moosburner M., Kosuri S., Yang L., Church G.M. CAS9 transcriptional activators for target specificity screening and paired nickases for cooperative genome engineering. Nat. Biotechnol. 2013;31:833–838. doi: 10.1038/nbt.2675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Liang P., Xie X., Zhi S., Sun H., Zhang X., Chen Y., Chen Y., Xiong Y., Ma W., Liu D. Genome-wide profiling of adenine base editor specificity by EndoV-seq. Nat. Commun. 2019;10:67. doi: 10.1038/s41467-018-07988-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tu M., Lin L., Cheng Y., He X., Sun H., Xie H., Fu J., Liu C., Li J., Chen D. A ‘new lease of life’: FnCpf1 possesses DNA cleavage activity for genome editing in human cells. Nucleic Acids Res. 2017;45:11295–11304. doi: 10.1093/nar/gkx783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zhang Y., Ge X., Yang F., Zhang L., Zheng J., Tan X., Jin Z.-B., Qu J., Gu F. Comparison of non-canonical PAMs for CRISPR/Cas9-mediated DNA cleavage in human cells. Sci. Rep. 2014;4:5405. doi: 10.1038/srep05405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wang Y., Wang B., Xie H., Ren Q., Liu X., Li F., Lv X., He X., Cheng C., Deng R. Efficient human genome editing using saCas9 ribonucleoprotein complexes. Biotechnol. J. 2019;14:e1800689. doi: 10.1002/biot.201800689. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.