Graphical abstract

Keywords: Transcription factor, Epigenetics, Single cell RNA-seq, Single cell methylome
Abstract
The brain is a highly complex organ consisting of numerous types of cells with ample diversity at the epigenetic level to achieve distinct gene expression profiles. During neuronal cell specification, transcription factors (TFs) form regulatory modules with chromatin remodeling proteins to initiate the cascade of epigenetic changes. Currently, little is known about brain epigenetic regulatory modules and how they regulate gene expression in a cell-type specific manner. To infer TFs involved in neuronal specification, we applied a recursive motif search approach on the differentially methylated regions identified from single-cell methylomes. The epigenetic transcription regulatory modules (ETRM), including EGR1 and MEF2C, were predicted and the co-expression of TFs in ETRMs were examined with RNA-seq data from single or sorted brain cells using a conditional probability matrix. Lastly, computational predications were validated with EGR1 ChIP-seq data. In addition, methylome and RNA-seq data generated from Egr1 knockout mice supported the essential role of EGR1 in brain epigenome programming, in particular for excitatory neurons. In summary, we demonstrated that brain single cell methylome and RNA-seq data can be integrated to gain a better understanding of how ETRMs control cell specification. The analytical pipeline implemented in this study is freely accessible in the Github repository (https://github.com/Gavin-Yinld/brain_TF).
1. Introduction
The importance of DNA methylation in neuronal differentiation [1], neural plasticity [2], [3], [4], [5], [6], and neural functioning [7], [8], [9] has been firmly established. During brain development, de novo DNA methylation occurs at the promoters of germ line-specific genes to repress pluripotency in progenitor cells, while methylation loss at the promoters activates neuron-specific genes [1]. Disorders in epigenetic machinery have been linked to many neurological diseases [10], [11], [12]. For instance, mutations in the DNA methyltransferase DNMT3B leads to defective brain development [13], and mutations in the methyl-cytosine binding protein MECP2 have been linked to Rett syndrome [14]. In addition, aberrant DNA methylation may lead to the premature activation of neuronal progenitor cells and, potentially, the development of brain tumors [15]. Despite our growing realization of neuroepigenetics, the epigenetic mechanism underlying brain cell specification remains largely unknown.
Transcription factors are known to be the master regulators of gene expression and play essential roles in cell-fate decision making. A number of databases, including TRANSFAC [16] and JASPAR [17], attempt to gather information on transcription factors together with their binding preferences in multiple species. On top of these databases, the Catalog of Inferred Sequence Binding Preferences (CIS-BP) [18] and HOCOMOCO [19] provide a large collection of TF binding motifs via the analyses of DNA binding domains in TFs and the consensus sequences predicted with large-scale ChIP-seq data. Recently, CIS-BP was expanded using the similarity regression method to quantify motif evolution with improved precision [20], and a comprehensive list of 1513 mouse transcription factors were documented in v2.0 of the CIS-BP database. Many of these transcription factors may interact with each other to form transcriptional regulatory modules essential for neuronal specification and diversity [21], [22], [23], [24]. For instance, our recent study demonstrated that EGR1, a transcription factor important for memory formation, can participate in brain methylome programming [25]. More specifically, EGR1 recruits a DNA demethylase, called TET1, to remove the methylation marks and activate downstream genes. However, it remains elusive how transcription factors in regulatory modules work together in a cell-type-specific manner.
Growing single cell “omics” data provide an opportunity to explore brain epigenetic regulatory modules with unprecedented resolution. Many different types of neurons have been determined with single cell RNA-seq [26], [27], [28], [29] and methylome sequencing [30], [31], [32], [33]. Each subtype of neurons has a distinct DNA methylation profile and, thus, some genomic loci may demonstrate a bipolar DNA methylation pattern, i.e., hypermethylated in one cell subset but hypomethylated in others [34]. The number of these bipolar methylated loci increased dramatically during the early stages of brain development in both human and mouse frontal cortices [24]. In addition, the development-related epigenetic changes tend to co-localize together in functional genomic regions critical for regulating gene expression [21]. It would be interesting to learn what transcription factors participate in the epigenetic control of these functional genomic regions and how they interplay with each other in the establishment of cell-type specific DNA methylation patterns.
In this study, we symmetrically determined transcription factors participating in the process of brain neuron specification. To predict neuronal cell-type specific epigenetic transcription regulatory modules, we started with over 3000 single-cell methylomes of sixteen neuronal subtypes and determined a set of transcription factors having motifs enriched in differentially methylated regions of neuronal subtypes. Epigenetic transcription regulatory modules were then inferred with a recursive motif search algorithm. The co-expression patterns of transcription factors in a module were further demonstrated together with cell-subtype specific markers using brain single-cell RNA-seq data. Finally, we focused on the Egr1 gene and validated the computational predictions with ChIP-seq, RNA-seq, and methylome sequencing data.
2. Results
2.1. Single-cell methylome analyses identified key TFs associated with brain cell specification
To determine what transcription factors are involved in the epigenetic regulation of neuronal cell specification, we started with brain single cell methylomes generated for 3377 neurons derived from the mouse frontal cortex [31] (Supplementary Table 1). In this publicly available dataset, an average of 1.4 million reads, covering around 4.7% of the genome, were obtained for each single neuron. 3377 neurons were clustered into 16 subpopulations according to their methylation profiles, including 10 excitatory neuron subtypes and 6 inhibitory neuron subtypes (Supplementary Table 2). For a given neuronal subtype, genomic regions with significantly lower methylation levels were defined as CG-DMRs (Differentially Methylated Regions) in the previous report [31]. Altogether, for the 16-neuron subtypes, a total of 575,524 genomic loci, covering 5.8% of the genome, were determined as CG-DMRs. 73.2% of these CG-DMRs were located more than 10 kb from the transcription start sites, suggesting that epigenetic regulation on distal enhancers is critical for neuronal cell specification.
For each neuronal subtype, we performed a recursive motif search to determine transcription factors with motifs enriched in CG-DMR genomic sequences. We used neuronal subtype mL2/3, which has the most CG-DMRs as an example to illustrate this procedure (Fig. 1A). The first iteration of the motif search identified the Egr1 gene as the “key” TF; this motif was the most significantly enriched in the 279,775 DMRs in the mL2/3 neurons that we started with. We then removed 40,171 CG-DMRs containing the motif for EGR1 and performed the second iteration of the motif search on the remaining (239,604) CG-DMRs. Following this procedure recursively, Mef2b, Fra1, Rfx2, and Oct genes were identified as “key” TFs in the second to the fifth iterations, respectively. The recursive search was terminated at the sixth iteration, where no TF motifs were found to have an enrichment p-value less than 1e-10 in the genomic sequences of the remaining 120,509 CG-DMRs (Fig. 1B). This recursive motif algorithm was expanded to the CG-DMRs of all 16 neuronal subtypes. A total of seventeen distinct “key” TFs were determined. Interestingly, these TFs can be classified into three groups according to motif enrichment in the DMRs of neuronal subtypes (Fig. 1C). Transcription factors involved in neurogenesis and early brain development, such as the Mef2, Atoh1, and Nf1 genes, have motifs that are enriched in both excitatory and inhibitory neuronal subtypes. Several neuronal-activity-induced transcription factors including Egr1, Atf3, and Junb (a subunit of Ap1) have motifs enriched in CG-DMRs in excitatory neuronal subtypes. The motifs of the Lhx3, Ap4, and Mafa genes are enriched in CG-DMRs for inhibitory neuronal subtypes. We selected the Egr, Mef2 and Maf genes as the representatives of the three groups of key TFs to further explore the characteristics of key TFs. Since multiple EGR, MEF2, and MAF family members are motif enriched, we used the ‘universalmotif’ R library (https://github.com/bjmt/universalmotif) to merge similar TF motifs belonging to the same family. From 2512 to 93,911 genomic loci hosting the motifs for the desired TFs were identified from the aforementioned 575,524 CG-DMRs. We determined the methylation profiles of these genomic loci across neuronal cell types, and observed an association between cell-subtype motif enrichment and the methylation patterns of genomic regions surrounding the binding motifs predicted for these key TFs. For instance, genomic loci hosting the motifs for EGR family members showed lower methylation levels in excitatory neurons than those for MAF family members, but higher methylation in inhibitory neurons (Fig. 1D). Compared to EGR and MAF families, MEF2 family members with motifs enriched in both excitatory and inhibitory neuronal subtypes showed a median methylation level between those of EGR and MAF in excitatory neurons but similar to that of MAF in inhibitory neurons (Fig. 1E). TF motif enrichment and the methylation profiles of their corresponding genomic loci suggest that the key TFs identified may perform their regulatory roles in a cell-type specific manner. Worthy of mention, we found that mIn1 subtype tends to be clustered with inhibitory instead of excitatory neurons (Fig. 1C). This is likely due to the clustering of single neurons in the previous study, which was based on mCH level instead of CG-DMRs [31].
Fig. 1.
Recursive motif analysis identified 17 key TFs. A) Recursive motif identification procedure. B) Results of recursive motif analysis on DMRs of mL23 neurons. C) Enrichment of key TFs identified from DMRs determined for 16 neuron types. D) Methylation level of genomic loci containing motif of EGR, MEF2, and MAF family members in excitatory and inhibitory neurons. E) Methylation level of genomic loci containing motif of EGR, MEF2, and MAF family members in sixteen neuronal subtypes.
2.2. Single-cell RNA-seq analyses revealed the co-expression of key TFs and marker genes in various cell types
To explore the cell-type specificity of key TFs, we made use of 11,886 single cell RNA-seq data from P60 mouse prefrontal cortex, which were assigned into eight major cell clusters according to the expression of cell type-specific markers [35] (see Methods). Among the 11,886 cells, 56.5% were assigned as excitatory neurons, 4.7% were assigned as inhibitory neurons, and the remaining cells were assigned as astrocytes (Astro), oligodendrocyte (Oligo), newly formed oligodendrocytes (NF_oligo), oligodendrocyte precursors (OPC), microglia, or endothelial cells (Endo), with the proportions ranging from 1.3% to 15.6% (Supplementary Table 3).
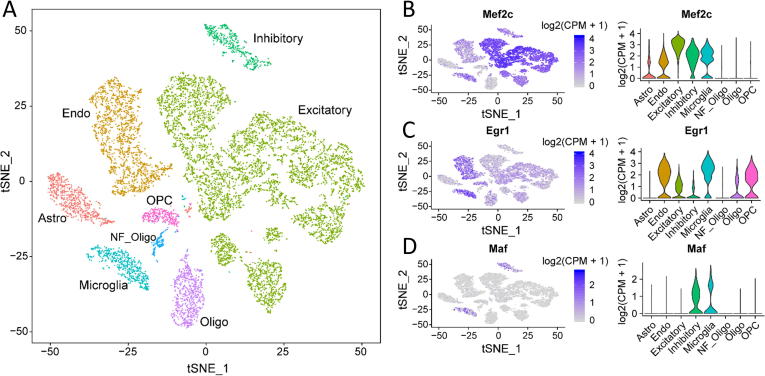
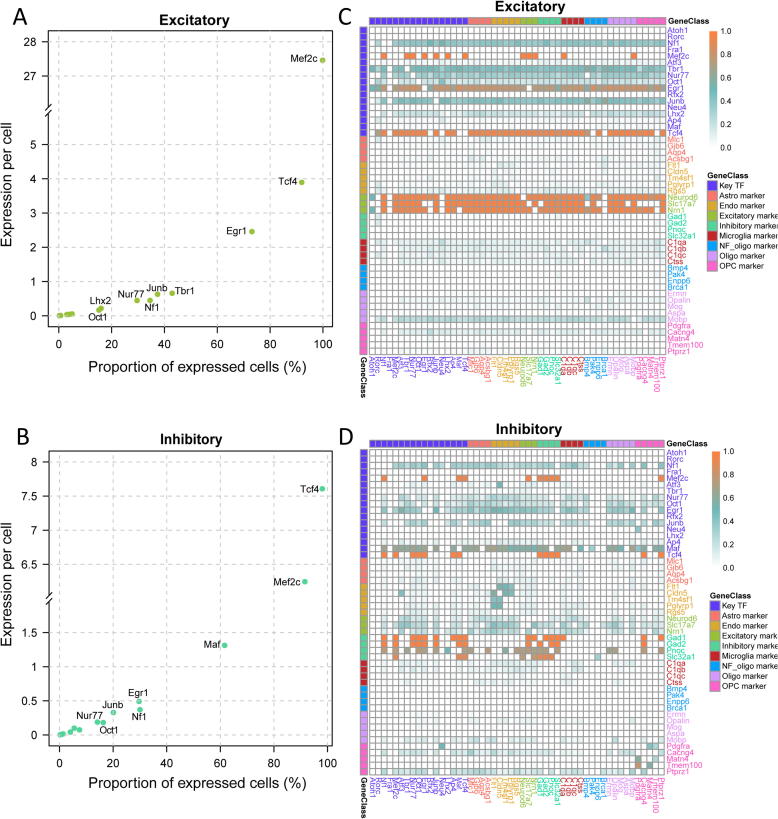
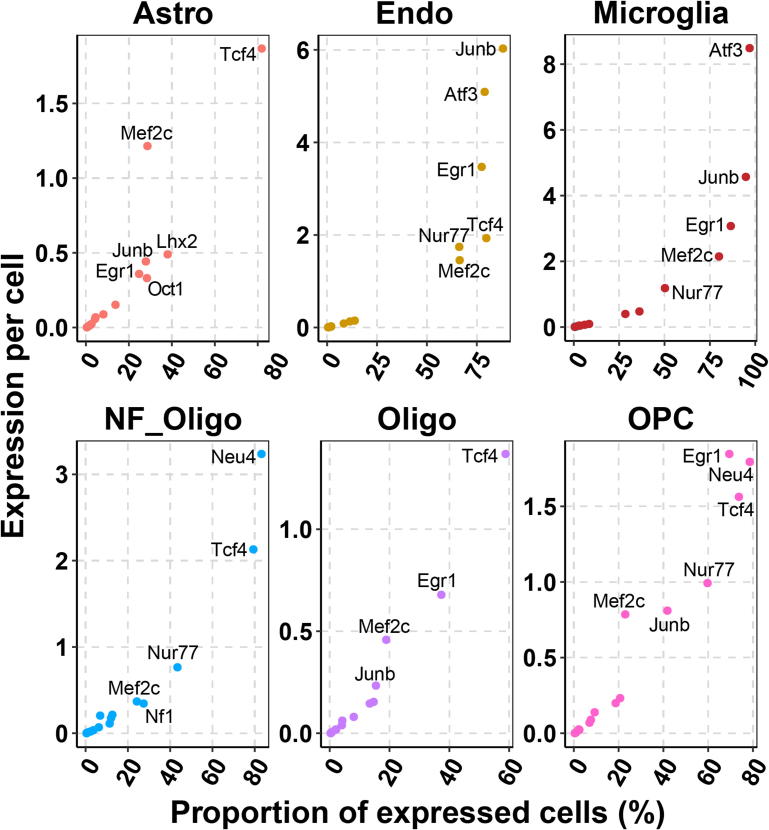
First, we followed the “Seurat” pipeline [36] to identify highly variable genes and demonstrated that the eight cell types were as shown in the previous report [35] (Fig. 2A). Next, we examined the expression profiles of key TFs in each cell type. While Mef2c expressed in both excitatory and inhibitory neurons (Fig. 2B), the Egr1 gene was expressed in excitatory neurons but was low in inhibitory neurons (Fig. 2C). In contrast, the Maf gene was expressed in inhibitory neurons but was depleted in excitatory neurons (Fig. 2D). We determined the expression level and the number of cells with key TF expression across single cells (Fig. 3A&B and Fig. S1). Approximately 99.6% excitatory and 91.3% inhibitory neurons have Mef2c expression. 72.9% of excitatory neurons have Egr1 expression and 61.4% of inhibitory neurons have Maf expression. Using the conditional probability matrix (see Methods section), we further investigated the association of gene expression among TFs and cell-type-specific markers reported previously [35] (Fig. 3C&D and Fig. S2). In excitatory neurons, Mef2c, Tcf4, and Egr1 showed co-expression patterns with Neurod6, which is a marker for excitatory neurons and is involved in the development and differentiation of the nervous system. More specifically, 99.7% of the excitatory neurons with expressed Neurod6 have Mef2c expression (Fig. 3C). On the other hand, 92.0% of the excitatory neurons with Mef2c expression also expressed Neurod6. In inhibitory neurons, highly expressed key TFs, such as Mef2c and Maf, showed co-expression patterns with inhibitory neuronal markers, including Gad2. Gad2 expression is present in 97.4% of the inhibitory neurons where Maf is expressed, and 59.9% of Gad2-expressing inhibitory neurons have Maf expression (Fig. 3D). These scRNA-seq results provide evidence for cell-type-specific expression of transcription factors. Thus, Mef2c may serve as a pan-neuron regulator and play roles in both excitatory and inhibitory neurons, while Egr1 and Maf may function in a neuron-subtype specific manner.
Fig. 2.
Expression of selective key TFs across brain cell types. A) Demonstration of eight cell clusters using t-SNE map. B) Expression of Mef2c in each cell cluster color-highlighted on t-SNE plots (left panel) and violin plot shows the expression level of Mef2c in each cell cluster (right panel). Gene expression profile of each single cell was normalized to counts per-million (CPM) and natural log transformed. C) Expression of Egr1 in each cell cluster color-highlighted on t-SNE plots (left panel) and violin plot shows the expression level of Egr1 in each cell cluster (right panel). D) Expression of Maf in each cell cluster color-highlighted on t-SNE plots (left panel) and violin plot shows the expression level of Maf in each cell cluster (right panel).
Fig. 3.
Key TFs co-expressed with marker genes in excitatory and inhibitory neurons. A and B) Summary of the expression of key TFs in excitatory (A) and inhibitory neurons (B). The expression count value obtained from Aritra et al. [35] were shown. C and D) Dependent expression relationship between key TFs and cell type specific expression markers in excitatory neurons (C) and inhibitory neurons (D). Each square in the heatmap represents the probability of TFA (y-axis) expressing under the condition of TFB (x-axis) expressing. The relationship between TF pairs with a p value over 0.05 in the hypergeometric test were set to 0.
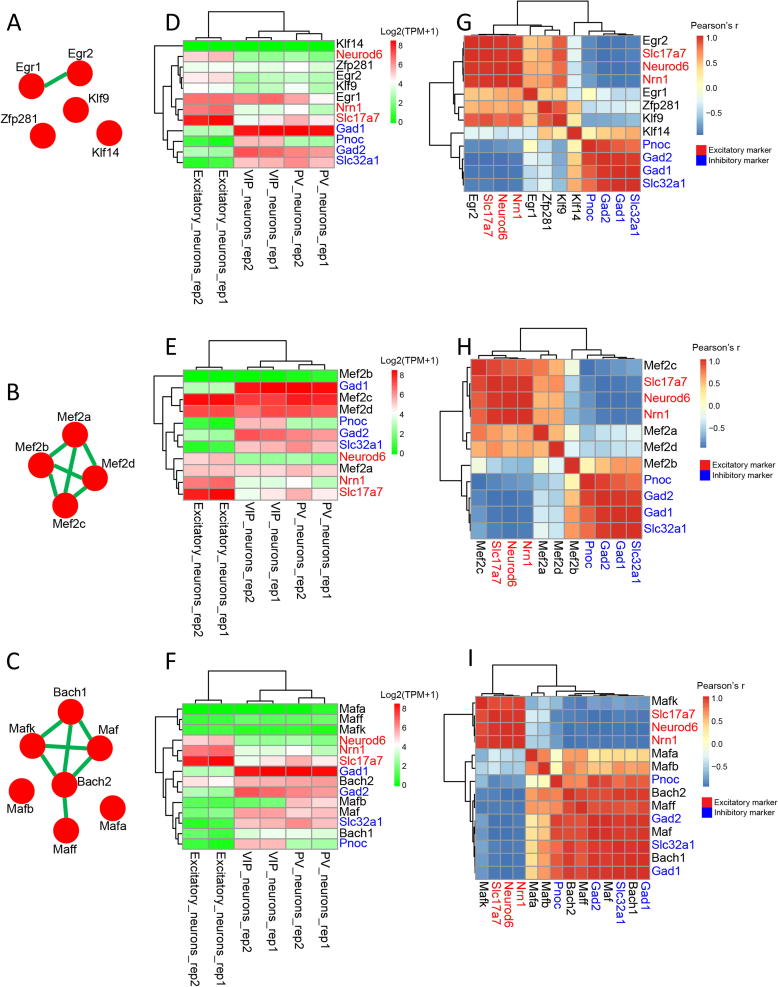
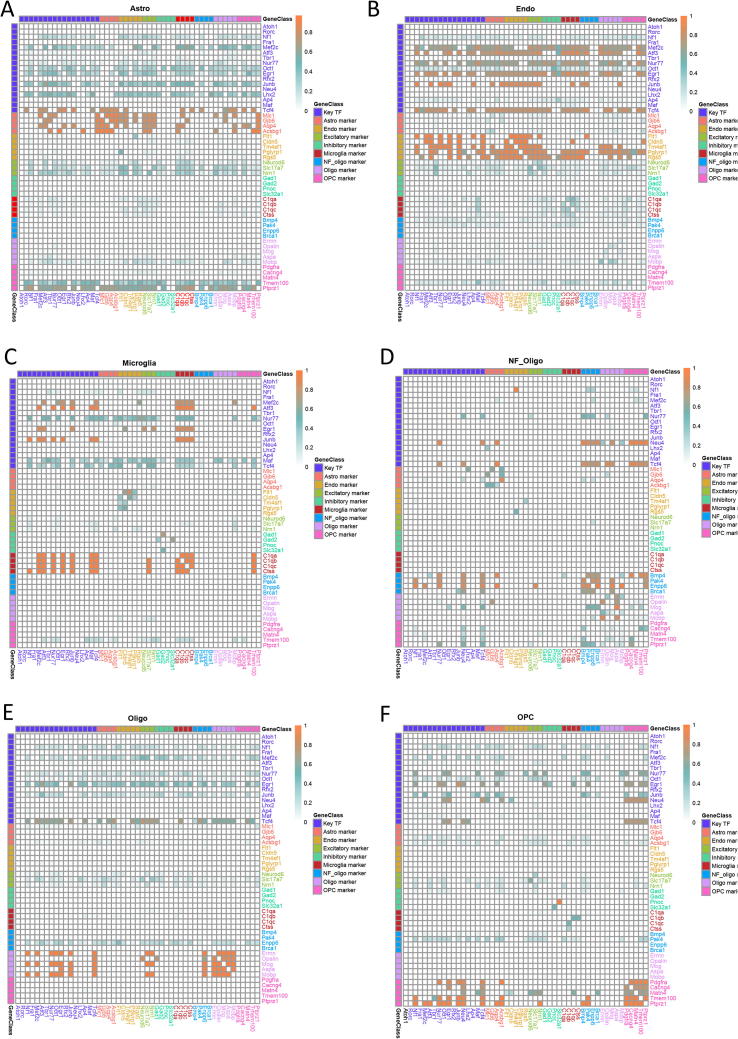
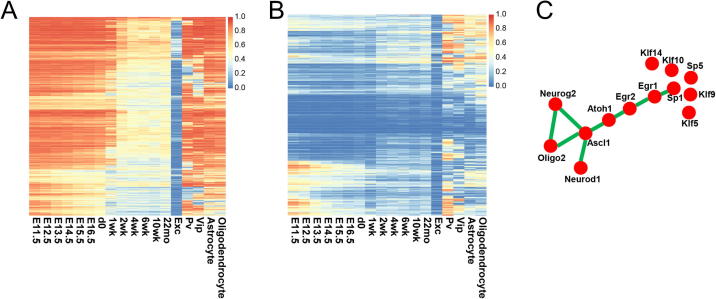
Recently, we implemented a pipeline to infer epigenetic transcriptional regulatory modules (ETRMs) associated with differentially methylated regions (DMRs) [22]. Using this pipeline, a recursive search algorithm was applied to identify co-enriched transcription factor motifs within specific DMRs for each of 16 neuronal clusters. Thus, an ETRM refers to a set of TFs with binding sites adjacent to a ‘key’ TF whose motif is the most significantly enriched within a subset of DMR. ETRM inferred from motif co-enrichment analysis is analogous to a protein–protein interaction network where the hub genes would be the key TFs in ETRMs. Using such an idea, motif co-enrichment networks for Egr1, Mef2c, and Maf were constructed for neuron subtypes including mL2/3 with the largest DMRs identified in excitatory neurons and mPv, which is the largest inhibitory neuronal subtype. In mL2/3 neurons, Egr2, Klf9, Klf14, and Zfp281 genes have motifs co-enriched in genomic loci containing the Egr1 motif (Fig. 4A). MEF2 members tended to have motifs co-enriched with the same TF family members (Fig. 4B). Similarly, in mPv neurons, MAF family members, including Bach1 and Bach2, had motifs co-enriched with Maf (Fig. 4C). In addition, most of these co-enriched TFs are documented in STRING database [37] to tightly interact with each other at protein level (Fig. 4B&C). Despite the low expression of some TFs in single neurons, we observed that transcription factors identified in an ETRM tended to co-express in single cells. For instance, Egr1, Klf9, and Zfp28 are co-expressed in excitatory neurons, while Maf, Mafb, and Bach1 are co-expressed in inhibitory neurons (Fig. S3A–C).
Fig. 4.
ETRMs of Egr1 in mL23, Mef2c in mL23, and Maf in mPv. A) ETRM predicted from Egr1 motif in DMRs of mL23 neurons. The green edge means that there is evidence in the STRING database to support interaction between the proteins coding from the connected two genes. B) ETRM predicted from the Mef2c motif in DMRs of mL23 neurons. C) ETRM predicted from the Maf motif in DMRs of mPv neurons. D-F) Expression of Egr1 ETRM (D), Mef2c ETRM (E), and Maf ETRM (F) in EXC, PV, and VIP neurons. G-I) Correlation between the expression of Egr1 ETRM (G), Mef2c ETRM (H), Maf ETRM (I) and excitatory/inhibitory markers in EXC, PV, and VIP neurons. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
2.3. RNA-seq analyses from sorted cells support the cell-subtype specific functions of key TFs
The current single cell RNA-seq technique has a limitation in its detection of transcripts with low expression. We expanded the co-expression analyses to include RNA-seq data generated for three types of sorted neuronal cells, including excitatory (EXC) neurons, parvalbumin (PV) expressing fast-spiking interneurons, and vasoactive intestinal peptide (VIP) expressing interneurons [38]. The TFs in Egr1 ETRM and Maf ETRM are primarily expressed in excitatory (Fig. 4D) and inhibitory neurons (Fig. 4F), respectively. On the other hand, the TFs in Mef2c ETRM are expressed in both excitatory and inhibitory neurons (Fig. 4E). Not surprisingly, strong expression correlations were observed between Egr1 ETRM and excitatory markers (Fig. 4G), Maf ETRM and inhibitory markers (Fig. 4I). Although most TFs in Mef2c ETRM are highly expressed in both excitatory and inhibitory neurons, some MEF2 family members show a higher correlation with excitatory markers (Fig. 4H). This indicates that MEF2 family members may have distinct functions in excitatory and inhibitory neurons.
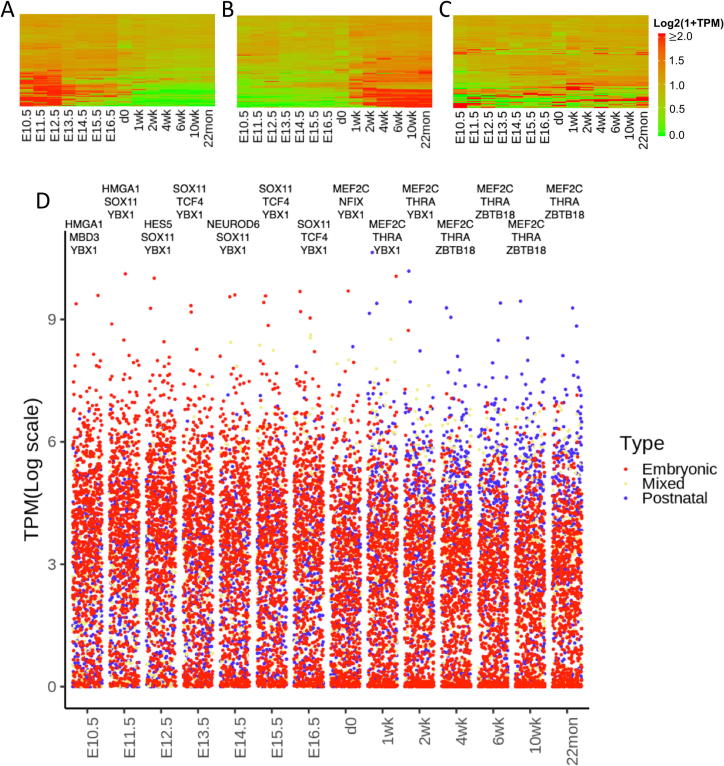
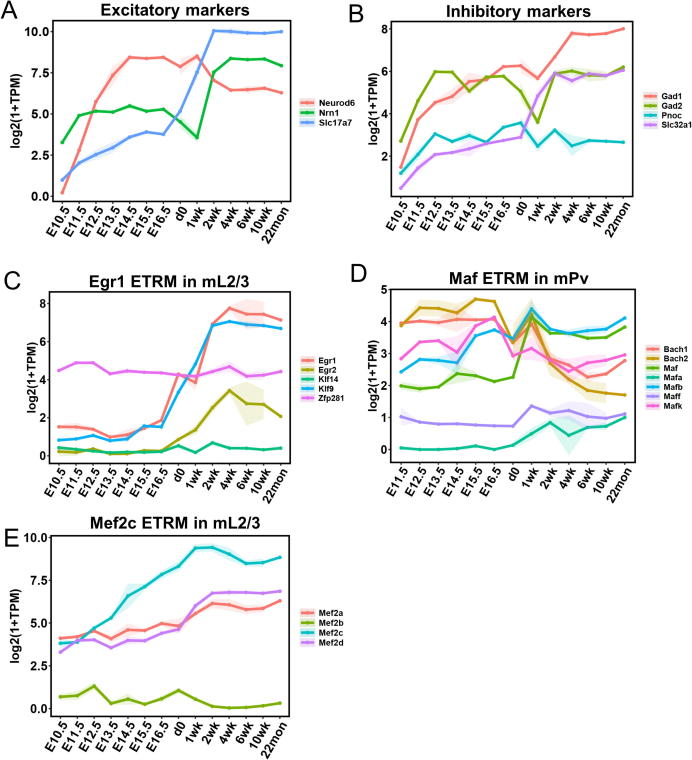
An important question to be addressed is whether the TFs in one ETRM would share a similar expression profile during brain development. To get an overview of developmental expression profiles, we collected mouse RNA-seq data from the forebrain, during embryonic stages (E10.5 to d0), and from postnatal frontal cortices in adult mice up to 22 months old (Supplementary Table 1). We first explored the expression of all the TFs in the mouse genome obtained from the CIS-BP database [20] during development and 1476 TFs were mapped to the RNA-seq data. We noticed that these TFs can be separated into three clusters: one being highly expressed in embryonic stages (Fig. S4A), one being highly expressed in postnatal stages (Fig. S4B), and the third showing a mixed pattern during development (Fig. S4C). These results reveal that many brain TFs have dynamic expression profiles and may function in specific stages during development. TFs highly expressed in embryonic stages may be key factors controlling neuronal stem cell proliferation and differentiation, while TFs highly expressed in postnatal stages may function in maturing or matured neurons. We showed that the top three TFs at each time point during development had the highest expression, for examples: Ybx1 from E10.5 to 2 week which is known for embryonic development [39], [40] and Mef2c from d0 to 22 month (Fig. S4D). Considering the exponential growing of glial cells after birth, some TFs with increased expression in postnatal stages could be glia-cell specific as well. In addition, most markers for excitatory and inhibitory neurons showed an increasing expression pattern during development (Fig. S5A&B). This trend was also observed for many cell-type specific ETRMs, such as Egr1, Egr2, and Klf9 in mL2/3 (Fig. S5C), MEF2 family members except for Mef2b in mL2/3 (Fig. S5E), and Maf and Mafb in mPv neurons (Fig. S5D). We also noticed that some TFs, such as Bach1, Bach2, and Mafk, showed a decreased expression pattern during development (Fig. S5D). This result suggests transient interactions among ETRM members and some family members may replace the others during development.
2.4. Experimental validation using EGR1 ChIP-seq, gene expression, and methylation profiles derived from Egr1KO mice
To validate the computational prediction of epigenetic regulation of brain cell specification, we made use of EGR1 ChIP-seq data generated from mouse frontal cortex, RNA-seq, and methylome data derived from Egr1 knockout (Egr1KO) mice [25]. We first predicted the ETRMs from EGR1 peaks identified in ChIP-seq data and overlapped EGR1 binding sites with the DMRs identified for mL2/3 neurons. Out of 12,015 EGR1 peaks obtained from ChIP-seq data, 3099 sites were located in the DMRs of mL2/3 neurons. Using CLARANS [41], these loci were first grouped into two clusters showing gradual hypo-methylation from embryonic to postnatal development (Fig. S6A), and hypo-methylation during embryonic development (Fig. S6B). Next, we predicted the co-enriched motifs in each cluster. The first cluster showed motif enrichment for EGR family members and KLF family members along with several TFs including Sp1, Oligo2, NeuroD1, etc. The second cluster showed motif enrichment for EGR and KLF family members only (Fig. S6C). This result is consistent with EGR1 ETRM predicted using DMRs alone (Fig. 4A) and the additional TFs identified in the first cluster are likely due to EGR1 peaks corresponding to the genomic loci bounded by the interacting proteins cross-linked together with EGR1 during the ChIP-seq procedure.
Next, to explore the influence of Egr1 loss on various neuronal cell types, we reanalyzed the methylome of Egr1KO cortex and focused on the methylation profiles of hyper-DMRs across 16 types of neurons. Comparing the methylomes derived from neuronal subtypes with controls, Egr1KO hyper-DMRs were found to be heavily methylated. According to their methylation profiles, these loci may be clustered into four groups (Fig. 5A). The first three groups have intermediate (group I), high (group II), and low (group III) methylation levels in the majority of methylomes examined, and have relatively constant methylation levels across sixteen neuronal subtypes (Fig. 5B). Interestingly, we found that 28.6% of Egr1KO hyper-DMRs (group IV) have lower methylation levels in excitatory neurons compared to those in inhibitory neurons (Fig. 5A&B). These results indicate that Egr1 could have significant functional differences between excitatory and inhibitory neurons.
Fig. 5.
Methylation profile of Egr1KO hyper-DMRs across sixteen neuronal cell types. A) Heatmap of a methylation profile of Egr1KO hyper-DMRs across sixteen neuronal cell types. Four co-methylated groups were identified by using the “cutree_rows” argument of “pheatmap” in R. B) Average methylation level of the four groups in Egr1KO hyper-DMRs. Shading shows the standard deviation.
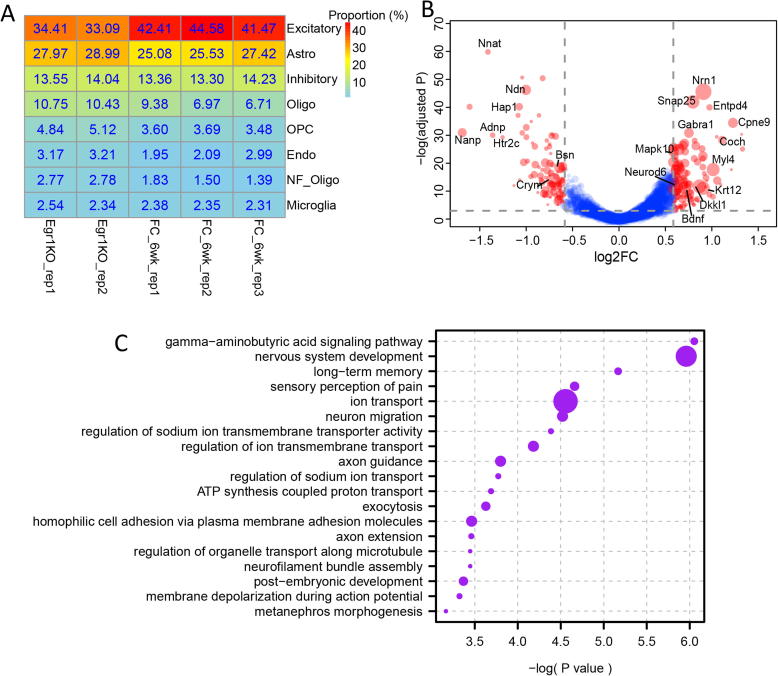
Finally, to further explore the influence that Egr1 knockout may have on brain cell specification, we made use of the expression profile of the Egr1 knockout (Egr1KO) mouse cortex [25], and estimated its cell-type composition from a single-cell RNA-seq-derived cell-type signature using the dampened weighted least squares algorithm [42]. This analysis was also performed on the expression profile of 6-week old mouse cortex [43] for comparison. As shown in Fig. 6A, compared to normal control, Egr1KO cortices were predicted to have a 9.1% reduction in excitatory neurons. Besides, Egr1 loss leads to the differential expression of gene markers for excitatory neurons including Nnat, Nrn1, and Snap25 (Fig. 6B). These genes are enriched in a pivotal biological process associated with brain functions, such as the “gamma-aminobutyric acid signaling pathway,” “nervous system development,” and “long-term memory” (Fig. 6C). In summary, these results confirmed the functional importance of EGR1 during brain development, particularly for the specification of excitatory neurons.
Fig. 6.
Excitatory neurons reduced after Egr1 knockout. A) Predicted cell-type compositions in Egr1 knockout and 6-week old mouse cortex. B) Volcano plot shows a comparison of the expression level of excitatory neuron marker genes in Egr1KO and 6-week old cortexes. The points colored in red represent the differentially expressed genes in Egr1KO compared to 6-weeks old cortex and the size of the points represents the fold change of expression in excitatory neurons compared to other cell types. C) Go enrichment analysis of the differentially expressed excitatory neuron markers in Egr1KO. The point size represents the number of genes enriched. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
3. Discussion
To our knowledge, this study is the first attempt to explore brain ETRMs via multi “omics” integration with single cell methylome and RNA-seq data from single cells, sorted neurons, and brain tissues at various developmental stages. To predict ETRMs, we exploited the recursive motif search approach [22] and implemented a novel expression-dependent single-cell analysis using a conditional probability matrix to demonstrate the cell type specificity of ETRMs. Additionally, EGR1 was selected as an example to demonstrate the functional importance of key TFs in a cell-type specific manner. Such validation may be extended to other key TFs if “omics” datasets available. The power of data integration is frequently limited by input quality. Single-cell methylomes often have low read depth leading to inaccuracy in cell classification and incomplete lists of cell type specific DMRs. A similar limitation also exists in detecting co-expressed TF pairs using single-cell RNA datasets, particularly for transcripts with low expression. In addition, the number of cell types and even the types of cells predicted with single-cell methylomes and single-cell RNA-seq data may not match each other. Despite these limitations, we have identified a number of ETRMs, which may serve as functional links between epigenetic regulatory loci to a specific type of neuron. We anticipate that the analytical procedure described in this study will facilitate “omics” data integration and that the predicted ETRMs will nurture hypotheses for future experimental design.
4. Methods
4.1. Datasets
The publicly available brain “omics” data used in this manuscript are summarized in Supplementary Table 1. All the analysis performed in this study were based on mouse genome GRCm38/mm10.
4.2. Analysis of single-cell methylomes
Single cell methylomes for 3377 neurons derived from mouse frontal cortex were downloaded. In the previous study [31], these 3377 neurons were clustered using an iterative, hierarchical and unsupervised clustering algorithm, BackSPIN [31]. Cells were split into two new clusters in each iteration, this procedure was performed recursively on each new cluster and terminated when no clusters met the splitting threshold. The clusters with highly similar mCH patterns were merged, and each cluster was annotated according to the depletion of mCH at known markers, including cortical glutamatergic or GABAergic neuron markers, cortical layer markers, or inhibitory neuron subtype markers. Finally, sixteen clusters were determined, including ten excitatory subtypes and six inhibitory subtypes [31] (Supplementary Table 2).
Specifically, in this work, single-cell methylomes assigned to the same cell type were merged to create synthetic methylomes for the sixteen neuronal cell types.
4.3. Recursive motif analysis to identify ETRMs
The recursive motif prediction was performed using the approach described in our previous study [22]. Briefly, the motif with the most significant p-value predicted by HOMER was selected as a key TF. In the case of ties involving two motifs sharing the same enrichment p-value, the motif with the higher frequency of target sequences was selected. Next, regions containing the key motif were identified using HOMER and the center of each region was shifted to the predicted binding site of the key TF for another round of motif search. Finally, regions containing the motif for the key TF were removed from the input dataset and the rest of the input sequences were used to identify the next most significant candidate motif. Such a motif searching process was performed recursively until no significant motif could be identified.
The 'universalmotif' R library was used to merge similar motifs. As described in the reference manual, four metrics were used: mean Pearson correlation coefficient, mean Euclidean distance, mean Sandelin-Wasserman similarity, and mean Kullback-Leilber divergence for similarity measures between two motifs. Means were used instead of just the similarity or distance metric to avoid the difference in results between comparisons of longer and shorter motifs.
4.4. Analysis of single-cell RNA-seq data
In the previous study [35], single cells from mouse prefrontal cortex were sequenced as 12 independent biological replicates. Cells with potential double droplets or having mitochondrial mRNA loads over 10% were filtered. Non-neuronal cells expressed less than 800 genes or neuronal cells expressed less than 1500 genes were also removed. Potential batch effect between samples were removed by CCA analysis [44]. In this study, we focused on the expression profile of 11,886 cells generated from the P60 mouse. The Seurat R package [36] was used to perform the single-cell RNA-seq dataset analysis. The top 2000 variable genes across cells were selected as features to perform linear dimensional reduction, and the top 10 principal components were used to generate the t-SNE (T-distributed Stochastic Neighbor Embedding) map. According to cell type annotation provide by Aritra et al. [35] (Supplementary Table 3), the cells were colored and labeled on the t-SNE map.
4.5. Transcription factor dependent analysis
To determine the relationship of co-expression between any two TFs (or genes), a conditional probability matrix was constructed for each cell type. NA, NB, and NAB denote the number of cells that express TFA, the number of cells that express TFB, and the number of cells that simultaneously express TFA and TFB, respectively. Then, the probability of TFB expressing under the condition of TFA expressing is:
Similarly, the probability of TFA expressing under the condition of TFB expressing is:
In addition to the significance of the co-expression between any two TFs, a hypergeometric test was used, in which the null hypothesis is that the target TFs are dependent upon each other. In the test, the testing statistic is the number of cells that simultaneously express TFA and TFB. Let N denote the total number of cells in a cell type. Then, under the null hypothesis, the testing statistic follows a hypergeometric distribution, that is
, wheredenote combinational formula.
In the test, the p-value was calculated by the cumulative probability of NAB. If the p-value is less than 0.05, the null hypothesis will be rejected and the co-expression between the target TFs will be determined.
4.6. Analysis of RNA-seq data
The RNA-seq datasets included samples from embryonic day 10.5 to postnatal day 0 and for 1-week, 2-week, 4-week, 6-week, 10 week, and 22-month, EXC neurons, PV neurons, and VIP neurons, which were downloaded from the GEO database. Adapters and bases of low quality were trimmed and the remaining reads were mapped to the mouse genome (mm10) by RSEM with Bowtie2 to achieve the expression level of each gene. For each time point or cell type, multiple replicates were merged and the average TPM (Transcripts Per Million) values from the replicates were calculated as the final value for the time points. Co-expression analysis in developmental stages was performed by using weighted correlation network analysis (WGCNA) [45], which automatically divides the gene expression matrix into smaller blocks and performs a two-level clustering. In the first step, genes that were weakly correlated were pre-clustered into different blocks using projective k-means. Next, for each block, a network analysis was performed by identifying clusters of highly correlated genes to estimate the cluster eigen-gene. Finally, clusters with a highly correlated eigen-gene were merged.
4.7. Analysis of ChIP-seq data
EGR1 ChIP-seq data with accession number GSE108768 were downloaded from the Gene Expression Omnibus (GEO) database, this dataset was generated for 6-week mouse frontal cortex. In this dataset, reproducible peaks on two biological replicates were achieved and the peaks overlapped with DMRs of mL2/3 were retained for downstream ETRM identification.
4.8. Estimation of cell-type composition from gene expression data
The DWLS package [42] was used to perform the decomposition analysis. A dampened weighted least squares algorithm was adopted to estimate the cell-type composition of bulk data from a single-cell RNA-seq-derived cell-type signature. First, the differentially expressed genes in each cell type compared to others cell types were defined as the markers for each cell type, using the given single-cell RNA-seq dataset with cell type information annotated. Then, a signature matrix with the lowest condition number was selected from the profile of markers. Lastly, a deconvolution was performed on the bulk RNA-seq data based on the dampened weighted least squares algorithm with the signature matrix as the reference.
Acknowledgments
Acknowledgements
This work was supported by the Center for One Health Research at the Virginia-Maryland, College of Veterinary Medicine, The Edward Via College of Osteopathic Medicine, and the Fralin Life Sciences Institute faculty development fund for H.X., and VT’s Open Access Subvention Fund; the Key Research Program of the Chinese Academy of Sciences (KFZD-SW-220-1 for X.L.), National Natural Science Foundation of China grants (31771416 for X.L.), the Second Tibetan Plateau Scientific Expedition and Research Program (STEP, 2019QZKK0501 for X.L.), and the CAS Light of West China Program (for X.L.). We recognize The Center for Engineered Health and the Virginia-Maryland College of Veterinary Medicine at Virginia Tech. We thank Dr. Janet Webster for English language editing.
Code availability
The analytical pipeline implemented in this study is freely accessible in the Github repository (https://github.com/Gavin-Yinld/brain_TF).
Author contributions
H.X. and X.L. conceived and designed the study; L.Y. and S.B. implemented the clustering and recursive motif finding procedure; L.Y., S.B., and J.H. conducted data analysis and integration; L.Y., S.B., J.F., and H.X. drafted the manuscript. All authors discussed the results and agreed on the contents of the manuscript.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.csbj.2020.04.007.
Contributor Information
Xuemei Lu, Email: xuemeilu@mail.kiz.ac.cn.
Hehuang Xie, Email: davidxie@vt.edu.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
Supplementary figure 1.
Supplementary figure 2.
Supplementary figure 3.
Supplementary figure 4.
Supplementary figure 5.
Supplementary figure 6.
References
- 1.Mohn F. Lineage-specific polycomb targets and de novo DNA methylation define restriction and potential of neuronal progenitors. Mol Cell. 2008;30:755–766. doi: 10.1016/j.molcel.2008.05.007. [DOI] [PubMed] [Google Scholar]
- 2.Martinowich K. DNA methylation-related chromatin remodeling in activity-dependent BDNF gene regulation. Science. 2003;302:890–893. doi: 10.1126/science.1090842. [DOI] [PubMed] [Google Scholar]
- 3.Ballas N., Grunseich C., Lu D.D., Speh J.C., Mandel G. REST and its corepressors mediate plasticity of neuronal gene chromatin throughout neurogenesis. Cell. 2005;121:645–657. doi: 10.1016/j.cell.2005.03.013. [DOI] [PubMed] [Google Scholar]
- 4.Setoguchi H. Methyl-CpG binding proteins are involved in restricting differentiation plasticity in neurons. J Neurosci Res. 2006;84:969–979. doi: 10.1002/jnr.21001. [DOI] [PubMed] [Google Scholar]
- 5.Kohyama J. Epigenetic regulation of neural cell differentiation plasticity in the adult mammalian brain. Proc Natl Acad Sci U S A. 2008;105:18012–18017. doi: 10.1073/pnas.0808417105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Liu J., Casaccia P. Epigenetic regulation of oligodendrocyte identity. Trends Neurosci. 2010;33:193–201. doi: 10.1016/j.tins.2010.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Nelson E.D., Kavalali E.T., Monteggia L.M. Activity-dependent suppression of miniature neurotransmission through the regulation of DNA methylation. J Neurosci. 2008;28:395–406. doi: 10.1523/JNEUROSCI.3796-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Meaney M.J., Ferguson-Smith A.C. Epigenetic regulation of the neural transcriptome: the meaning of the marks. Nat Neurosci. 2010;13:1313–1318. doi: 10.1038/nn1110-1313. [DOI] [PubMed] [Google Scholar]
- 9.Roth T.L., Roth E.D., Sweatt J.D. Epigenetic regulation of genes in learning and memory. Essays Biochem. 2010;48:263–274. doi: 10.1042/bse0480263. [DOI] [PubMed] [Google Scholar]
- 10.Jakovcevski M., Akbarian S. Epigenetic mechanisms in neurological disease. Nat Med. 2012;18:1194–1204. doi: 10.1038/nm.2828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gos M. Epigenetic mechanisms of gene expression regulation in neurological diseases. Acta Neurobiol Exp. 2013;73:19–37. doi: 10.55782/ane-2013-1919. [DOI] [PubMed] [Google Scholar]
- 12.Urdinguio R.G., Sanchez-Mut J.V., Esteller M. Epigenetic mechanisms in neurological diseases: genes, syndromes, and therapies. Lancet Neurol. 2009;8:1056–1072. doi: 10.1016/S1474-4422(09)70262-5. [DOI] [PubMed] [Google Scholar]
- 13.Hansen R.S. The DNMT3B DNA methyltransferase gene is mutated in the ICF immunodeficiency syndrome. Proc Natl Acad Sci U S A. 1999;96:14412–14417. doi: 10.1073/pnas.96.25.14412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Amir R.E. Rett syndrome is caused by mutations in X-linked MECP2, encoding methyl-CpG-binding protein 2. Nat Genet. 1999;23:185–188. doi: 10.1038/13810. [DOI] [PubMed] [Google Scholar]
- 15.Fanelli M. Loss of pericentromeric DNA methylation pattern in human glioblastoma is associated with altered DNA methyltransferases expression and involves the stem cell compartment. Oncogene. 2008;27:358–365. doi: 10.1038/sj.onc.1210642. [DOI] [PubMed] [Google Scholar]
- 16.Matys V. TRANSFAC and its module TRANSCompel: transcriptional gene regulation in eukaryotes. Nucleic Acids Res. 2006;34:D108–110. doi: 10.1093/nar/gkj143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Khan A. JASPAR 2018: update of the open-access database of transcription factor binding profiles and its web framework. Nucleic Acids Res. 2018;46:D1284. doi: 10.1093/nar/gkx1188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Weirauch M.T. Determination and inference of eukaryotic transcription factor sequence specificity. Cell. 2014;158:1431–1443. doi: 10.1016/j.cell.2014.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kulakovskiy I.V. HOCOMOCO: towards a complete collection of transcription factor binding models for human and mouse via large-scale ChIP-Seq analysis. Nucleic Acids Res. 2018;46:D252–D259. doi: 10.1093/nar/gkx1106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lambert S.A. Similarity regression predicts evolution of transcription factor sequence specificity. Nat Genet. 2019;51:981–989. doi: 10.1038/s41588-019-0411-1. [DOI] [PubMed] [Google Scholar]
- 21.He J. Retinal-input-induced epigenetic dynamics in the developing mouse dorsal lateral geniculate nucleus. Epigenetics Chromatin. 2019;12:13. doi: 10.1186/s13072-019-0257-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Banerjee S., Wei X., Xie H. Recursive motif analyses identify brain epigenetic transcription regulatory modules. Comput Struct Biotechnol J. 2019;17:507–515. doi: 10.1016/j.csbj.2019.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Banerjee S. Identifying transcriptional regulatory modules among different chromatin states in mouse neural stem cells. Front Genet. 2018;9:731. doi: 10.3389/fgene.2018.00731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sun M.A. Mammalian brain development is accompanied by a dramatic increase in bipolar DNA methylation. Sci Rep. 2016;6:32298. doi: 10.1038/srep32298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sun Z. EGR1 recruits TET1 to shape the brain methylome during development and upon neuronal activity. Nat Commun. 2019;10:3892. doi: 10.1038/s41467-019-11905-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Amit Zeisel A.B.M.-M. Cell types in the mouse cortex and hippocampus revealed by single-cell RNA-seq. Science. 2015 doi: 10.1126/science.aaa1934. [DOI] [PubMed] [Google Scholar]
- 27.Tasic B. Adult mouse cortical cell taxonomy revealed by single cell transcriptomics. Nat Neurosci. 2016;19:335–346. doi: 10.1038/nn.4216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tasic B. Shared and distinct transcriptomic cell types across neocortical areas. Nature. 2018;563:72–78. doi: 10.1038/s41586-018-0654-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hodge R.D. Conserved cell types with divergent features in human versus mouse cortex. Nature. 2019;573:61–68. doi: 10.1038/s41586-019-1506-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gravina S., Dong X., Yu B., Vijg J. Single-cell genome-wide bisulfite sequencing uncovers extensive heterogeneity in the mouse liver methylome. Genome Biol. 2016;17:150. doi: 10.1186/s13059-016-1011-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Luo C. Single-cell methylomes identify neuronal subtypes and regulatory elements in mammalian cortex. Science. 2017;357:600–604. doi: 10.1126/science.aan3351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Guo H. Single-cell methylome landscapes of mouse embryonic stem cells and early embryos analyzed using reduced representation bisulfite sequencing. Genome Res. 2013;23:2126–2135. doi: 10.1101/gr.161679.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Farlik M. Single-cell DNA methylome sequencing and bioinformatic inference of epigenomic cell-state dynamics. Cell Rep. 2015;10:1386–1397. doi: 10.1016/j.celrep.2015.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wu X., Sun M.-A., Zhu H., Xie H. Nonparametric Bayesian clustering to detect bipolar methylated genomic loci. BMC Bioinf. 2015;16 doi: 10.1186/s12859-014-0439-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bhattacherjee A. Cell type-specific transcriptional programs in mouse prefrontal cortex during adolescence and addiction. Nat Commun. 2019;10:4169. doi: 10.1038/s41467-019-12054-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Satija R., Farrell J.A., Gennert D., Schier A.F., Regev A. Spatial reconstruction of single-cell gene expression data. Nat Biotechnol. 2015;33:495–502. doi: 10.1038/nbt.3192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Szklarczyk D. STRING v11: protein-protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic Acids Res. 2019;47:D607–D613. doi: 10.1093/nar/gky1131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mo A. Epigenomic signatures of neuronal diversity in the mammalian brain. Neuron. 2015;86:1369–1384. doi: 10.1016/j.neuron.2015.05.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Uchiumi T. YB-1 is important for an early stage embryonic development – Neural tube formation and cell proliferation. J Biol Chem. 2006;281:40440–40449. doi: 10.1074/jbc.M605948200. [DOI] [PubMed] [Google Scholar]
- 40.Lu Z.H., Books J.T., Ley T.J. YB-1 is important for late-stage embryonic development, optimal cellular stress responses, and the prevention of premature senescence. Mol Cell Biol. 2005;25:4625–4637. doi: 10.1128/MCB.25.11.4625-4637.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ng R.T., Han J.W. CLARANS: a method for clustering objects for spatial data mining. IEEE Trans Knowl Data Eng. 2002;14:1003–1016. [Google Scholar]
- 42.Tsoucas D. Accurate estimation of cell-type composition from gene expression data. Nat Commun. 2019;10:2975. doi: 10.1038/s41467-019-10802-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lister R. Global epigenomic reconfiguration during mammalian brain development. Science. 2013;341:1237905. doi: 10.1126/science.1237905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Butler A., Hoffman P., Smibert P., Papalexi E., Satija R. Integrating single-cell transcriptomic data across different conditions, technologies, and species. Nat Biotechnol. 2018;36:411–420. doi: 10.1038/nbt.4096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Langfelder P., Horvath S. WGCNA: an R package for weighted correlation network analysis. BMC Bioinf. 2008;9:559. doi: 10.1186/1471-2105-9-559. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.