Abstract
Objectives
The streptozotocin (STZ) model is widely used in diabetes research. However, the cellular and molecular states of pancreatic endocrine cells in this model remain unclear. This study explored the molecular characteristics of islet cells treated with STZ and re-evaluated β-cell dysfunction and regeneration in the STZ model.
Methods
We performed single-cell RNA sequencing of pancreatic endocrine cells from STZ-treated mice. High-quality sequencing data from 2,999 cells were used to identify clusters via Louvain clustering analysis. Principal component analysis (PCA), t-distributed stochastic neighbor embedding (t-SNE), uniform manifold approximation and projection (UMAP), force-directed layout (FDL), and differential expression analysis were used to define the heterogeneity and transcriptomic changes in islet cells. In addition, qPCR and immunofluorescence staining were used to confirm findings from the sequencing data.
Results
Untreated β-cells were divided into two populations at the transcriptomic level, a large high-Glut2 expression (Glut2high) population and a small low-Glut2 expression (Glut2low) population. At the transcriptomic level, Glut2low β-cells in adult mice did not represent a developmentally immature state, although a fraction of genes associated with β-cell maturation and function were downregulated in Glut2low cells. After a single high-dose STZ treatment, most Glut2high cells were killed, but Glut2low cells survived and over time changed to a distinct cell state. We did not observe conversion of Glut2low to Glut2high β-cells up to 9 months after STZ treatment. In addition, we did not detect transcriptomic changes in the non-β endocrine cells or a direct trans-differentiation pathway from the α-cell lineage to the β-cell lineage in the STZ model.
Conclusions
We identified the heterogeneity of β-cells in both physiological and pathological conditions. However, we did not observe conversion of Glut2low to Glut2high β-cells, transcriptomic changes in the non-β endocrine cells, or direct trans-differentiation from the α-cell lineage to the β-cell lineage in the STZ model. Our results clearly define the states of islet cells treated with STZ and allow us to re-evaluate the STZ model widely used in diabetes studies.
Keywords: β-cell heterogeneity, Streptozotocin (STZ) model, Single-cell RNA-sequencing, Trans-differentiation, Diabetes
Highlight
-
•
β-cells show heterogeneity in both physiological and pathological conditions.
-
•
Glut2low β-cells survive from high dose of STZ, but over time change to a distinct cell state.
-
•
The expression profiles of non-β endocrine cells rarely change in the STZ model.
-
•
Glut2low β-cells do not represent a developmentally immature state.
1. Introduction
Diabetes mellitus is a metabolic disorder resulting from loss or dysfunction of insulin-producing β-cells in the pancreas or lack of insulin effect. Endogenous repair is a promising strategy to restore β-cell mass and relieve diabetes. Previous studies described various β-cell regeneration pathways, such as replication of pre-existent β-cells [1], intra-islet cell trans-differentiation [[2], [3], [4]], and β-cell neogenesis [5,6]. Studies of in vivo regeneration of β-cells require appropriate animal models [7,8].
The streptozotocin (STZ)-induced diabetic model is widely used in diabetes research. A single high dose of STZ results in near-total ablation of β-cells in rodents, leading to insulin deficiency, hyperglycemia, polydipsia, and polyuria, all of which mimic human type-1 diabetes [9]. However, rats that received low-dose STZ treatments combined with high-fat diets have been used as a model for type 2 diabetes [10,11]. Due to its ease of use and ability to mimic diabetic phenotypes, the STZ model has been used by several groups to study β-cell regeneration after acute β-cell injury [4,[12], [13], [14]]. One group observed δ-cell-dependent β-cell regeneration and diabetes recovery in juvenile mice after a single high dose of STZ [4]. Cheng et al. identified an intermediate cell type (vimentin+/MafB+) as β-cell progenitors following STZ-induced extreme β-cell ablation [12]. Grossman et al. studied the effect of insulin treatment in the STZ model and found that it could restore β-cell mass as well as normoglycemia after 120 days of glycemic control in female mice [13]. However, the molecular characteristics of each islet lineage after STZ treatment at the single-cell level remain undescribed, which hinders our understanding of the cell fate plasticity of endocrine lineages and β-cell state and regeneration in the STZ model.
In this study, we performed single-cell RNA sequencing (scRNA-seq) to investigate the single-cell transcriptomic profiles of all endocrine lineages over 9 months after a single high dose of STZ. We analyzed the characteristics of islet endocrine cells after STZ treatment to describe their cell heterogeneity, maturation, functional state, and trans-differentiation.
2. Materials and methods
2.1. Mice
Ngn3-Cre; Rosa-RFP [15] and Ins1-RFP [50] transgenic mice were used in this study. Male mice were weaned to regular chow at postnatal day 21 (P21). All of the procedures were approved by the Institutional Animal Care and Use Committee of Peking University.
2.2. In vivo STZ treatment
STZ was dissolved in 50 mM sodium citrate buffer (pH 4.5) and injected within 15 min of preparation. Two-month-old male mice were injected intraperitoneally with a single high dose of STZ (200 mg/kg, V900890, Sigma) after an overnight fast. The control mice were injected with an equal volume of sodium citrate buffer.
2.3. Insulin treatment and blood glucose monitoring
To prevent death in long-term experiments, the mice received subcutaneous insulin implants (LinBit Pr-1-B, LinShin Canada) when hyperglycemic (>400 mg/dL). The first insulin implant was administered 2 days after STZ injection. Non-fasted blood glucose levels were determined from the tail vein using a Contour Next blood glucose meter.
2.4. Insulin secretion assay
Eight-week-old male C57BL/6 mice were injected with STZ as previously described. Three or six days after STZ treatment, blood was collected from STZ-treated and control mice after fasting for 6 hours. Fasting serum insulin levels were determined using a Rat/Mouse Insulin ELISA Kit (EZRMI-13K, Millipore) according to the manufacturer's instructions.
2.5. Fluorescence-activated cell sorting (FACS)
The pancreas was perfused and digested using 0.5 mg/mL collagenase P (11213873001, Roche) as previously reported [16]. After centrifugation and removal of the supernatant, pancreatic tissue was dissociated into single cells via 0.25% trypsin-EDTA treatment for 5 min at 37 °C, and digestion was terminated with 0.4 volumes of fetal bovine serum (FBS). The cells were sorted using a BD FACS Aria SORP cell sorter.
2.6. Single-cell RNA-seq
Library preparation was conducted following the modified STRT-seq protocol [17,18]. Briefly, after cell sorting, single Ngn3-Cre; Rosa-RFP+ cells were transferred quickly into lysis buffer by a mouth pipette. Next, we reverse-transcribed the RNA and amplified the resulting cDNA over 18 cycles of PCR. The quality of the cDNA was assessed by qPCR of the housekeeping gene Gapdh. cDNA of good-quality single cells was pooled together and purified. Biotinylated pre-indexed primers were added to the cDNA during an additional 4 cycles of PCR. Approximately 300 ng of purified PCR product was sheared to approximately 300 bp by sonication. Sequencing libraries were prepared using a Kapa Hyper Prep Kit (KK8502, Kapa Biosystems).
2.7. Bulk cell reverse transcription quantitative PCR (RT-qPCR)
Ins1-RFP+ cells from the control and STZ-treated D12 Ins1-RFP mice were obtained by FACS. Total RNA from sorted cells was prepared using an RNAprep Pure Kit (for micro samples) (DP420, Tiangen). First-strand cDNA was synthesized using HiScript II Q RT SuperMix (R223, Vazyme). Real-time PCR was performed with 2X M5 HiPer SYBR Premix EsTaq (MF787-01, Mei5 Biotechnology).
The following primer sequences were used for RT-qPCR:
- Actb: forward 5′-TCCTGAGCGCAAGTACTCTGT-3′,
- reverse 5′-CTGATCCACATCTGCTGGAAG-3′;
- Gapdh: forward 5′-ATGGTGAAGGTCGGTGTGAAC-3′,
- reverse 5′-GCCTTGACTGTGCCGTTGAAT-3′;
- Glut2: forward 5′-GTGCTGCTGGATAAATTCGCC-3′,
- reverse 5′-ATTGCAGACCCAGTTGCTGA-3′;
- Ucn3: forward 5′-AAGCCTCTCCCACAAGTTCTA-3′,
- reverse 5′-GAGGTGCGTTTGGTTGTCATC-3′;
- Ins1: forward 5′-GCAAGCAGGTCATTGTTTCAAC-3′,
- reverse 5′-AAGCCTGGGTGGGTTTGG-3′;
- Nkx6.1: forward: 5′-AACACACCAGACCCACGTTCT-3′,
- reverse 5′-ATCCCCAGAGAATAGGCCAAG-3′;
- Pdx1: forward 5′-AACTTGAGCGTTCCAATACGGA-3′,
- reverse 5′-CAGCCGCCTTTCGTTATTCTTA-3′;
- Vdr: forward 5′-GGCTTCCACTTCAACGCTATG-3′,
- reverse 5′-ATGCTCCGCCTGAAGAAAC-3′;
- Etv1: forward 5′-TGCCTAGCTGCCACTCCAT-3′,
- reverse 5′-GCCTGGGACCCTTCTCAAAC-3′;
- Klf10: forward 5′-AGAAGAACCCACGGAAAT-3′,
- reverse 5′-GAGGAAGGCACAGCAAAG-3′;
- Tspan8: forward 5′-GGAGTTCCGTTTACCCAAAGA-3′,
- reverse 5′-CTCAATAACGGCCAGTCCAA-3′;
- Ins2: forward 5′-TGGCTTCTTCTACACACCCA-3′,
- reverse 5′-TCTAGTTGCAGTAGTTCTCCA-3′.
2.8. Immunofluorescence staining
Tissues were fixed in 4% paraformaldehyde overnight at 4 °C. For frozen sections, the tissues were cryoprotected in 30% sucrose, embedded in Optimal Cutting Temperature (OCT) media (Thermo Fisher, 6502), and frozen on dry ice. For paraffin sections, the tissues were dehydrated with 30–100% ethanol and embedded in paraffin. The sections (6 μm thick) were probed with primary antibodies against insulin (1:500, Abcam, ab7842), NKX6.1 (1:100, DSHB, F55A12-c), GLUT2 (1:50, Santa Cruz Biotechnology, sc-518022), and glucagon (1:200, Millipore, AB932) followed by incubation with Alexa Fluor 594 AffiniPure Donkey Anti-Guinea Pig IgG (H + L) (Jackson ImmunoResearch, 706-585-148) and Alexa Fluor 488 Donkey Anti-Mouse IgG (H + L) antibody (Thermo Fisher, A21202) as the secondary antibodies. Images were acquired using a Zeiss Axioimager M2 fluorescence microscope or Zeiss LSM 710 NLO and DuoScan System.
2.9. Quantification of gene expression from single-cell RNA-seq
Modified STRT-seq libraries were sequenced on an Illumina HiSeq 4000 System to generate 150-bp paired-end reads. Read1 included transcript sequences, and Read2 included an 8-bp cell-specific barcode and an 8-bp unique molecular identifier (UMI) sequence. The fastq file for each single-cell sample was split based on 8-bp cell-specific barcode sequences. Then 8-bp UMIs were added into the corresponding Read1. The polyA sequences in Read1 were trimmed. We aligned preprocessed Read1 reads to the mouse genome (mm10) with TopHat (v2.1.0) [19]. Mapped reads were annotated to genes using featureCounts (v1.5.3) [20]. The gene ID was appended as the XT tag in the bam file. After sorting and indexing the bam file with SAMtools (v.1.3.1) [21], we quantified the UMIs of each gene with UMI-tools (v0.5.0) using the following parameters: “count --per-gene --gene-tag = XT --method unique -I indexed_sorted_bam -S out_file” [22] and generated the UMI matrix. To normalize the sequencing depth, the UMI matrix was transformed to transcripts per 0.1 million (TP0.1M) (one UMI was considered as one transcript). To eliminate the influence of individual genes on the transcriptome, we ignored extremely highly expressed genes (Ins1, Ins2, Gcg, Sst, Ppy, Pyy, Iapp, and Yam1) in the calculations of the total transcripts. Single-cell samples were discarded if their mapped read count was less than 50,000 or if less than 1,500 genes were detected.
2.10. Cell type identification
To eliminate the index switching effect of the HiSeq 4000 System (5–10% of sample indexes were incorrectly assigned) [23], we subtracted 10% of the maximum TP0.1M value for each gene. Then we calculated a log2 (subtracted TP0.1M + 1) matrix to be used as the input for Seurat (v3.0.2) [24]. The top 2,000 variable genes were identified using the FindVariableFeatures function in the Seurat package. Cell cycle-related genes (including cell cycle regulators, such as cyclins and CDKs) were identified with hierarchical clustering and were excluded in downstream analyses. Principal component analysis (PCA) was performed with filtered variable genes using RunPCA in the Seurat package. The first 20 principal components (PCs) were selected for t-distributed stochastic neighbor embedding (t-SNE), uniform manifold approximation and projection (UMAP), and Louvain clustering using RunTSNE, RunUMAP, and FindNeighbors/FindClusters in the Seurat package, respectively. Cell types were assigned according to the marker gene expressions: β-cells (Ins1+; Ins2+), α-cells (Gcg+), δ-cells (Sst+), PP-cells (Ppy+), ductal cells (Spp1+), acinar cells (Cel+), and immune cells (Ptprc+).
2.11. β-cell heterogeneity analysis
The log2 (TP0.1M + 1) matrix was imported into Seurat. The top 1,000 highly variable genes were identified, and we retained genes detected in at least five samples. Cell cycle-related genes and index switching effect-related genes (including genes highly expressed in non-β-cells, such as the exocrine enzymes Cel and Ctrb1) were identified using hierarchical clustering and were excluded from further analyses. After PCA, we observed that the major variation between the control and STZ-treated β-cells was represented on PC1. Therefore, we performed hierarchical clustering on the top 200 high-loading genes of PC1, and divided the β-cells into three subpopulations. We performed differential expression analyses (group 1 vs group 2/3 and group 2 vs group 3) using FindMarkers in Seurat with the parameter “logfc.threshold = 0.6”. To reduce noise, we only retained differentially expressed genes co-expressed with at least 10 other differentially expressed genes (ρp > 0.2) [25]. We also performed hierarchical clustering on the retained differentially expressed genes to identify and exclude index switching effect-related genes. Gene ontology (GO) enrichment analyses were conducted using GOstats (v2.50.0) [26]. To evaluate β-cell state transitions, we performed PCA on the β-cells with filtered differentially expressed genes, and fit the principal curve on the PCA plot using princurve (v2.1.4) [27]. The lambda values from the princurve results were extracted and used as the pseudo-time values.
2.12. Comparison of immature β-cells and Glut2low cells
The P3, P12, and P21 β-cell identification pipeline was similar to the pipeline described in Section 2.10. PCA, UMAP, and forced-directed layout (FDL) were performed on P3, P12, P21, and adult β-cells with the top 2,000 highly variable genes (excluding cell cycle-related genes and index switching effect-related genes). To define subgroups in the P3, P12, and P21 β-cells, we performed hierarchical clustering on P3, P12, P21, and adult β-cells with genes differentially expressed among three groups of adult β-cells (Figure 2D). If a P3, P12, or P21 β-cell clustered with group 2 adult β-cells, we considered this β-cell to be a group 2 β-cell. To identify the maturation-related genes, we performed differential expression analysis between P3 and adult group 1 β-cells and filtered genes using the same criteria described in Section 2.11. We performed differential expression analysis between the P3 and adult group 1 β-cells using FindMarkers in Seurat with the parameter logfc.threshold = 0.6 and only retained genes co-expressed with at least 10 other differentially expressed genes (ρp > 0.2). Then we performed hierarchical clustering on the retained genes to identify and exclude index switching effect-related genes. The overlap between the maturation-related genes and STZ-related genes (genes in Figure 2D) is exhibited in the Venn diagram (Figure 3E).
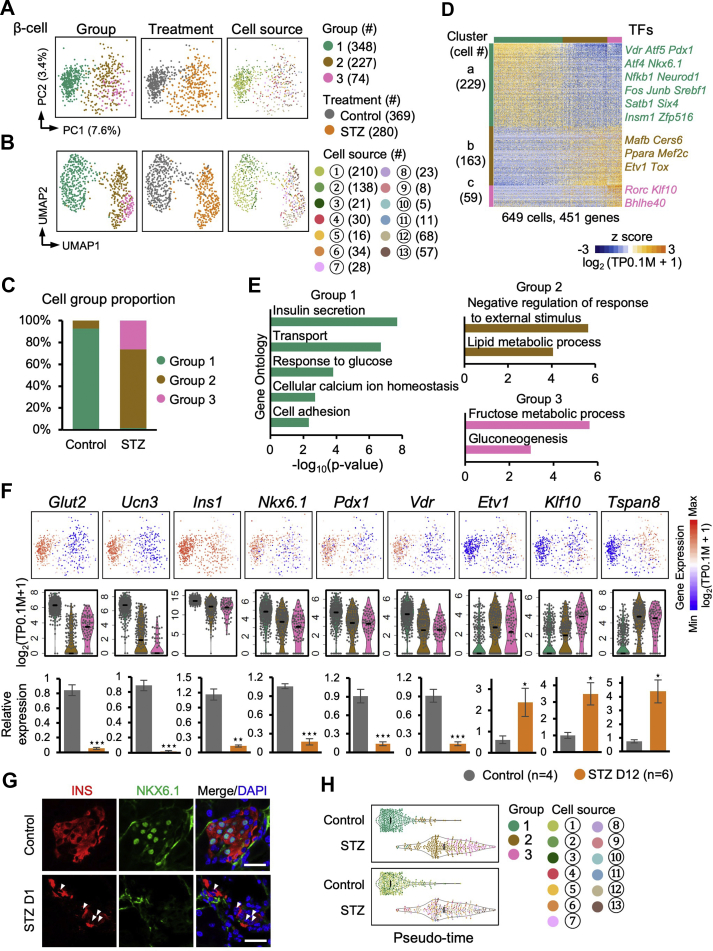
Figure 2.
β-cell heterogeneity.A and B: PCA and UMAP plots of the β-cells. Each dot represents a single cell sample. The colors denote the group (left), treatment condition (middle), and cell source (right). The circled numbers indicate the cell source labeled in Figure 1A. C: Bar plot showing the proportion of the three cell groups in the β-cells from the control and STZ-treated mice. D: Heatmap showing the expression patterns of genes differentially expressed among the three groups. Each column represents a single cell sample, and each row represents one gene. The gene count of each gene cluster is labeled on the left of the heatmap. Transcription factors (TFs) included in each gene cluster are labeled on the right of the heatmap. E: Selected GO terms enriched in the three clusters in (D). F: Expression levels of differently expressed genes are shown on the PCA plot (top) and violin plot (middle). The black line within each violin plot indicates the median of expression level. The expression levels of the indicated genes were verified via bulk cell RT-qPCR and normalized to β-actin (Actb) expression (bottom). Data are shown as means ± SEM. Student t-test: ∗ p-value < 0.05; ∗∗ p-value < 0.01; ∗∗∗ p-value < 0.001. G: Immunofluorescent staining of insulin and NKX6.1 in frozen sections from the control (top) and STZ-treated D1 (bottom) pancreatic tissues. The arrowheads indicate NKX6.1low β-cells in the STZ pancreas. Scale bars: 20 μm. H: The violin plots showing the pseudo-time value for each group and cell source. The circled numbers indicate the cell source labeled in Figure 1A. The black line within each violin plot indicates the median of the pseudo-time values.
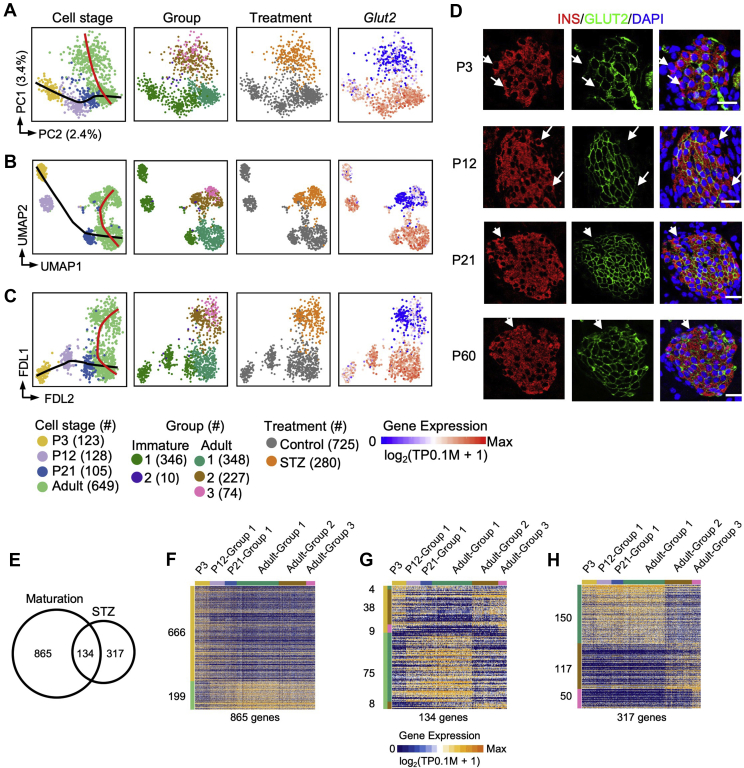
Figure 3.
Glut2lowcells represent a status distinct from immature β-cells.A, B, and C: PCA (A), UMAP (B), and FDL (C) plots of all of the Ngn3-Cre; Rosa-RFP+ β-cells. Each dot represents a single cell sample. The cells are colored by cell stage, cell type, treatment condition, and expression level of Glut2, respectively. Cell counts are indicated in brackets. The black and red curves indicate maturation and STZ-related trajectory, respectively. D: Immunofluorescent staining of insulin and GLUT2 from P3, P12, P21, and P60 pancreatic tissues. The arrows indicate insulin+GLUT2low β-cells. Scale bars: 20 μm. E: Venn plot showing the overlap between the maturation-related genes and the genes differentially expressed among groups 1–3 in Figure 2D. F–H: Heatmap showing the expression patterns of 865 maturation exclusively related genes (F), 134 maturation and STZ-related genes (G), and 317 STZ exclusively related genes (H) in Figure 3E among the Ngn3-Cre; Rosa-RFP+ β-cells. Each column represents a single cell sample, and each row represents one gene. The gene count of each gene cluster is labeled on the left of the heatmap.
For FDL analysis, we used FindNeighbors in the Seurat package to generate the shared nearest neighbor (SNN) matrix based on the PCA results. Then we built a graph using the graph.adjacency function in the igraph package (v1.2.4.1) [28] according to the SNN matrix. Finally, we generated the FDL results using the layout_with_fr function in the igraph package.
2.13. Heterogeneity analyses of α/δ/PP-cells
We applied the same pipeline in a β-cell heterogeneity analysis to explore the heterogeneity of α-cells, δ-cells, and PP-cells, respectively. We selected the top 1,000 highly variable genes, filtered out genes detected in less than 5 samples, and excluded cell cycle-related and index switching effect-related genes. The retained genes were used for PCA, UMAP, and hierarchical clustering. To evaluate whether the subgroups also existed in the α-cells, δ-cells, and PP-cells, we calculated the average silhouette widths [29] using the silhouette function in the cluster package (v2.0.5) [30] based on the hierarchical clustering. A relatively greater average silhouette width indicated an unambiguous clustering result. If a peak of the average silhouette width on N clusters was observed, the cells were expected to cluster into N subpopulations. In contrast, if no distinct peak of average silhouette width was observed, the cells were expected to be homogeneous.
3. Results
3.1. scRNA-seq of the STZ-treated pancreatic endocrine cells
To explore the transcriptomic states of individual pancreatic endocrine cells in the STZ-treated mice, we isolated cells from the STZ-treated mice via FACS and performedscRNA-seq using a modified STRT-seq (mSTRT-seq) approach [17,18]. We used the Ngn3-Cre; Rosa-RFP mice to enrich RFP+ pancreatic endocrine cells (Figure 1A–D). The mice were treated with a single high dose (200 mg/kg) of STZ and were assessed from day 6 (D6) to month 9 (M9) (Figure 1A). At D1 after STZ treatment, we observed an impaired islet structure, with many INS+ cells diminished (Supplementary Figure S1A). At D3 and D6, body weight significantly decreased, and the treated mice exhibited severe hyperglycemia and significantly reduced insulin concentrations compared with the control groups (Supplementary Figure S1B). These results indicated that the β-cells were significantly damaged in our STZ model. In this study, only mice with hyperglycemia at D2 after STZ treatment were used for subsequent experiments (Supplementary Figure S1C). To prevent the mice from dying from extreme hyperglycemia and study the cell states over a longer time period, we subcutaneously implanted an insulin capsule at D2 after STZ treatment [13]. We measured non-fasting blood glucose at 4 p.m. every three days and implanted insulin pellets when hyperglycemia (>400 mg/dL) was detected. The blood glucose level remained below 400 mg/dL after insulin implantation, but the mice without implantation remained hyperglycemic (>600 mg/dL) and subsequently died (Supplementary Figure S1D). However, two mice that did not receive an insulin supply but survived at M2 after STZ treatment were included in this study (Figure 1A). At D12, M5, and M9 after STZ treatment, we collected the pancreatic tissues to examine the morphology of the islets and observed few β-cells in the damaged islets (Supplementary Figure S1E and F).
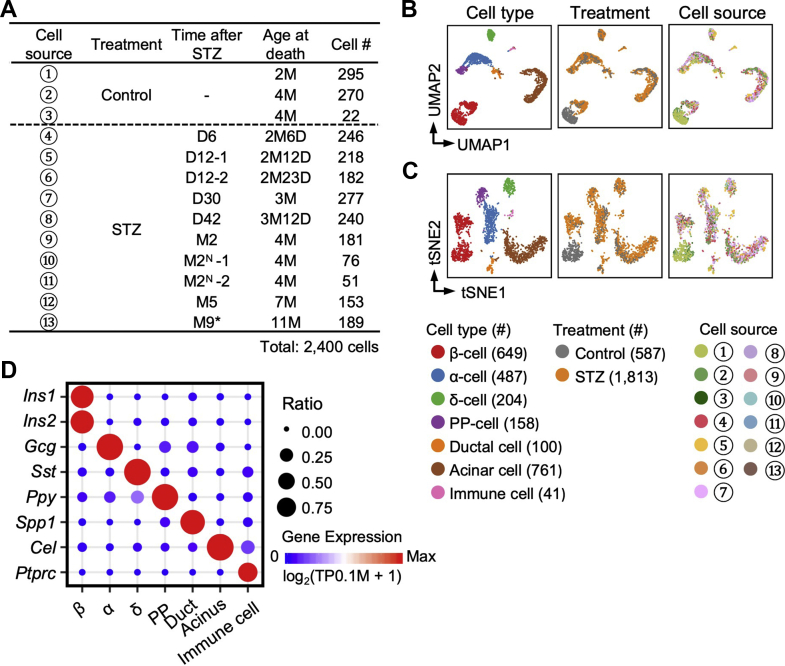
Figure 1.
Identification of cell types.A: Overview of 2,400 Ngn3-Cre; Rosa-RFP+ cells analyzed by scRNA-seq in this study. The cell # column shows the cell counts from the indicated treatment conditions. D (M): days (months) after STZ-treatment. Insulin was subcutaneously implanted at D2 after STZ treatment. N: no insulin implant. ∗:no insulin implant during M7-M9. B–C: The UMAP and t-SNE plots show the distribution of cells with different types and treatment conditions. Each dot represents a single cell sample. Cell counts are indicated in brackets. The circled numbers indicate the cell source labeled in (A). D: Expression levels of marker genes in each cell type. The point color from blue to red represents the expression level from low to high. The point size represents the detected ratio of the marker gene in the corresponding cell type.
In total, 2,400 Ngn3-Cre; Rosa-RFP+ mSTRT-seq samples passed quality control. On average, 9 × 104 unique molecular identifiers (UMIs) and 4,500 genes were detected in each single cell (Supplementary Figure S2A and B and Supplementary Table 1). Using UMAP [31] and t-SNE with Louvain cell clustering algorithms [24], we divided the cells into seven cell types based on their marker gene expressions: β-cells (Ins1+ and Ins2+), α-cells (Gcg+), δ-cells (Sst+), PP-cells (Ppy+), ductal cells (Spp1+), acinar cells (Cel+), and immune cells (Ptprc+) (Figure 1B–D and Supplementary Figure S2C). Ngn3 is also expressed in a fraction of the multipotent pancreatic progenitor cells and trunk cells (progenitors of ductal cells and endocrine progenitors) [32], which can also produce an exocrine population. These non-endocrine cells could be sorted from the single-cell suspension of the STZ-treated pancreas. For this reason, we obtained many non-endocrine cells from the Ngn3-Cre; Rosa-RFP mice. Notably, we collected at least 150 cells for scRNA-seq from each of the STZ-treated animals with an insulin supply. Because the cells from a range of time points were clustered together on the UMAP and t-SNE plots and did not show variations at the transcriptomic level (Figure 1B and C), we concluded that the cell states after STZ treatment were stable over time and that inter-individual variations of the transcriptomic profiles between different time points could be ignored in this analysis.
3.2. β-cell heterogeneity in the STZ model
Next we focused our analyses on β-cells. PCA and UMAP revealed three β-cell groups (Figure 2A and B). Group 1 included cells from the non-STZ-treated mice, group 2 contained cells from both the STZ-treated and non-STZ-treated mice, and group 3 exclusively included cells from the STZ-treated mice (Figure 2C). We identified 451 differentially expressed genes, including 23 transcription factors (TFs), among these three β-cell groups (Figure 2D and Supplementary Table 2). Cluster a genes were downregulated in group 2/3 cells and included key TFs important for maintaining β-cell identity, such as Nkx6.1, Pdx1, and Vdr [[33], [34], [35]], and genes associated with β-cell function, such as Glut2 (Slc2a2), Ucn3, and Ins1 (Figure 2F). Gene ontology (GO) enrichment analysis confirmed that cluster a genes were enriched for terms related to β-cell functions (Figure 2E and Supplementary Table 3). Cluster b genes were upregulated in the group 2/3 cells and enriched for the GO term of negative regulation of response to external stimulus (Figure 2E and Supplementary Table 4). The GO analysis also showed that fructose metabolism was upregulated in the group 3 β-cells (Figure 2E and Supplementary Table 5). Several genes, such as Fbp1, Fbp2, and Pdk1, have been reported to participate in fructose metabolism, resulting in reduced insulin secretion (Supplementary Tables 2 and 5) [[36], [37], [38]]. Our scRNA-seq analysis revealed that these genes were upregulated in the group 3 β-cells, suggesting that the group 3 β-cells were in a dysfunctional state regarding secreting insulin to regulate glucose homeostasis. Etv1 and Rorc, negative regulators of insulin secretion [39,40], were included in cluster b/c (Figure 2D), consistent with the lower expression of Ins1 in group 2/3 (Figure 2F), and suggesting weak insulin secretion in the group 2/3 cells. We selectively validated the downregulation of Glut2, Ucn3, Ins1, Nkx6.1, Pdx1, and Vdr and the upregulation of Etv1, Klf10, and Tspan8 in the sorted STZ-treated β-cells from Ins1-RFP transgenic mice (D12) [50] compared to the control using RT-qPCR (Figure 2F) and also confirmed the reduced expression of NKX6.1 in the STZ-treated mice via immunofluorescence staining (Figure 2G).
To analyze whether the group 2/3 β-cells could be restored to functional group 1 β-cells, we aligned the cells according to their positions along the group 1 to group 3 trajectory (Figure 2H). We found that the group 1 β-cells were quickly eliminated after STZ treatment, and the group 3 β-cells appeared over time (Figure 2H). However, only a few group 1 cells were detected in the STZ pancreas, independent of the presence of an insulin implant (Figure 2H and Supplementary Figure S3A). This finding indicated that the group 2 cells transformed into group 3 β-cells but did not restore their group 1 β-cell identity over a long period of time following STZ treatment. Taken together, our analyses revealed the heterogeneity of both the normal (group 1 and group 2 cells) and STZ-treated β-cells (group 2 and group 3 cells). In the STZ-treated animals, the group 1 β-cells were selectively killed, whereas the group 2 β-cells escaped STZ-mediated cell death but correspondingly altered their gene expression and morphed into a pathological state represented as group 3 β-cells.
It has been reported that a small fraction of β-cells locates to the islet edge and lacks expression of Glut2 and Ucn3 [41]. We re-analyzed the transcriptional profiles of the Ucn3- (Glut2low) and Ucn3+ (Glut2high) β-cells from this previous study [41]. We found that the 229 genes highly expressed in the group 1 cells and 163 genes highly expressed in the group 2 β-cells (no STZ treatment) were also generally highly expressed in the Ucn3+ (Glut2high) and Ucn3- (Glut2low) cells, respectively (Figure 2D and Supplementary Figure S3B and C). Therefore, based on this global similarity of gene expression patterns, we presumed that the group 2 β-cells included “virgin” β-cells identified by van der Meulen et al. [41].
3.3. Glut2low β-cells in the adult mice represented a state distinct from immature β-cells
Glut2low β-cells in the adult mice were considered immature β-cells because they displayed a low expression of the maturation marker gene Ucn3 (Figure 2F) [41,42]. To evaluate the maturation status of these Glut2low β-cells, we performed mSTRT-seq on the Ngn3-Cre; Rosa-RFP+ cells from the postnatal day 3 (P3), P12, and P21 mice (Supplementary Figure S4). Combining these datasets with adult Ngn3-Cre; Rosa-RFP+ datasets, we performed PCA, UMAP, and FDL analyses (Figure 3A–C). On the PCA plot, most of the β-cells were arranged along a path from P3 to adult Glut2high that reflected the β-cell maturation process (Figure 3A). Curiously, we observed a few Glut2low cells from the P3, P12, and P21 mice (Figure 3A), but at P12 and P21, not P3, the Glut2low cells were separated from the Glut2high cells on the developmental trajectory (Figure 3A). By performing immunostaining, we observed GLUT2low β-cells at the islet periphery at various developmental stages (Figure 3D). The distribution pattern of the GLUT2low β-cells was consistent with Van der Meulen's findings [41], which further indicated that the Glut2low β-cells (group 2) in the healthy mice identified by scRNA-seq were “virgin” β-cells. These findings clearly showed that Glut2low cells developed during the β-cell maturation process. However, STZ-treated adult β-cells (consisting of group 2/3 cells) were not located along this maturation path (as indicated by the black curve in Figure 3A). The UMAP and FDL analyses showed a similar result (Figure 3B and C). Moreover, the genes that were variably expressed during the maturation process displayed limited overlap with the genes variably expressed between the Glut2high and Glut2low β-cells from our STZ study (Figure 3E–H and Supplementary Table 6). The overlapping portion of the Venn diagram, including genes such as Ucn3 and Glut2 (Supplementary Table 6), was related to both β-cell maturation and β-1/2/3 cell transition processes.
3.4. The expression profiles of the non-β endocrine cells were not affected in the STZ model
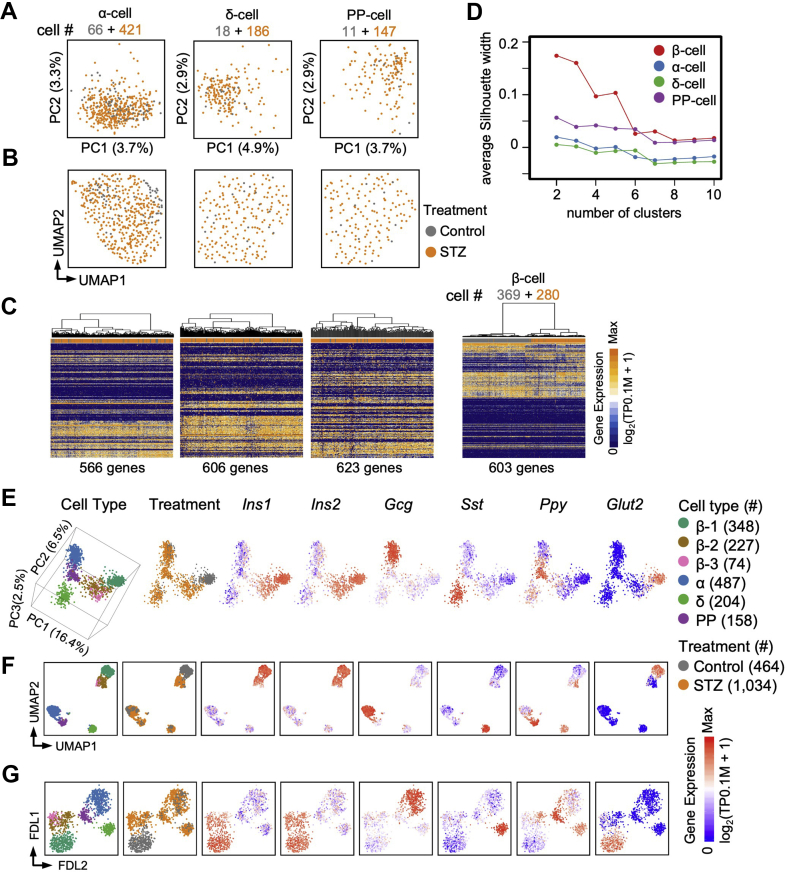
To investigate whether non-β endocrine cells were affected in the STZ model, we extended our analyses of the α-cells, δ-cells, and PP-cells. Although a small fraction of cells expressed at least two of the Ins1/Ins2, Gcg, Sst, and Ppy genes, these multi-hormone expressing cells were defined as a specific endocrine cell type based on their transcriptomic profiles (Figure 1B and C). We found that α-cells, δ-cells, and PP-cells from the control and STZ-treated mice overlapped on the PCA and UMAP plots and did not form distinct clusters associated with STZ treatment (Figure 4A and B). Additionally, hierarchical clustering based on highly variable genes could not distinguish non-β endocrine cells between the control and STZ-treated mice, indicating that the transcriptional statuses of the α-cells, δ-cells, and PP-cells were not affected by STZ treatment, unlike the β-cells (Figure 4C). Furthermore, the silhouette analysis [29] (Materials and Methods) suggested that in the control and STZ-treated mice, the α-cells, δ-cells, and PP-cells were homogeneous, whereas the β-cells from the non-treated control mice by themselves were heterogeneous (Supplementary Figure S5A and B), and after a high dose of STZ, the β-cells existed as three subpopulations (Figure 4D). Therefore, we concluded that STZ treatment did not affect the transcriptomic profiles of the non-β endocrine cells.
Figure 4.
Transcriptomic profile of non-β endocrine cells in the STZ model.A and B: PCA (A) and UMAP (B) of α-, δ-, and PP-cells from the Ngn3-Cre; Rosa-RFP pancreas. Each dot represents a single cell sample. The colors denote the treatment condition. C: Hierarchical clustering of highly variable genes on the α-, δ-, PP-, and β-cells. Each column represents a single cell sample, and each row represents one gene. D: Silhouette analyses of the hierarchical clustering of the α-, δ, PP-, and β-cells in (C). A relatively higher average silhouette width indicates an unambiguous clustering result. If a peak of the average silhouette width on N clusters is observed, the cells are expected to be clustered into N subpopulations. In contrast, if no distinct peak of the average silhouette width is observed, the cells are expected to be homogeneous. E-G: Three dimensions (3D) PCA (E), UMAP (F), and FDL (G) of the endocrine cells from the Ngn3-Cre; Rosa-RFP pancreas. Each dot represents a single cell. The colors denote cell type and treatment condition. Expression levels of Ins1, Ins2, Gcg, Sst, Ppy, and Glut2 are shown on the right.
It has been reported that, under normal physiological conditions, Glut2lowUcn3low “virgin” β-cells represent an intermediate stage in α to β trans-differentiation [41]. We therefore performed PCA, UMAP, and FDL analyses on endocrine lineages (Figure 4E–G and Supplementary Figure S5C and D). However, we did not detect a clear STZ-induced trans-differentiation trajectory from the α-cells to β-cells on the PCA, UMAP, and FDL plots (Figure 4E–G and Supplementary Figure S5C and D). Moreover, we performed hierarchical clustering analysis between the group 1 β-cells (Glut2high), group 2 β-cells (Glut2low), and the α-cells, but did not find that the group 2 β-cells were more similar to the α-cells at the transcriptomic level (Supplementary Figure S5E and F). These analyses suggest that trans-differentiation from the α-cells to Glut2low β-cells may not be a dominant mechanism in the maintenance of the small population of β-cells after STZ treatment.
4. Discussion
The STZ model is commonly used to study type-1 and type-2 diabetes [10,11]. However, precise characterization of the β-cell states in the STZ model has been challenging due to the rarity of β-cells and the model's impaired islet structure. In this study, we performed scRNA-seq on endocrine cells from reporter mice over a long period of time (D6 to M9) after STZ treatment. We found that the adult pancreas included two groups of β-cells that differed in their Glut2 expression (Glut2high and Glut2low). In our previous single-cell study, we detected several Glut2low β-cells [43]. However, due to a low number of sequenced cells, this small population of Glut2low β-cells could not be identified as a cell cluster in that study. We also confirmed the existence of Glut2low Ucn3low β-cells under normal physiological conditions, a population of cells previously shown to locate to the edge of the islets [41]. Although some genes related to β-cell functions were downregulated in the Glut2low Ucn3low cells, our transcriptomic analysis suggests that the Glut2low Ucn3low β-cells induced by STZ did not reflect immature β-cells, as they differentially expressed ∼300 genes that were not related to maturation (Figure 3E). Glut2 mediates STZ membrane transport, as a consequence, most of the Glut2high β-cells were damaged by STZ, but the Glut2low cells and possibly a small fraction of the Glut2high β-cells survived. However, these surviving cells gradually altered their states after STZ treatment despite exogenous insulin application. This result may suggest that normal functional β-cells or normal islet structures are necessary to maintain the status of Glut2low cells. In this study, we did not detect restoration of the Glut2low to Glut2high cells even 5–9 months after STZ-treatment.
Non-β endocrine cells have been reported to convert into insulin-secreting β-like cells under certain conditions [2,4,41,44,45]. Genetic inactivation of Dnmt1 and Arx in adult mice [44] or Pdx1-enforced expression in endocrine progenitors [45] could convert pancreatic α-cells into β-cells. Thorel et al. found that α-cells could transform into β-cells after near-total β-cell ablation in a diphtheria-toxin-induced model [2]. During trans-differentiation, α-cells gradually upregulated β-cell markers, such as Pdx1 and Nkx6.1, and eventually expressed insulin. Van der Meulen et al. also observed α- to β-cell trans-differentiation via a “virgin” β-cell state under normal conditions [41]. Moreover, an anti-malarial drug artemisinins was shown to drive the in vivo α-to β-cell fate transition by enhancing GABA signaling [46], although the main conclusion of that study has been challenged by others [47,48]. These findings encouraged us to investigate the process of cell fate trans-differentiation in the STZ model. If trans-differentiation from the non-β endocrine cells to the β-cells occurs in the STZ model, we would expect to observe heterogeneity in the non-β endocrine cell populations, which may reflect their state changes and reveal a defined cell fate transition trajectory via scRNA-seq analysis. However, we did not detect heterogeneity in the non-β endocrine cell populations or the previously described cell fate transition trajectory in this study (Figure 4). We acknowledge that the number of cells analyzed in this study may not be sufficient to detect cell trans-differentiation events in a small proportion of cells. A larger scale single-cell analysis combined with a lineage-tracing approach is required to investigate whether trans-differentiation events occur in the STZ model.
Many genes related to β-cell functions and diabetes development were differentially expressed in the STZ-treated β-cells compared to the functional Glut2high β-cells, indicating group 2/3 (Glut2low) cells were not fully functional in responding to glucose and insulin secretion. Group 3 cell upregulated genes Fbp1 and Fbp2 are primarily expressed in gluconeogenic organs, such as liver and muscle. Fbp1 negatively regulates insulin secretion, and Fbp2 is proposed to regulate indirect glycogen synthesis from lactate. Both genes were found to be upregulated in diabetic animal models [36] and diabetic subjects [38]. FBP inhibitors have been designed to improve insulin secretion, and several inhibitors, such as MB05032, have been reported to increase insulin secretion in the MIN6 β-cell line and in isolated mouse islets [49]. The transcriptomic signatures we observed in the group 3 cells may reveal other therapeutic targets to improve β-cell function. In addition, the group 2/3 (Glut2low) cells may represent a platform for screening factors and drugs that can reverse β-cell dysfunction via restoration of Glut2low to Glut2high cells as well as to study the molecular mechanisms related to this process. Our single-cell analyses have deepened our understanding of β-cell heterogeneity, the transcriptomic profiles of β-cell subsets, and the applicability of the STZ diabetes model.
Data availability
The RNA-seq data were submitted to the GEO repository under accession number GSE137909.
Funding
This study was supported by the National Key Research and Development Program of China (2019YFA0801500), the Ministry of Science and Technology of the People's Republic of China (973 Program 2015CB942800), the National Natural Science Foundation of China (31521004, 91753138), funding to C.-R.X., and funding provided by Boehringer Ingelheim Pharma GmbH.
Author contributions
C.-R.X. conceived the project. W.Z., J.Y., J.D.H., and C.-R.X. designed the study. Y.F., X.-X.Y., Y.Z., M.-Y.H., L.Y., and L.-C.L. conducted the research. W.-L.Q., W.Z., M.F., J.Y., J.D.H., and C.-R.X. analyzed the data. Y.F., W.-L.Q., J.D.H., and C.-R.X. wrote the manuscript. C.-R.X. is the guarantor of this work and, as such, had full access to the study data and takes responsibility for the integrity of the data and the accuracy of the data analysis.
Acknowledgments
We thank the members of the Xu laboratory for their advice and comments as well as the high-performance computing platform at the Peking-Tsinghua Center for Life Sciences. We thank Fei Wang and Yinghua Guo from the flow cytometry core at the National Center for Protein Sciences at Peking University for their technical assistance.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.molmet.2020.100982.
Contributor Information
Junqing Ye, Email: junqing.ye@boehringer-ingelheim.com.
Joerg D. Hoeck, Email: joerg.hoeck@boehringer-ingelheim.com.
Cheng-Ran Xu, Email: cxu@pku.edu.cn.
Conflict of interest
Weiyi Zhang, Michael Franti, Joerg Hoeck, and Junqing Ye are current employees of Boehringer Ingelheim Pharmaceuticals, Inc.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
Supplementary Figure S1. STZ model construction.A: Immunofluorescent staining of insulin and glucagon from the control (top) and STZ-treated D1 (bottom) pancreatic tissues. Scale bars: 50 μm. B: Fasting body weight, blood glucose level, and serum insulin of the control and STZ-treated D3 and D6 mice (no insulin implantation). n: Number of mice. C: Blood glucose level of each Ngn3-Cre; Rosa-RFP mouse (corresponding to Figure 1A) 2 days after STZ treatment. D: Blood glucose level and body weight of the STZ M5 and STZ M2NNgn3-Cre; Rosa-RFP mice before sacrifice. The STZ-treated M2N mouse had severe hyperglycemia starting 2 days after STZ treatment. E: Immunofluorescent staining of insulin and glucagon in paraffin sections from the normal, STZ D12, M5, and M9 pancreatic tissue. Scale bars: 50 μm ∗: no insulin implant during M7-M9. F: Proportion of β-cells in the islets shown in (E). Data are shown as means ± SEM. Supplementary Figure S2. Quality control of scRNA-seq samples.A and B: Bar plots showing UMI (A) and detected gene count (B) of the available single-cell samples. Each bar represents one sample. C: Projection of the expression level of markers to the UMAP plot in Figure 1B. The colored circles represent the corresponding cell type from Figure 1B. Supplementary Figure S3. Heterogeneously expressed genes in the β-cells.A: Violin plot showing the pseudo-time value for each group and cell source. The circled numbers indicate the cell source labeled in Figure 1A. The black line within each violin plot indicates the median of the pseudo-time values. B: Heatmap showing genes differentially expressed between group 1 and group 2 β-cells from Ngn3-Cre; Rosa-RFP control mice. C: Heatmap showing the expression patterns of the genes in (B) in bulk cell RNA-seq samples from Meulen et al. [41]. Supplementary Figure S4. mSTRT-seq reveals the β-cell maturation pathway.A: The t-SNE plots showing the cell types (top) and stage (bottom) of the P3, P12, and P21 Ngn3-Cre; Rosa-RFP+ cells. Each dot represents a single cell sample. Cell counts are indicated in brackets. B: Projection of the expression level of markers to the t-SNE plot in (A). C: PCA plot of β-cells from the P3, P12, P21, and normal adult Ngn3-Cre; Rosa-RFP mice. The adult Ngn3-Cre; Rosa-RFP mice include one 2M and two 4M mice, as indicated in Figure 1A. Supplementary Figure S5. Heterogeneity analysis of normal β-cells and α-cells at the transcriptomic level.A: PCA (left) and UMAP (right) of normal adult β-cells. Each dot represents a single cell sample. The colors denote the group. B: Silhouette analyses of the hierarchical clustering of normal adult β-cells. C: Three dimensions (3D) PCA (left), UMAP (middle), and FDL (right) on endocrine cells from Ngn3-Cre; Rosa-RFP pancreas. Each dot represents a single cell. Multiple hormone cells are distinguished by different colors. D: Table showing cell counts of the multiple hormone cells in the adult control and STZ-treated samples. E: PCA of the α-cells and group 1 and group 2 β-cells from the Ngn3-Cre; Rosa-RFP pancreas. Each dot represents a single cell sample. The colors denote the cell type and treatment condition. F: Hierarchical clustering of highly variable genes on the α-cells and group 1 and group 2 β-cells. Each column represents a single cell sample, and each row represents one gene.
References
- 1.Dor Y., Brown J., Martinez O.I., Melton D.A. Adult pancreatic beta-cells are formed by self-duplication rather than stem-cell differentiation. Nature. 2004;429(6987):41–46. doi: 10.1038/nature02520. [DOI] [PubMed] [Google Scholar]
- 2.Thorel F., Nepote V., Avril I., Kohno K., Desgraz R., Chera S. Conversion of adult pancreatic alpha-cells to beta-cells after extreme beta-cell loss. Nature. 2010;464(7292):1149–1154. doi: 10.1038/nature08894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chung C.H., Hao E., Piran R., Keinan E., Levine F. Pancreatic beta-cell neogenesis by direct conversion from mature alpha-cells. Stem Cells. 2010;28(9):1630–1638. doi: 10.1002/stem.482. [DOI] [PubMed] [Google Scholar]
- 4.Chera S., Baronnier D., Ghila L., Cigliola V., Jensen J.N., Gu G. Diabetes recovery by age-dependent conversion of pancreatic delta-cells into insulin producers. Nature. 2014;514(7523):503–507. doi: 10.1038/nature13633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Xu X., D'Hoker J., Stange G., Bonne S., De Leu N., Xiao X. Beta cells can be generated from endogenous progenitors in injured adult mouse pancreas. Cell. 2008;132(2):197–207. doi: 10.1016/j.cell.2007.12.015. [DOI] [PubMed] [Google Scholar]
- 6.Van de Casteele M., Leuckx G., Baeyens L., Cai Y., Yuchi Y., Coppens V. Neurogenin 3+ cells contribute to beta-cell neogenesis and proliferation in injured adult mouse pancreas. Cell Death and Disease. 2013;4:e523. doi: 10.1038/cddis.2013.52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ellis C., Ramzy A., Kieffer T.J. Regenerative medicine and cell-based approaches to restore pancreatic function. Nature Reviews Gastroenterology and Hepatology. 2017;14(10):612–628. doi: 10.1038/nrgastro.2017.93. [DOI] [PubMed] [Google Scholar]
- 8.Lysy P.A., Weir G.C., Bonner-Weir S. Making beta cells from adult cells within the pancreas. Current Diabetes Reports. 2013;13(5):695–703. doi: 10.1007/s11892-013-0400-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rakieten N., Rakieten M.L., Nadkarni M.R. Studies on the diabetogenic action of streptozotocin (NSC-37917) Cancer Chemotherapy Reports. 1963;29:91–98. [PubMed] [Google Scholar]
- 10.Reed M.J., Meszaros K., Entes L.J., Claypool M.D., Pinkett J.G., Gadbois T.M. A new rat model of type 2 diabetes: the fat-fed, streptozotocin-treated rat. Metabolism. 2000;49(11):1390–1394. doi: 10.1053/meta.2000.17721. [DOI] [PubMed] [Google Scholar]
- 11.Skovso S. Modeling type 2 diabetes in rats using high fat diet and streptozotocin. Journal of Diabetes Investigation. 2014;5(4):349–358. doi: 10.1111/jdi.12235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cheng Y., Kang H., Shen J., Hao H., Liu J., Guo Y. Beta-cell regeneration from vimentin+/MafB+ cells after STZ-induced extreme beta-cell ablation. Scientific Reports. 2015;5:11703. doi: 10.1038/srep11703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Grossman E.J., Lee D.D., Tao J., Wilson R.A., Park S.Y., Bell G.I. Glycemic control promotes pancreatic beta-cell regeneration in streptozotocin-induced diabetic mice. PloS One. 2010;5(1):e8749. doi: 10.1371/journal.pone.0008749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Guz Y., Nasir I., Teitelman G. Regeneration of pancreatic beta cells from intra-islet precursor cells in an experimental model of diabetes. Endocrinology. 2001;142(11):4956–4968. doi: 10.1210/endo.142.11.8501. [DOI] [PubMed] [Google Scholar]
- 15.Schonhoff S.E., Giel-Moloney M., Leiter A.B. Neurogenin 3-expressing progenitor cells in the gastrointestinal tract differentiate into both endocrine and non-endocrine cell types. Developmental Biology. 2004;270(2):443–454. doi: 10.1016/j.ydbio.2004.03.013. [DOI] [PubMed] [Google Scholar]
- 16.Li L.C., Yu X.X., Zhang Y.W., Feng Y., Qiu W.L., Xu C.R. Single-cell transcriptomic analyses of mouse pancreatic endocrine cells. Journal of Visualized Experiments. 2018;139 doi: 10.3791/58000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dong J., Hu Y., Fan X., Wu X., Mao Y., Hu B. Single-cell RNA-seq analysis unveils a prevalent epithelial/mesenchymal hybrid state during mouse organogenesis. Genome Biology. 2018;19(1):31. doi: 10.1186/s13059-018-1416-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Islam S., Kjallquist U., Moliner A., Zajac P., Fan J.B., Lonnerberg P. Characterization of the single-cell transcriptional landscape by highly multiplex RNA-seq. Genome Research. 2011;21(7):1160–1167. doi: 10.1101/gr.110882.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kim D., Pertea G., Trapnell C., Pimentel H., Kelley R., Salzberg S.L. TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biology. 2013;14(4):R36. doi: 10.1186/gb-2013-14-4-r36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Liao Y., Smyth G.K., Shi W. featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics. 2014;30(7):923–930. doi: 10.1093/bioinformatics/btt656. [DOI] [PubMed] [Google Scholar]
- 21.Li H., Handsaker B., Wysoker A., Fennell T., Ruan J., Homer N. The sequence alignment/map format and SAMtools. Bioinformatics. 2009;25(16):2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Smith T., Heger A., Sudbery I. UMI-tools: modeling sequencing errors in Unique Molecular Identifiers to improve quantification accuracy. Genome Research. 2017;27(3):491–499. doi: 10.1101/gr.209601.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sinha R., Stanley G., Gulati G.S., Ezran C., Travaglini K.J., Wei E. Index switching causes “spreading-of-signal” among multiplexed samples in Illumina HiSeq 4000 DNA sequencing. bioRxiv. 2017:125724. [Google Scholar]
- 24.Stuart T., Butler A., Hoffman P., Hafemeister C., Papalexi E., Mauck W.M., 3rd Comprehensive integration of single-cell data. Cell. 2019;177(7):1888–1902. doi: 10.1016/j.cell.2019.05.031. e1821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Quinn T.P., Richardson M.F., Lovell D., Crowley T.M. Propr: an R-package for identifying proportionally abundant features using compositional data analysis. Scientific Reports. 2017;7(1):16252. doi: 10.1038/s41598-017-16520-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Falcon S., Gentleman R. Using GOstats to test gene lists for GO term association. Bioinformatics. 2007;23(2):257–258. doi: 10.1093/bioinformatics/btl567. [DOI] [PubMed] [Google Scholar]
- 27.Hastie T., Stuetzle W. Principal curves. Jasa. 1989;84(406):502–516. [Google Scholar]
- 28.Csardi G., Nepusz T. The igraph software package for complex network research. InterJournal Complex Systems. 2006;1695(5):1–9. [Google Scholar]
- 29.Rousseeuw P.J. Silhouettes: a graphical aid to the interpretation and validation of cluster analysis. Journal of Computational and Applied Mathematics. 1987;20:53–65. [Google Scholar]
- 30.Maechler M., Rousseeuw P., Struyf A., Hubert M., Hornik K. 2016. Cluster: cluster analysis basics and extensions. R package version 2.0.5. [Google Scholar]
- 31.McInnes L., Healy J., Melville J. 2018. Umap: uniform manifold approximation and projection for dimension reduction. arXiv preprint arXiv:1802.03426. [Google Scholar]
- 32.Yu X.X., Qiu W.L., Yang L., Zhang Y., He M.Y., Li L.C. Defining multistep cell fate decision pathways during pancreatic development at single-cell resolution. The EMBO Journal. 2019;38(8) doi: 10.15252/embj.2018100164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Taylor B.L., Liu F.F., Sander M. Nkx6.1 is essential for maintaining the functional state of pancreatic beta cells. Cell Reports. 2013;4(6):1262–1275. doi: 10.1016/j.celrep.2013.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gao T., McKenna B., Li C., Reichert M., Nguyen J., Singh T. Pdx1 maintains beta cell identity and function by repressing an alpha cell program. Cell Metabolism. 2014;19(2):259–271. doi: 10.1016/j.cmet.2013.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bid H.K., Konwar R., Aggarwal C.G., Gautam S., Saxena M., Nayak V.L. Vitamin D receptor (FokI, BsmI and TaqI) gene polymorphisms and type 2 diabetes mellitus: a North Indian study. Indian Journal of Medical Sciences. 2009;63(5):187–194. [PubMed] [Google Scholar]
- 36.Brereton M.F., Rohm M., Shimomura K., Holland C., Tornovsky-Babeay S., Dadon D. Hyperglycaemia induces metabolic dysfunction and glycogen accumulation in pancreatic beta-cells. Nature Communications. 2016;7:13496. doi: 10.1038/ncomms13496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Westermeier F., Holyoak T., Asenjo J.L., Gatica R., Nualart F., Burbulis I. Gluconeogenic enzymes in beta-cells: pharmacological targets for improving insulin secretion. Trends in Endocrinology and Metabolism. 2019;30(8):520–531. doi: 10.1016/j.tem.2019.05.004. [DOI] [PubMed] [Google Scholar]
- 38.Kebede M., Favaloro J., Gunton J.E., Laybutt D.R., Shaw M., Wong N. Fructose-1,6-bisphosphatase overexpression in pancreatic beta-cells results in reduced insulin secretion: a new mechanism for fat-induced impairment of beta-cell function. Diabetes. 2008;57(7):1887–1895. doi: 10.2337/db07-1326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Suriben R., Kaihara K.A., Paolino M., Reichelt M., Kummerfeld S.K., Modrusan Z. Beta-cell insulin secretion requires the ubiquitin ligase COP1. Cell. 2015;163(6):1457–1467. doi: 10.1016/j.cell.2015.10.076. [DOI] [PubMed] [Google Scholar]
- 40.Taneera J., Mohammed A.K., Dhaiban S., Hamad M., Prasad R.B., Sulaiman N. RORB and RORC associate with human islet dysfunction and inhibit insulin secretion in INS-1 cells. Islets. 2019;11(1):10–20. doi: 10.1080/19382014.2019.1566684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.van der Meulen T., Mawla A.M., DiGruccio M.R., Adams M.W., Nies V., Dolleman S. Virgin beta cells persist throughout Life at a neogenic Niche within pancreatic islets. Cell Metabolism. 2017;25(4):911–926. doi: 10.1016/j.cmet.2017.03.017. e916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Blum B., Hrvatin S., Schuetz C., Bonal C., Rezania A., Melton D.A. Functional beta-cell maturation is marked by an increased glucose threshold and by expression of urocortin 3. Nature Biotechnology. 2012;30(3):261–264. doi: 10.1038/nbt.2141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Qiu W.L., Zhang Y.W., Feng Y., Li L.C., Yang L., Xu C.R. Deciphering pancreatic islet beta cell and alpha cell maturation pathways and characteristic features at the single-cell level. Cell Metabolism. 2017;25(5):1194–1205. doi: 10.1016/j.cmet.2017.04.003. e1194. [DOI] [PubMed] [Google Scholar]
- 44.Chakravarthy H., Gu X., Enge M., Dai X., Wang Y., Damond N. Converting adult pancreatic islet alpha cells into beta cells by targeting both Dnmt1 and Arx. Cell Metabolism. 2017;25(3):622–634. doi: 10.1016/j.cmet.2017.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yang Y.P., Thorel F., Boyer D.F., Herrera P.L., Wright C.V. Context-specific alpha- to-beta-cell reprogramming by forced Pdx1 expression. Genes & Development. 2011;25(16):1680–1685. doi: 10.1101/gad.16875711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Li J., Casteels T., Frogne T., Ingvorsen C., Honore C., Courtney M. Artemisinins target GABAA receptor signaling and impair alpha cell identity. Cell. 2017;168(1–2):86–100. doi: 10.1016/j.cell.2016.11.010. e115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ackermann A.M., Moss N.G., Kaestner K.H. GABA and artesunate do not induce pancreatic alpha-to-beta cell transdifferentiation in vivo. Cell Metabolism. 2018;28(5):787–792. doi: 10.1016/j.cmet.2018.07.002. e783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.van der Meulen T., Lee S., Noordeloos E., Donaldson C.J., Adams M.W., Noguchi G.M. Artemether does not turn alpha cells into beta cells. Cell Metabolism. 2018;27(1):218–225. doi: 10.1016/j.cmet.2017.10.002. e214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zhang Y., Xie Z., Zhou G., Zhang H., Lu J., Zhang W.J. Fructose-1,6-bisphosphatase regulates glucose-stimulated insulin secretion of mouse pancreatic beta-cells. Endocrinology. 2010;151(10):4688–4695. doi: 10.1210/en.2009-1185. [DOI] [PubMed] [Google Scholar]
- 50.Piccand J., Meunier A., Merle C., Jia Z., Barnier J.V., Gradwohl G. Pak3 promotes cell cycle exit and differentiation of β-cells in the embryonic pancreas and is necessary to maintain glucose homeostasis in adult mice. Diabetes. 2014;63:203–215. doi: 10.2337/db13-0384. PMID: 24163148 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figure S1. STZ model construction.A: Immunofluorescent staining of insulin and glucagon from the control (top) and STZ-treated D1 (bottom) pancreatic tissues. Scale bars: 50 μm. B: Fasting body weight, blood glucose level, and serum insulin of the control and STZ-treated D3 and D6 mice (no insulin implantation). n: Number of mice. C: Blood glucose level of each Ngn3-Cre; Rosa-RFP mouse (corresponding to Figure 1A) 2 days after STZ treatment. D: Blood glucose level and body weight of the STZ M5 and STZ M2NNgn3-Cre; Rosa-RFP mice before sacrifice. The STZ-treated M2N mouse had severe hyperglycemia starting 2 days after STZ treatment. E: Immunofluorescent staining of insulin and glucagon in paraffin sections from the normal, STZ D12, M5, and M9 pancreatic tissue. Scale bars: 50 μm ∗: no insulin implant during M7-M9. F: Proportion of β-cells in the islets shown in (E). Data are shown as means ± SEM. Supplementary Figure S2. Quality control of scRNA-seq samples.A and B: Bar plots showing UMI (A) and detected gene count (B) of the available single-cell samples. Each bar represents one sample. C: Projection of the expression level of markers to the UMAP plot in Figure 1B. The colored circles represent the corresponding cell type from Figure 1B. Supplementary Figure S3. Heterogeneously expressed genes in the β-cells.A: Violin plot showing the pseudo-time value for each group and cell source. The circled numbers indicate the cell source labeled in Figure 1A. The black line within each violin plot indicates the median of the pseudo-time values. B: Heatmap showing genes differentially expressed between group 1 and group 2 β-cells from Ngn3-Cre; Rosa-RFP control mice. C: Heatmap showing the expression patterns of the genes in (B) in bulk cell RNA-seq samples from Meulen et al. [41]. Supplementary Figure S4. mSTRT-seq reveals the β-cell maturation pathway.A: The t-SNE plots showing the cell types (top) and stage (bottom) of the P3, P12, and P21 Ngn3-Cre; Rosa-RFP+ cells. Each dot represents a single cell sample. Cell counts are indicated in brackets. B: Projection of the expression level of markers to the t-SNE plot in (A). C: PCA plot of β-cells from the P3, P12, P21, and normal adult Ngn3-Cre; Rosa-RFP mice. The adult Ngn3-Cre; Rosa-RFP mice include one 2M and two 4M mice, as indicated in Figure 1A. Supplementary Figure S5. Heterogeneity analysis of normal β-cells and α-cells at the transcriptomic level.A: PCA (left) and UMAP (right) of normal adult β-cells. Each dot represents a single cell sample. The colors denote the group. B: Silhouette analyses of the hierarchical clustering of normal adult β-cells. C: Three dimensions (3D) PCA (left), UMAP (middle), and FDL (right) on endocrine cells from Ngn3-Cre; Rosa-RFP pancreas. Each dot represents a single cell. Multiple hormone cells are distinguished by different colors. D: Table showing cell counts of the multiple hormone cells in the adult control and STZ-treated samples. E: PCA of the α-cells and group 1 and group 2 β-cells from the Ngn3-Cre; Rosa-RFP pancreas. Each dot represents a single cell sample. The colors denote the cell type and treatment condition. F: Hierarchical clustering of highly variable genes on the α-cells and group 1 and group 2 β-cells. Each column represents a single cell sample, and each row represents one gene.
Data Availability Statement
The RNA-seq data were submitted to the GEO repository under accession number GSE137909.