Abstract
Laccases (E.C. 1.10.3.2) are multicopper oxidases of great importance in the industry due to their non-specificity and high oxidative potential. Laccases are useful to bleach synthetic dyes, oxidize phenolic compounds and degrade pesticides, among others. Hence, the objective of this work was to optimize low cost culture media for recombinant (rPOXA 1B) laccase production from Pleurotus ostreatus in Pichia pastoris. To this end, low cost nitrogen sources were studied, such as malt extract, isolated soy protein and milk serum. Following, two central composite designs (CCD) were performed. In CCD-1 different concentrations of glucose USP (0–13.35 gL-1), protein isolated soy protein (5–25 gL-1), malt extract (3.5–17.5 gL-1) and (NH4)2SO4 (1.3–6.5 gL-1) were evaluated. In CCD-2 only different concentrations of glucose USP (7.9–22 gL-1) and isolated soy protein (15.9–44.9 gL-1) were evaluated. CCD-2 results led to a One Factor Experimental design (OFED) to evaluate higher isolated soy protein (20–80 gL-1) concentrations. In all designs, (CCD-1, CCD-2 and OFED) CuSO4 (0.16 gL-1) and chloramphenicol (0.1 gL-1) concentrations remained unchanged. For the OFED after sequential statistical optimization, an enzyme activity of 12,877.3 ± 481.2 UL−1 at 168 h was observed. rPOXA 1B activity increased 30.54 % in comparison with CCD-2 results. Final composition of optimized media was: 20 gL-1 glucose USP, 50 gL-1 isolated soy protein 90 % (w/w), 11.74 gL-1 malt extract, and 4.91 gL-1 (NH4)2SO4. With this culture media, it was possible to reduce culture media costs by 89.84 % in comparison with improved culture media previously described by our group.
Keywords: Biotechnology, Microbiology, Mycology, Microbial biotechnology, Enzymology, Laccase, Enzyme, Pichia pastoris, Isolated soy protein, Malt extract, Low-cost substrate
Biotechnology; Microbiology; Mycology; Microbial biotechnology; Enzymology; Laccase; Enzyme; Pichia pastoris; Isolated soy protein; Malt extract; Low-cost substrate
1. Introduction
Laccases (EC 1.10.3.2) or para-bezenediol: dioxygen oxidoreductases are monomeric multicopper enzymes [1], with an approximate molecular weight of 60–70 kDa [2]. They belong to a group of lignolytic enzymes that have the capacity to oxidize phenolic compounds and similar substrates, in addition to reducing simultaneously molecular O2 to H2O [3]. Because of laccases low specificity for their substrates [4], they are used in a great number of biotechnological applications; such as delignification of cellulose pulp, treatment of black liquor residue of cellulose pulping, degradation of aromatic polycyclic hydrocarbons, independent and simultaneous bioconversion of low density plastic and lignocellulosic material, soil bioremediation, wastewater treatment, discoloration and detoxification of industrial dyes, manufacture and synthesis of food processing material [5, 6, 7, 8, 9, 10, 11, 12].
Different white rot fungi such as Pleurotus ostreatus, Trametes versicolor, Rigidoporus lignosus, Trametes villosa, Rhizoctonia solani and Ganoderma lucidum [3, 13] have been reported to produce laccase. Specifically, P. ostreatus produces 11 types of laccases (isoenzymes) [3], of which POXA 1B has been widely studied for its great stability at alkaline pH and enzyme activity [14, 15]. However, enzyme production from the native organism is not sufficient to meet industrial demand, based fundamentally on fungi prolonged culture time [16].
Heterologous laccase production in hosts such as Trichoderma reesei [17], Pichia methanolica [18], Yarrowia lipolytica [19], Saccharomyces cerevisiae [20] and Pichia pastoris [15] has been a strategy to comply with industrial demands, due to achieved overexpressions through recombinant DNA technologies [21]. Never the less, among various heterologous hosts, Pichia pastoris a methylotrophic yeast, is known for recombinant protein production because of its robust secretion system, favouring protein processing [22]. Moreover, this yeast can reach high cell densities [23], it can be easily manipulated in the lab, expresses high levels of extracellular proteins and performs characteristic post-translational modifications of eukaryotes, such as glycosylations and disulfide bond formation, among others [24, 25].
Recombinant protein expression in P. pastoris is influenced by factors such as pH, temperature, media composition [26] and the promoter used for the genetic construction [27]. P. pastoris can grow in chemically defined basic media [25] and in complex media with biosynthetic precursors that can be catalyzed into various anabolic pathways, decreasing the need to produce it, which economizes metabolic energy and efficiency in the production of enzymes [28, 29]. For the design of culture media, it is necessary to know the nutritional requirements of the microorganism to be cultured [30], as well as the nutritional value for each component [31], for which different design methodologies and optimization of culture media have been employed. It is known that approximately 28 % of operation costs correspond to raw material [32, 33]. Therefore, research has been carried out with low cost media and it has been demonstrated that industrial grade compounds increases yields, productivities and enzyme production [27, 34].
Previous to this study, our group cloned P. ostreatus POXA 1B in P. pastoris [15] with an enzyme activity of 451.08 ± 6.46 UL−1 at 158 h, at shake flask scale in YPD media (2 % (w/v) D-glucose, 2 % (w/v) peptone, 1 % (w/v) yeast extract). Following the media was statistically improved (10 gL-1 D-glucose, 20 gL-1 peptone, 15 gL-1 yeast extract, 0.16 gL-1 CuSO4, 2.67 gL-1 (NH4)2SO4), obtaining at 168 h a laccase activity of 1,343.52 ± 40.30 UL−1 at shake-flask scale. Improved media was then assayed at 10 L bioreactor (with 6 L of working effective volume WEV) obtaining 3159.93 ± 498.90 UL−1 at 192 h [35]. Therefore, the objective of this work was to design and statistically optimize a low cost culture media that increased Pleurotus ostreatus rPOXA 1B activity in P. pastoris.
2. Materials and methods
2.1. Strain
P. pastoris X33/pGAPZαA-LaccPost-Stop strain containing the optimized synthetic gene coding for laccase POXA 1B under the control of the constitutive promoter of glyceraldehyde-3-phosphate dehydrogenase (pGAP) (E.C. 1.2.1.12), was used [15]. The microorganism is stored in YPD broth, supplemented with glycerol at 20 % (w/v) at -30 °C at the Working Cell Bank (WCB) [36], of Molecular Biotechnolgy Laboratory from the “Grupo de Biotecnología Ambiental e Industrial (GBAI), Departamento de Microbiología, Facultad de Ciencias, Pontificia Universidad Javeriana, Bogotá, D.C., Colombia”.
2.2. Strain reactivation and inoculum preparation
P. pastoris X33/pGAPZαA-LaccPost-Stop vials were recovered from the WCB and inoculated into a 50 mL Erlenmeyer flask containing 5 mL YPD broth and incubated for 24 h at 200 r.p.m., and 30 °C. Once the strain was re-activated the inoculum was prepared in previously improved media (10 gL-1 D-glucose, 20 gL-1 peptone, 15 gL-1 yeast extract, 0.16 gL-1 CuSO4, 2.67 gL-1 (NH4)2SO4) [35], incubated at 200 rpm and 30 °C until a minimal optical density reading of 1.00 at 600 nm (ODλ600) was reached. Assays performed in 250 mL Erlenmeyer flasks containing 150 mL effective work volume (EWV) were inoculated with 10 % (v/v) inoculum.
2.3. Low cost nitrogen sources
Three inexpensive nitrogen sources at different concentrations were selected by cost for evaluation: isolated soy protein (90 %), (Ciacomeq SAS), malt extract (Proquímicas JG SAS) and powdered milk (ChemiPharma) (Table 1, Supplementary Materials 1, 2 and 3). Taking into account nitrogen organic sources concentration was 35 gL-1 (20 gL-1 peptones, 15 gL-1 yeast extract) in previously improved media [35]; nitrogen sources for each treatment to be evaluated were adjusted to 35 gL-1. Each treatment was performed in triplicate in 250 mL Erlenmeyer flask containing 10 % (v/v) inoculum with WEV of 150 mL at 30 °C, for 168 h. Previously improved culture media was used as control [35]. Samples were collected at 0 and 168 h of culture and centrifuged for 10 min at 4,427 x g and 25 °C. Laccase activity (UL−1) was determined from supernatant [37].
Table 1.
Media composition with different nitrogen sources.
| Treatment | Carbon source/(gL−1) | Organic sources of nitrogen/(gL−1) | Inorganic sources of nitrogen/(gL−1) | Cofactor/(gL−1) | Other (gL−1) |
|---|---|---|---|---|---|
| Control | Glucose/(10) | Peptone/(20) Yeast extract/(15) | (NH4)2SO4/(2.67) | CuSO4/(0.16) | Chloramphenicol (0.1) |
| T1 | D-Glucose USP/(10) | Isolated soy protein/(20) | |||
| Malt extract/(15) | |||||
| T2 | Isolated soy protein/(35) | ||||
| T3 | Milk serum/(20) | ||||
| Malt extract/(15) |
For statistical analysis one factor ANOVA was employed, followed by a Tukey post-hoc test to evaluate significant differences among treatments at 168 h of culture. Normal distribution of the response variable (enzyme activity in UL−1) was previously verified using Kolmogorov-Smirnov test and variance homogeneity through Levene test. Statistical analysis was performed using SPSS Statistics® V. 21.0. (IBM).
2.4. Response surface methodology (RSM) for low cost culture media design
2.4.1. Central composite Design-1 (CCD-1)
To determine optimal concentrations for each pre-selected factor a first Central Composite Design-1 (CCD-1) was performed including a Central point (assay in triplicate) and two axial points (-1.5 and 1.5); axial points were calculated using a rotatable alpha (k = 2). Factors and levels included in the CCD-1 are illustrated in Table 2. Each treatment was performed in 250 mL Erlenmeyer flask with a 150 mL WEV, 10 % (v/v) inoculum incubated at 200 r.p.m., at 30 °C for 168 h. 2 mL of sample were collected at 0, 12, 72 and 168 h of culture and centrifuged at 4,427 x g and 25 °C. Residual reducing sugars (gL−1) [38], were measured in the supernatant, as well as laccase activity (UL−1) [37], and total protein concentration (mg mL−1), [39]. Enzyme activity (UL−1) and specific activity (Umg−1) were used as response variables at 72 and 168 h of culture.
Table 2.
CCD-1 composition, evaluated factors and their respective levels.
| Factor | Level |
||||
|---|---|---|---|---|---|
| -1.5 | -1 | Central Point | 1 | 1.5 | |
| Glucose USP (gL−1) | -0.05∗∗ | 3.3 | 6.65 | 10.0 | 13.35 |
| Isolated soy protein (gL−1) | 5.0 | 10.0 | 15.0 | 20.0 | 25.0 |
| Malt extract (gL−1) | 3.5 | 7.0 | 10.5 | 14.0 | 17.5 |
| (NH4)2SO4 (gL−1) | 1.3 | 2.6 | 3.9 | 5.2 | 6.5 |
| CuSO4 (gL−1)∗ | 0.16 | ||||
| Chloramphenicol (gL−1)∗ | 0.1 | ||||
These factors remained unchanged for all combinations.
This value was assumed as 0.0.
2.4.2. Central composite Design-2 (CCD-2)
To adjust the concentrations of significant CCD-1 factors a second Central Composite Design-2 (CCD-2) was performed including one central point, assayed in triplicate and two axial points (-1.5 and 1.5) that were evaluated in triplicate for 27 experiments; axial points were calculated using a rotatable alpha (k = 1.4142). Composition of each treatment proposed for this design is illustrated in Table 3. Assay conditions, sampling and response variables were similar to CCD-1.
Table 3.
CCD-2 composition, evaluated factors and their respective levels.
| Factor |
Level |
||||
|---|---|---|---|---|---|
| -1.5 |
-1 |
Central Point |
1 |
1.5 |
|
| D-Glucose USP (gL−1) |
7.9 |
10.0 |
15.0 |
20.0 |
22.1 |
| Isolated soy protein (gL−1) | 15.9 | 20.0 | 30.0 | 40.0 | 44.1 |
| Malt extract (gL−1)∗∗ | 11.74 | ||||
| (NH4)2SO4 (gL−1)∗∗ | 4.91 | ||||
| CuSO4 (gL−1)∗ | 0.161 | ||||
| Chloramphenicol (gL−1)∗ | 0.1 | ||||
These factors remained unchanged for all combinations.
These factors remained unchanged for all combinations and were derived from CCD-1 obtained results.
2.4.3. One Factor Experimental design (OFED)
For this design, isolated soy protein, the significant factor in CCD-2, was taken into account. A cubic model between 20 gL-1 and 80 gL-1 (-1 and +1 respectively) was used and provided 5 central points for a total of 10 treatments (combinations with 20, 50 and 80 gL-1) performed in duplicates. In this design, the factor Glucose (20 gL-1) remained unchanged as established in CCD-2 (Table 4). Assay conditions, sampling and response variables were similar to CCD-1 and CDD-2.
Table 4.
OFED composition, evaluated factors and their respective levels.
| Factor |
Levels |
||||||
|---|---|---|---|---|---|---|---|
| -1 |
Central Points |
1 |
|||||
| Isolated soy protein (gL−1) | 20.00 | 30.02 | 40.01 | 50.00 | 59.99 | 69.98 | 80.00 |
| Glucose USP (gL−1)∗∗∗ | 20 | ||||||
| Malt extract (gL−1)∗∗ | 11.74 | ||||||
| (NH4)2SO4 (gL−1)∗∗ | 4.91 | ||||||
| CuSO4 (gL−1)∗ | 0.16 | ||||||
| Chloramphenicol (gL−1)∗ | 0.1 | ||||||
These factors remained unchanged for all combinations.
These factors remained unchanged for all combinations and were derived from results obtained from CCD-1.
This factor remained unchanged for all combinations and was derived from results obtained in CCD-2.
2.4.4. CCD-1, CCD-2 and OFED statistical analyses
Factorial designs statistical analyses were performed using Design Expert® V9.0. Response variable normal distribution was evaluated using Kolmogorov-Smirnov test and variance homogeneity using Levene test.
2.5. Analytical techniques
2.5.1. Residual reducing sugar determination
Reducing sugar concentration obtained from sample supernatant was determined by 3,5-dinitrosalicylic acid assay (DNS), [38]. Standard curve was calculated from Eq. (1).
| (1) |
2.5.2. Enzyme activity assay
Laccase enzyme activity was performed using ABTS (2,2′-Azino-bis(3-ethylbenzothiazoline-6-sulfonic acid)) as substrate. In a spectrophotometric cuvette 100 μL of 20 mM ABTS was added in addition to centrifuged supernatant (from 2 to 20 μL, depending on the amount of enzyme present in the sample) to complete a volume of 1 mL with 0.1 M (pH 3.0 ± 0.2) citrate buffer. Absorbance change for 1 min at 420 nm was measured as resulting from ABTS oxidation [37]. One unit of enzyme (U) is defined as the quantity of enzyme capable of transforming 1 μmol ABTS substrate per minute, per liter, and was calculated based on the following equation:
| (2) |
Where: ΔE corresponds to final absorbance minus initial absorbance during 1 min of reaction, Vt total volume used in the reaction, ε ABTS molar extinction coefficient (M−1 cm−1) at 420 nm, d cuvette diameter in cm and Vs sample volume (enzyme) added to the reaction [39].
2.5.3. Total protein determination
Total protein concentration determination for each sample was obtained from direct absorbance at 280 nm using UV-Vis NanoDrop 2000 (Thermo Scientific) spectrophotometer [40].
2.5.4. Total carbon (TC, gL−1), total nitrogen (TN, gL−1) and initial carbon/nitrogen ratio (C/N)0 determination
Total organic carbon (TOC) analyzer, Shimadzu (TOC-L) was used to measure both TC and TN concentrations, this device develops a unique method of catalytic combustion oxidation and NDIR, according to Standard Method 5310B [41]. Standard solutions (0.1 gL-1) of each media components (separately) were prepared and then analyzed in the TOC device. Once TC and TN concentrations were detected, stoichiometric calculations were performed to determine the concentration in each culture medium (gL−1) and the initial (C/N)0 ratio.
2.5.5. Raw material cost analysis
Raw material costs employed for optimized low-cost media formulation were calculated, including 10 % contingencies and 19 % sales tax (VAT rate) at present for Colombia.
3. Results
3.1. Selection of low-cost nitrogen sources
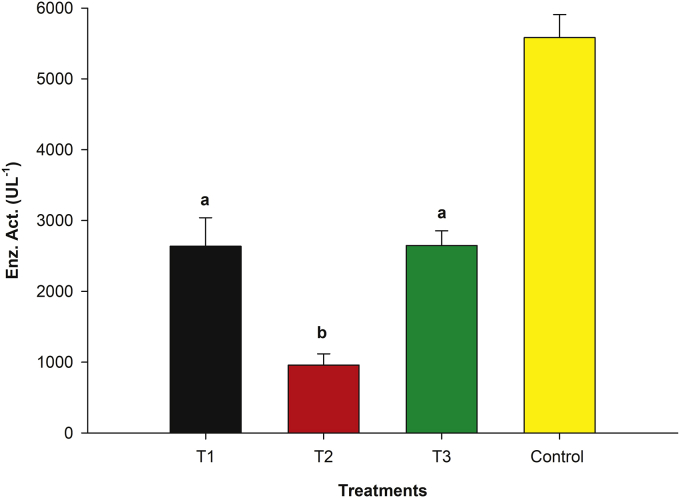
Treatments generated two groups which were significantly different (p < 0.001) in regard to laccase activity at 168 h of culture (Figure 1). Control media was not included in mean comparison and had a laccase activity of 5,583.33 ± 324.14 UL−1. Treatments T3 and T1 had maximal laccase activities of 2,636.57 ± 402.1 UL−1 and 2,645.83 ± 208.7 UL−1, respectively (group a mean comparison), with no significant differences (p > 0.05). Moreover, laccase activity for T2 (958.33 ± 158.11 UL−1), (group b), was considerably lower. Treatment T1 was the selected media for statistical optimization (10 gL-1 D-glucose USP, 20 gL-1 isolated soy protein, 15 gL-1 malt extract, 2.67 gL-1 (NH4)2SO4 and 0.16 gL-1 CuSO4.
Figure 1.
Laccase activity (UL−1) mean comparison obtained from different treatments. Each bar corresponds to laccase activity at 168 h of culture (mean of n = 3). An α = 0.05 was used.
3.2. Central composite Design-1 (CCD-1)
In CCD-1 quadratic model was significant at 72 h for the enzyme activity (UL−1) and specific enzyme activity (Umg−1); whereas at 168 h the lineal model was significant (Table 5). However, despite R-Squared values were low it was decided to continue working with specific activity (Umg−1) at 72 for having the least significant lack of fit and the second highest R-Squared.
Table 5.
CCD-1 ANOVA result analysis.
| Response variable | Source | Sum of Squares | df | Mean Square | F-Value | p-value Prob > F | R-Squared |
|---|---|---|---|---|---|---|---|
| Enz. Act. (UL−1) 72h | Quadratic Model | 1.98E+07 | 14 | 1.42E+06 | 3.26 | 0.0235∗ | 0.7917 |
| Lack of Fit | 5.12E+06 | 10 | 5.12E+05 | 10.78 | 0.0878 | ||
| Enz. Act. (UL−1) 168h | Linear Model | 1.05E+07 | 4 | 2.64E+06 | 4.77 | 0.0063∗ | 0.4645 |
| Lack of Fit | 1.05E+07 | 20 | 5.26E+05 | 0.64 | 0.7645 | ||
| Spec. Act. (Umg−1) 72h | Quadratic Model | 1.84E+05 | 14 | 1.31E+04 | 2.94 | 0.0345∗ | 0.7740 |
| Lack of Fit | 4.59E+04 | 10 | 4.59E+03 | 1.18 | 0.5418 | ||
| Spec. Act. (Umg−1) 168h | Linear Model | 7.56E+04 | 4 | 1.89E+04 | 4.65 | 0.0072∗ | 0.4580 |
| Lack of Fit | 8.54E+04 | 20 | 4.27E+03 | 2.10 | 0.3722 |
Significant at 95%.
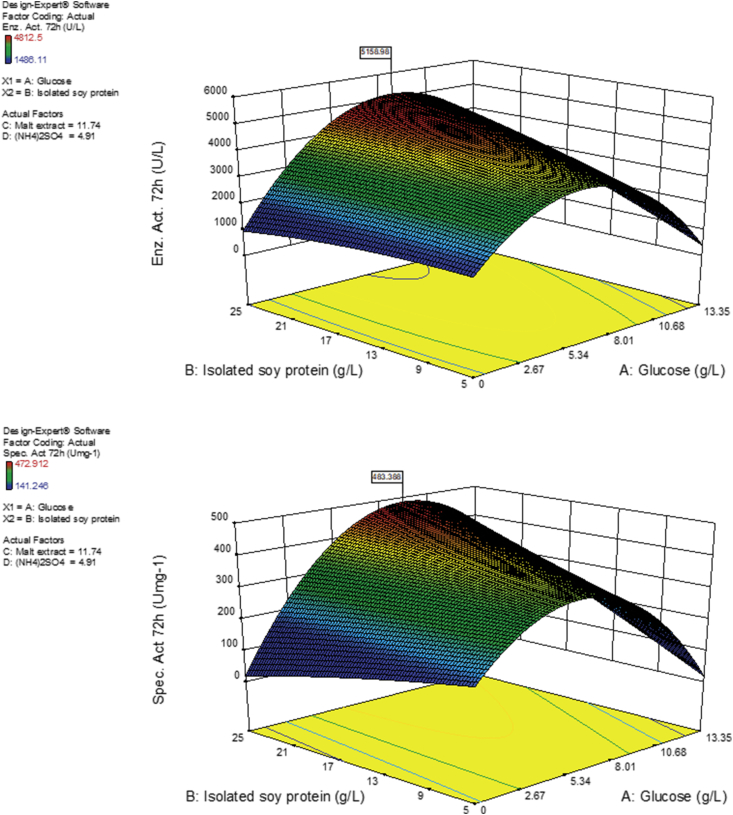
For both response variables (enzyme activity and specific enzyme activity) at both selected hours (72 or 168 h) and both models (quadratic and lineal), the factors isolated soy protein and glucose were significant between 89 and 99% (Table 6). For the rest of the factors and their combinations and squares in the quadratic model was below 85%. Interaction prediction of the two most significant variables (D-glucose USP and isolated soy protein) on specific activity (Umg−1) and laccase activity (UL−1) at 72 and 168 h, respectively is illustrated in Figure 2. It was observed; augmented response variable resulted from simultaneous glucose and isolated soy protein concentration, increase.
Table 6.
CCD-1 quadratic model individual factor significance, duplicates and combined (Response variable: specific activity at 72 h de culture).
| Spec. Act. (UL−1) 72 h | ||||||
|---|---|---|---|---|---|---|
| Source | Sum of |
df | Mean |
F |
p-value |
|
| Squares | Square | Value | Prob > F | |||
| Quadratic Model | 1.84E+05 | 14 | 1.31E+04 | 2.94 | 0.0345∗ | |
| A-Glucose | 1.35E+04 | 1 | 1.35E+04 | 3.02 | 0.1078∗∗∗ | |
| B- isolated soy protein | 2.54E+04 | 1 | 2.54E+04 | 5.68 | 0.0346∗ | |
| C-Malt extract | 6.59E+02 | 1 | 6.59E+02 | 0.15 | 0.7076 | |
| D-(NH4)2SO4 | 1.30E+04 | 1 | 1.30E+04 | 2.91 | 0.1140∗∗∗ | |
| AB | 1.75E+04 | 1 | 1.75E+04 | 3.91 | 0.0715∗∗ | |
| AC | 1.07E+04 | 1 | 1.07E+04 | 2.40 | 0.1473 | |
| AD | 3.71E+03 | 1 | 3.71E+03 | 0.83 | 0.3800 | |
| BC | 2.81E+02 | 1 | 2.81E+02 | 0.06 | 0.8063 | |
| BD | 9.04E+02 | 1 | 9.04E+02 | 0.20 | 0.6608 | |
| CD | 1.19E+02 | 1 | 1.19E+02 | 0.03 | 0.8729 | |
| A2 | 8.49E+04 | 1 | 8.49E+04 | 19.00 | 0.0009∗ | |
| B2 | 1.16E+02 | 1 | 1.16E+02 | 0.03 | 0.8746 | |
| C2 | 9.22E+01 | 1 | 9.22E+01 | 0.02 | 0.8882 | |
| D2 | 3.10E+03 | 1 | 3.10E+03 | 0.69 | 0.4209 | |
| Residual | 5.36E+04 | 12 | 4.47E+03 | |||
| Lack of Fit | 4.59E+04 | 10 | 4.59E+03 | 1.18 | 0.5418 | |
| Pure Error | 7.75E+03 | 2 | 3.87E+03 | |||
| Cor Total | 2.37E+05 | 26 | ||||
| R-Squared | 0.7740 | |||||
| Adj R-Squared | 0.5104 | |||||
| Adeq Precision | 7.2192 | |||||
96–99 % significance.
93–95 % significance.
89–90 % significance.
Figure 2.
CCD-1 contour graph. Effect of factor interaction between glucose (A) and isolated soy protein (B) on laccase activity (top) and specific activity (bottom).
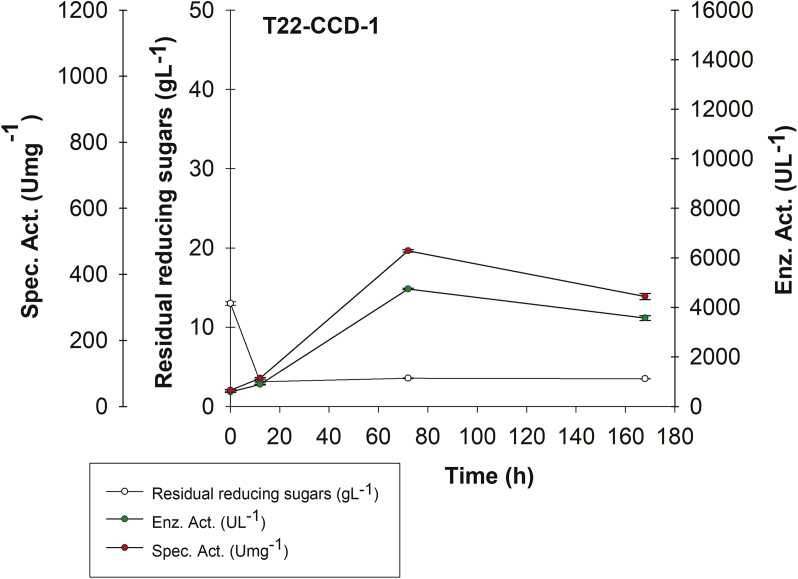
CCD-1 results at 72 h demonstrated the need to vary glucose USP and isolated soy protein concentrations (Table 6, Figure 2). To this end, for CCD-2 malt extract was defined at 11.74 gL-1 and ammonium sulphate at 4.91 gL-1 (CCD-1 optimal predicted concentrations, instead of including them as CCD-2 factors). However, to obtain an ampler surface navigation than that initially proposed in CCD-1 glucose USP (9.53 gL-1) and isolated soy protein (19.25 gL-1) concentrations had to be included in the increased range of CCD-2 concentrations (Table 3). This design highlighted T22 treatment, whose (C/N)0 ratio was 6.0 ± 0.03 (Figure 3, Supplementary Material 4) with an enzyme activity of 4,743.05 ± 29.46 UL−1 at 72 h. However, at 168 h laccase activity decreased to 3,569.44 ± 96.23 UL−1. This same behavior was also observed with obtained specific activity, which decreased from 471.32 ± 3.37 Umg−1 at 72h to 332.81 ± 9.11 Umg−1 at 168 h. On the other hand, reducing sugar concentrations considerably decreased during the first 12 h from 12.99 to 3.11 gL-1. However, there was no total consumption at 168 h, with approximately 3 gL-1 residual sugar concentration at 168 h (Figure 3).
Figure 3.
rPOXA 1B production kinetics. Central Composite Design-1 (CCD-1) for treatment T22.
3.3. Central composite Design-2 (CCD-2)
Enzyme activity and specific activity in the CCD-2 quadratic model was significant at 72 h and 168 h. However, lack of fit was significant for enzyme activity at 72 h, but not for specific activity at 168 h (Table 7, Supplementary Material 6). Therefore, the remaining of the analysis was based on specific activity (Umg−1) response variable.
Table 7.
CCD-2 ANOVA result analysis for specific activity (Umg−1) at 168 h.
| Source | Sum of |
df | Mean |
F |
p-value |
|
|---|---|---|---|---|---|---|
| Squares | Square | Value | Prob > F | |||
| Quadratic Model | 1.E+06 | 5 | 2.E+05 | 80.49 | < 0.0001 | |
| A-Glucose | 6.E+05 | 1 | 6.E+05 | 240.24 | < 0.0001 | |
| B-Isolated soy protein | 3.E+05 | 1 | 3.E+05 | 111.96 | < 0.0001 | |
| AB | 8.E+04 | 1 | 8.E+04 | 34.24 | < 0.0001 | |
| A2 | 2.E+02 | 1 | 2.E+02 | 0.08 | 0.7841 | |
| B2 | 3.E+04 | 1 | 3.E+04 | 10.61 | 0.0038 | |
| Residual | 5.E+04 | 21 | 2.E+03 | |||
| Lack of Fit | 1.E+04 | 3 | 4.E+03 | 1.89 | 0.1677 | |
| Pure Error | 4.E+04 | 18 | 2.E+03 | |||
| Cor Total | 1.E+06 | 26 | ||||
| R-Squared | 0.9504 | |||||
| Adj R-Squared | 0.9386 | |||||
| Pred R-Squared | 0.9193 | |||||
| Adeq Precision | 23.496 | |||||
In bold: model and significant factors (p < 0.05).
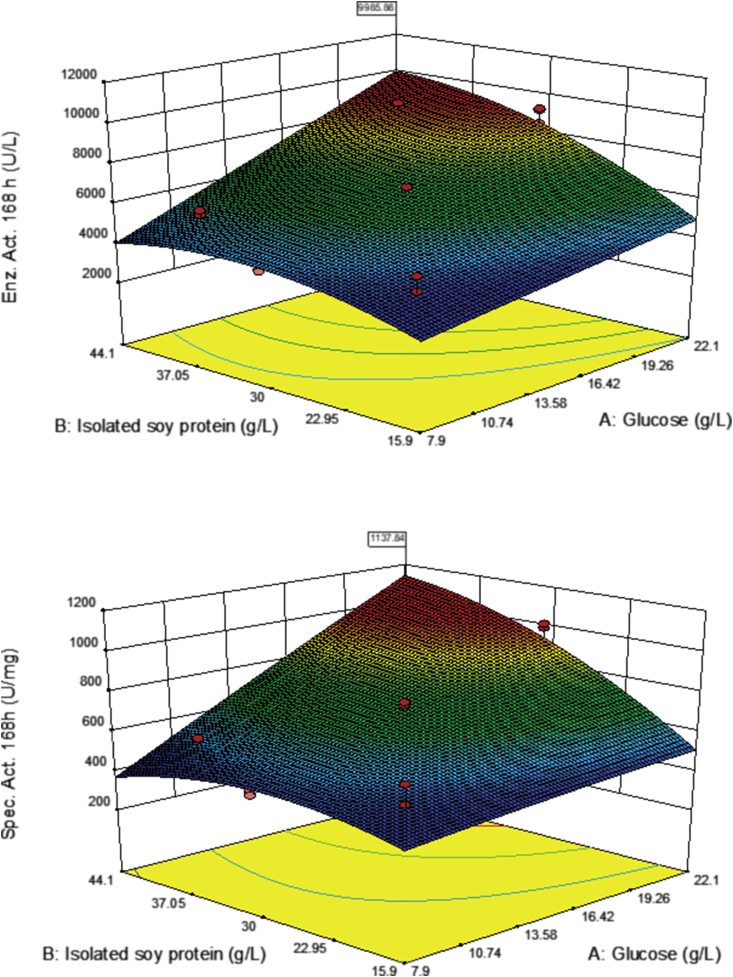
Both factors glucose and isolated soy protein had a positive influence on specific activity and laccase activity. As factor concentration was augmented response variables also increased (Figure 4).
Figure 4.
CCD-2 contour graph. Effect of factor interaction between glucose (A) and isolated soy protein (B) on laccase activity (top) and specific activity (bottom).
To obtain a laccase activity of 9,985.9 UL−1 and a specific activity of 1,137.8 Umg−1 (Figure 4) according to the model glucose optimal concentration had to be 20 gL-1 and isolated soy protein 39.87 gL-1. Nonetheless, increasing isolated soy protein could improve activity results further (Figure 5, Table 7). Isolated soy protein (Factor B2) was significant for the model (p = 0.0038) as depicted in Table 7. Moreover, interaction between factors AB was significant (p < 0.0001) although A2 was not (p = 0.7841). Therefore, in an attempt to optimize isolated soy protein concentration a One Factor Design was implemented.
Figure 5.
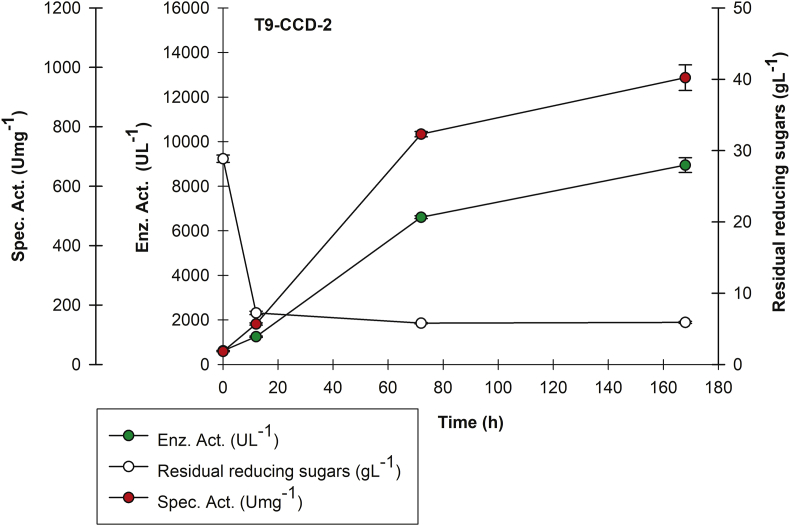
rPOXA 1B production kinetics. Central Composite Design-1 (CCD-2) for treatment T9.
This design highlighted T9 treatment, whose (C/N)0 ratio was 6.7 ± 0.02 (Figure 5) with obtained higher laccase activity and specific activity (Supplementary Material 5), at 72 h (6,608.80 ± 62.20 UL−1 and 725.21 ± 14.41 Umg−1), as well as 168 h with 8,944.44 ± 330.28 UL−1 and 971.92 ± 17.61 Umg−1. Results demonstrate CCD-2 exceeded T22 (CCD-1) for both sampling time (72 and 168 h). Again, during the first 12 h a decrease in reducing sugars was observed, although not completely with approximately 5 gL-1 residual sugars maintained until 168 h.
3.4. One Factor Experimental design (OFED)
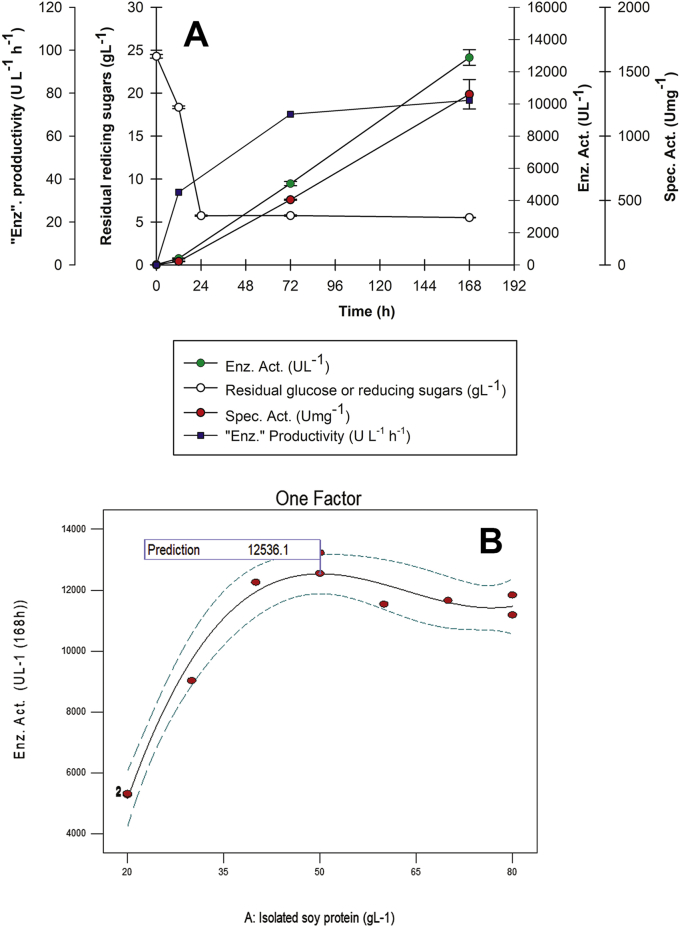
In this design the cubic model was significant (p < 0.0001) at 168 h of culture (Table 8) and treatment T4 with a (C/N)0 ratio of 5.4 ± 0.03 and 50 gL-1 isolated soy protein demonstrated the highest enzyme activity (12,877.31 ± 481.23 UL−1) and the highest specific activity (1,324.58 ± 114.19 Umg−1) at 168 h (Figure 6A), slightly exceeding model prediction which was 12,536.1 UL−1 with a desirability of 0.914 at 168 h (Figure 6B).
Table 8.
OFED ANOVA result analysis for enzyme activity (U L−1) at 168 h.
| Source | Sum of |
df | Mean |
F |
p-value |
|
|---|---|---|---|---|---|---|
| Squares | Square | Value | Prob > F | |||
| Model Cubic | 7.39E+07 | 3.00 | 2.46E+07 | 84.75 | <0.0001 | |
| A-isolated soy protein | 6.33 | 1.00 | 6.33 | 0.00 | 0.9964 | |
| Aˆ2 | 3.22E+07 | 1.00 | 3.22E+07 | 110.88 | <0.0001 | |
| Aˆ3 | 3.55E+06 | 1.00 | 3.55E+06 | 12.21 | 0.0129 | |
| Residual | 1.74E+06 | 6.00 | 2.91E+05 | |||
| Lack of Fit | 1.30E+06 | 3.00 | 4.33E+05 | 2.92 | 0.2014 | |
| Pure Error | 4.46E+05 | 3.00 | 1.49E+05 | |||
| Cor Total | 7.57E+07 | 9.00 | ||||
| R-Squared | 0.9769 | |||||
| Adj R-Squared | 0.9654 | |||||
| Pred R-Squared | 0.9394 | |||||
| Adeq Precision | 21.6448 | |||||
Bold signifies model and significant factors (p < 0.05).
Figure 6.
One Factor Design kinetics and prediction. A. rPOXA 1B production kinetics. B. One Factor design prediction.
3.5. Raw material cost analysis
Optimized low cost culture media had a cost of $ 1,227 USD/litre, which represents close to 89.84 % less in comparison with previously improved media ($12,084 USD/litre). Cost analysis is shown in Table 9 including 10 % contingencies and 19 % sales tax at present for Colombia [42].
Table 9.
Media cost comparison between previously improved media [35] vs. low cost optimized media.
| Previously improved media |
Low cost optimized media |
||||
|---|---|---|---|---|---|
| Reference |
[35] |
Reference |
Present study |
||
| Component | (g L−1) o L | Cost/L (USD) | Component | (g L−1) o L | Cost/L (USD) |
| Glucose (Merck) | 10 | $ 0.640 | Glucose USP (ChemiPharma) | 20 | $ 0.037 |
| Peptone (Oxoid) | 20 | $ 6.740 | Isolated soy protein (Ciacomeq SAS) | 50 | $ 0.200 |
| Yeast extract (Oxoid) | 15 | $ 1.485 | Malt extract (Proquímicas JG SAS) | 10.28 | $ 0.063 |
| CuSO4 (Scharlau) | 0.16 | $ 0.077 | CuSO4 (Scharlau) | 0.16 | $ 0.077 |
| (NH4)2SO4 (Merck) | 2.6 | $ 0.240 | (NH4)2SO4 (Merck) | 4.91 | $ 0.389 |
| Chloramphenicol (SIGMA) | 0.1 | $ 0.184 | Chloramphenicol (SIGMA) | 0.1 | $ 0.184 |
| H2O for dilution∗ | 1 | $ 0.001 | H2O for dilution∗ | 1 | $ 0.001 |
| Total | $ 9.367 | Total | $ 0.951 | ||
| 10 % contingencies | $ 0.937 | 10 % contingencies | $ 0.095 | ||
| 19 % sales tax | $ 1.780 | 19 % sales tax | $ 0.181 | ||
| Grand total | $ 12,084 | Grand total | $ 1.227 | ||
| U L−1 (168 h, 0.15 L) | 1,373.72 ± 0.37 | U L−1 (168 h, 0.15 L) | 12,877.31 ± 481.23 | ||
The cost of water for dissolving was obtained from the value of m3 of water established by the Waterworks Company for the industrial zone in Bogotá D.C., Colombia, July of 2018 [43].
4. Discussion
P. pastoris is a widely used host for expression of recombinant proteins. Culture media optimization design has been very successful with other organisms; however, it has not been widely explored for P. pastoris. In general, chemically defined media are the most used for recombinant protein production in P. pastoris, subjecting the microorganism to synthesize required metabolic intermediates, usually implying a slower growth decreasing recombinant protein expression due to metabolic burden [31, 44]. Hence, chemically defined media increase costs, and not always increasing primary and secondary metabolite production [31].
4.1. Low cost nitrogen source selection
It was observed enzyme activity was much lower when, isolated soy protein (90 %) was used as the only source of organic nitrogen source in culture media (T2, statistical group b), in comparison with T1 and T3 treatments, which were similar (statistical group a) (Figure 1). However, none of the proposed treatments exceeded control culture media enzyme activity.
Treatments T1 and T3, with higher enzyme activities, had in common malt extract as the organic source of nitrogen with 2.71 % w/w protein and 77.34 % w/w reducing sugars such as maltose, (Supplementary Material 1). Malt extract also contains vitamins and growth promoters [45], which were important for rPOXA 1B production (Figure 1). Even though, there were no differences in laccase activity between T1 and T3 (statistical group b), nitrogen source selection was performed taking into account the concentration of each of the evaluated nitrogen sources, since nitrogen metabolism is critical for the catabolism and anabolism of proteins, amino acids and other nitrogenous compounds [46]. T3 contained milk serum as the source of nitrogen, although rich in vitamins and minerals, only 11 % of its composition are proteins (Supplementary Material 2) in comparison with 90 % proteins in isolated soy protein in T1 media (Supplementary Material 3).
Proteins of vegetable origin, such as isolated soy protein (90 %) contain a high percentage of essential amino acids, such as glutamine [47, 48]. Glutamine is a source of carbon and nitrogen that partially increases the metabolic load caused by recombinant protein production [44], it increases growth rate, and is a precursor of other amino acids [31, 46]. In addition, it raises Krebs cycle (TCA) flux, increasing recombinant enzyme yield and productivity between 20 - 26 % and 15–27 %, respectively [44].
4.2. Response surface methodology (RSM) for low cost culture media design
4.2.1. Central composite Design-1 (CCD-1)
Starting from the composition of T1, optimization of the concentrations of selected organic nitrogen sources, inorganic nitrogen and glucose began. CCD-1 results (Figure 2), demonstrated treatment T22 at 72 h of culture achieved an enzyme activity and specific activity higher than that observed for T1 and T3 (Figure 1), yet similar to control results in low cost nitrogen source selection. Nonetheless, T22 enzyme activities and specific activities decreased at 168 h of culture (Figure 2), yet they were higher than those observed in T1 and T3 results (Figures 1 and 2).
Statistical analysis for the four response variables (Table 5), revealed the quadratic model was significant (p < 0.05) for enzyme activity and specific enzyme activity at 72 h of culture, whereas at 168 h of culture the linear model was significant instead (p < 0.05). Nonetheless, lack of fit for response variable: enzyme activity at 72 h was significant at 91 %, therefore this response variable was discarded. Although lack of fit for enzyme activity at 168 h and specific activities at 72 and 168 h were not significant (p > 0.05), specific activity at 72 h response variable was selected for having the highest R2 (0.7740) among all response variables, whose lack of fit was significant at least less than 63 % (Table 4).
Table 5 shows that an F-value of 2.94 implies the model was significant, and there was only a 3.45 % chance that an F-value this large could occur due to noise. The "Lack of Fit F-value" of 1.18 implied the Lack of Fit was not significant relative to pure error and there was a 54.18 % chance that a "Lack of Fit F-value" this large could occur due to noise, representing a good signal for the model. On the other hand, "Adeq Precision" (7.2192) measures the signal to noise ratio with a desirable ratio greater than 4, indicating that this quadratic model applied at specific activity at 72 h of culture, can be used to navigate the design space.
Table 6 also shows that isolated soy protein (factor B) and B2 had a significance between 96 and 99%, whereas factor A (glucose) and factor D ((NH4)2SO4) were significant between 89 and 90%. However, AB interaction was significant between 93 and 95%, while AD, BD and D2 were not significant (p > 0.05). Hence, it was decided to maintain unchanged the values recommended by the software for the factors: C-Malt extract (11.74 g L−1) and D-(NH4)2SO4 (4.91 g L−1) (Figure 3), and study in a new design the interaction between factors A and B.
Statistical analyses indicated isolated soy protein was a significant factor (p = 0.0346); thus, an increase in its concentration was proposed. Our results agree with those of other investigations, where use of organic sources increased protein production in comparison with inorganic sources of nitrogen [49, 50, 51]. Nevertheless, the combination of organic and inorganic nitrogen is a strategy to increase laccase activity [52], whose effectiveness could be due to nitrogen assimilation. Inorganic nitrogen is easily absorbed, while organic nitrogen supplies cells with growth factors and amino acids for cell metabolism and protein synthesis [50].
Even though ammonium sulphate did not have an important effect on enzyme activity, it has been reported cell density increases with media supplementation with ammonium ion (NH4+), as long as it does not exceed 600 mmol L−1 [53], since excess inhibits cell growth [31].
Additionally, other authors have reported protein expression under the control of the pGAP promoter varies according to carbon source [4, 45]. It has been reported glycerol as a source of carbon results in 30% less production in comparison with glucose [4, 13, 44, 54]. In contrast, other authors have reported higher levels of expression when glycerol is used as a source of carbon [16, 55], evidencing that intrinsic characteristics of the protein to be expressed also play a key role.
In this part of the study for rPOXA 1B specific activity glucose was a significant factor (p = 0.1078) between 89 - 90 % (Table 5), probably as a result of energy demand during microbial growth to synthesize rPOXA 1B, although the cell readjusts its metabolic flow distribution to increase ATP yield, this adjustment is limited [44].
Moreover, proteins whose expression positively correlate with the glycolytic flux tend to express in the same manner the enzymes that participate in the pathway [35]; which arises when proteins are expressed under the control of the pGAP promoter in the presence of carbon sources that stimulate glycolysis.
For biomass production P. pastoris detects the presence of nutrients through multiple signaling pathways [45]. One of the most important pathways in response to the presence of carbohydrates is the: Ras/PKA (Protein Kinase A, E.C. 2.7.11.11) signaling pathway. Supplementing with glucose activates Ras (a regulator of cell proliferation) that in turn activate synthesis of cAMP and PKA. Protein kinase A acts as a central regulator of metabolism of the cell's transcriptional state, carbohydrate storage, factors regulators of stress and ribosomal biogenesis [56, 57].
In comparison with S. cerevisiae, P. pastoris glucose absorption is limited due to the decreased number of hexose transporter genes [58]. Mattanovich et al., (2009) identified 14 supposed hexose transporters in P. pastoris. They found two had low affinity to transport hexose and they were homologous to some S. cerevisiae transporters, whereas two other transporters in P. pastoris with high affinity for hexose were homologous with some in Kluyveromyces lactis [59]. In contrast, in S. cerevisiae 20 HXT genes coding for hexose transporters of high and low affinity have been reported [60], which suggests higher rates of glucose absorption [45] in comparison with P. pastoris.
Regarding P. pastoris carbon metabolism it has been reported for recombinant strains Krebs cycle flow (TCA) increases 29 % [61], and part of the administrated glucose is metabolized through the phosphate pentose pathway (PPP), conclusion reached due to flow relationship analyzes that determine the origin of a specific metabolite [61, 62, 63]. Although it is not possible to determine with precision the percentage of oxidized glucose through the PPP, it is known an increase in the metabolic flux through the PPP has an influence on the biomass yield and heterologous protein production [61, 64]. Consequently, one could say that controlled consumption of glucose is a cellular strategy to conserve carbon and electron flow within the cell, which allows yeast to maintain a high rate of division [34]. Our results are in agreement with those reported by Vellanki et al., (2009), who demonstrated for recombinant protein expression in P. pastoris, glucose percentage should be higher than 1 % (w/v), [65].
It was observed enzyme activity (UL−1) and specific activity (Umg−1) and the desirability of the model were favoured when isolated soy protein concentration was increased above glucose's concentration (Figure 3), maintaining malt extract at 11.74 gL-1 and (NH4)2SO4 at 4.91 gL-1; which was determined after 100 optimization cycles navigating through the design surface.
According to the aforementioned, and due to the fact that glucose and isolated soy protein were the only significant factors in the design, it was decided to continue culture media optimization taking into account the model's predicted concentrations. Moreover, glucose USP (9.53 gL-1) and isolated soy protein (19.25 gL-1) were included within the amplified navigation surface in CCD-2, while malt extract and (NH4)2SO4 remained unchanged (Table 3).
4.2.2. Central composite Design-2 (CCD-2)
CCD-2 T9 treatment surpassed CCD-1 T22 in laccase activity, as well as specific activity at 72 h and 168 h of culture (T9, 6,608.80 ± 62.20 U L−1, 775.09 ± 8.75 U mg−1 at 72 h), (T9, 8,944.44 ± 330.29 U L−1, 965.12 ± 43.08 U mg−1 at 168 h), Figure 4. In Figure 4, it was observed too a change in slope was observed for enzyme activity and specific activity at 72 h. In addition, none of the response variables decreased, as was observed for CCD-1, suggesting (C/N)0 ratio did not influence the result, since relatively higher nitrogen concentrations and lower (C/N)0 ratios provide primary and secondary metabolite production [34, 66].
In this CCD-2 design the quadratic model (Table 7) was significant for laccase activity (p < 0.0001) and specific activity (p < 0.0001), however, R2 for specific activity was higher (R2 = 0.9504) and lack of fit was not significant (p = 0.1677). Therefore, it was decided to continue statistical analysis with this response variable (Supplementary Material 5). CCD-2 quadratic model's F-value of 80.49 implied the model was significant and there was only a 0.01% chance that an F-value this large could be due to noise. In this instace, A, B, AB and B2 were significant model terms. On the other hand, a lack of fit F-value of 1.89 implied it was not significant relative to pure error and that there was a 16.77 % chance that an F-value this large could occur due to noise. The Pred R-Squared of 0.9193 was in reasonable agreement with the Adj R-Squared of 0.9386 because differences between them were less than 0.2. An Adeq Precision ratio of 23.496 indicated this model could be used to navigate the design space (Table 7).
CCD-2 statistical results demonstrated AB interaction was significant (p < 0.0001), B2 was also significant (p = 0.0038), whereas A2 was not significant (p > 0.05). This connoted glucose concertation had to remain at 20 gL-1 and isolated soy protein concentration had to be increased to amplify the navigation surface. Thus, a One Factor Design was performed. This higher concentration study would theoretically allow increasing U L−1 and U mg−1 (Figure 4).
4.2.3. One Factor Experimental design (OFED)
The study showed there was only a 0.01 % chance an F-value this large could occur due to noise. In this instance, A2 and A3 were significant model terms. The lack of fit F-value of 2.92 implied that it was not significant relative to pure error, and there was a 20.14 % chance that an F-value this large could occur due to noise.
On the other hand, the Pred R-Squared of 0.9394 was in reasonable agreement with the Adj R-Squared of 0.9654, because difference between them was less than 0.2. Adeq Precision ratio of 21.645 indicated that this cubic model could be used to navigate the design space.
In Figure 6A it was observed OFED Treatment T4 surpassed CCD-2 T9 laccase activity, as well as specific activity reaching at 168 h an enzyme activity of 12,877.3 ± 481.2 U L−1 and 1,243.83 ± 24.1 Umg−1 specific activity. Additionally, no change in enzyme activity tendency was observed from 72 h onwards, as was observed for treatment T22 in CCD-1 and CCD-2 for T9, which could be indicating a (C/N)0 ratio of 5.4 ± 0.03 favoured rPOXA 1B production.
When comparing residual reducing sugars in CCD-1, CCD-2 and OFED (Figures 3 and 5 and 6A), it was observed between 12 and 24 h most of the sugars were consumed, with an approximate remnant of 5 gL-1 until the end of the culture. These results are different from those reported by Palma et al., (2012), who observed a reduction in glucose's absorption rate when the media had a nitrogen limitation in S. cerevisiae [67]. However, it is important to call to mind, P. pastoris possesses less hexose transporters and it can use glutamine present in isolated soy protein as a carbon source, which could influence P. pastoris glucose absorption. Additionally, it is important to consider glucose USP, malt extract (Supplementary Material 1) and isolated soy protein (0.126 g reducing sugars/10 g isolated soy protein, data not shown) have reducing compounds that could interfere with DNS assay used for residual reducing sugar evaluation [38].
As observed in Figure 6B, enzyme activity slightly decreased for isolated soy protein concentrations higher than 50 gL-1, suggesting under assay conditions media optimization had been reached. Henceforth, final media composition and optimal culture conditions were 20 gL-1 glucose USP (ChemiPharma), 50 gL-1 isolated soy protein (Ciacomeq SAS), 11.74 gL-1 malt extract (Proquímicas JG SAS), 4.91 gL-1 (NH4)2SO4 (Merck), 0.161 gL-1 CuSO4 (Merck), 0.1 gL-1 chloramphenicol (SIGMA).
4.2.4. Raw material cost analysis
One of the objectives of this study was to design a low-cost culture media. With the produced OFED media laccase activity increased by 30.54 % in comparison with results obtained from CCD-2 and 75.76 % concerning results obtained from 10 L bioreactor, where previously improved media was used [35]. Likewise, culture media cost was reduced (Table 9).
In conclusion, a low cost culture media with a (C/N)0 ratio of 5.4 ± 0.03 was optimized that attained an enzyme activity of 12,877.3 ± 481.2 UL−1 and 1,324.58 ± 114.19 Umg−1 specific activity at 168 h of culture, increasing produced laccase activity in CCD-1 and CCD-2 by 63.16 % and 30.54 %, respectively. Furthermore, results demonstrated that, isolated soy protein, malt extract and glucose USP are low cost substrates that increased P. pastoris rPOXA 1B activity at Erlenmeyer flask scale 28.55 fold in comparison with clone assay at Erlenmeyer flask scale [15]; 9.58 fold in comparison with previously improved media at Erlenmeyer flask scale [35], and 4.11 fold in comparison with previously improved media evaluated at 10 L bioreactor scale (6L WEV), [35]. Most important, considerable cost reduction was achieved.
Declarations
Author contribution statement
Leidy D. Ardila-Leal: Performed the experiments; Analyzed and interpreted the data; Wrote the paper.
María F. Alvarado-Ramírez: Performed the experiments; Analyzed and interpreted the data.
Ivonne S. Gutiérrez-Rojas, Balkys Quevedo-Hidalgo, Alejandro Pérez-Flórez, Aura M. Pedroza-Rodríguez: Conceived and designed the experiments; Analyzed and interpreted the data.
Raúl A. Poutou-Piñales: Conceived and designed the experiments; Analyzed and interpreted the data; Wrote the paper.
Funding statement
This work was supported by Grant ID: 00006337 (Optimización del medio de cultivo para producción de la lacasa recombinante POXA 1B de Pleurotus ostreatus en Pichia pastoris) from Pontificia Universidad Javeriana, Bogotá, D.C. Colombia and Grant ID: 00007885 (Estudio de la estabilidad a tiempo real del concentrado de la lacasa rPOXA 1B de Pleurotus ostreatus producida en Pichia pastoris) from Pontificia Universidad Javeriana, Bogotá, D.C. Colombia.
Competing interest statement
The authors declare no conflict of interest.
Additional information
No additional information is available for this paper.
Acknowledgements
Authors thank to María Lucía Gutiérrez, Ph.D. for English editing.
Appendix A. Supplementary data
The following is the supplementary data related to this article:
References
- 1.Strong P.J., Claus H. Laccase: a review of its past and its future in bioremediation. Crit. Rev. Environ. Sci. Technol. 2011;41:373–434. [Google Scholar]
- 2.Cannatelli M.D., Ragauskas A.J. Two decades of laccases. Adv. Sustain. Chem. Indu. 2017;17:122–140. doi: 10.1002/tcr.201600033. [DOI] [PubMed] [Google Scholar]
- 3.Rivera-Hoyos C.M. Fungal laccases. Fung. Biol. Rev. 2013;27:67–82. [Google Scholar]
- 4.Wang F. Influence of carbon source on the production of extracellular ligninolytic enzymes by Phanerochaete chrysosporium. BioRes. 2016;11:5676–5686. [Google Scholar]
- 5.Colao M.C., Lupino S., Garzillo A.M., Buonocore V., Ruzzi M. Heterologous expression of lcc1 gene from Trametes trogii in Pichia pastoris and characterization of the recombinant enzyme. Microb. Cell Factories. 2006;5:1–11. doi: 10.1186/1475-2859-5-31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gómez-Méndez L.D. Biodeterioration of plasma pretreated LDPE sheets by Pleurotus ostreatus. PloS One. 2018;13 doi: 10.1371/journal.pone.0203786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Morales-Álvarez E.D. Partial removal and detoxification of Malachite Green and Crystal Violet from laboratory artificially contaminated water by Pleurotus ostreatus. Univ. Sci. 2016;21:259–285. [Google Scholar]
- 8.Morales-Álvarez E.D. Malachite Green and Crystal Violet decolorization by Ganoderma lucidum and Pleurotus ostreatus supernatant and by rGlLCC1 and rPOXA 1B concentrates: molecular docking analysis. Appl. Biochem. Biotechnol. 2018;184:794–805. doi: 10.1007/s12010-017-2560-y. [DOI] [PubMed] [Google Scholar]
- 9.Moreno-Bayona D.A. Simultaneous bioconversion of lignocellulosic residues and oxodegradable polyethylene by Pleurotus ostreatus for biochar production, enriched with phosphate solubilizing bacteria for agricultural use. PloS One. 2019;14 doi: 10.1371/journal.pone.0217100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Piscitelli A., Pezzella C., Giardina P., Faraco V., Sannia G. Heterologous laccase production and its role in industrial applications. Bioeng. Bug. 2010;1:252–262. doi: 10.4161/bbug.1.4.11438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rivera-Hoyos C.M. Detoxification of pulping black liquor with Pleurotus ostreatus or recombinant Pichia pastoris followed by CuO/TiO2/visible photocatalysis. Sci. Rep. 2018;8:3503. doi: 10.1038/s41598-018-21597-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tonin F. Comparison of different microbial laccases as tools for industrial uses. N. Biotech. 2016;33:387–398. doi: 10.1016/j.nbt.2016.01.007. [DOI] [PubMed] [Google Scholar]
- 13.Nicolini C., Bragazzi N.L., Pechkova E., Bruzzese D., Cambria M.T. Recombinant laccase: I. Enzyme cloning and characterization. J. Cell. Biochem. 2013;114:599–605. doi: 10.1002/jcb.24397. [DOI] [PubMed] [Google Scholar]
- 14.Giardina P. Protein and gene structure of a blue laccase from Pleurotus ostreatus. Biochem. J. 1999;341:655–663. [PMC free article] [PubMed] [Google Scholar]
- 15.Rivera-Hoyos C.M. Computational analysis and low-scale constitutive expression of laccases synthetic genes GlLCC1 from Ganoderma lucidum and POXA 1B from Pleurotus ostreatus in Pichia pastoris. PloS One. 2015;10 doi: 10.1371/journal.pone.0116524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Flores C., Vidal C., Trejo-Hernández M., Galindo E., Serrano-Carreon L. Selection of Trichoderma strains capable of increasing laccase production by Pleurotus ostreatus and Agaricus bisporus in dual cultures. J. Appl. Microbiol. 2009;106:249–257. doi: 10.1111/j.1365-2672.2008.03998.x. [DOI] [PubMed] [Google Scholar]
- 17.Saloheimo M., Niku-Paavola M.-L. Heterologous production of a ligninolytic enzyme: expression of the phlebia Radiata laccase gene in Trichoderma reesei. Nat. Biotechnol. 1991;9:987–990. [Google Scholar]
- 18.Guo M., Lu F., Du L., Pu J., Bai D. Optimization of the expression of a laccase gene from Trametes versicolor in Pichia methanolica. Appl. Microbiol. Biotechnol. 2006;71:848–852. doi: 10.1007/s00253-005-0210-8. [DOI] [PubMed] [Google Scholar]
- 19.Theerachat M. Engineering and production of laccase from Trametes versicolor in the yeast Yarrowia lipolytica. Biores. Technol. 2012;125:267–274. doi: 10.1016/j.biortech.2012.07.117. [DOI] [PubMed] [Google Scholar]
- 20.Bleve G. Molecular cloning and heterologous expression of a laccase gene from Pleurotus eryngii in free and immobilized Saccharomyces cerevisiae cells. Appl. Microbiol. Biotechnol. 2008;79:731–741. doi: 10.1007/s00253-008-1479-1. [DOI] [PubMed] [Google Scholar]
- 21.Trono D. Recombinant enzymes in the food and pharmaceutical industries. In: Singh R.S., editor. Advances in Enzyme Technology. Elsevier; 2019. pp. 349–387. [Google Scholar]
- 22.Gidijala L., Uthoff S., van Kampen S.J., Steinbüchel A., Verhaert R.M.D. Presence of protein production enhancers results in significantly higher methanol-induced protein production in Pichia pastoris. Microb. Cell Factories. 2018;17:112. doi: 10.1186/s12934-018-0961-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Juturu V., Wu J.C. Heterologous protein expression in Pichia pastoris: latest research progress and applications. Chembiochem. 2018;19:7–21. doi: 10.1002/cbic.201700460. [DOI] [PubMed] [Google Scholar]
- 24.Cereghino G.P., Cereghino J., Ilgen C., Cregg J. Production of recombinant proteins in fermenter cultures of the yeast Pichia pastoris. Curr. Opin. Biotechnol. 2002;13:329–332. doi: 10.1016/s0958-1669(02)00330-0. [DOI] [PubMed] [Google Scholar]
- 25.Zhang A.-L. Recent advances on the GAP promoter derived expression system of Pichia pastoris. Mol. Biol. Rep. 2009;36:1611–1619. doi: 10.1007/s11033-008-9359-4. [DOI] [PubMed] [Google Scholar]
- 26.Batra J., Beri D., Mishra S. Response Surface Methodology based optimization of β-glucosidase production from Pichia pastoris. Appl. Biochem. Biotechnol. 2014;172:380–393. doi: 10.1007/s12010-013-0519-1. [DOI] [PubMed] [Google Scholar]
- 27.Mao R. Optimization of expression conditions for a novel NZ2114-derived antimicrobial peptide-MP1102 under the control of the GAP promoter in Pichia pastoris X-33. BMC Microbiol. 2015;15:57. doi: 10.1186/s12866-015-0389-5. 57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hahn-Hägerdal B. Role of cultivation media in the development of yeast strains for large scale industrial use. Microb. Cell Factories. 2005;4:31. doi: 10.1186/1475-2859-4-31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nocon J. Model based engineering of Pichia pastoris central metabolism enhances recombinant protein production. Metab. Eng. 2014;24:129–138. doi: 10.1016/j.ymben.2014.05.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gómez G., Batista C. Optimización de medios de cultivos para microorganismos, una valiosa estrategia para la producción de biopreparados de interés agrícola. Cultiv. Trop. 2006;27:17–24. [Google Scholar]
- 31.Matthews C.B., Kuo A., Love K.R., Love J.C. Development of a general defined medium for Pichia pastoris. Biotechnol. Bioeng. 2018;115:103–113. doi: 10.1002/bit.26440. [DOI] [PubMed] [Google Scholar]
- 32.Osma J.F., Toca-Herrera J.L., Rodríguez-Couto S. Cost analysis in laccase production. J. Environ. Manag. 2011;92:2907–2912. doi: 10.1016/j.jenvman.2011.06.052. [DOI] [PubMed] [Google Scholar]
- 33.Ravindran R., Jaiswal A.K. Microbial enzyme production using lignocellulosic food industry wastes as feedstock: a review. Bioengineering. 2016;3:1–22. doi: 10.3390/bioengineering3040030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zheng J. Development of an industrial medium and a novel fed-batch strategy for high-level expression of recombinant b-mananase by Pichia pastoris. Biores. Technol. 2012;118:257–264. doi: 10.1016/j.biortech.2012.05.065. [DOI] [PubMed] [Google Scholar]
- 35.Ardila-Leal L.D. Media improvement for 10 L bioreactor production of rPOXA 1B laccase by P. pastoris. 3 Biotech. 2019;9 doi: 10.1007/s13205-019-1979-y. Article 447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Poutou R.A., Amador E., Candelario M. Banco de células primario (BCP): caracterización y papel en la producción de proteínas recombinantes. Biotecnol. Apl. 1994;11:55–59. [Google Scholar]
- 37.Pezzella C. A step forward in laccase exploitation: recombinant production and evaluation of techno-economic feasibility of the process. J. Biotechnol. 2017;259:175–181. doi: 10.1016/j.jbiotec.2017.07.022. [DOI] [PubMed] [Google Scholar]
- 38.Miller G. Use of dinitrosalicilic acid reagent for determination of reducing sugar. Anal. Chem. 1959;31:426–428. [Google Scholar]
- 39.Amaral P.F.F., Ribeiro B.D. Assays of phenoloxidase activity. In: Vermelho A.B., Couri S., editors. Methods to Determine Enzymatic Activity. Bentham eBooks; 2013. pp. 195–207. [Google Scholar]
- 40.Desjardins P., Hansen J.B., Allen M. Microvolume protein concentration determination using the NanoDrop 2000c spectrophotometer. JoVE. 2009;33:3p. doi: 10.3791/1610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rice E.W. 23rd ed. Water Environment Federation; 2017. Standard Methods for the Examination of Water and Wastewater 2540 A; p. 277. [Google Scholar]
- 42.Tributario Nacional Estatuto. 2017. Art. 468. Tarifa general de inpuesto sobre las ventanas.http://estatuto.co/?e=645&w=libro-tercero - top [Google Scholar]
- 43.Empresa de Acueducto Alcantarillado y Aseo de Bogotá, Tarifas acueducto. https://http://www.acueducto.com.co/wps/html/resources/2018/tarifas2018/Tarifas_BOGOTA_2018_Revisadas-1.xls. 2018.
- 44.Heyland J., Fu J., Blank L., Schmid A. Carbon metabolism limits recombinant protein production in Pichia pastoris. Biotechnol. Bioeng. 2011;108:1942–1953. doi: 10.1002/bit.23114. [DOI] [PubMed] [Google Scholar]
- 45.Calık P. Recombinant protein production in Pichia pastoris under glyceraldehyde-3-phosphate dehydrogenase promoter: from carbon source metabolism to bioreactor operation parameters. Biochem. Eng. J. 2015;95:20–36. [Google Scholar]
- 46.Zhang W., Du G., Zhou J., Chen J. Regulation of sensing, transportation, and catabolism of nitrogen sources in Saccharomyces cerevisiae. Microbiol. Mol. Biol. Rev. 2018;82 doi: 10.1128/MMBR.00040-17. e00040-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Calderón De la Barca A.M., Ruiz-Salazar R.A., Jara-Marini M.E. Enzymatic hydrolysis and synthesis of soy protein to improve its amino acid composition and functional properties. Food Chem. Toxicol. 2000;65:246–253. [Google Scholar]
- 48.Lenders C.M. Evaluation of a novel food composition database that includes glutamine and other amino acids derived from gene sequencing data. Eur. J. Clin. Nutr. 2009;63:1433–1439. doi: 10.1038/ejcn.2009.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Choi D.B., Park E.Y. Enhanced production of mouse a-amylase by feeding combined nitrogen and carbon sources in fed-batch culture of recombinant Pichia pastoris. Process Biochem. 2006;41:390–397. [Google Scholar]
- 50.de Almeida A.F., aulk-Tornisielo S.M.T., Cano Carmona E. Influence of carbon and nitrogen sources on lipase production by a newly isolated Candida viswanathii strain. Ann. Microbiol. 2013;63:1225–1234. [Google Scholar]
- 51.Pritchett J., Baldwin S.A. The effect of nitrogen source on yield and glycosylation of a human cystatin C mutant expressed in Pichia pastoris. J. Ind. Microbiol. Biotechnol. 2004;31:553–558. doi: 10.1007/s10295-004-0181-2. [DOI] [PubMed] [Google Scholar]
- 52.Patel H., Gupte A., Guote S. Effect of different culture conditions and inducers on production of laccase by a Basidiomycete fungal isolate Pleurotus ostreatus HP-1 under solid state fermentation. BioRes. 2009;4:268–284. [Google Scholar]
- 53.Yu X.-W., Lu X., Zhao L.-S., Xu Y. Impact of NH4+ nitrogen source on the production of Rhizopus oryzae lipase in Pichia pastoris. Process Biochem. 2013;48:1462–1468. [Google Scholar]
- 54.Waterham H.R., Digan M.H., Koutz P.J., Lair S.V., Cregg J.M. Isolation of the Pichia pastoris glyceroldehyde-3-phosphate dehydrogenase gene and regulation and use of its promoter. Gene. 1997;186:37–44. doi: 10.1016/s0378-1119(96)00675-0. [DOI] [PubMed] [Google Scholar]
- 55.Várnai A. Expression of endoglucanases in Pichia pastoris under control of the GAP promoter. Microb. Cell Factories. 2014;13:57. doi: 10.1186/1475-2859-13-57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Peeters Ken. Fructose-1,6-bisphosphate couples glycolytic flux to activation of Ras. Nat. Commun. 2017;8:922. doi: 10.1038/s41467-017-01019-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Zaman S., Lippman S.I., Zhao X., Broach J.R. How Saccharomyces responds to nutrients. Annu. Rev. Genet. 2008;42:27–81. doi: 10.1146/annurev.genet.41.110306.130206. [DOI] [PubMed] [Google Scholar]
- 58.Peña D.A., Gasser B., Zanghellini J., Steiger M.G., Mattanovich D. Metabolic engineering of Pichia pastoris. Metab. Eng. 2018;50:2–15. doi: 10.1016/j.ymben.2018.04.017. [DOI] [PubMed] [Google Scholar]
- 59.Mattanovich D. Open access to sequence: browsing the Pichia pastoris genome. Microb. Cell Factories. 2009;8 doi: 10.1186/1475-2859-8-53. Art 53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Diderich J.A. Glucose uptake kinetics and transcription of HXT genes in chemostat cultures of Saccharomyces cerevisiae. J. Biol. Chem. 1999;274:15350–15359. doi: 10.1074/jbc.274.22.15350. [DOI] [PubMed] [Google Scholar]
- 61.Nocon J. Increasing pentose phosphate pathway flux enhances recombinant protein production in Pichia pastoris. Appl. Microbiol. Biotechnol. 2016;100:5955–5963. doi: 10.1007/s00253-016-7363-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Baumann K. A multi-level study of recombinant Pichia pastoris in different oxygen conditions. BMC Syst. Biol. 2010;4:131. doi: 10.1186/1752-0509-4-141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Zhang Z.-y. Pichia pastoris X33 GT2 release the glycerol repression on AOX1 and efficiently express heterologous proteins. China Biotechnol. 2017;37:38–45. [Google Scholar]
- 64.Nie Y. Impacts of high -galactosidase expression on central metabolism ofrecombinant Pichia pastoris GS115 using glucose as sole carbonsource via13C metabolic flux analysis. J. Biotechnol. 2014;187:124–134. doi: 10.1016/j.jbiotec.2014.07.011. [DOI] [PubMed] [Google Scholar]
- 65.Vellanki R.N., Potumarthi R., Mangamoori L.N. Constitutive expression and optimization of nutrients for streptokinase production by Pichia pastoris using statistical methods. Appl. Biochem. Biotechnol. 2009;158:25–40. doi: 10.1007/s12010-008-8315-z. [DOI] [PubMed] [Google Scholar]
- 66.Lopes V.R.O., Farias M.A., Belo I.M.P., Coelho M.A.Z. Nitrogen sources on TPOMW valorization through solid state fermentation performed by Yarrowia lipolytica. Braz. J. Chem. Eng. 2016;33:261–270. [Google Scholar]
- 67.Palma M., Madeira S.C., Mendes-Ferreira A., Sá-Correia I. Impact of assimilable nitrogen availability in glucose uptake kinetics in Saccharomyces cerevisiae during alcoholic fermentation. Microb. Cell Factories. 2012;11:99. doi: 10.1186/1475-2859-11-99. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.