Abstract
Background
As part of the Household Influenza Vaccine Evaluation (HIVE) study, acute respiratory infections (ARI) have been identified in children and adults from 2010 to 2018.
Methods
Annually, 890 to 1441 individuals were followed and contacted weekly to report ARIs. Specimens collected during illness were tested for human coronaviruses (HCoV) types OC43, 229E, HKU1, and NL63.
Results
In total, 993 HCoV infections were identified during the 8 years, with OC43 most commonly seen and 229E the least. HCoVs were detected in a limited time period, between December and April/May and peaked in January/February. Highest infection frequency was in children <5 years (18 per 100 person-years), with little variation in older age groups (range, 7 to 11 per 100 person-years). Overall, 9% of adult cases and 20% of cases in children were associated with medical consultation. Of the 993 infections, 260 were acquired from an infected household contact. The serial interval between index and household-acquired cases ranged from 3.2 to 3.6 days and the secondary infection risk ranged from 7.2% to 12.6% by type.
Conclusions
Coronaviruses are sharply seasonal. They appear, based on serial interval and secondary infection risk, to have similar transmission potential to influenza A(H3N2) in the same population.
Keywords: coronaviruses, seasonality, epidemiology, transmission, respiratory illness
In the HIVE study, acute respiratory infections were reported in children and adults from 2010 to 2018 and HCoVs identified. HCoV infection was sharply seasonal, occurring December–May. Transmission potential of HCoV was similar to influenza A(H3N2) in this population.
(See the Major Article by Nickbakhsh et al., on pages 17–25.)
Coronaviruses, long known to infect a wide variety of species, have been recognized as human respiratory pathogens for more than 60 years [1–3]. While animal coronaviruses have been associated with severe disease in their respective hosts, human coronaviruses (HCoV) were historically detected in mild respiratory illnesses [4–6]. Therefore, the identification of severe acute respiratory syndrome (SARS) as caused by a coronavirus of animal origin in 2002 was a surprise because of its epidemic behavior as well as its severity [7, 8]. Also a surprise was the ultimate interruption of SARS transmission by public health measures [9]. In 2012, another novel coronavirus of animal origin emerged as the causative agent of Middle East respiratory syndrome (MERS) [10]. Unlike SARS, human-to-human transmission of MERS has been limited and sporadic cases have continued to occur, primarily in Saudi Arabia among those with contact with camels [11]. The current outbreak of a novel coronavirus (SARS-CoV-2) has focused attention back to this often forgotten and little studied group of pathogens [12].
HCoVs were first recognized when specimens from individuals ill with respiratory illnesses were inoculated into organ cultures. One of these viruses is now known as OC (organ culture) 43 [13]. Another virus, 229E was first grown in primary human kidney culture [14]. Because of the difficulties in isolating them, much of the initial work in understanding their occurrence in was done by serology [15, 16]. Following renewed interest after the SARS epidemic, 2 additional coronaviruses, HKU1 and NL63, were identified in the 2000s, initially from persons with severe respiratory illnesses [17–20].
Availability of reverse-transcriptase polymerase chain reaction (RT-PCR) has now made it possible to detect infection easily for each of the 4 known human coronaviruses. However, as data from population-based studies using molecular methods have accumulated, minimal attention has been paid to these viruses, perhaps because there was so little known about their role in respiratory illnesses. The Household Influenza Vaccine Evaluation (HIVE) study is a longitudinal investigation of respiratory illnesses in households with children in the Ann Arbor, Michigan area that has been ongoing since 2010. Here we report on the occurrence of the 4 HCoV types over an 8-year period.
METHODS
Study Population
The complete methods of the HIVE cohort have been published previously [21]. Households were recruited from local households receiving primary care from the University of Michigan Health Care System. From 2010–2011 through 2015–2016, eligible households were those with 4 or more members at least 2 of whom were younger than 18 years. In subsequent years, eligible households contained 3 or more members including at least 1 child younger than 5 years (2016–2017) or younger than 10 years (2017–2018). Each year in the spring or summer, households attended an enrollment or reengagement visit to collect or update the demographic characteristics for each participant and household. Adult household members provided informed consent for themselves and their children, and children 7 years or older also provided verbal assent prior to participating. This study was reviewed and approved by the institutional review board at the University of Michigan Medical School.
Acute Respiratory Illness Surveillance
HIVE participants were instructed to report the onset of 2 or more acute respiratory illness (ARI) symptoms in anyone in the household, including: cough, fever or feverishness, nasal congestion, chills, headache, body aches, and sore throat. Weekly email and phone contacts were used to confirm the presence or absence of ARI in the household. Beginning in 2014–2015, a separate set of age-specific symptom qualifiers were provided for participants younger than 3 years, including: cough, fever or feverishness, nasal congestion, trouble breathing, fussiness, decreased appetite, and fatigue. Ill participants with 2 or more of the above symptoms attended an illness visit within 7 days of illness onset where study staff interviewed them about their illness and collected nasal and throat swabs (nasal only in children <3 years) combined in a single vial of viral transport media. Study staff followed-up with ill participants approximately a week after their illness visit to collect additional information about the course of the illness, including participants’ reports of whether they contacted a health care provider regarding their illness.
Prior to the 2014–2015 season, HIVE ARI surveillance periods were seasonal based on the likely influenza season. For the first 4 study years, the annual surveillance periods were 1 October 2010 through 30 April 2011, 1 December 2011 through 10 May 2012, 1 October 2012 through 13 May 2013, and 1 October 2013 through 30 April 2014. Surveillance resumed on 1 October 2014 and has been operating year round since then. New surveillance years are designated beginning 1 July and lasting until 30 June of the subsequent year.
Laboratory Testing
Respiratory specimens collected at illness visits were tested for detection of respiratory viruses, including the 4 seasonal coronavirus types (229E, OC43, HKU1, and NL63). For specimens collected prior to the 2016–2017 study year, testing was performed by singleplex RT-PCR using primers and probes developed by the CDC Division of Viral Diseases, Gastroenteritis, and Respiratory Viruses [22]. Specimens collected in the 2016–2017 and 2017–2018 years were tested using the FTD Respiratory Pathogen 33 multiplex PCR kit (Fast Track Diagnostics).
Statistical Analysis
Seasonal case counts of HCoV 229E, HKU1, NL63, and OC43 were used to assess annual patterning of each type; counts were also summed by month across all seasons combined to assess broad seasonality by type. Illnesses with multiple, coincident HCoV detections were only counted once in analyses of overall HCoV infections, but each HCoV type involved in coincident detection was also included in type-specific analyses.
Type-specific coronavirus incidence and corresponding 95% confidence intervals (CI) were calculated by age group (<5, 6–11, 12–17, 18–49, and ≥50 years) and by study year. Incidence was calculated as the number of type-specific coronavirus infections divided by the total participant time included during a specified time period. Because the majority of the HCoV season was captured even in study years without year-round surveillance, all participants were treated as contributing a full year of person time for each study year in which they were enrolled.
For each study year, we defined household index cases as the first person in a household to develop ARI with HCoV detection. Secondary cases were defined as those who developed ARI with HCoV detection within 14 days following illness in a household contact with the same type. The serial interval was calculated as the average number of days from the illness onset in a household index case to the illness onset in secondary cases. Secondary infection risks were calculated as the proportion of household contacts exposed to an index case who developed secondary infection. For the purpose of these transmission analyses, only the first introduction of HCoV to a household was considered.
We used latent class analysis to identify and compare classes of illness severity of HCoV infection across age groups and the 4 HCoV types (Supplementary Materials). For the latent items, we identified 7 participant-reported potential markers of illness severity including illness symptoms (cough, fever, fatigue, wheezing, and dyspnea), missed work or school, and medically attended illness (phone consultation, outpatient care, and inpatient care). A priori, we anticipated 3 latent classes of severity to represent mild, moderate, and severe illness; model diagnostics also confirmed that a 3-class model was most parsimonious. Latent class groups were then assigned to each HCoV infection, not including cases with multiple coincident HCoV types detected. Age-adjusted multinomial logistic regression models were then used to compare the odds of being assigned to each latent class between the 4 HCoV types.
RESULTS
Characteristics of the Study Population
The number of individuals under study ranged from 895 to 1441, and the number of households ranged from 209 to 340 over the 8 years (Table 1). Because of the requirement that children be present in the household, the studied population tended to be young, with relative absence of older adults. The numbers in the cohort varied from year to year but the age structure remained similar in all years, due to combined retention of existing households and recruitment of new households with young children.
Table 1.
Characteristics of the Study Population and Counts of Human Coronavirus Infections by Study Year: Household Influenza Vaccine Evaluation (HIVE) Cohort 2010–2018
| Season | ||||||||
|---|---|---|---|---|---|---|---|---|
| Characteristic | 2010–2011 | 2011–2012 | 2012–2013 | 2013–2014 | 2014–2015 | 2015–2016 | 2016–2017 | 2017–2018 |
| Surveillance period | 1 Oct–30 Apr | 1 Dec–10 May | 1 Oct–13 May | 1 Oct–30 Apr | 1 Oct–30 Jun | 1 Jul–30 Jun | 1 Jul–30 Jun | 1 Jul–30 Jun |
| Households enrolled, No. | 328 | 213 | 321 | 232 | 340 | 227 | 209 | 293 |
| Participants enrolled, No. | 1441 | 943 | 1426 | 1049 | 1431 | 996 | 895 | 1189 |
| Age groups, y, No. (%) | ||||||||
| 0–5 | 250 (17.4) | 131 (13.9) | 225 (15.8) | 133 (12.7) | 205 (14.3) | 97 (9.7) | 117 (13.1) | 220 (18.5) |
| 6–11 | 382 (26.5) | 278 (29.5) | 401 (28.1) | 316 (30.1) | 445 (31.1) | 327 (32.8) | 257 (28.7) | 298 (25.1) |
| 12–17 | 208 (14.4) | 141 (15.0) | 208 (14.6) | 169 (16.1) | 212 (14.8) | 173 (17.4) | 150 (16.8) | 166 (14.0) |
| 18–49 | 546 (37.9) | 353 (37.4) | 536 (37.6) | 385 (36.7) | 512 (35.8) | 351 (35.2) | 332 (37.1) | 457 (38.4) |
| ≥50 | 55 (3.8) | 40 (4.2) | 56 (3.9) | 46 (4.4) | 57 (4.0) | 48 (4.8) | 39 (4.4) | 48 (4.0) |
| ARI reports, No. | 1028 | 417 | 1227 | 706 | 1362 | 934 | 847 | 1401 |
| HCoV-positive cases, No. (% of all ARI cases) | ||||||||
| All typesa | 152 (14.8) | 59 (14.1) | 200 (16.3) | 105 (14.9) | 113 (8.3) | 109 (11.7) | 112 (13.2) | 143 (10.2) |
| 229E | 7 (0.7) | 20 (4.8) | 8 (0.7) | 45 (6.4) | 8 (0.6) | 3 (0.3) | 46 (5.4) | 12 (0.9) |
| HKU1 | 28 (2.7) | 19 (4.6) | 17 (1.4) | 30 (4.2) | 14 (1) | 36 (3.9) | 0 (0) | 49 (3.5) |
| NL63 | 62 (6) | 14 (3.4) | 75 (6.1) | 21 (3) | 37 (2.7) | 40 (4.3) | 11 (1.3) | 63 (4.5) |
| OC43 | 56 (5.4) | 13 (3.1) | 113 (9.2) | 16 (2.3) | 71 (5.2) | 34 (3.6) | 58 (6.8) | 26 (1.9) |
Abbreviations: ARI, acute respiratory infections; HCoV, human coronavirus.
aIndividual types sum to more than all types because coinfections were counted in each type, but only included as single cases in counts of all types.
The number of ARIs reported also varied from year to year, mainly related to the number of individuals on report but also the differential occurrence of influenza and other respiratory viruses (Table 1). Overall, from 2010–2011 through 2017–2018, 7992 ARI cases were reported to HIVE; 7469 (93%) had an illness visit with specimen collection and were tested for respiratory virus detection. The proportion of ARIs associated with coronavirus identification varied between 8.3% during the major influenza year 2014–2015 to 16.3% in 2012–2013. Codetection of multiple coronavirus types ranged from 0.7% of all coronavirus ARIs in 2010–2011 to 14.2% in 2014–2015.
Variation in Identifying the 4 Coronaviruses by Seasons and Years
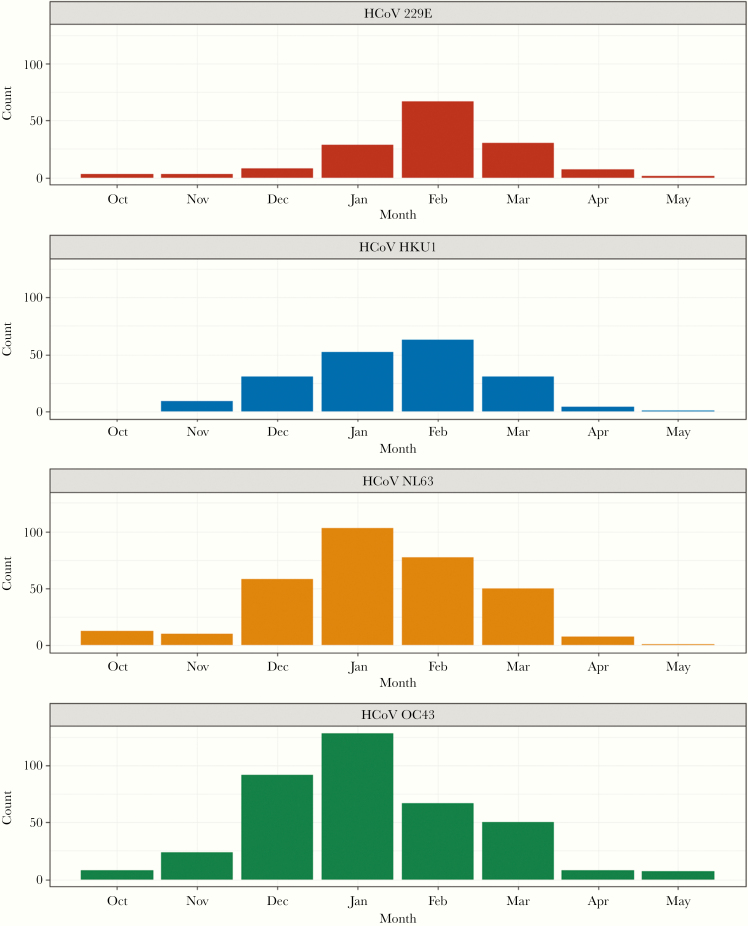
There was distinct seasonality in the identifications of the viruses. In the first 5 of the 8 years, no surveillance was conducted from June to September. When year-round surveillance was in place in the 2015–2016 through 2017–2018 study years, only 9 (2.5%) of the total 364 coronavirus-associated ARI occurred from June through September. Combined over the 8 years, the number of identifications for each virus increased in December, peaked in January or February, and began to decrease in March (Figure 1). The seasonal similarity between the 4 types is striking, with only the peak aggregate month differing between January and February.
Figure 1.
Counts of human coronavirus (HCoV) infections over 8 study years by type and month of illness onset: Household Influenza Vaccine Evaluation Study 2010–2018.
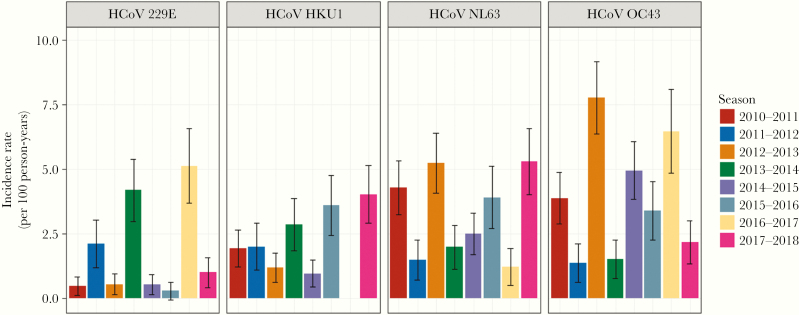
Identification of each of the 4 coronaviruses types occurred in each of the 8 study years, except for 2016–2017 when no HKU1 viruses were identified (Table 1 and Figure 2). In prior community studies [16, 23–25], there were suggestions of a cycling between 229E and OC43, with major years followed by lesser years. Such a pattern appears to be present for all types where relatively high incidence is not seen in consecutive years for a specific type. Likewise, combinations of cocirculating types were not found to be consistent from year to year.
Figure 2.
Incidence per 100 person-years of human coronavirus (HCoV) infections by type over 8 study years: Household Influenza Vaccine Evaluation Study 2010–2018. Error bars represent 95% confidence intervals.
Occurrence of Infections by Age
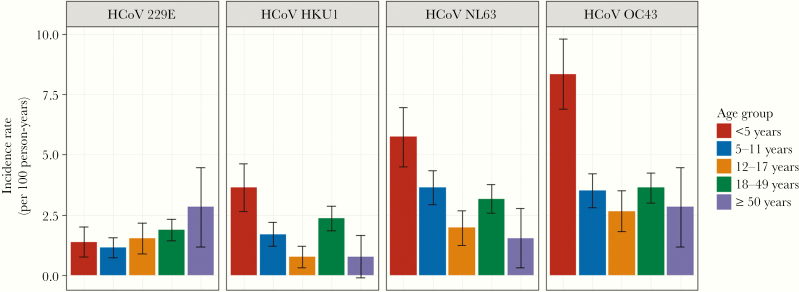
Infection rates for common respiratory pathogens are highest in young children and usually fall off with increasing age. With the exception of 229E, where incidence per 100 person-years was relatively low and did not vary by age, the incidence was consistently highest among children younger than 5 years (Figure 3). Among those older than 5 years, incidence was relatively flat with little consistent variation with increase in age.
Figure 3.
Incidence per 100 person-years of human coronavirus (HCoV) infections by type and age group over 8 study years: Household Influenza Vaccine Evaluation Study 2010–2018. Error bars represent 95% confidence intervals.
Illness Severity and Outcomes
Overall, 9% of adult cases and 20% of cases in children were associated with medical consultation. However, medical consultation only captures 1 facet of overall illness severity. Therefore, we used latent class analysis to categorize coronavirus case severity based on participant-reported symptoms and actions taken during the course of their illness (Supplementary Materials). Mild illnesses were primarily characterized by cough and fatigue, those with moderate illnesses were additionally likely to have fever and to miss school or work, and those with severe illness additionally were likely to have wheezing, dyspnea, or to seek care for their illness. Of all single coronavirus detections, 59% were categorized as mild illness by latent class methods. Moderate and severe illness were noted in 31% and 10% of coronavirus detections, respectively. HCoV illnesses in children younger than 5 years and adults older than 50 years were most likely to be classified as severe. In age-adjusted multinomial regression models (Table 2), 229E was associated with significantly higher odds of severe illness (versus mild) compared to the 3 other HCoV types (odds ratio [OR], 1.82; 95% CI, 1.01–3.27). On the other hand, NL63 was associated with significantly lower odds of severe illness (versus mild) compared to the other 3 types (OR, 0.59; 95% CI, .35–.99).
Table 2.
Odd Ratios and 95% Confidence Intervals From Multinomial Logistic Regression Models Predicting Illness Severity From Human Coronavirus (HCoV) Type and Age Group
| Severity Group, Odds Ratio (95% Confidence Interval)a | |||
|---|---|---|---|
| Mild | Moderate | Severe | |
| HCoV type | |||
| 229E | Ref | 1.51 (1.00–2.29) | 1.82 (1.01–3.27) |
| HKU1 | Ref | 1.13 (.77–1.66) | 1.51 (.88–2.59) |
| NL63 | Ref | 0.88 (.65–1.20) | 0.59 (.35–.99) |
| OC43 | Ref | 0.84 (.62–1.13) | 0.87 (.55–1.37) |
| Age group, y | |||
| 0–5 | Ref | 1.46 (1.04–2.04) | 2.06 (1.27–3.33) |
| 6–11 | Ref | 1.53 (1.11–2.11) | 1.02 (.60–1.72) |
| 12–17 | Ref | 0.99 (.62–1.61) | 0.71 (.31–1.62) |
| 18–49 | Ref | 0.55 (.40–.74) | 0.59 (.37–.94) |
| ≥50 | Ref | 0.63 (.24–1.61) | 1.23 (.40–3.80) |
aOdds ratios are relative to all other groups within category (ie, 229E vs not, 0–5 y vs not).
Household Transmission
Households present an ideal setting for determining the transmission characteristics of respiratory viruses. Of the 993 HCoVs identified, 260 occurred within 14 days of exposure to a household contact infected with the same type. The secondary attack risk was 7.2% for 229E, 8.6% for HKU1, 10.6% for OC43, and 12.6% for NL63. The average serial interval between the onset of illness in the household index case and onset of illness in secondary cases was similar across HCoV types (3.2 days for NL63 and HKU1, 3.3 days for OC43, and 3.6 days for 229E). Consistent with overall patterns of incidence by age (Figure 3), index cases were most frequently children, with the exception of 229E where adults aged 18 to 49 years were responsible for nearly half of the household introductions (Table 3).
Table 3.
Household Index Casesa by Human Coronavirus (HCoV) Type and Age Group
| HCoV Type, No. (Column %) | ||||
|---|---|---|---|---|
| 229E | HKU1 | NL63 | OC43 | |
| Age group, y | ||||
| 0–5 | 13 (11.5) | 38 (26.8) | 45 (20.7) | 82 (31.2) |
| 6–11 | 21 (18.6) | 35 (25.4) | 78 (36.9) | 65 (24.7) |
| 12–17 | 16 (14.2) | 9 (6.3) | 24 (11.1) | 24 (9.1) |
| 18–49 | 54 (47.8) | 56 (39.4) | 64 (29.5) | 83 (31.6) |
| ≥50 | 9 (8.0) | 3 (2.1) | 6 (2.8) | 9 (3.4) |
aOnly index cases from the first introduction of each coronavirus type to a household per season are included.
Discussion
While HCoVs have been recognized for many years to be a cause of common respiratory illnesses, documentation of their involvement has lagged behind that of other agents, such as the rhinoviruses; this includes frequency of infection, illnesses produced, and transmission characteristics. We might not even know about the existence of the 2 most recently identified viruses, NL63 and HKU1, if attention had not been directed to HCoV following the occurrence of SARS. The development of the RT-PCR test and the inclusion of the 4 types of HCoV in multiplex clinical testing panels, mainly used in severe illnesses such as pneumonia, have resulted in numerous reports of the involvement of these viruses in hospitalizations. While studies of hospitalized patients have established the role of HCoV in causing lower respiratory illness and the global circulation of these viruses, they are unable to shed light on the frequency or seasonality of these viruses in the broader community. There are a few exceptions that have followed HCoV infections over longer periods, often in special population groups. In particular, a recent study covered 9 years of Norwegian children hospitalized with respiratory infection and found that coronaviruses were involved in 9.1% of episodes [26]. The highest frequencies were in young children, similar to the infection rates demonstrated in the current study; also identical to the current study was the frequency of types involved, with OC43 the most common and 229E the least. While the authors did not emphasize seasonality, it was clear that most of the infections occurred in the winter months.
The regular appearance of all types of coronaviruses in the same months with minimal circulation in the summer was a clear finding of this study. Unlike the influenza viruses, which peak at different times during the colder season from year to year, and even the rhinoviruses, which are most common in the autumn and spring but are present year round, the coronaviruses are sharply seasonal with little spread after May until November or December. Our findings, on a larger scale, expand on the results of a 2-year study in childcare attendees suggesting a similar seasonal pattern [27]. It is of interest that the SARS coronavirus outbreaks occurred during the same time period, with the end of global outbreaks in the period of May–June 2003 [28]. Whether such a timetable applies to the current circulation of SARS-CoV-2 is impossible to determine at this time.
Another notable finding in this community study was the fact that, in those older than 4 years, HCoV infection rates were relatively consistent. Most agents causing lower respiratory infections typically have falling incidence with increasing age, sometimes with an increase in young adults [29]. However, it is clear that like other respiratory viruses, HCoV infects people of all ages and infection only produces relative immunity. The type-specificity of the protection is also not completely clear, although frequent cocirculation of types and codetection within individuals suggests that protection is likely not highly cross-reactive. As with SARS and MERS, SARS-CoV-2 has most frequently infected adults to date [30]. Whether the relative sparing of children has largely been driven by contact and exposure patterns or other factors remains unclear. However, the frequency of infection in adults suggests that there is likely little cross-protective immunity from prior seasonal HCoV infection.
The MERS coronavirus is limited in its potential for human to human transmission except in the nosocomial setting [11]. Although the SARS coronavirus more effectively transmitted human to human, transmission was interrupted by public health interventions [31]. The seasonal HCoV viruses are clearly different in their transmission characteristics given their continued seasonal circulation year after year. The serial interval between cases and the secondary infection risk in the HIVE cohort was similar to that previously found for A(H3N2) [32]. The secondary attack rates in the current study may reflect the relatively small household structure found in our region. A longitudinal study in Kenya found overall attack rates as high as 34.0% over 1 season in a study of households of sizes ranging up to 37 members [33].
The transition from pandemic to seasonal circulation is well established for influenza, wherein, following an influenza pandemic, the novel virus becomes the new seasonal virus, replacing the previous A subtype [34]. The 4 current HCoVs have been circulating for decades, and it is unclear how they initially emerged or whether they replaced previously circulating viruses [35]. Contrary to influenza, MERS and SARS have not become a fixed presence in seasonal respiratory illness circulation on a global scale. Only time will tell if SARS-CoV-2 will become a continuing presence in the seasonal HCoV landscape, continue with limited circulation as with MERS, or, like SARS, disappear from humans altogether.
Supplementary Data
Supplementary materials are available at The Journal of Infectious Diseases online. Consisting of data provided by the authors to benefit the reader, the posted materials are not copyedited and are the sole responsibility of the authors, so questions or comments should be addressed to the corresponding author.
Notes
Financial support. This work was supported by the National Institute of Allergy and Infectious Diseases (grant numbers R01 AI097150 and R56 AI097150).
Potential conflicts of interest. All authors: No reported conflicts of interest. All authors have submitted the ICMJE Form for Disclosure of Potential Conflicts of Interest. Conflicts that the editors consider relevant to the content of the manuscript have been disclosed.
References
- 1. Tyrrell DA, Bynoe ML. Cultivation of a novel type of common-cold virus in organ cultures. Br Med J 1965; 1(5448):1467–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Hamre D, Procknow JJ. A new virus isolated from the human respiratory tract. Proc Soc Exp Biol Med 1966; 121:190–3. [DOI] [PubMed] [Google Scholar]
- 3. McIntosh K, Dees JH, Becker WB, Kapikian AZ, Chanock RM. Recovery in tracheal organ cultures of novel viruses from patients with respiratory disease. Proc Natl Acad Sci U S A 1967; 57:933–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Hamre D, Beem M. Virologic studies of acute respiratory disease in young adults. V. Coronavirus 229E infections during six years of surveillance. Am J Epidemiol 1972; 96:94–106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. McIntosh K, Kapikian AZ, Turner HC, Hartley JW, Parrott RH, Chanock RM. Seroepidemiologic studies of coronavirus infection in adults and children. Am J Epidemiol 1970; 91:585–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Saif LJ. Animal coronaviruses: what can they teach us about the severe acute respiratory syndrome? 2004; 23:643–60. [DOI] [PubMed] [Google Scholar]
- 7. Drosten C, Günther S, Preiser W, et al. Identification of a novel coronavirus in patients with severe acute respiratory syndrome. N Engl J Med 2003; 348:1967–76. [DOI] [PubMed] [Google Scholar]
- 8. Peiris JS, Lai ST, Poon LL, et al. ; SARS Study Group Coronavirus as a possible cause of severe acute respiratory syndrome. Lancet 2003; 361:1319–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. World health Organization. Regional Office for the Western Pacific. SARS: how a global epidemic was stopped. Manila: WHO Regional Office for the Western Pacific, 2006. https://apps.who.int/iris/handle/10665/207501. Accessed 20 February 2020. [Google Scholar]
- 10. Zaki AM, van Boheemen S, Bestebroer TM, Osterhaus AD, Fouchier RA. Isolation of a novel coronavirus from a man with pneumonia in Saudi Arabia. N Engl J Med 2012; 367:1814–20. [DOI] [PubMed] [Google Scholar]
- 11. Hui DS, Azhar EI, Kim YJ, Memish ZA, Oh MD, Zumla A. Middle East respiratory syndrome coronavirus: risk factors and determinants of primary, household, and nosocomial transmission. Lancet Infect Dis 2018; 18:e217–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Zhu N, Zhang D, Wang W, et al. A novel coronavirus from patients with pneumonia in China, 2019. N Engl J Med 2020; 382:727–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Brucková M, McIntosh K, Kapikian AZ, Chanock RM. The adaptation of two human coronavirus strains (OC38 and OC43) to growth in cell monolayers. Proc Soc Exp Biol Med 1970; 135:431–5. [DOI] [PubMed] [Google Scholar]
- 14. Hamre D, Kindig DA, Mann J. Growth and intracellular development of a new respiratory virus. J Virol 1967; 1:810–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Cavallaro JJ, Monto AS. Community-wide outbreak of infection with a 229E-like coronavirus in Tecumseh, Michigan. J Infect Dis 1970; 122:272–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Monto AS, Lim SK. The Tecumseh study of respiratory illness. VI. Frequency of and relationship between outbreaks of coronavirus infection. J Infect Dis 1974; 129:271–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Guan Y, Zheng BJ, He YQ, et al. Isolation and characterization of viruses related to the SARS coronavirus from animals in southern China. Science 2003; 302:276–8. [DOI] [PubMed] [Google Scholar]
- 18. van der Hoek L, Pyrc K, Jebbink MF, et al. Identification of a new human coronavirus. Nat Med 2004; 10:368–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Fouchier RA, Hartwig NG, Bestebroer TM, et al. A previously undescribed coronavirus associated with respiratory disease in humans. Proc Natl Acad Sci U S A 2004; 101:6212–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Woo PC, Lau SK, Chu CM, et al. Characterization and complete genome sequence of a novel coronavirus, coronavirus HKU1, from patients with pneumonia. J Virol 2005; 79:884–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Monto AS, Malosh RE, Evans R, et al. ; HIVE Study Research Staff Data resource profile: Household Influenza Vaccine Evaluation (HIVE) Study. Int J Epidemiol 2019; 48:1040–1040g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Sakthivel SK, Whitaker B, Lu X, et al. Comparison of fast-track diagnostics respiratory pathogens multiplex real-time RT-PCR assay with in-house singleplex assays for comprehensive detection of human respiratory viruses. J Virol Methods 2012; 185:259–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Kaye HS, Dowdle WR. Seroepidemiologic survey of coronavirus (strain 229E) infections in a population of children. Am J Epidemiol 1975; 101:238–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Hendley JO, Fishburne HB, Gwaltney JM Jr. Coronavirus infections in working adults. Eight-year study with 229 E and OC 43. Am Rev Respir Dis 1972; 105:805–11. [DOI] [PubMed] [Google Scholar]
- 25. Hovi T, Kainulainen H, Ziola B, Salmi A. OC43 strain-related coronavirus antibodies in different age groups. J Med Virol 1979; 3:313–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Heimdal I, Moe N, Krokstad S, et al. Human coronavirus in hospitalized children with respiratory tract infections: a 9-year population-based study from Norway. J Infect Dis 2019; 219:1198–206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Fairchok MP, Martin ET, Chambers S, et al. Epidemiology of viral respiratory tract infections in a prospective cohort of infants and toddlers attending daycare. J Clin Virol 2010; 49:16–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Ofner-Agostini M, Wallington T, Henry B, et al. ; SARS Investigative Team Investigation of the second wave (phase 2) of severe acute respiratory syndrome (SARS) in Toronto, Canada. What happened? Can Commun Dis Rep 2008; 34:1–11. [PubMed] [Google Scholar]
- 29. Monto AS, Malosh RE, Petrie JG, Thompson MG, Ohmit SE. Frequency of acute respiratory illnesses and circulation of respiratory viruses in households with children over 3 surveillance seasons. J Infect Dis 2014; 210:1792–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Li Q, Guan X, Wu P, et al. Early transmission dynamics in Wuhan, China, of novel coronavirus-infected pneumonia. N Engl J Med 2020; 382:1199–207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Lipsitch M, Cohen T, Cooper B, et al. Transmission dynamics and control of severe acute respiratory syndrome. Science 2003; 300:1966–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Petrie JG, Ohmit SE, Cowling BJ, et al. Influenza transmission in a cohort of households with children: 2010-2011. PLoS One 2013; 8:e75339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Munywoki PK, Koech DC, Agoti CN, Cane PA, Medley GF, Nokes DJ. Continuous invasion by respiratory viruses observed in rural households during a respiratory syncytial virus seasonal outbreak in coastal Kenya. Clin Infect Dis 2018; 67:1559–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Monto AS, Fukuda K. Lessons from influenza pandemics of the last 100 years. Clin Infect Dis 2020; 70: 951–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Talbot HK, Shepherd BE, Crowe JE Jr, et al. The pediatric burden of human coronaviruses evaluated for twenty years. Pediatr Infect Dis J 2009; 28:682–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.