Abstract
A new type of pneumonia caused by a novel coronavirus SARS-CoV-2 outbreaks recently in China and spreads into many other countries. This disease, named as COVID-19, is similar to patients infected by SARS-CoV and MERS-CoV, and nearly 20% of patients developed severe condition. Cardiac injury is a prevalent complication of severe patients, exacerbating the disease severity in coronavirus disease 2019 (COVID-19) patients. Angiotensin-converting enzyme 2 (ACE2), the key host cellular receptor of SARS-CoV-2, has been identified in multiple organs, but its cellular distribution in human heart is not illuminated clearly. This study performed the first state-of-art single cell atlas of adult human heart, and revealed that pericytes with high expression of ACE2 might act as the target cardiac cell of SARS-CoV-2. The pericytes injury due to virus infection may result in capillary endothelial cells dysfunction, inducing microvascular dysfunction. And patients with basic heart failure disease showed increased ACE2 expression at both mRNA and protein levels, meaning that if infected by the virus these patients may have higher risk of heart attack and critically ill condition. The finding of this study explains the high rate of severe cases among COVID-19 patients with basic cardiovascular disease; and these results also perhaps provide important reference to clinical treatment of cardiac injury among severe patients infected by SARS-CoV-2.
Keywords: SARS-CoV-2, Heart injury, Heart failure, ACE2
1. Introduction
In December 2019, a new type of pneumonia outbreak caused by a novel coronavirus severe acute respiratory syndrome coronavirus (SARS-CoV)-2 was firstly reported in Wuhan, Hubei province, China.1 As of 29 February 2020, this disease, named coronavirus disease 2019 (COVID-19), had caused 79 968 laboratory-confirmed infectious cases, and 2873 death in China. And currently, this virus has recently spread into many other areas especially in East Asia and European Countries.
The clinical symptoms of COVID-19 are similar to that caused by SARS-CoV and the Middle East respiratory syndrome. COVID-19 holds relatively lower fatality rate (∼2%), with nearly 20% of patients developing into severe conditions like acute respiratory distress syndrome and multiple organ failure.2 Cardiac injury was observed among 23% of severe patients.3 Even in the general COVID-19 patient cohort, the creatinine kinase (CK) elevation (>200 U/L) was observed in 13% patients, most of whom did not have cytokine storm-induced systemic inflammatory response.2 Cardiac injury has been evidenced as a common complication, which exacerbates the disease severity among patients infected with SARS-CoV-2.
The genomic analysis revealed that SARS-CoV-2 shared an average identity of 82% with the SARS-CoVs that emerged in 32 countries or regions globally during 2002–3, and more than 89% identity with two bat-SARS-CoV-like viruses, bat-SL-CoVZC45 and bat-SL-CoVZXC21, which were isolated from Rhinolophus sinicus in Zhoushan, China in 2015–17.1 And SARS-CoV-2 was demonstrated to share the angiotensin-converting enzyme 2 (ACE2) as the host cellular receptor for virus spike (S) protein according to structural analysis and in vivo experiment.1 Therefore, the expression and distribution of ACE2 are key determinants for the entry of the virus. Meanwhile, in SARS-CoV, two more receptors DC-SIGN (encoded by CD209) and L-SIGN (encoded by CLEC4M) were also reported to enhance virus entry,4 but not evaluated in SARS-CoV-2. Previous studies have identified cell-specific expression pattern of ACE2 in multiple organs such as lung, ACE2 expression in heart, however, was not well studied due to lack of state-of-art adult human single-cell atlas.
In the present study, we investigated ACE2 expression in the adult human hearts from healthy and diseased individuals, to illuminate the potential capacity of heart infection by SARS-CoV-2.
2. Methods
Human heart tissues were obtained from abandon donors in our transplantation centre. Myocardial nuclei were isolated and purified using sucrose buffer. Single nucleus RNA sequencing was performed based on the 10× genomics chromium platform (10× genomics, USA) with Chromium Single Cell 3’ v3 Reagent, followed by sequencing using Hi-seq X-ten sequencer (Illumina, USA). The raw reads were processed by cellranger 3.0.0. And routine pipeline for single-cell data analysis was based on Seurat 3.1.1. After quality control (filter out cells with nFeature_RNA ≤ 800 or percentage of mitochondrial genes >5%), we selected the top 1500 variable genes to conduct Principal Component Analysis (PCA) and identified significant PCs according to the elbow plot. The PCA results were taken as input to achieve the two-dimensional visualizations using t-distributed stochastic neighbor embedding (t-SNE) plot. Cell types were identified by classical cell-specific marker genes. Cell–cell interaction weights were computed by the product of the fold change of ligands in sender-cell types and the fold change of the corresponding receptors in receiver-cell types.
Transcriptome analysis was based on an in-house RNA sequencing dataset containing 15 donor hearts and 40 failing explanted hearts, which were obtained from the heart transplantation centre of Fuwai Hospital. The total RNA was extracted from frozen tissue and used for library construction (strand-specific RNA library) followed by sequencing on Hi-seq X-ten sequencer (Illumina, USA). The gene expression was expressed as transcripts per million, and compared by Student’s t-test between two groups.
Proteomics analysis was based on Liquid chromatography-tandem mass spectrometry (LC-MS/MS) as our previously reported.5 Heart tissues from eight failing hearts and eight normal donors were prepared for comprehensive proteome profiling using TMT-labelled strategy. The protein level was expressed as fold change to normal donors, and compared by Student’s t-test between two groups.
Spatial structures of the receptor-binding domain (RBD) of SARS-CoV-2 spike protein and human ACE2 were predicted based on target-template alignment by using ProMod3 on SWISS MODEL server (https://swissmodel.expasy.org/). PyMOL software was used to read and analyse the RBD of the fetched docking complex.
The Institutional Review Board of our institution approved the protocol, which was in compliance with the Declaration of Helsinki. And all participants comprehensively understood the details of the study and provided written informed consent.
3. Results and discussion
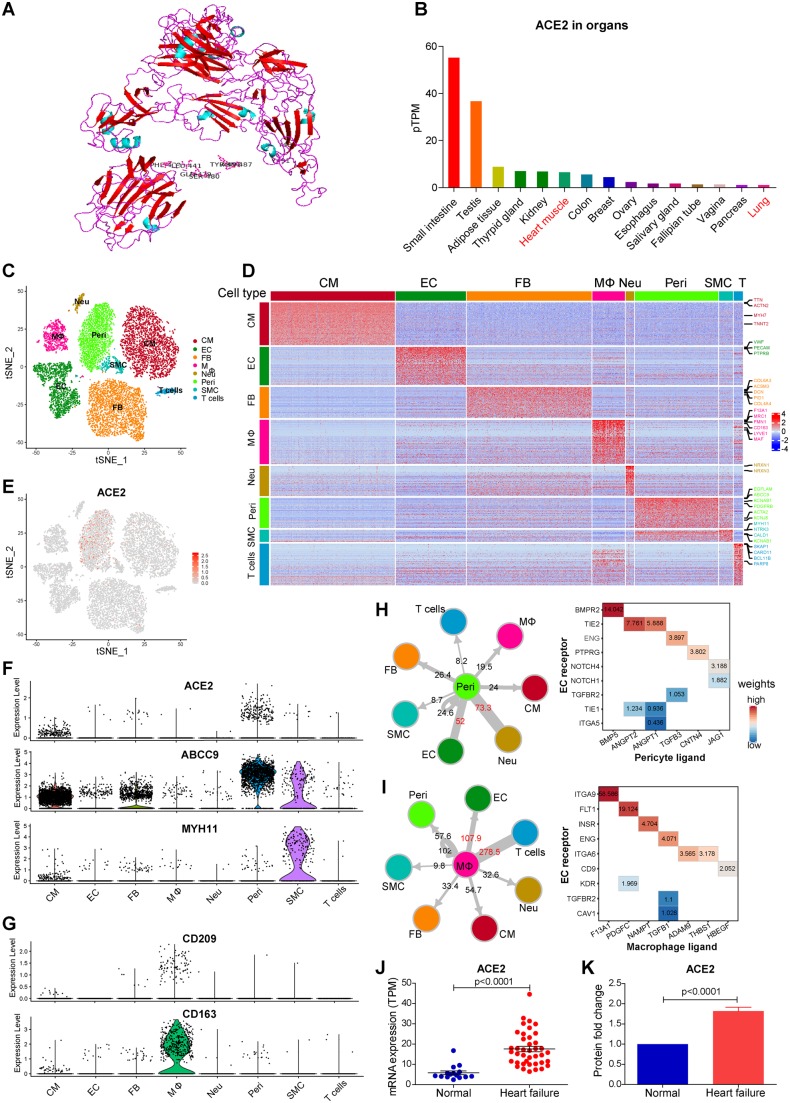
Based on modelling and docking, SARS-CoV-2 has an RBD that may bind well with human ACE2. The residues 441Leu, 472Phe, 479Gln, 480Ser, 487Asn, 491Tyr of mature spike protein appear to be critical for binding to the human ACE2 receptor and determining the host cellular range (Figure 1A).
Figure 1.
(A) The spatial and protein docking of human ACE2 protein and Spike protein of SARS-CoV-2. (B) ACE2 mRNA expression level across human organs by GTEx; (C) t-SNE plot of cell clusters in human hearts; (D). Heatmap of gene signatures of each cell type with marker genes labels; (E) Feature plot of ACE2 expression across cell types in the heart; (F) Violin plot reveals high expression of ACE2 in pericyte, and this cell type highly expresses ABCC9 (pericyte marker), but not MYH11 (SMC marker gene); (G) Violin plot reveals specifically high expression of CD209 in macrophage (co-expression with CD163); (H) Cell–cell interaction between pericyte (ligand) and other cell types (receptor) (left panel), and dominant cross-talks from pericyte to endothelial cell (EC) (right panel); (I) Cell–cell interaction between macrophage (ligand) and other cell types (receptor) (left panel), and dominant cross-talks from macrophage to EC (right panel); (J) ACE2 mRNA is increased in failing hearts (n = 40) compared to normal donors (n = 15) by RNA sequencing (P < 0.0001 by Student’s t-test); (K) ACE2 protein is increased in failing hearts (n = 8) compared to normal donors (n = 8) by proteomics (P < 0.0001 by Student’s t-test).
We screened the ACE2 mRNA expression in human organs based on the GTEx database (Figure 1B). Digestive tract intestine displayed the highest expression of ACE2, followed by testis and kidney, suggesting the vulnerability to virus in these organs, which could explain the positive detection of SARS-CoV-2 in patients’ faeces and urine.6 The expression of ACE2 in the heart is lower than that in the intestine and kidney, but higher than that in the lung which serves as a main target organ of this virus, indicating a potential infection susceptibility of human heart.
To further illuminate the cell constitution and cellular distribution of ACE2 in the human heart, we isolated myocardial nuclei and performed a single nuclear transcriptome analysis of the adult human hearts. At least eight major cell types were identified with distinct gene profiling and marker genes, including cardiomyocyte (CM, TTN, MYH7), endothelial cell (EC, VWF, PECAM), macrophage (MΦ, CD163, MRC1), fibroblast (FB, COL6A3, DCN), pericyte (Peri, ABCC9, KCNJ8), smooth muscle cell (SMC, MYH11, CALD1), T cell (SKAP1, BCL11B), and previously unknown neuron-like cell (Neu) identified by NRXN1 and NRXN1 (Figure 1C and D). Among these cell types, CMs took up only around 30% of the total nuclei, and fibroblast was the largest population (26.8%) among non-myocytes, followed by the EC. Remarkable, pericyte, a type of perivascular mural cells, took up >10% of total nuclei, which was substantially undervalued before. To the best of our knowledge, this is the first state-of-art comprehensive cell atlas of adult human myocardium.
Despite low expression in CMs, ACE2 was highly and specifically expressed in pericytes (Figure 1E). In addition, the expression of ABCC9 and PDGFRB and the absence of MYH11 help us to reaffirm the identity of the cells as pericytes instead of SMCs (Figure 1F). Different from SMCs that are located in the coronary arteries or arterioles, pericytes spread outside the endothelial cell of capillary and part of venules, which may play an essential role in myocardial micro-circulation. This result suggested that pericyte was a potential SARS-CoV-2 virus targeted host cell type in the human heart. We further screened another two related receptors of SARS-CoV, CD209, and CLEC4M. CD209 was specifically expressed in macrophage, and co-expressed with macrophage marker gene CD163, which may enhance the virus entry in the human heart (Figure 1G). CLEC4M was not enriched in human heart cells.
Virus-infected cells can be damaged, subsequently disturbing the micro-environment of the myocardium. To elucidate the potential mechanism of myocardial injury when the host cells are attacked by SARS-CoV-2, we conducted cell–cell interaction analysis centred on pericyte. Using the pericytes as ligand-secreting cells, we calculated the total number of interactions with each receptor-secreting cell type (Figure 1H). Two closest cell type crosstalking with pericytes were Neu and ECs. The former cell type was not identified and studied in the heart before. ECs showed extensive linkages with pericytes, with dominant role of angiopoietin ligands (ANGPT1/2)–Tie receptor (TIE2) interaction in normal heart, which could promote vascular EC survival and reduced vascular leakage. The ANGPT1/2-TIE1 crosstalk also exists but is weak between pericytes and ECs in the normal hearts. As the molecular balance between ANGPT1:ANGPT2 and TIE2:TIE1 is important in determining endothelial stability,7 it is suggested that pericytes play an important role in maintaining EC function in capillary vessels. The CD209 expressing cell macrophage also showed close interaction with ECs following T cells, mainly through ITGA9 and FLT1 (VEGFRA) receptor-mediated pathways (Figure 1I). Taken together, these results suggested that SARS-CoV-2 infection in the human heart might attack pericytes, and cause capillary ECs dysfunction, thus inducing micro-circulation disorder. This may explain the clinical manifestation of CK elevation among 13% of COVID-19 patients.2 And thus, medications to improve micro-circulation may alleviate heart injury in these patients.
COVID-19 patients with basic cardiovascular disease are more likely to progress to the severe conditions and even death. Hence, we further studied the expression of ACE2 in patients with heart muscle disease. RNA sequencing revealed that myocardial ACE2 expression was significantly increased in patients with heart failure (Figure 1J, fold change = 3.00, P < 0.0001), which was further validated at the protein level by proteomics profiling (Figure 1K, fold change = 1.82, P < 0.0001). This result indicated that heart failure patients were more susceptible to heart infection by SARS-CoV-2, and might develop further cardiac injury and even progress to critical condition.
4. Conclusions
ACE2 was highly expressed in pericytes of adult human hearts, which indicated an intrinsic susceptibility of heart to SARS-CoV-2 infection. Patients with basic heart failure disease exhibited increased expression of ACE2, and might have high possibility of heart attack and progressing to severe condition after infection. The finding of the present study may provide guidance to clinical treatment of cardiac injury of patients with COVID-19.
Funding
This work was supported by the National Natural Science Foundation of China (81872673), the National Key Research and Development Program of China (2017YFC1200203), and the COVID-19 research projects of Fudan University.
Conflict of interest: none declared.
References
- 1. Zhou P, Yang X-L, Wang X-G, Hu B, Zhang L, Zhang W, Si H-R, Zhu Y, Li B, Huang C-L, Chen H-D, Chen J, Luo Y, Guo H, Jiang R-D, Liu M-Q, Chen Y, Shen X-R, Wang X, Zheng X-S, Zhao K, Chen Q-J, Deng F, Liu L-L, Yan B, Zhan F-X, Wang Y-Y, Xiao G-F, Shi Z-L.. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature 2020;579:270–273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Guan W-J, Ni Z-Y, Hu Y, Liang W-H, Ou C-Q, He J-X, Liu L, Shan H, Lei C-L, Hui DSC, Du B, Li L-J, Zeng G, Yuen K-Y, Chen R-C, Tang C-L, Wang T, Chen P-Y, Xiang J, Li S-Y, Wang J-L, Liang Z-J, Peng Y-X, Wei L, Liu Y, Hu Y-H, Peng P, Wang J-M, Liu J-Y, Chen Z, Li G, Zheng Z-J, Qiu S-Q, Luo J, Ye C-J, Zhu S-Y, Zhong N-S; China Medical Treatment Expert Group for Covid-19. Clinical characteristics of coronavirus disease 2019 in China. N Engl J Med 2020; doi: 10.1056/NEJMoa2002032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Yang X, Yu Y, Xu J, Shu H, Ja X, Liu H, Wu Y, Zhang L, Yu Z, Fang M, Yu T, Wang Y, Pan S, Zou X, Yuan S, Shang Y.. Clinical course and outcomes of critically ill patients with SARS-CoV-2 pneumonia in Wuhan, China: a single-centered, retrospective, observational study. Lancet Respir Med 2020; doi: 10.1016/S2213-2600(20)30079-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Jeffers SA, Tusell SM, Gillim-Ross L, Hemmila EM, Achenbach JE, Babcock GJ, Thomas WD Jr, Thackray LB, Young MD, Mason RJ, Ambrosino DM, Wentworth DE, Demartini JC, Holmes KV.. CD209L (L-SIGN) is a receptor for severe acute respiratory syndrome coronavirus. Proc Natl Acad Sci USA 2004;101:15748–15753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Chen L, Yang F, Chen X, Rao M, Zhang NN, Chen K, Deng H, Song JP, Hu SS.. Comprehensive myocardial proteogenomics profiling reveals C/EBPalpha as the key factor in the lipid storage of ARVC. J Proteome Res 2017;16:2863–2876. [DOI] [PubMed] [Google Scholar]
- 6. Wang W, Xu Y, Gao R, Lu R, Han K, Wu G, Tan W.. Detection of SARS-CoV-2 in different types of clinical specimens. JAMA 2020; doi: 10.1001/jama.2020.3786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Bilimoria J, Singh H.. The Angiopoietin ligands and Tie receptors: potential diagnostic biomarkers of vascular disease. J Recept Signal Transduct Res 2019;39:187–193. [DOI] [PubMed] [Google Scholar]