Abstract
The tumor suppressor p53 transcriptionally activates target genes to suppress cellular proliferation during stress. p53 has also been implicated in the repression of the proto-oncogene Myc, but the mechanism has remained unclear. Here, we identify Pvt1b, a p53-dependent isoform of the long noncoding RNA (lncRNA) Pvt1, expressed 50 Kb downstream of Myc, which becomes induced by DNA damage or oncogenic signaling and accumulates near its site of transcription. We show that production of the Pvt1b RNA is necessary and sufficient to suppress Myc transcription in cis without altering the chromatin organization of the locus. Inhibition of Pvt1b increases Myc levels and transcriptional activity and promotes cellular proliferation. Furthermore, Pvt1b loss accelerates tumor growth, but not tumor progression, in an autochthonous mouse model of lung cancer. These findings demonstrate that Pvt1b acts at the intersection of the p53 and Myc transcriptional networks to reinforce the anti-proliferative activities of p53.
Graphical Abstract

eTOC
Olivero et al. identify the conserved lncRNA isoform Pvt1b as a locus-specific transcriptional regulator that serves to repress Myc transcription during the p53-mediated response to stress. Production of the Pvt1b RNA inhibits cellular proliferation and tumor growth, revealing tumor suppressor activities for this cancer-associated lncRNA.
Introduction
The p53 (also known as TP53) network is a central tumor suppressive mechanism in mammalian cells that is inactivated in the vast majority of human cancers (Vousden and Prives, 2009). In response to cellular stress induced by DNA damage or oncogenic signaling, p53 transcriptionally activates target genes to limit cellular proliferation or to permanently eliminate damaged cells (Vousden and Prives, 2009). Transcriptional activation by p53 relies on its binding to conserved p53 response elements (p53REs) in the promoters of target genes (Levine and Oren, 2009). p53 has also been implicated in the repression of cell cycle regulators (Engeland, 2018). One of the prominent targets of p53 repression is the Myelocytomasis (Myc) oncogene (Ho et al., 2005; Levy et al., 1993; Sachdeva et al., 2009), a global transcriptional amplifier that responds to mitogenic signals to promote cellular proliferation (Lin et al., 2012). Multiple models for how p53 negatively affects Myc levels have been proposed, including p53 binding to the Myc promoter to suppress histone acetylation, binding to a distal regulatory element to alter nucleosome positioning in the Myc promoter, or activating repressive Myc-targeting microRNAs (Ho et al., 2005; Porter et al., 2017; Sachdeva et al., 2009). However, the mechanism of p53-mediated Myc downregulation and its contribution to tumor suppression in vivo have remained unclear.
Long noncoding RNAs (lncRNAs) can modulate gene expression locally by accumulating near their sites of transcription (Kopp and Mendell, 2018). In dosage compensation, Xist and other lncRNAs expressed from the X-chromosome specifically repress genes across the entire X-chromosome through the recruitment of epigenetic regulators (Lee, 2012). Other cis-regulatory lncRNAs act in a more limited, locus-specific manner, such as the p53 target lincRNA-p21 proposed to promote the levels of its neighbor p21 (also known as Cdkn1a) by recruiting activating factors (Dimitrova et al., 2014). While studies of locus-specific cis-regulatory lncRNAs have revealed important roles in diverse biological processes (Dimitrova et al., 2014; Elling et al., 2018; Kotzin et al., 2016), characterization of the RNA molecule is often confounded by potential functional roles of DNA regulatory sequences in the lncRNA locus (Bassett et al., 2014; Engreitz et al., 2016; Groff et al., 2016). Defining the RNA-mediated regulation provides important opportunities for RNA-based therapeutics that can alter hardwired molecular interactions to change cellular responses.
Plasmacytoma variant 1 (Pvt1), a lncRNA expressed 50 Kb downstream of Myc, is altered in a large fraction of human cancers. Frequent translocations and viral integrations in the Pvt1 locus in lymphomas suggest important roles for Pvt1 in cancer progression (Cory et al., 1985; Graham and Adams, 1986; Graham et al., 1985). In addition, co-amplification of Myc and Pvt1 across multiple cancer types correlates with poor cancer patient prognosis, suggesting cooperation between the two genes during tumorigenesis (Cui et al., 2016; Tseng and Bagchi, 2015; Zeng et al., 2017). This pro-oncogenic cooperation between Myc and Pvt1 was recently confounded by the identification of a p53-binding site in the Pvt1 locus and by the description of the Pvt1 promoter as a transcriptional repressor of Myc (Cho et al., 2018; Porter et al., 2017). These studies suggested undefined roles for Pvt1 in cancer progression and a potential crosstalk between the tumor suppressor p53 pathway and the oncogenic Myc network.
In this study, we characterize Pvt1b, a p53-induced isoform of the lncRNA Pvt1, and we determine its contribution to Myc regulation and the p53 response to stress. We show that production of the Pvt1b RNA downstream of p53 represses Myc transcription and suppresses cellular proliferation during stress and in the early stages of tumorigenesis. The model presented here illuminates a role for the lncRNA isoform Pvt1b as a locus-specific transcriptional regulator that serves to enact selective gene repression downstream of the broad p53 transcriptional activation network.
Results
p53 suppresses Myc under conditions of genotoxic and oncogenic stress
To gain insight into the mechanism by which p53 causes suppression of Myc, we used multiple independent approaches to model the p53-dependent response to stress. To model the cellular response to genotoxic stress, we utilized wild-type (WT) mouse embryonic fibroblasts (MEFs) treated with the genotoxic agent Doxorubicin (Doxo) (Figure 1A). We observed that activation of the p53 transcriptional program following Doxo treatment for 24 hours resulted in 3-fold induction of the p53 target p21 and a concomitant reduction in Myc RNA and protein levels by 34±6% (p=0.008, Figure 1B) and 44±15% (p=0.0051, Figure 1C), respectively, consistent with previous findings (Ho et al., 2005; Porter et al., 2017). We also found that p53 activation by oncogenic stress, modeled by Tamoxifen (Tam)-CreER-dependent restoration of endogenous p53 expression in a murine lung adenocarcinoma cell line (K-rasLA2-G12D/+; p53LSL/LSL; Rosa26-CreERT2+, KPR) (Figure 1D) (Feldser et al., 2010), similarly led to a 70-fold activation of p21, a 34±7% repression of Myc RNA (p=0.0020, Figure 1E) and a 37±10% decrease in Myc protein (p=0.0028, Figure 1F). Myc repression by 39±5% was also observed in intestinal epithelium cells isolated from mice exposed to 6 Grays (Gy) of whole-body irradiation, which leads to a well-characterized p53-mediated response to genotoxic stress in vivo (p=0.0007, Figures 1G and 1H) (Clarke et al., 1994). Altogether, these results suggested that Myc repression is a general event downstream of p53 transcriptional activation.
Figure 1. p53 suppresses Myc in response to genotoxic and oncogenic stress.
(A) Schematic of the model system for studying p53-mediated response to genotoxic stress in WT MEFs untreated or treated with Doxo for 24 h. Activation of p53 by passaging or by genotoxic stress is represented by light and dark red nuclei, respectively.
(B) p21 and Myc RNA levels in cells from (A). Data show mean ± SEM (n=4, biological replicates), *p<0.05, **p<0.01, paired t test.
(C) Left Representative image and quantification of Myc protein levels from cells in (A). Hsp90 as a loading control. Right Bargraph of Myc protein levels showing mean±SEM (n=5, biological replicates), **p<0.01, paired t test.
(D) Schematic of the model system for studying p53-mediated response to oncogenic stress in KPR cells untreated or treated with Tam for 24 h. Activation of p53 by oncogenic stress is represented by red nucleus.
(E) p21 and Myc RNA levels in cells from (D). Data show mean ± SEM (n=6, biological replicates), ***p<0.001, paired t test.
(F) Left Representative image and quantification of Myc protein levels from cells in (D). Hsp90 as a loading control. Right Bargraph of Myc protein levels showing mean±SEM (n=5, biological replicates), **p<0.01, paired t test.
(G) Schematic of the model system for studying p53-mediated response in vivo in intestinal epithelial cells isolated from WT mice at 6 h post 6 Gy whole-body irradiation.
(H) p21 and Myc RNA levels from mice in (G). Data show mean ± SEM (n=3, biological replicates) **p<0.01, ***p<0.001, unpaired t test.
(I) Enrichment of p53 binding at the Pvt1-associated p53RE by ChIP-qPCR in Left Doxo-treated MEFs and Right Tam-treated KPR cells. Data show mean ± SEM (MEFs: n=4; KPR: n=3, biological replicates) *p<0.05, **p<0.01, paired t test.
(J) Myc RNA levels in p53-deficient or p53-proficient MEFs, untreated or treated with Doxo for 24 h. Data show mean ± SEM (n=3, biological replicates), ns = not significant, *p<0.05, paired t test.
(K) Left Representative image and quantification of Myc protein levels from cells in (J). Hsp90 as a loading control. Right Bargraph of Myc protein levels showing mean ± SEM (n=3, biological replicates), ns = not significant, *p<0.05, paired t test.
(L) Myc RNA levels in WT MEFs, untreated or treated with Doxo for the indicated times. Data show mean ± SEM (n=4, biological replicates), ***p<0.001, paired t test.
(M) Left Representative image and quantification of Myc protein levels from cells in (L). Hsp90 as a loading control. Right Bargraph of Myc protein levels showing mean ± SEM (n=4, biological replicates), **p<0.01, ***p<0.001, paired t test.
(N) Left Representative image and quantification of Myc protein levels following treatment with cycloheximide (Chx) for indicated times in WT MEFs, untreated or treated with Doxo for 8 h. Right Myc protein half-life (n=3, biological replicates), ns = not significant, paired t test.
In an effort to elucidate the mechanism by which p53 activation results in Myc repression, we examined whether p53 associates with the Myc locus. We observed that both in Doxo-treated MEFs and Tam-treated KPR cells, stress-dependent Myc repression was accompanied by binding of p53 to a distal p53RE, located 50 Kb downstream of Myc, which has previously been implicated in limiting Myc expression (Figure 1I)(Porter et al., 2017).
Consistent with p53 dependency, the changes in Myc RNA and protein levels were present in p53-proficient, but not p53-deficient MEFs (Figures 1J and 1K). Additionally, the decrease in Myc RNA levels was detectable as early as 4 hours following p53 activation and was coincident with the decrease in Myc protein levels, suggesting direct transcriptional modulation by p53 (Figures 1L and 1M). Inhibition of protein translation with Cycloheximide (Chx) revealed that Myc protein stability was not significantly affected by the presence of stress, suggesting that the decrease in Myc levels was not primarily due to post-translational regulation (Figure 1N).
Myc repression correlates with activation of a p53-dependent Pvt1 isoform, Pvt1b
We were intrigued that the distal p53RE was located within the gene body of the lncRNA Pvt1 (Figure 2A), which has previously been implicated as a p53 target (Barsotti et al., 2012). Considering lncRNAs can act in cis to regulate the transcription of neighboring genes, we examined whether Pvt1 played a role in restricting Myc expression during stress. We noted significant stress-dependent induction of an isoform of Pvt1, termed Pvt1b, initiated at a transcription start site located immediately downstream of the p53RE. We observed a 3.1±0.2-fold induction of Pvt1b in Doxo-treated MEFs (Figure 2B) and a 38±6-fold induction of Pvt1b in Tam-treated KPR cells (Figure 2C). Pvt1a, an isoform of Pvt1 initiated at exon 1a, was induced to a lesser extent in Doxo-treated MEFs (Figure 2B) and was not significantly induced by Tam in KPR cells (Figure 2C). Copy number calculations suggested that Pvt1b was induced from 20 to 210 copes per cell, while Pvt1a was expressed at 300-400 copies per cell (Figure S1A). Notably, activation of Pvt1b was coincident with Myc repression and occurred as early as 4 hours following Doxo treatment in MEFs (Figure S1B) or 6 hours following Tam treatment in KPR cells (Figure S1C), consistent with direct transcriptional regulation by p53. Similarly, Doxo-treated human fibroblasts exhibited a 2-fold decrease in MYC levels and an 8-fold increase of human PVT1B (Figure S1D). These findings indicated that the downregulation of Myc and the activation of a p53-dependent, stress-specific Pvt1 variant are conserved between mouse and human.
Figure 2. p53-dependent induction of the Pvt1 isoform, Pvt1b.
(A) Schematic of the mouse Myc-Pvt1 locus, highlighting exons 1a and 1b of Pvt1 and the location of the p53RE (green *).
(B, C) Isoform-specific and total Pvt1 RNA levels detected with primers located in indicated exons in (B) WT MEFs and (C) KPR cells, treated as indicated. Data show mean ± SEM (n=3, biological replicates), *p<0.05, **p<0.01, ***p<0.001, paired t test.
(D, E) RT-PCR detection of Pvt1a isoforms (a, blue), amplified with primers from exon 1a to exon 5, and Pvt1b isoforms (b, orange), amplified with primers from exon 1b to exon 5, in RNA isolated from (D) MEFs and (E) KPR cells, ladder (L).
(F) Genome browser tracks and Sashimi plots from TimeLapse-seq data in KPR cells, treated as indicated. Average number of splice junctions from 2 biological replicates from exon 1a to exon 2 (blue) and from exon 1b to exon 2 (orange) are indicated.
To further characterize the transcripts produced from the Pvt1 locus, we performed RT-PCR with forward primers located in either exon 1a or 1b and a reverse primer in exon 5. We found evidence for extensive alternative splicing and confirmed that variants containing exon 1b were induced by p53, while exon 1a-containing variants were constitutively expressed (Figures 2D and 2E). Despite the splicing heterogeneity, sequencing of nascent RNA revealed that stress-induced Pvt1b differed from constitutively expressed Pvt1a solely by the use of exon 1b versus exon 1a, and exhibited comparable splicing patterns to downstream exons (Figure 2F). We concluded that p53 activation during genotoxic and oncogenic stress initiated transcription in the Pvt1 locus from exon 1b, leading to the production of the p53-dependent isoform, Pvt1b, while Pvt1a represented a largely constitutively expressed isoform.
Stress-induced Myc repression occurs in the absence of promoter-enhancer contact reorganization
Previous work has shown that CRISPR-mediated transcriptional regulation of the Pvt1 promoter in p53-deficient cancer cells causes reorganization of the chromatin architecture in the locus and impacts the access of Myc to downstream enhancers (Cho et al., 2018). To test whether the stress-responsive, p53-dependent induction of Pvt1b was associated with changes of these chromatin contacts, we performed Chromosome Conformation Capture (3C) in MEFs and KPR cells. Using an anchor in the Myc promoter, we confirmed that the Myc promoter accessed multiple upstream and downstream enhancers, including previously described Pvt1 intragenic enhancers (Figures S2A and S2B) (Cho et al., 2018). However, we did not detect significant changes in the chromatin looping between the Myc promoter and Myc-associated enhancers during the p53-mediated stress response (Figures S2A and S2B). These results argue against a model where p53-dependent activation of Pvt1b leads to reorganization of the three-dimensional architecture of the locus.
Accumulation of Pvt1b in the chromatin surrounding the Pvt1-Myc locus
To gain insight into the potential regulatory function of Pvt1b, we performed single-molecule RNA Fluorescence in situ Hybridization (smRNA-FISH), which allows visualization of individual RNA molecules by utilizing multiple fluorescently-labeled probes per transcript. We designed four independent probesets to detect Pvt1 transcripts. Pvt1a- and Pvt1b-specific probesets (named Pvt1a (ex. 1a) and Pvt1b (ex. 1b)) were designed against the first exon of each isoform. While isoform-specific, the two probesets were not expected to detect single RNA molecules due to the low number of probes per transcript. The probeset Pvt1 (ex. 1a-10) was designed to detect both full-length Pvt1a and full-length Pvt1b at single-molecule resolution, while the Pvt1 (introns) probeset was specific to unspliced Pvt1 molecules. Finally, we designed a probeset to detect Myc intronic regions (Myc (intron)) and mark the site of Myc transcription. We observed that Pvt1a and Pvt1b exhibited a primarily 2- or 4-dot nuclear pattern in Etoposide (Etop)-treated MEFs, reflective of G1 or S/G2 stages of the cell cycle, respectively (Figures 3A and S3A). Pvt1a and Pvt1b formed larger clouds in Tam-treated KPR cells (Figures 3B and S3B), which have amplified the locus, as shown by DNA Fluorescence in situ Hybridization (DNA-FISH) (Figure S3C). By co-staining either Pvt1a or Pvt1b with total Pvt1, we concluded that both isoforms exhibited an identical localization pattern (Figures 3A, 3B, S3A and S3B). Notably, Pvt1a- and Pvt1b-containing foci co-localized with signals specific to the introns of nascent Myc (Figures 3C and 3D) as well as with nascent Pvt1 transcripts (Figures 3E and 3F). These results led us to conclude that, following transcription, Pvt1a and Pvt1b are retained on the chromatin surrounding the Pvt1-Myc locus. Subcellular fractionation analysis confirmed enrichment of both Pvt1 variants in the chromatin fraction (Figure 3G).
Figure 3. Accumulation of Pvt1 isoforms in the chromatin surrounding the Pvt1-Myc locus.
(A-F) smRNA-FISH with indicated probes in (A, C, E) WT MEFs, untreated or treated with Etop for 24 h and in (B, D, F) KPR cells untreated or treated with Tam for 24 h. DNA, DAPI. Note: Pvt1b is detectable in untreated, p53-proficient MEFs likely due to activation of the p53 pathway by passaging in primary cells but is undetectable in untreated, p53-deficient KPR cells.
(G) Pvt1a and Pvt1b RNA levels in Doxo-treated WT MEFs following subcellular fractionation (representative from n=2 biological replicates). Rn7s1 and Kcnq1ot1 used as controls for the cytoplasmic and chromatin fractions, respectively.
Pvt1b RNA represses Myc levels in cis
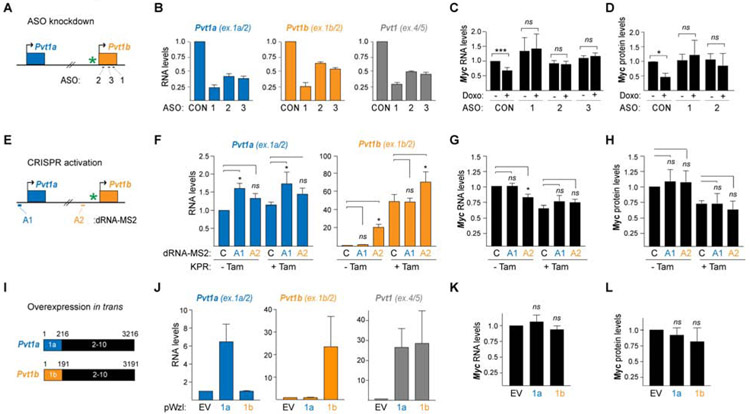
Based on the stress-dependent expression of Pvt1b and its local chromatin accumulation, we hypothesized that Pvt1b could be involved in Myc repression through an RNA-dependent mechanism. To directly test this hypothesis, we designed three independent antisense oligonucleotides (ASOs) specific to exon 1b (Figure 4A). We used a non-targeting ASO (CON) as a negative control. As ASOs lead to co-transcriptional RNA cleavage and degradation, ASO1, 2, and 3 significantly downregulated both Pvt1a and Pvt1b (Figure 4B).
Figure 4. Production of Pvt1b RNA suppresses Myc expression in cis.
(A) Schematic of ASO design. * denotes p53RE.
(B) Isoform-specific and total Pvt1 RNA levels in WT MEFs transfected with indicated control (CON) or Pvt1-targeting ASOs and harvested 24 h post Doxo treatment. Data are normalized to CON and show mean ± SEM (n=3, biological replicates).
(C) Myc RNA levels in cells from (B), untreated or treated with Doxo for 24 h. Data are normalized to CON-Doxo and show mean ± SEM (n=3, biological replicates), ***p<0.001, ns = not significant, paired t test.
(D) Quantification of Myc protein levels in cells from (B). Data are normalized to CON-Doxo and show mean ± SEM (n=3, biological replicates), *p<0.05, ns = not significant, paired t test.
(E) Schematic of CRISPRa dRNA design. * denotes p53RE.
(F) Pvt1a and Pvt1b RNA levels following Pvt1a (A1) or Pvt1b (A2) transcriptional activation in KPR cells, untreated or treated with Tam for 24 hours. Data are normalized to control dRNA (C) and show mean ± SEM (n=5, biological replicates), ns = not significant, *p<0.05, paired t test.
(G) Myc RNA levels from experiment in (F).
(H) Quantification of Myc protein levels in cells from (F). Data show mean ± SEM (n=3, biological replicates), ns = not significant, paired t test.
(I) Schematic of Pvt1a and Pvt1b overexpression constructs.
(J) Isoform-specific and total Pvt1 RNA levels in WT MEFs transiently overexpressing full length Pvt1a (1a) or Pvt1b (1b). Data are normalized to empty vector (EV) and show mean ± SEM (n=3, biological replicates), ns = not significant, paired t-test.
(K) Myc RNA levels from experiment in (J).
(L) Quantification of Myc protein levels in cells from (J). Data show mean ± SEM (n=3, biological replicates), ns = not significant, paired t test.
Next, we examined how Pvt1-targeting ASOs affected Myc expression levels. In untreated MEFs, Myc RNA and protein levels were not significantly altered in ASO compared to CON samples, indicating that knockdown of Pvt1 isoforms did not affect Myc regulation in the absence of stress, consistent with previous findings (Figures 4C, 4D and S4A) (Cho et al., 2018). As expected, upon treatment with Doxo, CON MEFs experienced a significant decrease in Myc RNA (Figure 4C) and protein levels (Figures 4D and S4A). On the other hand, we found that Pvt1-targeting ASOs completely rescued stress-induced downregulation of Myc RNA and protein (Figures 4C, 4D and S4A). These findings revealed that transcriptional activation of Pvt1b by p53 is required for Myc repression during stress. As a control, the absence of Myc downregulation was not due to altered association of p53 with the Pvt1b-associated p53RE (Figure S4B).
To test the sufficiency of Pvt1b in suppressing Myc, we employed the CRISPR-SAM (Synergistic Activation Mediator) system to activate the expression of endogenous Pvt1b in p53-deficient cells (Dahlman et al., 2015). CRISPR-SAM combines nuclease-proficient Cas9 with 15-nucleotide ‘dead RNAs’ (dRNAs), which are competent for Cas9 recruitment but do not support Cas9 nuclease activity. In CRISPR-SAM, the dRNA scaffold is extended by two MS2 binding loops (dRNA-MS2), which serve to recruit the MS2-binding protein (MBP) fused to the transcriptional activator domains of p65 and HSF1, allowing CRISPR activation (CRISPRa) of target genes (Dahlman et al., 2015). We designed A1 and A2 dRNA-MS2 targeting the promoters of Pvt1a and Pvt1b, respectively (Figure 4E). Compared to a non-targeting control (C), CRISPRa using A1 led to 1.6-fold induction of Pvt1a, without altering Pvt1b levels, while A2 resulted in a 20-fold activation of Pvt1b with no significant induction of Pvt1a (Figure 4F). Next, we examined the effect of activation of endogenous Pvt1a and Pvt1b on Myc levels. In support of our model, we found that CRISPRa of Pvt1b, but not Pvt1a, was sufficient to significantly repress Myc RNA in p53-deficient cells compared to control dRNA-expressing cells (p=0.023, Figure 4G). Activation of Pvt1b did not further downregulate Myc levels following p53 restoration, indicating that Pvt1b acted downstream of p53 (Figure 4G). On the other hand, activation of Pvt1b was not sufficient to suppress Myc protein levels, opening the possibility for Pvt1b-independent input at the post-transcriptional level (Figures S4C and 4H).
To distinguish between activity in cis versus in trans, we tested whether exogenous overexpression of Pvt1a and Pvt1b by transfection of cDNA constructs containing exons 1a-10 (1a) or 1b-10 (1b) affected Myc expression (Figure 4I). We observed a 6.5-fold overexpression of Pvt1a as well as a 23-fold overexpression of Pvt1b, which were comparable to CRISPRa-induced overexpression (Figure 4J). However, we found that exogenously delivered Pvt1a or Pvt1b did not significantly affect Myc RNA or protein levels, arguing against an effect in trans (Figures 4K, 4L and S4D). Altogether, these data supported a previously unappreciated role for Pvt1b, but not Pvt1a, in the repression of Myc in cis.
Genetic inhibition of Pvt1b reverses stress-induced Myc downregulation
To investigate the functional contribution of Pvt1b to the p53 tumor suppressor pathway, we developed a genetic approach to specifically inhibit Pvt1b expression by mutating the p53RE required for its expression. We targeted Cas9 to the Pvt1b p53RE by designing a guide RNA (ΔRE) adjacent to the GGG protospacer adjacent motif (PAM) site located in the central region of the p53 consensus binding motif (Figure 5A). A non-targeting gRNA (Con) was used as a negative control. We generated control (Con) and mutant (ΔRE) KPR population, MEF population, and KPR clonal cell lines, which contain numerous or clone-specific CRISPR/Cas9-induced mutations of the Pvt1b-associated p53RE. We confirmed mutagenesis of the p53RE by Sanger sequencing (Figures 5A, S5A and S5B) and showed by ChIP that ΔRE mutagenesis reduced p53 binding by 15-fold (Figure 5B). Importantly, by qRT-PCR, Pvt1b levels were significantly suppressed in ΔRE cells compared to controls (Figures 5C, S5C, S5D and S5G), and, by smRNA-FISH, we observed loss of Pvt1b-specific signal in Tam-treated ΔRE KPR cells compared to Tam-treated controls (Figures 5D and 5E). These observations led us to conclude that mutagenesis of the Pvt1b-associated p53RE led to efficient abrogation of stress-dependent Pvt1b activation.
Figure 5. Genetic inhibition of Pvt1b leads to increased Myc levels.
(A) Top Schematic of p53RE mutagenesis, indicating the PAM site (red box) and Cas9 cleavage site (red arrow). Bottom Mutant alleles, determined by Sanger sequencing.
(B) ChIP-qPCR analysis of p53 enrichment at Pvt1b-associated p53RE in indicated cells and treatments. Data show mean ± SEM (n=3, biological replicates) *p<0.05, paired t test.
(C) Pvt1a and Pvt1b RNA levels in indicated cells and treatments. Data show mean ± SEM (n=3, biological replicates), **p<0.01, ns = not significant, paired t test.
(D, E) smRNA-FISH of Pvt1b (ex.1b, red) co-localized with (D) total Pvt1 (ex1a-10, green) or (E) nascent Myc (intron, green) in indicated cells and treatments. DNA, DAPI.
(F) Myc RNA levels in indicated cells and treatments. Data show mean ± SEM (n=3, biological replicates), *p<0.05, ***p<0.001, ns = not significant, paired t test.
(G) Representative image and quantification of Myc protein levels in indicated cells and treatments. Hsp90 as a loading control.
(H) Quantification of Myc protein levels from experiments in (G). Data show mean ± SEM (n=6, biological replicates), *p<0.05, ***p<0.001, paired t test.
Next, we queried whether ΔRE mutagenesis led to isoform-specific inhibition. By qRT-PCR and smRNA-FISH, we found that Pvt1a RNA levels and localization pattern were not significantly altered in ΔRE KPR population and clonal cell lines compared to controls, indicating that mutation of the p53RE led to specific inhibition of Pvt1b in KPR cells (Figures 5C, 5D, S5C and S5D). On the other hand, mutagenesis of the p53RE in MEFs led to a significant reduction of Pvt1a (Figure S5G), consistent with our findings that Pvt1a expression has a p53-dependent component in this cell type (Figure 2B).
Finally, we examined by qRT-PCR and immunoblotting the effects of the ΔRE mutation and the resulting loss of Pvt1b expression on Myc levels during the cellular response to stress. In Con KPR population, KPR clonal, and MEF lines, exposure to oncogenic or genotoxic stress led to the expected significant decrease in Myc RNA (Figures 5F, S5E, S5F and S5H) and protein levels (Figures 5G, 5H, S5I and S5J). In contrast, exposure to stress in ΔRE KPR population, KPR clonal, and MEF lines did not lead to a significant decrease in Myc RNA levels compared to unstressed cells, consistent with the ASO data (Figures 5F, S5E, S5F and S5H). These results provided an independent, genetic confirmation that Pvt1b regulates Myc RNA levels downstream of p53.
Interestingly, while Myc protein levels were significantly elevated in ΔRE KPR+Tam and ΔRE MEF+Doxo lines compared to Con KPR+Tam and Con MEF+Doxo lines, respectively, the rescue was not complete (Figures 5G, 5H, S5I and S5J), consistent with the possibility of Pvt1b-independent regulatory input at the post-transcriptional level (Figure 4H).
Of note, mutagenesis of the Pvt1b-associated p53RE did not impact the long-range chromatin interactions in the locus, consistent with chromatin architecture not playing a significant role in p53-mediated Myc repression (Figure S6A).
Pvt1b suppresses Myc transcriptional activity and cellular proliferation in vitro
By analyzing the effects of the ΔRE mutation on gene expression in total RNA from untreated and Doxo-treated ΔRE and Con MEFs, we confirmed that Myc is a target of Pvt1b regulation in response to stress (Figure 6A). Next, to test whether Pvt1b acted at the transcriptional or post-transcriptional level, we sequenced nascent RNA from untreated and Tam-treated ΔRE and Con KPR cells (Schofield et al., 2018). We found that nascent Myc transcripts were significantly upregulated in ΔRE+Tam compared to Con+Tam KPR cells, indicative of transcriptional regulation (Figures 6B and 6C). These data revealed that Pvt1b production promotes transcriptional suppression of Myc.
Figure 6. Pvt1b suppresses Myc transcription and proliferative function.
(A, B) Butterfly plot depicting the fold change (logFC) in gene expression of indicated samples relative to statistical significance (−log10(p-value), MEF: n=3; KPR: n=2, biological replicates). Gene expression profiling was performed by (A) RNAseq of polyA-selected RNA isolated from Con or ΔRE gRNA-expressing MEFs, untreated or treated with Doxo for 24 hours or (B) TimeLapse-seq of ribosomal cDNA-depleted s4U-labeled RNA isolated from Con or ΔRE gRNA-expressing KPR cells, untreated or treated with Tam for 16 hours. Total Pvt1 (blue) and Myc (red) are labeled.
(C) Top Genome browser tracks depicting the Myc-Pvt1 locus and Bottom Detail of the Myc locus from TT-TimeLapse-seq.
(D, E) Cumulative frequency distribution plot of differential expression for a set of curated Myc target genes and a matched set of control genes from analyses in (A, B).
(F) Population doublings in Con or ΔRE gRNA-expressing KPR cells, untreated or treated with Tam over indicated timecourse. Data show mean ± SEM (n=3, biological replicates), **p<0.01, unpaired t test.
(G) Representative images of colony formation assay of Tam-treated KPR cells, infected with Con or ΔRE gRNAs. Numbers show mean ± SEM (n=3, biological replicates), **p<0.01, unpaired t test.
Next, we queried how the changes in Myc RNA levels affected the Myc transcriptional program by examining the consequence of Pvt1b loss on a curated set of 196 Myc target genes (Gene Set Enrichment Analysis, HALLMARK_MYC_TARGETS_V1 (Liberzon et al., 2015)). We plotted the cumulative frequency distribution of the fold change of Myc target genes in ΔRE cells relative to Con cells in the presence of stress (logFC [ΔRE/Con+stress]). Compared to a randomly generated set of control genes expressed at comparable levels, we found a significant increase in the levels of Myc targets in MEFs and KPR cells (Figures 6D and E). We concluded that Myc derepression by ΔRE mutagenesis led to a small but significant increase in the transcriptional activity of Myc.
Considering Myc target genes include factors that promote cellular growth, we compared the proliferation of mutant cells compared to controls. It has previously been shown that Tam-mediated p53 restoration in KPR cells leads to a permanent cell cycle arrest, called senescence (Feldser et al., 2010). While loss of Pvt1b expression did not overcome senescence, it led to a significant increase in cellular proliferation and colony formation compared to control cells (Figures 6F and 6G). As a control, the ΔRE mutation did not impact Myc levels and proliferation in p53-deficient cells, ruling out off target effects (Figures S6B and 6F ). These data suggested that Pvt1b mediates specific aspects of p53 function to suppress the proliferative potential of cells in vitro.
Tumor-specific inhibition of Pvt1b promotes tumor growth in vivo
Inactivation of p53 in the K-rasLSL-G12D/+(K) autochthonous mouse model of lung cancer has been shown to increase tumor burden and promote tumor progression from benign to aggressive disease (DuPage et al., 2009; Jackson et al., 2005; Jackson et al., 2001). To elucidate whether Pvt1b mediated some aspects of p53 function, we performed tumor-specific mutagenesis of the Pvt1b-associated p53RE (Figure 7A). We built a bifunctional lentiviral construct (U6-gRNA PGK-Cre, UGPC) for co-expression of the ΔRE gRNA (UGPC-ΔRE) and Cre recombinase, required for Cas9 targeting and tumor initiation, respectively (DuPage et al., 2009). Expression of Cas9 in a tumor-specific manner was achieved by crossing the K model to Rosa26-Cas9LSL (C) mice to generate KC animals (Platt et al., 2014). As a negative control, we used a non-targeting control (UGPC-Con). As a positive control, we used a previously described gRNA that targets the open reading frame of p53 (UGPC-p53KO) (Xue et al., 2014). Sanger sequencing confirmed successful mutagenesis of the Pvt1b-associated p53RE in UGPC-ΔRE-infected animals (Figure 7B).
Figure 7. Tumor-specific editing in a lung cancer model reveals a role for Pvt1b in suppressing tumor growth, but not progression.
(A) Schematic of tumor-specific gene editing in KC and KPC lung cancer mouse models.
(B) Mutant ΔRE alleles, determined by Sanger sequencing of bulk DNA isolated from tumor-bearing lungs.
(C) H&E staining of lung sections of KC mice infected with indicated gRNAs and analyzed at 16 weeks post tumor initiation (pti). Scale bars as indicated.
(D) Quantification of tumor grade in mice described in (C). The number of tumors analyzed from n=5 mice is indicated for each group.
(E) Quantification of tumor burden in mice described in (C). Dots represent individual animals and bargraph shows mean ± SEM (n=7 mice), ***p<0.001, **p<0.01, ns = not significant, unpaired t test.
(F) Representative images of immunohistochemistry for the mitotic marker pHH3 in lung sections from (C). Scale bars as indicated.
(G) Quantification of images in (F). Data show mean ± SEM of n=13-15 tumors from n=5 mice, **p<0.01, Mann-Whitney test.
(H) Quantification of tumor burden in KC and KPC mice infected with indicated gRNAs and analyzed at 12 weeks pti. Dots represent individual animals and bargraph shows mean ± SEM (KC: n=6 mice, KPC: n=3 mice), *p<0.05, ns=not significant, unpaired t-test.
We next examined hematoxylin and eosin (H&E) sections of lungs from mice infected with UGPC-Con, -p53KO and -ΔRE virus and sacrificed at 16 weeks post tumor initiation. In the K model, progression of atypical adenomatous hyperplasia (AAH, grade 1) and lung adenoma (grade 2) to adenocarcinoma (grade 3) and invasive adenocarcinoma (grade 4) is promoted by loss of p53 function (Jackson et al., 2005). Indeed, histopathological analysis revealed that all of the tumors (53/53 tumors) in UGPC-Con-infected animals manifested grade 1 features (Figures 7C and 7D). In contrast, 70% of UGPC-p53KO-expressing tumors (39/56 tumors) were marked by atypical nuclei, desmoplasia, and transition to a poorly differentiated phenotype and were classified as grade 2 or 3 (Figures 7C and 7D) (DuPage et al., 2008). Based on these data, we estimated that a large portion of the tumors underwent successful CRISPR/Cas9 editing in vivo. Editing of the Pvt1b-associated p53RE resulted in tumors with histopathological features comparable to controls and only 3% of tumors (2/67 tumors) in UGPC-ΔRE-infected animals were classified grade 2 or 3, suggesting that tumor progression was not accelerated by Pvt1b inhibition (Figures 7C and 7D). We concluded that Pvt1b does not likely mediate the ability of p53 to restrain tumor progression from benign hyperplasia to advanced disease.
On the other hand, quantification of the tumor area relative to the total lung area revealed that the tumor burden in UGPC-ΔRE-infected animals (21±4%) was significantly increased compared to the burden of control mice (12±2%) (p=0.0040, Figure 7E). Notably, the tumor burden in p53RE-edited mice was comparable to the tumor burden in UGPC-p53KO-infected mice (26±3%) (Figure 7E). These findings suggested that Pvt1b mediated in large part the growth-restrictive functions downstream of p53, particularly during the pre-malignant stages of the disease. As a control for potential off-target effects of Cas9 expression and CRISPR editing, we used two independent sgRNAs (sg1 and sg2) to target the p53RE in intron 1 of an unrelated lncRNA, Gm26542, for which we had evidence for direct p53 regulation (Figures S7A, S7B and S7C). In contrast to Pvt1b, inhibition of Gm26542 did not affect proliferation in Tam-treated KPR cells in vitro (Figure S7D) and did not significantly alter the tumor burden in KC mice in vivo (Figure S7E).
The increase in tumor burden in UGPC-ΔRE-infected animals compared to UGPC-Con mice was not due to decreased apoptosis as there was no evidence for Cleaved Caspase 3 (CC3) immunohistochemistry (IHC) staining in lung sections. Instead, the increase in tumor burden could be attributed to enhanced proliferation, as manifested by the significantly greater number of phosphorylated histone H3 (pHH3)-positive mitotic cells in Pvt1b-deficient tumors from UGPC-ΔRE-infected animals compared to tumors from UGPC-Con-infected mice (p=0.0026, Figures 7F and 7G).
Finally, to investigate whether Pvt1b acted downstream or independent of p53, we performed an epistasis experiment. We generated cohorts of either KC or K-rasLSL-G12D/+; p53FL/FL; Rosa26-Cas9LSL/LSL (KPC) animals, which have genetically engineered Cre-inducible loss-of-function alleles of p53. We analyzed tumor burden at 12 weeks post tumor initiation with UGPC-Con or -ΔRE virus. Consistent with our findings above, we observed a significant increase in the tumor burden of UGPC-ΔRE-infected mice compared to UGPC-Con-infected KC animals (p=0.0035, Figure 7H). In contrast, we found that the tumor burden was not significantly different between UGPC-ΔRE and UGPC-Con-infected KPC animals (Figure 7H). Moreover, there was no statistically significant difference between the tumor burden of KC mice infected with UGPC-ΔRE and KPC mice infected with UGPC-Con (Figure 7H). Altogether, these results revealed that Pvt1b and p53 enhance the expansion of pre-malignant tumors through a common pathway.
Discussion
Our study provides new mechanistic insights into the function of the lncRNA Pvt1 in the context of the p53 tumor suppressor pathway. We identify a conserved isoform of Pvt1, Pvt1b, which is directly activated by p53 in response to genotoxic and oncogenic stress. Our data reveal that production of Pvt1b functions as a p53-dependent mechanism that is wired into the Myc-Pvt1 locus to directly and swiftly down-regulate Myc transcription during stress. This appears to be the primary mechanism underlying stress-induced Myc reduction at the transcriptional level, although our data are also consistent with Pvt1b-independent regulation at the post-transcriptional level.
Functionally, we observed that Pvt1b activation leads to restricted Myc levels and transcriptional activity and suppressed cellular proliferation. Furthermore, using an autochthonous mouse model of lung cancer, we determined that Pvt1b acts downstream of p53 during the early stages of cancer development to limit tumor growth. Strikingly, in this respect, epistasis analysis suggested that Pvt1b acts as the primary mediator of p53. On the other hand, we found that Pvt1b is not involved in other aspects of p53 function, such as promoting senescence or limiting tumor progression to advanced disease. Altogether, these analyses define the specific contributions of Pvt1b downstream of p53, pointing to growth limiting and tumor suppressive functions of Pvt1b in the context of cancer. These conclusions contrast the common classification of Pvt1 as an oncogene, which is based on extensive correlative evidence linking Pvt1 aberrations with increased invasive capacities of cancer cells and poor patient survival (Guan et al., 2007; Guo et al., 2018; Kong et al., 2015; Riquelme et al., 2014; Tseng et al., 2014; Zeng et al., 2017; Zhang et al., 2019; Zhao et al., 2018; Zheng et al., 2016; Zhu et al., 2017). On the other hand, our data are consistent with recent reports of tumor suppressive elements in the Pvt1 locus (Barsotti et al., 2012; Cho et al., 2018; Porter et al., 2017).
Our findings shed light on a subset of genomic aberrations reported across a variety of malignancies, which represent translocations between the first exon of Pvt1a fused to various 3’ gene partners (Iwakawa et al., 2013; Kim et al., 2014; Nagoshi et al., 2012; Northcott et al., 2012). Such rearrangements would be expected to separate the Myc locus from Pvt1b, providing cells with a proliferative advantage due to the inability of p53 to suppress Myc levels during early stages of tumor development. On the other hand, the proposed tumor suppressive role of Pvt1b is at odds with the common amplification of the Pvt1 locus in cancer (Guan et al., 2007; Riquelme et al., 2014). We propose that amplification of other elements, such as the Pvt1a transcript or Pvt1-associated Myc enhancers may be the drivers of oncogenic activities in this setting, as proposed by others (Tseng et al., 2014; Cho et al., 2018). Alternatively, these alterations might be occurring following p53 inactivation, which would preclude Pvt1b expression.
Mechanistically, we provide direct evidence for a role of Pvt1b RNA production in Myc regulation. Antisense-mediated depletion experiments reveal that Pvt1b is required for stress-induced Myc inhibition, whereas epigenetic activation from the endogenous locus shows that Pvt1b is sufficient to repress Myc in the absence of stress or a functional p53 pathway. While ASO-based knockdown and CRISPR-guided epigenetic experiments cannot formally differentiate between the mature Pvt1b molecules or the production of nascent Pvt1b transcripts as the mediator of Myc repression, our data support an RNA-based mechanism.
This conclusion differs from the recent finding that the Pvt1a promoter suppresses Myc levels in an RNA-independent manner (Cho et al., 2018). The discrepancy can potentially be explained by the previous focus on the constitutive Pvt1 isoform, by the use of p53-deficient cell lines, or by the use of ineffective ASOs (Cho et al., 2018). Alternatively, we propose that the two tumor suppressive activities in the Pvt1 locus, one p53- and RNA-dependent and the other p53- and RNA-independent, may co-exist and operate in distinct cellular contexts. Our findings also do not contradict studies that have implicated Pvt1a or circular Pvt1 isoforms as oncogenes via diverse mechanisms, such as oncoprotein stabilization or competition for miRNA binding (Tseng et al., 2014; Xu et al., 2017; Zhao et al., 2018). Indeed, the complexity of the Pvt1 locus highlights the need for further rigorous dissection of the various alternative start site- and splice-variants.
It is important to note that Pvt1b mediates a repressive event downstream of p53, which is a well-characterized transcriptional activator. Considered in the context of the previously characterized p53-dependent cis-regulatory lincRNA-p21 (Dimitrova et al., 2014), it appears that transcription factors use lncRNAs to either enhance their inherent activity or to allow reverse regulation within local circuits. LncRNAs which accumulate at their sites of transcription, such as Pvt1b, are poised to act as modulators of gene expression in a locus-specific manner. Indeed, Pvt1b activation leads to Myc repression within four hours of exposure to genotoxic stress, which is comparable to the kinetics of activation of p53 target genes. We propose that production and/or chromatin accumulation of p53-induced Pvt1b transcripts act in cis during the cellular response to stress to rapidly influence the transcriptional environment at the Myc promoter. Thus, locus-specific transcriptional regulation by lncRNAs may provide additional tools within a transcriptional program that allow dynamic and swift responses to cellular challenges. As the mechanisms of more p53-dependent lncRNAs are revealed, we can gain new insight into how regulatory RNAs contribute to the cellular responses to stress mediated by p53. Although future work will determine the functional elements of Pvt1b transcripts, the widespread importance of this regulatory circuit in normal and transformed cells in vitro and in vivo suggests the possibility of controlling Myc levels in cancer by modulating Pvt1b activity.
STAR METHODS
CONTACT FOR REAGENT AND RESOURCE SHARING
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Nadya Dimitrova (nadya.dimitrova@yale.edu). Plasmids generated in this study have been deposited to Addgene.
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Mouse strains
All animal work was conducted in accordance with a protocol approved by the Yale University Institutional Animal Care and Use Committee. K-rasLSL-G12D/+(K) and p53FL/FL (P) mice were previously described (Jackson et al., 2005; Jackson et al., 2001) and obtained from the laboratory of T. Jacks (MIT). Rosa26-Cas9LSL/LSL (C) mice were previously described (Platt et al., 2014) and purchased from Jackson Laboratories (026556). Wild-type (WT) C57BL/6J mice were purchased from Jackson Laboratories (000664). For irradiation experiments, 4-8 months-old mice were irradiated with 6 Gy of whole body irradiation and sacrificed 6 hours post irradiation. For tumor studies, 3-6 months-old mice were used. Experiments were performed blind to gender and with an equal distribution of males and females in each experimental group.
Cell culture and drug treatments
WT MEFs were isolated from embryos at E13.5 from timed matings of WT C57BL/6J animals. All MEF experiments were performed at passages 2-10. KPR8 lung adenocarcinoma cell line of the genotype K-rasG12D/+; p53LSL/LSL; Rosa-CreERT2 was previously established from spontaneously arising primary tumors isolated from K-rasLA2-G12D/+; p53LSL/LSL; Rosa-CreERT2 mice, as previously described (Feldser et al., 2010). p53-restorable p53LSL/LSL; Rosa-CreERT2 MEFs were previously described (Ventura et al., 2007). Genotypes and Tam-mediated restoration of p53 expression were validated by genotyping and by qRT-PCR and immunoblotting, respectively. Puromycin-sensitive KPR8 (KPR) and p53-restorable MEF clones were generated by transient transfection with a guide RNA targeting the ORF of puromycin to inactivate the puromycin-resistance gene expressed from the Stop cassette, cloned downstream of a U6 promoter in a BRD004 lentiviral construct (a gift from the Broad Institute, MIT) that co-expresses spCas9 and GFP. Normal human fetal lung fibroblasts were purchased from the NIA Aging Cell Culture Repository (TIG-1, NG06173). Primary MEFs and human fibroblasts were maintained in DMEM (Gibco) supplemented with 15% FBS (F0926, Sigma-Aldrich), 50 U/ml pen/strep (Gibco), 2 mM L-glutamine (Gibco), 0.1 mM non-essential amino acids (Gibco), and 0.055 mM β-mercaptoethanol (Gibco). Cancer cells and 293 viral packaging cells were cultured in DMEM supplemented with 10% FBS, 50 U/ml pen/strep, 2 mM L-glutamine, and 0.1 mM non-essential amino acids. All cell cultures were maintained at 37°C in a humidified incubator with 5% CO2. Viral titering was performed in 3TZ cells, a derivative of 3T3 cells, expressing a LSL-LacZ transgene (generously provided by the laboratory of T. Jacks, MIT).
To delete the loxP-STOP-loxP (LSL) cassette preventing p53 expression, cells were treated with 0.5 μM 4-hydroxytamoxifen (Tam, Cayman Chemical Company). To induce DNA damage, cells were treated with 0.5 μM doxorubicin (Doxo, Sigma-Aldrich) or 50 μM etoposide (Etop, Millipore Sigma) for smRNA-FISH studies. To assess protein stability, cells were treated with 50 μg/ml cycloheximide (Chx, Sigma-Aldrich) for the indicated times.
Constructs
Mutagenesis of p53REs in cultured cells was performed with a gRNA targeting the p53RE of Pvt1b (gΔRE) or Gm26542 (g1 or g2), cloned downstream of a U6 promoter in BRD001 or BRD004 lentiviral constructs (gifts from the Broad Institute, MIT) that co-express spCas9 and either an IRES-driven puromycin-resistance gene or GFP, respectively. Control gRNA targeting dTomato (Con) was used as a negative control. Tumor-specific mutagenesis of p53REs in vivo was performed with gRNAs cloned downstream of a U6 promoter in UGPC (U6-gRNA-PGK-Cre) lentiviral vector. UGPC-Con targeting dTomato was used as a negative control. UGPC-p53KO targeting the ORF of p53 was used as a positive control (Xue et al., 2014). For CRISPRa experiments, a lentiviral vector (lenti-SAM-Hygro) was constructed to co-express nuclease-proficient spCas9, a U6-driven 15-mer ‘dead RNA’ (dRNA) extended by two MS2 loops (dRNA-MS2) (Dahlman et al., 2015), the transcriptional activator domains p65 and HSF1 fused to the MS2-binding protein (MBP), and a hygromycin-resistance gene. All sgRNA and dRNA sequences used in this study can be found in Table S1.
Lentivirus was produced in 293 cells by co-transfecting the lentiviral constructs with pCMV-dR8.2 dvpr (Addgene plasmid #8455) and pCMV-VSV-G (Addgene plasmid #8454) viral packaging constructs. Viral containing supernatants supplemented with 4 μg/ml polybrene (Millipore Sigma) were used to infect WT MEFs and KPR cells by 2-3 consecutive lentiviral infections, delivered at 24 hour-intervals. Following infections, cells were selected with 5 μg/ml (KPR) or 2 μg/ml (MEFs) puromycin (Sigma-Aldrich) or 800 μg/ml hygromycin (Roche). UGPC lentivirus was prepared as above, concentrated by ultracentrifugation, and titered by infecting 3TZ cells and determining the number of viral particles based on the fraction of LacZ-positive cells as previously described (DuPage et al., 2009).
Mutagenesis of the Pvt1b and Gm26542 p53REs was confirmed by PCR amplification of the region, subsequent cloning into pCR-Blunt II-TOPO® vector (Invitrogen) and Sanger sequencing.
For overexpression experiments, full-length Pvt1a (exon 1a-10) and Pvt1b (exon 1b-10) cDNAs were synthesized as gene blocks and cloned into pWZL Hygro retroviral vector (Addgene plasmid #18750). 5 μg of empty vector, Pvt1a-, or Pvt1b-expressing constructs were transfected into 1-3x106 WT MEFs using the Amaxa Mouse/Rat Hepatocyte Nucleofector Kit (Lonza, VPL-1004) and the Nucleofector 2b Device (Lonza). Analysis was performed at 48 hours post transfection.
METHOD DETAILS
RNA isolation and qRT-PCR
For RNA-seq and qRT-PCR analysis, RNA was isolated with the RNeasy Mini Kit (Qiagen) and 0.5-1 μg of total RNA was reverse transcribed using the High Capacity cDNA Reverse Transcription Kit (Applied Biosystems). SYBR Green PCR master mix (Kapa Biosystems) was used for quantitative PCR in triplicate reactions with primers listed in Table S1. Relative RNA expression levels were calculated using the ddCt method compared to GAPDH and normalized to control samples.
Immunoblotting
Cells were collected, counted, and lysed in 2×Laemmli buffer (100 mM Tris-HCl pH6.8, 200 mM DTT, 3% SDS, 20% glycerol) at 0.5-1x104 cells/μl. Samples were heated at 95°C for 7 minutes and passed through an insulin syringe. Protein from 1x105 cells was separated on 10% SDS-polyacrylamide gels and transferred to nitrocellulose membranes (Bio-Rad). After blocking (5% milk, PBST), membranes were incubated overnight at 4°C in primary antibody, then 1hr at RT in secondary antibody. The following antibodies were used: anti-c-Myc (1:1000, clone Y69, ab32072, Abcam), anti-Hsp90 (1:2500, 610419, BD Transduction Laboratories), anti-Hsp90 (1:1000, 4877S, Cell Signaling Technology), goat anti-mouse secondary antibody (1:50000, 1706516, Bio-Rad), and donkey anti-rabbit secondary antibody (1:50000, 711-035-152, Jackson ImmunoResearch). Protein bands were visualized using Amersham ECL Prime Western Blotting Detection Reagent (GE Healthcare). Quantification of Myc and Hsp90 protein levels was performed using the rectangle selection and measure tools in FIJI and Myc levels plotted relative to Hsp90 levels and normalized to negative control in relevant graphs. For cycloheximide experiments, Myc levels were normalized to negative control and half-life of Myc protein was determined using Prism8 software.
Chromatin immunoprecipitation (ChIP)
Cells were harvested by trypsinization, counted, washed once in PBS and crosslinked in 1% methanol-free formaldehyde (Thermo Scientific) diluted in PBS for 10 min at RT. The reaction was stopped by adding glycine to a final concentration of 100 mM and placing the samples on ice for 5 min. Cells were washed twice in cold PBS and the pellet was frozen and stored at −80°C.
5-10x106 nuclei were isolated by incubating the thawed cell pellet in Cell lysis buffer (20 mM Tris-HCl, pH 8.0, 85 mM KCl, 0.5% NP-40), supplemented with protease inhibitors (1 mM PMSF and Mini Complete Protease Inhibitor Cocktail Tablet, Roche) on ice for 10 min. After centrifugation, the supernatant was removed and the nuclei were resuspended in Nuclei lysis buffer (50 mM Tris-HCl, pH 8.0, 10 mM EDTA, 1% SDS supplemented with protease inhibitors) and incubated for 10 min on ice. Next, chromatin was sonicated to 300-500 bp fragment size in an ice-water slurry for 10 cycles (15” ON, 30” OFF) using a Bioruptor sonicator (Diagenode). Sonicated lysates were centrifuged at 13K rpm for 20 min and diluted in ChIP dilution buffer (0.01% SDS, 1.1% Triton- X100, 1.1 mM EDTA, 20 mM Tris-HCl, pH 8.0, 167 mM NaCl, supplemented with protease inhibitors). Input aliquots were saved at this point. The sonicated chromatin was precleared with beads (PureProteome Protein G Magnetic Beads, Millipore Sigma) and used to set up chromatin immunoprecipitations with a p53 antibody (P53-CM5P-L, Leica), or control IgG (ab46540, Abcam) and incubated overnight at 4°C on a rotator. Beads (PureProteome Protein G Magnetic Beads, Millipore Sigma) were blocked overnight in 1% BSA in PBS supplemented with 20 μg salmon sperm DNA (Invitrogen) per immunoprecipitation reaction. The next day, the blocked beads were added to the immunoprecipitation reactions and samples were incubated on the rotator for an additional hour. Beads were washed once in each of the following washes for 5 min at 4°C on the rot ator: Low salt wash (0.1% SDS, 1 % Triton-X100, 2 mM EDTA, 20 mM Tris-HCl pH 8.0, 150 mM NaCl supplemented with protease inhibitors), High salt wash (0.1% SDS, 1% Triton-X100, 2 mM EDTA, 20 mM Tris-HCl, pH 8.0, 500 mM NaCl), LiCl wash (0.25 M LiCl, 1% NP-40, 1% Na deoxycholate, 1 mM EDTA, 20 mM Tris-HCl, pH 8.0), and TE wash (10 mM Tris-HCl, pH 8.0, 1 mM EDTA).
After completely removing any remaining liquid from the washes, beads were resuspended in Elution buffer (50 mM Tris-HCl, pH 8.0, 10 mM EDTA, pH 8.0, 1% SDS) and incubated at 65°C for 15 min with frequent vortexing to prevent settling. After elution, the beads were pelleted, and the supernatant was transferred to a new tube and incubated overnight at 65°C to reverse the crosslinking. The next day, samples were treated with RNaseA or 2 hours at 37°C, followed by a proteinase K (Roche) treatment for 30 min at 55°C. The DNA was purified by phenol-chloroform extraction and EtOH precipitation. The DNA pellet was air dried, resuspended in 200 μl H2O and used for quantitative PCR analysis (ChIP-qPCR) using primers listed in Table S1.
Single-molecule FISH (smRNA-FISH)
Quasar570 (Q570)- and Quasar670 (Q670)-conjugated Stellaris FISH probes are listed in Table S1 (Stellaris, LGC Biosciences). smRNA-FISH was performed according to the manufacturer recommendations. Briefly, cells were grown on coverslips and fixed for 10 min in 4% methanol-free formaldehyde (Thermo Scientific) at RT, followed by PBS washes. Cells were dehydrated overnight at 4°C in 70% EtOH (diluted in DEPC-H2O) and stored in 70% EtOH for up to a week at 4°C. Coverslips were transferred to a hybridization chamber and equilibrated for 5 min in Wash Buffer A (Stellaris, LGC Biosciences) prepared with formamide (Millipore Sigma) according to manufacturer’s instructions. Cells were incubated overnight at 30°C with the indicated probes diluted 1:50 in Hybridization solution (Stellaris, LGC Biosciences) prepared with formamide according to manufacturer’s instructions. The next day, cells were washed 2 times for 30 min at 30°C in Wash Buffer A, incubate d in Wash Buffer B (Stellaris, LGC Biosciences) for 5 min at RT, and mounted in antifade reagent (Vectashield Mounting medium with DAPI, Vector Laboratories). The following probesets were used: Pvt1b (ex. 1b) detecting Pvt1b isoform with 10 probes spanning exon 1b, labeled with Q670 and false-colored in red; Pvt1a (ex.1a) detecting Pvt1a isoform with 11 probes spanning exon 1a, labeled with Q670 and false-colored in red; Pvt1 (ex. 1a-10) detecting total Pvt1 with 48 probes spanning exons 1a-10, labeled with Q570 and false-colored in green; Pvt1 (introns) detecting nascent Pvt1a with 31 probes spanning intron 1 upstream of exon 1b, labeled with Q670 and false-colored in red; and Myc (intron) detecting nascent Myc with 33 probes spanning intron 1 of Myc, labeled with Q570 and false-colored in green. Pvt1a (ex. 1a) and Pvt1b (ex. 1b) probesets do not detect at the single molecule level. Images were captured using an Axio Imager 2 microscope system (Zeiss) with a PlanApo 63x 1.4 oil DIC objective lens (Zeiss). For KPR cells, z-stacks of 12 planes at 0.5 μm steps were acquired and used to generate maximum intensity projections. For WT MEFs, single plane images were acquired. All images were edited using Adobe Photoshop.
DNA-Fluorescence in situ hybridization (FISH)
DNA-FISH was performed as previously described (Chaumeil et al., 2008). To generate probes, the following BAC clones were used: RP23-55F11 (Myc) and RP24-301E22 (Chr 6) (BACPAC Resources). BAC DNA was purified with a Nucleobond Xtra BAC kit (Takara Bio USA) and nick translated with a nick translation system (Invitrogen) and Alexa Fluor® 488-5-UTP or Alexa Fluor® 594-5-UTP (Invitrogen) following manufacturer instructions. Final probes were ethanol precipitated with 7.5M ammonium acetate and stored in sterile TE at −20°C.
20 ng of nick-translated probe was precipitated with 3 μg of salmon sperm DNA (Invitrogen) and 1 μg of mouse COT1 DNA (Invitrogen) using 1/10th volume of sodium acetate (3M, pH 5.5) and 2.5 volumes of ethanol. Probes were stored overnight at −20°C, then centrifuged at 13K rpm for 30 min at 4°C, washed twice with 70% ethanol, and air dried. Pellets were resuspended in formamide (Millipore Sigma), incubated at 37°C for at least 10 min, and denatured for 7 min at 75°C. After denaturing, an equal volume of 2X hybridization buffer (4X SSC, 20% w/v dextran sulfate, 2 mg/mL BSA, 40 mM RVC) was added and probe-DNA mixtures were pre-annealed for 30 min to 1 hour at 37°C.
Cells were plated on coverslips and fixed in 4% paraformaldehyde in PBS for 10 min at RT, followed by PBS washes. Cells were permeabilized in 0.5% Triton X-100 in PBS for 6 min, washed twice with 70% ethanol and stored in 70% ethanol at −20°C. Cover slips were dehydrated in an ethanol series (80%, 90%, 100%), air dried, and incubated in RNase A diluted in 2X SSC (100 μg/mL) for 1 hour at 37°C. Cover slips were washed three times with 2X SSC for 5 min and incubated in 50 μg/mL pepsin diluted into prewarmed 0.01M HCl for 3 min at 37°C, followed by two 5 min PBS washes and one in 1X PBS/MgCL2. After washing, cover slips were incubated in 1% formaldehyde (Thermo Scientific) in 1X PBS/MgCL2 for 10 min at RT. Cover slips were next washed in PBS for 5 min and dehydrated in an ethanol series (70%/90%/100%) and air dried. Cover slips were then denatured in prewarmed 50% formamide in 2X SSC for 30 min at 80°C, dehydrated in an ice-cold ethanol series (70%/90%/100%), and incubated with denatured probe DNA overnight at 42°C in a dark chamber humidified with 50% formamide in 2X SSC. Following incubation, cover slips were washed three times with prewarmed 50% formamide in 2X SSC at 42°C for 5 min and three times with prewarmed 2X SSC at 42°C for 5 min. Cover slips were mounted on slides with antifade mounting medium with DAPI (Vector Laboratories) and sealed with nail polish. Single plane images were captured using an Axio Imager 2 microscope system (Zeiss) with a PlanApo 63x 1.4 oil DIC objective lens (Zeiss).
Subcellular fractionation
Subcellular fractionation was performed as previously described (Conrad and Orom, 2017) with slight modifications. Briefly, cells were harvested by trypsinization, rinsed once in PBS and re-suspended in 1 mM EDTA in PBS. 1x106 cells were set aside for whole cell (WC) RNA isolation using TRIzol (Invitrogen) following the manufacturer’s protocol. 3 x106 cells were lysed in 0.4 mL cell lysis buffer (10 mM TrisHCl pH 7.5, 0.15% NP-40, 150 mM NaCl, 100 U/mL RNase-IN (Promega)) for 5 min on ice. Lysate was layered on a sucrose cushion (24% w/v sucrose, 150 mM NaCl, 10 mM TrisHCl pH 7.5,100 U/mL RNase-IN) and centrifuged for 10 min at 3,500g, yielding the cleared cytoplasmic fraction (supernatant) and pelleted nuclei. Nuclear pellets were washed once in PBS supplemented with 1 mM EDTA, re-suspended in 0.25 mL glycerol buffer (50% glycerol, 20 mM Tris-HCl pH 7.5, 75 mM NaCl, 0.5 mM EDTA, 0.85 mM DTT, 100 U/mL RNase-IN), and lysed by the immediate addition of an equal volume nuclear lysis buffer (10 mM HEPES pH 7.6, 7.5 mM MgCl2, 0.2 mM EDTA, 300 mM NaCl, 1% NP-40, 1 mM DTT, 1M Urea, 100 U/mL RNase-IN) with 2 min incubation on ice. Centrifugation for 2 min at 18,800g yielded the nucleoplasmic and chromatin- associated fractions in the supernatant and pellet, respectively. Chromatin pellets were washed once in 1 mM EDTA in PBS and solubilized in 1 mL TRIzol reagent by syringing. RNA was extracted from the cytoplasmic and nucleoplasmic fractions using TRIzol-LS (Invitrogen) and from the chromatin-associated fraction using TRIzol following the manufacturer’s protocols. Subcellular RNA enrichment patterns were determined by qRT-PCR, normalizing fraction Ct values to WC Ct values. Cytoplasmically-enriched RNA Rn7s1 and chromatin-enriched RNA Kcnq1ot1 served as fractionation quality controls. Primer sequences can be found in Table S1.
Antisense knockdown
1 μM Pvt1-targeting (ASO1, ASO2, and ASO3) or control (CON) antisense LNA Gapmers (Exiqon, Qiagen) were transfected into 1-3x106 MEFs using the Amaxa Mouse/Rat Hepatocyte Nucleofector Kit (Lonza, VPL-1004) and the Nucleofector 2b Device (Lonza). Knockdown of Pvt1 variants and the corresponding effects on p21 and Myc expression were assayed at 72 hours post-transfection by qRT-PCR following the indicated treatments. The sequences of all ASOs are listed in Table S1.
Chromosome Conformation Capture (3C)
Chromosome conformation capture was performed as described previously with minor modifications (Hagege et al., 2007). Briefly, cells were harvested by trypsinization, counted, washed once in PBS and 5-10x106 cells were crosslinked in 1% methanol-free formaldehyde (Thermo Scientific) diluted in PBS for 10 min at RT. The reaction was stopped by adding 1.425 ml of 1 M glycine. Cell pellets were frozen in a bath of dry ice covered in 100% EtOH and stored at −80°C, or were processed immediately. Cells were lysed in 5 ml cell lysis buffer (20 mM TrisHCl pH8.0, 85 nM KCl, 0.5% NP-40, 5 mM MgCl2, 0.1 mM EGTA) including 1x complete protease inhibitor (Roche). Cell nuclei were resuspended in 0.5 ml of 1.2x Cutsmart restriction buffer (New England Biolabs) and SDS was added to each tube to a final concentration of 0.3%. Following extraction with 2% Triton X-100, chromatin was digested overnight at 37°C with 400-800 U BamHI-HF (New England Biolabs). Ligations were performed in a total reaction volume of 6.125 mL of 1.15x ligation buffer (10x Ligation Buffer: 600 mM Tris-HCl pH7.5, 50 mM DTT, 50 mM MgCl2, 10 mM ATP (New England Biolabs)) using 100 U of T4 DNA ligase (New England Biolabs) with incubation at 16°C for 4 h, followed by further incubation at RT for 30 min. Reversal of crosslinking was performed by adding 300 μg proteinase K (Roche) followed by incubation at 65°C overnight. DNA was extracted with phenol-chloroform followed by EtOH precipitation. The efficiency of restriction enzyme digestion was examined using qRT-PCR with primer sets spanning BamHI sites. The concentrations of 3C libraries were determined by qRT-PCR and compared to a genomic DNA reference of known concentration. Samples were subsequently diluted to a concentration of 20 ng/μl and a total of 50 ng was used for each qRT-PCR reaction. Interaction frequencies were determined using a unidirectional primer strategy with an anchor designed against the promoter of Myc (A1) and were normalized to a control region in the Myc-Pvt1 locus. The primer sequences can be found in Table S1.
RNA-seq
Total RNA was isolated in three biological replicates. PolyA selection and cDNA library preparation was performed using TruSeq Stranded mRNA Library Prep (Illumina). Paired-end 100 bp sequencing was performed on an Illumina HiSeq 4000 instrument. RNA-seq read files were merged from technical replicates and mapped to the mm10 genome assembly using Tophat (ver 2.0.14) (Trapnell et al., 2009) with gencode (vM10) annotation used as the transcriptome index. Additional transcripts were assembled using stringtie (1.2.4) (Pertea et al., 2015) and reads within exon sequences counted using HTSeq (HTSeq-0.6.1) counts (Anders et al., 2015). The differential expression analysis was performed with EdgeR (3.22.3) (using general linear model settings for biological triplicates with blocked matrix model for paired comparisons) (Robinson et al., 2010). For analysis of Myc targets, the Hallmark Gene Set in the Molecular Signature Database (Broad Institute) (Liberzon et al., 2015) was used and compared to randomly selected and expression matched genes with statistical significance of differential expression determined with a Kolmogorov-Smirnov test.
Transcriptome-wide TimeLapse-seq
At approximately 60% cellular confluence, media was spiked with a final concentration of 100 μM s4U (Alfa Aesar) and grown in the dark for 1 hour. Cells were rinsed once with PBS, scraped from plates, suspended in 1 mL TRIzol (Invitrogen), and frozen overnight at −80°C. Total RNA was purified and treated with TimeLapse chemistry essentially as described (Schofield et al., 2018) with minor modifications. Briefly, following chloroform extraction and isopropanol precipitation (supplemented with 1 mM DTT) genomic DNA was depleted by treating with TURBO DNase (Invitrogen) and total RNA was extracted with acidic phenol:chloroform:isoamyl alcohol and EtOH precipitation. Isolated total RNA was mixed with 600 mM TFEA, 1 mM EDTA and 100 mM sodium acetate, pH 5.2 in water. A solution of 10 mM NaIO4 was added and the reaction mixture was incubated at 45°C for 1 hr. Ch emically treated RNA was purified using Agencourt RNAclean XP beads (1 equivalent volume, Beckman Coulter) according to manufacturer's instructions. Purified material was then incubated in a reducing buffer (10 mM DTT, 100 mM NaCl, 10 mM Tris pH 7.4, 1 mM EDTA) at 37°C for 30 min, followed by a second RNAclean bead purification. For each sample, 10 ng of total RNA input was used to prepare sequencing libraries from the Clontech SMARTer Stranded Total RNA-Seq kit Pico Input (Takara Bio USA) with ribosomal cDNA depletion. Paired-end 100 bp sequencing was performed on an Illumina HiSeq 4000 instrument.
TT-TimeLapse-seq
At approximately 60% cellular confluence, media was spiked with s4U (1 μM final, Alfa Aesar) and cells were grown in the dark for 5 min. Total RNA and DNA isolation were performed as described above. Total RNA (50 μg) was biotinylated with MTSEA biotin-XX (Biotium), isolation and streptavidin enrichment essentially as described (Schofield et al., 2018). Enriched RNA was chemically treated as described above. Library construction and sequencing were performed essentially as described above.
TimeLapse-seq mutational analysis
Filtering and alignment to the mouse GRCm38.p5 were performed essentially as described previously (Schofield et al., 2018). Briefly, reads were filtered to remove duplicate sequences with FastUniq (Xu et al., 2012), trimmed of adaptor sequences with Cutadapt v1.16 (Martin, 2011) and aligned to GRCm38 using HISAT2 v2.1.0 (Kim et al., 2015) (with default parameters except -mp 4,2). Reads aligning to transcripts were quantified with HTSeq (Anders et al., 2015) htseq-count. SAMtools v1.5 (Li et al., 2009) was used to collect only read pairs with a mapping quality greater than 2 and concordant alignment (sam FLAG = 147/99 or 83/163). Mutation calling was performed essentially as described previously (Schofield et al., 2018). Briefly, T-to-C mutations were only considered if they met several conditions. Mutations must have a base quality score greater than 40 and be more than 3 nucleotides from the read’s end. Sites of likely single-nucleotide polymorphisms (SNPs) and alignment artifacts were identified with bcftools or from sites of high mutation levels in the non-s4U treated controls and were not considered in mutation calling. Browser tracks were made using STAR v2.5.3a (Dobin et al., 2013) and normalized across samples using scale factors calculated using RNA-seq reads using edgeR (Robinson et al., 2010) (calcNormFactors using method = ‘upperquartile’).
Differential expression analysis
Differential expression analysis of transcriptome-wide TimeLapse-seq and TT-TimeLapse-seq data was performed with DESeq2 (Love et al., 2014) essentially as described previously (Schofield et al., 2018). DESeq2 expression analysis was performed on TT-TimeLapse-seq and transcriptome-wide TimeLapse-seq data to determine changes in transcriptional activity and mRNA expression, respectively.
Growth curve and colony assay
To generate growth curves, Con-, ΔRE-, sg1-, or sg2-expressing KPR cells were grown in the presence or absence of Tam. Population doublings over indicated time course were plotted as the average of three independent experiments. For colony assays, 4x105 Con- or ΔRE - expressing KPR cells were plated in the presence of Tam in 6 cm dishes and monitored for colony formation. Plates were washed with PBS, fixed in 0.5% Crystal Violet; 25% MeOH for 10 minutes and washed in ddH2O. The average of three biological replicates is shown.
Tumor studies
Lung tumorigenesis was initiated in cohorts of KC and KPC mice as described in (DuPage et al., 2009) by intratracheal infection with 1x105 pfu UGPC lentiviruses . Mice were analyzed at 12 or 16 weeks post tumor initiation. For histological analyses, lungs were inflated with 4% paraformaldehyde, and fixed overnight in 4% paraformaldehyde, prior to dehydration in 70% ethanol. Fixed lungs were embedded in paraffin, sectioned, and stained with hematoxylin and eosin (H&E). Tumor burden scored as tumor area relative to total lung area was determined using the freehand selection tool and Measure feature in ImageJ on images acquired with an Axio Imager 2 microscope system (Zeiss) with a PlanApo 10x 0.3 objective lens (Zeiss). Tumor grade was scored as previously described (DuPage et al., 2009; Nikitin et al., 2004).
Immunohistochemistry
Immunohistochemistry on paraffin sections was performed using the ABC Vectastain kit (Vector Labs) with an antibody to pHH3 Serine 10 (9701S, Cell Signaling Technologies). The staining was visualized with DAB (Vector Labs) and slides were counterstained with hematoxylin.
QUANTIFICATION AND STATISTICAL ANALYSIS
In relevant figures, figure legends convey the statistical details of experiments including statistical tests used and type and number (n) of biological replicates, while asterisks define degree of significance as described. All Student’s t-tests and Mann-Whitney U-tests were analyzed in two sided. All sequencing data were aligned to the mouse genome (GRCm38/mm10). All statistical analyses were performed and graphics were generated using Prism8 software.
DATA AND SOFTWARE AVAILABILITY
All software used in this study is listed in the Key Resources Table. Data generated in this study are available through Gene Expression Omnibus (GEO) under accession number GEO: GSE126940. Graphical abstract was created using graphics from www.Biorender.com.
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Rabbit monoclonal (Y69) anti-c-myc | Abcam | Cat#ab32072 |
| Mouse monoclonal anti-hsp90 | BD Transduction Laboratories | Cat#610419 |
| Rabbit monoclonal anti-hsp90 | Cell Signaling Technologies | Cat#4877S |
| Rabbit polyclonal anti-p53 | Leica | Cat#P53-CM5P-L |
| Rabbit Anti-Mouse IgG (H&L) | Abcam | Cat#ab46540 |
| Rabbit polyclonal anti-pHH3 Serine 10 | Cell Signaling Technologies | Cat#9701S |
| Goat Anti-Mouse IgG (H + L)-HRP Conjugate | Bio-Rad | Cat#1706516 |
| Peroxidase AffiniPure Donkey Anti-Rabbit IgG (H+L) | Jackson ImmunoResearch | Cat#711-035-152 |
| Bacterial and Virus Strains | ||
| Stable Competent E.coli (High Efficiency) | New England Biolabs | Cat#C3040H |
| 5-alpha Competent E.coli (High Efficiency) | New England Biolabs | Cat#C2987U |
| Chemicals, Peptides, and Recombinant Proteins | ||
| 4-hydroxy tamoxifen | Cayman Chemical Company | Cat#17308-10 |
| Doxorubicin hydrochloride | Sigma-Aldrich | Cat#D1515-10MG |
| Etoposide | Millipore Sigma | Cat#E1383-25MG |
| Cycloheximide | Sigma-Aldrich | Cat#C4859-1ML |
| Hexadimethrine bromide (polybrene) | Millipore Sigma | Cat#107689-10G |
| Puromycin dihydrochloride | Sigma-Aldrich | Cat#58-58-2 |
| Hygromycin B | Roche | Cat#10843555001 |
| DMEM, high glucose, pyruvate | Gibco | Cat#11995065 |
| Fetal Bovine Serum | Sigma-Aldrich | Cat#F0926-500ML |
| L-Glutamine | Gibco | Cat#25030-081 |
| Pen-strep | Gibco | Cat#15140-122 |
| NEAA | Gibco | Cat#11140-050 |
| 2-mercaptoethanol | Gibco | Cat#21985023 |
| SYBR Fast Master Mix | Kapa Biosystems | Cat#kk4602 |
| Nitrocellulose membranes | Bio-Rad | Cat#1620112 |
| ECL Prime Western Blotting detection reagent | GE Healthcare | Cat#RPN2232 |
| Methanol-free formaldehyde | Thermo Scientific | Cat#28908 |
| cOmplete, Mini Protease Inhibitor Cocktail | Roche | Cat#4693124001 |
| PureProteome Protein G Magnetic Beads | Millipore Sigma | Cat#LSKMAGG02 |
| Salmon Sperm DNA | Invitrogen | Cat#15632011 |
| Mouse Cot-1 DNA | Invitrogen | Cat#18440016 |
| Proteinase K | Roche | Cat#03115879001 |
| Formamide | Millipore Sigma | Cat#F9037-100ML |
| Stellaris® RNA FISH Hybridization Buffer | LGC Biosciences | Cat#SMF-HB1-10 |
| Stellaris® RNA FISH Wash Buffer A | LGC Biosciences | Cat#SMF-WA1-60 |
| Stellaris® RNA FISH Wash Buffer B | LGC Biosciences | Cat#SMF-WB1-20 |
| VECTASHIELD® Antifade Mounting Medium with DAPI | Vector Laboratories | Cat#H-1200 |
| Alexa Fluor® 488-5-UTP | Invitrogen | Cat#C11397 |
| Alexa Fluor® 594-5-UTP | Invitrogen | Cat#C11400 |
| TRIzol Reagent | Invitrogen | Cat#15-596-018 |
| TRIzol-LS Reagent | Invitrogen | Cat#10296028 |
| RNasin Plus RNase inhibitor | Promega | Cat#N2615 |
| CutSmart® Buffer | New England Biolabs | Cat#B7204S |
| BamHI-HF® | New England Biolabs | Cat#R3136L |
| T4 DNA Ligase | New England Biolabs | Cat#M0202L |
| Adenosine 5'-Triphosphate (ATP) | New England Biolabs | Cat#P0756L |
| 4-thiouridine (s4U) | Alfa Aesar | Cat#AAJ60679MC |
| TURBO DNase | Invitrogen | Cat#AM2238 |
| Agencourt RNAclean XP beads | Beckman Coulter | Cat#A63987 |
| MTSEA biotin-XX | Biotium | Cat#900661 |
| Critical Commercial Assays | ||
| Mouse/Hepatocyte Nucleofector Kit | Lonza | Cat#VPL1004 |
| RNeasy Mini Kit | Qiagen | Cat#Q74106 |
| High Capacity cDNA Reverse Transcription Kit | Applied Biosystems | Cat#Q74106 |
| Zero Blunt TOPO PCR Cloning Kit | Invitrogen | Cat#450245 |
| Nucleobond Xtra BAC kit | Takara Bio USA | Cat#740436.10 |
| Nick Translation System | Invitrogen | Cat#18160-010 |
| TruSeq Stranded mRNA Library Prep | Illumina | Cat#20020594 |
| Clontech SMARTer Stranded Total RNA-Seq kit Pico Input Mammalian | Takara Bio USA | Cat#634411 |
| Vectastain Elite ABC-HRP Kit (Peroxidase, rabbit IgG) | Vector Laboratories | Cat#PK-6101 |
| DAB Peroxidase (HRP) Substrate Kit | Vector Laboratories | Cat#SK-4100 |
| Deposited Data | ||
| Raw and analyzed data of mouse cell line | This study | GEO: GSE126940 |
| Experimental Models: Cell Lines | ||
| Mouse: Primary WT embryonic fibroblasts (MEFs) | This paper | N/A |
| Mouse: K-rasG12D/+; p53LSL/LSL; Rosa-CreERT2 (KPR8) cell line | Feldser et al., 2010 | N/A |
| Mouse: p53LSL/LSL; Rosa-CreERT2 (p53-restorable MEFs) | Ventura et al., 2007 | N/A |
| Mouse: 3TZ (3T3 derivative) cell line | T. Jacks, MIT | N/A |
| Human: fetal lung fibroblasts (TIG-1) | NIA Aging Cell Culture Repository | Cat#NG06173 |
| Experimental Models: Organisms/Strains | ||
| Mouse: Wild-type (WT) C57BL/6J | Jackson Laboratories | Cat#000664 |
| Mouse: K-rasLSL-G12D/+ (K) | Jackson et al., 2001; Jackson Laboratories | Cat#008179 |
| Mouse: p53FL/FL (P) | Jackson et al., 2005; Jackson Laboratories | Cat#008462 |
| Mouse: Rosa26-Cas9LSL/LSL (C) | Platt et al., 2014; Jackson Laboratories | Cat#026556 |
| Oligonucleotides | ||
| qRT-PCR primers, see Table S1 | This paper | N/A |
| PCR primers, see Table S1 | This paper | N/A |
| smRNA FISH probes, see Table S1 | This paper | see Table S1 |
| 3C qPCR primers, see Table S1 | This paper | N/A |
| ChIP qPCR primers, see Table S1 | This paper | N/A |
| ASO sequences, see Table S1 | This paper | see Table S1 |
| sgRNA sequences, see Table S1 | This paper; Xue et al., 2014 | N/A |
| dRNA sequences, see Table S1 | This paper | N/A |
| Recombinant DNA | ||
| pCMV-dR8.2 dvpr | Stewart et al., 2003 | Addgene #8455 |
| pCMV-VSV-G | Stewart et al., 2003 | Addgene #8454 |
| pWZL Hygro | S. Lowe, unpublished | Addgene #18750 |
| BRD001 | Broad Institute | N/A |
| BRD004 | Broad Institute | N/A |
| UGPC | This paper | N/A |
| lenti-SAM-hygro | This paper | N/A |
| Myc BAC | BACPAC Resources Center | Cat#RP23-55F11 |
| Chr 6 BAC | BACPAC Resources Center | Cat#RP24-301E22 |
| Software and Algorithms | ||
| GraphPad Prism, version 8.2.1 for MacOS | N/A | www.graphpad.com |
| Biorender | N/A | www.biorender.com |
| Tophat (v2.0.14) | Trapnell et al., 2009 | http://ccb.jhu.edu/software/tophat/index.shtml |
| stringtie (v1.2.4) | Pertea et al., 2015 | https://ccb.jhu.edu/software/stringtie/ |
| HTSeq (v0.6.1) | Anders et al., 2015 | https://htseq.readthedocs.io/en/release_0.11.1/ |
| EdgeR (v3.22.3) | Robinson et al., 2010 | https://bioconductor.org/packages/release/bioc/html/edgeR.html |
| FastUniq | Xu et al., 2012 | http://sourceforge.net/projects/fastuniq/ |
| Cutadapt (v1.16) | Martin, 2011 | https://cutadapt.readthedocs.io/en/stable/ |
| HISAT2 (v2.1.0) | Kim et al., 2015 | https://ccb.jhu.edu/software/hisat2/index.shtml |
| SAMtools (v1.5) | Li et al., 2009 | http://samtools.sourceforge.net |
| STAR (v2.5.3a) | Dobin et al., 2013 | https://github.com/alexdobin/STAR |
| DEseq2 | Love et al., 2014 | https://bioconductor.org/packages/release/bioc/html/DESeq2.html |
Supplementary Material
Table S1. Oligonucleotide, probe, sgRNA and dRNA sequences used in this study, Related to Figures 1-5 and S1-S7.
Highlights.
Pvt1b is a p53-dependent lncRNA isoform, induced by genotoxic and oncogenic stress
Production of Pvt1b RNA represses Myc transcription in cis
Pvt1b suppresses Myc transcriptional program and cellular proliferation
Pvt1b limits tumor growth but not tumor progression in a mouse lung tumor model
Acknowledgements
We are grateful to David Feldser for providing KPR cell lines. We thank YCGA, the Keck DNA Sequencing Facility and the Yale Mouse Research Pathology Histology Service. This work was supported in part by the LCRF (ND), IRG-ACS 58-012-58 (ND), the V Foundation (ND), the Pew-Stewart Foundation (ND), NIH R37CA230580 (ND), and by a Career Development Award funded by SPORE in Lung Cancer (Yale University, NIH P50CA196530). CO and ET were supported by NIH T32GM007223.
Footnotes
Declaration of interests
The authors declare no competing interests.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Anders S, Pyl PT, and Huber W (2015). HTSeq--a Python framework to work with high-throughput sequencing data. Bioinformatics 31, 166–169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barsotti AM, Beckerman R, Laptenko O, Huppi K, Caplen NJ, and Prives C (2012). p53-Dependent induction of PVT1 and miR-1204. J Biol Chem 287, 2509–2519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bassett AR, Akhtar A, Barlow DP, Bird AP, Brockdorff N, Duboule D, Ephrussi A, Ferguson-Smith AC, Gingeras TR, Haerty W, et al. (2014). Considerations when investigating lncRNA function in vivo. Elife 3, e03058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaumeil J, Augui S, Chow JC, and Heard E (2008). Combined immunofluorescence, RNA fluorescent in situ hybridization, and DNA fluorescent in situ hybridization to study chromatin changes, transcriptional activity, nuclear organization, and X-chromosome inactivation. Methods Mol Biol 463, 297–308. [DOI] [PubMed] [Google Scholar]
- Cho SW, Xu J, Sun R, Mumbach MR, Carter AC, Chen YG, Yost KE, Kim J, He J, Nevins SA, et al. (2018). Promoter of lncRNA Gene PVT1 Is a Tumor-Suppressor DNA Boundary Element. Cell 173, 1398–1412 e1322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clarke AR, Gledhill S, Hooper ML, Bird CC, and Wyllie AH (1994). p53 dependence of early apoptotic and proliferative responses within the mouse intestinal epithelium following gamma-irradiation. Oncogene 9, 1767–1773. [PubMed] [Google Scholar]
- Conrad T, and Orom UA (2017). Cellular Fractionation and Isolation of Chromatin-Associated RNA. Methods Mol Biol 1468, 1–9. [DOI] [PubMed] [Google Scholar]
- Cory S, Graham M, Webb E, Corcoran L, and Adams JM (1985). Variant (6;15) translocations in murine plasmacytomas involve a chromosome 15 locus at least 72 kb from the c-myc oncogene. EMBO J 4, 675–681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui M, You L, Ren X, Zhao W, Liao Q, and Zhao Y (2016). Long non-coding RNA PVT1 and cancer. Biochem Biophys Res Commun 471, 10–14. [DOI] [PubMed] [Google Scholar]
- Dahlman JE, Abudayyeh OO, Joung J, Gootenberg JS, Zhang F, and Konermann S (2015). Orthogonal gene knockout and activation with a catalytically active Cas9 nuclease. Nat Biotechnol 33, 1159–1161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dimitrova N, Zamudio JR, Jong RM, Soukup D, Resnick R, Sarma K, Ward AJ, Raj A, Lee JT, Sharp PA, et al. (2014). LincRNA-p21 activates p21 in cis to promote Polycomb target gene expression and to enforce the G1/S checkpoint. Mol Cell 54, 777–790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, Batut P, Chaisson M, and Gingeras TR (2013). STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29, 15–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DuPage M, Dooley AL, and Jacks T (2009). Conditional mouse lung cancer models using adenoviral or lentiviral delivery of Cre recombinase. Nat Protoc 4, 1064–1072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elling R, Robinson EK, Shapleigh B, Liapis SC, Covarrubias S, Katzman S, Groff AF, Jiang Z, Agarwal S, Motwani M, et al. (2018). Genetic Models Reveal cis and trans Immune-Regulatory Activities for lincRNA-Cox2. Cell Rep 25, 1511–1524 e1516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engeland K (2018). Cell cycle arrest through indirect transcriptional repression by p53: I have a DREAM. Cell Death Differ 25, 114–132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engreitz JM, Haines JE, Perez EM, Munson G, Chen J, Kane M, McDonel PE, Guttman M, and Lander ES (2016). Local regulation of gene expression by lncRNA promoters, transcription and splicing. Nature 539, 452–455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feldser DM, Kostova KK, Winslow MM, Taylor SE, Cashman C, Whittaker CA, Sanchez-Rivera FJ, Resnick R, Bronson R, Hemann MT, et al. (2010). Stage-specific sensitivity to p53 restoration during lung cancer progression. Nature 468, 572–575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graham M, and Adams JM (1986). Chromosome 8 breakpoint far 3' of the c-myc oncogene in a Burkitt's lymphoma 2;8 variant translocation is equivalent to the murine pvt-1 locus. EMBO J 5, 2845–2851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graham M, Adams JM, and Cory S (1985). Murine T lymphomas with retroviral inserts in the chromosomal 15 locus for plasmacytoma variant translocations. Nature 314, 740–743. [DOI] [PubMed] [Google Scholar]
- Groff AF, Sanchez-Gomez DB, Soruco MML, Gerhardinger C, Barutcu AR, Li E, Elcavage L, Plana O, Sanchez LV, Lee JC, et al. (2016). In Vivo Characterization of Linc-p21 Reveals Functional cis-Regulatory DNA Elements. Cell Rep 16, 2178–2186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guan Y, Kuo WL, Stilwell JL, Takano H, Lapuk AV, Fridlyand J, Mao JH, Yu M, Miller MA, Santos JL, et al. (2007). Amplification of PVT1 contributes to the pathophysiology of ovarian and breast cancer. Clin Cancer Res 13, 5745–5755. [DOI] [PubMed] [Google Scholar]
- Guo J, Hao C, Wang C, and Li L (2018). Long noncoding RNA PVT1 modulates hepatocellular carcinoma cell proliferation and apoptosis by recruiting EZH2. Cancer Cell Int 18, 98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagege H, Klous P, Braem C, Splinter E, Dekker J, Cathala G, de Laat W, and Forne T (2007). Quantitative analysis of chromosome conformation capture assays (3C-qPCR). Nat Protoc 2, 1722–1733. [DOI] [PubMed] [Google Scholar]
- Ho JS, Ma W, Mao DY, and Benchimol S (2005). p53-Dependent transcriptional repression of c-myc is required for G1 cell cycle arrest. Mol Cell Biol 25, 7423–7431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwakawa R, Takenaka M, Kohno T, Shimada Y, Totoki Y, Shibata T, Tsuta K, Nishikawa R, Noguchi M, Sato-Otsubo A, et al. (2013). Genome-wide identification of genes with amplification and/or fusion in small cell lung cancer. Genes Chromosomes Cancer 52, 802–816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson EL, Olive KP, Tuveson DA, Bronson R, Crowley D, Brown M, and Jacks T (2005). The differential effects of mutant p53 alleles on advanced murine lung cancer. Cancer Res 65, 10280–10288. [DOI] [PubMed] [Google Scholar]
- Jackson EL, Willis N, Mercer K, Bronson RT, Crowley D, Montoya R, Jacks T, and Tuveson DA (2001). Analysis of lung tumor initiation and progression using conditional expression of oncogenic K-ras. Genes Dev 15, 3243–3248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim D, Langmead B, and Salzberg SL (2015). HISAT: a fast spliced aligner with low memory requirements. Nat Methods 12, 357–360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim HP, Cho GA, Han SW, Shin JY, Jeong EG, Song SH, Lee WC, Lee KH, Bang D, Seo JS, et al. (2014). Novel fusion transcripts in human gastric cancer revealed by transcriptome analysis. Oncogene 33, 5434–5441. [DOI] [PubMed] [Google Scholar]
- Kong R, Zhang EB, Yin DD, You LH, Xu TP, Chen WM, Xia R, Wan L, Sun M, Wang ZX, et al. (2015). Long noncoding RNA PVT1 indicates a poor prognosis of gastric cancer and promotes cell proliferation through epigenetically regulating p15 and p16. Mol Cancer 14, 82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kopp F, and Mendell JT (2018). Functional Classification and Experimental Dissection of Long Noncoding RNAs. Cell 172, 393–407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kotzin JJ, Spencer SP, McCright SJ, Kumar DBU, Collet MA, Mowel WK, Elliott EN, Uyar A, Makiya MA, Dunagin MC, et al. (2016). The long non-coding RNA Morrbid regulates Bim and short-lived myeloid cell lifespan. Nature 537, 239–243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JT (2012). Epigenetic regulation by long noncoding RNAs. Science 338, 1435–1439. [DOI] [PubMed] [Google Scholar]
- Levine AJ, and Oren M (2009). The first 30 years of p53: growing ever more complex. Nat Rev Cancer 9, 749–758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levy N, Yonish-Rouach E, Oren M, and Kimchi A (1993). Complementation by wild-type p53 of interleukin-6 effects on M1 cells: induction of cell cycle exit and cooperativity with c-myc suppression. Mol Cell Biol 13, 7942–7952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R, and Genome Project Data Processing, S. (2009). The Sequence Alignment/Map format and SAMtools. Bioinformatics 25, 2078–2079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liberzon A, Birger C, Thorvaldsdottir H, Ghandi M, Mesirov JP, and Tamayo P (2015). The Molecular Signatures Database (MSigDB) hallmark gene set collection. Cell Syst 1, 417–425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagoshi H, Taki T, Hanamura I, Nitta M, Otsuki T, Nishida K, Okuda K, Sakamoto N, Kobayashi S, Yamamoto-Sugitani M, et al. (2012). Frequent PVT1 rearrangement and novel chimeric genes PVT1-NBEA and PVT1-WWOX occur in multiple myeloma with 8q24 abnormality. Cancer Res 72, 4954–4962. [DOI] [PubMed] [Google Scholar]
- Nikitin AY, Alcaraz A, Anver MR, Bronson RT, Cardiff RD, Dixon D, Fraire AE, Gabrielson EW, Gunning WT, Haines DC, et al. (2004). Classification of proliferative pulmonary lesions of the mouse: recommendations of the mouse models of human cancers consortium. Cancer Res 64, 2307–2316. [DOI] [PubMed] [Google Scholar]
- Northcott PA, Shih DJ, Peacock J, Garzia L, Morrissy AS, Zichner T, Stutz AM, Korshunov A, Reimand J, Schumacher SE, et al. (2012). Subgroup-specific structural variation across 1,000 medulloblastoma genomes. Nature 488, 49–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Platt RJ, Chen S, Zhou Y, Yim MJ, Swiech L, Kempton HR, Dahlman JE, Parnas O, Eisenhaure TM, Jovanovic M, et al. (2014). CRISPR-Cas9 knockin mice for genome editing and cancer modeling. Cell 159, 440–455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porter JR, Fisher BE, Baranello L, Liu JC, Kambach DM, Nie Z, Koh WS, Luo J, Stommel JM, Levens D, et al. (2017). Global Inhibition with Specific Activation: How p53 and MYC Redistribute the Transcriptome in the DNA Double-Strand Break Response. Mol Cell 67, 1013–1025 e1019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riquelme E, Suraokar MB, Rodriguez J, Mino B, Lin HY, Rice DC, Tsao A, and Wistuba II, (2014). Frequent coamplification and cooperation between C-MYC and PVT1 oncogenes promote malignant pleural mesothelioma. J Thorac Oncol 9, 998–1007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson MD, McCarthy DJ, and Smyth GK (2010). edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26, 139–140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sachdeva M, Zhu S, Wu F, Wu H, Walia V, Kumar S, Elble R, Watabe K, and Mo YY (2009). p53 represses c-Myc through induction of the tumor suppressor miR-145. Proc Natl Acad Sci U S A 106, 3207–3212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schofield JA, Duffy EE, Kiefer L, Sullivan MC, and Simon MD (2018). TimeLapse-seq: adding a temporal dimension to RNA sequencing through nucleoside recoding. Nat Methods 15, 221–225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell C, Pachter L, and Salzberg SL (2009). TopHat: discovering splice junctions with RNA-Seq. Bioinformatics 25, 1105–1111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tseng YY, and Bagchi A (2015). The PVT1-MYC duet in cancer. Mol Cell Oncol 2, e974467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tseng YY, Moriarity BS, Gong W, Akiyama R, Tiwari A, Kawakami H, Ronning P, Reuland B, Guenther K, Beadnell TC, et al. (2014). PVT1 dependence in cancer with MYC copy-number increase. Nature 512, 82–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ventura A, Kirsch DG, McLaughlin ME, Tuveson DA, Grimm J, Lintault L, Newman J, Reczek EE, Weissleder R, and Jacks T (2007). Restoration of p53 function leads to tumour regression in vivo. Nature 445, 661–665. [DOI] [PubMed] [Google Scholar]
- Vousden KH, and Prives C (2009). Blinded by the Light: The Growing Complexity of p53. Cell 137, 413–431. [DOI] [PubMed] [Google Scholar]
- Xu H, Luo X, Qian J, Pang X, Song J, Qian G, Chen J, and Chen S (2012). FastUniq: a fast de novo duplicates removal tool for paired short reads. PLoS One 7, e52249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu MD, Wang Y, Weng W, Wei P, Qi P, Zhang Q, Tan C, Ni SJ, Dong L, Yang Y, et al. (2017). A Positive Feedback Loop of lncRNA-PVT1 and FOXM1 Facilitates Gastric Cancer Growth and Invasion. Clin Cancer Res 23, 2071–2080. [DOI] [PubMed] [Google Scholar]
- Zeng Y, Wang T, Liu Y, Su Z, Lu P, Chen X, and Hu D (2017). LncRNA PVT1 as an effective biomarker for cancer diagnosis and detection based on transcriptome data and meta-analysis. Oncotarget 8, 75455–75466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Yang G, and Luo Y (2019). Long non-coding RNA PVT1 promotes glioma cell proliferation and invasion by targeting miR-200a. Exp Ther Med 17, 1337–1345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao L, Kong H, Sun H, Chen Z, Chen B, and Zhou M (2018). LncRNA-PVT1 promotes pancreatic cancer cells proliferation and migration through acting as a molecular sponge to regulate miR-448. J Cell Physiol 233, 4044–4055. [DOI] [PubMed] [Google Scholar]
- Zheng X, Hu H, and Li S (2016). High expression of lncRNA PVT1 promotes invasion by inducing epithelial-to-mesenchymal transition in esophageal cancer. Oncol Lett 12, 2357–2362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu S, Shuai P, Yang C, Zhang Y, Zhong S, Liu X, Chen K, Ran Q, Yang H, and Zhou Y (2017). Prognostic value of long non-coding RNA PVT1 as a novel biomarker in various cancers: a meta-analysis. Oncotarget 8, 113174–113184. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1. Oligonucleotide, probe, sgRNA and dRNA sequences used in this study, Related to Figures 1-5 and S1-S7.
Data Availability Statement
All software used in this study is listed in the Key Resources Table. Data generated in this study are available through Gene Expression Omnibus (GEO) under accession number GEO: GSE126940. Graphical abstract was created using graphics from www.Biorender.com.
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Rabbit monoclonal (Y69) anti-c-myc | Abcam | Cat#ab32072 |
| Mouse monoclonal anti-hsp90 | BD Transduction Laboratories | Cat#610419 |
| Rabbit monoclonal anti-hsp90 | Cell Signaling Technologies | Cat#4877S |
| Rabbit polyclonal anti-p53 | Leica | Cat#P53-CM5P-L |
| Rabbit Anti-Mouse IgG (H&L) | Abcam | Cat#ab46540 |
| Rabbit polyclonal anti-pHH3 Serine 10 | Cell Signaling Technologies | Cat#9701S |
| Goat Anti-Mouse IgG (H + L)-HRP Conjugate | Bio-Rad | Cat#1706516 |
| Peroxidase AffiniPure Donkey Anti-Rabbit IgG (H+L) | Jackson ImmunoResearch | Cat#711-035-152 |
| Bacterial and Virus Strains | ||
| Stable Competent E.coli (High Efficiency) | New England Biolabs | Cat#C3040H |
| 5-alpha Competent E.coli (High Efficiency) | New England Biolabs | Cat#C2987U |
| Chemicals, Peptides, and Recombinant Proteins | ||
| 4-hydroxy tamoxifen | Cayman Chemical Company | Cat#17308-10 |
| Doxorubicin hydrochloride | Sigma-Aldrich | Cat#D1515-10MG |
| Etoposide | Millipore Sigma | Cat#E1383-25MG |
| Cycloheximide | Sigma-Aldrich | Cat#C4859-1ML |
| Hexadimethrine bromide (polybrene) | Millipore Sigma | Cat#107689-10G |
| Puromycin dihydrochloride | Sigma-Aldrich | Cat#58-58-2 |
| Hygromycin B | Roche | Cat#10843555001 |
| DMEM, high glucose, pyruvate | Gibco | Cat#11995065 |
| Fetal Bovine Serum | Sigma-Aldrich | Cat#F0926-500ML |
| L-Glutamine | Gibco | Cat#25030-081 |
| Pen-strep | Gibco | Cat#15140-122 |
| NEAA | Gibco | Cat#11140-050 |
| 2-mercaptoethanol | Gibco | Cat#21985023 |
| SYBR Fast Master Mix | Kapa Biosystems | Cat#kk4602 |
| Nitrocellulose membranes | Bio-Rad | Cat#1620112 |
| ECL Prime Western Blotting detection reagent | GE Healthcare | Cat#RPN2232 |
| Methanol-free formaldehyde | Thermo Scientific | Cat#28908 |
| cOmplete, Mini Protease Inhibitor Cocktail | Roche | Cat#4693124001 |
| PureProteome Protein G Magnetic Beads | Millipore Sigma | Cat#LSKMAGG02 |
| Salmon Sperm DNA | Invitrogen | Cat#15632011 |
| Mouse Cot-1 DNA | Invitrogen | Cat#18440016 |
| Proteinase K | Roche | Cat#03115879001 |
| Formamide | Millipore Sigma | Cat#F9037-100ML |
| Stellaris® RNA FISH Hybridization Buffer | LGC Biosciences | Cat#SMF-HB1-10 |
| Stellaris® RNA FISH Wash Buffer A | LGC Biosciences | Cat#SMF-WA1-60 |
| Stellaris® RNA FISH Wash Buffer B | LGC Biosciences | Cat#SMF-WB1-20 |
| VECTASHIELD® Antifade Mounting Medium with DAPI | Vector Laboratories | Cat#H-1200 |
| Alexa Fluor® 488-5-UTP | Invitrogen | Cat#C11397 |
| Alexa Fluor® 594-5-UTP | Invitrogen | Cat#C11400 |
| TRIzol Reagent | Invitrogen | Cat#15-596-018 |
| TRIzol-LS Reagent | Invitrogen | Cat#10296028 |
| RNasin Plus RNase inhibitor | Promega | Cat#N2615 |
| CutSmart® Buffer | New England Biolabs | Cat#B7204S |
| BamHI-HF® | New England Biolabs | Cat#R3136L |
| T4 DNA Ligase | New England Biolabs | Cat#M0202L |
| Adenosine 5'-Triphosphate (ATP) | New England Biolabs | Cat#P0756L |
| 4-thiouridine (s4U) | Alfa Aesar | Cat#AAJ60679MC |
| TURBO DNase | Invitrogen | Cat#AM2238 |
| Agencourt RNAclean XP beads | Beckman Coulter | Cat#A63987 |
| MTSEA biotin-XX | Biotium | Cat#900661 |
| Critical Commercial Assays | ||
| Mouse/Hepatocyte Nucleofector Kit | Lonza | Cat#VPL1004 |
| RNeasy Mini Kit | Qiagen | Cat#Q74106 |
| High Capacity cDNA Reverse Transcription Kit | Applied Biosystems | Cat#Q74106 |
| Zero Blunt TOPO PCR Cloning Kit | Invitrogen | Cat#450245 |
| Nucleobond Xtra BAC kit | Takara Bio USA | Cat#740436.10 |
| Nick Translation System | Invitrogen | Cat#18160-010 |
| TruSeq Stranded mRNA Library Prep | Illumina | Cat#20020594 |
| Clontech SMARTer Stranded Total RNA-Seq kit Pico Input Mammalian | Takara Bio USA | Cat#634411 |
| Vectastain Elite ABC-HRP Kit (Peroxidase, rabbit IgG) | Vector Laboratories | Cat#PK-6101 |
| DAB Peroxidase (HRP) Substrate Kit | Vector Laboratories | Cat#SK-4100 |
| Deposited Data | ||
| Raw and analyzed data of mouse cell line | This study | GEO: GSE126940 |
| Experimental Models: Cell Lines | ||
| Mouse: Primary WT embryonic fibroblasts (MEFs) | This paper | N/A |
| Mouse: K-rasG12D/+; p53LSL/LSL; Rosa-CreERT2 (KPR8) cell line | Feldser et al., 2010 | N/A |
| Mouse: p53LSL/LSL; Rosa-CreERT2 (p53-restorable MEFs) | Ventura et al., 2007 | N/A |
| Mouse: 3TZ (3T3 derivative) cell line | T. Jacks, MIT | N/A |
| Human: fetal lung fibroblasts (TIG-1) | NIA Aging Cell Culture Repository | Cat#NG06173 |
| Experimental Models: Organisms/Strains | ||
| Mouse: Wild-type (WT) C57BL/6J | Jackson Laboratories | Cat#000664 |
| Mouse: K-rasLSL-G12D/+ (K) | Jackson et al., 2001; Jackson Laboratories | Cat#008179 |
| Mouse: p53FL/FL (P) | Jackson et al., 2005; Jackson Laboratories | Cat#008462 |
| Mouse: Rosa26-Cas9LSL/LSL (C) | Platt et al., 2014; Jackson Laboratories | Cat#026556 |
| Oligonucleotides | ||
| qRT-PCR primers, see Table S1 | This paper | N/A |
| PCR primers, see Table S1 | This paper | N/A |
| smRNA FISH probes, see Table S1 | This paper | see Table S1 |
| 3C qPCR primers, see Table S1 | This paper | N/A |
| ChIP qPCR primers, see Table S1 | This paper | N/A |
| ASO sequences, see Table S1 | This paper | see Table S1 |
| sgRNA sequences, see Table S1 | This paper; Xue et al., 2014 | N/A |
| dRNA sequences, see Table S1 | This paper | N/A |
| Recombinant DNA | ||
| pCMV-dR8.2 dvpr | Stewart et al., 2003 | Addgene #8455 |
| pCMV-VSV-G | Stewart et al., 2003 | Addgene #8454 |
| pWZL Hygro | S. Lowe, unpublished | Addgene #18750 |
| BRD001 | Broad Institute | N/A |
| BRD004 | Broad Institute | N/A |
| UGPC | This paper | N/A |
| lenti-SAM-hygro | This paper | N/A |
| Myc BAC | BACPAC Resources Center | Cat#RP23-55F11 |
| Chr 6 BAC | BACPAC Resources Center | Cat#RP24-301E22 |
| Software and Algorithms | ||
| GraphPad Prism, version 8.2.1 for MacOS | N/A | www.graphpad.com |
| Biorender | N/A | www.biorender.com |
| Tophat (v2.0.14) | Trapnell et al., 2009 | http://ccb.jhu.edu/software/tophat/index.shtml |
| stringtie (v1.2.4) | Pertea et al., 2015 | https://ccb.jhu.edu/software/stringtie/ |
| HTSeq (v0.6.1) | Anders et al., 2015 | https://htseq.readthedocs.io/en/release_0.11.1/ |
| EdgeR (v3.22.3) | Robinson et al., 2010 | https://bioconductor.org/packages/release/bioc/html/edgeR.html |
| FastUniq | Xu et al., 2012 | http://sourceforge.net/projects/fastuniq/ |
| Cutadapt (v1.16) | Martin, 2011 | https://cutadapt.readthedocs.io/en/stable/ |
| HISAT2 (v2.1.0) | Kim et al., 2015 | https://ccb.jhu.edu/software/hisat2/index.shtml |
| SAMtools (v1.5) | Li et al., 2009 | http://samtools.sourceforge.net |
| STAR (v2.5.3a) | Dobin et al., 2013 | https://github.com/alexdobin/STAR |
| DEseq2 | Love et al., 2014 | https://bioconductor.org/packages/release/bioc/html/DESeq2.html |