Abstract
Objectives
Many countries use faecal immunochemical testing (FIT) to screen for colorectal cancer. There is increasing evidence that faecal microbiota play a crucial role in colorectal cancer carcinogenesis. We assessed the possibility of measuring faecal microbial features in FIT as potential future biomarkers in colorectal cancer screening.
Methods
Bacterial stability over time and the possibility of bacterial contamination were evaluated using quantitative polymerase chain reaction analysis. Positive FIT samples (n = 200) of an average-risk screening cohort were subsequently analysed for universal 16S, and bacteria. Escherichia coli (E. coli), Fusobacterium nucleatum (F. nucleatum), Bacteroidetes and Faecalibacterium prausnitzii (F. prausnitzii) by qPCR. The results were compared with colonoscopy findings.
Results
Faecal microbiota in FIT were stably measured up to six days for E. coli (p = 0.53), F. nucleatum (p = 0.30), Bacteroidetes (p = 0.05) and F. prausnitzii (p = 0.62). Overall presence of bacterial contamination in FIT controls was low. Total bacterial load (i.e. 16S) was significantly higher in patients with colorectal cancer and high-grade dysplasia (p = 0.006). For the individual bacteria tested, no association was found with colonic lesions.
Conclusions
These results show that the faecal microbial content can be measured in FIT samples and remains stable for six days. Total bacterial load was higher in colorectal cancer and high-grade dysplasia. These results pave the way for further research to determine the potential role of microbiota assessment in FIT screening.
Keywords: Microbiome, colorectal cancer, screening, faecal occult blood test
Key summary
What is the established knowledge on this subject?
Faecal immunochemical tests are used worldwide in colorectal cancer screening.
There is increasing evidence that the gut microbiota play a crucial role in colorectal cancer carcinogenesis.
What are the significant and/or new findings of this study?
The gut microbiota can be measured in faecal immunochemical test samples.
Individual microbial features remain relatively stable in faecal immunochemical test samples for up to six days
Screenees with high-grade dysplasia and colorectal cancer have a higher load of total universal 16S in their faecal immunochemical test samples.
Introduction
Colorectal cancer (CRC) is a major cause of cancer-related morbidity and mortality.1 The aetiology of CRC is complex and not yet completely understood. There is increasing attention for the gut microbiota and its role in colorectal carcinogenesis.2–4 It is estimated that at least 20%, perhaps more, of the cancer burden worldwide can be attributed to microbial agents.5
An association between CRC and specific faecal bacteria has already been reported a long time ago.6 In a small Dutch study of 12 patients with Streptococcus bovis bacteraemia, CRC was diagnosed in eight and gastric cancer in one patient.7 CRC appears to have a complex aetiology with potential aetiological contribution of multiple bacterial species playing different roles.3,4,8 Most gut bacteria cannot easily be cultivated, yet sequencing of bacterial DNA following polymerase chain reaction (PCR) allows for the identification of the composition of the faecal microbiota. Evidence to date suggests that inflammatory processes triggered by enterotoxigenic bacteria can contribute to CRC development by facilitating DNA damage in intestinal epithelial cells.4,9 The ensuing accumulation of genetic lesions can contribute to oncogenesis along the adenoma–carcinoma sequence. Several studies have shown that the bacterial composition of malignant lesions differs from that of surrounding normal tissue.4,10,11 While most previous research has focused on the unravelling of the complex microbial composition in CRC and the role of the gut microbiota in the pathogenesis of CRC, it is of interest to see whether altered bacterial presence may be valuable in improving screening strategies.
In the past decade, an increasing number of countries have embarked on CRC screening. Many of those use faecal immunochemical tests (FITs) as their screening method.12 FITs rely on the measurement of trace amounts of blood from neoplastic lesions. However, not all lesions bleed (e.g. serrated adenomas), and conversely, occult blood can be detected in faecal samples of healthy individuals.13 In spite of high participation rates and a relatively high sensitivity for CRC of 75–85% depending on the cut-off used, the sensitivity of FIT for detection of advanced adenomas is much lower and generally ranges below 50%.14,15 For this reason there is an urgent need for additional markers to increase FIT sensitivity without losing its specificity, as the latter is of crucial importance in a screening setting. Investigation of faecal bacterial features could present one such possible additional marker.16 Hence, it would be of great interest to detect bacterial features in the test materials of FIT screenees, which would preclude additional material collection from screenees. Therefore, the aim of our study was to evaluate the possibility of measuring faecal microbiota in FIT in relation to endoscopic findings, to evaluate their stability over time and to assess the effect of potential bacterial contaminants in downstream PCR analysis. For this proof of principle, we have selected four different bacterial markers for quantitative PCR (qPCR) analysis: suspected driver bacteria of the Enterobacteriaceae (Escherichia coli; E. coli), Bacteroidetes-species, the most often associated CRC bacterium Fusobacterium nucleatum (F. nucleatum) and the anti-inflammatory Clostridiaceae Faecalibacterium prausnitzii (F. prausnitzii), which was described to be less prevalent in CRC patients.17–24 In addition, taxonomic profiling was carried out for six pooled samples from patients with different endoscopy outcomes to evaluate the feasibility of using FIT fluid for future 16S rRNA gene sequencing purposes.
Methods
Patients, FIT screening and data collection
Details about the design of this ongoing population-based FIT CRC screening programme have been described previously.25 In short, demographic data of all individuals between 50 and 74 years old living in the southwest of The Netherlands were randomly obtained from municipal population registers and were invited for FIT screening biennially. At present, four rounds of FIT screening have taken place. For this study only FIT samples of the end of the third and beginning of the fourth screening round were used, aiming for a total of 200 FITs to be included. Recruitment of these third and fourth screening rounds took place between February 2013 and August 2014. In the third screening round all invitees received the OC-sensor (Eiken, Japan). In the fourth screening round invitees were randomized between the OC-sensor and FOB-Gold (Sentinel, Italy). Participants were instructed to send the FIT sample within one day after collection and to keep the FIT sample in the refrigerator until sending it to the laboratory. A cut-off of ≥10 µg Hb/g faeces was used to refer the screenee for a colonoscopy within four weeks. All colonoscopies were performed by gastroenterologists with an experience based on at least 1000 colonoscopies. All lesions were evaluated by trained gastrointestinal pathologists according to the Vienna criteria.26 Advanced adenomas were defined as an adenoma with a diameter ≥10 mm, and/or with a ≥25% villous component, and/or high-grade dysplasia (HGD). Advanced neoplasia included advanced adenoma and CRC, with the most advanced lesion used for analysis. Serrated polyps were defined as serrated adenomas (with our without dysplasia) and hyperplastic polyps. For this study only FIT-positive screenees were included.
Bacterial quantitative analysis
After occult blood measurement, FIT samples were stored at −20℃ until analysis. DNA was isolated from FIT liquid by Wizard Genomic DNA Purification kit (Promega, Leiden, The Netherlands) with modifications. Information on primers, PCR and qPCR analyses can be found in Supplementary Material file 1 online.
Microbial stability and contamination in FIT
For analysis of the stability of the microbial content of FIT over time, seven FITs from stool samples with (n = 2) and without blood (n = 5) of healthy volunteers were collected and stored at −20℃ immediately, or after 24, 48, 72, 96, 120 and 144 h in order to mimic FIT transit time. DNA was isolated from all samples upon thawing and the presence of E. coli, F. nucleatum, Bacteroidetes, F. prausnitzii and universal bacterial 16S was detected by PCR and qPCR as described above. F. nucleatum was below detection level in all FIT samples of our healthy donors and could not be included in the analysis. To test for unintentional bacterial contamination, FIT controls with and without blood but not containing faeces underwent the same procedure for comparison with FIT with blood and faecal material.
16S rRNA gene sequencing pilot
To determine the feasibility of using FIT material for 16S rRNA gene sequencing, a pilot was conducted using six different pooled DNA samples from patients with the following conditions: 1) no endoscopic findings, 2) tubular adenoma, 3) HGD, 4) CRC, 5) sessile adenoma and 6) hyperplastic polyp. All samples were shipped on dry ice to the Macrogen Institute in Seoul, Korea. The V3-4 region of the 16S rRNA gene was amplified and sequenced on the Illumina MiSeq platform. The 16S rRNA gene sequencing data was subsequently processed using the SILVA database for taxonomic profiling at genus level, allowing for the global assessment of the bacterial presence of our selected markers in FIT material.
Statistical analysis
Descriptive data were reported as proportions or means with the standard deviation. For non-normally distributed data the median and interquartile range (IQR) were given. Chi-square tests were used to analyse categorical data; continuous data were analysed using Student’s t-tests or one-way analysis of variance. Linear regression analysis was used to assess bacterial load and transit time. Correction for multiple testing was done according to Bonferroni resulting in a two-sided p-value of <0.01 that was considered to be statistically significant. Statistical analysis was performed using IBM SPSS version 21.0.
Results
FIT screenees
A total of 200 samples from FIT positive screenees were collected. Of these, 20 samples had to be discarded for various reasons (e.g. because multiple samples from the same screenee were included, because a sample was misclassified or because pathology outcome was missing), resulting in 180 samples available for analysis of microbial content. These included 119 OC-sensor tests (66%) and 61 FOB-Gold tests (34%). Of those, 56% were male with a median age of 64 years (IQR 58–69 years). Median faecal Hb concentration was 21 µg Hb/g faeces (IQR 13–55 µg Hb/g faeces). All screenees included in this study underwent complete colonoscopy and in 31% (n = 55) patients advanced neoplasia was detected, of whom five were diagnosed with CRC. All colonoscopy findings are described in Table 1.
Table 1.
Primer sequences used in this study.
| Bacterium | Sequence (5'–3') | Product size | Reference | |
|---|---|---|---|---|
| Universal 16S | Forward | CGGTGAATACGTTCCCGG | 145 | 1, 9–12 |
| Reverse | TACGGCTACCTTGTTACGACTT | |||
| Fusobacterium nucleatum | Forward | CTTAGGAATGAGACAGAGATG | 140 | 2, 13 |
| Reverse | TGATGGTAACATACGAAAGG | |||
| Escherichia coli | Forward | CATGCCGCGTGTATGAAGAA | 96 | 3, 4, 14 |
| Reverse | CGGGTAACGTCAATGAGCAAA | |||
| Bacteroidetes | Forward | GGTGTCGGCTTAAGTGCCAT | 140 | 5, 6, 15 |
| Reverse | CGGACGTAAGGGCCGTGC | |||
| Faecalibacterium prausnitzii | Forward | GATGGCCTCGCGTCCGATTAG | 198 |
Stability microbiota in FIT over time and bacterial contaminants
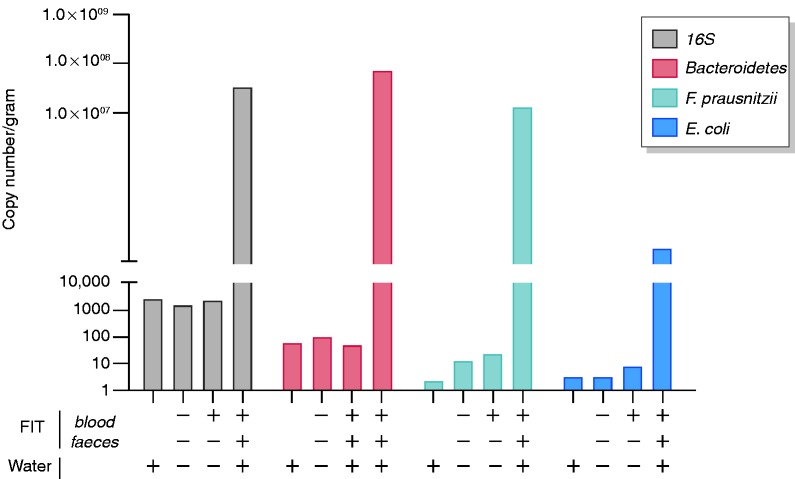
Transit time of the FIT from screenee to the laboratory could potentially affect the microbial composition detected. Although growth of anaerobic bacteria is not expected, and FIT buffer contains bacteriostatic sodiumazide, degradation of bacterial DNA might occur. Therefore, we first analysed the stability of the bacterial composition in FIT. Universal bacterial 16S, Bacteroidetes, F. prausnitzii and E. coli DNA was consistently detected by qPCR, with no significant loss in detection levels for up to 144 h in FIT positive (Figure 1(a)) and FIT negative (Figure 1(b) and (c)) samples. Microbial contamination did not influence our results as indicated by relatively low levels of bacterial contamination detected in FIT controls (Figure 2). The 16S and individual bacterial marker levels in FIT fluid without faecal content were similar to water controls and several times lower than FIT fluid containing faecal material.
Figure 1.
Stability of bacterial composition over time for faecal immunochemical testing (FIT) positive (n = 2) (a) and FIT negative specimens (n = 5) (b) and (c). This has been depicted for universal 16S and specific markers including Bacteroidetes, Faecalibacterium prausnitzii and Escherichia coli. Bacterial composition remained stable up to at least 144 h.
Figure 2.
Quantitative polymerase chain reaction (qPCR) assessment of unintentional bacterial contamination. Faecal immunochemical testing controls in either the presence or the absence of occult blood and/or faecal material were tested for 16S, Bacteroidetes, Faecalibacterium prausnitzii and Escherichia coli. Water controls were concurrently processed for qPCR analysis.
The average time between faecal sampling by the screenee and analysis at the laboratory (i.e. transit time) was one day (IQR 1–2 days), with 91% of FITs arriving at the laboratory within two days after sampling. For all screenees the correlation between absolute copy number of the four bacteria and transit time were evaluated (Figure 3). No significant decrease in faecal microbiota was seen in up to six days for E. coli (p = 0.53), F. nucleatum (p = 0.30), Bacteroidetes (p = 0.05) and F. prausnitzii (p = 0.62; Figure 3).
Figure 3.
Transit time (interval between faecal sampling and arrival of the faecal immunochemical testing (FIT) specimen at the laboratory) and absolute copy number/gram faecal immunochemical test for Escherichia coli (a), Fusobacterium nucleatum (b), Bacteroidetes (c), Faecalibacterium prausnitzii (d).
Microbiota in FIT and findings at colonoscopy
For all samples, copy number per gram (copy nr./g) FIT liquid was calculated for the total number of bacteria (i.e. 16S) and the four predefined bacteria. A significant difference was seen for 16S, with increasing abundance of total bacterial content in screenees with high-grade dysplasia and CRC (p = 0.006; Figure 4(a)). Significant differences between the groups were seen for the copy nr./g E. coli (p = 0.05; Figure 4(b)). Post hoc testing revealed that in particular patients with tubular and villous adenoma showed lower levels of E. coli, albeit not significant (p = 0.07). For F. nucleatum, F. prausnitzii and Bacteroidetes, no association was observed between the presence of the bacteria and any particular lesion (Supplementary Figure 1). No significant association between amounts of bacteria and presence of advanced neoplasia was observed (Supplementary Figure 2).
Figure 4.
Copy number per gram faecal immunochemical testing liquid for 16S (a) and Escherichia coli (b) according to colonoscopy outcomes.
nr.: number; SP: serrated polyp; TA: tubular adenoma; TVA: (tubulo)villous adenoma; HGD: high grade dysplasia; CRC: colorectal cancer
To correct for potential differences in amount of faecal matter in the FIT, the bacteria were also calculated relative to the total bacterial presence as determined by universal 16S (copy nr./g of 16S). No significant differences were found when evaluating FIT microbiota according to all colonoscopy findings, including CRC, for E. coli (p = 0.97), F. nucleatum (p = 0.98), Bacteroidetes (p = 0.15) and F. prausnitzii (p = 0.91; Figure 5). In addition, no significant differences in microbiota were found between screenees with and without advanced neoplasia (E. coli p = 0.30; F. nucleatum p = 0.55; Bacteroidetes p = 0.12; F. prausnitzii p = 0.93; Figure 6). When evaluating FIT microbiota according to location of the most advanced lesion (i.e. distal vs. proximal), again no significant differences were seen for all four bacteria (Supplementary Figure 3).
Figure 5.
Absolute copy number per 16S and most advanced colonoscopy finding* for Escherichia coli (a), Fusobacterium nucleatum (b), Bacteroidetes (c) and Faecalibacterium prausnitzii (d).
nr.: number; *SP: serrated polyp; TA: tubular adenoma; TVA: (tubulo)villous adenoma; HGD: high grade dysplasia; CRC: colorectal cancer
Figure 6.
Absolute copy number (nr.) per 16S for screenees with no advanced neoplasia (No AN) and advanced neoplasia (AN) for Escherichia coli (a), Fusobacterium nucleatum (b), Bacteroidetes (c), Faecalibacterium prausnitzii (d).
16S rRNA gene sequencing of pooled samples
Although it is well known that the overall bacterial abundance in FIT material is relatively low compared with stool specimens, 16S rRNA gene sequencing data were generated for all six pooled DNA samples from patients with respectively: 1) no endoscopic findings, 2) tubular adenoma, 3) HGD, 4) CRC, 5) sessile adenoma and 6) hyperplastic polyp. Taxonomic classification indicated that Bacteroidetes in addition to genus Faecalibacterium (with regard to F .prausnitzii) and family Enterobacteriaceae (with regard to E. coli) were present in all samples (Supplementary Figure 4). Genus Fusobacterium (with regard to F. nucleatum) was detected in the pooled DNA samples from tubular adenomas, suggesting that this lower abundant bacterial marker is present in FIT material. More importantly, these findings confirm the ability to use FIT specimens for qPCR analysis as well as future 16S rRNA sequencing purposes.
Discussion
Our results show that faecal microbial DNA can be isolated from FIT samples and remains stable for up to six days. The inclusion of FIT controls in qPCR analysis allowed the assessment of bacterial contamination which appeared to be of minimal impact as the detected levels were similar to water controls. When the qPCR findings were put against the endoscopic findings, screenees with HGD and CRC had a higher load of total universal 16S. With respect to specific microbial features, no relation was found between numbers of specific bacteria and colonoscopy findings relative to total 16S, except that numbers of E. coli were reduced in patients with tubular and villous adenoma. With regard to location of the lesion, no differences were found between a lesion in the distal or proximal colon and number of faecal bacteria observed.
E. coli and Bacteroidetes species have been suggested to promote inflammation, driving the colorectal epithelium to a carcinogenic state.18,20,21 This state of inflammation and dysbiosis gives room for opportunistic bacteria, such as F. nucleatum, to further induce carcinogenesis, whereas anti-inflammatory bacteria such as F. prausnitzii may be ‘crowded out’.4,21 Consequently various bacteria take part in the process of carcinogenesis, with many of these bacteria being variably present during carcinogenesis.4 This could explain why lower concentrations of E. coli were found in tubular and villous adenomas, although screenees with normal colonoscopy and those with CRC had similar concentrations. Our findings did not support a role for E. coli and Bacteroidetes as additional biomarkers in FIT samples to identify FIT-positive screenees at risk of carrying advanced adenomas. Previous studies have suggested a role for F. nucleatum in CRC, in particular, the detection of sessile serrated lesions, with the mucus cap on these lesions suggested as a cause for the high levels of F. nucleatum.19 Additional detection of sessile serrated lesions would be especially valuable in FIT screening, as FIT is known to have a poor sensitivity for these lesions.27 However, our results did not show any association between F. nucleatum and hyperplastic polyps or serrated lesions compared with other neoplasia or a normal colon (p = 0.82; data not shown). This could be because F. nucleatum is not sensitive enough by itself as a biomarker in a screening setting due to overabundance in healthy subjects.28
We found that faecal microbial DNA remained stable over six days, which is in line with findings from a previous study comparing different collection methods of faeces, including FIT.29 Furthermore, one other study has looked specifically at isolating bacterial DNA from FIT samples.30 Its findings are in line with ours showing that faecal material contained in FIT sampling is sufficient to perform microbiota characterization and is representative of bacterial findings in a full stool sample.
Most other studies regarding the role of the microbiome in colorectal carcinogenesis have looked specifically at the microbiome at and around the tumour-site and it is conceivable that a faecal sample obtained by FIT is not representative of onsite mucosal dysbiosis.31,32 However, microbiota on mucosa retrieved during colonoscopy or surgery could be influenced by the bowel preparation that all patients undergo prior to the intervention. Furthermore, most of these studies had a case–control design and were thus prone to overestimation of diagnostic performance.33 To date, a small number of studies have looked at the faecal microbiome in FIT screenees, showing a difference in overall faecal microbiome between healthy patients and patients with colorectal adenomas.34,35 Two of these studies analysed the microbiota in full stool samples and not in the FIT samples themselves, making comparison with our data complex. However, a full stool sample may ask for a considerable effort from the screenee, making the design undesirable in a screening setting as it might hamper participation rates. Baxter et al. used 16S sequencing of stool samples to identify a microbiota-based model to predict colonic lesions and subsequently showed that this model also worked on DNA isolated from FITs indicating that FIT fluid may indeed provide additional biomarkers for CRC detection.30,35 While 16S rRNA gene sequencing data may allow a more in depth analysis of the microbiome as seen in patients with different degrees of intestinal malignancy, its use for diagnostic purposes of individual patients may be cumbersome. Furthermore, in our pilot study, it was not possible to retrieve detailed taxonomic information on species level as, with a limited sequencing depth, 16S rRNA gene sequencing did not pick up specific markers at species level such as F. nucleatum. For implementation in diagnostic laboratories, it might be preferable to find microbial biomarkers which may be identified in FIT by readily available PCR techniques rather than 16S or metagenomic sequencing efforts. However, with current literature not agreeing on the actual absence or presence of bacteria featured in colonic lesions, identifying the right biomarker to investigate is key.
The strengths of our study include the fact that all FIT samples were retrieved from a population-based CRC screening cohort, consisting of average risk screenees, resulting in a high external validity. Also, as gut microbiota were measured in FIT samples, no additional stool samples were required from the participants. It is the first study comparing microbial features between previously untreated patients across the adenoma-to-carcinoma range, including all the different stages of malignancy. Furthermore, we included FIT samples that tested positive for occult blood for both lesions and non-lesions, precluding the possibility of a bias introduced by potential microbe–blood interactions.36 In order to appreciate our results, some limitations also need to be addressed. At present, the exact pathway and role of the gut micriobiota are unknown. Since no known common suspects have been consistently identified, we have selected four bacteria for this qPCR study, but the inclusion of other bacteria could be of more interest in the future. As only FIT-positive subjects underwent colonoscopy, it was not possible to evaluate prime indicators of diagnostic performance, including sensitivity, specificity and the area under the receiver-operating curve. However, we considered analysis of only positive FITs justified as, in the end, this is the population for whom identificational biomarkers would be of benefit to avoid unnecessary endoscopic screening. Furthermore, we used the most advanced lesion detected during colonoscopy, while screenees sometimes have more than one lesion. The presence of multiple lesions could theoretically lead to our findings being an underestimation of the relation between faecal microbiome and colonic neoplasia, although for screening purposes subjects at highest risk (i.e. with advanced neoplasia) are of most interest. Another important limitation is the small sample size and the absence of negative controls in the 16S rRNA gene sequencing pilot. PCR analysis indicates that, while orders of multitude lower, contaminants will be present during isolation and amplification of DNA from FIT fluids. Although the markers of choice were detectable in the pooled samples, which shows their feasibility to use for qPCR purposes, future studies should incorporate negative controls to confirm the detection of biological relevant signals and to control for bacterial contamination.
In conclusion, our results illustrate that the gut microbial markers can be stably measured in FIT samples in CRC screening, with a higher total bacterial load for CRC and high-grade dysplasia. The need to increase FIT sensitivity, especially for advanced adenomas, remains of evident importance and further studies should be conducted to determine the role of microbiota, and preferably specific biomarkers, in FIT.
Supplemental Material
Supplemental material, UEG890732 Supplemental Material for First steps towards combining faecal immunochemical testing with the gut microbiome in colorectal cancer screening by Esmée J Grobbee, Suk Yee Lam, Gwenny M Fuhler, Blerdi Blakaj, Sergey R Konstantinov, Marco J Bruno, Maikel P Peppelenbosch, Ernst J Kuipers and Manon CW Spaander in United European Gastroenterology Journal
Acknowledgements
We would like to acknowledge Erasmus MC MRACE for the funding of this study. The funding source had no involvement in the study design, collection of data, analysis and interpretation of the data, in the writing of the report, nor in the decision to submit the paper.
Author contribution
Conceived idea for the study: EJG, EJK, GMF, MPP, MCWS; EJG and GMF designed and conceptualized the study; supervised execution of the study was done by MCWS and MPP; samples were analysed by BB, SRK and GMF. Responsible for data entry were EJG, SYL, BB, SRK and GMF; analysis and interpretation of data was done by EJG, SYL and GMF. EJG drafted the manuscript. SYL, GMF, BB, SRK, MJB, MPP, EJK, MCWS provided critical revision of the manuscript for important intellectual content.
Declaration of conflicting interests
The authors declare that there is no conflict of interest.
Ethics approval
The study was approved by the Medical Ethical Committee of the Erasmus MC, Rotterdam (reference number: MEC-2014-212) on 6 May 2014. The study protocol conforms to the ethical guidelines of the 1975 Declaration of Helsinki as reflected in a prior approval by the institution’s human research committee.
Informed consent
All participants gave written informed consent to participate in CRC screening.
Funding
This work was supported by Erasmus MC MRACE (a population-based study on F. nucleatum and CRC) (grant no 105565).
ORCID iD
Esmée J Grobbee https://orcid.org/0000-0002-2943-174X
References
- 1.Kuipers EJ, Grady WM, Lieberman D, et al. Colorectal cancer. Nat Rev Dis Primers 2015; 1: 15065. [DOI] [PMC free article] [PubMed]
- 2.Narayanan V, Peppelenbosch MP, Konstantinov SR. Human fecal microbiome-based biomarkers for colorectal cancer. Cancer Prev Res (Phila) 2014; 7: 1108–1111. [DOI] [PubMed] [Google Scholar]
- 3.Gagniere J, Raisch J, Veziant J, et al. Gut microbiota imbalance and colorectal cancer. World J Gastroenterol 2016; 22: 501–518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tjalsma H, Boleij A, Marchesi JR, et al. A bacterial driver-passenger model for colorectal cancer: Beyond the usual suspects. Nat Rev Microbiol 2012; 10: 575–582. [DOI] [PubMed] [Google Scholar]
- 5.Zur Hausen H. The search for infectious causes of human cancers: Where and why (Nobel lecture). Angew Chem Int Ed Engl 2009; 48: 5798–5808. [DOI] [PubMed] [Google Scholar]
- 6.Klein RS, Recco RA, Catalano MT, et al. Association of Streptococcus bovis with carcinoma of the colon. N Engl J Med 1977; 297: 800–802. [DOI] [PubMed] [Google Scholar]
- 7.Kuipers EJ, de Jong A. Gastrointestinal disorders and Streptococcus bovis bacteremia. Ned Tijdschr Geneeskd 1990; 134: 1337–1339. [PubMed] [Google Scholar]
- 8.Sears CL, Garrett WS. Microbes, microbiota, and colon cancer. Cell Host Microbe 2014; 15: 317–328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bhatt AP, Redinbo MR, Bultman SJ. The role of the microbiome in cancer development and therapy. CA Cancer J Clin 2017; 67: 326–344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Flanagan L, Schmid J, Ebert M, et al. Fusobacterium nucleatum associates with stages of colorectal neoplasia development, colorectal cancer and disease outcome. Eur J Clin Microbiol Infect Dis 2014; 33: 1381–1390. [DOI] [PubMed] [Google Scholar]
- 11.Marchesi JR, Dutilh BE, Hall N, et al. Towards the human colorectal cancer microbiome. PLoS One 2011; 6: e20447–e20447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Schreuders EH, Ruco A, Rabeneck L, et al. Colorectal cancer screening: A global overview of existing programmes. Gut 2015; 64: 1637–1649. [DOI] [PubMed] [Google Scholar]
- 13.Van Doorn SC, Stegeman I, Stroobants AK, et al. Fecal immunochemical testing results and characteristics of colonic lesions. Endoscopy 2015; 47: 1011–1017. [DOI] [PubMed] [Google Scholar]
- 14.Lee JK, Liles EG, Bent S, et al. Accuracy of fecal immunochemical tests for colorectal cancer: Systematic review and meta-analysis. Ann Intern Med 2014; 160: 171–171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Imperiale TF, Gruber RN, Stump TE, et al. Performance characteristics of fecal immunochemical tests for colorectal cancer and advanced adenomatous polyps: A systematic review and meta-analysis. Ann Intern Med 2019; 170: 319–329. [DOI] [PubMed] [Google Scholar]
- 16.Gudra D, Shoaie S, Fridmanis D, et al. A widely used sampling device in colorectal cancer screening programmes allows for large-scale microbiome studies. Gut 2019; 68: 1723–1725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lopez-Siles M, Martinez-Medina M, Suris-Valls R, et al. Changes in the abundance of Faecalibacterium prausnitzii phylogroups I and II in the intestinal mucosa of inflammatory bowel disease and patients with colorectal cancer. Inflamm Bowel Dis 2016; 22: 28–41. [DOI] [PubMed] [Google Scholar]
- 18.Liang Q, Chiu J, Chen Y, et al. Fecal bacteria act as novel biomarkers for noninvasive diagnosis of colorectal cancer. Clin Cancer Res 2017; 23: 2061–2070. [DOI] [PubMed] [Google Scholar]
- 19.Ito M, Kanno S, Nosho K, et al. Association of Fusobacterium nucleatum with clinical and molecular features in colorectal serrated pathway. Int J Cancer 2015; 137: 1258–1268. [DOI] [PubMed] [Google Scholar]
- 20.Leung A, Tsoi H, Yu J. Fusobacterium and Escherichia: Models of colorectal cancer driven by microbiota and the utility of microbiota in colorectal cancer screening. Expert Rev Gastroenterol Hepatol 2015; 9: 651–657. [DOI] [PubMed] [Google Scholar]
- 21.Sears CL, Pardoll DM. Perspective: Alpha-bugs, their microbial partners, and the link to colon cancer. J Infect Dis 2011; 203: 306–311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yu J, Feng Q, Wong SH, et al. Metagenomic analysis of faecal microbiome as a tool towards targeted non-invasive biomarkers for colorectal cancer. Gut 2017; 66: 70–78. [DOI] [PubMed] [Google Scholar]
- 23.Wirbel J, Pyl PT, Kartal E, et al. Meta-analysis of fecal metagenomes reveals global microbial signatures that are specific for colorectal cancer. Nat Med 2019; 25: 679–689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Balamurugan R, Rajendiran E, George S, et al. Real-time polymerase chain reaction quantification of specific butyrate-producing bacteria, Desulfovibrio and Enterococcus faecalis in the feces of patients with colorectal cancer. J Gastroenterol Hepatol 2008; 23: 1298–1303. [DOI] [PubMed] [Google Scholar]
- 25.Grobbee EJ, van der Vlugt M, van Vuuren AJ, et al. A randomised comparison of two faecal immunochemical tests in population-based colorectal cancer screening. Gut 2017; 66: 1975–1982. [DOI] [PubMed] [Google Scholar]
- 26.Schlemper RJ, Riddell RH, Kato Y, et al. The Vienna classification of gastrointestinal epithelial neoplasia. Gut 2000; 47: 251–255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Imperiale TF, Ransohoff DF, Itzkowitz SH, et al. Multitarget stool DNA testing for colorectal-cancer screening. N Engl J Med 2014; 370: 1287–1297. [DOI] [PubMed] [Google Scholar]
- 28.Kostic AD, Chun E, Robertson L, et al. Fusobacterium nucleatum potentiates intestinal tumorigenesis and modulates the tumor-immune microenvironment. Cell Host Microbe 2013; 14: 207–215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Vogtmann E, Chen J, Amir A, et al. Comparison of collection methods for fecal samples in microbiome studies. Am J Epidemiol 2017; 185: 115–123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Baxter NT, Koumpouras CC, Rogers MA, et al. DNA from fecal immunochemical test can replace stool for detection of colonic lesions using a microbiota-based model. Microbiome 2016; 4: 59–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zoetendal EG, von Wright A, Vilpponen-Salmela T, et al. Mucosa-associated bacteria in the human gastrointestinal tract are uniformly distributed along the colon and differ from the community recovered from feces. Appl Environ Microbiol 2002; 68: 3401–3407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chen W, Liu F, Ling Z, et al. Human intestinal lumen and mucosa-associated microbiota in patients with colorectal cancer. PLoS One 2012; 7: e39743–e39743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Deeks JJ, Bossuyt PM and Gatsonis C. Cochrane handbook for systematic reviews of diagnostic test accuracy, version 1.0.0. The Cochrane Collaboration, 2009, http://srdta.cochrane.org/.
- 34.Goedert JJ, Gong Y, Hua X, et al. Fecal microbiota characteristics of patients with colorectal adenoma detected by screening: A population-based study. EBioMedicine 2015; 2: 597–603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Baxter NT, Ruffin MTt, Rogers MA, et al. Microbiota-based model improves the sensitivity of fecal immunochemical test for detecting colonic lesions. Genome Med 2016; 8: 37–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Schroedl W, Kleessen B, Jaekel L, et al. Influence of the gut microbiota on blood acute-phase proteins. Scand J Immunol 2014; 79: 299–304. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental material, UEG890732 Supplemental Material for First steps towards combining faecal immunochemical testing with the gut microbiome in colorectal cancer screening by Esmée J Grobbee, Suk Yee Lam, Gwenny M Fuhler, Blerdi Blakaj, Sergey R Konstantinov, Marco J Bruno, Maikel P Peppelenbosch, Ernst J Kuipers and Manon CW Spaander in United European Gastroenterology Journal