Summary
Trained immunity confers a sustained augmented response of innate immune cells to a secondary challenge, via a process dependent on metabolic and transcriptional reprogramming. Because of its previous associations with metabolic and transcriptional memory, as well as the importance of H3 histone lysine 4 monomethylation (H3K4me1) to innate immune memory, we hypothesize that the Set7 methyltransferase has an important role in trained immunity induced by β-glucan. Using pharmacological studies of human primary monocytes, we identify trained immunity-specific immunometabolic pathways regulated by Set7, including a previously unreported H3K4me1-dependent plasticity in the induction of oxidative phosphorylation. Recapitulation of β-glucan training in vivo additionally identifies Set7-dependent changes in gene expression previously associated with the modulation of myelopoiesis progenitors in trained immunity. By revealing Set7 as a key regulator of trained immunity, these findings provide mechanistic insight into sustained metabolic changes and underscore the importance of characterizing regulatory circuits of innate immune memory.
Keywords: trained immunity, Set7, methylation, β-glucan, oxidative phosphorylation, immunometabolism, inflammation, monocyte, macrophage
Graphical Abstract
Highlights
-
•
Set7 regulates enhanced cytokine production in trained immunity in vitro
-
•
Set7 knockout mice are unable to mount trained immunity against endotoxin challenge
-
•
Set7 modulates cellular respiration in β-glucan-trained macrophages
-
•
Set7-dependent histone methylation regulates MDH2 and SDHB in trained cells
Using a combination of pharmacological and genetic approaches, Keating et al. show that the Set7 methyltransferase is a regulator of trained immunity induced by β-glucan. Activation of Set7 increases oxidative phosphorylation in trained cells via histone lysine methylation at gene enhancers of key enzymes of the TCA cycle.
Introduction
A series of recent discoveries has uncovered how cells of the innate immune system such as monocytes and macrophages undergo functional reprogramming to mount a de facto immune memory of an infectious or inflammatory injury by a process called trained immunity, which facilitates augmented responses to subsequent pathogenic encounters (Netea et al., 2020). In the context of infections or vaccination, trained immunity provides beneficial heterologous effects by the enhanced cytokine response to stimulation with non-related pathogens. Prototypical stimuli of trained immunity include the fungal cell wall component β-glucan (Quintin et al., 2012) and the bacillus Calmette-Guérin (BCG) vaccine (Kleinnijenhuis et al., 2012). Recent attention has also turned to endogenous drivers of inflammation as inducers of trained immunity (Bekkering et al., 2014, van der Valk et al., 2016). These stimuli shape innate immunological memories by reprogramming metabolic and transcriptional profiles (Arts et al., 2016a, Cheng et al., 2014).
Posttranslational methylation of proteins conveys information to cellular pathways, including those that regulate gene expression. In vitro experiments with pan-methyltransferase inhibitors revealed the pivotal importance of this chemical modification for trained macrophages (Cheng et al., 2014, Quintin et al., 2012). Changes in histone lysine methyl modifications (H3 histones monomethylated [H3K4me1] or trimethylated [H3K4me3] at lysine 4) underlie β-glucan-induced trained immunity (Novakovic et al., 2016). Signaling factors derived from local tissue environments play key roles in determining macrophage fate, and evidence points to the role of enhancer elements in shaping specialized macrophage populations (Denisenko et al., 2017, Gosselin et al., 2014). H3K4me1 is a chromatin signature of enhancers (Heintzman et al., 2007). This modification was shown to persist at decommissioned distal elements, indicating that H3K4me1 provides a mechanism for epigenetic memory in trained immunity in macrophages (Saeed et al., 2014). Despite this, the mechanisms linking immunological signals induced by microbial stimuli or vaccines to chromatin-dependent changes in trained immunity are unclear. Moreover, the identities of the chromatin-modifying enzymes critical to these processes remain obscure.
One enzyme that writes the H3K4me1 modification to transcriptionally activating or poised genomic regions is the Set7 lysine methyltransferase (Wang et al., 2001) (also called Set9 [Nishioka et al., 2002], Set7/9 [Tamura et al., 2018], or KMT7 [Allis et al., 2007], and encoded by SETD7). Set7 writes a persistent H3K4me1 signature pertaining to vascular endothelial inflammatory signaling (Brasacchio et al., 2009). The importance of Set7 for mediating H3K4me1 signatures at enhancers associated with endothelial gene expression was demonstrated using an unbiased epigenome-wide approach (Keating et al., 2014). Although Set7 has not been studied in the specific context of trained immunity, our previous analysis of macrophages trained with β-glucan identified elevated levels of SETD7 expression (Quintin et al., 2012).
The current study explored the role of Set7 in β-glucan-induced trained immunity. Using genetic and pharmacological approaches, we demonstrate that Set7 is critical for the induction of trained immunity in vitro and in vivo. We identify a role for Set7 in immunometabolic pathways, including the induction of oxidative phosphorylation (OXPHOS). Characterization of epigenetic networks is key to a deeper understanding of regulatory mechanisms supporting trained immunity, and could identify strategies to modulate pro-inflammatory circuits of the innate immune system.
Results
Set7 Expression and Activity Are Increased in Human Primary Monocytes and Macrophages Stimulated with β-Glucan
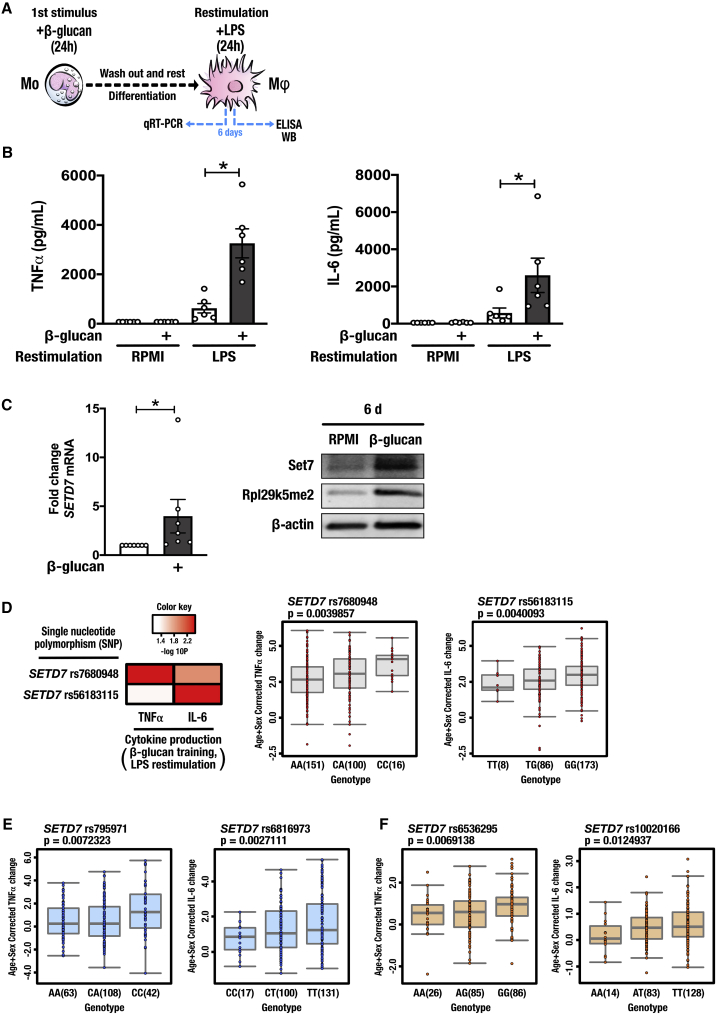
To investigate the role of Set7, we adopted a previously described in vitro model of trained immunity using the fungal cell wall component β-glucan (Cheng et al., 2014). Adherent human primary monocytes were incubated with culture medium or β-glucan (1 μg/mL) for 24 h. Cells were washed and incubated in normal culture conditions for a further 5 days, during which time they differentiated into macrophages. On day 6, the cells were restimulated with the Toll-like receptor 4 ligand lipopolysaccharide (LPS) (10 ng/mL) for 24 h and pro-inflammatory cytokine production was measured (Figure 1A). Tumor necrosis factor alpha (TNFα) and IL-6 were measured as functional readouts of trained immunity (Arts et al., 2016a, Cheng et al., 2014). Cells stimulated with β-glucan exhibited enhanced TNFα and IL-6 production following LPS restimulation (Figure 1B). We validated our previous transcriptome data showing that SETD7 mRNA expression was significantly increased on day 6 of the in vitro training protocol in macrophages trained with β-glucan. Currently, there is no gold standard test for Set7 activity in primary cells. However, a recent study identified the ribosomal protein Rpl29, a component of the 60S ribosomal subunit, as a non-histone methylation substrate of Set7 (Hamidi et al., 2018), and dimethylated Rpl29 (Rpl29k5me2) was shown to serve as a reliable biomarker for Set7 activity in cancer cells. We observed that Rpl29k5me2 levels correlated with Set7 protein expression in macrophages trained with β-glucan (Figure 1C).
Figure 1.
Set7 Is Associated with Trained Immunity Induced by β-Glucan
(A) Graphical outline of in vitro training methods. Adherent monocytes (Mo) were stimulated with 1 μg/mL β-glucan or standard culture medium (RPMI) for 24 h (first stimulus), allowed to differentiate to macrophages (Mϕ) for 5 days, and restimulated for 24 h with LPS or RPMI on day 6.
(B) Production of pro-inflammatory cytokines TNFα and IL-6 by trained macrophages following restimulation (n = 6 healthy volunteers per group).
(C) Expression of SETD7 mRNA prior to restimulation (6 d) (n = 7 healthy volunteers per group). Representative western blot analysis of differentiated macrophages performed on day 6, prior to restimulation. Set7 encoded by the SETD7 gene; Rpl29k5me2 as a marker for Set7 activity; β-actin was used as loading control.
(D) Single-nucleotide polymorphisms (SNPs) within SETD7 suggestively associated with trained responses to β-glucan in peripheral blood mononuclear cells (n = 267). Age- and sex-corrected TNFα and IL-6 changes are shown as boxplots for rs7680948 and rs56183115, respectively.
(E) SNPs near SETD7 suggestively associated with trained responses to the bacillus Calmette-Guérin vaccine in peripheral blood mononuclear cells. Age- and sex-corrected TNFα and IL-6 changes are shown as boxplots for rs795971 (n = 213) and rs6816973 (n = 248), respectively.
(F) SNPs near SETD7 suggestively associated with trained responses to the oxidized low-density lipoprotein in peripheral blood mononuclear cells. Age- and sex-corrected TNFα and IL-6 changes are shown as boxplots for rs6536295 (n = 197) and rs10020166 (n = 225), respectively.
Data are represented as mean ± SEM. ∗p < 0.05, Wilcoxon signed-rank test.
To further investigate the association of Set7 to trained immunity, we conducted a genetic study of peripheral blood mononuclear cells (PBMCs) isolated from 267 healthy volunteers of Western European ancestry. Adherent PBMCs from all volunteers were incubated with trained with culture medium or β-glucan (1 μg/mL) for 24 h as described above. We tested for associations among common single-nucleotide polymorphisms (SNPs) and variation in the magnitude of β-glucan-trained cytokine responses of individual subjects and identified two SNPs suggestively associated (p < 9.99 × 10−3) with adaptive changes in cytokine production mapped within SETD7. Intronic variants rs7680948 (located in intron 4 of SETD7 [p < 0.004]) and rs56183115 (located in intron 3 of SETD7 [p = 0.004]) were associated with the potentiation of TNFα and IL-6 production, respectively, upon induction of trained immunity by β-glucan (Figure 1D). To investigate Set7 in trained immunity induced by other stimuli, we tested for associations among SNPs near SETD7 and variation in the magnitude of cytokine responses of individuals trained with BCG and oxidized low-density lipoprotein (oxLDL). We identified numerous SNPs suggestively associated (p < 9.99 × 10−3) with adaptive changes in pro-inflammatory cytokine production mapped within 250 kb of SETD7. Variants rs795971 (located approximately 250 kb upstream of SETD7 [p = 0.007]) and rs6816973 (located approximately 160 kb upstream of SETD7 [p < 0.003]) were associated with the potentiation of TNFα and IL-6 production, respectively, upon induction of trained immunity by BCG (Figure 1E). In addition, we identified that variants rs6536295 (located approximately 3 kb upstream of SETD7 [p < 0.007]) and rs10020166 (located approximately 15 kb upstream of SETD7 [p = 0.01]) were associated with the production of TNFα and IL-6, respectively, upon induction of trained immunity by oxLDL (Figure 1F).
Pharmacological Inhibition of Set7 Attenuates β-Glucan-Induced Trained Immunity In Vitro
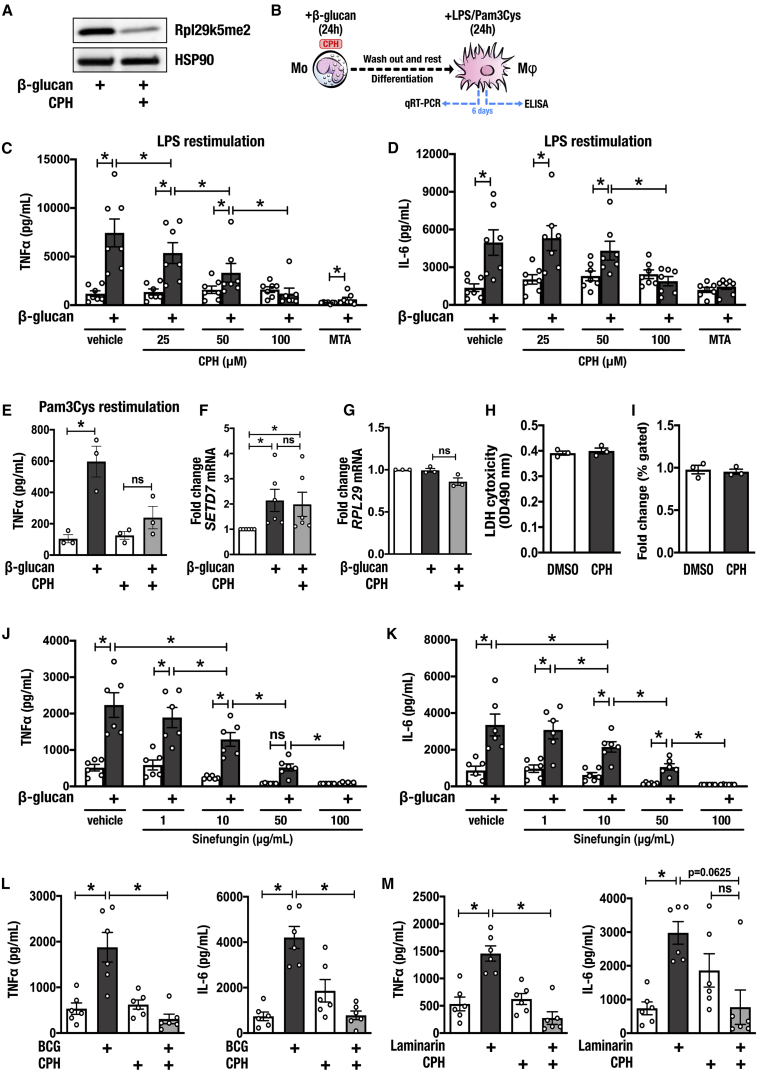
Recent studies demonstrated specific inhibition of Set7 activity by cyproheptadine (CPH) in human breast cancer cells (Takemoto et al., 2016). To test whether CPH similarly inhibits Set7 in primary cells, we measured Rpl29K5me2 levels of macrophages trained with β-glucan and observed a strong reduction of this posttranslational modification following 24-h incubation with 100 μM CPH (Figure 2A). To characterize the effects of CPH on trained immunity, we conducted in vitro training experiments on monocytes exposed to a range of CPH concentrations (Figure 2B). When co-incubated with β-glucan for only the first 24 h of the training protocol, CPH dose-dependently attenuated the heightened responsiveness of trained cells to restimulation with LPS (Figures 2C and 2D). This effect was most pronounced at the highest concentration of CPH tested (100 μM); however, a significant reduction in TNFα production was observed after co-incubation with 25 μM CPH.
Figure 2.
Pharmacological Inhibition of Set7 Dose-Dependently Attenuates the Pro-inflammatory Cytokine Response of Trained Immunity In Vitro
(A) Western blot analysis of β-glucan-trained macrophages incubated with cyproheptadine (CPH) for 24 h. HSP90 was used as a loading control.
(B) Graphical overview of in vitro training methods. Adherent monocytes (Mo) were stimulated with β-glucan or RPMI culture medium for 24 h in the presence of CPH or DMSO vehicle control, allowed to differentiate to macrophages (Mϕ), and restimulated for 24 h with LPS, Pam3Cys, or RPMI on day 6.
(C and D) Production of (C) TNFα and (D) IL-6 by β-glucan-trained macrophages incubated with CPH or 5′-methylthioadenosine (MTA) for the first 24 h of in vitro training and restimulated with LPS (n = 7 healthy volunteers).
(E) Production of TNFα by β-glucan-trained macrophages incubated with CPH for the first 24 h of in vitro training and restimulated with Pam3Cys (n = 3 healthy volunteers).
(F) Expression of SETD7 mRNA on day 6 by cells trained with β-glucan in the presence of 100 μM CPH (n = 6 healthy volunteers; open bar represents DMSO vehicle control).
(G) Expression of RPL29 mRNA at 24 h by cells trained with β-glucan in the presence of 100 μM CPH (n = 3 healthy volunteers; open bar represents DMSO vehicle control).
(H) Analysis of lactate dehydrogenase (LDH) as a measure of cytotoxicity in cells incubated with 100 μM CPH for 24 h (n = 3 healthy volunteers).
(I) Analysis of viability and apoptosis with Annexin V and PI staining in cells incubated with 100 μM CPH versus DMSO vehicle controls for 24 h (n = 3 healthy volunteers). Fold change difference between CPH and vehicle controls.
(J and K) Production of (J) TNFα and (K) IL-6 by β-glucan-trained macrophages incubated with sinefungin for the first 24 h of in vitro training and restimulated with LPS (n = 6 healthy volunteers).
(L) Production of TNFα and IL-6 by bacillus Calmette-Guérin (BCG)-trained macrophages incubated with 100 μM CPH for the first 24 h of in vitro training and restimulated with LPS (n = 6 healthy volunteers).
(M) Production of TNFα and IL-6 by laminarin-trained macrophages incubated with 100 μM CPH for the first 24 h of in vitro training and restimulated with LPS (n = 6 healthy volunteers).
Data are represented as mean ± SEM. ∗p < 0.05, Wilcoxon signed-rank test or t test where appropriate.
See also Figures S1 and S2.
Similar inhibition of β-glucan-induced trained immunity by CPH (100 μM) was observed when the cells were restimulated with the Toll-like receptor 2 agonist Pam3Cys (10 μg/mL) (Figure 2E). Exposure to this concentration of CPH did not alter the mRNA expression of SETD7 (measured on day 6, prior to restimulation), indicating that the inhibitory effect on trained immunity occurred at the level of Set7 activity (Figure 2F, open bar represents DMSO vehicle controls). Similarly, RPL29 mRNA expression was unaffected by 100 μM CPH (Figure 2G, open bar represents DMSO vehicle controls). The attenuating effect of CPH on cytokine production was not due to cytotoxicity of the compound (Figure 2H) or induction of apoptosis (Figure 2I). We excluded the possibility that CPH inhibits trained immunity via its antihistamine properties by performing experiments using an alternative antihistamine inhibitor, diphenhydramide, which did not alter trained immunity induced by β-glucan (Figure S1).
To validate the observation of Set7-dependent regulation of cytokine production, we also tested the Set7 inhibitor sinefungin (Sasaki et al., 2016). Sinefungin dose-dependently inhibited the heightened production of TNFα and IL-6 by trained cells following restimulation with LPS (Figures 2J and 2K). To test the broader role of Set7 as a key regulator of trained immunity, we assessed the effects of CPH on trained cytokine production by other compounds previously shown to induce trained immunity. Indeed, incubation with 100 μM CPH also inhibited the induction of trained immunity by BCG (Figure 2L). Similarly, the dectin-1 ligand laminarin was previously demonstrated to induce trained immunity (Petit et al., 2019). Here, we show that the augmented TNFα production exhibited by macrophages trained with laminarin was inhibited by CPH. A similar trend was observed for IL-6 production, with no difference observed between cells that were incubated with laminarin and CPH, or laminarin alone (Figure 2M).
To understand the mechanism of Set7-dependent cytokine production in trained immunity, we assessed the TNF and IL6 promoters by chromatin immunoprecipitation (ChIP) on day 6. We did not observe significant increases in H3K4me1 enrichment at either promoter in cells trained with β-glucan. A similar pattern of H3K4me1 enrichment was observed for cells trained in the presence of 100 μM CPH (Figure S2), suggesting that Set7 does not directly regulate TNF and IL6 expression via promoter histone methylation.
Together, these findings identify an important role for Set7 in trained immunity in vitro.
Set7 Regulates Trained Immunity In Vivo
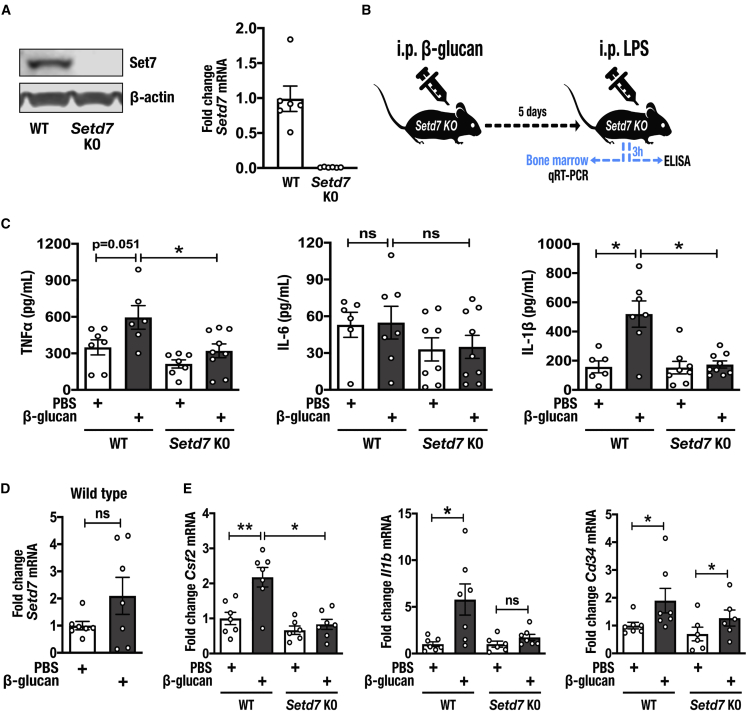
Using murine models, we and others (Cheng et al., 2014, Garcia-Valtanen et al., 2017) have described the specific contribution of β-glucan to the activation of trained immunity in vivo. Wild-type mice that received β-glucan injections exhibited enhanced cytokine production by innate immune cells in response to a secondary challenge or infection (Arts et al., 2016a). We adopted a similar approach to test the role of Set7 in trained immunity in vivo. We generated a Set7 constitutive knockout (Setd7 KO) mouse model by the deletion of Setd7 exon 2 region (unpublished data). Deletion of exon 2, which encodes the first MORN (membrane occupation and recognition nexus) repeat, results in a frameshift and inactivation of Set7. Western blot and gene expression analyses confirmed that Set7 was absent in bone marrow (BM) of the homozygous Setd7 KO mice (Figure 3A). Wild-type and Setd7 KO mice were systemically administered a single intraperitoneal 1-mg dose of β-glucan as described previously (Cheng et al., 2014). Control mice were injected with endotoxin-free phosphate-buffered saline (PBS). Five days after β-glucan administration, the mice were challenged with intraperitoneal injections of 10 μg of LPS, and after 3 h the serum levels of cytokines were quantified (Figure 3B). Contrasting the augmented pro-inflammatory cytokine production in wild-type mice administered β-glucan, mice lacking functional Set7 were unable to mount trained immunity against endotoxin challenge with regard to TNFα and IL-1β production. However, the effects of Set7 deletion on the trained production of IL-6 in vivo are less clear, because IL-6 production was already at high levels in wild-type mice that received PBS (Figure 3C).
Figure 3.
Set7 Regulates Trained Immunity Induced by β-Glucan In Vivo
(A) Representative western blot of Set7 protein expression in the bone marrow of wild-type (WT) and Setd7 KO mice. β-Actin was used as loading control. Expression of Setd7 mRNA in the bone marrow of WT and Setd7 KO mice (n = 7 mice per group).
(B) Schematic overview of in vivo induction of trained immunity by β-glucan.
(C) Plasma levels of TNFα, IL-6, and IL-1β in WT and Setd7 KO mice trained with PBS or β-glucan on day 1 and administered LPS on day 6 (n = 6–9 mice per group).
(D) Day 6 analysis of Setd7 mRNA expression in the bone marrow of WT mice administered PBS or β-glucan (n = 7 mice per group).
(E) Bone marrow mRNA expression of Csf2, Il1b, and Cd34 in WT and Setd7 KO mice trained with PBS or β-glucan on day 1 and administered LPS on day 6 (n = 6–7 mice per group).
Data are represented as mean ± SEM. ∗p < 0.05, ∗∗p < 0.01, Mann-Whitney test.
The modulation of myeloid progenitors is an integral component of trained immunity (Christ et al., 2018, Kaufmann et al., 2018, Mitroulis et al., 2018), which can explain the sustained activation of the innate immune system beyond the short life span of circulating myeloid cells. A recent study showed that administration of β-glucan to mice resulted in the expansion and polarization of hematopoietic stem and progenitor cells (HSPCs) toward myelopoiesis, which was associated with elevated signaling by innate immune mediators such as IL-1β and granulocyte-macrophage colony-stimulating factor (GM-CSF), as well as changes in lipid and glucose metabolism (Mitroulis et al., 2018). To investigate the role of Set7 in these adaptations, we analyzed Setd7 mRNA in the BM of wild-type mice and observed a trend to increase expression in mice administered β-glucan (Figure 3D). We replicated key findings of the previous study (Mitroulis et al., 2018), albeit in whole bone marrow, and found that β-glucan-dependent transcriptional induction of Csf2 (GM-CSF) and Il1b was significantly reduced in Setd7 KO mice. We also observed a reduction in the expression of the surrogate marker for HSPCs Cd34 in Set7 null mice; however, the expression of this gene remained elevated relative to Set7-null mice that received PBS injections (Figure 3E). These data demonstrate that Set7 regulates the in vivo pro-inflammatory cytokine response to induction of trained immunity by β-glucan and indicate an important role in hematopoietic adaptations that support the sustained phenotype.
Set7 Regulates Key Metabolic Changes in Macrophages Trained with β-Glucan
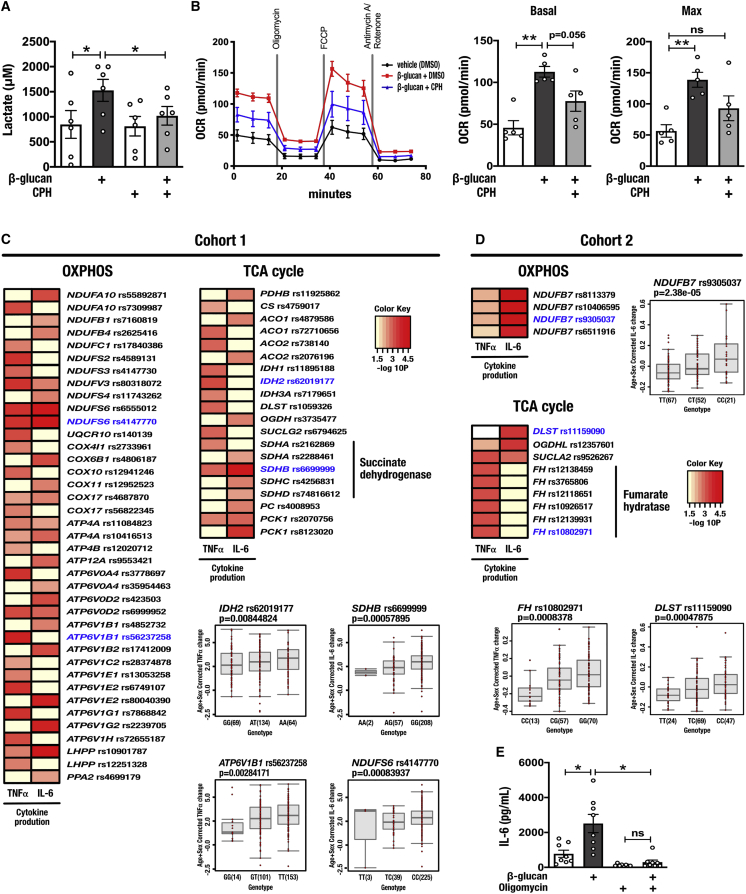
Trained immunity induced by β-glucan or BCG is characterized by metabolic reprogramming including increased glycolysis and intracellular accumulation of fumarate and mevalonate (Arts et al., 2016a, Bekkering et al., 2018). To explore the potential role of Set7 in the hallmark glycolytic metabolism of trained immunity, we measured extracellular lactate levels in day 6 macrophages trained with β-glucan. We observed a significant increase in these levels in β-glucan-trained cells that was abolished by coincubation with 100 μM CPH for the first 24 h of in vitro training (Figure 4A). In addition to changes in glycolysis, previous studies found that β-glucan training is accompanied by the repression of OXPHOS. In contrast to those findings (Cheng et al., 2014), we observed increased oxygen consumption at day 6 by cells trained with 1 μg/mL β-glucan (Figure 4B). By performing parallel respirometry experiments with the Seahorse XF Extracellular Flux Analyzer and the Oxygraph-2k from Oroboros (the instrument used for measuring oxygen consumption by Cheng et al., 2014), we found that the stimulatory dose of β-glucan can explain the discrepancy between the current (increased oxygen consumption) and previously reported findings (decreased oxygen consumption) (Cheng et al., 2014). Specifically, we observed, using the Oxygraph-2k, training with 1 μg/mL β-glucan stimulated oxygen consumption measured on day 6. On the other hand, the stimulatory dose of 10 μg/mL described by Cheng et al. led to an overall reduction of oxygen consumption by trained macrophages (Figures S3A and S3B). To confirm these observations and to rule out donor-specific variation as the cause, we trained cells from the same set of donors with 1 μg/mL β-glucan or 10 μg/mL β-glucan and analyzed them in parallel using the Seahorse system. While both stimulatory doses showed a trend to increase the extracellular acidification rate, training with 1 μg/mL β-glucan increased oxygen consumption, whereas training with 10 μg/mL β-glucan reduced oxygen consumption for each donor (Figures S3C and S3D). Next, we sought to determine whether Set7 was mechanistically involved in the upregulation of OXPHOS by cells trained with β-glucan. Paralleling its attenuating effect on cytokine production, CPH blunted the increase in oxygen consumption induced by β-glucan (Figure 4B; open bars represent DMSO vehicle controls).
Figure 4.
Plasticity in the Induction of Oxidative Phosphorylation Is Important for the Pro-inflammatory Cytokine Production by Cells Trained by β-Glucan
(A) Lactate production by β-glucan-trained macrophages incubated with CPH for the first 24 h of in vitro training (n = 6 healthy volunteers).
(B) Oxygen consumption analysis (seahorse) of macrophages 5 days after incubation with β-glucan and co-incubation with CPH. Basal and maximum oxygen consumption rates (OCR) are indicated (n = 5 healthy volunteers; open bars represent DMSO vehicle controls).
(C and D) Heatmap of the p values of association between SNPs mapped to genes involved in oxidative phosphorylation (OXPHOS) and the tricarboxylic acid (TCA) cycle and the magnitude of cytokine production capacity by PBMCs trained with β-glucan in vitro isolated from (C) 300BCG (cohort 1) and (D) 200FG (cohort 2). The color legend for the heatmap indicates the range of p values from QTL mapping. Boxplots show the genotype-stratified cytokine levels for the OXPHOS and TCA cycle loci (cohort 1, n = 238 healthy volunteers for TNFα, n = 251 healthy volunteers for IL-6; cohort 2, n = 119 healthy individuals).
(E) Production of IL-6 by β-glucan-trained macrophages incubated with 1 μM oligomycin for the first 24 h of in vitro training and restimulated with LPS (n = 8 healthy volunteers).
Data are represented as mean ± SEM. ∗p < 0.05, ∗∗p < 0.01, Wilcoxon signed-rank test or Mann-Whitney test where appropriate.
See also Figure S3.
To understand the significance of this change in oxygen consumption for the induction of trained immunity, we investigated the effect of genetic variation on individual responses to β-glucan. Drawing from our genetic study of PBMCs isolated from 267 healthy volunteers (cohort 1; 300BCG), we tested for associations among common SNPs (minor allele frequency >5%) and variation in the magnitude of β-glucan-trained TNFα and IL-6 responses of individual subjects. Although genome-wide significant cytokine quantitative trait loci (cQTLs) were not observed, we identified numerous SNPs suggestively associated (p < 9.99 × 10−3) with adaptive changes in pro-inflammatory cytokine production mapped within 250 kb of genes related to OXPHOS as well as genes encoding key tricarboxylic acid (TCA) cycle enzymes. Variation in genes encoding isocitrate dehydrogenase enzymes was associated with the potentiation of TNFα production upon training with β-glucan. Similarly, variation in genes that encode subunits of the succinate dehydrogenase (SDH) complex, which oxidizes succinate to fumarate as a key component of the TCA cycle and ubiquinone to ubiquinol as complex II in the mitochondrial electron transport chain, was associated with IL-6 production. Other OXPHOS genes implicated by this study include those encoding various subunits of NADH:ubiquinone oxidoreductase (complex I of mitochondrial electron transport chain) (Figure 4C).
We validated the cytokine potentiating importance of plasticity in OXPHOS using a genetic study of PBMCs isolated from 119 healthy volunteers of Western European ancestry from the 200 Functional Genomics cohort (cohort 2; 200FG) of the Human Functional Genomics Study (https://www.humanfunctionalgenomics.org). In this second cohort, we observed a suggestive association (p < 9.99 × 10−3) between IL-6 production and variation in the gene encoding mitochondrial electron transport chain complex I subunit NDUFB7 (rs9305037 p = 2.38 × 10−5). Variation in fumarate hydratase (FH), which catalyzes the conversion of fumarate to malate in the TCA cycle, was associated with TNFα production in PBMCs trained with β-glucan. On the other hand, variation in dihydrolipoamide s-succinyltransferase (DLST), a component of the TCA cycle oxoglutarate dehydrogenase complex that catalyzes the conversion of 2-oxoglutarate to succinyl-CoA, was associated with variable IL-6 production (Figure 4D). To determine the physiological significance of this metabolic pathway to cytokine production in trained immunity, we inhibited OXPHOS with the FoF1-ATP synthase inhibitor oligomycin during restimulation with LPS, and observed a significant reduction in the IL-6 production capacity of cells trained with β-glucan (Figure 4E). These data collectively demonstrate that increased OXPHOS is necessary for induction of the trained phenotype by β-glucan and indicate a regulatory role for Set7 in this metabolic adaptation.
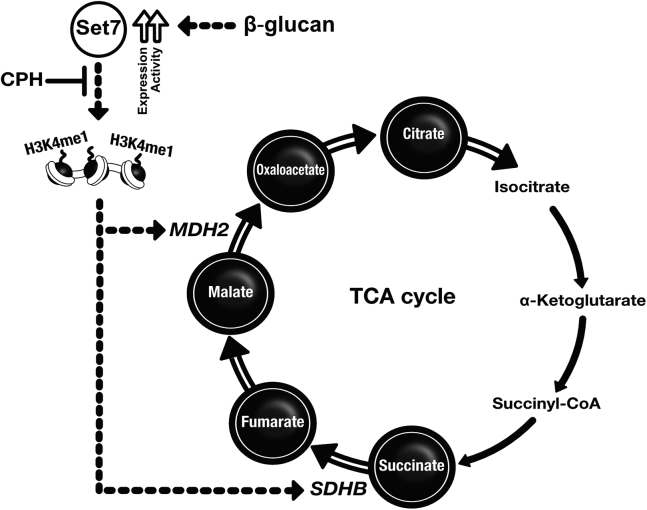
Set7 Regulates TCA Cycle Metabolite Production in Macrophages Trained with β-Glucan
Previous studies reported induction of TCA cycle metabolites succinate, fumarate, and malate in monocytes and macrophages trained with β-glucan (Arts et al., 2016a). We were therefore prompted to investigate the role of Set7 in these metabolic changes. Metabolite measurements revealed small, yet significant (fumarate, malate) differences between naive and trained cells after 24-h incubation with β-glucan. However, on day 6, the intracellular metabolism of β-glucan-trained cells was clearly distinguishable from untrained cells, with major differences observed for succinate, fumarate, malate, oxaloacetate, and citrate. Incubation with 100 μM CPH for the first 24 h of the in vitro training protocol significantly inhibited the production of fumarate, and malate. By contrast, levels of succinate, oxaloacetate, and citrate were largely unaffected by CPH at either time point (Figure 5A; open bars represent DMSO vehicle controls).
Figure 5.
Inhibition of Set7 Regulates Metabolic Changes in Macrophages Trained with β-Glucan
(A) Key TCA cycle metabolite concentrations in primary human monocytes/macrophages measured 24 h and 5 days after incubation with β-glucan and co-incubation with CPH (n = 6 healthy volunteers; open bars represent DMSO vehicle controls).
(B) Schematic overview of the TCA cycle and succinate dehydrogenase (SDH) complex. Enzymes analyzed for gene expression in (C) and (D) are indicated.
(C) Expression analysis of genes encoding enzymes involved in the TCA cycle by primary human monocytes/macrophages measured 24 h and 5 days after incubation with β-glucan and co-incubation with 100 μM CPH (n = 6–8 healthy volunteers; open bars represent DMSO vehicle controls).
(D) Expression analysis of genes encoding SDH subunits in primary human monocytes/macrophages measured 24 h and 5 days after incubation with β-glucan and co-incubation with 100 μM CPH (n = 6–8 healthy volunteers; open bars represent DMSO vehicle controls).
(E) Bone marrow mRNA expression of Mdh2, Sdhb, Fh1, and Suclg1 in WT and Setd7 KO mice trained with PBS or β-glucan on day 1 and administered LPS on day 6 (n = 6–7 mice per group).
Data are represented as mean ± SEM. ∗p < 0.05, ∗∗p < 0.01, Wilcoxon signed-rank test or Mann-Whitney test where appropriate.
To understand mechanisms underlying the immunometabolic modulatory effects of CPH, we measured the expression of genes encoding key enzymes involved in the regulation of the TCA cycle and OXPHOS (Figure 5B). Significant upregulation of SUCLG1, FH, and MDH2 expression immediately following 24 h of stimulation with β-glucan was maintained or further elevated until at least day 6. A similar pattern, although not statistically significant at 24 h, was observed for CS expression. Importantly, incubation with CPH for the initial 24-h training period inhibited the β-glucan-induced activation of FH at the 24-h time point. Contrasting the upregulation of MDH2 by β-glucan, expression of this gene in cells incubated with CPH during the initial 24-h training period was not significantly different from the control group at either time point (Figure 5C; open bars represent DMSO vehicle controls). Key to our observations of CPH-responsive β-glucan-induced fumarate accumulation is the activity of SDH. To explore the role of Set7 in regulating this complex, we measured the expression of genes encoding individual SDH subunits. We observed significant transcriptional upregulation of SDHB, SDHC, and SDHD following 24 h of stimulation with β-glucan. For SDHB and SDHD, the expression was increased on day 6. Incubation with CPH during the initial 24-h training period revealed a tendency to prevent the β-glucan-induced transcriptional activation of SDHB at the 24-h and 6-day time points. In contrast to the upregulation of SDHB by β-glucan, expression of this gene in cells incubated with CPH during the initial 24-h training period was not significantly different from the control group at either time point (Figure 5D; open bars represent DMSO vehicle controls). To determine the importance of Set7 for the control of MDH2 and SDHB in trained immunity in vivo, we analyzed the expression of these genes in BM of wild-type and Setd7 KO mice. Similar to our results for human cells, we observed a significant increase in the expression of Mdh2 and Sdhb in wild-type mice that received β-glucan injections, which were not observed in Setd7 KO mice. In contrast, we did not observe significant changes to Fh1 or Suclg1 expression (Figure 5E). These observations indicate a role for Set7 in sustained metabolic gene changes associated with trained immunity via the persistent activation of MDH2 and SDHB.
Histone Methylation at Distal Enhancers Support Metabolic Reprogramming of Macrophages Trained with β-Glucan
At the level of gene regulation, trained immunity induced by β-glucan is characterized by changes to chromatin modifications such as H3K4me1 (Novakovic et al., 2016, Saeed et al., 2014) that modulate transcriptional programs. In eukaryotes, H3K4me1 is predominantly enriched at enhancers—distal regulatory elements that, via chromatin remodeling events and looping of DNA, are brought into close proximity to physically interact with target promoters to fine-tune spatially and temporally restricted gene expression patterns. Three-dimensional chromatin structures, known as topologically associating domains (TADs), divide the genome into regions enriched in chromosomal looping contacts (Fanucchi and Mhlanga, 2019). Studies of chromatin architecture have begun to define the regulatory importance of TAD formation for the epigenetic priming and transcriptional memory of β-glucan-trained innate immune gene promoters (Fanucchi et al., 2019). Because previous studies demonstrated that metabolic changes occur upstream of enhanced cytokine production in trained immunity, we reasoned that H3K4me1 at enhancers associated with the regulation of metabolic genes could underpin the necessity of Set7 for the trained phenotype induced by β-glucan.
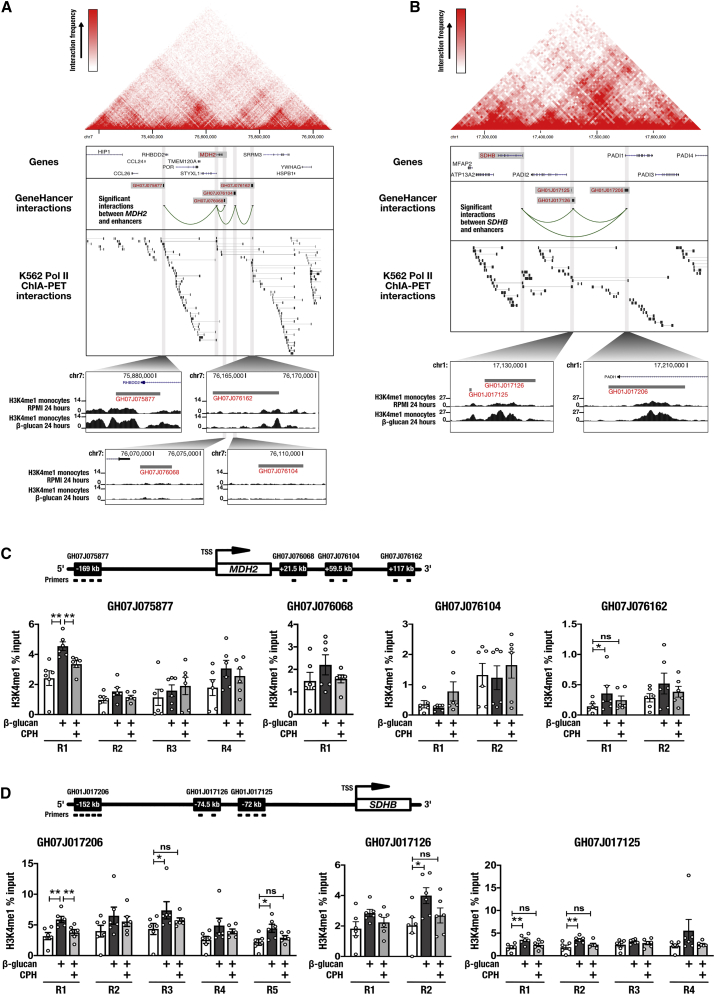
Drawing from the GeneHancer database of human enhancers and their inferred target genes (Fishilevich et al., 2017) as well as publicly accessible Pol II ChIA-PET data generated from K562 cells (GEO sample accession: GSM970213), we identified four enhancer regions that interact with MDH2. The most distal region (GeneHancer ID: GH07J075877) is approximately 3.1 kb in size and is located −169 kb upstream of MDH2, spanning the promoter of RHBDD2. Three additional MDH2 TAD enhancers, located in closer proximity downstream of MDH2, were identified as GH07J076068 (3.4 kb, +22.3 kb from the MDH2 transcription start site [TSS]), GH07J076104 (5.1 kb, +59.5 kb from TSS), and GH07J076162 (4.9 kb, +117.1 kb from TSS). Analysis of publicly accessible data derived from sequencing of immunoprecipitated chromatin (ChIP-seq) generated by the Blueprint Consortium (Adams et al., 2012) indicated that GH07J075877 and GH07J076162 became enriched for H3K4me1 in monocytes following stimulation with β-glucan for 24 h. By contrast, H3K4me1 at GH07J076068 and GH07J076104 was unaffected by β-glucan training (Figure 6A). The same approach identified three distal enhancers that interact within the SDHB TAD in K562 cells. GH07J017206 is approximately 3.1 kb in size and is located −152 kb upstream of the SDHB TSS, residing at the first exon of PADI1. GH07J017126 is approximately 3.7 kb in size and is located −74.7 kb upstream of SDHB. Adjacent to this second regulatory element is a relatively smaller (approximately 1 kb) enhancer located −71.9 kb upstream of SDHB (GH07J017125). Analysis of ChIP-seq data revealed specific enrichment for H3K4me1 at these SDHB TAD enhancers following 24-h stimulation of monocytes with β-glucan (Figure 6B).
Figure 6.
H3K4me1 Changes at Distal Enhancers Regulating MDH2 and SDHB Support Metabolic Reprogramming of Macrophages Trained with β-Glucan
(A) Gene-enhancer interactions within the topologically associating domain (TAD) surrounding MDH2 derived from ChIA-PET interactions in K562 cells (upper panel). ChIP-seq-derived H3K4me1 maps of these enhancer regions in monocytes stimulated with β-glucan for 24 h (lower panels).
(B) Gene-enhancer interactions within the TAD surrounding SDHB derived from ChIA-PET interactions in K562 cells (upper panel). ChIP-seq-derived H3K4me1 maps of these enhancer regions in monocytes stimulated with β-glucan for 24 h (lower panels).
(C) Levels of H3K4me1 at enhancer sites associated with the transcriptional regulation of MDH2 in primary human macrophages measured 5 days after incubation with β-glucan and co-incubation with 100 μM CPH (n = 6 healthy volunteers; open bars represent DMSO vehicle controls).
(D) Levels of H3K4me1 at enhancer sites associated with the transcriptional regulation of SDHB in primary human macrophages measured 5 days after incubation with β-glucan and co-incubation with 100 μM CPH (n = 6 healthy volunteers; open bars represent DMSO vehicle controls).
Data are represented as mean ± SEM. ∗p < 0.05, ∗∗p < 0.01, Wilcoxon signed-rank test.
See also Figures S4, S5, and S6.
We assessed the chromatin at enhancers within the MDH2 domain on day 6 of the in vitro training protocol by ChIP and observed H3K4me1 enrichment in response to β-glucan at three discrete regions of GH07J075877, reaching statistical significance at region 1 (R1). The addition of 100 μM CPH for the first 24 h of in vitro training attenuated this enrichment at R1 as well as at R2 and R4 on day 6. A similar pattern of β-glucan-induced and CPH-responsive H3K4me1 enrichment was also observed at GH07J076162, but not at GH07J076104 as predicted by ChIP-seq. However, contrasting the ChIP-seq data, we observed a trend toward H3K4me1 enrichment at GH07J076068 in cells trained with β-glucan (Figure 6C; open bars represent DMSO vehicle controls). Stimulation of monocytes with β-glucan also led to the sustained enrichment of H3K4me1 at enhancers within the SDHB TAD in day 6 macrophages. This was most pronounced at GH07J017206, where three regions (R1, R3, and R5) exhibited significant increases in H3K4me1 in response to β-glucan. Moreover, incubation with CPH for the first 24 h of in vitro training sufficiently attenuated this chromatin modification. Significant H3K4me1 enrichment was also observed at GH07J017126 (R2) and GH07J017125 (R1 and R2), reflecting an overall pattern of β-glucan-induced and CPH-responsive acquisition of H3K4me1 at SDHB enhancers (Figure 6D; open bars represent DMSO vehicle controls). To investigate the evolutionary conservation of enhancers regulating MDH2 and SDHB, we compared the DNA sequence similarity across different species. For enhancers regulating MDH2, we observed a high level of conservation in mice for GH07J075877. In contrast, GH07J076068, GH07J076104, and GH07J076162 were poorly conserved (Figure S4). For enhancers regulating SDHB, we observed that GH01J017125 is partially conserved in mice. GH01J017126 is only conserved in primates, whereas GH01J017206 is conserved in all species analyzed (Figure S5). In contrast to enhancers of MDH2 and SDHB, and consistent with gene expression data (Figures 5C and 5D), β-glucan did not induce significant changes to H3K4me1 at GH11J112155 and GH02J084740, which are associated with SDHD (+74.9 kb from TSS) and SUCLG1 (−281.5 kb from TSS) regulation, respectively (Figure S6). Together, these data reveal chromatin-dependent mechanisms of TCA cycle gene regulation in trained immunity and strongly implicate Set7 in this epigenetic process.
Discussion
Several experimental observations led us to investigate the role of Set7 in trained immunity. Set7 had been strongly implicated in transcriptional memory and specifically the persistent activation of the RELA gene by vascular endothelial cells in response to transient stimulation with a high concentration of glucose (Brasacchio et al., 2009, El-Osta et al., 2008), pointing to a role in the clinical phenomenon of metabolic memory in diabetes (Keating et al., 2018). With regard to innate immune cells, analysis of TNFα-stimulated THP-1s revealed that a large proportion of NFκB-dependent genes were attenuated by Set7 knockdown (Li et al., 2008). The second consideration is that the H3K4me1 chromatin-templated enzymatic product of Set7 is a characteristic feature of transcriptionally permissive regulatory elements such as enhancers (Heintzman et al., 2007). We previously demonstrated the persistence of H3K4me1 at decommissioned enhancers in trained cells, supporting the notion that H3K4me1 provides a mechanism for epigenetic memory induced by β-glucan (Saeed et al., 2014). The third argument for Set7 in trained immunity came from our previous transcriptome profiling studies of macrophages trained with β-glucan that identified specific activation of SETD7 expression (Quintin et al., 2012). Indeed, the inhibition of Set7 for the initial 24-h training period is sufficient to attenuate the augmentation in pro-inflammatory cytokine production, suggesting that Set7 plays an early role in establishing changes that underpin trained immunity.
The inability of Setd7 KO mice to mount a β-glucan-induced amplified response to a secondary challenge of systemic inflammation highlights the importance of Set7 for trained immunity in vivo. Because mature monocytes have a short life span in the circulation relative to the duration of trained immunity, recent attention has turned to mechanistic processes acting at the level of myeloid precursors. Administration of β-glucan to mice induced a bias of HSPCs toward myelopoiesis and activated transcriptional and metabolic pathways associated with trained immunity at the expense of pathways associated with lymphopoiesis. Enhanced myelopoiesis was associated with elevated signaling by innate immune mediators such as IL-1β and GM-CSF (Mitroulis et al., 2018). Our analysis of total bone marrow validated the gene expression signature of IL1b and Csf2 in animals administered β-glucan and revealed a striking inhibition of this effect in mice lacking functional Set7. Similarly, trained immunity is induced in myeloid progenitor cells with skewing toward myelopoiesis by high-cholesterol Western-type diet (WD) in mice. Involvement of the IL-1 pathway in this process argues for a role for Set7 in the sustained atherogenic inflammation associated with WD-induced trained immunity (Christ et al., 2018).
Specific metabolic pathways distinguish and support the spectrum of macrophage phenotypes and activation states. For trained immunity induced by β-glucan (Cheng et al., 2014) and BCG (Arts et al., 2016b), the process is characteristically reliant on enhanced glycolysis. Indeed, the attenuation of lactate production by CPH in cells trained with β-glucan indicates a role for Set7 in the glycolytic metabolism. Most remarkable are our observations supporting a role for Set7 in the upregulation of OXPHOS in β-glucan-trained cells. Previous studies indicated that brief exposure to a high concentration of β-glucan causes a classical Warburg shift to glycolysis at the expense of OXPHOS (Cheng et al., 2014), contrasting trained immunity induced by BCG (Arts et al., 2016b), which induces upregulation of both energy systems. Using measurements of metabolic flux and functional genomics, we underscore the importance of OXPHOS for the trained macrophage phenotype. Findings presented by the current study suggest that the disparity in terms of the role of OXPHOS arises from the stimulatory dose of β-glucan: a β-glucan concentration of 1 μg/mL induces both glycolysis and OXPHOS, whereas a concentration of 10 μg/mL induces glycolysis but inhibits OXPHOS (Cheng et al., 2014).
Accumulation of succinate, fumarate, and malate in trained cells was previously interpreted as anaplerotic repurposing of TCA cycle metabolism. The importance of these metabolic changes is highlighted by the observation that fumarate can partly recapitulate the training effects of β-glucan (Arts et al., 2016a). Our current data indicate that the amplified TCA cycle in fact remains functional. Moreover, we show that the inhibitory effect of CPH during the first 24 h of in vitro training is sufficient to attenuate the augmented metabolite profiles in day 6 macrophages. Genes encoding important TCA cycle components were transcriptionally activated by β-glucan in a sustained and CPH-sensitive manner. This observation is particularly revealing for SDHB, which functions in the TCA cycle as well as OXPHOS.
Emerging evidence points toward specific chromatin structural organization in the transcriptional memory of β-glucan-trained innate immune gene promoters (Fanucchi et al., 2019). We propose that chromatin modifications associated with specific TAD formation contribute to the persistent transcriptional activation of key metabolic genes that support trained immunity. We observed H3K4me1 enrichment at distal enhancers that form regulatory domains with MDH2 and SDHB, respectively, and are predicted to be involved in their transcriptional regulation. Inhibition of this enrichment by CPH during the first 24 h of in vitro training suggests a memory function for Set7-mediated H3K4me1 within the respective TADs of MDH2 and SDHB. Our data largely mirror the H3K4me1 enrichment patterns of primary monocytes stimulated with β-glucan for 24 h. In contrast, we could not identify similar regulatory elements for FH.
While there is limited understanding of the factors that regulate structural genomic interactions, a central tenet of promoter-enhancer communication is the specific binding of transcription factors, which can be constrained to regulatory elements by chromatin modifications such as H3K4me1 (Stadhouders et al., 2019). In addition to H3 histones, Set7 controls the stability and function of several transcription factors by post-translational lysine methylation (Keating and El-Osta, 2013). One such transcription factor substrate is YY1, an important structural regulator of enhancer-promoter loops (Weintraub et al., 2017). Methylation of YY1 at K173 and K411 by Set7 modulates DNA binding and YY1-regulated gene transcription in HeLa cells (Zhang et al., 2016). While it is unclear whether similar processes govern innate immune transcriptional plasticity, several of the enhancers investigated in the current study harbor binding motifs for YY1 (data not shown).
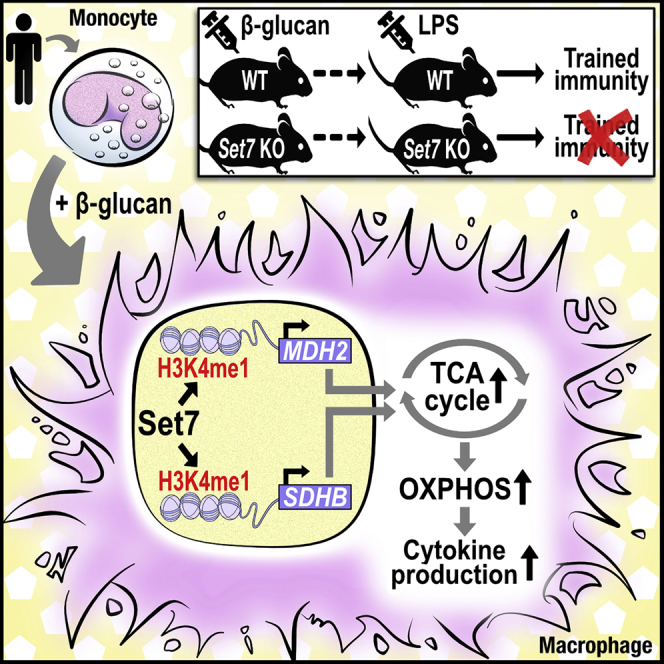
Set7 is a crucial component of the cellular machinery responsible for the establishment and maintenance of metabolic and transcriptional memory programs that support the trained immunity phenotype induced by β-glucan. At least part of this role involves the regulation of OXPHOS at the level of chromatin modification and gene expression, with potentially important consequences for specific TCA cycle metabolites and downstream signaling events, including metabolite-dependent chromatin-modifying reactions (Arts et al., 2016a, Keating and El-Osta, 2015; Figure 7). Our study is limited by our focus on the role of Set7 in one aspect of β-glucan-mediated metabolic gene regulation. Further elucidation of this key regulatory circuitry will provide important mechanistic and physiological insights into trained immunity, and lead to the identification of strategies to enhance or dampen inflammatory responses in certain clinical settings.
Figure 7.
Set7 Regulates Metabolic Changes in Cells Trained with β-Glucan
β-Glucan stimulation activates Set7 to write the H3K4me1 modification to distal enhancers associated with sustained MDH2 and SDHB gene expression leading to increases in TCA cycle metabolites and increased oxidative phosphorylation important for enhanced cytokine production in trained immunity.
STAR★Methods
Key Resources Table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Rabbit monoclonal anti-mono-methyl-Histone H3 (Lys4) | Cell Signaling Technology | Cat#5326; RRID: AB_10695148 |
| Rabbit monoclonal anti-di-methyl-Rpl29 (Lys5) | Cell Signaling Technology | Cat#19495; RRID: AB_2798819 |
| Rabbit polyclonal anti-Set7 | Cell Signaling Technology | Cat#2813; RRID: AB_823636 |
| Rabbit polyclonal anti-HSP90 | Cell Signaling Technology | Cat#4874; RRID: AB_2121214 |
| Rabbit polyclonal anti-β-actin | Sigma-Aldrich | Cat#A2066; RRID: AB_476693 |
| Mouse monoclonal anti-β-actin | Cell Signaling Technology | Cat#3700; RRID: AB_2242334 |
| swine-anti-rabbit Ig polyclonal secondary HRP antibody | Dako | Cat#P0217; RRID: AB_2728719 |
| Chemicals, Peptides, and Recombinant Proteins | ||
| β-glucan (b1,3-(D)-glucan | Professor David Williams, College of Medicine, Johnson City, USA | N/A |
| Lipopolysaccharide | Sigma-Aldrich | Cat#L2880 From E.coli serotype 055:B5 |
| Percoll | Sigma-Aldrich | Cat#P1644 |
| Ficoll-Paque | GE Healthcare | Cat#17-1440-03 |
| Roswell Park Memorial Institute medium (RPMI) | Invitrogen | Cat#22406031 |
| iScript reverse transcriptase | Bio-Rad | Cat#1708840 |
| TRIzol reagent | Life Technologies | Cat#15596018 |
| SYBR Green | Applied Biosciences | Cat#4368708 |
| 16% Formaldehyde | Fisher Scientific | Cat#28908 |
| Cyproheptadine | Selleckchem | Cat#S2044 |
| Sinefungin | Sigma-Aldrich | Cat#S8559 |
| Bacillus Calmette–Guérin vaccine | Statens Serum Institut, Copenhagen, Denmark | N/A |
| Laminarin | Sigma-Aldrich | Cat#L9634 |
| Diphenhydramide hydrochloride | Sigma-Aldrich | Cat#D3630 |
| Critical Commercial Assays | ||
| Pierce BCA protein assay kit | ThermoFisher Scientific | Cat#23225 |
| Human TNFα DuoSet ELISA | R&D systems | Cat#DY210 |
| Human IL-6 DuoSet ELISA | R&D systems | Cat#DY206 |
| Mouse TNFα Quantikine ELISA | R&D systems | Cat#MTA00B |
| Mouse IL-6 Quantikine ELISA | R&D systems | Cat#M6000B |
| Mouse IL-1β Quantikine ELISA | R&D systems | Cat#MLB00C |
| Lactate Fluorometric Assay kit | Biovision | Cat#K607 |
| MinElute PCR purification column | QIAGEN | Cat#28006 |
| iScript cDNA synthesis kit | Bio-Rad | Cat#1708891 |
| Cytox 96 assay | Promega | Cat#G1780 |
| Succinate colorimetric assay kit | Sigma-Aldrich | Cat#MAK184 |
| Fumarate colorimetric assay kit | Sigma-Aldrich | Cat#MAK060 |
| Malate colorimetric assay kit | Sigma-Aldrich | Cat#MAK067 |
| Oxaloacetate colorimetric assay kit | Sigma-Aldrich | Cat#MAK070 |
| Citrate colorimetric assay kit | Sigma-Aldrich | Cat#MAK057 |
| Experimental Models: Organisms/Strains | ||
| 300BCG cohort (Human Functional Genomics Project) | N/A | https://www.humanfunctionalgenomics.org |
| 200FG cohort (Human Functional Genomics Project) | N/A | https://www.humanfunctionalgenomics.org |
| Setd7 knockout mouse | Professor Assam El-Osta, Monash University, Melbourne | N/A |
| Oligonucleotides | ||
| See Table S1 | This paper | N/A |
| Software and Algorithms | ||
| GraphPad Prism 8.12 | Graphpad software | https://www.graphpad.com |
| R statistical programming | N/A | RRID:SCR_001905 |
Resource Availability
Lead Contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Niels P. Riksen (niels.riksen@radboudumc.nl).
Materials Availability
This study did not generate new unique reagents.
Data and Code Availability
This study did not generate any unique datasets or code.
Experimental Model and Subject Details
Human subjects
With regard to the in vitro studies, buffy coats from male and female healthy donors were obtained after written informed consent (Sanquin blood bank, Nijmegen, the Netherlands). Cells were isolated and experiments conducted on the same day.
Human cohorts
The 300BCG cohort consists of 267 healthy males and females of Western European ancestry. The second cohort consists of 119 healthy individuals of Western European ancestry from the 200 Functional Genomics cohort (2011/399) of the Human Functional Genomics Project. The 300BCG cohort and 200FG cohort studies were approved by the local ethics committee (CMO regio Arnhem-Nijmegen, number NL58553.091.16 and number 2011-399, respectively). Inclusion of volunteers and experiments were conducted according to the principles expressed in the Declaration of Helsinki. All volunteers gave written informed consent before any material was taken.
Mice
The Setd7 knockout (KO) mice were generated by genOway (Lyon, France). The Setd7 targeting vector was designed within the first MORN domain of exon 2 containing two loxP sites (unpublished data). Insertion of loxP sites was introduced 1.6kb upstream of exon 2 by the integration of a loxP-flanked neomycine cassette. A targeting vector containing loxP sites flanking exon 2 of the Setd7 gene was integrated by homologous recombination in mouse embryonic stem cells. Recombinant clones were injected into mouse C57BL/6J strain blastocysts and implanted into pseudopregnant females. Once the construct was integrated into C57BL/6J background mice, it was crossed with a CMV promoter-driven Cre recombinase mouse to create a constitutive Setd7 KO mouse. Wild-type and Setd7 KO mice housed under specific pathogen-free conditions were used at the age of 9-11 weeks. Food and water was provided ad libitum. All animal studies were approved by the Alfred Medical Research and Education Precinct (AMREP) Animal Ethics Committee under guidelines laid down by the National Health and Medical Research Council (NHMRC) of Australia.
Method Details
Cells and reagents
Human peripheral blood mononuclear cells (PBMCs) were isolated from healthy volunteers by density-gradient centrifugation over Ficoll-Paque (GE Healthcare). Percoll isolation of monocytes was performed as previously described (Arts et al., 2016a). Cells were cultured in RPMI 1640 Dutch-modified culture medium (RPMI medium, Invitrogen) supplemented with 10 μg/mL gentamicin (Centraform), 2 mM Glutamax (Invitrogen), 1 mM pyruvate (Invitrogen), and 10% pooled human serum. Stimuli and inhibitors used were Escherichia coli lipopolysaccharide (LPS; serotype 055:B5, Sigma-Aldrich, 10 ng/mL), and Pam3Cys (EMC microcollections, L2000, 10 μg/mL), Laminarin (Sigma), BCG (Statens Serum Institut, Copenhagen, Denmark), cyproheptadine (Selleckchem), sinefungin (Sigma), and diphenhydramide (Sigma).
In vitro training and pharmacological inhibition
β-1,3-(D)-glucan (β-glucan) was kindly provided by Professor David Williams (College of Medicine, Johnson City, USA). For isolation of cell wall β-glucans C. albicans was cultivated in 25 mL of YPD (1% yeast extract, 2% dextrose, 2% peptone) for 48 hours at 30°C. The cells were harvested by centrifugation at 5,000x g for 5 minutes and pellet washed once with dH2O. The washed cell pellets were then frozen at −20°C overnight. Prior to extracting the cell wall β-glucans, the cell pellets were subjected to repeated freeze-thaw cycles (3X) to lyse the cells. Cell pellets were then extracted with a base/acid isolation approach. The supernatant contained the water soluble mannans. Glucans are water insoluble and were harvested by centrifugation and washing in dH2O prior to lyophilization. The structure and purity of the β-glucans was determined by solution, high field one and two-dimensional Nuclear Magnetic Resonance Spectroscopy (1 and 2-D NMR).
Adherent monocytes were trained as described previously (Bekkering et al., 2014). Cells were incubated with β-glucan (1 μg/mL), Laminarin (1 mg/mL), or BCG (5 μg/mL) for 24 hours, washed with warm phosphate buffered saline (PBS) and incubated in normal culture medium at 37°C, 5% CO2. For pharmacological inhibition experiments, cells were pre-incubated with cyproheptadine (25-100 μM), sinefungin (1, 10, 50, and 100 μg/mL), or diphenhydramide (10, 50, 100 μg/mlL), for 1 hour prior to stimulation. Following 5 days in culture, cells were restimulated with medium alone, 10 ng/mL LPS, or 10 μg/mL Pam3Cys for 24 hours at 37°C, 5% CO2. Cytokine production was measured in supernatants by enzyme-linked immunosorbent assay (ELISA) according to the manufacturer’s instructions (R&D Systems).
Cytokine measurement
Cytokine production in supernatants and plasmas was determined using commercial enzyme-linked immunosorbent assay kits for TNFα, IL-6, IL-1β (R&D Systems, MN, USA), IL-10 (Sanquin) according to the instructions of the manufacturers.
Annexin V/PI staining and lactate dehydrogenase (LDH) measurements for cell viability
Apoptosis and cell viability of monocytes after 24h exposure to 100 μM CPH or vehicle control was evaluated with Annexin V-FITC (Biovision) and Propidium Iodide ECD (Biovision) fluorescence with cytoFLEX flow cytometer (Beckman Coulter) and analyzed with Kaluza 2.1 (Beckman Coulter). Analysis of LDH as a measure of cytotoxicity in cells incubated with CPH for 24 hours was assessed in the supernatants by using a Cytotox 96 kit (Promega).
Quantitative RT-PCR
Total RNA was isolated from human primary macrophages and total bone marrow of mice using TRIzol reagent according to the manufacturer’s instructions. 0.5-1 μg of total RNA was used to synthesize cDNA with the SuperScript III First-Strand Synthesis System (Thermo Fisher Scientific) according to the manufacturer’s protocol. Quantitative RT-PCR was performed using an Applied Biosciences StepOne PLUS qRT-PCR machine using SYBR Green (Invitrogen). All reactions were performed for at least 6 biological replicates and the values expressed as fold increase in mRNA levels relative to those in non-trained cells. 18s (human) or H3f3a (mouse) was used as a housekeeping gene. qRT-PCR primers are listed in Table S1.
Western blot analysis of primary human cells
For protein expression analysis of primary human cells, approximately 1 × 106 monocytes exposed for 24 hours to 100 μM CPH and 1 × 106 macrophages were lysed with 100 μL of lysis buffer (1M Tris pH 7.4), 5M NaCl, 0.5M EDTA, 10% NP-40, 0.5M NaF, 2.5% sodium deoxycholate, PhosSTOP (Roche), cOmplete (Roche)) prior to stimulation on day 6. The homogenate was frozen, then thawed and centrifuged at 4°C for 10 min at 15,000 x g, and the supernatant was taken for analysis. The western blot was performed using a Trans Turbo Blot System (Bio-Rad) according to the manufacturer’s instructions. Protein was loaded and separated on SDS-PAGE using 4%–15% gradient precast gels and transferred to nitrocellulose membranes using the semi-dry method (Bio-Rad). Rabbit polyclonal primary antibodies were used for both Set7 (1:1000, 2813, Cell Signaling Technology) and Rpl29k5me2 (1:1000, 19495, Cell Signaling Techonolgy. Swine-anti-rabbit polyclonal secondary HRP antibody (1:5000, P0217, Dako) was used to detect Set7 and Rpl29k5me2 protein expressions. β-actin was detected on the blots using rabbit polyclonal primary antibody (1:1000, A2066, Sigma-Aldrich) and swine-anti-rabbit polyclonal secondary HRP antibody (1:5000, P0217, Dako). HSP90 was detected on the blots using rabbit polyclonal primary antibody (1:1000, 4874, Cell Signaling Technology) and swine-anti-rabbit polyclonal secondary HRP antibody (1:5000, P0217, Dako). Blots were developed with ECL (GE Healthcare) according to the manufacturer’s instructions.
Mouse experiments
For the in vivo study of trained immunity, mice were injected intraperitoneally with 1 mg of β-glucan in 200 μL of endotoxin-free phosphate-buffered saline (PBS). Intraperitoneal injections of PBS were performed as control. Five days after β-glucan administration, the mice were injected intraperitoneally with 10 μg of LPS from E. coli 055:B5 (Sigma) as a secondary challenge. Mice were euthanized at 3 hours after the LPS challenge.
To isolate bone marrow cells (BMCs), mouse femurs and tibias were collected, trimmed and flushed with DPBS (GIBCO) using a 20 mL syringe with a 25 gauge needle to release BMCs. Bone marrow suspensions were gently harvested on 40 μm nylon mesh strainer (Falcon) in 50 mL conical tubes. After centrifugation (5 min, 350 x g, 4°C), the cells were suspended with RBC lysis buffer (155 mM NH4Cl, 10 mM KHCO3, 0.1mM EDTA) to remove erythrocytes.
Protein analyses were performed as described previously (Okabe et al., 2012). Briefly, approximately 5 × 106 BMCs were lysed with 250 μL of buffer C (20 mM HEPES-KOH (pH 7.5), 25% Glycerol, 520 mM KCl, 5 mM MgCl2, 0.1 mM EDTA, 1mM DTT, 0.5 mM PMSF, 0.2% NP-40 and proteinase inhibitor cocktail) for 15 min at 4°C, then centrifuged for 15 min at 15,000 x g, 4°C. The supernatant was collected for analysis. Primary antibodies were used for both Set7 (1:2,000, 2813, Cell Signaling Technology) and β-actin (1:10,000, 3700, Cell Signaling Technology). IRDye 800CW Donkey anti-mouse IgG and IRDye 680RD Donkey anti-rabbit IgG secondary antibodies (1:10,000 each, LI-COR) were used to detect Set7 and β-actin protein signals simultaneously.
Metabolic analysis
Approximately 1 × 107 monocytes were trained with β-glucan (1 μg/mL) in 10 cm Petri dishes (Greiner) in 10 mL medium volumes for 24 hours, washed with warm PBS and incubated in normal culture medium at 37°C, 5% CO2. Following 5 days in culture, cells were detached with versene solution (ThermoFisher Scientific) and 1 × 105 cells were plated to overnight-calibrated cartridges in assay medium (RPMI with 0.6 mM glutamine, 5 mM glucose and 1 mM pyruvate [pH adjusted to 7.4]) and incubated for 1 hour in a non-CO2-corrected incubator at 37°C. Oxygen consumption rate (OCR) was measured using a Cell Mito Stress Kit (for OCR) or a glycolysis stress test kit in an XFp Analyzer (Seahorse Bioscience), with final concentrations of 1 μM oligomycin, 1 μM FCCP, and 0.5 μM rotenone/antimycin A.
Oxygen consumption measurement
Culture medium was collected from cells treated with either RPMI or β-glucan (1 μg/mL or 10 μg/mL). After stimulation, the cells were trypsinized, washed, and resuspended in the collected culture medium. Cell suspensions containing 1 × 106 cells were then used for cellular O2 consumption analysis. Oxygen consumption was measured at 37°C using polarographic oxygen sensors in a two-chamber Oxygraph (OROBOROS Instruments, Innsbruck, Austria). DatLab software (Oroboros) was used for data acquisition (2 s time interval) and analysis (Gnaiger, 2001). First, basal respiration (baseline oxygen consumption) was measured. Next, leak respiration was determined by addition of 2.5 μM of the specific complex V inhibitor oligomycin A (OLI). Then, maximal electron transport chain complex (ETC) capacity (maximum oxygen consumption) was quantified by applying increasing concentrations of the mitochondrial uncoupler FCCP (0.25 to 20 μM final maximal concentration). Finally, minimal (non-mitochondrial) respiration was assessed by addition of the specific complex I inhibitor rotenone (ROT; 100 nM) and the complex III inhibitor antimycin A (AA; 2.5 μM).
Metabolite measurements
Cells were trained with β-glucan (1 μg/mL) with and without CPH (100 μM) as described above, using DMSO as a vehicle control. Metabolite concentrations measured from at least 1 × 106 trained monocytes for succinate, fumarate, malate, oxaloacetate, and citrate were determined using a commercial colorimetric assay kit (Sigma) according to the manufacturer’s instructions.
Genetic analysis
We conducted in vitro β-glucan, BCG and oxLDL training of adherent PBMCs from 267 healthy individuals of Western European ancestry from the 300BCG cohort (NL58553.091.16). DNA samples of these individuals were genotyped using the commercially available SNP chip, Infinium Global Screening Array MD v1.0 from Illumina. Genotype information on approximately 4 million single-nucleotide polymorphisms (SNPs) was obtained upon imputation (MAF > 5% and R2 > 0.3 for imputation quality). First, raw cytokine levels were log-transformed and the ratio between trained and non-trained cytokine levels were taken as the change of cytokine levels. The cytokine changes were mapped to genotype data using a linear regression model with age and sex as covariates. Genetic outliers (n = 15) and samples stimulated with low β-glucan (< 1 μg/mL) were removed before QTL mapping.
We also conducted in vitro β-glucan training of adherent PBMCs in a second cohort of 119 healthy individuals of Western European ancestry from the 200 Functional Genomics cohort (2011/399) of the Human Functional Genomics Project (www.humanfunctionalgenomics.org). Genotype information on approximately 4 million single-nucleotide polymorphisms (SNPs) was obtained using Illumina HumanOmniExpressExome SNP chip upon imputation. Only SNPs with a minor allele frequency of ≥ 5% that passed standard quality filters were included in the analysis. Raw cytokine levels were log-transformed and the ratio between trained and non-trained cytokine levels was used to quantify the trained immunity response. They were subsequently mapped to genotype data using a linear regression model with age and sex as co-variates (Li et al., 2016).
ChIP-seq and ChIA-PET analysis
This study makes use of H3K4me1 ChIP-seq datasets generated by the Blueprint Consortium (Adams et al., 2012). This study makes use of ChIA-PET data (accession number GSM970213). In the graphical display of the ChIA-PET data, the paired end tags (PETs) or chromosomal interactions are represented by two blocks for each end of the contact, connected by a horizontal line. The number of PETs in a cluster reflects the strength of the chromosomal interaction. The pre-processed datasets were visualized using the UCSC genome browser with the GRCh37/hg19 assembly (Kent et al., 2002). Enhancers were identified from the GeneHancer database of human regulatory elements (Fishilevich et al., 2017). Enhancers with the highest annotation-derived confidence score were selected. The study makes use of Hi-C data from the K562 cell line (accession GSE63525). Hi-C maps were generated at 5kb resolution using the 3D genome browser with the GRCh37/hg19 assembly (Wang et al., 2018).
Chromatin immunoprecipitation
Trained monocytes on day 6 were cross-linked in methanol free 1% formaldehyde, followed by sonication and immunoprecipitation using antibodies against H3K4me1 (Cell Signaling Technology). Immunoprecipitated chromatin was processed further for qRT-PCR analysis using the MiniElute DNA purification kit (QIAGEN). Primers used in the reaction are listed in Table S1. Samples were analyzed with a comparative Ct method on the StepOne PLUS qPCR machine (Applied Biosystems) using SYBR green (Invitrogen) in accordance with the manufacturer’s instructions.
Quantification and Statistical Analysis
Statistical parameters including the exact value of n, the definition of center, dispersion and precision measures (mean ± SEM), and statistical significance are reported in the figures and figure legends. Statistical analysis was performed using GraphPad Prism 8.12 (GraphPad Inc.). Analysis of human qPCR, ELISA and cellular assays was performed using Wilcoxon signed-rank test, t test or non-parametric Mann-Whitney tests, as appropriate. Analysis of mouse data used Mann-Whitney tests for comparisons between groups. R-package Matrix-eQTL was used for cytokine QTL mapping. A p value < 0.05 (∗) was considered statistically significant, (∗∗) p < 0.01. Data are shown as mean ± SEM.
Acknowledgments
The authors thank all volunteers who participated in the 200FG and 300BCG cohorts. The authors acknowledge Jori Wagenaars and Merel Adjobo-Hermans from the Department of Biochemistry, Radboud University Medical Center, Nijmegen, the Netherlands, for their experimental support. M.G.N. is supported by a European Research Council (ERC) Advanced Grant (833247) and a Spinoza grant of the Netherlands Organisation for Scientific Research. N.P.R. is recipient of a grant of the European Research Area Cardiovascular Disease (ERA-CVD) Joint Transnational Call 2018, which is supported by the Dutch Heart Foundation (JTC2018, project MEMORY; 2018T093). N.P.R., L.A.B.J., and M.G.N. received funding from the European Union Horizon 2020 Research and Innovation Program under Grant Agreement 667837, and the IN-CONTROL grant from the Heart Foundation Netherlands (CVON2012-03 and CVON2018-27). L.A.B.J. was supported by a Competitiveness Operational Programme grant of the Romanian Ministry of European Funds (HINT, ID P_37_762; MySMIS 103587). Y.L. was supported by the Radboud University Medical Center Hypatia Grant (2018). M.O. was supported by a VENI grant (016.176.006) from the Netherlands Organisation for Scientific Research (NWO). This study makes use of data generated by the Blueprint Consortium. A full list of the investigators who contributed to the generation of the data is available from http://www.blueprint-epigenome.eu. Funding for the project was provided by the European Union’s Seventh Framework Programme (FP7/2007-2013) under Grant Agreement 282510 BLUEPRINT.
Author Contributions
Conceptualization, S.T.K., L.G., L.A.B.J., M.G.N., and N.P.R.; Methodology, S.T.K., L.G., C.D.C.C.v.d.H., J.C.d.S., H.R., J.H.v.P., L.H., M.P.N., J.O., E.P.B., W.J.H.K., M.G.N., and N.P.R.; Formal Analysis, S.T.K., L.G., C.D.C.C.v.d.H., H.R., J.C.d.S., S.F., J.O., H.K., J.H.v.P., V.M., Y.L., E.P.B., and W.J.H.K.; Investigation, S.T.K., L.G., C.D.C.C.v.d.H., H.R., J.C.d.S., S.F., J.O., K.H., J.H.v.P., L.H., M.P.N., L.C.J.d.B., V.A.C.M.K., S.J.C.F.M.M., V.P.M., J.D.-A., M.O., E.P.B., and W.J.H.K.; Writing—Original Draft, S.T.K.; Writing—Review and Editing, S.T.K., L.G., C.D.C.C.v.d.H., J.C.d.S., S.F., J.O., J.H.v.P., M.P.N., V.M., L.C.J.d.B., V.A.C.M.K., S.J.C.F.M.M., V.P.M., J.D.-A., M.O., E.P.B., W.J.H.K., L.A.B.J., M.G.N., and N.P.R.; Funding Acquisition, L.A.B.J., M.G.N., and N.P.R.; Supervision, J.O., M.M., A.E.-O., L.A.B.J., M.G.N., and N.P.R.
Declaration of Interests
W.J.H.K. is a scientific advisor of Khondrion (Nijmegen, the Netherlands) and of Fortify Therapeutics. These subject matter experts had no involvement in the data collection, analysis and interpretation, writing of the manuscript, and the decision to submit the manuscript for publication.
Published: April 21, 2020
Footnotes
Supplemental Information can be found online at https://doi.org/10.1016/j.celrep.2020.107548.
Supplemental Information
References
- Adams D., Altucci L., Antonarakis S.E., Ballesteros J., Beck S., Bird A., Bock C., Boehm B., Campo E., Caricasole A. BLUEPRINT to decode the epigenetic signature written in blood. Nat. Biotechnol. 2012;30:224–226. doi: 10.1038/nbt.2153. [DOI] [PubMed] [Google Scholar]
- Allis C.D., Berger S.L., Cote J., Dent S., Jenuwien T., Kouzarides T., Pillus L., Reinberg D., Shi Y., Shiekhattar R. New nomenclature for chromatin-modifying enzymes. Cell. 2007;131:633–636. doi: 10.1016/j.cell.2007.10.039. [DOI] [PubMed] [Google Scholar]
- Arts R.J., Novakovic B., Ter Horst R., Carvalho A., Bekkering S., Lachmandas E., Rodrigues F., Silvestre R., Cheng S.C., Wang S.Y. Glutaminolysis and fumarate accumulation integrate immunometabolic and epigenetic programs in trained immunity. Cell Metab. 2016;24:807–819. doi: 10.1016/j.cmet.2016.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arts R.J.W., Carvalho A., La Rocca C., Palma C., Rodrigues F., Silvestre R., Kleinnijenhuis J., Lachmandas E., Gonçalves L.G., Belinha A. Immunometabolic pathways in BCG-induced trained immunity. Cell Rep. 2016;17:2562–2571. doi: 10.1016/j.celrep.2016.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bekkering S., Quintin J., Joosten L.A., van der Meer J.W., Netea M.G., Riksen N.P. Oxidized low-density lipoprotein induces long-term proinflammatory cytokine production and foam cell formation via epigenetic reprogramming of monocytes. Arterioscler. Thromb. Vasc. Biol. 2014;34:1731–1738. doi: 10.1161/ATVBAHA.114.303887. [DOI] [PubMed] [Google Scholar]
- Bekkering S., Arts R.J.W., Novakovic B., Kourtzelis I., van der Heijden C., Li Y., Popa C.D., Ter Horst R., van Tuijl J., Netea-Maier R.T. Metabolic induction of trained immunity through the mevalonate pathway. Cell. 2018;172:135–146.e9. doi: 10.1016/j.cell.2017.11.025. [DOI] [PubMed] [Google Scholar]
- Brasacchio D., Okabe J., Tikellis C., Balcerczyk A., George P., Baker E.K., Calkin A.C., Brownlee M., Cooper M.E., El-Osta A. Hyperglycemia induces a dynamic cooperativity of histone methylase and demethylase enzymes associated with gene-activating epigenetic marks that coexist on the lysine tail. Diabetes. 2009;58:1229–1236. doi: 10.2337/db08-1666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng S.C., Quintin J., Cramer R.A., Shepardson K.M., Saeed S., Kumar V., Giamarellos-Bourboulis E.J., Martens J.H., Rao N.A., Aghajanirefah A. mTOR- and HIF-1α-mediated aerobic glycolysis as metabolic basis for trained immunity. Science. 2014;345:1250684. doi: 10.1126/science.1250684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christ A., Günther P., Lauterbach M.A.R., Duewell P., Biswas D., Pelka K., Scholz C.J., Oosting M., Haendler K., Baßler K. Western diet triggers NLRP3-dependent innate immune reprogramming. Cell. 2018;172:162–175.e14. doi: 10.1016/j.cell.2017.12.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denisenko E., Guler R., Mhlanga M.M., Suzuki H., Brombacher F., Schmeier S. Genome-wide profiling of transcribed enhancers during macrophage activation. Epigenetics Chromatin. 2017;10:50. doi: 10.1186/s13072-017-0158-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- El-Osta A., Brasacchio D., Yao D., Pocai A., Jones P.L., Roeder R.G., Cooper M.E., Brownlee M. Transient high glucose causes persistent epigenetic changes and altered gene expression during subsequent normoglycemia. J. Exp. Med. 2008;205:2409–2417. doi: 10.1084/jem.20081188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fanucchi S., Mhlanga M.M. Lnc-ing trained immunity to chromatin architecture. Front. Cell Dev. Biol. 2019;7:2. doi: 10.3389/fcell.2019.00002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fanucchi S., Fok E.T., Dalla E., Shibayama Y., Börner K., Chang E.Y., Stoychev S., Imakaev M., Grimm D., Wang K.C. Immune genes are primed for robust transcription by proximal long noncoding RNAs located in nuclear compartments. Nat. Genet. 2019;51:138–150. doi: 10.1038/s41588-018-0298-2. [DOI] [PubMed] [Google Scholar]
- Fishilevich S., Nudel R., Rappaport N., Hadar R., Plaschkes I., Iny Stein T., Rosen N., Kohn A., Twik M., Safran M. GeneHancer: genome-wide integration of enhancers and target genes in GeneCards. Database (Oxford) 2017;2017:bax028. doi: 10.1093/database/bax028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Valtanen P., Guzman-Genuino R.M., Williams D.L., Hayball J.D., Diener K.R. Evaluation of trained immunity by β-1, 3 (d)-glucan on murine monocytes in vitro and duration of response in vivo. Immunol. Cell Biol. 2017;95:601–610. doi: 10.1038/icb.2017.13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gnaiger E. Bioenergetics at low oxygen: dependence of respiration and phosphorylation on oxygen and adenosine diphosphate supply. Respir. Physiol. 2001;128:277–297. doi: 10.1016/s0034-5687(01)00307-3. [DOI] [PubMed] [Google Scholar]
- Gosselin D., Link V.M., Romanoski C.E., Fonseca G.J., Eichenfield D.Z., Spann N.J., Stender J.D., Chun H.B., Garner H., Geissmann F., Glass C.K. Environment drives selection and function of enhancers controlling tissue-specific macrophage identities. Cell. 2014;159:1327–1340. doi: 10.1016/j.cell.2014.11.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamidi T., Singh A.K., Veland N., Vemulapalli V., Chen J., Hardikar S., Bao J., Fry C.J., Yang V., Lee K.A. Identification of Rpl29 as a major substrate of the lysine methyltransferase Set7/9. J. Biol. Chem. 2018;293:12770–12780. doi: 10.1074/jbc.RA118.002890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heintzman N.D., Stuart R.K., Hon G., Fu Y., Ching C.W., Hawkins R.D., Barrera L.O., Van Calcar S., Qu C., Ching K.A. Distinct and predictive chromatin signatures of transcriptional promoters and enhancers in the human genome. Nat. Genet. 2007;39:311–318. doi: 10.1038/ng1966. [DOI] [PubMed] [Google Scholar]
- Kaufmann E., Sanz J., Dunn J.L., Khan N., Mendonça L.E., Pacis A., Tzelepis F., Pernet E., Dumaine A., Grenier J.C. BCG educates hematopoietic stem cells to generate protective innate immunity against tuberculosis. Cell. 2018;172:176–190.e19. doi: 10.1016/j.cell.2017.12.031. [DOI] [PubMed] [Google Scholar]
- Keating S.T., El-Osta A. Transcriptional regulation by the Set7 lysine methyltransferase. Epigenetics. 2013;8:361–372. doi: 10.4161/epi.24234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keating S.T., El-Osta A. Epigenetics and metabolism. Circ. Res. 2015;116:715–736. doi: 10.1161/CIRCRESAHA.116.303936. [DOI] [PubMed] [Google Scholar]
- Keating S.T., Ziemann M., Okabe J., Khan A.W., Balcerczyk A., El-Osta A. Deep sequencing reveals novel Set7 networks. Cell. Mol. Life Sci. 2014;71:4471–4486. doi: 10.1007/s00018-014-1651-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keating S.T., van Diepen J.A., Riksen N.P., El-Osta A. Epigenetics in diabetic nephropathy, immunity and metabolism. Diabetologia. 2018;61:6–20. doi: 10.1007/s00125-017-4490-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kent W.J., Sugnet C.W., Furey T.S., Roskin K.M., Pringle T.H., Zahler A.M., Haussler D. The human genome browser at UCSC. Genome Res. 2002;12:996–1006. doi: 10.1101/gr.229102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleinnijenhuis J., Quintin J., Preijers F., Joosten L.A., Ifrim D.C., Saeed S., Jacobs C., van Loenhout J., de Jong D., Stunnenberg H.G. Bacille Calmette-Guerin induces NOD2-dependent nonspecific protection from reinfection via epigenetic reprogramming of monocytes. Proc. Natl. Acad. Sci. USA. 2012;109:17537–17542. doi: 10.1073/pnas.1202870109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y., Reddy M.A., Miao F., Shanmugam N., Yee J.K., Hawkins D., Ren B., Natarajan R. Role of the histone H3 lysine 4 methyltransferase, SET7/9, in the regulation of NF-kappaB-dependent inflammatory genes. Relevance to diabetes and inflammation. J. Biol. Chem. 2008;283:26771–26781. doi: 10.1074/jbc.M802800200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y., Oosting M., Deelen P., Ricaño-Ponce I., Smeekens S., Jaeger M., Matzaraki V., Swertz M.A., Xavier R.J., Franke L. Inter-individual variability and genetic influences on cytokine responses to bacteria and fungi. Nat. Med. 2016;22:952–960. doi: 10.1038/nm.4139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitroulis I., Ruppova K., Wang B., Chen L.S., Grzybek M., Grinenko T., Eugster A., Troullinaki M., Palladini A., Kourtzelis I. Modulation of myelopoiesis progenitors is an integral component of trained immunity. Cell. 2018;172:147–161.e12. doi: 10.1016/j.cell.2017.11.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Netea M.G., Domínguez-Andrés J., Barreiro L.B., Chavakis T., Divangahi M., Fuchs E., Joosten L.A.B., van der Meer J.W.M., Mhlanga M.M., Mulder W.J.M. Defining trained immunity and its role in health and disease. Nat. Rev. Immunol. 2020 doi: 10.1038/s41577-020-0285-6. Published online March 4, 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishioka K., Chuikov S., Sarma K., Erdjument-Bromage H., Allis C.D., Tempst P., Reinberg D. Set9, a novel histone H3 methyltransferase that facilitates transcription by precluding histone tail modifications required for heterochromatin formation. Genes Dev. 2002;16:479–489. doi: 10.1101/gad.967202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novakovic B., Habibi E., Wang S.Y., Arts R.J.W., Davar R., Megchelenbrink W., Kim B., Kuznetsova T., Kox M., Zwaag J. beta-Glucan reverses the epigenetic state of LPS-induced immunological tolerance. Cell. 2016;167:1354–1368.e14. doi: 10.1016/j.cell.2016.09.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okabe J., Orlowski C., Balcerczyk A., Tikellis C., Thomas M.C., Cooper M.E., El-Osta A. Distinguishing hyperglycemic changes by Set7 in vascular endothelial cells. Circ. Res. 2012;110:1067–1076. doi: 10.1161/CIRCRESAHA.112.266171. [DOI] [PubMed] [Google Scholar]
- Petit J., Embregts C.W.E., Forlenza M., Wiegertjes G.F. Evidence of trained immunity in a fish: conserved features in carp macrophages. J. Immunol. 2019;203:216–224. doi: 10.4049/jimmunol.1900137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quintin J., Saeed S., Martens J.H.A., Giamarellos-Bourboulis E.J., Ifrim D.C., Logie C., Jacobs L., Jansen T., Kullberg B.J., Wijmenga C. Candida albicans infection affords protection against reinfection via functional reprogramming of monocytes. Cell Host Microbe. 2012;12:223–232. doi: 10.1016/j.chom.2012.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saeed S., Quintin J., Kerstens H.H., Rao N.A., Aghajanirefah A., Matarese F., Cheng S.C., Ratter J., Berentsen K., van der Ent M.A. Epigenetic programming of monocyte-to-macrophage differentiation and trained innate immunity. Science. 2014;345:1251086. doi: 10.1126/science.1251086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasaki K., Doi S., Nakashima A., Irifuku T., Yamada K., Kokoroishi K., Ueno T., Doi T., Hida E., Arihiro K. Inhibition of SET domain-containing lysine methyltransferase 7/9 ameliorates renal fibrosis. J. Am. Soc. Nephrol. 2016;27:203–215. doi: 10.1681/ASN.2014090850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stadhouders R., Filion G.J., Graf T. Transcription factors and 3D genome conformation in cell-fate decisions. Nature. 2019;569:345–354. doi: 10.1038/s41586-019-1182-7. [DOI] [PubMed] [Google Scholar]
- Takemoto Y., Ito A., Niwa H., Okamura M., Fujiwara T., Hirano T., Handa N., Umehara T., Sonoda T., Ogawa K. Identification of cyproheptadine as an inhibitor of SET domain containing lysine methyltransferase 7/9 (Set7/9) that regulates estrogen-dependent transcription. J. Med. Chem. 2016;59:3650–3660. doi: 10.1021/acs.jmedchem.5b01732. [DOI] [PubMed] [Google Scholar]
- Tamura R., Doi S., Nakashima A., Sasaki K., Maeda K., Ueno T., Masaki T. Inhibition of the H3K4 methyltransferase SET7/9 ameliorates peritoneal fibrosis. PLoS ONE. 2018;13:e0196844. doi: 10.1371/journal.pone.0196844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Valk F.M., Bekkering S., Kroon J., Yeang C., Van den Bossche J., van Buul J.D., Ravandi A., Nederveen A.J., Verberne H.J., Scipione C. Oxidized phospholipids on lipoprotein(a) elicit arterial wall inflammation and an inflammatory monocyte response in humans. Circulation. 2016;134:611–624. doi: 10.1161/CIRCULATIONAHA.116.020838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H., Cao R., Xia L., Erdjument-Bromage H., Borchers C., Tempst P., Zhang Y. Purification and functional characterization of a histone H3-lysine 4-specific methyltransferase. Mol. Cell. 2001;8:1207–1217. doi: 10.1016/s1097-2765(01)00405-1. [DOI] [PubMed] [Google Scholar]
- Wang Y., Song F., Zhang B., Zhang L., Xu J., Kuang D., Li D., Choudhary M.N.K., Li Y., Hu M. The 3D Genome Browser: a web-based browser for visualizing 3D genome organization and long-range chromatin interactions. Genome Biol. 2018;19:151. doi: 10.1186/s13059-018-1519-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weintraub A.S., Li C.H., Zamudio A.V., Sigova A.A., Hannett N.M., Day D.S., Abraham B.J., Cohen M.A., Nabet B., Buckley D.L. YY1 is a structural regulator of enhancer-promoter loops. Cell. 2017;171:1573–1588.e28. doi: 10.1016/j.cell.2017.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang W.J., Wu X.N., Shi T.T., Xu H.T., Yi J., Shen H.F., Huang M.F., Shu X.Y., Wang F.F., Peng B.L. Regulation of transcription factor Yin Yang 1 by SET7/9-mediated Lysine methylation. Sci. Rep. 2016;6:21718. doi: 10.1038/srep21718. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
This study did not generate any unique datasets or code.