Abstract
Huwe1 is a highly conserved member of the HECT E3 ubiquitin ligase family. Here, we explore the growing importance of Huwe1 in nervous system development, function and disease. We discuss extensive progress made in deciphering how Huwe1 regulates neural progenitor proliferation and differentiation, cell migration, and axon development. We highlight recent evidence indicating that Huwe1 regulates inhibitory neurotransmission. In covering these topics, we focus on findings made using both vertebrate and invertebrate in vivo model systems. Finally, we discuss extensive human genetic studies that strongly implicate HUWE1 in intellectual disability, and heighten the importance of continuing to unravel how Huwe1 affects the nervous system.
Keywords: Huwe1, EEL-1, Ubiquitin ligase, HECT, Neuron, Neural progenitor, Neurotransmission, Axon, Transcription factor, Intellectual disability
Introduction
HECT, UBA and WWE domain containing protein 1 (Huwe1) is an E3 ubiquitin ligase that is highly conserved across the animal kingdom [1–4] (Fig. 1a). While rodent, zebrafish and fly orthologs are also generally referred to as Huwe1, the C. elegans ortholog is called Enhancer of EFL-1 (EEL-1) based on its discovery in genetic screens. Early studies found that Huwe1 was involved in regulating tumor cell proliferation/suppression, apoptosis, responses to DNA damage and embryogenesis [2–8]. Many of these initial studies refer to Huwe1 by other names including Mule, ArfBP1, Lasu1, HectH9 or Ureb1. More recently, Huwe1 was shown to have important roles in the immune system and myogenesis [9–11].
Fig. 1.
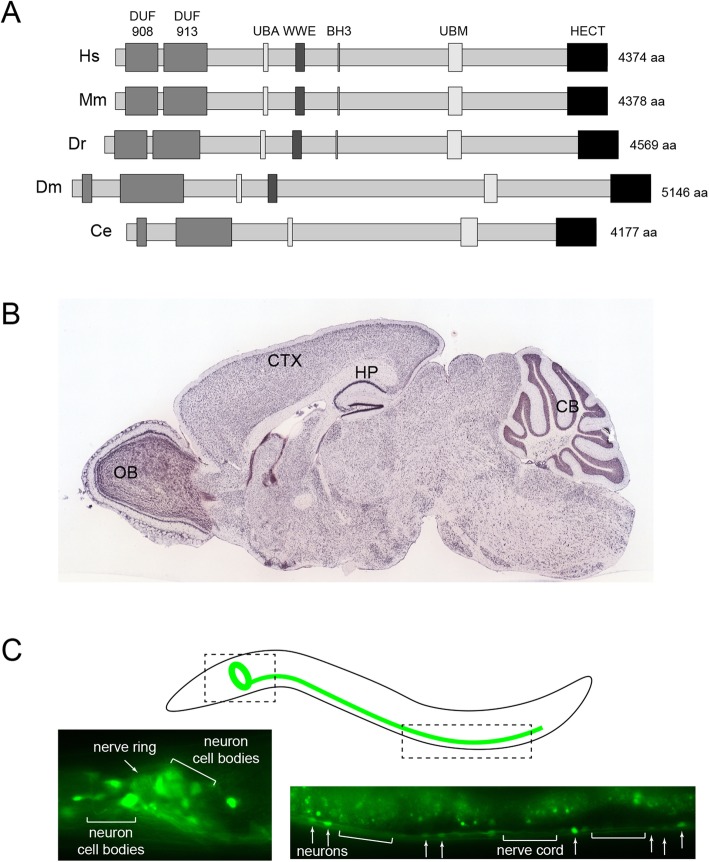
HUWE1 is evolutionarily conserved and broadly expressed in animal nervous systems. a Domain architecture of Huwe1 orthologs from Homo sapiens (Hs), Mus musculus (Mm), Danio rerio (Dr), Drosophila melanogaster (Dm) and Caenorhabditis elegans (Ce). b In situ hybridization of Huwe1 in mouse brain. Note Huwe1 is expressed throughout the brain with high expression in olfactory bulb (OB), cerebral cortex (CTX), hippocampus (HP) and cerebellum (CB). ceel-1 promoter driving GFP expression in C. elegans. eel-1 is widely expressed in the nerve ring and head neurons (left), and ventral nerve cord (right; bars = nerve cord; arrows = motor neuron cell bodies). Schematic denotes locations of images shown and axon bundles of the nerve ring and ventral nerve cord. DUF = conserved Domain of Unknown Function, UBA = Ubiquitin Associated domain, WWE = domain with conserved tryptophan (W) and glutamate (E) residues, UBM = Ubiquitin-Binding Motif (previously known as DUF4414), HECT = Homologous to the E6-AP Carboxyl Terminus (HECT)-type E3 ubiquitin ligase domain. For B, image credit: Allen Institute© 2007 Allen Institute for Brain Science. Allen Mouse Brain Atlas. Available from: https://mouse.brain-map.org/experiment/show/70445293. For C, images reprinted from Cell Reports, Vol. 19, Opperman, Mulcahy, Giles, Risley, Birnbaum, Tulgren, Dawson-Scully, Zhen, Grill, The HECT Family Ubiquitin Ligase EEL-1 Regulates Neuronal Function and Development, 822–835, Copyright 2017, with permission from Elsevier
A growing body of work over the last 10 years, spanning multiple model systems, has shown that Huwe1 is important in nervous system development and function. Extensive human genetic studies have provided compelling evidence implicating Huwe1 in multiple neurodevelopmental disorders, including both non-syndromic and syndromic forms of X-linked intellectual disability (ID). In this review, we discuss how Huwe1 contributes to nervous system development, function and disease. We hope that exploring this literature encourages further studies on Huwe1 in the nervous system, and highlights valuable directions for future investigation from both a basic science and disease perspective.
Huwe1 protein composition and nervous system expression
Huwe1 is a ubiquitin ligase in the HECT family, a group of E3 ubiquitin ligases that function as single subunit enzymes [5, 12]. A striking feature of Huwe1 is its enormous size — well over 4000 amino acids (Fig. 1a). Huwe1 has numerous protein domains including a catalytic ubiquitin ligase domain, several domains involved in ubiquitin binding, and multiple conserved domains of unknown function (Fig. 1a and Fig. 4). The best characterized domain is the HECT domain, which is located at the C-terminus and is the catalytic E3 ubiquitin ligase domain that conjugates ubiquitin to substrates [13, 14]. Other domains involved in ubiquitination include a UBA domain that binds polyubiquitin chains, a WWE domain that mediates protein-protein interactions during ubiquitination, and a ubiquitin binding motif (UBM) that contains several repeated sequences and binds ubiquitin (previously called DUF4414). There is also an extremely small Bcl-2 homology domain 3 (BH3) that is a protein-protein interaction region, and only present in vertebrate isoforms of Huwe1. Two studies have solved the structure of the HECT domain [13, 14]. Interestingly, the Huwe1 HECT domain forms a dimer, which leads to autoinhibition of catalytic ubiquitin ligase activity [14]. At present, the functional roles of many domains in Huwe1 remain unknown, and the biochemical mechanisms that allow for substrate specificity are unclear. The full complement of proteins that interact with Huwe1 and potential non-ubiquitin ligase mechanisms of Huwe1 function also remain largely unexplored.
Fig. 4.
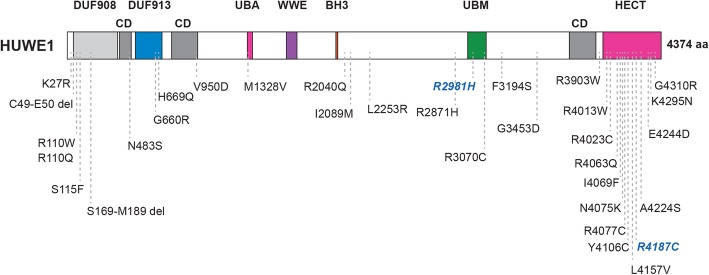
Mutations in HUWE1 associated with non-syndromic and syndromic forms of X-linked Intellectual Disability. Schematic of human HUWE1 and amino acid changes resulting from ID-associated mutations in HUWE1. ID-associated mutations occur broadly across the HUWE1 protein sequence with two hotspots, the HECT domain and DUF908 domain. Blue italics highlight two ID-associated mutations that were shown to result in loss-of-function when tested using in vivo assays of neuron function in C. elegans. Note three Conserved Domains (CD) of unknown function are annotated
Interest in Huwe1 function in the nervous system becomes apparent when one simply considers how broadly expressed this molecule is across animal nervous systems. In both humans and rodents, Huwe1 is expressed throughout the adult brain with particularly strong expression in the olfactory bulb, superficial layers of the cortex, hippocampus and cerebellum (Fig. 1b) [15–17]. In adult C. elegans, the Huwe1 ortholog EEL-1 is expressed broadly throughout the nervous system. This includes sensory neurons, the nerve ring (a central bundle of hundreds of axons), and the motor neurons of the nerve cord (Fig. 1c) [18]. The broad expression of Huwe1 in adult mammalian and C. elegans nervous systems is consistent with Huwe1 having widespread, important roles in the nervous system, which we discuss further.
Huwe1 ubiquitin ligase activity in neural progenitor proliferation and differentiation
Mammalian nervous systems are composed of millions to billions of neurons. Waves of cell proliferation in various regions of the developing brain are critical to achieve these large numbers. Two particularly important regions for brain growth are the ventricular zone (VZ) of the neocortex (Fig. 2a) and the external granule layer (EGL) of the cerebellum (Fig. 2b). Proliferation of neural stem cells and neural progenitors takes place in deep layers of the neocortex. As precursors differentiate into neurons they migrate to the cortical plate (CP). In the cerebellum, proliferation of neural progenitors occurs in the superficial EGL and differentiating neurons migrate deeper to the internal granule layer (IGL). Interestingly, Huwe1 expression is lowest in regions where proliferation of neural stems cells and progenitors occurs and expression gradually increases away from these regions where neurons are differentiating and migrating [15]. Consistent with these expression patterns, both cell-based and in vivo approaches have shown that Huwe1 plays prominent roles in early development of the nervous system.
Fig. 2.
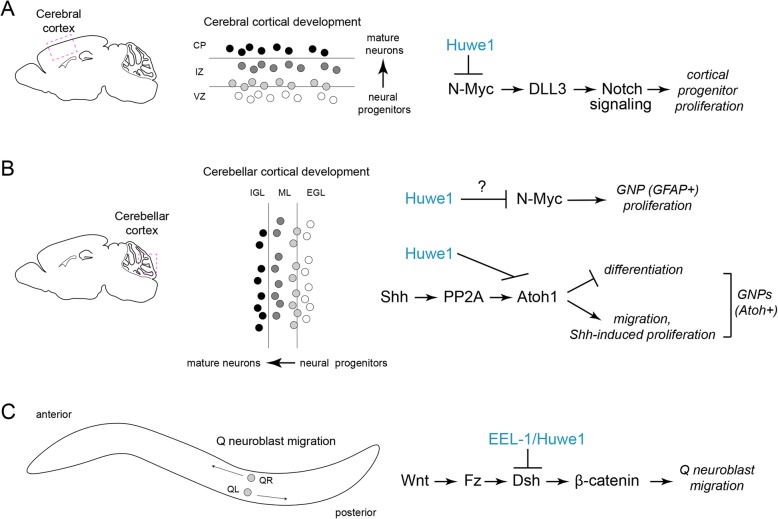
HUWE1 regulates neural development. (a and b) Shown to the left are diagrams of sagittal section of mouse brain. Boxes indicate a portion of (a) cerebral cortex and (b) cerebellar cortex. Center is a schematic of Huwe1 expression in neural progenitors that are proliferating, differentiating and migrating during development in (a) cerebral cortex and (b) cerebellar cortex (white = low expression, black = high expression). To the right are molecular pathways Huwe1 regulates to affect progenitor development in (a) cerebral cortex and (b) cerebellar cortex. Note that in the cerebellum Huwe1 potentially affects two pathways in different GNP populations. c Shown is a schematic of Q neuroblast migration in C. elegans which is mediated by Wnt signaling. To the left is the molecular mechanism by which EEL-1 and HUWE1 influence Wnt signaling in neuroblast migration and mammalian cell-based assays
The first cell-based experiments focused on differentiation of cultured embryonic stem (ES) cells into a neuronal lineage [15]. Both germline and conditional knock out of Huwe1 impaired ES cell differentiation. In cultured mouse brain slice, Huwe1 knockdown and knockout increased proliferation of cortical progenitors and reduced differentiation to neurons [15]. A subsequent in vivo study conditionally deleted the Huwe1 ubiquitin ligase domain broadly in neurons [19]. Huwe1 conditional knockouts (cKO) displayed neonatal lethality within 24 h of birth. Increased numbers of neural progenitors were observed in the developing cortex of Huwe1 cKO animals further solidifying the concept that Huwe1 inhibits neural progenitor proliferation. Consistent with this, overexpression of Huwe1 in cultured cortical slices caused the opposite phenotype, fewer dividing cells. Huwe1 ubiquitin ligase activity also functions later in development, as Huwe1 ubiquitin ligase cKOs displayed defects in differentiation of neurons in the cortex [19]. Taken together, these studies indicate that Huwe1 ubiquitin ligase activity inhibits proliferation of neural progenitors early in development, and encourages neuronal differentiation during cortical development. These functions of Huwe1 are required for proper patterning of the cortex.
Because Huwe1 cKO in neurons throughout the brain resulted in lethality [19], it was plausible Huwe1 had potential roles in nervous system development outside the cortex. This was tested when Lasorella and colleagues conditionally deleted Huwe1 ubiquitin ligase activity in granule neuron progenitors (GNPs) and radial glia of the cerebellum using the GFAP promoter to express Cre [20]. 50% of these Huwe1 cKO mice died within 4 weeks of birth. Survivors developed ataxia consistent with cerebellar dysfunction. Similar to results in cortex, cerebellar GNPs lacking Huwe1 displayed expanded proliferation resulting in increased GNP numbers, increased EGL size, and reduced numbers of mature neurons in the IGL [20]. Abnormalities in cerebellar glia were also observed.
GNPs of the developing cerebellum are not a homogenous population of cells. As a result, when Huwe1 was conditionally knocked out in a subset of GNPs that expressed the Atoh1 transcription factor, GNP differentiation was impaired [21]. Proliferation of Atoh1-positive GNPs could be affected in Huwe1 cKOs, but only when proliferation was increased in culture using the mitogenic stimulant Sonic Hedge Hog (Shh). Thus, compelling evidence from cell culture, brain slice, and multiple intact developing brain regions indicates that Huwe1 ubiquitin ligase activity is critical in regulating the switch from proliferation to differentiation in neural progenitors.
Huwe1 affects multiple transcriptional programs to influence proliferation and differentiation
Let us now comment on the mechanisms by which Huwe1 regulates neural progenitor proliferation and differentiation early in development. As noted above, knockout strategies in which the HECT ubiquitin ligase domain was conditionally deleted indicated that Huwe1 ubiquitin ligase activity is required to switch neural progenitors from a proliferative state towards differentiation in the developing nervous system. Genetic and biochemical results indicated that this occurs because Huwe1 can ubiquitinate and inhibit two transcription factors, N-Myc and Atoh1 [15, 19, 21]. Consistent with this, N-Myc and Atoh1 are expressed in proliferating neural stem cells and progenitors, are down regulated as cells differentiate and can affect neural progenitor proliferation and/or differentiation.
Affinity purification proteomics with a human neuroblastoma cell line initially identified Huwe1 as an N-Myc binding protein [15]. Several further in vitro and cell-based biochemical experiments indicated that Huwe1 polyubiquitinates N-Myc resulting in degradation by the 26S proteasome. Experiments in ES cells, cultured brain slice and intact brain showed that Huwe1 inhibits N-Myc to affect proliferation and differentiation of neural stem cells and progenitors. For example, Huwe1 knockout in ES cells impairs differentiation towards neural cell fates, which is rescued by N-Myc knockdown [15]. Consistent with this, levels of the N-Myc transcriptional target Cyclin D2 are elevated when Huwe1 is impaired. Similar results in cultured brain slice indicate that increased proliferation and decreased differentiation of neural progenitors caused by Huwe1 knockdown is rescued by N-Myc knockdown [15]. In developing mouse brain, Huwe1 cKO in cortex resulted in increased neuronal progenitor proliferation that was suppressed by N-Myc knockdown [19]. In both the cortex and cerebellum, loss of Huwe1 function increased N-Myc expression, and altered the N-Myc transcriptional target Cyclin D2 [19, 20]. These results indicate that Huwe1 inhibits N-Myc to affect cortical progenitor proliferation. Effects of Huwe1 on N-Myc might also influence cerebellar progenitor proliferation, but this remains unclear.
Another principle gene affected by Huwe1 inhibition of N-Myc is the Notch ligand DLL3 [19]. DLL3 was discovered as a possible downstream effector of N-Myc by transcriptomics with a large number of gliomas that had differing levels of Huwe1 expression. DLL3 was confirmed as a transcriptional target of N-Myc, as N-Myc binds the DDL3 promoter and ectopic N-Myc expression increases DLL3 levels and Notch signaling. Consistent with this, Huwe1 degradation of N-Myc influences DLL3 [19]. For example, Huwe1 cKO animals display increased DLL3 mRNA and Notch1 signaling in the brain. In cultured brain slice, Huwe1 cKO causes increased neural progenitor proliferation which is suppressed by knocking down DLL3 or N-Myc. Collectively, this is strong evidence that Huwe1 directly inhibits N-Myc thereby affecting DLL3 transcription and, ultimately, neural progenitor proliferation and differentiation in the developing cortex (Fig. 2a).
In the cerebellum, Huwe1 inhibition of N-Myc might affect proliferation of GNPs that express N-Myc [20], but this remains untested. A subset of cerebellar GNPs express the Atoh1 transcription factor. Huwe1 ubiquitinates and degrades Atoh1, and robustly affects development of GNPs that express this transcription factor [21]. This discovery initially stemmed from affinity purification proteomics using HEK 293 cells, which identified Huwe1 as an Atoh1 binding protein. Cell-based experiments showed that Huwe1 knockdown and overexpression affect Atoh1 ubiquitination and stability. Further supporting these findings, Huwe1 cKO resulted in increased Atoh1 protein levels in GNPs in culture and in vivo.
Overexpression of Atoh1 in cultured, purified GNPs impairs differentiation, but does not affect progenitor proliferation [22]. Consistent with Huwe1 being a negative regulator of Atoh1, Huwe1 cKOs where the Atoh1 promoter was used to drive Cre expression mimicked Atoh1 overexpression phenotypes with decreased differentiation of cultured GNPs, but no effect on GNP proliferation [21]. In cultured cerebellar brain slice, Huwe1 knockdown results in impaired differentiation of GNPs. This effect was strongly suppressed when Huwe1 and Atoh1 were simultaneously knocked down [21]. Further experiments evaluating GNP differentiation in vivo using conditional knockout alleles for both Huwe1 and Atoh1 will be valuable in further evaluating this genetic relationship. Nonetheless, existing results with knockdown approaches taken together with extensive, high-quality biochemical studies support the conclusion that Huwe1 ubiquitinates and inhibits Atoh1 to affect differentiation of a subset of GNPs in the cerebellum.
Interestingly, Huwe1 can inhibit proliferation of GNPs expressing Atoh1, but this only occurs when GNPs are deprived of and re-treated with Shh to stimulate proliferation [21]. This likely occurs because Shh signaling through PP2A affects dephosphorylation of Atoh1 at two serine residues. Atoh1 dephosphorylation then prevents Huwe1 ubiquitination and degradation. Thus, Huwe1 and Shh-PP2A signaling display differing effects on Atoh1 stability, which influences proliferation of Atoh1 expressing GNPs (Fig. 2b).
Work on cochlear development also found that Huwe1 ubiquitinates and inhibits Atoh1 to regulate hair cell differentiation [23]. This independent study reproduced proteomic and biochemical findings to confirm that Huwe1 binds and ubiquitinates Atoh1. Interestingly, Casein Kinase 1 (CK1) phosphorylation of Atoh1 affected ubiquitination by Huwe1. This provides further evidence that phosphorylation of Atoh1 affects Huwe1-mediated degradation, and demonstrates that Huwe1 inhibits Atoh1 to affect cellular differentiation in multiple cell types.
In summary, Huwe1 restricts genetic programs required to switch from proliferation to differentiation in multiple types of neural progenitors. This occurs by Huwe1 ubiquitin ligase activity triggering proteasomal degradation and inhibition of the N-Myc and Atoh1 transcription factors. This compelling body of work on Huwe1 in early nervous system development has several future research directions poised to be particularly fruitful. 1) Determining how Huwe1 is regulated in the nervous system. 2) Identifying Huwe1 binding proteins that are not ubiquitination substrates, but potentially function with this enormous molecule. 3) Evaluating if further Huwe1 ubiquitination substrates, that are novel or previously identified in cell-based studies, have functional relevance in early nervous system development.
There is one final point regarding nervous system disease that is worth making here. Genetic programs involved in early development of the nervous system are often hijacked in brain cancers. Indeed, genetic changes in HUWE1 are implicated in patients with glioblastoma and medulloblastoma [19, 21]. Whether HUWE1 is involved in neuroblastoma remains unknown. These intriguing initial links to cancer now encourage further research aimed at exploring how broadly and frequently HUWE1 mutations occur in different brain cancers.
Huwe1 and adult stem cell proliferation
Huwe1 can also affect neural stem cell proliferation after completion of the developmental program. In the adult hippocampus, neural stem cells in the dentate gyrus must maintain a balance between proliferation and quiescence. Too much proliferation and the stem cell compartment accumulates DNA damage and becomes exhausted; an excess of quiescence results in a dearth of new neurons. When Huwe1 was conditionally knocked out in adult animals using tamoxifen-inducible Cre, stem cell proliferation increased and differentiation decreased [24]. Experiments that varied the timing of cell cycle labelling relative to the induction of Huwe1 cKO revealed that Huwe1 has a prominent role in promoting quiescence of stem cells that are in a proliferative state. Experiments with Huwe1 cKO after 5 months indicated that defects in neural stem cell quiescence caused by loss of Huwe1 function result in long-term defects in proliferation. Thus, Huwe1 is needed to restrain proliferation, ensure a return to quiescence, and promote long-term maintenance of the adult neural stem cell compartment.
Huwe1 affects adult stem cell proliferation by ubiquitinating and degrading the transcription factor Ascl1 [24]. AscI was initially identified as a Huwe1 ubiquitination substrate using affinity purification proteomics from purified hippocampal stem cells. Ascl1 polyubiquitination was inhibited by Huwe1 knockdown and Ascl1 stability was increased when Huwe1 was conditionally knocked out in cultured neural stem cells. Huwe1 polyubiquitination of Ascl in neural stem cells was independently verified [25]. Genetic outcomes support these biochemical and proteomic findings, as Huwe1 cKO in adulthood resulted in increased Ascl1 expression and increased numbers of Ascl1 expressing neural progenitors in the hippocampus [24]. This was accompanied by reduced differentiation of stem cells into neural progenitors. Consistent with Huwe1 inhibiting Ascl1, the Ascl1 transcriptional targets Cyclin D1 and D2 were elevated in stem cells lacking Huwe1.
These results suggest that Huwe1 constrains Ascl1 transcription factor levels in neural stem cells to restrict proliferation and promote quiescence in the hippocampus of adult mice. Going forward, genetic experiments in which Huwe1 and Ascl1 are both eliminated will be important to solidify this intriguing discovery.
Huwe1 is a conserved regulator of neuronal migration
It can be challenging to dissect how genetic perturbations affect differentiation and migration of maturing neurons because problems with differentiation can indirectly influence migration. Nonetheless, because genetic manipulation of Huwe1 caused consistent, conserved effects on migration, this warrants commentary. Furthermore, studies using C. elegans have provided valuable insight into the molecular mechanisms Huwe1 influences to specifically affect neuroblast migration.
Huwe1 effects on neural migration have been most extensively studied by evaluating granule neuron migration in the cerebellum [20]. Normally, large numbers of differentiating granule neurons migrate from the EGL through the molecular layer (ML) to the IGL (Fig. 2b). In Huwe1 cKOs, few cells reach the IGL, and most cells are trapped in the ML or EGL layers. It is important to note that Huwe1 cKO in cerebellum also affects the organization of Bergmann glial cells [20]. Bergmann glia provide a structural scaffold and cues for migrating granule neurons in the developing cerebellum. Thus, granule neuron migration defects in Huwe1 cKOs are potentially caused by a combination of differentiation defects in GNPs and abnormalities in Bergmann glia. Experiments with cultured brain slice delved into the molecular mechanism by which Huwe1 affects granule neuron migration in the cerebellum [21]. Huwe1 knockdown in developing cerebellum dramatically impaired migration of GNPs from the EGL to the IGL. These defects were mimicked by overexpression of Atoh1, a transcription factor and Huwe1 ubiquitination substrate. Importantly, double knockdown of Huwe1 and Atoh1 suppressed migration defects caused by Huwe1 single knockdown. These results indicate that Huwe1 inhibits Atoh1 to promote GNP migration in cerebellum (Fig. 2b).
C. elegans has also yielded important insight into how Huwe1 regulates neuroblast migration during early nervous system development. As noted earlier, C. elegans has a single, highly conserved Huwe1 ortholog called EEL-1. In vivo genetic results indicate EEL-1 has a pronounced role in Wnt-mediated migration of Q neuroblasts [26]. Because Wnt signaling regulates Q neuroblast migration, when Wnt secretion is reduced in vps-29 mutants the left Q neuroblast (QL) progeny migrate incorrectly to the anterior. This genetic background was used to identify regulators of Wnt signaling. An RNAi screen against a swath of ubiquitin machinery found that eel-1 knockdown suppressed migration defects in vsp-29 mutants. Cell-specific knockdown confirmed that EEL-1 functions cell autonomously in Q neuroblasts. These results indicated that EEL-1 likely functions downstream of Wnt signaling. Further genetic results showed that EEL-1 functions upstream of the β-catenin BAR-1, a Wnt signaling effector. Biochemical experiments using HEK 293 cells demonstrated that Huwe1 ubiquitinates Dishevelled, and Wnt stimulates this ubiquitination [26]. Proteomic and coimmunoprecipitation results indicated endogenous Huwe1 binds two Dishevelled isoforms (Dvl2 and Dvl3) in cultured murine embryonic fibroblasts. Similar to findings on Huwe1 ubiquitination of Atoh1 during cochlear development [23], CK1 affects Huwe1 ubiquitination of Dishevelled [26]. When Huwe1 was knocked down and evaluated in cell-based assays for Wnt signaling, outcomes indicated that Huwe1 can act as a conserved inhibitor of Wnt signaling. Interestingly, Huwe1 ubiquitination does not trigger proteasomal degradation of Dishevelled, but rather blocks its activation thereby preventing downstream signaling to β-catenin. These findings demonstrate that Huwe1 is a conserved inhibitor of Wnt signaling that influences neuroblast migration in C. elegans in vivo (Fig. 2c).
Going forward, it will be important to know if Huwe1 affects Wnt signaling to influence neural migration in other organisms. Since Wnts affect axon polarity, axon guidance, synapse formation and synaptic plasticity [27], it could be valuable to explore whether Huwe1 plays a role in these events. This is encouraged by findings from Drosophila (discussed below) that show Huwe1 can affect axon development [28], a possible interaction between Huwe1 and another ubiquitin ligase that affects Wnt signaling [29], and evidence that Huwe1 affects Wnt signaling during intestinal oncogenesis [30]. Studies spanning rodent and C. elegans model systems also bring to the forefront several questions about Huwe1 and cell migration. Can it be more definitively determined if Huwe1 effects on migration are a secondary consequence of altered progenitor differentiation, the result of Huwe1 function in glia, or a combination of both? How widespread a role does Huwe1 have in neural migration? Finally, can future experiments provide evidence that Huwe1 directly effects cell migration signaling in mammals in vivo? Experiments from C. elegans suggest that this last possibility might occur in other systems.
Huwe1 influences axon development
Thus far, we have detailed the extensive progress made in understanding Huwe1 function and mechanisms that impact early nervous system development. We now pivot to discuss a growing body of evidence that indicates Huwe1 has other roles later in development and in neuron function. We begin with the effects of Huwe1 on axon development.
Initial studies on cultured ES cells differentiated into neurons [15] and in cerebellum [20] suggested that Huwe1 KO impairs neurite and parallel fiber axon growth. While this hinted at a possible role for Huwe1 in axon development, these defects could simply arise from impaired differentiation. However, findings from other systems discussed below suggest the question of whether Huwe1 affects axon outgrowth might warrant further more direct evaluation.
The first clear indication that Huwe1 can influence axon development arose from experiments in Drosophila. Flies overexpressing exogenous human HUWE1 displayed increased terminal axon branching of dorsal cluster neurons [28]. Genetic results indicated HUWE1 causes excess axon branching by negatively regulating Wnt signaling most likely via effects on Dishevelled. This is similar to results showing that mammalian Huwe1 and C. elegans EEL-1 inhibit Wnt signaling by effecting Dishevelled [26]. Despite this interesting finding, it is notable that we still know nothing about how Huwe1 loss of function affects nervous system development and function in flies. This remains rich ground for future experiments, particularly given the rapid, powerful genetics of the Drosophila system.
Subsequent studies in C. elegans combined eel-1 (lf) mutants with enhancer genetics to explore how EEL-1 affects axon development. The sensitizing genetic background used involved mutants for components of a highly conserved, atypical Skp/Cullin/F-box (SCF) ubiquitin ligase complex that consists of the RPM-1 E3 ubiquitin ligase and the F-box protein FSN-1 [31–34]. The RPM-1/FSN-1 ubiquitin ligase complex is an important regulator of axon termination in C. elegans [35, 36], and functions as both a ubiquitin ligase and a signaling hub to affect numerous downstream signaling pathways [31, 37–42]. In two different eel-1 (lf) mutants, low levels of axon termination defects were observed in two types of neurons, the SAB motor neurons and the mechanosensory neurons that sense gentle touch [18]. Interestingly, eel-1; fsn-1 double mutants had strong enhancer effects indicating that EEL-1 and FSN-1 function in parallel pathways to regulate axon termination. rpm-1 (lf) mutants have more severe, higher frequency axon termination defects than fsn-1 mutants, because RPM-1 functions as both a ubiquitin ligase and a signaling hub. rpm-1; eel-1 double mutants showed no increase in axon termination defects indicating that EEL-1 and RPM-1 function in the same pathway to regulate axon termination.
Mammalian cell-based experiments have shown that Huwe1 and the human RPM-1 ortholog, called Protein associated with Myc (PAM) or Myc binding protein 2 (MYCBP2), can interact in non-neuronal cell lines [43]. However, there is no evidence that this occurs in neurons or in C. elegans, and EEL-1 was not noted in recent comprehensive proteomic studies with RPM-1 [42]. Like HUWE1, human PAM binds MYC [44], but the domain in PAM required for this interaction is not conserved in C. elegans RPM-1 [31]. Moreover, axon termination defects in eel-1 (lf) mutants were not suppressed by null alleles of mml-1, the C. elegans MYC ortholog. Thus, it seems unlikely that RPM-1 and EEL-1 function in the same pathway to regulate axon termination by affecting MYC. At present, a molecular explanation for why EEL-1 and RPM-1 function in the same pathway remains absent, but this will hopefully become more clear as proteomic and genetic studies continue with both molecules.
Overall, experiments from both flies and worms indicate that Huwe1 is a conserved regulator of axon development. However, the use of sensitizing genetic backgrounds in C. elegans and effects of Huwe1 on axon branching in a subset of neurons in flies suggest these are likely to be less prominent Huwe1 functions. Because Huwe1 inhibits Wnt signaling to affect neuroblast migration in worms and axon branching in flies, it is possible other genetic programs Huwe1 affects to regulate differentiation or migration might influence axon termination or branching later in development. Whether this is the case or not awaits further experiments more temporally and anatomically focused on axon development.
Huwe1 affects inhibitory GABAergic and Glycinergic neuron function
High levels of Huwe1 expression persist in neurons broadly across the adult nervous systems of humans, mice and C. elegans [16–18] (Fig. 1B, C). In both mouse and human, Huwe1 expression is particularly high in superficial cortex, hippocampus and cerebellum. One possible explanation for these observations is that Huwe1 expression must be maintained in mature neurons to prevent cell proliferation. Alternatively, widespread expression might occur because Huwe1 has other roles in neuron development or function beyond its effects on neural progenitor proliferation, differentiation and migration during early nervous system development. Further support for this second possibility came from the observation that Huwe1 is present in synaptosomes isolated from mouse brain [45]. Consistent with this, imaging in live C. elegans found that EEL-1/Huwe1 localizes to the presynaptic terminals of inhibitory GABAergic neurons [18]. In cultured mouse spinal cord neurons, Huwe1 is present at or near postsynaptic terminals [46]. These observations indicate that in both worms and mammals Huwe1 is present at synapses where it would be capable of influencing neuron function.
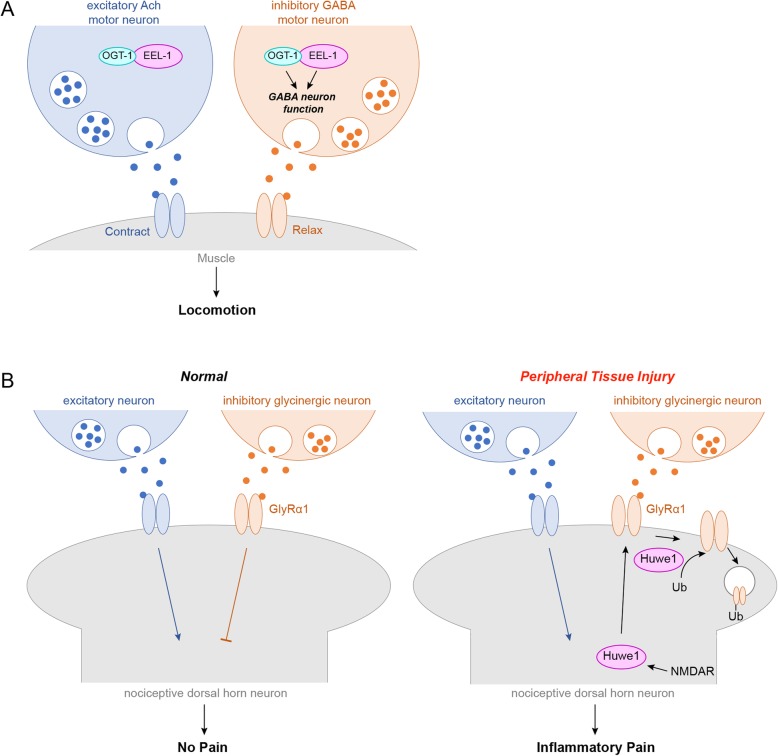
The first functional evidence indicating that Huwe1 affects neurotransmission and neuronal function emerged from in vivo studies using C. elegans [18]. This relied upon both genetic loss- and gain-of-function approaches, pharmacological manipulation, and electrophysiological readouts using the inhibitory GABAergic motor neurons. The motor circuit of C. elegans controls locomotion and is composed of opposing cholinergic and GABAergic motor neurons that excite or inhibit body wall muscles, respectively (Fig. 3a). The circuit can be manipulated and studied pharmacologically using aldicarb, an acetylcholine esterase inhibitor. Aldicarb blocks acetylcholine (Ach) clearance from neuromuscular junctions leading to excess cholinergic signaling, increased muscle contraction, and eventually paralysis. There are two general principles that emerge from a large number of studies with aldicarb [47]. 1) Mutants with defective Ach function have slower build-up of Ach leading to slower paralysis, or resistance to aldicarb. 2) In contrast, mutants with impaired inhibitory GABA neuron function have reduced inhibition onto muscles. As a result, aldicarb treatment and Ach accumulation acts more quickly resulting in more rapid paralysis, or hypersensitivity to aldicarb. eel-1 null mutants are hypersensitive to aldicarb compared to wild-type animals suggesting that EEL-1 affects GABA neuron function [18]. EEL-1 overexpression results in the opposing phenotype with slower paralysis in response to aldicarb, suggesting that GABA neuron function is increased. Electrophysiological recordings confirmed that eel-1 mutants have robust defects in GABAergic presynaptic transmission, but normal cholinergic transmission. Genetic results also demonstrated that EEL-1 functions cell autonomously in GABA neurons to regulate presynaptic transmission, consistent with EEL-1 localization at GABAergic presynaptic terminals. Thus, EEL-1 is expressed in both cholinergic excitatory motor neurons and inhibitory GABAergic motor neurons, but predominantly influences GABAergic transmission (Fig. 3a). Importantly, effects of EEL-1 on synapse formation in GABA neurons was ruled out indicating defects in GABAergic transmission in eel-1 mutants most likely stem from direct effects on presynaptic transmission.
Fig. 3.
Huwe1 regulates inhibitory neurotransmission. a Schematic of a C. elegans motor circuit showing that EEL-1/HUWE1 is expressed in excitatory cholinergic and inhibitory GABAergic motor neurons, but preferentially affects GABAergic transmission. EEL-1 forms a complex with OGT-1 and functions parallel to OGT-1 to regulate GABA neuron function. b Schematic of excitatory and inhibitory connections to a mouse dorsal horn neuron in the spinal cord. Normally (left), glycinergic transmission inhibits excitatory input to nociceptive neurons. After peripheral tissue damage (right), Huwe1 is required to reduce glycinergic inhibitory transmission that stimulates inflammatory pain. This requires NMDAR-dependent recruitment of Huwe1, which ubiquitinates GlyRα1 and triggers GlyRα1 endocytosis (right). For A, diagram originally published in the Journal of Biological Chemistry. Giles, Desbois, Opperman, Tavora, Maroni, Grill. J. Biol. Chem. 2019; 294: 6843–6856.© Giles, Desbois, Opperman, Tavora, Maroni, Grill
There is some evidence EEL-1 has a more modest role in synapse formation [18]. Although electrophysiological results indicate synaptic transmission is not impaired in the cholinergic motor neurons of eel-1 mutants, mild defects in synapse formation were observed in these neurons. While GABA neurons formed synapses normally in eel-1 mutants, the use of an fsn-1 sensitizing genetic background revealed a role for EEL-1 in synapse formation in GABA neurons. Similar to genetic outcomes with axon termination, eel-1 and rpm-1 function in the same pathway to regulate synapse formation in both cholinergic and GABAergic motor neurons. Thus, EEL-1 plays a prominent role in GABAergic presynaptic transmission, and has less pronounced effects on synapse formation.
Initial efforts to decipher how EEL-1 regulates GABAergic transmission relied upon in vivo affinity purification proteomics using C. elegans [48]. Importantly, proteomics was facilitated by the widespread expression of EEL-1 across the worm nervous system, despite the preferential effects of EEL-1 on GABAergic transmission. O-linked β-N-acetylglucosamine transferase 1 (OGT-1) was identified as a prominent EEL-1 binding protein, and this interaction was confirmed by coimmunoprecipitation from C. elegans. OGT-1 is a highly conserved glycosyltransferase that post-translationally modifies many cytosolic and nuclear proteins [49, 50]. OGT is broadly expressed in the nervous system of mammals including high expression in the brain, and it is localized to synapses where it modifies synaptic proteins [51–53]. In C. elegans, OGT-1 was localized to presynaptic terminals of GABA motor neurons which is a reasonable subcellular location for OGT-1 to bind and function with EEL-1 [48]. Results with automated aldicarb assays showed that ogt-1 mutants were hypersensitive to aldicarb suggesting that OGT-1 regulates GABA neuron function similar to EEL-1. Analysis of ogt-1; eel-1 double mutants, and transgenic rescue experiments showed that OGT-1 and EEL-1 function cell autonomously in parallel to regulate GABA neuron function, but do not affect synapse formation in these neurons [48]. The most likely explanation for these findings is that OGT-1 and EEL-1 work together in a complex to affect GABAergic transmission (Fig. 3a). While EEL-1 ubiquitin ligase activity is required for GABA neuron function, outcomes from genetic, proteomic and biochemical experiments suggest that OGT-1 is unlikely to be an EEL-1 ubiquitination substrate [48]. A better understanding of the relationship between EEL-1 and OGT-1, and how the EEL-1/OGT-1 protein complex affects GABA neuron function now awaits further study. Importantly, more work is encouraged by the finding that human OGT binds HUWE1 indicating that this interaction is conserved [48].
Interestingly, transgenic rescue experiments demonstrated differing roles for the catalytic activity of EEL-1 and OGT-1 in GABA neuron function. While EEL-1 ubiquitin ligase activity was required for GABA neuron function, OGT-1 glycosyltransferase activity was dispensable [48]. Thus, EEL-1 and OGT-1 likely converge on a single molecular target or cellular process that affects GABA neuron function, but this cannot simply be the result of both ubiquitination and glycosylation activity. Rather, OGT-1 is likely to have a novel molecular function as part of a complex with EEL-1. The nature of this molecular mechanism remains unknown, but could result from lesser studied roles of OGT as a scaffold protein [49, 54].
It is unknown if Huwe1 regulates GABAergic transmission in mammals, but recent work indicates that Huwe1 affects another type of inhibitory neurotransmission, glycinergic transmission [46]. Cell-based experiments with HEK 293 cells showed that Huwe1 can directly ubiquitinate Glycine receptor α1 (GlyRα1). Huwe1 is present in cultured dorsal horn neurons where it localizes to a small percentage of postsynaptic terminals. Ubiquitination of GlyRα1, which is stimulated by excitation with NMDA, is blocked when Huwe1 is knocked down in spinal dorsal horn. The cellular effects of Huwe1 on GlyRα1 were determined by examining endocytosis in cultured spinal neurons [46]. Huwe1 knockdown reduced activity-induced surface labeling and internalization of GlyRα1 indicating that Huwe1 is required for GlyRα1 endocytosis.
To address the functional effects of Huwe1 on glycinergic transmission, Huwe1 was knocked down and electrophysiology was performed on spinal cord slice. Huwe1 knockdown resulted in failure to decrease inhibitory glycinergic transmission following an inflammatory challenge. Consistent with this, Huwe1 knockdown blunted pain responses to inflammation. It is notable that Huwe1 knockdown did not affect baseline glycinergic transmission in the absence of an inflammatory challenge. This could be because Huwe1 only modifies glycinergic transmission in the context of inflammation, or because Huwe1 knockdown does not completely abolish Huwe1 function. These results demonstrate that after tissue injury Huwe1 ubiquitinates GlyRα1 to decrease inhibitory glycinergic neurotransmission and facilitate pain sensation (Fig. 3b). Experiments with Huwe1 null animals will be important in further assessing Huwe1 effects on glycinergic transmission in the spinal cord. Nonetheless, existing findings suggest that Huwe1 could be an important new molecular target for treating inflammatory pain.
Thus, results from C. elegans and rodent model systems indicate that Huwe1 has a conserved role in regulating inhibitory neurotransmission. While much remains unknown about how EEL-1 regulates GABAergic transmission, initial outcomes from in vivo proteomics and genetics indicates that EEL-1 forms a complex with OGT-1, and functions parallel to OGT-1 to regulate GABA neuron function. Continued exploration of the proteomic and genetic space surrounding EEL-1 using invertebrate models like C. elegans is ideally positioned to further reveal how EEL-1 regulates GABAergic transmission. The discovery that Huwe1 regulates inhibitory glycinergic neurotransmission in spinal cord following tissue injury is another prominent step forward. These findings across systems prompt an important question: Does Huwe1 regulate GABAergic and/or glycinergic inhibitory neurotransmission in the brain?
Huwe1 in mitochondrial function and degradation
We now turn to emerging evidence that Huwe1 can effect mitochondria. This includes regulation of mitochondrial function and fragmentation, as well as mitochondrial turnover via mitophagy. Huwe1 effects on mitochondria could have important implications in the nervous system where mitochondria provide energy and calcium buffering for neuron function, and altered turnover of mitochondria is implicated in nervous system disease.
Mitofusins are key regulators of mitochondrial fusion and function, affect neurodegeneration, and are causally implicated in Charcot-Marie-Tooth (CMT) Disease which features peripheral axon degeneration [55–58]. Huwe1 regulates Mitofusin 2 (Mfn2), the Mitofusin isoform that is involved in CMT and neurodegeneration [59]. This discovery stemmed from affinity purification proteomics that identified Huwe1 as an Mfn2 binding protein in cell lines. Cell-based experiments using HEK 239 cells showed this interaction involves the BH3 domain of Huwe1, and is dependent upon serine phosphorylation of Mfn2. Huwe1 knockdown prevented degradation of Mfn2 and mitochondrial fragmentation in response to pharmacologically-induced stress in the U2OS non-neuronal cell line. Consistent with Huwe1 ubiquitinating and inhibiting Mfn2, Huwe1 knockdown results in increased U2OS cell death, which is suppressed by simultaneous Mfn2 knockdown [59]. A subsequent study using S2 insect cells showed that steric acid signaling opposes Huwe1 effects on Mfn2 thereby influencing mitochondrial fragmentation [60].
Huwe1 can also affect mitophagy, a specific form of autophagy that allows removal of damaged mitochondria. Mitophagy is regulated by the pro-autophagy molecule autophagy/beclin-1 regulator-1 (AMBRA1). Affinity purification proteomics using Hela cells stimulated to undergo mitophagy identified Huwe1 as a an AMBRA1 binding protein [61]. Mitophagy induced by mitochondrial membrane targeting of AMBRA1 was rescued by Huwe1 knockdown. This result suggests that AMBRA1 functions through Huwe1 to influence mitophagy. This is further supported by the observation that Huwe1 ubiquitination of Mfn2 was increased by coexpression of membrane targeted AMBRA1. These results indicate that AMBRA1 functions with Huwe1 to regulate Mfn2 degradation and mitophagy. Interestingly, Huwe1 also ubiquitinates AMBRA1 leading to AMBRA1 phosphorylation and induction of mitophagy. This suggests that Mfn2 and mitophagy are affected by a complex signaling relationship between Huwe1 and AMBRA1.
While these are important discoveries, effects of Huwe1 on mitochondria and mitophagy have only been shown in non-neuronal cell lines to date. Whether Huwe1 degrades Mfn2 to affect mitochondrial function, fragmentation and mitophagy in the nervous system remains unknown. However, if this is the case, it would have intriguing implications for nervous system health and disease.
HUWE1 copy number variations and non-syndromic intellectual disability
The first genetic links between human HUWE1 and neurodevelopmental disorders emerged with the discovery that X-chromosome microduplications that lead to copy number variations (CNV) in HUWE1 are associated with non-syndromic, X-linked ID [62–64]. One of these studies also identified missense mutations in HUWE1 associated with non-syndromic X-linked ID [64], which we discuss later. Since these early discoveries, a plethora of human genetic studies have linked HUWE1 genetic changes with ID. While genetic changes in HUWE1 are associated with other neurodevelopmental conditions, such as seizure/epilepsy, autism and schizophrenia, we primarily focus on ID because it has the most compelling links to HUWE1. Patients with HUWE1 genetic changes do have developmental abnormalities outside of the nervous system (for example slowed growth and altered facial features) that we do not comment on here. We believe a discussion of these topics is better left to a review with a greater clinical focus. Nonetheless, we hope our review encourages further studies and commentary aimed at determining whether particular genetic changes in HUWE1 represent one or many molecularly-defined syndromes. This already appears to be well under way in one case, that of Juberg-Marsidi-Brooks syndrome [65].
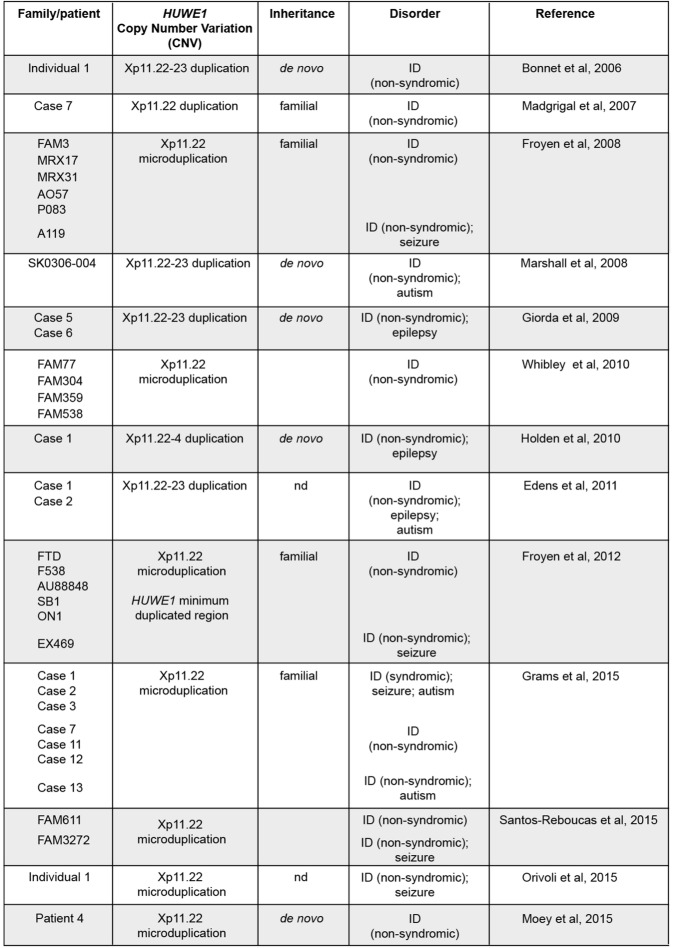
Microduplications of chromosome Xp11.22 which result in CNV in HUWE1 were initially identified independently by two groups using microarray-comparative hybridization approaches [63, 64] (Table 1). These two studies showed that HUWE1 is a common gene present in all Xp11.22 microduplications identified from 7 families. Moreover, in one study microduplications containing HUWE1 were shown to segregate with ID in 6 of these families and were not identified in control samples [64]. Importantly, this study also performed qPCR and showed that HUWE1 expression levels were elevated in these patient samples, consistent with an overexpression/gain-of-function effect on HUWE1. Subsequent studies that used exome or next generation sequencing identified HUWE1 CNVs in numerous other families with non-syndromic ID [66–75] (Table 1).
Table 1.
HUWE1 copy number variations associated with non-syndromic ID.
One particular study was critical in establishing the link between HUWE1 CNV and ID [71]. Froyen and colleagues identified 6 families with Xp11.22 microduplications containing HUWE1, showed that HUWE1 was the only common gene duplicated, and demonstrated that HUWE1 expression was increased in individuals with ID. Importantly, one of these Xp11.22 microduplications did not include HSD17B10, the gene adjacent to HUWE1. Similar findings ruled out two microRNAs, miRNA98 and let7f-2, that are also often present with HUWE1 in Xp11.22 microduplications. Through a compelling collective effort, the field now has data from 35 different families/individuals that convincingly argues increased HUWE1 function is likely to cause non-syndromic ID (Table 1).
Individuals with HUWE1 CNVs sometimes have other neurodevelopmental conditions that accompany ID. The most compelling links are to epilepsy or seizure [64, 67, 69–74]. Autism-like behaviors have also been documented [66, 70, 72]. While seizure/epilepsy and autism are observed in some individuals with HUWE1 CNVs, the genetic links between HUWE1 and these neurodevelopmental conditions are much less clear and less consistent than ID. Thus, increased HUWE1 function might contribute to these outcomes, but it is also possible that they are caused by other genes present in duplications that contain HUWE1.
The concept that increased HUWE1 function leads to ID is further supported by outcomes from both vertebrate and invertebrate animal models, which show that increased HUWE1 function is detrimental to the nervous system. For example, overexpression of HUWE1 in cortical neurons results in reduced proliferation of neural progenitors [19]. In Drosophila, overexpression of HUWE1 results in altered axon branching in some types of neurons [28]. Finally, work in C. elegans showed that overexpressing HUWE1 in GABAergic neurons alters their function [18]. Thus, findings from three different in vivo model systems indicate that increased HUWE1 function can adversely affect neuron function and development. Despite this progress, we still know relatively little about how increasing HUWE1 function affects different brain regions and different types of neurons in vivo. Moreover, which HUWE1 signaling mechanisms are the most important to phenotypes caused by HUWE1 overexpression remains entirely unclear.
HUWE1 mutations in syndromic and non-syndromic intellectual disability
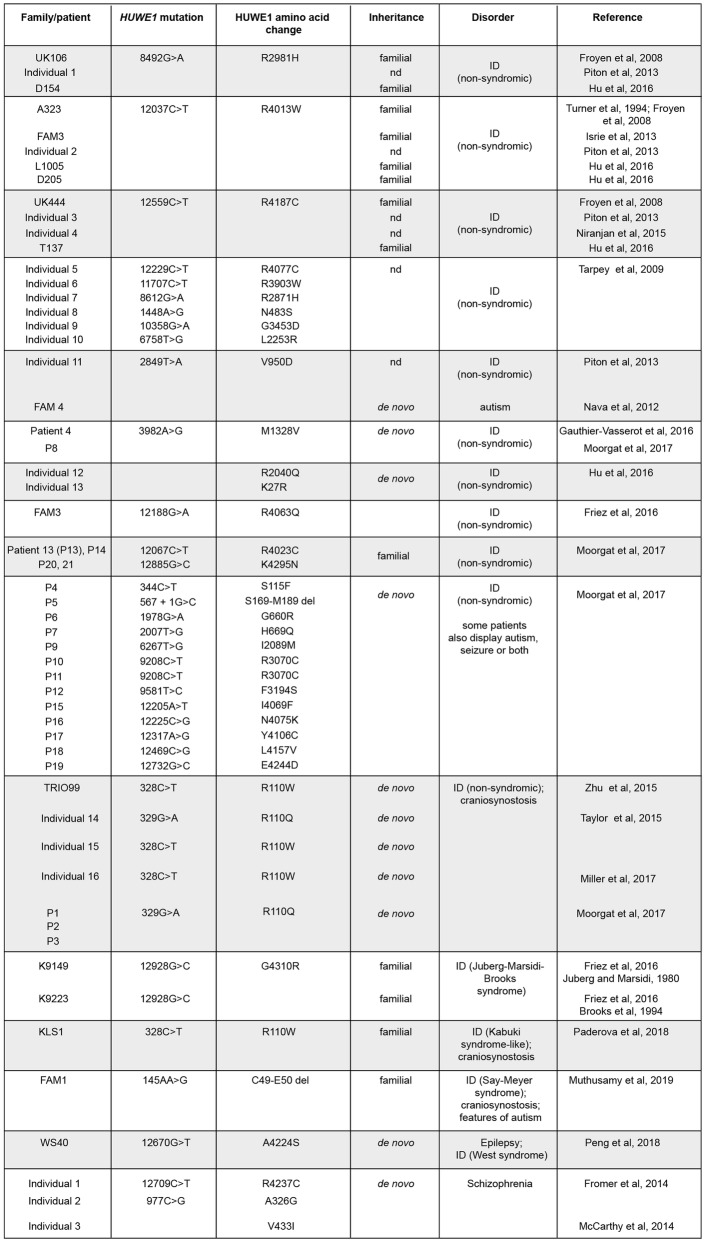
A growing number of human genetic studies have also identified HUWE1 missense mutations that are associated with both syndromic and non-syndromic forms of X-linked ID. The first HUWE1 mutations identified in three families with non-syndromic ID were R4013W, R2981H and R4187C [64, 76] (Table 2, Fig. 4). All three arginine residues (and motifs containing them) are highly conserved from C. elegans through humans, and two residues are in the HECT ubiquitin ligase domain. Experiments using C. elegans showed two of these exact disorder-associated mutations engineered into EEL-1/HUWE1 fail to rescue GABA neuron defects in eel-1 (lf) mutants [18] (Fig. 4, blue italics). In contrast, wild-type EEL-1 does rescue eel-1 (lf) defects. Thus, results from in vivo experiments demonstrate that two ID-associated mutations impair EEL-1 function, and therefore are likely to impair HUWE1 as well. We note that these point mutations are probably not HUWE1 null alleles for two reasons. 1) Huwe1 knockout mice display neonatal lethality [19, 20]. 2) More damaging large deletion alleles have not been identified for HUWE1 despite large numbers of human genetic studies [71].
Table 2.
HUWE1 missense mutations associated with neurodevelopmental disorders including non-syndromic and syndromic ID
Independent studies identified numerous other missense mutations in HUWE1 associated with non-syndromic ID that show familial inheritance or occur de novo in individuals [65, 77–83] (Table 2; Fig. 4). To date, a total of 31 genetic changes in HUWE1 isolated from 49 different families or individuals with non-syndromic ID have been identified (Table 2; Fig. 4). Importantly, several mutations (R4013W, R2981H, R4187C, R110Q/W and M1328V) were identified in multiple different families/individuals (Table 2). This high quality, extensive progress with human genetics makes a strong case that mutations in HUWE1 are likely to cause non-syndromic ID.
There are other important points to be made based on the impressive scope of these studies. First, mutations in HUWE1 associated with non-syndromic ID cause amino acid changes that are broadly distributed across the entire HUWE1 protein (Fig. 4). Mutations affect annotated domains of unknown function (DUF 908, DUF 913), unannotated evolutionarily conserved domains (CD) of unknown function, the UBA domain, the UBM domain, and the HECT catalytic ubiquitin ligase domain (Fig. 4). With that said, there are two domains in HUWE1 that are the most frequently mutated, and possible ID hot spots. These are the N-terminal DUF 908 domain (5 mutations) and, even more so, the C-terminal HECT domain (13 mutations) (Fig. 4). These findings suggest that HUWE1 ubiquitin ligase activity is likely to be involved in ID, and other domains harboring ID-associated mutations are likely to be important for HUWE1 function or structure.
Comparison of ID-associated mutations with the HECT domain crystal structure revealed further valuable insight. Two mutations, E4244D and K4295N, affect residues required for HECT domain structure [13] (Fig. 4). Another mutation, R4013W, is adjacent to K4014, another structurally important residue [13]. The L4157V mutation affects a residue that is required for binding to the E2 enzyme, suggesting it could affect substrate ubiquitination [13]. Mutations occur in two domains that bind ubiquitin [84, 85], the UBA (M1328V) and UBM (R2981H, R3070C) domains (Fig. 4). While the structural and functional roles of the UBA and UBM domains in HUWE1 remain unclear, it is possible mutations in these domains might affect binding to ubiquitin thereby influencing the efficiency of substrate ubiquitination or the stability of ubiquitin modifications on substrates. No mutations were found in a small region (D3951 to L3992) that mediates dimerization and inhibition of the HUWE1 HECT domain [14] (Fig. 4). Collectively, these observations suggest that ID-associated mutations are likely to impair HUWE1 function.
Other neurodevelopmental conditions can show comorbidity with non-syndromic ID in some individuals with HUWE1 mutations. For example, a small number of studies focused on craniosynostosis identified mutations in HUWE1, and also observed non-syndromic ID in these individuals (Table 2) [86–88]. In all cases, R110Q/W mutations occurred, which suggests that this residue might be particularly relevant in craniosynostosis. Alternatively, because most studies focused on non-syndromic ID might not have evaluated craniosynostosis, this could be a core, undiagnosed feature of disease associated with mutations in HUWE1. Indeed, one study examining non-syndromic ID identified several individuals with R110Q mutations and noted that they have craniosynostosis (Table 2) [83]. This study also observed seizures and autism as comorbidities with non-syndromic ID in some individuals with HUWE1 mutations (Table 2) [83].
Importantly, HUWE1 mutations have also emerged in syndromic forms of ID. Friez and colleagues used next generation exome sequencing to revisit the genetic basis of two syndromic forms of ID, Juberg-Marsidi and Brooks syndromes [65] (Table 2). Identical HUWE1 mutations, G4310R, were identified in patients from both of these syndromes. This led the authors to reclassify this as a single syndromic form of ID, Juberg-Marsidi-Brooks (JMB) syndrome, that has the same molecular genetic basis. Cell-based experiments using cells derived from these individuals showed that HUWE1 ubiquitination substrates were increased [65]. Thus, the G4310R lesion is likely to result in loss of HUWE1 function, but is likely to be a hypomorphic allele for reasons discussed above. Another possible link between mutations in HUWE1 and syndromic ID involves a patient with a Kabuki-like syndrome (Table 2) [89]. Interestingly, this individual has the same mutation, R110W, that was identified in several individuals with craniosynostosis and non-syndromic ID (Table 2) [86–88]. Finally, a splicing mutation that deletes two amino acids from HUWE1 was found in a family that potentially has Say-Meyer syndrome, another syndromic form of ID (Table 2) [90]. Taken as a whole, these studies suggest that mutations in HUWE1 potentially result in several types of syndromic ID, including a potential causal link to JMB. While this is important progress, identification of HUWE1 mutations in more families with JMB, and other syndromic forms of ID, would be valuable.
HUWE1 missense mutations can be associated with other neurodevelopmental conditions. For example, a variant in HUWE1 was found in an individual with West syndrome, a form of early infantile epileptic encephalopathy that has accompanying ID (Table 2) [91]. De novo mutations in HUWE1 were found in individuals with schizophrenia [92, 93] and possibly autism [94] (Table 2). Notably, this single patient with autism has the same mutation, V950D, that was identified in a different individual with non-syndromic ID (Table 2) [79]. Thus, ID and autism could be comorbidities associated with mutations in HUWE1, or certain HUWE1 mutations could be risk factors for these conditions such that ID and autism can occur separately or together. Answers to these questions now await further more extensive behavioral and clinical studies on individuals with HUWE1 mutations.
Further links between HUWE1 and ID have emerged from identification of proteins that interact with HUWE1, and studies that evaluated HUWE1 RNA expression. Proteomics identified the glycosyltransferase OGT as a conserved binding protein for both C. elegans EEL-1/HUWE1 and human HUWE1 [48]. OGT was found to function with EEL-1 to regulate inhibitory GABA neuron function. Similar to HUWE1, human genetic studies have identified OGT mutations in multiple families with non-syndromic ID [80, 95–98]. To date, 7 different mutations in OGT have been found in 11 individuals with ID. Familial inheritance indicates that OGT mutations segregate with ID, providing further support for the involvement of OGT in ID [96–98].
There is one last intriguing link between HUWE1 and ID. Findings from mice and Drosophila showed that HUWE1 mRNA is a prominent, conserved target of the RNA binding protein FMRP, and FMRP affects HUWE1 RNA levels [99, 100]. This is a potentially important observation as mutations in FMRP cause Fragile X syndrome, a highly prevalent syndromic form of ID. Whether HUWE1 is a relevant target of FMRP in regard to pathology, behavioral abnormalities or cognitive impairment in Fragile X syndrome remains unclear, but is certainly an interesting possibility given mounting evidence linking HUWE1 genetic changes to ID.
In summary, a compelling body of human genetic studies and work using both mammalian and invertebrate model systems support several overarching concepts regarding HUWE1 and neurodevelopmental disorders. 1) There is extensive evidence that both CNVs and mutations in HUWE1 likely cause non-syndromic ID. Thus, both gain- and loss-of-function genetic changes in HUWE1 are likely to result in ID. This is similar to other prominent players in ID, such as MECP2 and SHANK3 [101]. 2) Mutations in HUWE1 are potentially associated with several syndromic forms of ID with the most substantive links to JMB syndrome. 3) Further evidence linking HUWE1 to ID has emerged from protein interaction and RNA expression studies. This suggests that HUWE1 could be part of signaling networks involved in ID.
Conclusion
Over the past decade, HUWE1 has emerged as a relevant player in several neurodevelopmental disorders with the most prominent links to ID. As a result, several future research directions have become particularly compelling. It will be imperative that we comprehensively unravel the mechanisms by which HUWE1 regulates nervous system function and development in order to identify potential therapeutic targets and strategies for molecular intervention. In vivo invertebrate and vertebrate models are likely to be invaluable in this process. It is critical to further investigate how precise HUWE1 genetic changes that occur in ID affect neuron function and development. Finally, we hope that continued human genetic and clinical studies will allow for more comprehensive, precise molecular genetic classification of HUWE1 neurodevelopmental disorders. While identification of the molecular genetic basis of JMB is a valuable start, much important work remains to be done.
Acknowledgements
We thank Dr. Damon Page for helpful discussions.
Authors’ contributions
ACG and BG wrote and edited the manuscript.
Authors’ information
Department of Neuroscience, The Scripps Research Institute, Jupiter, Florida, 33458, USA.
Funding
BG was supported by National Institutes of Health Grant R01 NS072129.
Availability of data and materials
Not applicable.
Ethics approval and consent to participate
Not applicable.
Consent for publication
For materials that have been reproduced, informed consent was obtained from authors and publishers. In all cases, reproduction guidelines have been followed.
Competing interests
The authors declare no competing financial interests.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Huibregtse JM, Scheffner M, Beaudenon S, Howley PM. A family of proteins structurally and functionally related to the E6-AP ubiquitin-protein ligase. Proc Natl Acad Sci U S A. 1995;92:2563–2567. doi: 10.1073/pnas.92.7.2563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zhong Q, Gao W, Du F, Wang X. Mule/ARF-BP1, a BH3-only E3 ubiquitin ligase, catalyzes the polyubiquitination of Mcl-1 and regulates apoptosis. Cell. 2005;121:1085–1095. doi: 10.1016/j.cell.2005.06.009. [DOI] [PubMed] [Google Scholar]
- 3.Chen D, Kon N, Li M, Zhang W, Qin J, Gu W. ARF-BP1/mule is a critical mediator of the ARF tumor suppressor. Cell. 2005;121:1071–1083. doi: 10.1016/j.cell.2005.03.037. [DOI] [PubMed] [Google Scholar]
- 4.Page BD, Diede SJ, Tenlen JR, Ferguson EL. EEL-1, a Hect E3 ubiquitin ligase, controls asymmetry and persistence of the SKN-1 transcription factor in the early C. elegans embryo. Development. 2007;134:2303–2314. doi: 10.1242/dev.02855. [DOI] [PubMed] [Google Scholar]
- 5.Kao SH, Wu HT, Wu KJ. Ubiquitination by HUWE1 in tumorigenesis and beyond. J Biomed Sci. 2018;25:67. doi: 10.1186/s12929-018-0470-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Adhikary S, Marinoni F, Hock A, Hulleman E, Popov N, Beier R, Bernard S, Quarto M, Capra M, Goettig S, et al. The ubiquitin ligase HectH9 regulates transcriptional activation by Myc and is essential for tumor cell proliferation. Cell. 2005;123:409–421. doi: 10.1016/j.cell.2005.08.016. [DOI] [PubMed] [Google Scholar]
- 7.Liu Z, Oughtred R, Wing SS. Characterization of E3Histone, a novel testis ubiquitin protein ligase which ubiquitinates histones. Mol Cell Biol. 2005;25:2819–2831. doi: 10.1128/MCB.25.7.2819-2831.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hall JR, Kow E, Nevis KR, Lu CK, Luce KS, Zhong Q, Cook JG. Cdc6 stability is regulated by the Huwe1 ubiquitin ligase after DNA damage. Mol Biol Cell. 2007;18:3340–3350. doi: 10.1091/mbc.e07-02-0173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hao Z, Duncan GS, Su YW, Li WY, Silvester J, Hong C, You H, Brenner D, Gorrini C, Haight J, et al. The E3 ubiquitin ligase mule acts through the ATM-p53 axis to maintain B lymphocyte homeostasis. J Exp Med. 2012;209:173–186. doi: 10.1084/jem.20111363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hao Z, Sheng Y, Duncan GS, Li WY, Dominguez C, Sylvester J, Su YW, Lin GH, Snow BE, Brenner D, et al. K48-linked KLF4 ubiquitination by E3 ligase mule controls T-cell proliferation and cell cycle progression. Nat Commun. 2017;8:14003. doi: 10.1038/ncomms14003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Li L, Martinez SS, Hu W, Liu Z, Tjian R. A specific E3 ligase/deubiquitinase pair modulates TBP protein levels during muscle differentiation. Elife. 2015;4:e08536. doi: 10.7554/eLife.08536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rotin D, Kumar S. Physiological functions of the HECT family of ubiquitin ligases. Nat Rev Mol Cell Biol. 2009;10:398–409. doi: 10.1038/nrm2690. [DOI] [PubMed] [Google Scholar]
- 13.Pandya RK, Partridge JR, Love KR, Schwartz TU, Ploegh HL. A structural element within the HUWE1 HECT domain modulates self-ubiquitination and substrate ubiquitination activities. J Biol Chem. 2010;285:5664–5673. doi: 10.1074/jbc.M109.051805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sander B, Xu W, Eilers M, Popov N, Lorenz S. A conformational switch regulates the ubiquitin ligase HUWE1. Elife. 2017;6. [DOI] [PMC free article] [PubMed]
- 15.Zhao X, Heng JI, Guardavaccaro D, Jiang R, Pagano M, Guillemot F, Iavarone A, Lasorella A. The HECT-domain ubiquitin ligase Huwe1 controls neural differentiation and proliferation by destabilizing the N-Myc oncoprotein. Nat Cell Biol. 2008;10:643–653. doi: 10.1038/ncb1727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lein ES, Hawrylycz MJ, Ao N, Ayres M, Bensinger A, Bernard A, Boe AF, Boguski MS, Brockway KS, Byrnes EJ, et al. Genome-wide atlas of gene expression in the adult mouse brain. Nature. 2007;445:168–176. doi: 10.1038/nature05453. [DOI] [PubMed] [Google Scholar]
- 17.Hawrylycz MJ, Lein ES, Guillozet-Bongaarts AL, Shen EH, Ng L, Miller JA, van de Lagemaat LN, Smith KA, Ebbert A, Riley ZL, et al. An anatomically comprehensive atlas of the adult human brain transcriptome. Nature. 2012;489:391–399. doi: 10.1038/nature11405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Opperman KJ, Mulcahy B, Giles AC, Risley MG, Birnbaum RL, Tulgren ED, Dawson-Scully K, Zhen M, Grill B. The HECT family ubiquitin ligase EEL-1 regulates neuronal function and development. Cell Rep. 2017;19:822–835. doi: 10.1016/j.celrep.2017.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhao X, D DA, Lim WK, Brahmachary M, Carro MS, Ludwig T, Cardo CC, Guillemot F, Aldape K, Califano A, et al: The N-Myc-DLL3 cascade is suppressed by the ubiquitin ligase Huwe1 to inhibit proliferation and promote neurogenesis in the developing brain. Dev Cell 2009, 17:210–221. [DOI] [PMC free article] [PubMed]
- 20.D'Arca D, Zhao X, Xu W, Ramirez-Martinez NC, Iavarone A, Lasorella A. Huwe1 ubiquitin ligase is essential to synchronize neuronal and glial differentiation in the developing cerebellum. Proc Natl Acad Sci U S A. 2010;107:5875–5880. doi: 10.1073/pnas.0912874107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Forget A, Bihannic L, Cigna SM, Lefevre C, Remke M, Barnat M, Dodier S, Shirvani H, Mercier A, Mensah A, et al. Shh signaling protects atoh1 from degradation mediated by the e3 ubiquitin ligase huwe1 in neural precursors. Dev Cell. 2014;29:649–661. doi: 10.1016/j.devcel.2014.05.014. [DOI] [PubMed] [Google Scholar]
- 22.Ayrault O, Zhao H, Zindy F, Qu C, Sherr CJ, Roussel MF. Atoh1 inhibits neuronal differentiation and collaborates with Gli1 to generate medulloblastoma-initiating cells. Cancer Res. 2010;70:5618–5627. doi: 10.1158/0008-5472.CAN-09-3740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cheng YF, Tong M, Edge AS. Destabilization of Atoh1 by E3 ubiquitin ligase Huwe1 and casein kinase 1 is essential for Normal sensory hair cell development. J Biol Chem. 2016;291:21096–21109. doi: 10.1074/jbc.M116.722124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Urban N, van den Berg DL, Forget A, Andersen J, Demmers JA, Hunt C, Ayrault O, Guillemot F. Return to quiescence of mouse neural stem cells by degradation of a proactivation protein. Science. 2016;353:292–295. doi: 10.1126/science.aaf4802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gillotin S, Davies JD, Philpott A. Subcellular localisation modulates ubiquitylation and degradation of Ascl1. Sci Rep. 2018;8:4625. doi: 10.1038/s41598-018-23056-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.de Groot RE, Ganji RS, Bernatik O, Lloyd-Lewis B, Seipel K, Sedova K, Zdrahal Z, Dhople VM, Dale TC, Korswagen HC, Bryja V: Huwe1-mediated ubiquitylation of dishevelled defines a negative feedback loop in the wnt signaling pathway. Sci Signal 2014, 7:ra26. [DOI] [PubMed]
- 27.Park M, Shen K. WNTs in synapse formation and neuronal circuitry. EMBO J. 2012;31:2697–2704. doi: 10.1038/emboj.2012.145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Vandewalle J, Langen M, Zschaetzsch M, Nijhof B, Kramer JM, Brems H, Bauters M, Lauwers E, Srahna M, Marynen P, et al. Ubiquitin ligase HUWE1 regulates axon branching through the Wnt/beta-catenin pathway in a Drosophila model for intellectual disability. PLoS One. 2013;8:e81791. doi: 10.1371/journal.pone.0081791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Callow MG, Tran H, Phu L, Lau T, Lee J, Sandoval WN, Liu PS, Bheddah S, Tao J, Lill JR, et al. Ubiquitin ligase RNF146 regulates tankyrase and Axin to promote Wnt signaling. PLoS One. 2011;6:e22595. doi: 10.1371/journal.pone.0022595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Dominguez-Brauer C, Khatun R, Elia AJ, Thu KL, Ramachandran P, Baniasadi SP, Hao Z, Jones LD, Haight J, Sheng Y, Mak TW. E3 ubiquitin ligase mule targets beta-catenin under conditions of hyperactive Wnt signaling. Proc Natl Acad Sci U S A. 2017;114:E1148–E1157. doi: 10.1073/pnas.1621355114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Grill B, Murphey RK, Borgen MA. The PHR proteins: intracellular signaling hubs in neuronal development and axon degeneration. Neural Dev. 2016;11:8. doi: 10.1186/s13064-016-0063-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sharma J, Baker ST, Turgeon SM, Gurney AM, Opperman KJ, Grill B. Identification of a peptide inhibitor of the RPM-1.FSN-1 ubiquitin ligase complex. J Biol Chem. 2014;289:34654–34666. doi: 10.1074/jbc.M114.614065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Baker ST, Grill B. Defining minimal binding regions in regulator of presynaptic morphology 1 (RPM-1) using Caenorhabditis elegans neurons reveals differential signaling complexes. J Biol Chem. 2017;292:2519–2530. doi: 10.1074/jbc.M116.748004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Desbois M, Crawley O, Evans PR, Baker ST, Masuho I, Yasuda R, Grill B. PAM forms an atypical SCF ubiquitin ligase complex that ubiquitinates and degrades NMNAT2. J Biol Chem. 2018;293:13897–13909. doi: 10.1074/jbc.RA118.002176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Schaefer AM, Hadwiger GD, Nonet ML. rpm-1, a conserved neuronal gene that regulates targeting and synaptogenesis in C. elegans. Neuron. 2000;26:345–356. doi: 10.1016/S0896-6273(00)81168-X. [DOI] [PubMed] [Google Scholar]
- 36.Opperman KJ, Grill B. RPM-1 is localized to distinct subcellular compartments and regulates axon length in GABAergic motor neurons. Neural Dev. 2014;9:10. doi: 10.1186/1749-8104-9-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Nakata K, Abrams B, Grill B, Goncharov A, Huang X, Chisholm AD, Jin Y. Regulation of a DLK-1 and p38 MAP kinase pathway by the ubiquitin ligase RPM-1 is required for presynaptic development. Cell. 2005;120:407–420. doi: 10.1016/j.cell.2004.12.017. [DOI] [PubMed] [Google Scholar]
- 38.Grill B, Bienvenut WV, Brown HM, Ackley BD, Quadroni M, Jin Y. C. elegans RPM-1 regulates axon termination and synaptogenesis through the Rab GEF GLO-4 and the Rab GTPase GLO-1. Neuron. 2007;55:587–601. doi: 10.1016/j.neuron.2007.07.009. [DOI] [PubMed] [Google Scholar]
- 39.Grill B, Chen L, Tulgren ED, Baker ST, Bienvenut W, Anderson M, Quadroni M, Jin Y, Garner CC. RAE-1, a novel PHR binding protein, is required for axon termination and synapse formation in Caenorhabditis elegans. J Neurosci. 2012;32:2628–2636. doi: 10.1523/JNEUROSCI.2901-11.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tulgren ED, Turgeon SM, Opperman KJ, Grill B. The Nesprin family member ANC-1 regulates synapse formation and axon termination by functioning in a pathway with RPM-1 and beta-catenin. PLoS Genet. 2014;10:e1004481. doi: 10.1371/journal.pgen.1004481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Baker ST, Opperman KJ, Tulgren ED, Turgeon SM, Bienvenut W, Grill B. RPM-1 uses both ubiquitin ligase and phosphatase-based mechanisms to regulate DLK-1 during neuronal development. PLoS Genet. 2014;10:e1004297. doi: 10.1371/journal.pgen.1004297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Crawley O, Opperman KJ, Desbois M, Adrados I, Borgen MA, Giles AC, Duckett DR, Grill B. Autophagy is inhibited by ubiquitin ligase activity in the nervous system. Nat Commun. 2019;10:5017. doi: 10.1038/s41467-019-12804-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yin L, Joshi S, Wu N, Tong X, Lazar MA. E3 ligases Arf-bp1 and pam mediate lithium-stimulated degradation of the circadian heme receptor rev-erb alpha. Proc Natl Acad Sci U S A. 2010;107:11614–11619. doi: 10.1073/pnas.1000438107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Guo Q, Xie J, Dang CV, Liu ET, Bishop JM. Identification of a large Myc-binding protein that contains RCC1-like repeats. Proc Natl Acad Sci U S A. 1998;95:9172–9177. doi: 10.1073/pnas.95.16.9172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tai HC, Besche H, Goldberg AL, Schuman EM. Characterization of the brain 26S proteasome and its interacting proteins. Front Mol Neurosci. 2010;3. [DOI] [PMC free article] [PubMed]
- 46.Zhang ZY, Guo Z, Li HL, He YT, Duan XL, Suo ZW, Yang X, Hu XD. Ubiquitination and inhibition of glycine receptor by HUWE1 in spinal cord dorsal horn. Neuropharmacology. 2019;148:358–365. doi: 10.1016/j.neuropharm.2019.02.002. [DOI] [PubMed] [Google Scholar]
- 47.Mahoney TR, Luo S, Nonet ML. Analysis of synaptic transmission in Caenorhabditis elegans using an aldicarb-sensitivity assay. Nat Protoc. 2006;1:1772–1777. doi: 10.1038/nprot.2006.281. [DOI] [PubMed] [Google Scholar]
- 48.Giles AC, Desbois M, Opperman KJ, Tavora R, Maroni MJ, Grill B. A complex containing the O-GlcNAc transferase OGT-1 and the ubiquitin ligase EEL-1 regulates GABA neuron function. J Biol Chem. 2019;294:6843–6856. doi: 10.1074/jbc.RA119.007406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Levine ZG, Walker S. The biochemistry of O-GlcNAc Transferase: which functions make it essential in mammalian cells? Annu Rev Biochem. 2016;85:631–657. doi: 10.1146/annurev-biochem-060713-035344. [DOI] [PubMed] [Google Scholar]
- 50.Hart GW. Nutrient regulation of signaling and transcription. J Biol Chem. 2019;294:2211–2231. doi: 10.1074/jbc.AW119.003226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kreppel LK, Blomberg MA, Hart GW. Dynamic glycosylation of nuclear and cytosolic proteins. Cloning and characterization of a unique O-GlcNAc transferase with multiple tetratricopeptide repeats. J Biol Chem. 1997;272:9308–9315. doi: 10.1074/jbc.272.14.9308. [DOI] [PubMed] [Google Scholar]
- 52.Cole RN, Hart GW. Cytosolic O-glycosylation is abundant in nerve terminals. J Neurochem. 2001;79:1080–1089. doi: 10.1046/j.1471-4159.2001.00655.x. [DOI] [PubMed] [Google Scholar]
- 53.Vosseller K, Trinidad JC, Chalkley RJ, Specht CG, Thalhammer A, Lynn AJ, Snedecor JO, Guan S, Medzihradszky KF, Maltby DA, et al. O-linked N-acetylglucosamine proteomics of postsynaptic density preparations using lectin weak affinity chromatography and mass spectrometry. Mol Cell Proteomics. 2006;5:923–934. doi: 10.1074/mcp.T500040-MCP200. [DOI] [PubMed] [Google Scholar]
- 54.Yang X, Zhang F, Kudlow JE. Recruitment of O-GlcNAc transferase to promoters by corepressor mSin3A: coupling protein O-GlcNAcylation to transcriptional repression. Cell. 2002;110:69–80. doi: 10.1016/S0092-8674(02)00810-3. [DOI] [PubMed] [Google Scholar]
- 55.Escobar-Henriques M, Joaquim M. Mitofusins: disease gatekeepers and hubs in mitochondrial quality control by E3 ligases. Front Physiol. 2019;10:517. doi: 10.3389/fphys.2019.00517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zuchner S, Mersiyanova IV, Muglia M, Bissar-Tadmouri N, Rochelle J, Dadali EL, Zappia M, Nelis E, Patitucci A, Senderek J, et al. Mutations in the mitochondrial GTPase mitofusin 2 cause Charcot-Marie-tooth neuropathy type 2A. Nat Genet. 2004;36:449–451. doi: 10.1038/ng1341. [DOI] [PubMed] [Google Scholar]
- 57.Chen H, McCaffery JM, Chan DC. Mitochondrial fusion protects against neurodegeneration in the cerebellum. Cell. 2007;130:548–562. doi: 10.1016/j.cell.2007.06.026. [DOI] [PubMed] [Google Scholar]
- 58.Detmer SA, Chan DC. Complementation between mouse Mfn1 and Mfn2 protects mitochondrial fusion defects caused by CMT2A disease mutations. J Cell Biol. 2007;176:405–414. doi: 10.1083/jcb.200611080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Leboucher GP, Tsai YC, Yang M, Shaw KC, Zhou M, Veenstra TD, Glickman MH, Weissman AM. Stress-induced phosphorylation and proteasomal degradation of mitofusin 2 facilitates mitochondrial fragmentation and apoptosis. Mol Cell. 2012;47:547–557. doi: 10.1016/j.molcel.2012.05.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Senyilmaz D, Virtue S, Xu X, Tan CY, Griffin JL, Miller AK, Vidal-Puig A, Teleman AA. Regulation of mitochondrial morphology and function by stearoylation of TFR1. Nature. 2015. [DOI] [PMC free article] [PubMed]
- 61.Di Rita A, Peschiaroli A, P DA, Strobbe D, Hu Z, Gruber J, Nygaard M, Lambrughi M, Melino G, Papaleo E, et al: HUWE1 E3 ligase promotes PINK1/PARKIN-independent mitophagy by regulating AMBRA1 activation via IKKalpha. Nat Commun 2018, 9:3755. [DOI] [PMC free article] [PubMed]
- 62.Bonnet C, Gregoire MJ, Brochet K, Raffo E, Leheup B, Jonveaux P. Pure de-novo 5 Mb duplication at Xp11.22-p11.23 in a male: phenotypic and molecular characterization. J Hum Genet. 2006;51:815–821. doi: 10.1007/s10038-006-0023-3. [DOI] [PubMed] [Google Scholar]
- 63.Madrigal I, Rodriguez-Revenga L, Armengol L, Gonzalez E, Rodriguez B, Badenas C, Sanchez A, Martinez F, Guitart M, Fernandez I, et al. X-chromosome tiling path array detection of copy number variants in patients with chromosome X-linked mental retardation. BMC Genomics. 2007;8:443. doi: 10.1186/1471-2164-8-443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Froyen G, Corbett M, Vandewalle J, Jarvela I, Lawrence O, Meldrum C, Bauters M, Govaerts K, Vandeleur L, Van Esch H, et al. Submicroscopic duplications of the hydroxysteroid dehydrogenase HSD17B10 and the E3 ubiquitin ligase HUWE1 are associated with mental retardation. Am J Hum Genet. 2008;82:432–443. doi: 10.1016/j.ajhg.2007.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Friez MJ, Brooks SS, Stevenson RE, Field M, Basehore MJ, Ades LC, Sebold C, McGee S, Saxon S, Skinner C, et al. HUWE1 mutations in Juberg-Marsidi and Brooks syndromes: the results of an X-chromosome exome sequencing study. BMJ Open. 2016;6:e009537. doi: 10.1136/bmjopen-2015-009537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Marshall CR, Noor A, Vincent JB, Lionel AC, Feuk L, Skaug J, Shago M, Moessner R, Pinto D, Ren Y, et al. Structural variation of chromosomes in autism spectrum disorder. Am J Hum Genet. 2008;82:477–488. doi: 10.1016/j.ajhg.2007.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Giorda R, Bonaglia MC, Beri S, Fichera M, Novara F, Magini P, Urquhart J, Sharkey FH, Zucca C, Grasso R, et al. Complex segmental duplications mediate a recurrent dup(X)(p11.22-p11.23) associated with mental retardation, speech delay, and EEG anomalies in males and females. Am J Hum Genet. 2009;85:394–400. doi: 10.1016/j.ajhg.2009.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Whibley AC, Plagnol V, Tarpey PS, Abidi F, Fullston T, Choma MK, Boucher CA, Shepherd L, Willatt L, Parkin G, et al. Fine-scale survey of X chromosome copy number variants and indels underlying intellectual disability. Am J Hum Genet. 2010;87:173–188. doi: 10.1016/j.ajhg.2010.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Holden ST, Clarkson A, Thomas NS, Abbott K, James MR, Willatt L. A de novo duplication of Xp11.22-p11.4 in a girl with intellectual disability, structural brain anomalies, and preferential inactivation of the normal X chromosome. Am J Med Genet A. 2010;152a:1735–1740. doi: 10.1002/ajmg.a.33457. [DOI] [PubMed] [Google Scholar]
- 70.Edens AC, Lyons MJ, Duron RM, Dupont BR, Holden KR. Autism in two females with duplications involving Xp11.22-p11.23. Dev Med Child Neurol. 2011;53:463–466. doi: 10.1111/j.1469-8749.2010.03909.x. [DOI] [PubMed] [Google Scholar]
- 71.Froyen G, Belet S, Martinez F, Santos-Reboucas CB, Declercq M, Verbeeck J, Donckers L, Berland S, Mayo S, Rosello M, et al. Copy-number gains of HUWE1 due to replication- and recombination-based rearrangements. Am J Hum Genet. 2012;91:252–264. doi: 10.1016/j.ajhg.2012.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Grams SE, Argiropoulos B, Lines M, Chakraborty P, McGowan-Jordan J, Geraghty MT, Tsang M, Eswara M, Tezcan K, Adams KL, et al. Genotype-phenotype characterization in 13 individuals with chromosome Xp11.22 duplications. Am J Med Genet A. 2015. [DOI] [PubMed]
- 73.Santos-Reboucas CB, de Almeida LG, Belet S, Dos Santos SR, Ribeiro MG, da Silva AF, Medina-Acosta E, Dos Santos JM, Goncalves AP, Bahia PR, et al. Novel microduplications at Xp11.22 including HUWE1: clinical and molecular insights into these genomic rearrangements associated with intellectual disability. J Hum Genet. 2015;60:207–211. doi: 10.1038/jhg.2015.1. [DOI] [PubMed] [Google Scholar]
- 74.Orivoli S, Pavlidis E, Cantalupo G, Pezzella M, Zara F, Garavelli L, Pisani F, Piccolo B. Xp11.22 Microduplications including HUWE1: Case Report and Literature Review. Neuropediatrics. 2015. [DOI] [PubMed]
- 75.Moey C, Hinze SJ, Brueton L, Morton J, DJ MM, Kamien B, Barnett CP, Brunetti-Pierri N, Nicholl J, Gecz J, Shoubridge C. Xp11.2 microduplications including IQSEC2, TSPYL2 and KDM5C genes in patients with neurodevelopmental disorders. Eur J Hum Genet. 2015. [DOI] [PMC free article] [PubMed]
- 76.Turner G, Gedeon A, Mulley J. X-linked mental retardation with heterozygous expression and macrocephaly: pericentromeric gene localization. Am J Med Genet. 1994;51:575–580. doi: 10.1002/ajmg.1320510456. [DOI] [PubMed] [Google Scholar]
- 77.Tarpey PS, Smith R, Pleasance E, Whibley A, Edkins S, Hardy C, O'Meara S, Latimer C, Dicks E, Menzies A, et al. A systematic, large-scale resequencing screen of X-chromosome coding exons in mental retardation. Nat Genet. 2009;41:535–543. doi: 10.1038/ng.367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Isrie M, Kalscheuer VM, Holvoet M, Fieremans N, Van Esch H, Devriendt K. HUWE1 mutation explains phenotypic severity in a case of familial idiopathic intellectual disability. Eur J Med Genet. 2013. [DOI] [PubMed]
- 79.Piton A, Redin C, Mandel JL. XLID-causing mutations and associated genes challenged in light of data from large-scale human exome sequencing. Am J Hum Genet. 2013;93:368–383. doi: 10.1016/j.ajhg.2013.06.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Niranjan TS, Skinner C, May M, Turner T, Rose R, Stevenson R, Schwartz CE, Wang T. Affected kindred analysis of human X chromosome exomes to identify novel X-linked intellectual disability genes. PLoS One. 2015;10:e0116454. doi: 10.1371/journal.pone.0116454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Hu H, Haas SA, Chelly J, Van Esch H, Raynaud M, de Brouwer AP, Weinert S, Froyen G, Frints SG, Laumonnier F, et al. X-exome sequencing of 405 unresolved families identifies seven novel intellectual disability genes. Mol Psychiatry. 2016;21:133–148. doi: 10.1038/mp.2014.193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Gauthier-Vasserot A, Thauvin-Robinet C, Bruel AL, Duffourd Y, St-Onge J, Jouan T, Riviere JB, Heron D, Donadieu J, Bellanne-Chantelot C, et al. Application of whole-exome sequencing to unravel the molecular basis of undiagnosed syndromic congenital neutropenia with intellectual disability. Am J Med Genet A. 2016. [DOI] [PubMed]
- 83.Moortgat S, Berland S, Aukrust I, Maystadt I, Baker L, Benoit V, Caro-Llopis A, Cooper NS, Debray FG, Faivre L, et al. HUWE1 variants cause dominant X-linked intellectual disability: a clinical study of 21 patients. Eur J Hum Genet. 2017. [DOI] [PMC free article] [PubMed]
- 84.Dikic I, Wakatsuki S, Walters KJ. Ubiquitin-binding domains - from structures to functions. Nat Rev Mol Cell Biol. 2009;10:659–671. doi: 10.1038/nrm2767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Su V, Lau AF. Ubiquitin-like and ubiquitin-associated domain proteins: significance in proteasomal degradation. Cell Mol Life Sci. 2009;66:2819–2833. doi: 10.1007/s00018-009-0048-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Zhu X, Petrovski S, Xie P, Ruzzo EK, Lu YF, McSweeney KM, Ben-Zeev B, Nissenkorn A, Anikster Y, Oz-Levi D, et al. Whole-exome sequencing in undiagnosed genetic diseases: interpreting 119 trios. Genet Med. 2015;17:774–781. doi: 10.1038/gim.2014.191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Taylor JC, Martin HC, Lise S, Broxholme J, Cazier JB, Rimmer A, Kanapin A, Lunter G, Fiddy S, Allan C, et al. Factors influencing success of clinical genome sequencing across a broad spectrum of disorders. Nat Genet. 2015;47:717–726. doi: 10.1038/ng.3304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Miller KA, Twigg SR, McGowan SJ, Phipps JM, Fenwick AL, Johnson D, Wall SA, Noons P, Rees KE, Tidey EA, et al. Diagnostic value of exome and whole genome sequencing in craniosynostosis. J Med Genet. 2017;54:260–268. doi: 10.1136/jmedgenet-2016-104215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Paderova J, Drabova J, Holubova A, Vlckova M, Havlovicova M, Gregorova A, Pourova R, Romankova V, Moslerova V, Geryk J, et al. Under the mask of kabuki syndrome: elucidation of genetic-and phenotypic heterogeneity in patients with kabuki-like phenotype. Eur J Med Genet. 2018;61:315–321. doi: 10.1016/j.ejmg.2018.01.005. [DOI] [PubMed] [Google Scholar]
- 90.Muthusamy B, Nguyen TT, Bandari AK, Basheer S, Selvan LDN, Chandel D, Manoj J, Gayen S, Seshagiri S, Chandra Girimaji S, Pandey A. Exome sequencing reveals a novel splice site variant in HUWE1 gene in patients with suspected say-Meyer syndrome. Eur J Med Genet. 2019. [DOI] [PMC free article] [PubMed]
- 91.Peng J, Wang Y, He F, Chen C, Wu LW, Yang LF, Ma YP, Zhang W, Shi ZQ, Chen C, et al. Novel west syndrome candidate genes in a Chinese cohort. CNS Neurosci Ther. 2018;24:1196–1206. doi: 10.1111/cns.12860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Fromer M, Pocklington AJ, Kavanagh DH, Williams HJ, Dwyer S, Gormley P, Georgieva L, Rees E, Palta P, Ruderfer DM, et al. De novo mutations in schizophrenia implicate synaptic networks. Nature. 2014;506:179–184. doi: 10.1038/nature12929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.McCarthy SE, Gillis J, Kramer M, Lihm J, Yoon S, Berstein Y, Mistry M, Pavlidis P, Solomon R, Ghiban E, et al. De novo mutations in schizophrenia implicate chromatin remodeling and support a genetic overlap with autism and intellectual disability. Mol Psychiatry. 2014. [DOI] [PMC free article] [PubMed]
- 94.Nava C, Lamari F, Heron D, Mignot C, Rastetter A, Keren B, Cohen D, Faudet A, Bouteiller D, Gilleron M, et al. Analysis of the chromosome X exome in patients with autism spectrum disorders identified novel candidate genes, including TMLHE. Transl Psychiatry. 2012;2:e179. doi: 10.1038/tp.2012.102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Bouazzi H, Lesca G, Trujillo C, Alwasiyah MK, Munnich A. Nonsyndromic X-linked intellectual deficiency in three brothers with a novel MED12 missense mutation [c.5922G>T (p.Glu1974His)] Clin Case Rep. 2015;3:604–609. doi: 10.1002/ccr3.301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Vaidyanathan K, Niranjan T, Selvan N, Teo CF, May M, Patel S, Weatherly B, Skinner C, Opitz J, Carey J, et al. Identification and characterization of a missense mutation in the O-GlcNAc Transferase gene that segregates with X-linked intellectual disability. J Biol Chem. 2017. [DOI] [PMC free article] [PubMed]
- 97.Willems AP, Gundogdu M, Kempers MJE, Giltay JC, Pfundt R, Elferink M, Loza BF, Fuijkschot J, Ferenbach AT, van Gassen KLI, et al. Mutations in N-acetylglucosamine (O-GlcNAc) transferase in patients with X-linked intellectual disability. J Biol Chem. 2017;292:12621–12631. doi: 10.1074/jbc.M117.790097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Selvan N, George S, Serajee FJ, Shaw M, Hobson L, Kalscheuer V, Prasad N, Levy SE, Taylor J, Aftimos S, et al. O-GlcNAc transferase missense mutations linked to X-linked intellectual disability deregulate genes involved in cell fate determination and signaling. J Biol Chem. 2018;293:10810–10824. doi: 10.1074/jbc.RA118.002583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Darnell JC, Van Driesche SJ, Zhang C, Hung KY, Mele A, Fraser CE, Stone EF, Chen C, Fak JJ, Chi SW, et al. FMRP stalls ribosomal translocation on mRNAs linked to synaptic function and autism. Cell. 2011;146:247–261. doi: 10.1016/j.cell.2011.06.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Greenblatt EJ, Spradling AC. Fragile X mental retardation 1 gene enhances the translation of large autism-related proteins. Science. 2018;361:709–712. doi: 10.1126/science.aas9963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Zoghbi HY, Bear MF. Synaptic dysfunction in neurodevelopmental disorders associated with autism and intellectual disabilities. Cold Spring Harb Perspect Biol. 2012;4. [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Not applicable.