SUMMARY
Cellular translation surveillance rescues ribosomes that stall on problematic mRNAs. During translation surveillance, endonucleolytic cleavage of the problematic mRNA is a critical step in rescuing stalled ribosomes. Here we identify NONU-1 as a factor required for translation surveillance pathways including no-go and nonstop mRNA decay. We show that (1) NONU-1 reduces nonstop and no-go mRNA levels; (2) NONU-1 contains an Smr RNase domain required for mRNA decay; (3) the domain architecture and catalytic residues of NONU-1 are conserved throughout metazoans and eukaryotes, respectively; and (4) NONU-1 is required for the formation of mRNA cleavage fragments in the vicinity of stalled ribosomes. We extend our results in C. elegans to homologous factors in S. cerevisiae, showing the evolutionarily conserved function of NONU-1. Our work establishes the identity of a factor critical to translation surveillance and will inform mechanistic studies at the intersection of translation and mRNA decay.
Graphical Abstract
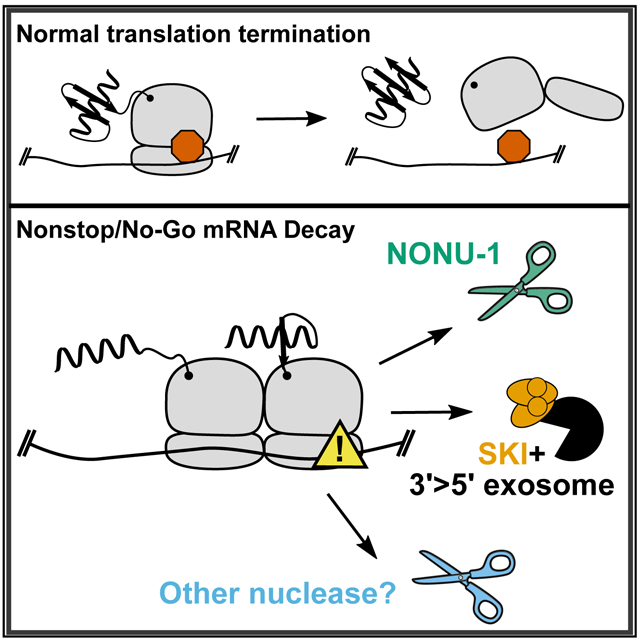
In Brief
Glover et al. report that NONU-1 is an endoribonuclease required for some mRNA cleavages after ribosome stalling during nonstop and no-go decay. In addition, NONU-1-independent mechanisms contribute to target mRNA degradation. Identification of NONU-1 should create a more complete picture of the molecular events underpinning relief from ribosome stalls.
INTRODUCTION
Numerous mechanisms exist to protect cells from the negative effects of errors in gene expression. Among these are translation surveillance pathways in which a ribosome identifies an early stop codon (nonsense-mediated mRNA decay [NMD]), a lack of stop codons (nonstop decay), or a block during translation elongation (no-go decay). Central to both nonstop and no-go decay is the process of ribosome stalling. Recent work has also shown that ribosomes stall during NMD, effectively funneling NMD targets into nonstop decay (Hashimoto et al., 2017; Arribere and Fire, 2018). Despite substantial mechanistic insight into translation surveillance pathways (reviewed by Joazeiro, 2017), how ribosomal stalling communicates with mRNA decay machinery remains a central unsolved question.
Mounting evidence points to endonucleolytic cleavage of the mRNA in the vicinity of stalled ribosomes as an important early event in translation surveillance (e.g., Doma and Parker, 2006; Guydosh and Green, 2014; Ikeuchi et al., 2019; D’Orazio et al., 2019; Navickas et al., 2020; Schaeffer and van Hoof, 2011). Subsequent to mRNA cleavage, target mRNAs are eventually cleared in part by 3′ > 5′ degradation facilitated by the SKI RNA helicase in conjunction with the exosome (Doma and Parker 2006; van Hoof et al., 2002; Hashimoto et al., 2017; Arribere and Fire, 2018). Knowledge of the identities and functions of factors that interface between translation and mRNA decay will illuminate a critical junction in gene expression and regulation. Identification of nuclease(s) at this junction would therefore significantly advance our understanding of translation, surveillance, and targeted mRNA decay.
Here we identify a mutation that blocks nonstop and no-go mRNA decay in C. elegans. The mutation identifies a conserved gene, nonu-1, whose structure predicts that it encodes a conserved nuclease component of translation surveillance. NONU-1 contains an Smr domain with the IF3-C fold previously implicated in processing RNA. Homologs of NONU-1 include the recently identified S. cerevisiae CUE2 and the uncharacterized YPL199C, which we show function redundantly in nonstop mRNA decay. Our results identify a critical component of the translation surveillance machinery in two model organisms and suggest why this factor has been recalcitrant to discovery in S. cerevisiae.
RESULTS
nonu-1 Encodes a Conserved Factor Required for Nonstop mRNA Decay
We previously developed a phenotypic reporter in C. elegans that allowed us to identify nonstop mRNA decay factors via reverse and forward genetics (Figure 1A; Arribere and Fire, 2018). Briefly, the reporter was constructed using the unc-54 locus, as expression and function of this gene have been extensively studied (Brenner, 1974; Epstein et al., 1974; Dibb et al., 1985, 1989; Moerman et al., 1982; Bejsovec and Anderson, 1988; Anderson and Brenner, 1984) and unc-54 has been used in previous suppressor screens (Hodgkin et al., 1989). The nonstop reporter has GFP integrated at the C terminus of UNC-54, a ribosomal skipping T2A sequence between GFP and the 3′ UTR, and all stop codons removed from the 3′ UTR. The T2A sequence is a viral-derived peptide that cotranslationally releases the upstream protein and allows UNC-54::GFP to escape nonstop protein decay (so-called ribosome quality control; Bengtson and Joazeiro, 2010; Shao et al., 2013; Shen et al., 2015). We hereafter refer to the unc-54::gfp::t2a::nonstop reporter as unc-54(nonstop). Animals with the unc-54(nonstop) reporter deficient in nonstop mRNA decay exhibit derepression of the locus, as evidenced by increased GFP fluorescence, mRNA expression, and egg laying (unc-54 encodes a muscle myosin required in the vulva for egg laying) (Figure 1B; Arribere and Fire, 2018). Although our initial screen successfully identified C. elegans’ skih-2 and ttc-37 (homologs of S. cerevisiae SKI2 and SKI3, respectively), it did not identify a factor that could function as an endonuclease.
Figure 1. NONU-1 and Its Homologs Have a Conserved Function in Nonstop Decay.
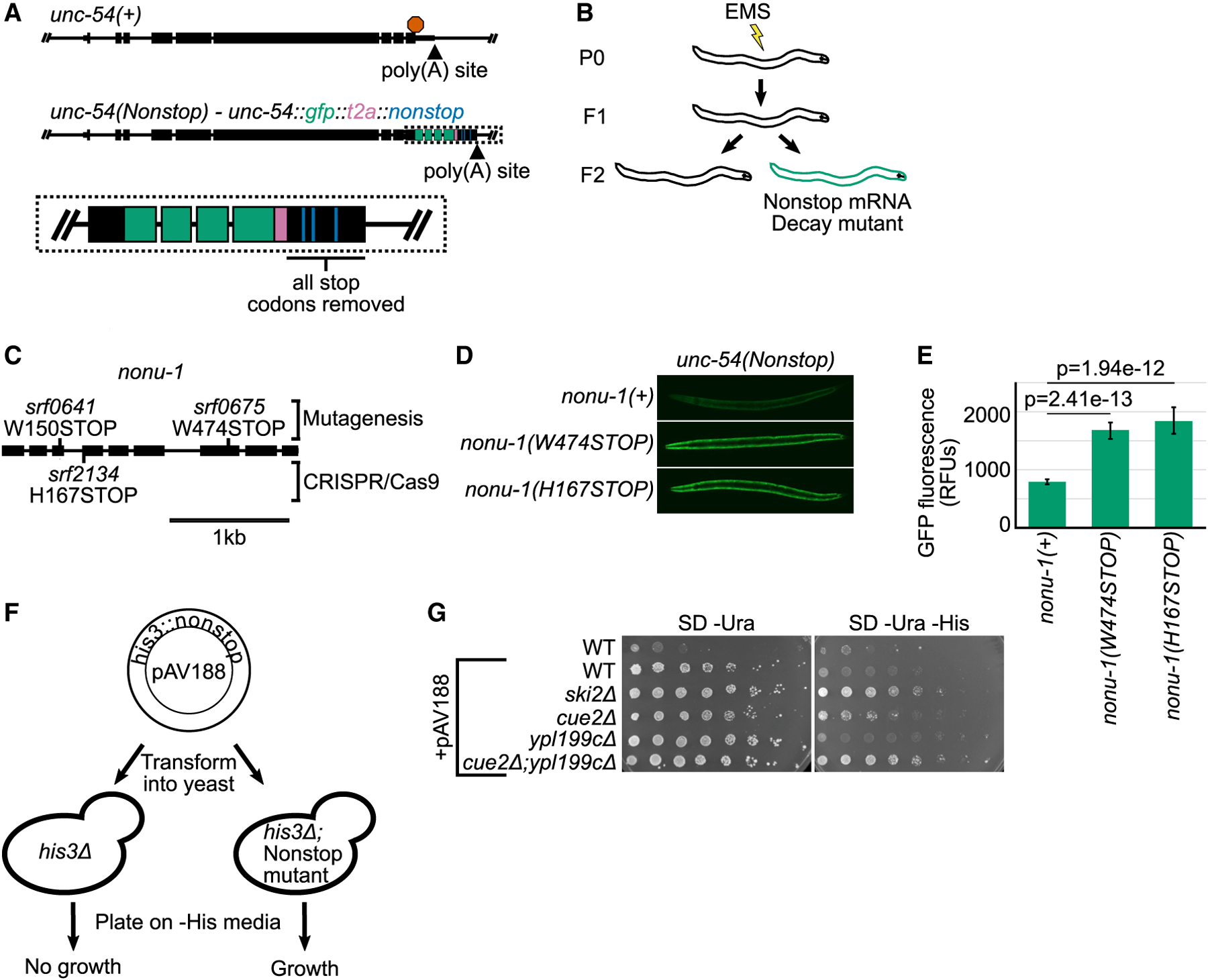
(A) Gene diagrams showing annotated exons (black rectangles) at the wild-type unc-54 locus (top) and the unc-54(nonstop) reporter (bottom). Red octagon indicates the stop codon; triangles indicate poly(A) sites. Inset shows zoom in with GFP (green), T2A (pink), and re-coded stop codons (blue).
(B) Cartoon of the genetic screen to identify nonstop mRNA decay mutants.
(C) Gene diagram showing annotation of the nonu-1 locus. Mutations made via EMS and CRISPR/Cas9 are above and below the gene, respectively.
(D) Images of GFP expression in the indicated strains. See STAR Methods.
(E) Quantification of GFP expression for strains shown in (D). Each bar represents average images of at least five different animals; 95% confidence interval shown as error bars. p values from Student’s t test comparing mutants with nonu-1(+). RFUs, relative fluorescence units.
(F) Diagram of the S. cerevisiae nonstop decay assay. See text for details.
(G) The indicated strains were transformed with the reporter plasmid, and strains were serially diluted and plated on the indicated media. Pictures are representative of experimental triplicates.
We repeated the genetic screen and isolated an additional 36 mutants. We genetically mapped the causative locus in each strain by backcrossing to a polymorphic strain (also called Hawaiian variant mapping; Figure S1; Arribere and Fire, 2018; Doitsidou et al., 2010). The majority of mutants mapped to loci homologous to known nonstop mRNA decay factors in other systems. However, two strains (WJA0675 and WJA0641) harbored mutations that mapped to an area lacking obvious known nonstop mRNA decay components (Figure S1). Visual inspection revealed that strain WJA0675 contained a Trp > STOP mutation in predicted ORF f26a1.13, and strain WJA0641 contained a Trp > STOP mutation in the neighboring ORF f26a1.14 (Figure S2A). Our subsequent analyses showed that f26a1.13 and f26a1.14 are a single gene that is required for nonstop mRNA decay (Figure S2; Lee et al., 2018), and we hereafter refer to this gene as nonu-1 (nonu [nonstop nuclease]). Homology searches with the encoded NONU-1 protein identified homologous proteins in diverse eukaryotes, but no homolog known to function in nonstop mRNA decay. During review of this manuscript, one homolog of NONU-1 in S. cerevisiae (CUE2) was identified as a factor involved in no-go mRNA decay (D’Orazio et al., 2019).
S. cerevisiae Homologs of NONU-1 Are Required for Nonstop mRNA Decay
A previous genetic screen in S. cerevisiae failed to identify a homolog of NONU-1 (Wilson et al., 2007). We performed a homology search and identified two candidate homologs in S. cerevisiae: YPL199C and CUE2. Of the two, CUE2 was recently identified as a factor involved in no-go mRNA decay (D’Orazio et al., 2019), but the relationship of YPL199C to CUE2 and nonu-1 has not been studied. To determine whether CUE2 and/or YPL199C function in nonstop decay, we assayed the ability of mutant strains to derepress a his3::nonstop reporter previously used to identify and study factors required for nonstop decay in S. cerevisiae (Wilson et al., 2007; van Hoof et al., 2002). When transformed into a his3Δ strain, the his3:: nonstop reporter allows the selective growth of nonstop decay mutants (Figure 1F).
Consistent with previous work, we observed substantial derepression of the reporter in a ski2 mutant (Figure 1G). In either a cue2Δ or ypl199cΔ mutant, we observed suppression of the his3::nonstop reporter. The magnitude of the suppression was significantly less than that conferred by a ski2 mutation but was reproducible across independent isolates and technical replicates. The small magnitude of growth suppression compared with other mutants (e.g., ski2) may have precluded either gene from being identified in a previous genetic screen (Wilson et al., 2007). Analysis of a cue2Δ ypl199cΔ double-mutant strain revealed an even greater suppression of nonstop decay than either single mutant, pointing to a functional redundancy that likely precluded detection from prior loss-of-function screens. We conclude that NONU-1 and its homologs in S. cerevisiae have a conserved function in nonstop decay. Although CUE2 and YPL199C each had a consistent effect on the his3::nonstop reporter, we note that the magnitude of this effect was below that of other factors (i.e., SKI2), suggesting multiple independent mechanisms exist to repress nonstop mRNAs.
Domain Architecture of NONU-1
To gain insight into NONU-1 function, we examined the domain structure of the protein and its metazoan orthologs and found that they contain several conserved domains (from N terminus to C terminus; Figures 2A and S3). The NONU-1 protein family is characterized by the following:
Figure 2. NONU-1 Contains a Conserved Smr Domain Required for Nonstop mRNA Decay.
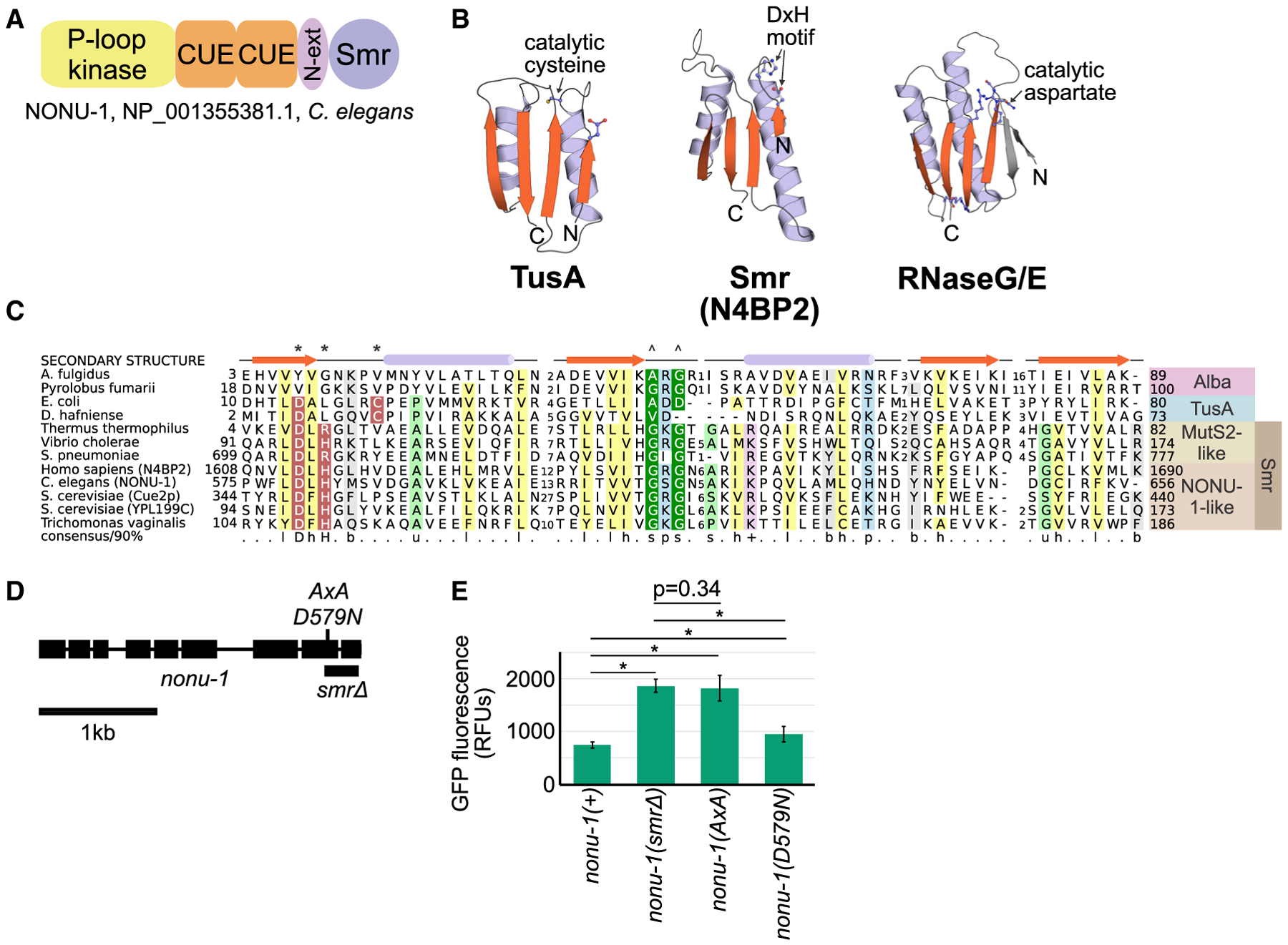
(A) Protein domain schematic of NONU-1. See text for details.
(B) Structures of representatives of the IF3-C fold domain, including the Smr domain from the human homolog of NONU-1, N4BP2. See also Figure S3C.
(C) Multiple sequence alignment of the TusA-Alba-Smr assemblage with protein secondary structure diagram and conserved amino acids highlighted. Asterisk and caret indicate amino acids involved in catalysis and substrate recognition, respectively, in TusA- or MutS2-like proteins. See also Figure S3A.
(D) Gene diagram showing the nonu-1 locus and mutations generated by CRISPR/Cas9.
(E) Quantification of GFP expression for the indicated strains (as in Figure 1E). p values from Student’s t test; *p < 10e-6. Compare with Figure 1E. See also Figure S3.
An N-terminal basic region similar to a ribosome-binding motif at the N terminus of the ribosomal protein S26AE. This basic stretch is only observed in the chordate versions of NONU-1 and is thus not pictured in Figure 2A. The basic stretch suggests that NONU-1 might interact directly with ribosomes.
A domain of the P loop kinase superfamily belonging to the polynucleotide kinase (PNK) clade. These kinase domains are known to phosphorylate RNA/DNA ends (Leipe et al., 2003; Burroughs and Aravind, 2016). The P loop kinase domain suggests NONU-1 may modify nonstop mRNAs or their degradation products.
Two ubiquitin-binding coupling of ubiquitin to ER degradation (CUE) domains of the UBA-like fold (Kang et al., 2003). Ub chains are an important signal for ribosomal stalling and suggest a mechanism of specificity for NONU-1 recruitment to stalled ribosomes (Ikeuchi et al., 2019; Simms et al., 2017; Garzia et al., 2017; Juszkiewicz et al., 2018; Saito et al., 2015).
An Smr domain, homologous in structure and sequence to domains known to bind, cleave, or process RNA (Figures 2B, 2C, and S3A; Aravind et al., 2003). Smr domains of some proteins function as an endoribonuclease (Bhandari et al., 2011; Zhou et al., 2017; Wu et al., 2016), and the Smr domain of CUE2 was recently identified as being critical for no-go mRNA decay in S. cerevisiae (D’Orazio et al., 2019). The NONU-1 Smr domain co-occurs with a highly charged, small helical extension that likely represents an extension of the Smr domain (“N-ext,” also known as “DUF1771”). The existence of a domain known to function as an endoribonuclease makes NONU-1 a prime candidate for an endonuclease involved in translation surveillance.
The combination of these domains characterizes the NONU-1 family of proteins found throughout metazoans and choanoflagellates as an endoribonuclease with a conserved role in diverse cell types and organisms.
Catalysis by the Smr Domain of NONU-1 Is Required for Nonstop mRNA Decay
Given the above observations, we investigated whether the Smr domain is important for NONU-1 function in nonstop mRNA decay. We used CRISPR/Cas9 to delete the Smr domain, generating nonu-1(srf0780), which we hereafter refer to as nonu-1(smrΔ) (Figure 2D). Expression, splicing, and stability of the nonu-1 transcript was not grossly perturbed in nonu-1(smrΔ) as assayed using RNA sequencing (RNA-seq). When combined with the unc-54(nonstop) reporter, nonu-1(smrΔ) conferred derepression of GFP expression comparable with the nonu-1 premature stop codon mutations originally isolated in the genetic screen (Figure 2E). We thus conclude that the NONU-1 Smr domain is required for nonstop mRNA decay.
We analyzed the sequence and structure of the NONU-1 Smr domain to better understand its potential catalytic mechanism. Sequence alignment of NONU-1 homologs across eukaryotes identified a highly conserved aspartate-x-histidine (DxH, where x is typically a hydrophobic amino acid) motif (Figures 2C and S3A) shared with two related endoribonucleases (Bhandari et al., 2011; Zhou et al., 2017). The DxH motif occupies a position in the Smr domain similar to the location of active site residues of other catalytic versions of the IF3-C fold (Figure 2B) and was also identified as being critical for CUE2 function in S. cerevisiae (D’Orazio et al., 2019). Given the defining DxH motif, we investigated if this motif is required for NONU-1’s role in nonstop decay. Alanine substitutions at this location (DxH > AxA) exhibited a defect in nonstop decay comparable with the nonu-1(smrΔ) as well as nonu-1 premature stop codon mutations (Figures 1E and 2E). Taken together, these observations are consistent with the idea that the DxH motif contributes to the active site of the NONU-1 endoribonuclease.
It was recently reported that endonucleolytic cleavage during no-go decay occurs via an unknown metal-independent nuclease resulting in a 3′ phosphate and 5′ hydroxl (Navickas et al., 2020). Although the catalytic mechanism of Smr is unknown, the emerging picture of Smr endoribonuclease activity is consistent with what is known about cleavage during no-go/nonstop. First, in vitro RNA cleavage with Smr domain-containing proteins is inhibited by metals (Zhou et al., 2017). Second, metal-dependent nucleases typically require negatively charged amino acids (Asp or Glu) to chelate the positively charged metal. The one residue in NONU-1 that could conceivably function in this manner is Asp579 of the DxH motif, but the isosteric mutation nonu-1(D579N) was still functional (Figure 2E). Functional substitution of Asp579 with an asparagine is most readily consistent with a metal-independent role for this residue in catalysis.
Our results are consistent with the model that NONU-1 is an endoribonuclease that acts during nonstop mRNA decay, with metal-independent cleavage being carried out by the highly conserved DxH motif within the Smr domain.
NONU-1 Reduces Nonstop mRNA Levels
The unc-54(nonstop) reporter contains a T2A “self-cleaving” peptide after the open reading frame that allows the nascent peptide to leave the ribosome and escape repression from ribosome quality control (Arribere and Fire, 2018). That we identified nonu-1 as a phenotypic suppressor of the unc-54(nonstop) reporter suggests that nonu-1 acts in nonstop mRNA decay rather than in ribosome quality control (Figure 3A). Consistent with this, by RNA-seq, we detected a 2.4-fold increase of the unc-54(nonstop) reporter mRNA in nonu-1(smrΔ) (Figures 3B and S4A). We thus conclude that nonu-1 acts to reduce nonstop mRNA levels.
Figure 3. NONU-1 Reduces mRNA Levels of Nonstop and No-Go Reporters.
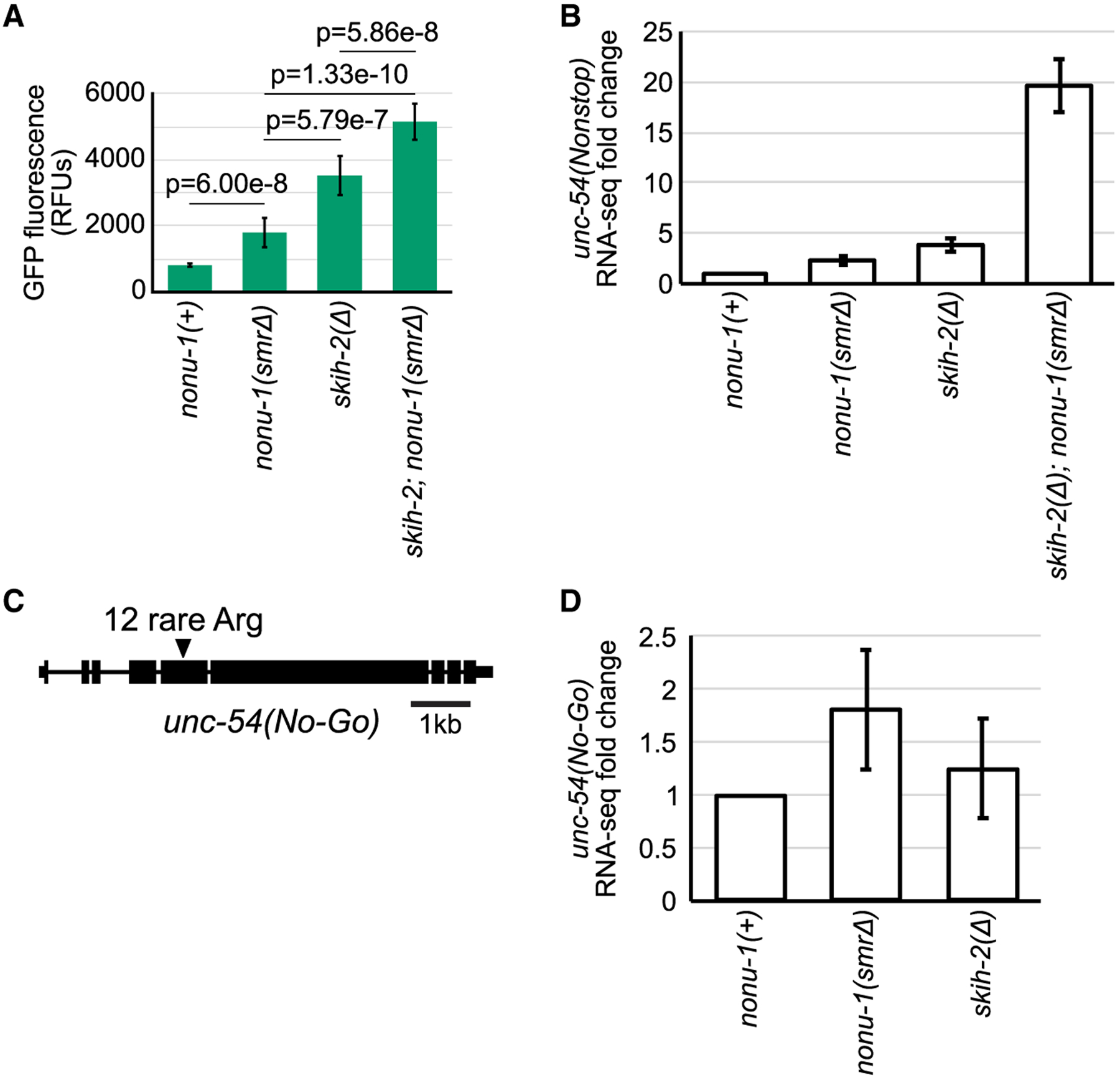
(A) Quantification of GFP expression for the indicated strains (as in Figure 1E). p values from Student’s t test.
(B) RNA-seq shown as average fold change of unc-54(nonstop) relative to nonu-1(+) with 95% confidence interval shown as error bars.
(C) Gene diagram of unc-54 no-go decay reporter. Triangle indicates an insertion of 12 rare arginine codons (allele srf0788).
(D) RNA-seq for unc-54(no-go) with 95% confidence interval as error bars. See also Figure S4.
We compared the phenotypic effect on unc-54(nonstop) conferred by mutations in nonu-1 relative to mutations in other nonstop mRNA decay components. The phenotype of nonu-1 mutants was distinct from skih-2-null mutants, as assayed by the level of GFP fluorescence and unc-54(nonstop) mRNA expression (Figures 3A and 3B). Interestingly, the skih-2 nonu-1 double mutant exhibited even greater nonstop suppression than either single mutant alone, as assayed by nonstop mRNA levels, nonstop protein levels, and suppression of unc-54’s egg-laying phenotype (Figures 3A, 3B, and S4B). This result is consistent with the idea that phenotypic suppression by skih-2 and nonu-1 do not strictly depend on each other. It is unclear how much of the SKI complex’s repressive effect in nonstop mRNA decay is a direct result of accelerated mRNA decay versus other effects (e.g., on mRNA translation initiation and/or recycling; Searfoss and Wickner, 2000; Searfoss et al., 2001; Schmidt et al., 2016).
Because the above analysis of unc-54 mRNA expression was done using RNA-seq, we were able to address the question of whether skih-2 and/or nonu-1 are required for normal expression of endogenous mRNAs. There are endogenous mRNAs targeted by the nonstop decay pathway in other organisms (e.g., Sparks and Dieckmann, 1998). Although we were able to detect mRNAs that increased in either skih-2 or nonu-1 (Tables S2 and S3), our subsequent analyses support the idea that these mRNAs change as a result of secondary effects (Figure S4; Hendriks et al., 2014).
NONU-1 Acts in No-Go mRNA Decay
In addition to nonstop decay, endonucleolytic cleavage of the mRNA is thought to be an important step in no-go decay (Doma and Parker, 2006). No-go decay results from blocks in translation elongation such as rare codons, polybasic amino acid stretches, and RNA structures (Doma and Parker, 2006). We generated a no-go decay reporter in C. elegans by inserting12 rare arginine codons in-frame in the unc-54 gene (unc-54[no-go]) (Figure 3C). We observed 2-fold derepression of the unc-54(no-go) mRNA in nonu-1(smrΔ)-mutant animals (Figure 3D). Thus NONU-1 is required for repression during no-go decay, and this function resides in the Smr domain. This result points to NONU-1 as a general player in translation surveillance.
We and others have recently shown that nonstop mRNA decay components including SKI and PELO act in NMD after a committed step of mRNA degradation (Hashimoto et al., 2017; Arribere and Fire, 2018). As with skih-2 and pelo-1, we failed to detect derepression of endogenous NMD targets using RNA-seq in nonu-1 mutants. Once there is additional information on the biochemistry, function, and relationship of NONU-1 to other translation surveillance events, it will become possible to directly test the hypothesis that NONU-1 functions in NMD.
NONU-1 Is Required for Some RNA Cleavages in the Vicinity of Stalled Ribosomes
A simple model to explain NONU-1’s function in translational surveillance is that it acts as an endonuclease after ribosome stalling. To test this model, we decided to characterize cleavage fragments during nonstop and no-go decay.
We first set out to characterize the role of NONU-1 in mRNA degradation during nonstop mRNA decay. Unfortunately, limitations preclude some techniques from being used to examine cleavage products in the unc-54(nonstop) mRNA reporter: the unc-54(nonstop) mRNA is huge (>6 kb), making northern analysis of small cleavage differences difficult, and the A/T-rich nature of the unc-54 3′ UTR causes short Ribo-seq reads to be lost during PCR (Arribere and Fire, 2018). For this reason we turned to a technique (3′RACE 0[3′ rapid amplification of cDNA ends]) that allows longer read lengths and 3′ end identification with single-nucleotide precision. To enhance the stability of cleavage products for detection, we performed these analyses in a skih-2 pelo-1 mutant background, which slows 3′> 5′ decay.
Our initial analysis of 3′ RACE reads mapping to the unc-54(nonstop) 3′ UTR revealed no major nonu-1-dependent differences (Figure 4A). Interestingly, we observed a population of 3′ends coinciding with the position of known ribosomal stalls (Arribere and Fire, 2018). The lack of a nonu-1-dependence to these cleavages points to the existence of nonu-1-independent decay mechanisms active during nonstop mRNA decay.
Figure 4. nonu-1-Dependent mRNA Cleavage Occurs in the Vicinity of Stalled Ribosomes.
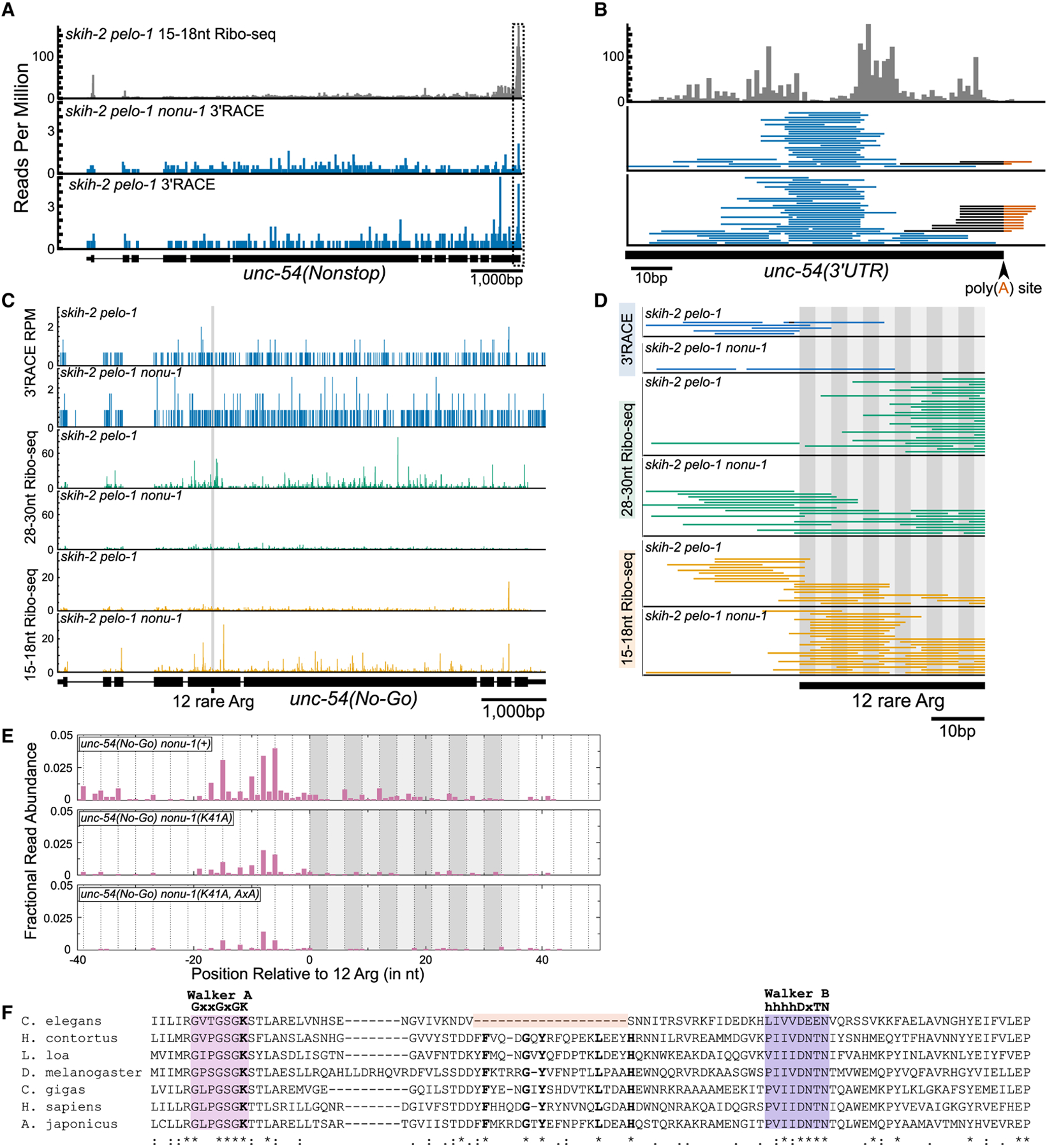
(A) 15–18 nt Ribo-seq reads from (Arribere and Fire, 2018) on top (gray) and 3′ RACE data from this study on bottom (blue). The 5′ ends of Ribo-seq and 3′ ends of 3′ RACE reads were mapped to unc-54(nonstop), with read counts displayed per million uniquely mapping reads (RPM).
(B) Zoomed-in view of the boxed area in (A). Reads aligned to the poly(A) junction are shown with the sequence mapping to the 3′ UTR in black and poly(A) sequence in red. The 3′ ends of Ribo-seq reads are shown.
(C) 3′ RACE reads (top panel; 3′ ends, blue), 28–30 nt Ribo-seq reads (middle panel; 5′ ends, green), and 15–18 nt Ribo-seq reads (bottom panel; 5′ ends, orange).
(D) Zoomed-in view from (C) showing region upstream of and including the 12 rare arginines (alternating gray boxes).
(E) 5′ RACE reads mapped to unc-54(no-go). The 5′ ends of reads are displayed as fractional read abundance (STAR Methods).
(F) Multiple sequence alignment of the P loop kinase domain of NONU-1 and its homologs. See text.
A caveat of the above analysis is that any cleavages occurring in the ~ 80 nt poly(A) tail (Lima et al., 2017) of unc-54 would generate unmappable reads that would be lost. Consistent with this idea, a manual inspection revealed several unmapped reads that matched the unc-54 3′ UTR and contained untem-plated adenosines at the annotated poly(A) site (Figure 4B). Although we observed a general decrease in such reads in the nonu-1(AxA) mutant, the limited number of such reads makes it difficult to draw strong conclusions.
Given the difficulties in detecting poly(A)-internal cleavage events, we decided to analyze nonu-1’s role in no-go mRNA decay. We performed two variations on Ribo-seq: 15–18 nt Ribo-seq, which captures the products of terminally stalled ribosomes at cleavage sites, and 28–30 nt Ribo-seq, which captures some elongating ribosomes (Ingolia et al., 2009; Guydosh and Green, 2014; Arribere and Fire, 2018). The unc-54(no-go) reporter contains AGG and CGG arginine codons in a non-random order so as to allow unique mapping of short RNA fragments at and around the stretch of 12 arginines.
Expression analysis showed active translation throughout the unc-54(no-go) reporter, with no substantial difference in 28–30 nt Ribo-seq reads for the thousands of bases before and after the stall (Figure 4C). Consistent with this, we failed to observe a large discrete peak in either the 3′RACE or 15–18 nt Ribo-seq data. Taken together, these datasets are consistent with the idea that ribosomal stalling by arginine codons is relatively inefficient in C. elegans, with most ribosomes translating through the 12 consecutive arginines. We note this is in contrast to S. cerevisiae, in which even two rare arginine (CGA) codons are sufficient to induce stalling (Letzring et al., 2010).
Next we closely analyzed reads in the vicinity of the arginine stall. In a skih-2 pelo-1 background, we observed an absence of 28–30 nt Ribo-seq reads just upstream of the stall, coincident with the appearance of 15–18 nt Ribo-seq and 3′RACE reads in this region (Figure 4D). A simple model to explain this observation is that some elongating ribosomes stall and experience cleavage. This effect was lost in the nonu-1 mutant; instead 28–30 nt Ribo-seq reads accumulated upstream of the stall, and 3′ RACE and 15–18 nt Ribo-seq reads were absent. We note that the low read counts in this region are consistent with the idea that stalling and cleavage is a relatively inefficient event. Nevertheless, the data from the three techniques fit a model in which nonu-1 facilitates mRNA cleavage after ribosomal stalling.
We next turned our attention to detection of the downstream fragment associated with cleavage. Using a gene-specific 5′RACE protocol, we detected 5′ ends upstream of the rare arginine stretch in the same region in which we observed nonu-1-dependent 3′ends via 3′RACE and 15–18 nt Ribo-seq (Figure 4E). Also consistent with the 3′RACE and 15–18 nt Ribo-seq data, we failed to detect a single discrete site, instead observing a distribution of 5′ ends spanning up to ~ 18 nt upstream of the arginines. The detection of both 5′ and 3′ ends just upstream of the rare arginine stretch is consistent with endonucleolytic cleavage in this region. We note that our 5′RACE protocol was 5′ hydroxyl dependent, adding further evidence to support the idea that the captured ends are the product of a metal-independent cleavage reaction.
We also analyzed the effect of nonu-1 mutations on the presence of 5′ ends. First we characterized the effect of mutation of a highly conserved lysine (K41) present in the Walker A motif of the NONU-1 PNK domain known to be essential for kinase activity (Wang et al., 2002, 2012). In a model in which the kinase domain phosphorylates the 5′ OH end for subsequent degradation, one would expect an increase in 5′ ends at ribosome stall sites in a PNK domain mutant. Curiously, this was not the case; instead there was a slight reduction in the abundance of 5′ ends in the nonu-1(K41A) mutant (Figure 4E). Inspection of the PNK domain of C. elegans NONU-1 revealed that it contains a large (19 amino acid) in-frame deletion spanning a region of the protein including five residues that are otherwise universally conserved in animals (Figure 4F). Further analysis revealed this deletion to be present throughout the Caenorhabditis lineage but not in other nematodes. We thus conclude that the Caenorhabditis lineage suffered a deletion possibly inactivating the PNK domain of NONU-1, complicating an analysis of PNK domain function in this system.
We also tested the effect of the nonu-1(AxA) mutant on the abundance of 5′ ends (nonu-1(K41A, AxA) mutant). We saw a slight reduction in the abundance of 5′ ends, consistent with a role for NONU-1 in their generation (Figure 4E). However, we noted a persistent low level of reads in the nonu-1(K41A, AxA) mutant, thus demonstrating that not all 5′ ends in this region depend on the DxH motif of nonu-1 for their generation. This result demonstrates that although nonu-1 is important for cleavage in the vicinity of stalled ribosomes, at least some cleavages persist in its absence, consistent with other results (e.g., Figures 4A–4D) and data from other labs in other systems pointing to multiple nucleases (Ikeuchi et al., 2019; Navickas et al., 2020).
Evolution of NONU-1 and Smr-Domain Proteins
Given that previous studies have only partly examined the evolution of Smr domains in eukaryotes (Liu et al., 2013), we conducted an in-depth sequence and structure analysis of the Smr domain. Smr is an IF3-C fold domain that also includes the nucleic acid-binding Alba, and the tRNA thiotransfer-catalyzing TusA (Figures 2B, 2C, S3A, and S3B). One unifying sequence feature of this assemblage of IF3-C fold domains is a strongly conserved sxs motif (where s is a small residue, with the second s typically a glycine) in the extended loop region between the second strand and the second helix thought to be involved in substrate binding (Figures 2B, 2C, and S3A; Guo et al., 2014). Among the catalytically active versions of the Smr-TusA-Alba assemblage, a conserved aspartate is observed at the C-terminal end of the initial core strand (Figures 2B, 2C, and S3A). This aspartate is near a histidine in NONU-1 and forms the conserved DxH motif. More distant branches of the IF3-C fold include domains that bind, cleave, or process RNA, including the RNaseG/E nucleases (Fukui et al., 2008), the synaptojanin/calcineurin domain phosphoesterases and nucleases (Burroughs and Aravind, 2016), the Schlafen domain endoribonucleases (Makarova et al., 2001; Li et al., 2018; Yang et al., 2018), and the RtcA RNA end cyclases (Figure S3B; Palm et al., 2000).
At least one NONU-1-like protein is traceable to the last eukaryotic common ancestor and is in practically all major lineages, suggesting a widespread and ancient role for Smr domain surveillance endonucleases in eukaryotes. NONU-1 homologs show a diversity of domain architectures across eukaryotes, including fusions to RNA-binding (CCCH, PWI), 2′−3′ cyclic phosphoesterase (2H), and ubiquitin-binding and conjugation (UIM, UBL, and RING) domains (Figure S3C). We also noted multiple instances of rapidly evolving lineage-specific expansions of NONU-1 homologs. These include multiple paralogs (25 or more) with distinct domain architectures in nematodes of the Caenorhabditis lineage. Some of these have predicted signal peptides, suggesting that they are secreted (e.g., in Caenorhabditis remanei and Entamoeba). The combination of lineage-specific expansions, rapid evolution, and secretion is a hallmark of proteins deployed as effectors in defensive or offensive roles in biological conflicts (Krishnan et al., 2018; Zhang et al., 2016; Lespinet et al., 2002). In light of this, we hypothesize that several eukaryotic Smr proteins, especially the expanded versions, might function beyond translation surveillance as effectors deployed against viral or parasitic RNA. This is consistent with the discovery of a comparable role for the structurally related Schlafen domain in tRNA processing and retroviral RNA restriction (Li et al., 2012; Yang et al., 2018), as well as numerous studies showing that cellular RNA processing and translation surveillance factors have antiviral functions (Toh-E et al., 1978; Garcia et al., 2014; Balistreri et al., 2014).
DISCUSSION
The Role of NONU-1 in Translation Surveillance
NONU-1 and its homologs in S. cerevisiae are the first factors with a nuclease domain to be identified as required for nonstop/no-go mRNA decay. The Smr domain is conserved throughout eukaryotes, and its function in translation surveillance is conserved between S. cerevisiae and C. elegans, establishing NONU-1 as an ancient factor critical to ribosome rescue and mRNA decay. Our identification and characterization of NONU-1 sheds light on the poorly understood intersection of translation and mRNA decay and sets the stage for a more complete molecular understanding of ribosome rescue.
Our analyses support a model in which NONU-1 is required for some mRNA cleavages upstream of ribosomal stalls. This is consistent with the idea that NONU-1 acts on a trailing ribosome that collides with a stalled ribosome, lending support to the idea of ribosomal collisions as an important signal in translational surveillance (Simms et al., 2017). We also note this role of NONU-1 is conserved to S. cerevisiae, where the NONU-1 homolog Cue2p is required for cleavage upstream of a ribosomal stall (D’Orazio et al., 2019).
Our analysis also revealed a population of nonu-1-independent 15–18 nt Ribo-seq reads internal to the stall (Figures 4C and 4D). Arginine-internal ribosomal stalls have not been reported in S. cerevisiae, perhaps because stalling at CGA is so efficient (Letzring et al., 2010), and because the repetitive nature of the (CGA)12 reporter commonly used prevents read mapping internal to the stall. Additional work will help illuminate the diversity of ribosome stalling events and downstream cellular responses.
Interestingly, we note that NONU-1 is not required for full repression of the mRNA targets of translation surveillance. Even in presumed nonu-1-null mutants, we observed substantial repression of nonstop/no-go targets that could be relieved with other suppressors (e.g., skih-2). Two simple, non-mutually exclusive models are as follows: (1) NONU-1 may function redundantly with other endonucleases in translation surveillance. Recent work in S. cerevisiae points to the existence of at least two nucleases active during no-go decay, though their identities remain unknown (Ikeuchi et al., 2019; Navickas et al., 2020). Our analyses here corroborate this idea, as we observed some cleavages that persisted in nonu-1 mutants (Figure 4). (2) There may be cleavage-independent mechanisms that repress the mRNA targets of translation surveillance. Whether through additional nucleases or cleavage-independent mechanisms, our work supports a redundancy in translation surveillance that ensures robust repression of its targets and efficient rescue of stalled ribosomes.
Our results support the idea that NONU-1 acts in translation surveillance largely independently of the SKI complex. We observed multiplicative effects in skih-2 nonu-1 double mutants, and in both C. elegans and S. cerevisiae, we observed a greater derepression of nonstop reporters in skih-2/ski2Δ mutants relative to nonu-1/cue2Δ/ypl199cΔ mutants. This is surprising given the prevailing model in the field that SKI accelerates 3′ > 5′ decay after endonucleolytic cleavage. One possibility is that NONU-1 may function redundantly with other endonucleases to create SKI substrates. Another possibility is that SKI’s role in surveillance is misunderstood. Although it is widely known that one function of the SKI complex is to destabilize the upstream (5′) mRNA fragment during translation surveillance (e.g., Doma and Parker, 2006; Hashimoto et al., 2017), it is unclear if this effect is responsible for the phenotypic suppression of nonstop reporters by SKI. Alternative models include functional suppression by SKI’s effects on ribosome recycling, initiation, or mRNA extraction from the ribosome (Searfoss and Wickner, 2000; Searfoss et al., 2001; Zinoviev et al., 2020), which is also consistent with recent structural data showing a direct role for SKI on the ribosome and near the 5′ ends of ORFs (Schmidt et al., 2016).
In S. cerevisiae, endonucleolytic cleavage during no-go creates a 5′ hydroxyl that is phosphorylated by Rlg1/Trl1 to facilitate 5′ > 3′ digestion by Xrn1 (Navickas et al., 2020). Although Rlg1/Trl1 is widely conserved in several eukaryotic lineages, including fungi, plants, alveolates, and kinetoplastids (Burroughs and Aravind, 2016), its absence in the animal/choanoflagellate lineage raises the question of what protein carries out this RNA repair. Interestingly, the animal/choanoflagellate NONU-1 homologs have acquired a P loop kinase domain that is related to but distinct from the kinase domain of Rlg1/Trl1. The acquisition of a P loop kinase domain in NONU-1 in organisms that have lost Rlg1/Trl1 suggests a model in which NONU-1 phosphorylates its own cleavage products to facilitate degradation by Xrn1. Although we were unable to test this model in C. elegans, we expect that it will inform efforts to understand the fate of cleaved fragments in other animal systems such as humans.
STAR⋆METHODS
Detailed methods are provided in the online version of this paper and include the following:
LEAD CONTACT AND MATERIALS AVAILABILITY
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Joshua Arribere (jarriber@ucsc.edu). All C. elegans strains, S. cerevisiae strains, and plasmids generated in this study are available on request from the Lead Contact without restriction.
EXPERIMENTAL MODEL AND SUBJECT DETAILS
C. elegans growth and propagation
Strains were derived from N2 background (VC2010 [Thompson et al., 2013]). Hermaphroditic animals were grown at 22C on NGM plates using OP50 as a food source per standard C. elegans husbandry (Brenner 1974). Some strains were provided by the CGC, which is funded by NIH Office of Research Infrastructure Programs (P40 OD010440). A full list of strains, sources, and constructions is available in Table S1.
S. cerevisiae husbandry
S. cerevisiae strains were grown on YPAD media at 30C. A full list of strains, sources, and constructions is available in Table S1. All mutations were verified by at least two independent PCR primers.
METHOD DETAILS
EMS mutagenesis
EMS mutagenesis was performed essentially as described (Arribere and Fire, 2018). Briefly, a large population of unc-54(cc4092) was washed with M9 and resuspended in a final volume of 4ml M9. EMS was added to a final concentration of ~ 1mM and animals incubated for 4 h at room temperature with rotation. Animals were washed and allowed to recover overnight on plates with OP50. The next day animals were washed and eggs isolated via sodium hypochlorite treatment. 100–200 eggs were placed on a single small NGM plate and allowed to develop. Plates were screened for individuals with increased GFP fluorescence at the F2/F3 generation. Only a single isolate was kept per small NGM plate, ensuring independence of mutations identified.
Mutation mapping
We crossed each isolated suppressor to Hawaiian unc-54(cc4112) males (expressing an UNC-54::mCherry fusion engineered by CRISPR/Cas9). Cross progeny were picked to new plates to self fertilize. From among the F2, we picked ~ 30 GFP+ progeny to a new plate and allowed the animals to self-fertilize and starve. Upon starvation, animals were washed off the plate with 1mL EN50, and further washed with EN50 to remove residual E. coli. Genomic DNA was extracted after proteinase K treatment and resuspended in 50uL TE pH7.4. 50ng genomic DNA was used as input for Nextera (Tn5) sequencing libraries. Libraries were sequenced at the Vincent J. Coates Genomics Sequencing Laboratory at UC Berkeley.
Reads were mapped to the C. elegans genome using bowtie2 (version 2.3.4.1). Reads were assigned using GATK (McKenna et al., 2010) and a previously published dataset of Hawaiian SNPs (Thompson et al., 2013). The fraction of reads that were assignable to Hawaiian or N2 animals was calculated across the genome, and linkage was identified by portions of the genome that went to 0% Hw. Regions of linkage were then manually inspected to identify candidate lesions/loci.
CRISPR/Cas9 mutagenesis
CRISPR/Cas9 was performed as previously described (Arribere et al., 2014). A full list of gRNAs is available in Table S1, and exact sequences of mutant alleles is provided alongside C. elegans strains in Table S1. Multiple genetic isolates of each mutation were obtained and observed to have identical phenotypes.
Microscopy and image quantification
L4 worms were anesthetized in 2uL 1mM levamisole in a microscope well slide with a 0.15mm coverslip. A Zeiss AxioZoom microscope was used with a 1.0x objective and a GFP fluorescence light source to acquire all images. The unc-54(cc4092); skih-2(cc2854) strain was used to set parameters (exposure time 330ms., shift 50%, zoom 80%) and the same parameters were used for all images. Five representative worms were imaged for each strain. All comparisons shown are between images obtained during the same imaging session.
We used FIJI to define the area of the worm, subtract the background, and determine mean pixel intensity for the area of each worm. A mean fluorescence intensity was calculated for each strain based upon quantification of 5 representative images per strain. We calculated two standard deviations above and below the mean to obtain a 95% confidence interval.
RNA-seq and analysis
25–50 day 1 adults were picked from a blank plate into S-basal solution and washed to remove E. coli. Animals were dissolved in trizol and total RNA extracted. Ribosomal RNA was depleted from 250 ng of total RNA using an NEBNext rRNA Depletion Kit (Human/Mouse/Rat) and libraries were constructed using an NEBNext Ultra RNA Library Prep Kit for Illumina sequencing. Libraries were sequenced at the University of California, Santa Cruz using the Illumina NextSeq platform.
RNA-seq reads were trimmed with cutadapt and mapped using STAR (version 2.5.0a) to the C. elegans genome (WBCel235) with the unc-54 locus modified to match the unc-54(cc4092) allele. Reads that mapped within the annotated bounds of a protein coding gene were assigned to that gene. Multiply-mapping reads or reads that could not be unambiguously assigned to a gene (e.g., due to overlapping genes) were discarded. Read counts were median-normalized using DESeq (Anders and Huber, 2010).
For differential expression of endogenous mRNAs in skih-2 and nonu-1, genes with mRNAs that increased in biological duplicates (skih-2) or triplicates (nonu-1) were identified with DESeq. mRNA levels were deemed significantly different if they exhibited an adjusted p value < 0.05 (skih-2) or < 0.001 (nonu-1). Varying these cutoffs changed the number genes identified as skih-2 or nonu-1 targets, but did not alter our conclusions.
S. cerevisiae Nonstop Decay assay
Cells were transformed with the his3::nonstop reporter plasmid (pAV188) via lithium acetate transformation and plated on selective media (SD-Ura). Two Ura+ transformants were taken for each strain, and results were reproducible across these independent isolates. Cells were subsequently grown on SD-Ura plates and in SD-Ura liquid media to maintain the plasmid. For the his3::nonstop reporter assay, 100mL liquid cultures were grown overnight and harvested at mid-log phase (OD600 ~ 0.5). Cells were pelleted and resuspended in 3mL media. The OD600 was measured and the same OD600 was used as the starting number of cells for all strains. Cells were serially diluted 1:6 and plated on selective media (SD-Ura and SD-Ura-His). Plates were photographed after 4 days of growth at 30C.
Sequence analysis
Domain sequence similarity searches were performed using PSI-BLAST program (Altschul et al., 1997) against the non-redundant (nr) database housed at the NCBI and the HHpred program (Zimmermann et al., 2018) against pfam and pdb databases (Finn et al., 2016; Burley et al., 2019). Structure similarity searches were performed using the DALI server (Holm and Sander, 1995). Multiple sequence alignments were built using the Kalign2 program, with manual adjustments based on profile-profile and high-scoring pair sequence similarity search results (Lassmann et al., 2009). Domain architectures Smr domain-containing proteins were elucidated by first running rpsblast searches against a PSSM library constructed from the pfam profile database (Marchler-Bauer and Bryant 2004). Regions lacking any annotation were then used as seeds in further rounds of iterative similarity searches. Phylogenetic analyses were carried out using approximate-maximum-likelihood as enacted by the FastTree 2.1 program with default parameters for amino acid sequences (Price et al., 2010).
3′ RACE
Strains for 3′RACE in Figures 4A and 4B were grown up as populations of mixed developmental stages (L1-adult); animals in Figures 4C and 4D were grown as populations of synchronized L4/young adult animals. Animals were passed through a 5% sucrose cushion, washed with N50 to remove E. coli, and snap frozen in liquid nitrogen. Frozen animal pellets were dissolved in trizol and total RNA was extracted and stored in TE pH7.4. 1ug of RNA was treated with T4 PNK (NEB) to remove 3′phosphates. RNA was then extracted with phenol chloroform and precipitated into TE pH7.4. AF-JA-34 was ligated onto RNA 3′ends with T4 RNA Ligase 1 (NEB). Adaptor cleanup was performed with 5′ deadenylase (NEB) and RecJ (NEB), followed by RNA extraction as before. RNA was fragmented with 2x AF buffer (10mM Na2CO3, 90mM NaHCO3, 0.5mM EDTA) and then ran on a 15% polyacrylamide gel. AF-JA-126 was used for reverse transcription on gel purified RNA with Superscript II RT (Thermo Fisher). RNA was hydrolyzed with 1N NaOH and the remaining cDNA product was run on 15% polyacrylamide gel. Gel purified cDNA was circularized with circligase (Lucigen) and then used for PCR. Libraries were agarose gel purified and sequenced at the Vincent J. Coates Genomics Sequencing Laboratory at UC Berkeley.
Ribo-Seq
Strains for Ribo-seq were harvested at the L4/young adult stage. Animals were passed through a 5% sucrose cushion and washed in N50 to remove E. coli, and snap frozen in liquid nitrogen. Frozen animal pellets were ground in PLB (20mM Tris pH8.0, 140mM KCl, 1.5mM MgCl2, 1% Triton) with 0.1mg/mL cycloheximide and liquid nitrogen with a mortar and pestle. Ground powder was mixed with PLB and 100ug/mL cycloheximide and clarified via a 10’ spin at 10,000 rcf. RNA in the supernatant was quantified with a nanodrop and OD units were used to calculate the amount of RNaseI to use (total OD × 0.3). RNA was treated with RNaseI (Ambion) and loaded onto a 10%–50% sucrose gradient. Gradients were spun in an SW41 Ti rotor in an ultracentrifuge at 35,000 rpm for 4.5hrs. Monosomes were collected on a fractionator and digested with proteinase K. Monosome RNA was cleaned up by acid phenol chloroform extraction and stored in TE pH7.4. 2ug was ran on 15% polyacrylamide gel and size-selected for 15–18nt or 28–30nt footprints. Gel purified RNA was treated with T4 PNK (NEB) to remove 3′ phosphates. The remaining library preparation was as per 3′ RACE, except no fragmentation was performed.
5′RACE
Strains for 5′RACE were grown up as populations of mixed developmental stages (L1-adult). Animals were passed through a 5% sucrose cushion, washed with N50 to remove E. coli, and snap frozen in liquid nitrogen. Frozen animal pellets were dissolved in trizol and total RNA was extracted and stored in TE pH7.4. 400 pmole of 5′adaptor (/5Phos/ACACGACGCTCTTCCGATCT [barcode] GC rNrNrNrNrNrNrNrN/3Phos/) was ligated to free 5′ OH ends of 1ug RNA with RtcB ligase (NEB). We used distinct 5′ adaptors (JA-AA-313, JA-AA-314, JA-AA-315, and JA-AA-316) for each sample, each with an internal barcode of 6nts that allowed us to sort reads into their respective samples after sequencing. This approach normalized for sample-specific differences in the downstream enzymatic steps (RT, PCR) and the UMI allowed us to collapse PCR duplicates after sequencing. JA-AA-275 binds downstream of the 12 rare arginines in the unc-54(No-Go) reporter and was used for reverse transcription with Superscript II RT (Thermo Fisher). Two rounds of nested PCR were performed with oligos upstream of the RT oligo to reduce products not derived from unc-54, first with PCR primers JA-AA-276 and JA-AA-277, and then with primers for illumina barcoding. Biological and technical replicates with 5′adaptors flipped yielded similar data to that shown in Figure 4E. Libraries were gel purified and sequenced at the Vincent J. Coates Genomics Sequencing Laboratory at UC Berkeley.
Fractional Read Abundance (Figure 4E) was calculated as follows: reads were assigned to samples based on the six nucleotide barcode, and then mapped and collapsed based on common UMIs allowing for up to two mutations in their UMI (this was found to be necessary due to bottlenecking in the PCR which coupled with the error rate of PCR/sequencing led to extraneous UMIs one nucleotide mismatched from a far more abundant UMI). The “fractional read abundance” is the read abundance at each position divided by the total number of unique (UMI-collapsed) reads across all three samples. This metric allows for detection of differences in unc-54 expression across samples. This metric was also found to be reproducible across biological and technical replicates.
QUANTIFICATION AND STATISTICAL ANALYSIS
Statistical details for experiments can be found in Figure Legends and STAR Methods. Briefly, p values were determined by Student’s t test when data were normally distributed (Figures 1E, 2E, 3A, and S4B), and by Mann Whitney U when data were not normally distributed (Figure S4C). All 95% confidence intervals were mean ± two standard deviations from the mean.
DATA AND CODE AVAILABILITY
The accession number for the data reported in this paper is SRA: PRJNA548154.
Supplementary Material
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Chemicals, Peptides, and Recombinant Proteins | ||
| Ethyl Methanesulfonate | VWR/Alfa Aesar | Cat#AAAA12938–06 |
| Proteinase K | Sigma Aldrich | Cat#3115879001 |
| Tn5 | Gift from Chris Vollmers | N/A |
| (−)-Tetramisole Hydrochloride (Levamisole) | Sigma Aldrich | Cat#L9756–5G |
| TRIzol Reagent | Thermo Fisher | Cat#15596–026 |
| T4 Polynucleotide Kinase | NEB | Cat#M0201L |
| T4 RNA Ligase 1 | NEB | Cat#M0204L |
| 5′ Deadenylase | NEB | Cat#M0331S |
| RecJ | NEB | Cat#M0264L |
| Superscript II Reverse Transcriptase | Thermo Fisher | Cat#18064014 |
| CircLigase ssDNA Ligase | Lucigen | Cat#76081–608 |
| Cycloheximide | Sigma Aldrich | Cat#C1988–1G |
| RNase I | Ambion | Cat#AM2294 |
| RtcB Ligase | NEB | Cat#M0458S |
| Critical Commercial Assays | ||
| NEBNext rRNA Depletion Kit (human/mouse/rat) | NEB | Cat#E6310 |
| NEBNext Ultra RNA Library Prep Kit | NEB | Cat#E7530 |
| Deposited Data | ||
| All sequencing fastq data (SRA: PRJNA548154) | This paper | https://www.ncbi.nlm.nih.gov/bioproject/?term=PRJNA548154 |
| Experimental Models: Organisms/Strains | ||
| See Table S1 for all C. elegans strains | Table S1 | N/A |
| See Table S1 for all S. cerevisiae strains | Table S1 | N/A |
| Oligonucleotides | ||
| See Table S1 for all oligonucleotides | Table S1 | N/A |
| Recombinant DNA | ||
| gRNA sequences in pRB1017 backbone, see Table S1 for oligonucleotides used for cloning | Table S1 | N/A |
| pAV188 (his3::nonstop) | Gift from Ambro van Hoof (van Hoof et al., 2002) | N/A |
| pDD162 (Cas9) | Addgene | #47549 |
| Software and Algorithms | ||
| FIJI | Schindelin etal., 2012 | https://fiji.sc |
| Bowtie2 (version 2.3.4.1) | Langmead and Salzberg, 2012 | http://bowtie-bio.sourceforge.net/bowtie2/index.shtml |
| GATK (version 3.5) | McKenna et al., 2010 | https://gatk.broadinstitute.org/hc/en-us/ |
| Cutadapt (version 1.15) | Martin, 2011 | https://cutadapt.readthedocs.io/en/stable/ |
| STAR (version 2.5.0a) | Dobin et al., 2013 | https://github.com/alexdobin/STAR |
| DESeq (version 1.34.1) | Anders and Huber, 2010 | https://bioconductor.org/packages/release/bioc/html/DESeq.html |
| PSI-BLAST | Altschul et al., 1997 | https://blast.ncbi.nlm.nih.gov/Blast.cgi?CMD=Web&PAGE=Proteins&PROGRAM=blastp&RUN_PSIBLAST=on |
| DALI Server | Holm and Sander, 1995 | http://ekhidna2.biocenter.helsinki.fi/dali/ |
| Kalign2 (version 2.04) | Lassmann et al., 2009 | https://anaconda.org/bioconda/kalign2 |
| FastTree 2.1 | Price et al., 2010 | http://www.microbesonline.org/fasttree/ |
| HHpred | Zimmermann et al., 2018 | https://toolkit.tuebingen.mpg.de/tools/hhpred |
Highlights.
NONU-1 represses nonstop and no-go mRNA decay targets
The Smr RNase domain of NONU-1 is required for mRNA repression
NONU-1-dependent mRNA cleavages occur in the vicinity of stalled ribosomes
NONU-1 function and catalytic residues are conserved throughout eukaryotes
ACKNOWLEDGMENTS
We thank Manny Ares, Grant Hartzog, and their respective labs for providing yeast strains, reagents, and expertise; Ben Abrams and the University of California, Santa Cruz (UCSC), Microscopy Facility for imaging and quantification; Beth Shapiro, Beth Nelson, the UCSC Paleogenomics Lab, and the Vincent J. Coates Genomics Sequencing Facility for deep sequencing; Ambro van Hoof for sharing pAV188; Susan Strome and lab for reagents and microscopy; and Chris Vollmers for Tn5. We thank Manny Ares, Susan Strome, Al Zahler, and Sara Dubbury for comments on the manuscript. This work was supported in part by the Intramural Research Program of the National Library of Medicine at the National Institutes of Health (NIH) (A.M.B. and L.A.), the National Science Foundation (NSF) Graduate Research Fellowship (NSF DGE-1842400 to M.L.G.), an R01 grant from the National Institute of General Medical Sciences (NIGMS) (1R01GM131012-01 to J.A.A.), a Searle Scholars award (J.A.A.), and start-up funds from UCSC (J.A.A.).
Footnotes
SUPPLEMENTAL INFORMATION
Supplemental Information can be found online at https://doi.org/10.1016/j.celrep.2020.03.023.
DECLARATION OF INTERESTS
The authors declare no competing interests.
REFERENCES
- Altschul SF, Madden TL, Schäffer AA, Zhang J, Zhang Z, Miller W, and Lipman DJ (1997). Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25, 3389–3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anders S, and Huber W (2010). Differential expression analysis for sequence count data. Genome Biol. 11, R106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson P, and Brenner S (1984). A selection for myosin heavy chain mutants in the nematode Caenorhabditis elegans. Proc. Natl. Acad. Sci. U S A 81, 4470–4474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aravind L, Iyer LM, and Anantharaman V (2003). The two faces of Alba: the evolutionary connection between proteins participating in chromatin structure and RNA metabolism. Genome Biol. 4, R64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arribere JA, and Fire AZ (2018). Nonsense mRNA suppression via nonstop decay. eLife 7, e33292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arribere JA, Bell RT, Fu BXH, Artiles KL, Hartman PS, and Fire AZ (2014). Efficient marker-free recovery of custom genetic modifications with CRISPR/Cas9 in Caenorhabditis elegans. Genetics 198, 837–846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balistreri G, Horvath P, Schweingruber C, Zünd D, McInerney G, Merits A, Mühlemann O, Azzalin C, and Helenius A (2014). The host nonsense-mediated mRNA decay pathway restricts mammalian RNA virus replication. Cell Host Microbe 16, 403–411. [DOI] [PubMed] [Google Scholar]
- Bejsovec A, and Anderson P (1988). Myosin heavy-chain mutations that disrupt Caenorhabditis elegans thick filament assembly. Genes Dev. 2, 1307–1317. [DOI] [PubMed] [Google Scholar]
- Bengtson MH, and Joazeiro CAP (2010). Role of a ribosome-associated E3 ubiquitin ligase in protein quality control. Nature 467, 470–473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhandari D, Guha K, Bhaduri N, and Saha P (2011). Ubiquitination of mRNA cycling sequence binding protein from Leishmania donovani (LdCSBP) modulates the RNA endonuclease activity of its Smr domain. FEBS Lett. 585, 809–813. [DOI] [PubMed] [Google Scholar]
- Brenner S (1974). The genetics of Caenorhabditis elegans. Genetics 77, 71–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burley SK, Berman HM, Bhikadiya C, Bi C, Chen L, Di Costanzo L, Christie C, Dalenberg K, Duarte JM, Dutta S, et al. (2019). RCSB Protein Data Bank: biological macromolecular structures enabling research and education in fundamental biology, biomedicine, biotechnology and energy. Nucleic Acids Res. 47 (D1), D464–D474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burroughs AM, and Aravind L (2016). RNA damage in biological conflicts and the diversity of responding RNA repair systems. Nucleic Acids Res. 44, 8525–8555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D’Orazio KN, Wu CC-C, Sinha N, Loll-Krippleber R, Brown GW, and Green R (2019). The endonuclease Cue2 cleaves mRNAs at stalled ribosomes during no go decay. eLife 8, e49117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dibb NJ, Brown DM, Karn J, Moerman DG, Bolten SL, and Waterston RH (1985). Sequence analysis of mutations that affect the synthesis, assembly and enzymatic activity of the unc-54 myosin heavy chain of Caenorhabditis elegans. J. Mol. Biol 183, 543–551. [DOI] [PubMed] [Google Scholar]
- Dibb NJ, Maruyama IN, Krause M, and Karn J (1989). Sequence analysis of the complete Caenorhabditis elegans myosin heavy chain gene family. J. Mol. Biol 205, 603–613. [DOI] [PubMed] [Google Scholar]
- Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, Batut P, Chaisson M, and Gingeras TR (2013). STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29, 15–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doitsidou M, Poole RJ, Sarin S, Bigelow H, and Hobert O (2010). C. elegans mutant identification with a one-step whole-genome-sequencing and SNP mapping strategy. PLoS ONE 5, e15435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doma MK, and Parker R (2006). Endonucleolytic cleavage of eukaryotic mRNAs with stalls in translation elongation. Nature 440, 561–564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Epstein HF, Waterston RH, and Brenner S (1974). A mutant affecting the heavy chain of myosin in Caenorhabditis elegans. J. Mol. Biol 90, 291–300. [DOI] [PubMed] [Google Scholar]
- Finn RD, Coggill P, Eberhardt RY, Eddy SR, Mistry J, Mitchell AL, Potter SC, Punta M, Qureshi M, Sangrador-Vegas A, et al. (2016). The Pfam protein families database: towards a more sustainable future. Nucleic Acids Res. 44 (D1), D279–D285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukui K, Nakagawa N, Kitamura Y, Nishida Y, Masui R, and Kuramitsu S (2008). Crystal structure of MutS2 endonuclease domain and the mechanism of homologous recombination suppression. J. Biol. Chem 283, 33417–33427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia D, Garcia S, and Voinnet O (2014). Nonsense-mediated decay serves as a general viral restriction mechanism in plants. Cell Host Microbe 16, 391–402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garzia A, Jafarnejad SM, Meyer C, Chapat C, Gogakos T, Morozov P, Amiri M, Shapiro M, Molina H, Tuschl T, and Sonenberg N (2017). The E3 ubiquitin ligase and RNA-binding protein ZNF598 orchestrates ribosome quality control of premature polyadenylated mRNAs. Nat. Commun 8, 16056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo L, Ding J, Guo R, Hou Y, Wang D-C, and Huang L (2014). Biochemical and structural insights into RNA binding by Ssh10b, a member of the highly conserved Sac10b protein family in Archaea. J. Biol. Chem 289, 1478–1490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guydosh NR, and Green R (2014). Dom34 rescues ribosomes in 3′ untranslated regions. Cell 156, 950–962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hashimoto Y, Takahashi M, Sakota E, and Nakamura Y (2017). NonstopmRNA decay machinery is involved in the clearance of mRNA 5′-fragments produced by RNAi and NMD in Drosophila melanogaster cells. Biochem. Biophys. Res. Commun 484, 1–7. [DOI] [PubMed] [Google Scholar]
- Hendriks G-J, Gaidatzis D, Aeschimann F, and Großhans H (2014). Extensive oscillatory gene expression during C. elegans larval development. Mol. Cell 53, 380–392. [DOI] [PubMed] [Google Scholar]
- Hodgkin J, Papp A, Pulak R, Ambros V, and Anderson P (1989). A new kind of informational suppression in the nematode Caenorhabditis elegans. Genetics 123, 301–313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holm L, and Sander C (1995). Dali: a network tool for protein structure comparison. Trends Biochem. Sci 20, 478–480. [DOI] [PubMed] [Google Scholar]
- Ikeuchi K, Tesina P, Matsuo Y, Sugiyama T, Cheng J, Saeki Y, Tanaka K, Becker T, Beckmann R, and Inada T (2019). Collided ribosomes form a unique structural interface to induce Hel2-driven quality control pathways. EMBO J. 38, e100276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ingolia NT, Ghaemmaghami S, Newman JRS, and Weissman JS (2009). Genome-wide analysis in vivo of translation with nucleotide resolution using ribosome profiling. Science 324, 218–223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joazeiro CAP (2017). Ribosomal stalling during translation: providing substrates for ribosome-associated protein quality control. Annu. Rev. Cell Dev. Biol 33, 343–368. [DOI] [PubMed] [Google Scholar]
- Juszkiewicz S, Chandrasekaran V, Lin Z, Kraatz S, Ramakrishnan V, and Hegde RS (2018). ZNF598 is a quality control sensor of collided ribosomes. Mol. Cell 72, 469–481.e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang RS, Daniels CM, Francis SA, Shih SC, Salerno WJ, Hicke L, and Radhakrishnan I (2003). Solution structure of a CUE-ubiquitin complex reveals a conserved mode of ubiquitin binding. Cell 113, 621–630. [DOI] [PubMed] [Google Scholar]
- Krishnan A, Iyer LM, Holland SJ, Boehm T, and Aravind L (2018). Diversification of AID/APOBEC-like deaminases in metazoa: multiplicity of clades and widespread roles in immunity. Proc. Natl. Acad. Sci. U S A 115, E3201–E3210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B, and Salzberg SL (2012). Fast gapped-read alignment with Bowtie 2. Nat. Methods 9, 357–359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lassmann T, Frings O, and Sonnhammer ELL (2009). Kalign2: high-performance multiple alignment of protein and nucleotide sequences allowing external features. Nucleic Acids Res. 37, 858–865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee RYN, Howe KL, Harris TW, Arnaboldi V, Cain S, Chan J, Chen WJ, Davis P, Gao S, Grove C, et al. (2018). WormBase 2017: molting into a new stage. Nucleic Acids Res. 46 (D1), D869–D874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leipe DD, Koonin EV, and Aravind L (2003). Evolution and classification of P-loop kinases and related proteins. J. Mol. Biol 333, 781–815. [DOI] [PubMed] [Google Scholar]
- Lespinet O, Wolf YI, Koonin EV, and Aravind L (2002). The role of line-age-specific gene family expansion in the evolution of eukaryotes. Genome Res. 12, 1048–1059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Letzring DP, Dean KM, and Grayhack EJ (2010). Control of translation efficiency in yeast by codon-anticodon interactions. RNA 16, 2516–2528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li M, Kao E, Gao X, Sandig H, Limmer K, Pavon-Eternod M, Jones TE, Landry S, Pan T, Weitzman MD, and David M (2012). Codon-usage-based inhibition of HIV protein synthesis by human schlafen 11. Nature 491, 125–128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li M, Kao E, Malone D, Gao X, Wang JYJ, and David M (2018). DNA damage-induced cell death relies on SLFN11-dependent cleavage of distinct type II tRNAs. Nat. Struct. Mol. Biol 25, 1047–1058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lima SA, Chipman LB, Nicholson AL, Chen Y-H, Yee BA, Yeo GW, Coller J, and Pasquinelli AE (2017). Short poly(A) tails are a conserved feature of highly expressed genes. Nat. Struct. Mol. Biol 24, 1057–1063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu S, Melonek J, Boykin LM, Small I, and Howell KA (2013). PPRSMRs: ancient proteins with enigmatic functions. RNA Biol. 10, 1501–1510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makarova KS, Aravind L, Wolf YI, Tatusov RL, Minton KW, Koonin EV, and Daly MJ (2001). Genome of the extremely radiation-resistant bacterium Deinococcus radiodurans viewed from the perspective of comparative genomics. Microbiol. Mol. Biol. Rev 65, 44–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marchler-Bauer A, and Bryant SH (2004). CD-Search: protein domain annotations on the fly. Nucleic Acids Res. 32, W327–W331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin M (2011). Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet. J 17, 10–12. [Google Scholar]
- McKenna A, Hanna M, Banks E, Sivachenko A, Cibulskis K, Kernytsky A, Garimella K, Altshuler D, Gabriel S, Daly M, and DePristo MA (2010). The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 20, 1297–1303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moerman DG, Plurad S, Waterston RH, and Baillie DL (1982). Mutations in the unc-54 myosin heavy chain gene of Caenorhabditis elegans that alter contractility but not muscle structure. Cell 29, 773–781. [DOI] [PubMed] [Google Scholar]
- Navickas A, Chamois S, Saint-Fort R, Henri J, Torchet C, and Benard L (2020). No-go decay mRNA cleavage in the ribosome exit tunnel produces 5′-OH ends phosphorylated by Trl1. Nat. Commun 11, 122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palm GJ, Billy E, Filipowicz W, and Wlodawer A (2000). Crystal structure of RNA 3′-terminal phosphate cyclase, a ubiquitous enzyme with unusual topology. Structure 8, 13–23. [DOI] [PubMed] [Google Scholar]
- Price MN, Dehal PS, and Arkin AP (2010). FastTree 2—approximately maximum-likelihood trees for large alignments. PLoS ONE 5, e9490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saito K, Horikawa W, and Ito K (2015). Inhibiting K63 polyubiquitination abolishes no-go type stalled translation surveillance in Saccharomyces cerevisiae. PLoS Genet. 11, e1005197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaeffer D, and van Hoof A (2011). Different nuclease requirements for exosome-mediated degradation of normal and nonstop mRNAs. Proc. Natl. Acad. Sci. U S A 108, 2366–2371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schindelin J, Arganda-Carreras I, Frise E, Kaynig V, Longair M, Pietzsch T, Preibisch S, Rueden C, Saalfeld S, Schmid B, et al. (2012). Fiji: an open-source platform for biological-image analysis. Nat. Methods 9, 676–682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt C, Kowalinski E, Shanmuganathan V, Defenouillère Q, Braunger K, Heuer A, Pech M, Namane A, Berninghausen O, Fromont-Racine M, et al. (2016). The cryo-EM structure of a ribosome-Ski2-Ski3-Ski8 helicase complex. Science 354, 1431–1433. [DOI] [PubMed] [Google Scholar]
- Searfoss AM, and Wickner RB (2000). 3′ poly(A) is dispensable for translation. Proc. Natl. Acad. Sci. U S A 97, 9133–9137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Searfoss A, Dever TE, and Wickner R (2001). Linking the 3′ poly(A) tail to the subunit joining step of translation initiation: relations of Pab1p, eukaryotic translation initiation factor 5b (Fun12p), and Ski2p-Slh1p. Mol. Cell. Biol 21, 4900–4908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shao S, von der Malsburg K, and Hegde RS (2013). Listerin-dependent nascent protein ubiquitination relies on ribosome subunit dissociation. Mol. Cell 50, 637–648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen PS, Park J, Qin Y, Li X, Parsawar K, Larson MH, Cox J, Cheng Y, Lambowitz AM, Weissman JS, et al. (2015). Protein synthesis. Rqc2p and 60S ribosomal subunits mediate mRNA-independent elongation of nascent chains. Science 347, 75–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simms CL, Yan LL, and Zaher HS (2017). Ribosome collision is critical for quality control during no-go decay. Mol. Cell 68, 361–373.e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sparks KA, and Dieckmann CL (1998). Regulation of poly(A) site choice of several yeast mRNAs. Nucleic Acids Res. 26, 4676–4687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson O, Edgley M, Strasbourger P, Flibotte S, Ewing B, Adair R, Au V, Chaudhry I, Fernando L, Hutter H, et al. (2013). The million mutation project: a new approach to genetics in Caenorhabditis elegans. Genome Res. 23, 1749–1762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toh-E A, Guerry P, and Wickner RB (1978). Chromosomal superkiller mutants of Saccharomyces cerevisiae. J. Bacteriol 136, 1002–1007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Hoof A, Frischmeyer PA, Dietz HC, and Parker R (2002). Exosome-mediated recognition and degradation of mRNAs lacking a termination codon. Science 295, 2262–2264. [DOI] [PubMed] [Google Scholar]
- Wang LK, Lima CD, and Shuman S (2002). Structure and mechanism of T4 polynucleotide kinase: an RNA repair enzyme. EMBO J. 21, 3873–3880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang LK, Das U, Smith P, and Shuman S (2012). Structure and mechanism of the polynucleotide kinase component of the bacterial Pnkp-Hen1 RNA repair system. RNA 18, 2277–2286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson MA, Meaux S, and van Hoof A (2007). A genomic screen in yeast reveals novel aspects of nonstop mRNA metabolism. Genetics 177, 773–784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu W, Liu S, Ruwe H, Zhang D, Melonek J, Zhu Y, Hu X, Gusewski S, Yin P, Small ID, et al. (2016). SOT1, a pentatricopeptide repeat protein with a small MutS-related domain, is required for correct processing of plastid 23S-4.5S rRNA precursors in Arabidopsis thaliana. Plant J. 85, 607–621. 10.1111/tpj.13126. [DOI] [PubMed] [Google Scholar]
- Yang J-Y, Deng X-Y, Li Y-S, Ma X-C, Feng J-X, Yu B, Chen Y, Luo Y-L, Wang X, Chen M-L, et al. (2018). Structure of Schlafen13 reveals a new class of tRNA/rRNA- targeting RNase engaged in translational control. Nat. Commun 9, 1165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang D, Burroughs AM, Vidal ND, Iyer LM, and Aravind L (2016). Transposons to toxins: the provenance, architecture and diversification of a widespread class of eukaryotic effectors. Nucleic Acids Res. 44, 3513–3533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou W, Lu Q, Li Q, Wang L, Ding S, Zhang A, Wen X, Zhang L, and Lu C (2017). PPR-SMR protein SOT1 has RNA endonuclease activity. Proc. Natl. Acad. Sci. U S A 114, E1554–E1563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimmermann L, Stephens A, Nam S-Z, Rau D, Kübler J, Lozajic M, Gabler F, Söding J, Lupas AN, and Alva V (2018). A completely reimplemented MPI bioinformatics toolkit with a new HHpred server at its core. J. Mol. Biol 430, 2237–2243. [DOI] [PubMed] [Google Scholar]
- Zinoviev A, Ayupov RK, Abaeva IS, Hellen CUT, and Pestova TV (2020). Extraction of mRNA from stalled ribosomes by the Ski complex. Mol. Cell 77, 1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The accession number for the data reported in this paper is SRA: PRJNA548154.


