Abstract
Strigolactones (SLs) are a class of plant hormones involved in several biological processes that are of great agricultural concern. While initiating plant–fungal symbiosis, SLs also trigger germination of parasitic plants that pose a major threat to farming. In vascular plants, SLs control shoot branching, which is linked to crop yield. SL research has been a fascinating field that has produced a variety of different signaling models, reflecting a complex picture of hormone perception. Here, we review recent developments in the SL field and the crystal structures that gave rise to various models of receptor activation. We also highlight the increasing number of discovered SL molecules, reflecting the existence of cross-kingdom SL communication.
Strigolactones Have Many Functions in the Plant World
SLs are a class of terpenoid-derived compounds that were first discovered as (+)-strigol, stimulating the germination of seeds from the parasitic plant Striga [1]. Since the description of (+)-strigol, an astonishing functional diversity of SLs across plants has emerged and a multitude of different SL molecules has been found (Figures 1 and 2). In 2005, SLs were reported to stimulate hyphal branching in arbuscular mycorrhizal (AM) fungi to initiate a symbiotic relationship with their host plants [2]. It is thought that AM symbiosis (see Glossary) originated ~460 million years ago and coincided with the first appearance of bryophyte-like land plants [3]. The moss Physcomitrella patens uses SLs to control colony expansion, a process that is reminiscent of the quorum-sensing mechanism involved in bacterial growth regulation [4]. However, the bioactive SLs in bryophytes have not yet been clearly identified. SLs were later identified as endogenous hormones that regulate shoot branching in vascular plants [5,6]. Regulating the number of branches in a plant is imperative for controlling plant architecture and crucial for the efficient production of biomass. Therefore, increased shoot branching is associated with higher crop yields [7]. At the same time, SLs trigger germination of the parasitic plants of the Orobanchaceae [1], which cause major agricultural damage, especially to farmers in developing nations of Africa and Asia [8,9].
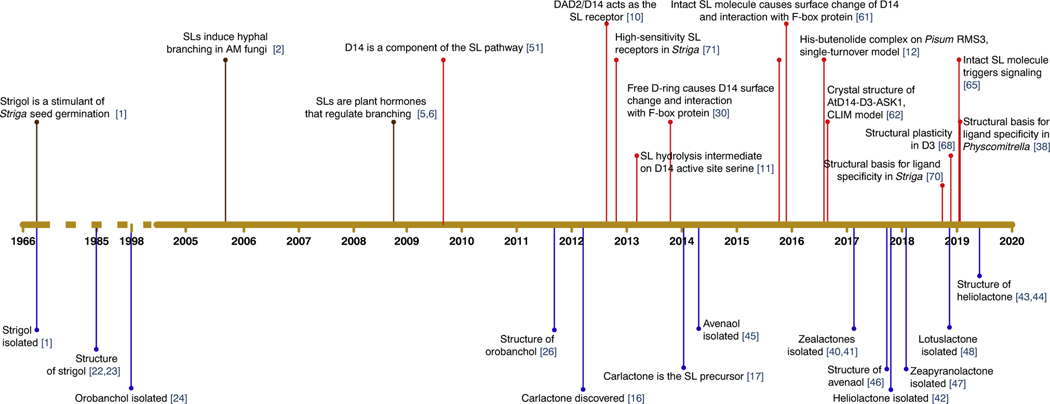
Figure 1. History of Discoveries in the Strigolactone (SL) Field.
Brown lines indicate key discoveries of the biological functions of SLs. Red lines indicate discoveries concerning the signaling mechanism. Blue lines indicate discoveries of SL molecules. Based on [1,2,5,6,10–12,16,17,22–24,26,30,38,40–48,51,61,62,65,68,70,71]. Abbreviations: AM, arbuscular mycorrhiza; CLIM, covalently linked intermediate molecule.
Figure 2. Chemical Structure of Strigolactones (SLs).
Canonical SLs feature a complete A, B, C, D ring structure and are split into two different families resulting from different conformations between the B and C ring. The 2′R conformation of the D-ring is conserved in all SLs (blue). (A) Strigol-type SLs have a conserved β orientation of the C-ring within the B-/C-ring junction (red). Modifications on the A-ring are variable (magenta). (B) Orobanchol-type SLs have a conserved α orientation of the C-ring within the B-/C-ring junction (red). Note the variety of the nature of the A-ring (magenta), as an epoxide in fabacol, a benzene in solanacol, and a cycloheptadiene in medicaol. (C) Noncanonical SLs feature an intact D-ring connected to the rest of the molecule in the same conformation as in canonical SLs (blue). However, the remaining ring system can adopt a different chemistry. (D) The synthetic SL analog GR24.
SLs are perceived by dual-functional receptor/hydrolase proteins with slow substrate turnover [10–12], a feature that adds to the challenge of elucidating how exactly the signal is transduced at the molecular level [13,14]. SL research has become a rapidly advancing field that has seen no less than six signaling models in the past 6 years, accompanied by several crystal structures. Capturing the intact SL molecule inside the receptor remains a major challenge to structural biologists [15]. Here, we review the published crystal structures and embed them in the newest results from the SL field. We discuss the currently known SL molecules together with the evolution of receptor proteins and emphasize the importance of the yet unknown SLs for signaling.
The Growing Family of SL Molecules
A multitude of different SL molecules has emerged in different species. SLs are derived from β-carotene, which is isomerized from a trans to a cis configuration by the β-carotene isomerase DWARF 27 (D27). Further steps include the creation of 9-cis-aldehyde by the enzyme CAROTENOID CLEAVAGE DIOXYGENASE 7 (CCD7) and the production of carlactone by the enzyme CAROTENOID CLEAVAGE DIOXYGENASE 8 (CCD8) from 9-cis-aldehyde [16]. CCD8 has high stereospecificity and, therefore, determines the stereochemistry of all carlactone-derived SLs [17]. SLs include a tricyclic ABC part that is connected to a butenolide D-ring by an enol ether bridge (Figure 2A). In natural SLs, the connection to the D-ring is conserved in the 2′R configuration. The 2′R configured enol ether bridge and the D-ring are required for biological activity [18,19]. There are two different configurations between the B and the C-ring, which ultimately created two different SL families: strigol types and orobanchol types (Figure 2A,B). While rice and tomato strictly produce orobanchol-type SLs, tobacco has both types of SL [20,21]. Determining the correct stereochemical centers has been a difficult task in SL characterization. Although strigol was isolated in 1966, it took until 1985 to determine its full structure [22,23]. Orobanchol was first isolated in 1998 [24] and its structure was initially incorrectly assigned [25] and later corrected [26]. The compound alectrol, which was isolated together with orobanchol, was later identified as the SL orobanchyl acetate [27].
SLs with a 2′S configuration between the C and D-ring are called ‘non-natural’ SLs. To our best knowledge, non-natural SLs do not exist in nature but are a product of racemic chemical synthesis [19,28]. When tested in germination assays using seeds of parasitic plants, (+)-strigol had higher activity on seeds of Striga asiatica compared with (−)-strigol; however, when tested on Alectra vogelii seeds, (−)-strigol activity was greater than (+)-strigol activity [29]. Rice D14 has been observed to degrade the synthetic SL analog (+)-GR24 but not (−)-ent-GR24 [30]. However, binding of both GR24 enantiomers to Arabidopsis D14 was reported [31]. Non-natural SL isomers have been shown to bind to the karrikin [32] receptor KAI2 [33], although at high concentrations [18]. This has led to growing acceptance of the idea that non-natural SLs are somewhat chemical mimics of an as yet undiscovered KAI2 ligand (KL) [34–36], the unknown intrinsic signaling molecule that is perceived by the karrikin receptor KAI2. In addition, GR24 samples may contain contalactone, a recently discovered contamination and by-product that is obtained during the last step of GR24 synthesis. Although present at low concentrations in GR24 samples, contalactone is hydrolyzed by Arabidopsis D14 and KAI2 [37]. The non-natural SL (−)-5-deoxystrigol binds to several receptors from the moss Physcomitrella patens in vitro. Since these proteins do not bind karrikin, it is likely that (−)-5-deoxystrigol acts a chemical mimic of at least one of the actual SLs in Physcomitrella [38]. Unfortunately, the identification of the native SLs in Physcomitrella [4] could not be reproduced by another group [39] or when using newer mass spectrometry (MS) equipment [21].
Thus far, the identification of SL molecules from different species has been based on the analyses of exudates, which contain the SLs that have been secreted. However, endogenous SLs might be different from secreted SLs and, thus, so could the perception of these molecules by the endogenous SL receptors.
In contrast to non-natural SLs, SLs that lack the A, B, or C ring but at the same time retain the enol-ether-D-ring moiety are called ‘noncanonical’ SLs (Figure 2C). This also includes the SL precursor carlactone, which is able to suppress the high-tillering phenotype of the SL-deficient rice mutants d10, d27, and htd-1 and can induce germination of Striga hermonthica seeds, although not as effectively as GR24 [16]. Carlactone has also been found in exudates of Physcomitrella patens [21]. Noncanonical SLs have been found in maize and sunflower, and were named zealactone [40,41] and heliolactone [42], respectively. The stereochemical confirmation of heliolactone was recently confirmed by its total synthesis [43,44]. Black oat (Avena strigosa) produces the noncanonical SL avenaol, which was isolated in 2014 [45] and synthesized and structurally confirmed in 2017 [46]. Most recently, zeapyranolactone [47], another non-canonical maize SL, and lotuslactone [48], a noncanonical SL isolated from Lotus japonicus, were added to the collection. The chemical origin of noncanonical SLs appears to be methyl carlactonoate and, therefore, it would in fact be more appropriate to refer to these SLs as ‘methyl lactonoates’, rather than calling them lactones [21]. Both canonical and noncanonical SLs can elicit germination of the parasitic plants Striga and Orobanche at different concentrations [21,49,50]. We speculate that the specialization among species in production of different noncanonical SLs might reflect the trade-off between AM symbiosis and the need to avoid stimulation of parasitic plants.
The Structure Puzzle of SL Perception
The SL receptor D14 was first identified as a component of the SL signaling pathway from an SL-insensitive mutant of rice, d14 [51]. Studies of the Petunia hybrida homolog DECREASED APICAL DOMINANCE 2 (DAD2) provided evidence that DAD2 is an SL receptor and an α/β hydrolase [10]. Subsequent research confirmed D14 as SL receptors in Oryza sativa [11,30] and also established the homologs D14 in Arabidopsis thaliana [52], and RAMOSUS 3 (RMS3) in Pisum sativum [12] as SL receptors. Upon SL binding, D14 forms a complex with the F-box protein D3 and the transcriptional repressor D53. SLs thereby induce D53 degradation by the proteasome, abrogating the repressing activity of D53 on the SL signaling pathway [53]. The entire process takes place within 15–30 min after exposure to SL [53–56].
Several different crystal structures of D14 have been solved, potentially depicting several different steps of the SL perception and hydrolysis mechanism. In 2012, the first crystal structure of an SL receptor, the protein DAD2 from Petunia hybrida [10], showed that DAD2 folds into a α/β hydrolase architecture [57] and features a four-helix lid domain as well as a putative hydrophobic binding pocket for the SL molecule. α/β hydrolases are a large protein superfamily that includes a variety of different plant proteins, including the gibberellin receptor GID1 [58]. In 2013, three different groups published the crystal structure of rice D14 [11,30,59]. Many SL ligand-binding studies use the chemical GR24, a racemic mixture of a synthetic SL analog [60] (Figure 2D). In one structure, the D-ring, a product of the GR24 hydrolysis reaction, was found to be located in the lid of D14 (Figure 3A), resulting in an altered protein surface structure [30]. The authors proposed that the altered surface was required for the interaction of D14 with the F-box protein D3 and, therefore, for SL signaling. The crystal structure from another group revealed electron density close to the active site serine instead (Figure 3C), which was interpreted as a reaction intermediate after the nucleophilic attack of the serine on the GR24 molecule [11]. Both structures support the concept of SL hydrolysis but do not show any conformational change in the protein structure upon SL binding. Two years later, the same group that provided the crystal structure showing the hydrolysis intermediate at the serine, presented a model in which the change of the D14 protein surface is not caused by the D-ring but by an intact GR24 molecule instead [61]. The authors explained the lack of electron density for most of the ligand molecule (Figure 3D) by its low occupancy in the crystal and by the presence of several reaction intermediates [61]. We would like to offer an alternative interpretation of the electron density. The X-ray data provide a conclusive fit when modeled with the same hydrolysis intermediate at the serine as published 2 years earlier [11] (Figure 3E).
Figure 3. Crystal Structures Containing Parts of the Strigolactone (SL) Analog GR24.
In each of (Ai–Hi), the position and electron difference maps of ligands in the D14 substrate-binding pocket are shown. In each of (Aii–Hii), the electron density of ligands after refinement of the ligands is illustrated. Difference electron density maps are Fo-Fc electron density maps at 3.0 σ calculated from ligand-deleted models. Maps after refinement are 2Fo-Fc electron density maps at 1.0 σ. Ligands were modeled with Coot [80], X-ray data were refined with Phenix [81], and visualizations were made with CCP4mg [82]. (A) Protein Data Bank (PDB) structure 3WIO of rice D14 with the hydroxy D-ring GR24 hydrolysis product [30]. (B) PDB structure 6BRT of rice D14 with the hydroxy D-ring GR24 hydrolysis product [68]. (C) PDB structure 4IHA of rice D14 in complex with a GR24 hydrolysis intermediate (open lactone) covalently linked to the active site Ser147 [11]. (D) PDB structure 5DJ5 of rice D14 in complex with an intact GR24 molecule [61]. (E) PDB structure 5DJ5 with an alternative interpretation of the electron density, showing the same GR24 hydrolysis intermediate as in (C). (F) PDB structure 5HZG of the AtD14-D3-ASK1 complex with electron density interpreted as a covalently linked intermediate molecule (CLIM) [62]. (G) PDB structure 5HZG of the AtD14-D3-ASK1 complex with electron density interpreted as an iodide ion [15]. (H) PDB structure 5HZG with an alternative interpretation of the electron density, showing a histidine-butenolide complex.
In 2016, two other reaction intermediates during SL perception were reported: a study of the D14 homolog in pea, RMS3 [12], and a publication about the crystal structure of the AtD14-OsD3-
AtASK1 complex reported at the same time the mass of the D-ring covalently attached to the histidine of the active site. In addition to MS data, the latter study included crystallographic results showing electron density stretching from the serine to the histidine of the active site (Figure 3F), representing a ligand named ‘covalently linked intermediate molecule’ (CLIM) [62]. The authors concluded that this intermediate, simultaneously attached to these two amino acids, might trigger the conformational change seen in D14 when bound to D3. Whether any of these modified active site residues directly causes the allosteric activation of D14 or whether it is triggered by D3 binding and only requires stabilization by the modification remain open questions. The interpretation of the crystallographic data picturing CLIM has been challenged by another report, which suggested that the atoms representing CLIM are not present in the crystal but that the electron density instead represents an iodide ion [15], because the element was present at high concentrations in the crystallization condition [62]. Our own refinements of the X-ray data using iodide resulted in positive electron density, indicating that the iodide is not an entirely satisfying fit (Figure 3G), and we propose a third possible interpretation for the identity of the ligand. A conclusive fit of the electron density was achieved by modeling a D-ring that is only attached to the histidine (the histidine-butenolide complex) (Figure 3H), which is a modification that has so far been reported by three different groups [12,38,62]. CLIM is part of the crystal structure of the AtD14-OsD3-AtASK1 complex, which revealed the interfaces between the binding partners and finally shed light on the conformational change occurring in D14 when bound to the F-box protein D3. Binding between AtD14 and D3 requires a structural change in helix αT1 in D14. When D14 is bound to MAX2, helix αT1 in D14 has extended and now terminates at residue G158, which is, before D3 binding, located in helix αT2 in the unbound D14 structure. D14 with the glycine mutated to a glutamate (AtD14 G158E) is still able to hydrolyze SL but cannot bind to D3 and fails to initiate SL signaling [62]. Whether this position strictly requires a glycine or if certain side chains or charges would also be tolerated, is an interesting question. The authors make a good point by suggesting that the flexibility of the G158 residue is likely essential for the formation of a π-turn structure at the end of the alpha helix [62], a common structural motif at the C-terminal end of helices that is often caused by glycine residues due to their missing side chain and extraordinary backbone flexibility [63,64].
A recent report challenged the idea of hydrolysis-mediated conformational changes in general and instead presented a model in which the intact SL molecule causes the active D14 signaling state. The study includes a mutant that substituted the so far experimentally neglected third amino acid of the catalytic center of D14, the aspartic acid, with an alanine residue (AtD14 D218A). This protein variant was able to bind, but unable to hydrolyze, SL and complemented the atd14 hyperbranching phenotype [65]. The authors proposed that this aspartate is part of an interface that binds to the signaling repressor D53, which raises the question why signaling is not impaired when this residue is mutated. After polyubiquitination and degradation of D53, the aspartate-containing loop flips back to complete the catalytic triad, hydrolyzing SL. This would mean that SL hydrolysis is not required for signaling and that it occurs after D53 has been degraded and SL signaling has taken place. So why does SL hydrolysis happen at all? The rationale behind this concept is that the SL molecule needs to be destroyed to stop it from getting involved in further signaling. Indeed, AtD14 D218A-expressing plants are very sensitive to exogenously applied SL. However, it appears that this final step would only be necessary if D14 does not get degraded with or after the rest of the complex. Nevertheless, SL-dependent degradation of D14 has been demonstrated by others [66,67], and it occurs within minutes of the SL signal [53–56].
An important outstanding question is why the intact SL molecule is not present in the structure of the AtD14-D3-ASK1 complex [62] if it indeed triggers the signaling state. A possible explanation is that the SL might have spontaneously hydrolyzed before it could be observed [65]. By using excess amounts of SL, it was also demonstrated that D14 is not a single-turnover enzyme, unlike a previous study that reported product inhibition of the homolog in P. sativum, RMS3 [12]. Future investigations will hopefully reveal whether this previously reported single-turnover mechanism of RMS3 is associated with the fluorescent SL analogs used in the study or if the SL receptors in different species have different turnover efficiencies that include single-substrate turnover enzymes.
Can the hydrolase activity of D14 be regulated? In 2018, a publication showed that the F-box protein D3, the rice homolog of Arabidopsis MAX2, can adopt two different conformational states via its C-terminal α-helix [68]. The C-terminal helix is highly dynamic and can dislodge from the rest of the LRR domain. The dislodged helix binds to the prehydrolysis, open-conformation D14 protein and arrests D14 to prevent premature SL hydrolysis before the D53 substrate is loaded and polyubiquitinated. D53 binding catalytically reactivates D3-bound D14, allowing it to carry out SL hydrolysis. The resulting intermediate stabilizes the closed conformation of D14, forcing the C-terminal helix of D3 back into the rest of the LRR domain, allowing D14 to be ubiquitinated. The crystal structure of D14 in complex with the C-terminal helix of D3 revealed a slight distortion of the lid domain of D14, which might explain how D3 arrests D14 in its catalytically inactive state. However, it does not provide an obvious explanation for why binding of the C-terminal helix to D14 is SL dependent. In addition, the D14-D3-helix fusion protein is catalytically less active than D14 itself, but it is not completely inactive [68]. Since crystallization usually occurs over a period of days, it is difficult to assess whether the structure shows the pre- or postcatalytic state. However, the authors did find electron density in the hydrophobic pocket and assigned it to be the free D-ring (Figure 3B), suggesting that GR24 hydrolysis did happen at some point during crystallization.
What Makes an SL Receptor Specific?
How is specificity within the ligand-binding pocket in D14 proteins achieved? D14 and the karrikin receptor KAI2 share a similar overall fold. However, there are differences that likely contribute to ligand specificity. The substrate-binding pocket in D14 is larger than that in KAI2, as shown in Arabidopsis and rice [11,59], as well as in S. hermonthica [69,70]. While presuming that the sheer size of the hydrophobic pocket determines ligand specificity probably overly simplifies the situation, several studies have suggested that the volume of the pocket is one important factor in ligand type and affinity. A striking example is the crystal structure of the Striga SL receptor ShHTL7. The ligand pocket size of ShHTL7 is large compared with other SL receptors and ShHTL7 has picomolar sensitivity to the synthetic SL analog GR24 [71]. It perceives a variety of SLs, such as strigol, 5-deoxystrigol, sorgolactone, and 4-deoxyorobanchol. The latter is interesting considering that Striga has low sensitivity for orobanchol and 4-deoxyorobanchol [72,73]. The finding that ShHTL7 has an extraordinarily large SL-binding pocket was confirmed by another group that solved the crystal structures of three different SL receptors from Striga [70]. Compared with its homologs, ShHTL7 has smaller residues that build an enlarged binding pocket. In addition, it appears that the pocket size can be regulated by other structural features within the protein architecture. Ligand preference between SL and karrikin by homologous Striga receptors is dependent on a hydrogen bond in the cap domain of the protein. A tyrosine to phenylalanine substitution and the subsequent loss of a hydrogen bond in the loop connecting two helices in the protein lid lets those helices slide away from the rest of the lid, resulting in a larger binding pocket. An analogous study reported a similar mechanism of pocket size regulation in D14/KAI2 proteins from Physcomitrella patens [38]. However, the size-regulating feature appears to be a hydrogen bond located in the loop connecting helices αT2 and αT3. P. patens and S. hermonthica appear to have independently evolved similar mechanisms to control the ligand pocket sizes in their SL receptors by using different parts of the protein architecture.
While the volume of the pocket appears to help discriminate between SL molecules and other ligands, the structural contributors that distinguish different SL molecules remain unclear.
The Catalytic Triad: More Than Just Hydrolysis
What is the role of the catalytic triad residues in protein architecture and substrate binding? The 2012 study of the SL receptor DAD2 [10] included a crystal structure of the catalytic DAD2 S96A mutant. In this structure, when compared with wild-type DAD2, the main chain of the F27 residue, which borders the side of the ligand binding cavity, has slightly moved away from the active site. This is likely the result of the loss of a hydrogen bond between the oxygen in S96 and the main chain nitrogen in F27. In addition, the side chain of F27 has less-defined electron density and higher temperature factors, indicating an increase in the flexibility of this residue. Differential scanning fluorimetry (DSF) experiments, which determined the melting points of the proteins, showed slightly lower melting points of both active site mutants, S96A and H246A, compared with wild-type DAD2 [10]. Similar DSF results have been obtained for the pea homolog RMS3, where the S96A mutant protein displayed a slightly lower melting point compared with wild-type RMS3 [12]. An examination of Arabidopsis D14 using DSF showed the same trend: Both active site mutants, AtD14 S97A and AtD14 H246A, had a slightly lower melting point compared with wild-type AtD14 [65]. These results confirm and extend what was observed in the crystal structures of DAD2. The catalytic triad residues, at least the serine and histidine, appear to contribute to the stability of the ligand-binding pocket. In addition, it has been reported that a substitution of the rice D14 active site histidine to an alanine impaired SL hydrolysis, but that the protein was still able to bind GR24 [30]. The architecture of the ligand-binding pocket appears to prefer a serine residue as the nucleophile. Intuitively, substituting the catalytic serine with a cysteine should increase enzymatic activity by introducing a stronger nucleophile [74,75]. However, the opposite has been demonstrated [12,65], thus the binding cavity might be unable to use a different nucleophile. A recent study that included an aspartate to alanine substitution in the active site of Arabidopsis D14 [65] allowed further investigation of SL binding and hydrolysis. AtD14 D218A not only complemented the atd14 hyperbranching phenotype, but was also more sensitive to GR24, which the authors explained by a lack of postresponse removal of SL due to the eliminated catalytic activity [65]. Their DSF experiments, which investigated whether destabilization of D14 takes place upon SL binding, showed that the substitution had made the protein unstable even without presenceof SL. The reason for the instability is unclear, but considering the location of the aspartic acid, it might hint at an altered architecture of the ligand-binding pocket. Taking into account the crystal structures of wild-type DAD2 and DAD2 S96A, as well as the DSF experiments using SL receptors with active site mutations from DAD2, RMS3, and AtD14, it appears that the active site contributes to the stability and rigidity of the ligand-binding pocket. In wild-type D14, the histidine-butenolide formation might interrupt the interaction between the active site histidine and aspartate and thereby stabilize the alternative loop conformation bound to D3/MAX2 (Figure 4). We speculate that the reported D218A mutation in D14 [65] might have created a constitutionally active state of D14 and a protein that can undergo the conformational change and bind to D3/MAX2 without SL hydrolysis, because the side chain of the aspartate has been removed. This might be an alternative explanation for the increased sensitivity for GR24 that the authors have observed. Hopefully, future crystallographic studies of D14 D218A will clarify whether, and to what extent, the ligand-binding site has been altered by the mutation.
Figure 4. Model of Strigolactone (SL) Signaling.
(A) The SL receptor D14 has a preformed binding pocket [10,11,59] for the SL molecule that comprises a tricyclic ABC part connected to the D-ring (red). (B) After binding of the intact SL molecule [65,68], the F-box protein D3 (MAX2) binds to D14 with its dislodged C-terminal helix, arresting D14 in a catalytically inactive state [68]. (C) Binding of the transcriptional repressor D53 to D3 causes D3 to retrieve its dislodged helix, restoring the catalytic activity of D14 [68]. D14 hydrolyzes the SL molecule, obtaining a covalent butenolide modification at the active site histidine (His) [12,38,62]. (D) During or after binding of the D3-D53 complex, the transcriptional repressor D53 is polyubiquitinated [53,54]. (E) D53 is degraded, initiating the SL gene response [53,54]. (F) D14 is then degraded [66,67] and D3 recycled.
Concluding Remarks and Outlook
Significant success has been achieved over the past few years in both the discovery of new SL molecules and the elucidation of more details in the signaling pathway, as recently highlighted in several excellent reviews [21,76–78]. The molecular basis of strigol and orobanchol-type SL discrimination in Striga and Orobanche species has not yet been investigated and it would be exciting to see how SL receptors in Orobanche are different from those already discovered. It will also be interesting to see how future studies on S. asiatica fit into this picture, given that its genome contains a high number of 17 potential SL receptors [79]. The role of noncanonical SLs is not fully understood. Although only less than ten noncanonical SLs have been found so far, there are likely many more and a complete picture of how noncanonical SLs are part of an SL communication key for specific symbiotic interaction and an evasive strategy against detection by parasitic plants is still missing (see Outstanding Questions).
Outstanding Questions.
At what point during signaling does SL hydrolysis occur?
How stable is the histidine-butenolide complex in vivo and how fast does polyubiquitination of D53 take place after it is formed?
Does D14 get recycled, degraded, or both?
What is the exact contribution of the active site residues to the architecture of the substrate-binding site in D14 and to SL binding?
Are the SL receptors in symbiotic fungi structurally similar to the SL receptors in plants?
How does the number of SL molecules reflect the diversity of the soil microbiome and the need to evade parasitic plants?
Can SLs be used in agriculture to remodel symbiotic relationships between plants and soil microbes to promote plant growth?
It has been established that Arabidopsis D14 is not a single-turnover enzyme [65]. The same has been reported for rice D14 [68]. Future research will hopefully clarify whether the Pisum homolog RMS3 is indeed a single-turnover enzyme [12] or if certain fluorescent SL derivatives cause product inhibition. Recent findings from two different groups [65,68] suggest that the SL molecule stays intact during the initial complex formation between D14, D3/MAX2, and D53. However, the lack of an intact SL molecule in the crystal structure of the AtD14-D3-ASK1 complex [62] and the repeated documentation of the histidine-butenolide intermediate [12,38,62] suggest that SL hydrolysis happens at some point before the complex dissociates. Although the CLIM molecule [62] is a logical link between the D-ring associated with the active site serine and histidine individually, alternative interpretations of the crystallographic data are available [15], including in this review (Figure 3H). The understanding of D14 catalytic triad mutants is not complete, a gap that will hopefully be filled with more crystal structures that complement the already existing DAD2 S96A structure [10] and clarify how other single mutations affect SL binding or destabilize D14. Finally, the interface between rice D14 and the dislodged D3 helix and the resulting distortion of the D14 substrate-binding pocket result from a crystal structure, in which the F-box helix was fused to D14 [68]. This arrangement might limit the number of possible interfaces and compete with crystallization contacts. A crystal structure of rice D14 in complex with full-length D3 would hopefully be able to verify how the interfaces compare with the AtD14-D3 interaction.
Structural biology might be both a source of confusion and the solution for the SL field. Despite the coexistence of several models and controversy, the contribution of X-ray crystallography to SL signaling models has been enormous and is likely to provide answers to many of the still outstanding questions.
Highlights.
SLs are a class of plant hormones that are involved in agriculturally important processes, such as shoot branching, arbuscular mycorrhizal symbiosis, and germination of parasitic plants.
An increasing number of different SLs have been identified.
Structural biology has been a treasure trove for SL research, but has also been a source of confusion.
Recent studies show that different species have independently developed similar features to regulate the architecture of their SL receptors.
New findings suggest that the intact SL molecule is part of a catalytically arrested D14-D3/MAX2 interaction but that a histidine-butenolide complex stabilizes the complex with the transcriptional repressor.
Acknowledgments
Our studies of plant hormones have been supported by NIH grants R01 GM52413 and R01 GM094428. We are currently supported by NIH grant R35 GM122604. Apologies are offered to those colleagues whose work was not cited due to space constrains. J.C. is an investigator of the Howard Hughes Medical Institute.
Glossary
- α/β hydrolases
a protein superfamily of enzymes with a typical α/β sheet architecture that contains eight β-strands and six α-helices. The superfamily includes lipases, esterases, proteases, and other hydrolases
- Arbuscular mycorrhizal (AM) symbiosis
a symbiotic relationship between plants and fungi of the phylum Glomeromycota
- Butenolide
a lactone with a four-carbon heterocyclic ring. The D-ring of an SL molecule is a butenolide
- Catalytic triad
a group of three amino acids in the active sites of many enzymes. The triad usually comprises a nucleophile, base, and acid. In D14, these roles are taken by the active site serine, histidine, and aspartate, respectively
- Covalently linked intermediate molecule (CLIM)
a proposed SL hydrolysis intermediate simultaneously linked to the serine and histidine residues of the active site of the D14 protein, triggering or stabilizing the interaction between D14 and the F-box protein D3
- Dual-functional receptor/hydrolase
a protein that is both a receptor and an enzyme. The protein binds and ultimately destroys its ligand by hydrolyzing it.
- F-box protein
a component of the multiprotein E3 ubiquitin ligase complex that carries out polyubiquitination of proteins, targeting them for degradation
- GR24
a synthetic SL analog that is used in many studies because its production is cheaper than that of actual SL molecules
- Karrikin
a chemical found in the smoke of burning plant material, triggering the germination of dormant seeds in the soil after a wildfire. Karrikins are perceived by the protein KAI2, a close homolog of D14
- Noncanonical SLs
SLs that do not have a complete tricyclic ABC part
- Non-natural SLs
SLs with the ‘wrong’ (2’S) stereochemical configuration between the C and the D-ring
References
- 1.Cook CE. et al. (1966) Germination of witchweed (Striga lutea Lour.): isolation and properties of a potent stimulant. Science 154, 1189–1190 [DOI] [PubMed] [Google Scholar]
- 2.Akiyama K. et al. (2005) Plant sesquiterpenes induce hyphal branching in arbuscular mycorrhizal fungi. Nature 435, 824–827 [DOI] [PubMed] [Google Scholar]
- 3.Remy W. et al. (1994) Four hundred-million-year-old vesicular arbuscular mycorrhizae. Proc. Natl. Acad. Sci. U. S. A 91, 11841–11843 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Proust H. et al. (2011) Strigolactones regulate protonema branching and act as a quorum sensing-like signal in the moss Physcomitrella patens. Development 138, 1531–1539 [DOI] [PubMed] [Google Scholar]
- 5.Gomez-Roldan V. et al. (2008) Strigolactone inhibition of shoot branching. Nature 455, 189–194 [DOI] [PubMed] [Google Scholar]
- 6.Umehara M. et al. (2008) Inhibition of shoot branching by new terpenoid plant hormones. Nature 455, 195–200 [DOI] [PubMed] [Google Scholar]
- 7.Xing Y and Zhang Q. (2010) Genetic and molecular bases of rice yield. Annu. Rev. Plant Biol 61, 421–442 [DOI] [PubMed] [Google Scholar]
- 8.Westwood JH. et al. (2010) The evolution of parasitism in plants. Trends Plant Sci. 15, 227–235 [DOI] [PubMed] [Google Scholar]
- 9.Berner D. et al. (1995) Striga research and control. A perspective from Africa. Plant Dis. 79, 652–660 [Google Scholar]
- 10.Hamiaux C. et al. (2012) DAD2 is an alpha/beta hydrolase likely to be involved in the perception of the plant branching hormone, strigolactone. Curr. Biol 22, 2032–2036 [DOI] [PubMed] [Google Scholar]
- 11.Zhao LH. et al. (2013) Crystal structures of two phytohormone signal-transducing alpha/beta hydrolases: karrikin-signaling KAI2 and strigolactone-signaling DWARF14. Cell Res. 23, 436–439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.de Saint Germain A. et al. (2016) An histidine covalent receptor and butenolide complex mediates strigolactone perception. Nat. Chem. Biol 12, 787–794 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Janssen BJ and Snowden KC. (2012) Strigolactone and karrikin signal perception: receptors, enzymes, or both? Front. Plant Sci 3, 296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Snowden KC and Janssen BJ. (2016) Structural biology: signal locked in. Nature 536, 402–404 [DOI] [PubMed] [Google Scholar]
- 15.Carlsson GH. et al. (2018) The elusive ligand complexes of the DWARF14 strigolactone receptor. J. Exp. Bot 69, 2345–2354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Alder A. et al. (2012) The path from beta-carotene to carlactone, a strigolactone-like plant hormone. Science 335, 1348–1351 [DOI] [PubMed] [Google Scholar]
- 17.Seto Y. et al. (2014) Carlactone is an endogenous biosynthetic precursor for strigolactones. Proc. Natl. Acad. Sci. U. S. A 111, 1640–1645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Scaffidi A. et al. (2014) Strigolactone hormones and their stereoisomers signal through two related receptor proteins to induce different physiological responses in Arabidopsis. Plant Physiol. 165, 1221–1232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Flematti GR. et al. (2016) Stereospecificity in strigolactone biosynthesis and perception. Planta 243, 1361–1373 [DOI] [PubMed] [Google Scholar]
- 20.Xie X. et al. (2013) Confirming stereochemical structures of strigolactones produced by rice and tobacco. Mol. Plant 6, 153–163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yoneyama K. et al. (2018) Which are the major players, canonical or non-canonical strigolactones? J. Exp. Bot 69, 2231–2239 [DOI] [PubMed] [Google Scholar]
- 22.Cook CE. et al. (1972) Germination stimulants. II. Structure of strigol, a potent seed germination stimulant for witchweed (Striga lutea). J. Am. Chem. Soc 94, 6198–6199 [Google Scholar]
- 23.Brooks DW. et al. (1985) The absolute structure of (+)-strigol. J. Org. Chem 50, 3779–3781 [Google Scholar]
- 24.Yokota T. et al. (1998) Alectrol and orobanchol, germination stimulants for Orobanche minor, from its host red clover. J. Phytochem 49, 1967–1973 [Google Scholar]
- 25.Mori K. et al. (1999) Structure and synthesis of orobanchol, the germination stimulant for Orobanche minor. Tetrahedron Lett. 40, 943–946 [Google Scholar]
- 26.Ueno K. et al. (2011) Ent-2’-epi-Orobanchol and its acetate, as germination stimulants for Striga gesnerioides seeds isolated from cowpea and red clover. J. Agric. Food Chem 59, 10485–10490 [DOI] [PubMed] [Google Scholar]
- 27.Xie X. et al. (2008) Isolation and identification of alectrol as (+)orobanchyl acetate, a germination stimulant for root parasitic plants. Phytochemistry 69, 427–431 [DOI] [PubMed] [Google Scholar]
- 28.Zwanenburg B and Pospisil T. (2013) Structure and activity of strigolactones: new plant hormones with a rich future. Mol. Plant 6, 38–62 [DOI] [PubMed] [Google Scholar]
- 29.Hauck C and Schildknecht H. (1990) Separation of enantiomers of the germination stimulant strigol on cellulose triacetate and determination of their biological activity. J. Plant Physiol 136, 126–128 [Google Scholar]
- 30.Nakamura H. et al. (2013) Molecular mechanism of strigolactone perception by DWARF14. Nat. Commun 4, 2613. [DOI] [PubMed] [Google Scholar]
- 31.Waters MT. et al. (2015) A Selaginella moellendorffii ortholog of KARRIKIN INSENSITIVE2 functions in Arabidopsis development but cannot mediate responses to karrikins or strigolactones. Plant Cell 27, 1925–1944 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Flematti GR. et al. (2004) A compound from smoke that promotes seed germination. Science 305, 977. [DOI] [PubMed] [Google Scholar]
- 33.Guo Y. et al. (2013) Smoke-derived karrikin perception by the alpha/beta-hydrolase KAI2 from Arabidopsis. Proc. Natl. Acad. Sci. U. S. A 110, 8284–8289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Conn CE and Nelson DC. (2015) Evidence that KARRIKININSENSITIVE2 (KAI2) receptors may perceive an unknown signal that is not karrikin or strigolactone. Front. Plant Sci 6, 1219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Flematti GR. et al. (2013) Karrikin and cyanohydrin smoke signals provide clues to new endogenous plant signaling compounds. Mol. Plant 6, 29–37 [DOI] [PubMed] [Google Scholar]
- 36.Morffy N. et al. (2016) Smoke and hormone mirrors: action and evolution of karrikin and strigolactone signaling. Trends Genet. 32, 176–188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.de Saint Germain A. et al. (2019) Contalactone, a contaminant formed during chemical synthesis of the strigolactone reference GR24 is also a strigolactone mimic. Phytochemistry 168, 112112 [DOI] [PubMed] [Google Scholar]
- 38.Bürger M. et al. (2019) Structural basis of karrikin and non-natural strigolactone perception in Physcomitrella patens. Cell Rep. 26, 855–865 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Decker EL. et al. (2017) Strigolactone biosynthesis is evolutionarily conserved, regulated by phosphate starvation and contributes to resistance against phytopathogenic fungi in a moss, Physcomitrella patens. New Phytol. 216, 455–468 [DOI] [PubMed] [Google Scholar]
- 40.Charnikhova TV. et al. (2017) Zealactones. Novel natural strigolactones from maize. Phytochemistry 137, 123–131 [DOI] [PubMed] [Google Scholar]
- 41.Xie X. et al. (2017) Methyl zealactonoate, a novel germination stimulant for root parasitic weeds produced by maize. J. Pestic. Sci 42, 58–61 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ueno K. et al. (2014) Heliolactone, a non-sesquiterpene lactone germination stimulant for root parasitic weeds from sunflower. Phytochemistry 108, 122–128 [DOI] [PubMed] [Google Scholar]
- 43.Woo S and McErlean CSP (2019) Total synthesis and stereochemical confirmation of heliolactone. Org. Lett 21, 4215–4218 [DOI] [PubMed] [Google Scholar]
- 44.Yoshimura M. et al. (2019) Total synthesis and biological evaluation of heliolactone. Helv. Chim. Acta 102, e1900211 [Google Scholar]
- 45.Kim HI. et al. (2014) Avenaol, a germination stimulant for root parasitic plants from Avena strigosa. Phytochemistry 103, 85–88 [DOI] [PubMed] [Google Scholar]
- 46.Yasui M. et al. (2017) Total synthesis of avenaol. Nat. Commun 8, 674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Charnikhova TV. et al. (2018) Zeapyranolactone − a novel strigolactone from maize. Phytochem. Lett 24, 172–178 [Google Scholar]
- 48.Xie X. et al. (2018) Lotuslactone, a non-canonical strigolactone from Lotus japonicus. Phytochemistry 157, 200–205 [DOI] [PubMed] [Google Scholar]
- 49.Kim HI. et al. (2010) Structure–activity relationship of naturally occurring strigolactones in Orobanche minor seed germination stimulation. J. Pestic. Sci 35, 344–347 [Google Scholar]
- 50.Kisugi T. et al. (2013) Strigone, isolation and identification as a natural strigolactone from Houttuynia cordata. Phytochemistry 87, 60–64 [DOI] [PubMed] [Google Scholar]
- 51.Arite T. et al. (2009) d14, a strigolactone-insensitive mutant of rice, shows an accelerated outgrowth of tillers. Plant Cell Physiol. 50, 1416–1424 [DOI] [PubMed] [Google Scholar]
- 52.Waters MT. et al. (2012) Specialisation within the DWARF14 protein family confers distinct responses to karrikins and strigolactones in Arabidopsis. Development 139, 1285–1295 [DOI] [PubMed] [Google Scholar]
- 53.Zhou F. et al. (2013) D14-SCF(D3)-dependent degradation of D53 regulates strigolactone signalling. Nature 504, 406–410 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Jiang L. et al. (2013) DWARF 53 acts as a repressor of strigolactone signalling in rice. Nature 504, 401–405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Soundappan I. et al. (2015) SMAX1-LIKE/D53 family members enable distinct MAX2-dependent responses to strigolactones and karrikins in Arabidopsis. Plant Cell 27, 3143–3159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wang L. et al. (2015) Strigolactone signaling in Arabidopsis regulates shoot development by targeting D53-like SMXL repressor proteins for ubiquitination and degradation. Plant Cell 27, 3128–3142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ollis DL. et al. (1992) The alpha/beta hydrolase fold. Protein Eng. 5, 197–211 [DOI] [PubMed] [Google Scholar]
- 58.Mindrebo JT. et al. (2016) Unveiling the functional diversity of the alpha/beta hydrolase superfamily in the plant kingdom. Curr. Opin. Struct. Biol 41, 233–246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kagiyama M. et al. (2013) Structures of D14 and D14L in the strigolactone and karrikin signaling pathways. Genes Cells 18, 147–160 [DOI] [PubMed] [Google Scholar]
- 60.Mangnus EM. et al. (1992) Improved synthesis of strigol analog GR24 and evaluation of the biological activity of its diastereomers. J. Agric. Food Chem 40, 1230–1235 [Google Scholar]
- 61.Zhao LH. et al. (2015) Destabilization of strigolactone receptor DWARF14 by binding of ligand and E3-ligase signaling effector DWARF3. Cell Res. 25, 1219–1236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Yao R. et al. (2016) DWARF14 is a non-canonical hormone receptor for strigolactone. Nature 536, 469–473 [DOI] [PubMed] [Google Scholar]
- 63.Richardson JS and Richardson DC. (1988) Amino acid preferences for specific locations at the ends of alpha helices. Science 240, 1648–1652 [DOI] [PubMed] [Google Scholar]
- 64.Aurora R. et al. (1994) Rules for alpha-helix termination by glycine. Science 264, 1126–1130 [DOI] [PubMed] [Google Scholar]
- 65.Seto Y. et al. (2019) Strigolactone perception and deactivation by a hydrolase receptor DWARF14. Nat. Commun 10, 191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Chevalier F. et al. (2014) Strigolactone promotes degradation of DWARF14, an alpha/beta hydrolase essential for strigolactone signaling in Arabidopsis. Plant Cell 26, 1134–1150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Hu Q. et al. (2017) DWARF14, a receptor covalently linked with the active form of strigolactones, undergoes strigolactone-dependent degradation in rice. Front. Plant Sci 8, 1935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Shabek N. et al. (2018) Structural plasticity of D3-D14 ubiquitin ligase in strigolactone signalling. Nature 563, 652–656 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Xu Y. et al. (2016) Structural basis of unique ligand specificity of KAI2–like protein from parasitic weed Striga hermonthica. Sci. Rep 6, 31386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Xu Y. et al. (2018) Structural analysis of HTL and D14 proteins reveals the basis for ligand selectivity in Striga. Nat. Commun 9, 3947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Toh S. et al. (2015) Structure-function analysis identifies highly sensitive strigolactone receptors in Striga. Science 350, 203–207 [DOI] [PubMed] [Google Scholar]
- 72.Nomura S. et al. (2013) Structural requirements of strigolactones for germination induction and inhibition of Striga gesnerioides seeds. Plant Cell Rep. 32, 829–838 [DOI] [PubMed] [Google Scholar]
- 73.Cardoso C. et al. (2014) Differential activity of Striga hermonthica seed germination stimulants and Gigaspora rosea hyphal branching factors in rice and their contribution to underground communication. PLoS ONE 9, e104201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Brotzel F and Mayr H. (2007) Nucleophilicities of amino acids and peptides. Org. Biomol. Chem 5, 3814–3820 [DOI] [PubMed] [Google Scholar]
- 75.Bischoff R and Schluter H. (2012) Amino acids: chemistry, functionality and selected non-enzymatic post-translational modifications. J. Proteome 75, 2275–2296 [DOI] [PubMed] [Google Scholar]
- 76.Lumba S. et al. (2017) The perception of strigolactones in vascular plants. Nat. Chem. Biol 13, 599–606 [DOI] [PubMed] [Google Scholar]
- 77.Machin DC. et al. (2019) Fellowship of the rings: a saga of strigolactones and other small signals. New Phytol. Published online August 23, 2019. 10.1111/nph.16135 [DOI] [PubMed] [Google Scholar]
- 78.Waters MT. et al. (2017) Strigolactone signaling and evolution. Annu. Rev. Plant Biol 68, 291–322 [DOI] [PubMed] [Google Scholar]
- 79.Yoshida S. et al. (2019) Genome sequence of Striga asiatica provides insight into the evolution of plant parasitism. Curr. Biol 29, 3041–3052 e4 [DOI] [PubMed] [Google Scholar]
- 80.Emsley P. et al. (2010) Features and development of Coot. Acta Crystallogr. D Biol. Crystallogr 66, 486–501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Adams PD. et al. (2010) PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr 66, 213–221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.McNicholas S. et al. (2011) Presenting your structures: the CCP4mg molecular-graphics software. Acta Crystallogr. D Biol.Crystallogr 67, 386–394 [DOI] [PMC free article] [PubMed] [Google Scholar]