Abstract
Objectives:
To establish the efficacy of pairing readers randomly and evaluate the merits of developing optimal pairing methodologies.
Materials and Methods:
Sensitivity, specificity, and proportion correct were computed for three different case-sets that were independently read by 16 radiologists. Performance of radiologists as single readers was compared with expected double reading performance. We theoretically evaluated all possible pairing methodologies. Bootstrap resampling methods were used for statistical analyses.
Results:
Significant improvements in expected performance for double vs. single reading (i.e., delta performance) were shown for all performance measures and case-sets (p ≤ .003), with overall delta performance across all theoretically possible pairing schemes (n=10,395) ranging between .05 and .08. Delta performance for the 20 best pairing schemes was significant (p < .001) and ranged between .07 and .10. Delta performance for 20 random pairing schemes was also significant (p ≤ .003) and ranged between .05 and .08. Delta performance for the 20 worst pairing schemes ranged between .03 and .06, reaching significance in delta proportion correct (p ≤ .021) for all three case-sets and in delta specificity for two case-sets (p < .033) but not for a third case-set (p = .131), and not reaching significance in delta sensitivity for any of the three case-sets (.098 ≥ p ≥ .067).
Conclusion:
Significant benefits accrue from double reading, and whilst random reader pairing achieves most double reading benefits, a strategic pairing approach may maximise the benefits of double reading.
Keywords: Radiologists, Digital Mammography, Observer Variation, Double Reading, Breast Cancer
Introduction
Independent double reading in mammography, which involves two radiologists reading the same mammogram, relies on the principle that if a cancer is missed by one radiologist there is a chance that it will be picked up by a second radiologist, thus leading to an improved cancer detection rate. It can also potentially reduce recall rates by having an arbitrator consider mammograms where one radiologist thinks a cancer may be present, whilst another records the findings as normal or benign. Previous studies reported that compared to single reading, double reading can reduce recall rates by about 25 to 32% (1, 2), with an increase in sensitivity of about 10 to 15% and an insignificant decrease in specificity of about 0.1–1.8% (3, 4). Double reading has also been shown to outperform single reading with computer-aided detection (CAD) (5–7). In Australia and New Zealand, independent double reading is the standard practice in breast cancer-screening programmes (8), and in circumstances when discordant findings between paired readers occur, concordance is achieved by consensus discussion or by third-reader arbitration process, though there can be circumstances where greatly increasing sensitivity is very important, in which case an “or” rule should be used, or where greatly increasing specificity is very important, in which case an “and” rule should be used (9). Despite the diagnostic accuracy improvement achieved through independent double reading, single reading is still commonly practised in a number of countries such as the United States of America.
In the couple of decades since the introduction of double reading in regions such as Australia, New Zealand and Europe (6), there have been no attempts to optimise the pairing of readers. Several studies have focussed on the efficacy of independent double reading compared to single reading and CAD by assessing recall rates, sensitivity, and cost-effectiveness (10–12), and two studies have examined whether changing the order of reading, where the second reader reviews a batch of images in a reverse order to that of the first, would improve cancer detection accuracy with double reading (13, 14). Countries employing double reading generally pair readers randomly or out of convenience, and there are no data on efforts to look at the feasibility and impact of optimally pairing readers when compared with single reading, nor has it been evaluated if non-strategic random pairing limits the benefits of double compared to single reading.
It is an open question whether methodologies could ever be developed that will enable clinics to determine the best possible pairing scheme for the readers at hand. It is important to note that, of course, the best pair could routinely outperform all other pairs. However, it is implausible that the best pair will handle all cases. Instead, we are interested in improving the entire double reading system, and so the question remains: how should readers be paired to maximize performance across all pairs?
In this study, we exhaustively explored all possible ways of pairing a large group of readers and evaluated the average double reading performance for each pairing scheme. We also assessed whether non-strategic random pairing captures most double reading benefits that could be obtained by an optimal pairing scheme (notwithstanding the current lack-of-knowledge in how to prospectively determine the best possible pairing scheme). Our results should be of value to countries involved or planning to be involved in breast cancer screening programs.
Materials and Methods
Data were collected through the Breast Screen Reader Assessment Strategy (BREAST) (http://sydney.edu.au/health-sciences/breastaustralia/). Institutional ethics approval was obtained from the human research ethics committees at the University of Sydney and the University of California, Santa Barbara. Informed consent was obtained from all participants prior to their readings to use their de-identified responses for the purpose of research.
Image case sets
Three case-sets offered by the BREAST platform were used, here referred to as Set 1, 2 and 3. The cases were selected by an expert breast radiologist overseeing training, quality control and clinical policies of a state’s breast screening program. Each set contained 60 digitally-acquired mammographic cases, with 40 normal and 20 cancer cases of different presentations (masses, calcifications, asymmetries, architectural distortions), each consisting of cranial-caudal and medio-lateral oblique projections of left and right breasts. The 40 normal cases were a mixture of “pristine” normal cases and cases with benign findings. Normality was confirmed by two independent screen readings, a negative follow-up screening mammogram after 2 years, and a review by two expert radiologists. The 20 cancer cases were biopsy-proven. All cases had prior images obtained 2 years apart from the current image, and all images were anonymized. With regard to mammographic density, the cases were chosen to reflect a normal screening caseload.
Participants
Data were collected from 16 board certified radiologists who completed all three case sets. The self-reported demographic details of radiologists at the time of each test are summarised in Table 1.
Table 1:
Demographics of the 16 radiologists at the time of each test
| Parameter | Case set | First Quartile |
Mean | Third Quartile |
Interquartile Range |
|---|---|---|---|---|---|
| Age (years) | Set 1 | 36.0 | 45.5 | 57.75 | 21.75 |
| Set 2 | 38.25 | 47.5 | 54.75 | 16.5 | |
| Set 3 | 36.75 | 47.0 | 57.75 | 21.0 | |
| No of years since qualification as radiologist | Set 1 | 6.25 | 11.5 | 20 | 13.75 |
| Set 2 | 3.25 | 11.5 | 20 | 16.75 | |
| Set 3 | 6.25 | 13.5 | 20 | 13.75 | |
| No of years reading mammograms | Set 1 | 3.25 | 7.0 | 17.5 | 14.25 |
| Set 2 | 1.75 | 5.0 | 10.75 | 9.0 | |
| Set 3 | 3.25 | 8.0 | 17.5 | 14.25 | |
| Annual Volume | Set 1 | 1620 | 3600 | 6300 | 4680 |
| Set 2 | 1260 | 1980 | 2700 | 1440 | |
| Set 3 | 1620 | 3600 | 6300 | 4680 | |
| No of hours per week | Set 1 | 7.5 | 10.25 | 16.75 | 9.25 |
| Set 2 | 4.0 | 13.0 | 18.0 | 14.0 | |
| Set 3 | 7.5 | 13.0 | 18.37 | 10.88 | |
Reading Environment
Readings were conducted in rooms to match the radiologic reporting environment. Ambient lighting ranged from 12 to 20 lux at the position of the reader, and the walls were painted a light matte colour with minimum specular reflection. The images were displayed in random sequence on two 5-megapixel reporting monitors (MFGD 5621; Barco, Kortrijk, Belgium and Radiforce G51; Eizo, Ishikawa, Japan) linked to their own specific video card (BarcoMed 5MP2FH; Barco and Matrox MED5MP-DVI; Dorval, Quebec, Canada respectively). Monitors were calibrated to the Digital Imaging and Communication in Medicine Gray-Scale Standard Display Function (DICOM GSDF) and displayed maximum luminance, minimum luminance and contrast ratio within 5% of 475 cd/m2, 1.3 cd/m2, and 365:1 respectively. Readers had unlimited time to complete a case set and had access to a range of post-processing tools including zooming, panning and windowing. Figure 1: Experimental setup and reading environment.
Figure 1:
Experimental setup and reading environment.
Reading process
Radiologists marked the location of perceived lesions and reported their confidence using the RANZCR (Royal Australian and New Zealand College of Radiologists) rating system as follows: (2) Benign, (3) Equivocal, (4) Suspicious, and (5) Malignant. A rating of (1) indicated no significant abnormality, and the next case was displayed. A demonstration of the software was given before commencing the test. Radiologists were allowed to mark as many lesions as they observed for all cases. Radiologists had no prior knowledge about the nature of abnormalities nor the proportion of abnormal cases in each set.
Data Analysis
Performance for each case-set based on sensitivity, specificity, and proportion correct was computed for individual readers and pairs of readers, and the single reading performance was then compared to the average performance of readers when grouped in pairs. Each normal case was considered correctly identified (true negative; TN=1) if the reader’s highest rating for the case was 1 or 2, whereas each cancer case was considered correctly identified (true positive; TP=1) if the reader’s highest rating for the case was 3, 4, or 5.
Sensitivity was defined as the proportion of cancer cases that were correctly identified: ΣTP / number of cancer cases. Specificity was defined as the proportion of normal cases that were correctly identified: ΣTN / number of normal cases. Proportion correct was defined as the overall proportion of cases that were correctly identified: (ΣTP + ΣTN) / total number of cases.
An important component of this study was the exhaustive exploration of all possible ways that a cohort of readers can be paired. A cohort of readers consisted of 12 of the 16 radiologists, with the 4 remaining radiologists used for arbitration. We explored all possible cohorts of 12 readers, and all possible pairing schemes within each cohort.
Given 16 radiologists, how many cohorts of 12 readers can we form? Using combinatorics, there are n choose distinct ways of creating a cohort of 12 readers (with the 4 remaining radiologists used for arbitration). Given a cohort of 12 readers, how many pairing schemes can we form? For any ‘k’ even number of readers, there are 1×3×5.,.x(k-1) distinct ways of dividing k readers into k/2 non-overlapping pairs of 2. Hence, for a cohort of size k=12, there are 1×3×5×7×9×11 = 10,395 distinct pairing schemes, where each pairing scheme consists of 6 non-overlapping pairs of 2.
For each case that a pair was discordant (because one reader’s highest rating was 1 or 2, whereas the other reader’s highest rating was 3, 4, or 5), we arbitrated the case using each of the 4 radiologists who were not in the current cohort and recorded the average performance for the case across all 4 arbitrators as follows: If all 4 arbitrators were correct, the decision outcome for the discordant pair was recorded as TP=1 if it was a cancer case or TN=1 if it was a normal case. If 3 of the 4 arbitrators were correct, the decision outcome for the discordant pair was recorded as TP=0.75 if it was a cancer case or TN=0.75 if it was a normal case. If 2 of the 4 arbitrators were correct, the decision outcome for the discordant pair was recorded as TP=0.5 if it was a cancer case or TN=0.5 if it was a normal case. If 1 of the 4 arbitrators was correct, the decision outcome for the discordant pair was recorded as TP=0.25 if it was a cancer case or TN=0.25 if it was a normal case. If none of the 4 arbitrators were correct, the decision outcome for the discordant pair was recorded as TP=0 if it was a cancer case or TN=0 if it was a normal case.
These TP and TN values for cases that the pair was discordant were used in the respective summations to compute the pair’s expected sensitivity, specificity, and proportion correct across all cases as defined above. This scoring technique using all 4 arbitrators for each case and assigning fractional decision outcomes, instead of randomly selecting a third reader every time that a pair was discordant, allowed us to implement a non-stochastic exploration of all possible pairing schemes and identify the best vs. worst pairing schemes from each cohort.
We computed delta performance measures for each pairing scheme by subtracting the cohort’s average single reader performance from the expected performance of the average pair in the scheme. Bootstrap resampling methods (15) for group decision analyses (16) were used to estimate uncertainties in the obtained performance measures due to reader and case variability in the original data, and to assess whether the obtained delta performance measures were statistically significant. Bootstrap resampling was used because it fits almost any statistical analysis employing random sampling techniques, and it is a more valid alternative to statistical inference for hypothesis testing, where parametric inference is unfeasible (17).
In bootstrap resampling, each bootstrap run uses the same amount of data as the original analyses (i.e., 16 bootstrap readers and 60 bootstrap cases per case-set), but parts of the original data are absent from the bootstrap run whereas other parts of the original data are overrepresented in the bootstrap run. This is accomplished for each bootstrap run by sampling readers and cases from the original with replacement. Thus, for each of 1,000 bootstrap runs, we repeated all original analyses for each case set using a bootstrap set of 16 random readers and a bootstrap set of 60 random cases consisting of 20 random cancer cases and 40 random normal cases. Using the bootstrap readers and cases for each run, we explored all 1,820 possible bootstrap cohorts and computed the delta performances of all 10,395 possible pairing schemes within each bootstrap cohort. These bootstrap runs were used to obtain p values and 95% confidence intervals for each reported performance measure. All p values < .05 were statistically significant after controlling for multiple comparisons (i.e., 12 comparisons per case set) using the false discovery rate procedure (18).
Results
Results obtained using all 10,395 pairing schemes from each cohort
Figure 2 shows the proportion of pairing schemes across all cohorts that the average pair outperforms the average single reader in the cohort for each measure and case set. The figure shows that the proportion of pairing schemes that outperform single readers is close to 1 for all measures and case sets, except for specificity in Set 1 where the proportion of pairing schemes across all cohorts that the average pair outperforms the average single reader in the cohort is .98.
Figure 2:
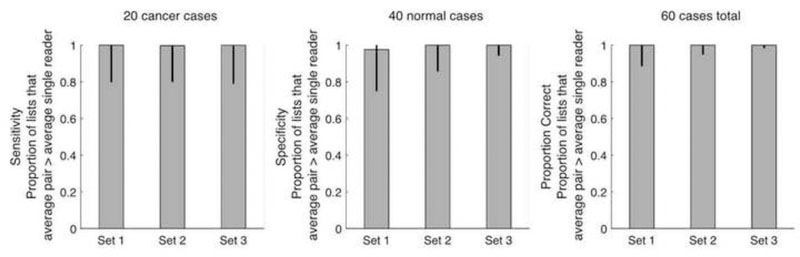
Proportion of pairing schemes across all cohorts that the average pair outperforms the average single reader in the cohort for each measure and case set. Error bars mark 95% bootstrap confidence intervals.
Figure 3 shows the average expected performance for each measure and case set across all 10,395 pairing schemes from each cohort (grey bars) and across the average single reader from each cohort (yellow bars). The raw performance values shown in Figure 3 were used to construct the overall delta performance values shown in Figure 4. All bootstrap resampling p values shown in Figure 4 are < .05, which means that all overall delta performance measures in all three case sets are statistically significantly greater than zero.
Figure 3:
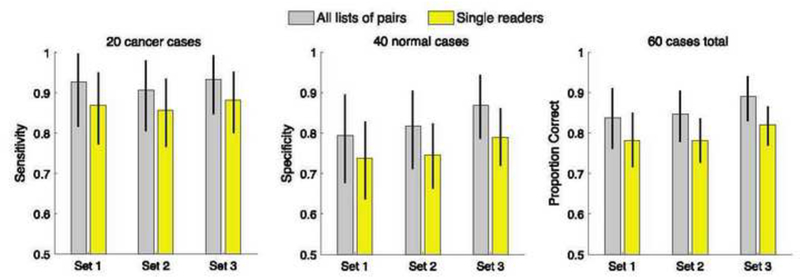
Average expected performance for each measure and case set across all 10,395 pairing schemes from each cohort and across the average single reader from each cohort. Error bars mark 95% bootstrap confidence intervals.
Figure 4:
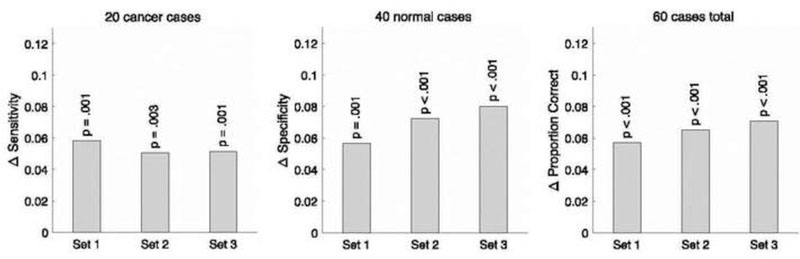
Average difference in expected performance (i.e., Δ performance) for each measure and case set between the average pair in the pairing scheme and the average single reader in the cohort across all 10,395 pairing schemes from each cohort. Statistically significant p values (p < .05) using bootstrap resampling methods were obtained for Δ sensitivity, Δ specificity, and Δ proportion correct in all three case sets.
Results obtained by subselecting the 20 best, 20 random and 20 worst pairing schemes from each cohort.
The overall results described above were obtained using all 10,395 pairing schemes from each cohort. To follow, we break down the results by sub-selecting the 20 best, 20 random, and 20 worst pairing schemes from each cohort to see the possible range of double-reading benefits. Figure 5 shows the average expected performance for each measure and case set across the 20 best pairing schemes from each cohort (blue bars), across 20 random pairing schemes from each cohort (green bars), across the 20 worst pairing schemes from each cohort (red bars), and across the average single reader from each cohort (yellow bars).
Figure 5:
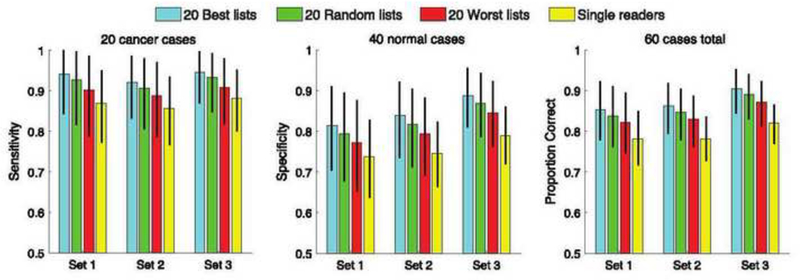
Average expected performance for each measure and case set across the 20 best pairing schemes from each cohort, across 20 random pairing schemes from each cohort, across the 20 worst pairing schemes from each cohort, and across the average single reader from each cohort. Error bars mark 95% bootstrap confidence intervals.
The raw performance values shown in Figure 5 were used to construct the delta performance values shown in Figure 6. The bootstrap resampling p values shown in Figure 6 are < .05 across the 20 best pairing schemes from each cohort and across 20 random pairing schemes from each cohort, which means that the delta performance measures in all three case sets are statistically significantly greater than zero, not only across the 20 best pairing schemes from each cohort, but also across 20 random pairing schemes from each cohort. On the other hand, while the delta performance values in all three case sets are all positive across the 20 worst pairing schemes from each cohort, not all bootstrap resampling p values are statistically significant.
Figure 6:
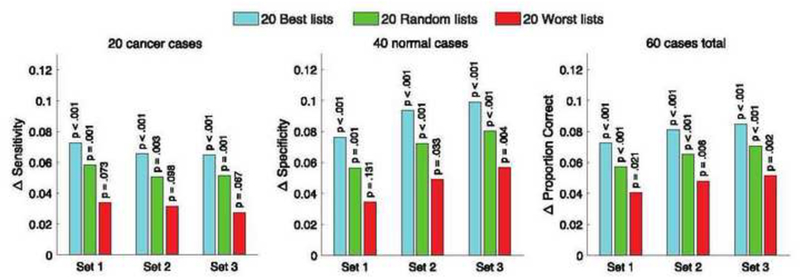
Average difference in expected performance (i.e., Δ performance) for each measure and case set between the average pair in the pairing scheme and the average single reader in the cohort across the 20 best pairing schemes from each cohort, across 20 random pairing schemes from each cohort, and across the 20 worst pairing schemes from each cohort. Statistically significant p values (p < .05) using bootstrap resampling methods were obtained for Δ sensitivity, Δ specificity, and Δ proportion correct in all three case sets across the 20 best pairing schemes in each cohort and across 20 random pairing schemes in each cohort; as opposed to across the 20 worst pairing schemes in each cohort where some p values were not statistically significant.
Discussion
Inherent human flaws have been identified as a major contributor of suboptimal cancer detection performance in mammography (6). One intervention introduced to improve diagnostic performance is independent double reading, which facilitates the interpretation of images by two individuals with different perceptual, cognitive, and decision-making expertise, allowing for arbitration when there is disagreement. However, most screening settings pair readers randomly or out of convenience with little thought given to how reader pairing would affect diagnostic accuracy.
In this study, we exhaustively evaluated every possible way of dividing a group of readers into distinct pairs and measured the overall impact of double reading for each theoretically possible pairing scheme. The results showed that whichever performance metric was used (sensitivity, specificity, or proportion correct) and whichever case set was tested (all radiologists completed three different case sets), statistically significant improvements in diagnostic accuracy were shown with paired compared to single readings for almost all pairing schemes. These findings suggest that double reading is an important mechanism to reduce errors in mammography interpretation and improve early detection of breast cancer.
Due to the paucity of research on optimising the pairing of readers, further analyses were performed to explore double reading performance when radiologists are paired in best, random and worst possible ways. From our results, all delta performance measures were statistically significantly greater than zero in all case sets, not only when readers were paired in best possible ways, but also when readers were paired randomly. These results highlight that even with a simple random pairing approach, important clinical benefits accrue with respective improvements in sensitivity and specificity of about 5% and 7% compared to single readers.
Interestingly, the delta proportion correct between the worst pairing schemes and single readers were also shown to be statistically significantly greater than zero in all case sets. However, no statistically significant improvement was observed in sensitivity for the worst pairing schemes when compared against the single readers’ sensitivity. This means that there might not be any benefits of double reading in mammography in improving sensitivity when readers get paired in worst possible ways. These findings suggest that using permanent pairings out of convenience run a small risk of obtaining poor performance compared to regularly pairing readers randomly.
Previous studies that explored double reading in screening mammography did not report the criteria for pairing readers (10, 11, 13, 14). Thus, it is unclear whether pairing readers based on some specific criteria, for example by pre-assessing each individual’s diagnostic accuracy and then matching readers with similar diagnostic accuracy (19), would yield near-optimal performance of double reading. We found that although random reader pairing yields significant benefits, it is theoretically possible to strategically pair readers so as to achieve better diagnostic performance overall. However, our data is not rich enough to identify criteria that can be used to prospectively determine best reader pairings and optimise the entire double reading system. Thus, further research with a large radiologists’ cohort is needed to potentially identify criteria that can be used to optimise reader pairings. Given that our exhaustive exploration of all possible pairing schemes showed only marginal benefits for best reader pairings compared to random reader pairings, it is logical that until criteria that optimise reader pairing are identified, regularly randomizing readers into pairs may capture most double reading benefits theoretically achieved through strategic pairing approaches.
Our findings are important firstly to those countries having a paired reader approach since it highlights the possibility that permanently pairing readers out of convenience presents a small possibility of greatly limiting the benefits of paired readings. Furthermore we suggest that strategic pairing approaches could potentially be developed by taking into account factors that are likely to affect individual reader cancer detection performance and matching readers so that they complement each other. Secondly, to those regions without paired readings, the work highlights that single reading is less efficacious than paired reading even when simple random reader pairing is used.
In conclusion, this study shows that compared to single reading, independent double reading in digital mammography improves the overall breast cancer detection performance by statistically significant and clinically important margins. The results also highlight that whilst a random pairing approach may be beneficial, it may be important to explore some sophisticated optimal pairing strategies so that the benefits of double reading can be maximized. Some observations noted with the worst pairing schemes suggest that identifying characteristics of reader pairs associated with high diagnostic performance may eliminate the chances of pairing readers who perform very poorly, and should be explored in future studies.
Acknowledgments
Funding
This research was supported by the National Geospatial-Intelligence Agency (HM04761610003), by the Institute for Collaborative Biotechnologies through funding from the U.S. Army Research Office (W911NF-09–0001), and by the SAGE Center for the Study of the Mind, UCSB. The content of the information does not necessarily reflect the position or the policy of the U.S. Government, and no official endorsement should be inferred. This research was supported by a grant (IN-15–029) from the National Breast Cancer Foundation (Australia). We also acknowledge the support of the BreastScreen Reader Assessment Strategy, Cancer Institute NSW, the New Zealand Ministry of Health, and the Royal Australian and New Zealand College of Radiology.
List of abbreviations
- CAD
computer-aided detection
- BREAST
Breast Screen Reader Assessment Strategy
- DICOM GSDF
Digital Imaging and Communication in Medicine Gray-Scale Standard Display Function
- RANZCR
Royal Australian and New Zealand College of Radiologists
- TN
true negative
- TP
true positive
- ΣTP
Summation of true positives
- ΣTN
Summation of true negatives
References
- 1.Ciatto S, Ambrogetti D, Bonardi R, Catarzi S, Risso G, Rosselli Del Turco M, et al. Second reading of screening mammograms increases cancer detection and recall rates. Results in the Florence screening programme. Journal of medical screening. 2005;12(2):103–6. [DOI] [PubMed] [Google Scholar]
- 2.Duijm LE, Groenewoud JH, Hendriks JH, de Koning HJ. Independent Double Reading of Screening Mammograms in the Netherlands: Effect of Arbitration Following Reader Disagreements 1. Radiology. 2004;231(2):564–70. [DOI] [PubMed] [Google Scholar]
- 3.Anderson E, Muir B, Walsh J, Kirkpatrick A. The efficacy of double reading mammograms in breast screening. Clinical radiology. 1994;49(4):248–51. [DOI] [PubMed] [Google Scholar]
- 4.Thurfjell EL, Lernevall KA, Taube A. Benefit of independent double reading in a population-based mammography screening program. Radiology. 1994;191(1):241–4. [DOI] [PubMed] [Google Scholar]
- 5.Azavedo E, Zackrisson S, Mejàre I, Heibert Arnlind M. Is single reading with computer-aided detection (CAD) as good as double reading in mammography screening? A systematic review. BMC Medical Imaging. 2012;12:22-. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ekpo EU, Alakhras M, Brennan P. Errors in Mammography Cannot be Solved Through Technology Alone. Asian Pac J Cancer Prev. 2018;19( 2):291–301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Taylor P, Potts HW. Computer aids and human second reading as interventions in screening mammography: two systematic reviews to compare effects on cancer detection and recall rate. European journal of cancer (Oxford, England : 1990). 2008;44(6):798–807. [DOI] [PubMed] [Google Scholar]
- 8.Australia B BreastScreen Australia National Accreditation Guidelines. BreastScreen Australia Quality Improvement Program [Internet]:[43 p.]. 2008. [Google Scholar]
- 9.Zou KH, Bhagwat JG, Carrino JA. Statistical combination schemes of repeated diagnostic test data. Academic radiology. 2006;13(5):566–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Posso M, Carles M, Rué M, Puig T, Bonfill X. Cost-Effectiveness of Double Reading versus Single Reading of Mammograms in a Breast Cancer Screening Programme. PLoS One. 2016;11(7):e0159806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gromet M Comparison of Computer-Aided Detection to Double Reading of Screening Mammograms: Review of 231,221 Mammograms. American Journal of Roentgenology. 2008;190(4):854–9. [DOI] [PubMed] [Google Scholar]
- 12.Houssami N, Macaskill P, Bernardi D, Caumo F, Pellegrini M, Brunelli S, et al. Breast screening using 2D-mammography or integrating digital breast tomosynthesis (3D-mammography) for single-reading or double-reading – Evidence to guide future screening strategies. European Journal of Cancer. 2014;50(10):1799–807. [DOI] [PubMed] [Google Scholar]
- 13.Taylor-Phillips S, Wallis MG, Parsons H, Dunn J, Stallard N, Campbell H, et al. Changing case Order to Optimise patterns of Performance in mammography Screening (CO-OPS): study protocol for a randomized controlled trial. Trials. 2014;15:17-. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Taylor-Phillips S, Wallis MG, Jenkinson D, et al. Effect of using the same vs different order for second readings of screening mammograms on rates of breast cancer detection: A randomized clinical trial. JAMA. 2016;315(18):1956–65. [DOI] [PubMed] [Google Scholar]
- 15.Efron B, Tibshirani RJ. An introduction to the bootstrap. 1993. [Google Scholar]
- 16.Juni MZ, Eckstein MP. The wisdom of crowds for visual search. Proceedings of the National Academy of Sciences. 2017;114(21):E4306–E15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Varian H Bootstrap Tutorial. Mathematica Journal. 2005;9:768–75. [Google Scholar]
- 18.Benjamini Y, Hochberg Y. Controlling the False Discovery Rate: A Practical and Powerful Approach to Multiple Testing. Journal of the Royal Statistical Society Series B (Methodological). 1995;57(1):289–300. [Google Scholar]
- 19.Kurvers RH, Herzog SM, Hertwig R, Krause J, Carney PA, Bogart A, et al. Boosting medical diagnostics by pooling independent judgments. Proc Natl Acad Sci U S A. 2016;113(31):8777–82. [DOI] [PMC free article] [PubMed] [Google Scholar]


