Abstract
Background
The presence of high-abundance drug-resistant HIV-1 jeopardizes success of antiretroviral therapy (ART). Despite numerous investigations, the clinical impact of low-abundance drug-resistant HIV-1 variants (LA-DRVs) at levels <15%–25% of the virus population in antiretroviral (ARV) drug-naive individuals remains controversial.
Methods
We systematically reviewed 103 studies assessing prevalence, detection methods, technical and clinical detection cutoffs, and clinical significance of LA-DRVs in antiretroviral drug-naive adults.
Results
In total, 14 919 ARV drug-naive individuals were included. Prevalence of LA-DRVs (ie, proportion of individuals harboring LA-DRVs) was 0%–100%. Technical detection cutoffs showed a 4 log range (0.001%–10%); 42/103 (40.8%) studies investigating the impact of LA-DRVs on ART; 25 studies included only individuals on first-line nonnucleoside reverse transcriptase inhibitor-based ART regimens. Eleven of those 25 studies (44.0%) reported a significantly association between preexisting LA-DRVs and risk of virological failure whereas 14/25 (56.0%) did not.
Conclusions
Comparability of the 103 studies is hampered by high heterogeneity of the studies’ designs and use of different methods to detect LA-DRVs. Thus, evaluating clinical impact of LA-DRVs on first-line ART remains challenging. We, the WHO HIVResNet working group, defined central areas of future investigations to guide further efforts to implement ultrasensitive resistance testing in routine settings.
Keywords: HIV-1, antiretroviral therapy, HIV-1 drug resistance, low-abundance drug-resistant HIV-1 variants, antitretroviral drug-naive individuals, next-generation sequencing, minority variants
We systematically assessed 103 studies on the prevalence of low-abundance drug-resistant HIV-1 variants and their impact on antiretroviral therapy, which remains uncertain. Important topics were defined to guide further efforts to implement ultrasensitive resistance testing in routine settings.
Since its introduction, antiretroviral therapy (ART) has greatly reduced global mortality rates and lengthened the lifespan of people living with human immunodeficiency virus (HIV) [1, 2]. However, despite its potency, the efficacy of ART in suppressing viral replication can be jeopardized by the presence of drug resistance [3]. Recognizing the potential consequences of drug resistance in achieving HIV epidemic control, the World Health Organization’s Global Action Plan on HIV Drug Resistance 2017–2021 defines areas where improved collective efforts are needed to strengthen the monitoring, prevention, and response to HIV drug resistance [4]. Using the framework of the Global Action Plan, WHO HIVResNet—a network of HIV drug resistance experts coordinated by WHO—developed a prioritized list of research gaps [5] that, once addressed, will enhance the ability to monitor resistance and interpret its impact on ART outcomes. These priority areas include the need for: (1) an improved understanding of optimal methods to detect low-abundance drug-resistant HIV-1 variants (LA-DRVs), often referred to as minority variants; (2) defining the appropriate technical and clinical detection thresholds for various types of assays; and (3) characterizing the clinical relevance of LA-DRVs in ARV drug-naive people initiating treatment with drugs to which LA-DRVs are present at varying levels of the viral population (or quasispecies) [5].
In HIV-1 infected, antiretroviral drug-naive individuals, LA-DRVs may arise due to de novo mutagenesis as a result of error-prone replication or by transmission from an HIV-1–infected antiretroviral drug-treated person [6]. In clinical practice, DRVs are commonly detected by population, Sanger-based sequencing of the HIV-1 pol gene [7]. LA-DRVs present at less than 15%–25% of the circulating viral population may not be detected by population sequencing [8, 9]. However, with the advent of more sensitive methods such as next-generation sequencing (NGS), virus variants present at low frequencies within the virus population of individuals can be detected. Several techniques to detect LA-DRVs have been developed. Some of the earliest methods included particularly allele-specific real-time polymerase chain reaction (AS-PCR) [10]. All of these techniques have different thresholds or cutoffs for detecting low-abundance variants, and specific strengths and limitations [11]. More recently, NGS has revolutionized the detection of LA-DRVs and is increasingly used for genotypic HIV-1 drug resistance testing worldwide [12, 13].
It has been shown that LA-DRVs can be detected in individuals acutely or recently infected with HIV-1 [14–19], and their transmission has been documented [20, 21], although these are probably very rare events. Although several reports indicate that the presence of drug-resistant HIV-1 at high abundance may affect future efficacy of ART, the clinical importance of LA-DRVs at time of treatment initiation remains uncertain, with some but not all studies reporting an association between LA-DRVs and suboptimal treatment outcomes. In this systematic review, we generate an up-to-date assessment of the prevalence, detection methods, technical and clinical detection cutoffs, and the clinical significance of LA-DRVs in antiretroviral drug-naive adults. This review is important to guide further efforts to implement resistance testing in routine settings for patient care and surveillance, in an evolving antiretroviral drug landscape.
METHODS
Search Strategy
We searched PubMed’s MEDLINE database for publications using 8 search strings constructed for Medline. Our search included every related article added to PubMed since its creation through 31 May 2019. Articles that contained all or some of the string words in the title or abstract were screened for inclusion. The search strings used in different combinations were: “HIV drug resistance,” “minority variants,” “minority mutations,” “minority quasispecies,” “low frequency variants,” “low-abundance variants,” “allele-specific,” “deep sequencing,” and “next-generation sequencing.”
Inclusion Criteria for Eligible Studies
A study was included if it mentioned the detection, prevalence (ie, the proportion of individuals harboring LA-DRVs), and/or clinical impact of LA-DRVs in ARV drug-naive adults. Infections caused by any HIV-1 subtype were included; only articles written in English were considered.
Exclusion Criteria for Noneligible Studies
Studies including only ARV drug-experienced individuals (ie, long- or short-term ART, the latter particularly applied for prevention of mother-to-child transmission of HIV). In addition, reviews, brief communications, conference proceedings, abstracts, or posters were excluded because of the very limited information that they provided or because of duplication of the information in full articles. All duplicate publications from the different search string results were removed.
RESULTS
Summary of Study Characteristics
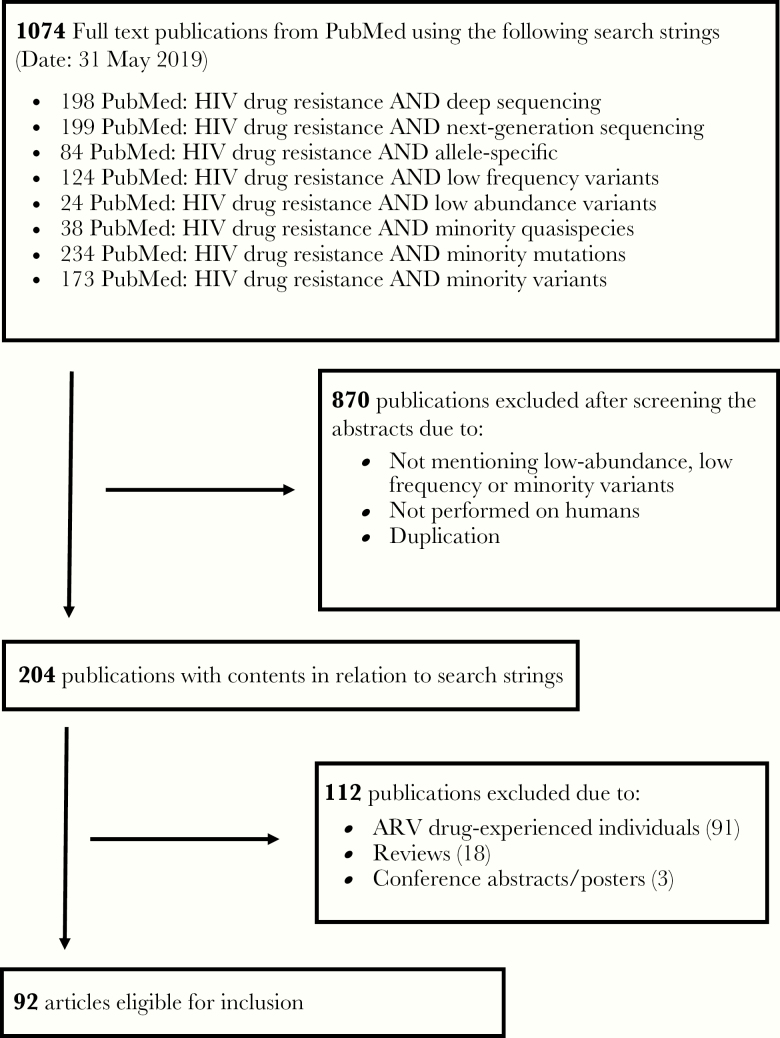
Applying the 8 search strings to PubMed, we found 1074 publications, the majority of which were duplicates. Of the 1074 publications, 204 were retained with contents matching the search strings. Ninety-two publications met inclusion criteria (Figure 1) [14–105]. Eight publications reported on 2–3 studies each, thus in total 103 studies were included in the analysis (Supplementary Table 1). Across the studies, numbers of participants ranged from 1 to 1148, with a median of 65 (interquartile range, 27–162) individuals (Table 1). Most participants were sampled between the years 2001 and 2014. The earliest specimens were collected in 1994 [20, 21] and the latest in 2016 [40, 41, 67, 93, 96]. Forty-two (40.8%) studies did not differentiate between acute and chronic HIV-1 infection. Seventeen (16.5%) studies reported on acute/recent HIV-1 infection and 44 (42.7%) studies focused (mainly) on chronic HIV-1 infection. Plasma was the most frequent specimen type (97 studies, 94.2%). Most studies (68, 66.0%) were conducted in high-income countries (mainly in Europe and the United States), with comparably fewer (30, 29.1%) conducted in low- and middle-income countries: sub-Saharan Africa (19, 18.4%), Asia (8, 7.8%), and Middle/South America (3, 2.9%).
Figure 1.
Summary of study search and selection procedure. Abbreviations: ART, antiretroviral therapy; HIV, human immunodeficiency virus.
Table 1.
Summary of All Publications on Detection, Prevalence, and/or Clinical Impact of Low-Abundance Drug-Resistant HIV-1 Variants in ARV Drug-Naive, Adult Individuals (92 Publications, 103 Studies)
| Parameter | Studies, n (%) |
|---|---|
| Number of participants, median (min–max) | 65 (1–1148) |
| Stage of HIV-1 infection at time of inclusion | |
| Acute/recent (Mainly) chronic Not specified | 17 (16.5) 44 (42.7) 42 (40.8) |
| Type of specimen used for LA-DRVs detection | |
| Plasma PBMCs Serum Dried blood spot CSF Virus isolate | 97 (94.2) 10 (9.7) 4 (3.9) 1 (1.0) 1 (1.0) 1 (1.0) |
| Geographic area | |
| Europe North America Africa Asia Latin America Worldwide Europe/North America Not specified | 40 (38.8) 21 (20.4) 19 (18.4) 12 (11.7) 4 (3.9) 4 (3.9) 2 (1.9) 1 (1.0) |
| Type of study | |
| Prevalence Prevalence and clinical impact of LA-DRVs Case-control studies Case reports Meta-analysis | 61 (59.2) 30 (29.1) 9 (8.7) 2 (1.9) 1 (1.0) |
| Detection method of LA-DRVs | |
| 454 pyrosequencing AS-PCR Illumina NGS Cloning + sequencing HIV-SNaPshot OLA Pyrosequencing (Pyro-Mark) DEEPGEN NGS RCA SGA SMRTS (PacBio) | 34 (33.0) 34 (33.0) 25 (24.3) 3 (2.9) 2 (1.9) 2 (1.9) 2 (1.9) 1 (1.0) 1 (1.0) 1 (1.0) 1 (1.0) |
| Inclusion/exclusion of individuals harboring DRVs detected by population sequencing | |
| DRVs not excluded Certain DRVs excluded Any DRVs excluded Presence of DRVs as inclusion criteria No population sequencing performed/no data shown | 32 (31.1) 15 (14.6.) 9 (8.7) 4 (3.9) 25 (24.3) |
Abbreviations: ART, antiretroviral therapy; AS-PCR, allele-specific polymerase chain reaction; CSF, cerebral spinal fluid; DRV, drug-resistant HIV-1 variant; HIV, human immunodeficiency virus; LA, low abundance; NGS, next-generation sequencing; OLA, oligonucleotide ligation assay; PBMCs, peripheral blood mononuclear cells; RCA, rolling-circle amplification; SGA, single genome amplification; SMRTS, single molecule real-time sequencing.
Sixty-one (59.2%) studies reported solely on the prevalence of LA-DRVs in ARV drug-naive individuals and did not assess their impact on treatment outcomes, while 30 (29.1%) different prevalence studies also investigated the impact of LA-DRVs on ART clinical outcomes (Table 1). Of the remaining 12 studies, 9 (8.7%) were case-control studies [38, 53, 57, 60, 70, 84, 88, 99, 105], 2 (1.9%) were case reports [25, 100], and 1 (1.0%) was a meta-analysis [65]; all reported on the association between LA-DRVs and treatment outcomes (Supplementary Table 1).
Methods Used in the Detection of Low-Abundance Drug-Resistant HIV-1 Variants
A variety of different methods were used to detect LA-DRVs in the 103 studies. Most studies used AS-PCR (34, 33.0%) and 454 pyrosequencing (34, 33.0%) or Illumina (25, 24.3%) NGS platforms (Table 1 and Supplementary Table 1). Prior to 2005, when the first NGS platform became available, AS-PCR was the most frequently used method. Despite having the highest sensitivity, a major limitation of point mutation assays, such as AS-PCR, is that they can only detect 1 single point mutation at a time [10] and their ability to detect alternative polymorphisms at the codon of interest is reduced [106].
Generally, each method may be affected by polymorphisms associated with drug resistance, which may skew the sensitivity of primers and probes used in the assay [106]. Other important issues, particularly when applying NGS assays, are experimental challenges during sample preparation, for example loss during DNA or RNA extraction or contaminations, and errors introduced during reverse transcription or amplifications, for example nucleotide misincorporation, resampling, biases due to primer/probe mismatches, or in vitro recombination [107, 108]. Some of them can be addressed by quantifying input cDNA copy numbers as done, for instance, in the study by Mbunkah et al [67], or by using primer IDs [36, 109, 110]. Details of the technical strengths and limitations of the methods used in the studies are shown in Table 2.
Table 2.
Detection Methods for Low-Abundance Drug-Resistant HIV-1 Variants Used in the Included Publications
| Detection Method | Technical Strengths | Technical Limitations | Technical Cutoff Applied, % Min–Max |
|---|---|---|---|
| 454 pyrosequencing | Good sensitivity and specificity; long reads at short run times | Homopolymer errors; relatively high insertion/deletion rate; low throughput; costly reagents, (not available anymore) | 0.02–5 |
| AS-PCR | High sensitivity and specificity; fairly labor-intensive; easy interpretation of results | Only particular mutations of interest can be detected; false-positive results at lower limits; polymorphisms at primer binding sites can reduce assay’s sensitivity/specificity; varying sensitivity/specificity for different mutations due to virus- and assay- related issues | 0.001–2 |
| Cloning + sequencing | High sensitivity; not susceptible to primer polymorphisms; genetic linkage is possible if single genome amplification is applied | Time and labor intensive | 0.5–10; depending on sequenced clones |
| Illumina NGS | High-throughput data with low error rates; high sensitivity; relatively cheap | Fairly laborious with long run times | 1–3 |
| OLA | High sensitivity and specificity; fairly labor intensive; easy interpretation of results | Only particular mutations of interest can be detected; false-positive results at lower limits; polymorphisms can reduce sensitivity | 2 |
| RCA | High sensitivity and specificity; fairly labor intensive; easy interpretation of results | Only particular mutations of interest can be detected; false-positive results at lower limits; polymorphisms can reduce sensitivity | 1 |
| SGA | Risk of nucleotide misincorporation or template switching introduced during PCR amplification is reduced; genetic linkage is possible | Very labor intensive and costly; much time involved in determining the appropriate dilution to use | 2 |
| SMRTS (Pacific Biosciences) | Long reads; low error rate due to circular consensus sequencing | Fairly laborious; high input amount of DNA required | 1 |
Abbreviations: AS-PCR, allele-specific polymerase chain reaction; HIV, human immunodeficiency virus; NGS, next-generation sequencing; OLA, oligonucleotide ligation assay; RCA, rolling-circle amplification; SGA, single-genome amplification; SMRTS, single-molecule real-time sequencing.
The technical sensitivity of each method dictates the lower limit of detection to be used with it. Studies that used AS-PCR had a minimum limit of detection (technical cutoff) of 0.001% and a maximum of 1%, with 0.01% used most often (Table 2). Studies using NGS technologies (454 pyrosequencing, Illumina NGS, or single-molecule real-time sequencing by Pacific Biosciences) had minimum technical cutoffs of 0.02% and 1%, respectively. A 1% technical cutoff was most commonly applied with those methods. Sources of errors in genotypic resistance assays, including errors introduced by reverse transcription, multiple rounds of amplification (including PCR recombination) followed by sequencing, may affect the technical cutoffs [108]. Lowering technical cutoff values below 1% for most NGS-based assays could give rise to false positives due to these inherent errors from the assay [111]. The high heterogeneity in limits of detection not only between the different methods used but also within the same methods argues for future interlaboratory studies [11, 94, 112].
Prevalence of Low-Abundance Drug Resistant HIV-1 Variants
Ninety-one (88.3%) studies of ARV drug-naive adults reported DRVs prevalence data. The reverse transcriptase region was the most commonly studied part of the HIV-1 genome. Sixty-one of the 91 studies (67.0%) were conducted in high-income countries, 14 (15.4%) in low- and middle-income countries, and 14 (15.4%) in upper middle-income countries, as classified by the World Bank [113]. For 2 (2.2%) studies, the countries of origin were not specified. The drug-resistant mutations K103N, Y181C, and M184V were the most commonly reported mutations at varying detection thresholds (Table 3 and Supplementary Table 1). The K103N, Y181C, and M184V mutations as LA-DRVs were detected in a median of 2.0%, 0.2%, and 0.5% of ARV drug-naive adults, respectively. The prevalence of the K103N, Y181C, and M184V mutations as LA-DRVs in ARV drug-naive individuals reached up to 33.0%, 10.0%, and 41.9%, respectively (Table 3 and Supplementary Table 1).
Table 3.
Prevalence of Low-Abundance Drug-Resistant HIV-1 Mutations K103N, Y181C, and M184V in the 91 Prevalence Studies
| K103N | Y181C | M184V | |
|---|---|---|---|
| Studies reporting the LA-DRV, No. (%) | 68 (74.7) | 57 (62.6) | 58 (63.7) |
| Number of participants/study, median (IQR; min–max) | 55 (29–151; 4–995) | 56 (26–133; 4–442) | 53 (21–123; 5–833) |
| Proportion of individuals harboring the LA-DRV, median % (IQR; min–max) | 2 (0–5.5; 0–33.3) | 0.2 (0–3; 0–10) | 0.5 (0–7.3; 0–41.9) |
Details are provided in Supplementary Table 1.
Most studies provided information for individuals harboring the K103N, Y181C, and M184V mutations at high abundance. These individuals were not included in this analysis.
Abbreviations: HIV, human immunodeficiency virus; IQR, interquartile range; LA-DRV, low-abundance drug-resistant HIV-1 variant.
Taken together, the prevalence of these LA-DRVs was highly variable in different studies of ARV drug-naive individuals. Several issues complicate the comparison of prevalence data between studies. Considerable heterogeneity was present with respect to time of sampling, detection methods used to characterize LA-DRVs, the thresholds applied in their detection, and to a lesser extent the study participants’ inclusion/exclusion criteria. Consequently, highly variable prevalence estimates of LA-DRVs are reported across the different studies, even those performed in the same country. Another complicating factor was the presence of DRVs as revealed by genotypic resistance testing methods based on population sequencing. Ten of 91 (11.0%) studies included only individuals without any DRVs detected by population-based sequencing, 15 (16.5%) excluded specific DRVs detected by population-based sequencing, and 37 (40.7%) allowed the presence of DRVs at high abundance. In 5 of 91 (5.5%) studies, the presence of DRVs detected by population-based sequencing was an inclusion criteria, and in 24 (26.4%) studies, genotypic resistance testing by population-based sequencing was not performed or the data were not shown (Table 1). Nevertheless, it was generally true that the use of more sensitive detection assays led to the reporting of higher prevalence estimates of DRVs.
Impact of Low-Abundance Drug-Resistant HIV-1 Variants on the Outcome of Antiretroviral Therapy
LA-DRVs have been suggested to have an impact on ART outcomes in antiretroviral drug-naive individuals [10]. We found 42/103 (40.8%) studies investigating the impact of LA-DRVs on first-line ART in ARV drug-naive adults. Of note, the presence of DRVs detected by standard population sequencing was not an exclusion criterion in most of the studies (Supplementary Table 1). Four studies described single individuals on nonnucleoside reverse transcriptase inhibitor (NNRTI)-based ART regimens in whom preexisting low-abundance NNRTI-resistant variants were rapidly selected and became the predominant variant during virological failure (Supplementary Table 1) [25, 27, 87, 100]. Another study showed the selection of preexisting low-abundance protease inhibitor (PI)-resistant variants during virological failure in 3 individuals receiving ritonavir-boosted protease inhibitor (PI/r)-based regimens [90].
Besides these 5 case reports, 1 meta-analysis [65], 9 case-control studies, and 27 prevalence studies investigated the clinical relevance of LA-DRVs (Supplementary Table 1). Four of these 37 studies (10.8%) investigated the impact of preexisting RTI- and/or PI LA-DRVs on first-line PI/r-based ART and did not find any impact of LA-DRVs on clinical outcome [17, 59, 60, 85]. In 8 of the 37 studies (21.6%), individuals received various first-line ART regimens, mainly NNRTI- or PI/r-based therapy. No impact of preexisting LA-DRVs on treatment outcomes was reported in these studies [15, 18, 29, 31, 44, 54, 71, 95]. So far, no study has reported the potential impact of integrase strand transfer inhibitor (INSTI) LA-DRVs on INSTI-based ART regimens in ARV drug-naive individuals.
The potential impact of LA-DRVs on first-line NNRTI-based ART regimens is very controversially discussed. Twenty-five of the 37 studies (67.6%) included only individuals on first-line NNRTI-based ART, regimens considered to have a relatively low genetic barrier to resistance. Case reports are not included in this subset of studies. We assessed the quality of these 25 studies using 18 criteria based on the recommendations by the Strengthening of Reporting of Observational Studies in Epidemiology (STROBE) statement covering information on study design, participants’ characteristics, methods, and results (Table 4 and Supplementary Table 2) [114]. A majority of the 25 studies reached high scores showing the high quality of these studies. Nevertheless, the comparability of those studies is hampered by the high heterogeneity of the studies’ designs (Supplementary Table 2) with respect to several factors: (1) some used AS-PCR, hence reporting only 1 or a few DRVs, while others used NGS and reported a selection of DRVs, including some studies reporting all nucleoside reverse transcriptase inhibitor (NRTI)- and NNRTI-DRVs; (2) a few studies focused solely on NNRTI-DRVs, while others included all RTI-DRVs; (3) a wide range of technical cutoffs was used, that is 0.001%–10.0%; (4) most studies excluded individuals with preexisting high-abundance DRVs, while others did not; (5) the number of participants was highly variable, ranging from 5 to 489; (6) the criteria and definitions used to determine the impact of LA-DRVs on clinical outcome were highly diverse; and (7) the clinical and epidemiological characteristics of the participants varied substantially, for example year of sampling and stage of HIV-1 infection at time of study enrolment (Supplementary Table 2).
Table 4.
Categorization of the 25 Studies Evaluating the Impact of NNRTI Low-Abundance Drug-Resistant HIV-1 Variants Based on Recommendations by the Strengthening of Reporting of Observational Studies in Epidemiology Statement [114]
| Reference | Study Design | Participants | Methods | Results | Sum | ||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Country | Sampling time period | Duration of follow-up | Definition of virological failurea | Number of participants | Age | Sex | Ethnicity | Route of HIV-1 infection | HIV-1 subtype | Cd4+ T-cell count at baseline | Viral load at baseline | Stage of HIV-1 infection | Details on ART regimens | GRT prior to study initiation | Technical cutoff | Clinical cutoff/mutational load | GRT at virological failure | ||
| Johnson et al 2008 [57] | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 1 | 1 | 0 | 1 | 11 |
| Metzner et al 2009 [70] | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 0 | 1 | 1 | 0 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 14 |
| Simen et al 2009 [115] | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 0 | 1 | 1 | 1 | 0 | 1 | 1 | 0 | 0 | 13 |
| Balduin et al 2009 [24] | 1 | 1 | 0 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 0 | 14 |
| Geretti et al 2009 [46] | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 0 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 14 |
| Paredes et al 2010 [84] | 1 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 15 |
| Goodman et al 2011 [48] | 1 | 0 | 1 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 1 | 0 | 1 | 1 | 1 | 1 | 1 | 11 |
| Li et al 2011 [65] | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 15 |
| Messiaen et al 2012 [68] | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 0 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | NA | 13 |
| Bansode et al 2013 [26] | 1 | 1 | 1 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 1 | 1 | 1 | 1 | 0 | NA | 10 |
| Mohamed et al 2014 [75] | 1 | 0 | 1 | 1 | 1 | 0 | 1 | 0 | 0 | 1 | 0 | 1 | 0 | 1 | 1 | 1 | 0 | 1 | 11 |
| Metzner et al 2014 [72] | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 17 |
| Neogi et al 2014 [79] | 1 | 0 | 1 | 1 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 1 | 1 | 0 | NA | 8 |
| Nicot et al 2015 [80] | 1 | 1 | 1 | 0 | 1 | 1 | 1 | 0 | 0 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | NA | 12 |
| Cozzi-Lepri et al 2015 [38] | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 1 | 1 | 0 | 16 |
| Zoufaly et al 2015 [105] | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 17 |
| Porter et al 2015 [88] | 0 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 1 | 0 | 1 | 1 | 1 | 0 | NA | 12 |
| Van Eygen et al 2016 [99] | 0 | 0 | 1 | 1 | 1 | 0 | 1 | 0 | 0 | 1 | 0 | 1 | 1 | 1 | 1 | 1 | 0 | NA | 10 |
| Mzingwane et al 2016 [77] | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 0 | 0 | 1 | 0 | 1 | 1 | 0 | 1 | 0 | 0 | 11 |
| Ávila-Ríos et al 2016 [23] | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 0 | 1 | 1 | 1 | 0 | 1 | 1 | 1 | 14 | |
| Raymond et al 2018 [89] | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 15 |
| Inzaule et al 2018 [53] | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 0 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 0 | 14 |
| Hassan et al 2019 [50] | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 0 | 1 | 1 | 0 | 0 | 1 | 0 | 1 | 0 | 1 | 12 |
| Derache et al 2019 [41] | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 0 | 0 | 1 | 1 | 0 | 1 | 0 | 1 | 1 | 0 | 12 |
| Su et al 2019 [96] | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 16 |
| Sum | 23 | 19 | 24 | 23 | 25 | 18 | 21 | 10 | 7 | 13 | 20 | 20 | 17 | 23 | 21 | 25 | 8 | 10 | |
1 = information is provided; 0 = information is not provided; NA = not applicable, eg, not all studies evaluated the impact of LA-DRVs on rate of virological failure but on, eg, time to viral load suppression. Details are given in Supplementary Table 2.
Abbreviations: ART, antiretroviral therapy; GRT, routine genotypic resistance test; HIV, human immunodeficiency virus; NNRTI, nonnucleoside reverse transcriptase inhibitor.
aDefinition of virological failure or other virological outcome parameters.
Fourteen of those 25 studies (56.0%) showed no association between preexisting LA-DRVs and risk of virological failure in individuals receiving NNRTI-based first-line ART: 3 case-control studies [88, 99, 105] and 11 prevalence studies [24, 26, 50, 68, 72, 75, 77, 79, 80, 89, 96]. Eleven of the 25 studies (44.0%) reported a higher risk of virological failure if low-abundance NNRTI-DRVs were present prior to treatment initiation (1 meta-analysis [65], 5 case-control studies [38, 53, 57, 70, 84], and 4 prevalence studies [23, 41, 46, 48, 115]; Supplementary Table 2).
In a multicohort European case-control study that included ARV drug-naive individuals (76 cases and 184 controls), the presence of preexisting low-abundance NNRTI-DRVs more than doubled the risk of virological failure in individuals on first-line NNRTI-based ART (odds ratio, 2.75; 95% confidence interval [CI], 1.35–5.60; P = .005) compared to individuals with no NNRTI LA-DRVs detected [38]. In a pooled analysis of 10 studies with a total of 985 ARV drug-naive individuals, 138 (14%) of whom carried either NNRTI or NRTI LA-DRVs, the detection of LA-DRVs at treatment initiation was associated with more than twice the risk of virological failure early after therapy initiation with NNRTI-based ART compared to individuals in whom no LA-DRVs were detected [65]. Among the few studies in low- and middle-income countries assessing the clinical impact of LA-DRVs on NNRTI-based ART, 1 study conducted in Mexico including 264 ARV drug-naive individuals initiating treatment reported an association of LA-DRVs with virological failure [23].
Sensitivity thresholds (for identifying cases) and tradeoffs with specificity (ability to identify controls) with respect to the clinical relevance of LA-DRVs have been debated. Sensitivity thresholds were evaluated in more recent studies. After a median follow-up of 8 months after ART initiation, Ávila-Ríos et al observed a variable increased risk of viral nonsuppression at 6 months depending on the sensitivity threshold used [23]. Findings were statistically significant at sensitivity thresholds of 20% (P = .019), 10% (P = .0064), and 5% (P = .015), but not at 2% (P = .074), suggesting an optimal threshold for NNRTI LA-DRVs of 5%. Inzaule et al also reported an optimal sensitivity threshold for NNRTI LA-DRVs of 5%, but not lower [53]: lowering the threshold from 20% through 1% results in improved sensitivity (ability to identify cases) but at a cost of reduced specificity (ability to identify controls). The adjusted odds ratio for virological failure was 9.2 (95% CI, 4.2–20.1) at a detection threshold of 20%, but changed as the threshold was lowered: 6.8 (95% CI, 3.3–13.9) at the 10% threshold, 7.6 (95% CI, 3.4–17.1) at the 5% threshold, and 4.5 (95% CI, 2.0–10.2) at the 1% threshold [53]. A very recent study by Derache et al showed a significantly higher risk of virological failure in the presence of LA-DRVs using a threshold of 5% [41].
An alternative predictor of clinical outcome that has been proposed is the absolute copy number of a particular viral mutant (mutational load), rather than the proportion. A few studies have investigated the relationship between mutational load and its impact on virological outcome. A threshold of 2000 copies/mL of variants with K103N prior to treatment initiation was shown to predict virological failure in a retrospective analysis investigating the effects of low levels of the K103N mutation present at treatment initiation [48]. In a pooled analysis of studies involving ARV drug-naive individuals initiating NNRTI-based regimens, a dose-dependent increased risk of virological failure of first-line ART was observed, although copy numbers of 10–99 per mL plasma or frequencies of <0.5% of low-abundance NNRTI-DRVs were already significantly associated with an increased risk of virological failure of first-line ART [65]. A dose-effect relationship between the mutational load and virological failure was also observed in the multicohort Europe-wide case-control study showing a significantly higher risk of virological failure at mutational loads ≥1000 copies per mL [38]. Inzaule et al also performed a sensitivity analysis based on mutational load and reported that the association between LA-DRVs and virological failure was significant only at a higher copy number (≥1000 copies per mL) [53].
Evaluating the clinical impact of LA-DRVs on first-line ART remains challenging. Most studies enrolled small numbers of participants and were often substudies or subanalyses where LA-DRVs were not the primary focus. Substantial variation in study design and use of different methods to detect LA-DRVs were also observed. The definitions of virological failure also varied and in most studies neither coadministered NRTIs nor the viral and mutational loads were considered. A meta-analysis of these studies would be inappropriate due to these biases, coupled with the fact that some of these studies also included individuals with preexisting DRVs detected by routine genotypic drug resistance assays.
CONCLUSIONS
The prevalence of LA-DRVs has been reported in antiretroviral drug-naive individuals across the globe at varying levels. In the past decade, different technologies have evolved and new ones have been developed, making detection of LA-DRVs in individuals easier in terms of costs and sensitivity. The application of these novel platforms to routine HIV drug resistance genotyping has the potential to be revolutionary. This review documents a considerable range in the lower limit of detection of LA-DRVs for different assays, from <0.01% as seen with AS-PCR to 1%–5% for ultradeep sequencing assays and other methods [54].
LA-DRVs have been shown to be clinically relevant, especially prior to the initiation of a first-line NNRTI-based regimen. Of note, not each ARV drug-naive individual harboring LA-DRVs experiences virological failure. Furthermore, in the case of virological failure, the preexisting LA-DRVs are not necessarily the selected variants. The clinical impact of LA-DRVs on response to regimens that are based on other drug classes generally remains even more uncertain, as reflected by the latest recommendations on HIV-1 drug resistance testing by the International Antiviral Society USA [116]. Although just a single study in this review reported low-abundance INSTI-DRVs at a prevalence of 2.4% [52], we believe that the relevance of LA-DRVs with potential impact on INSTI-containing regimens will depend on a number of factors including: (1) the prevalence of transmitted INSTI resistance, which remains low but which may increase in the future; and (2) the overall genetic barrier of INSTI regimens.
Important questions concerning LA-DRVs remain to be addressed, such as defining a clinically relevant threshold or cutoff at which they are associated with an increased risk of virological failure (Box 1). A large prospective study investigating the impact of LA-DRVs on different ART regimens and in different clinical settings in different countries could shed light on this widely debated, timely, and clinically relevant question, but such a study will be challenging and costly to set up. Future studies should assess the effect of mutational load on virological outcomes, identify which LA-DRVs are clinically relevant, the impact of linked mutations, and the time it takes to virological failure.
Box 1. Outstanding questions about LA-DRVs.
At what threshold are LA-DRVs associated with increased risk of virological failure for regimens based on NRTIs, NNRTIs, PIs, and INSTIs?
What is the role of the mutational load, ie, the total amount of particular LA-DRVs?
Which drug resistance mutations are clinically relevant in the context of LA-DRVs?
Are there any linkages or interplays between the mutations/variants and could this have any effects?
How much time does it take until virological failure occurs due to the outgrowth of LA-DRVs?
For which ART combination might the detection of LA-DRVs be beneficial?
Which viral and host factors contribute to virological failure due to the outgrowth of LA-DRVs? Vice versa, which factors prevent virological failure in the presence of LA-DRVs?
Abbreviation: ART, antiretroviral therapy; INSTI, integrase strand transfer inhibitor; LA-DRV, low-abundance drug-resistant human immunodeficiency virus-1 variants; NNRTI, nonnucleoside reverse transcriptase inhibitor; NRTI, nucleoside reverse transcriptase inhibitor; PI, protease inhibitor.
Standardization of assays and bioinformatics procedures in this field is also important [117]. Because it is crucial to understand the point at which LA-DRVs may become clinically significant, defining a clinically relevant threshold for HIV drug resistance testing will be equally valuable [23, 53]. Therefore, more studies are needed to determine a threshold, below which the risk of treatment failure decreases. Because cutoffs may depend on several factors such as viral load and the relative fitness cost of specific mutations, defining a single cutoff for all DRVs is unlikely to be possible. Consequently, clinical cutoffs specific for each mutation and treatment regimen may be required. In the near future, the clinical impact of LA-DRVs on treatment outcome will possibly depend on the genetic barrier of the remaining current drugs to resistance and the potency of emerging drugs. Furthermore, wider implementation of some of the sensitive technologies for detection of LA-DRVs in low- and middle-income countries is also encouraged.
Supplementary Data
Supplementary materials are available at The Journal of Infectious Diseases online. Consisting of data provided by the authors to benefit the reader, the posted materials are not copyedited and are the sole responsibility of the authors, so questions or comments should be addressed to the corresponding author.
Notes
Author contributions. S. B., R. P., N. P., M. R. J., and K. J. M. conceptualized the review. H. A. M. and K. J. M. searched the literature, selected studies, extracted the data, and wrote the manuscript. All authors made contributions to writing and review of the manuscript and also approved the final manuscript.
Disclaimer. The funding sources had no role in the design of this study, the data analyses, or the interpretation of the data.
Financial support. This work was supported by the Swiss Federal Commission for Scholarships (H. A. M.); the Hartmann Müller Foundation for Medical Research (grant number 1899 to K. J. M.); the Ensemble pour une Solidarité Thérapeutique Hospitalière En Réseau (ESTHER 2017 project to H. A. M. and K. J. M.); the European Union Erasmus Mundus Programme (S. I.); Amsterdam Institute for Global Health and Development (S. I. and T. F. RdW); and the Wellcome Trust (R. L. H.).
Potential conflicts of interest. G. H. reports grants from Eunice Kennedy Shriver National Institutes of Child Health and Human Development during the conduct of the study. R. P. reports travel grants and honoraria from ViiV HealthCare, Merck Sharpe & Dohme, and Gilead Sciences outside the submitted work. K. J. M. reports grants travel grants and honoraria from ViiV and Gilead Sciences outside the submitted work; and the University of Zurich received an unrestricted research grant from Gilead Science for studies that K. J. M. serves as principal investigator, unrelated to the submitted work. All other authors declare no potential conflict of interest.
All authors have submitted the ICMJE Form for Disclosure of Potential Conflicts of Interest. Conflicts that the editors consider relevant to the content of the manuscript have been disclosed.
References
- 1. Palella FJ Jr, Delaney KM, Moorman AC, et al. . Declining morbidity and mortality among patients with advanced human immunodeficiency virus infection. HIV Outpatient Study Investigators. N Engl J Med 1998; 338:853–60. [DOI] [PubMed] [Google Scholar]
- 2. Antiretroviral Therapy Cohort Collaboration. Survival of HIV-positive patients starting antiretroviral therapy between 1996 and 2013: a collaborative analysis of cohort studies. Lancet HIV 2017; 4:e349–e56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. World Health Organization. HIV drug resistance report 2017 http://apps.who.int/iris/bitstream/handle/10665/255896/9789241512831-eng.pdf;jsessionid=8E49187FF0167858B46AD727759E2A4A?sequence=1. Accessed 12 December 2018.
- 4. World Health Organization. Global action plan on HIV drug resistance 2017–2021 https://www.who.int/hiv/pub/drugresistance/hivdr-action-plan-2017-2021/en/. Accessed 12 December 2018.
- 5. World Health Organization. WHO HIVResNet meeting report: Johannesburg, South Africa, 11–12 November 2017, 2018. https://apps.who.int/iris/handle/10665/273133. Accessed 9 December 2018. [Google Scholar]
- 6. World Health Organization. WHO HIV drug resistance report 2012 http://apps.who.int/iris/bitstream/handle/10665/75183/9789241503938_eng.pdf?sequence=1. Accessed 12 December 2018.
- 7. Hirsch MS, Günthard HF, Schapiro JM, et al. ; International AIDS Society-USA Antiretroviral drug resistance testing in adult HIV-1 infection: 2008 recommendations of an International AIDS Society-USA panel. Clin Infect Dis 2008; 47:266–85. [DOI] [PubMed] [Google Scholar]
- 8. Günthard HF, Wong JK, Ignacio CC, Havlir DV, Richman DD. Comparative performance of high-density oligonucleotide sequencing and dideoxynucleotide sequencing of HIV type 1 pol from clinical samples. AIDS Res Hum Retroviruses 1998; 14:869–76. [DOI] [PubMed] [Google Scholar]
- 9. Schuurman R, Demeter L, Reichelderfer P, Tijnagel J, de Groot T, Boucher C. Worldwide evaluation of DNA sequencing approaches for identification of drug resistance mutations in the human immunodeficiency virus type 1 reverse transcriptase. J Clin Microbiol 1999; 37:2291–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Metzner KJ. Detection and significance of minority quasispecies of drug-resistant HIV-1. J HIV Ther 2006; 11:74–81. [PubMed] [Google Scholar]
- 11. Halvas EK, Aldrovandi GM, Balfe P, et al. . Blinded, multicenter comparison of methods to detect a drug-resistant mutant of human immunodeficiency virus type 1 at low frequency. J Clin Microbiol 2006; 44:2612–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Gibson RM, Meyer AM, Winner D, et al. . Sensitive deep-sequencing-based HIV-1 genotyping assay to simultaneously determine susceptibility to protease, reverse transcriptase, integrase, and maturation inhibitors, as well as HIV-1 coreceptor tropism. Antimicrob Agents Chemother 2014; 58:2167–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Chimukangara B, Samuel R, Naidoo K, de Oliveira T. Primary HIV-1 drug resistant minority variants. AIDS Rev 2017; 19:89–96. [PubMed] [Google Scholar]
- 14. Metzner KJ, Rauch P, Walter H, et al. . Detection of minor populations of drug-resistant HIV-1 in acute seroconverters. AIDS 2005; 19:1819–25. [DOI] [PubMed] [Google Scholar]
- 15. Peuchant O, Thiébaut R, Capdepont S, et al. ; ANRS CO3 Aquitaine Cohort Transmission of HIV-1 minority-resistant variants and response to first-line antiretroviral therapy. AIDS 2008; 22:1417–23. [DOI] [PubMed] [Google Scholar]
- 16. Toni TA, Asahchop EL, Moisi D, et al. . Detection of human immunodeficiency virus (HIV) type 1 M184V and K103N minority variants in patients with primary HIV infection. Antimicrob Agents Chemother 2009; 53:1670–2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Metzner KJ, Rauch P, von Wyl V, et al. . Efficient suppression of minority drug-resistant HIV type 1 (HIV-1) variants present at primary HIV-1 infection by ritonavir-boosted protease inhibitor-containing antiretroviral therapy. J Infect Dis 2010; 201:1063–71. [DOI] [PubMed] [Google Scholar]
- 18. Gianella S, Delport W, Pacold ME, et al. . Detection of minority resistance during early HIV-1 infection: natural variation and spurious detection rather than transmission and evolution of multiple viral variants. J Virol 2011; 85:8359–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Nicot F, Saliou A, Raymond S, et al. . Minority variants associated with resistance to HIV-1 nonnucleoside reverse transcriptase inhibitors during primary infection. J Clin Virol 2012; 55:107–13. [DOI] [PubMed] [Google Scholar]
- 20. Metzner KJ, Scherrer AU, Preiswerk B, et al. ; Swiss HIV Cohort Study Origin of minority drug-resistant HIV-1 variants in primary HIV-1 infection. J Infect Dis 2013; 208:1102–12. [DOI] [PubMed] [Google Scholar]
- 21. Lipscomb JT, Switzer WM, Li JF, Masciotra S, Owen SM, Johnson JA. HIV reverse-transcriptase drug resistance mutations during early infection reveal greater transmission diversity than in envelope sequences. J Infect Dis 2014; 210:1827–37. [DOI] [PubMed] [Google Scholar]
- 22. Avidor B, Girshengorn S, Matus N, et al. . Evaluation of a benchtop HIV ultradeep pyrosequencing drug resistance assay in the clinical laboratory. J Clin Microbiol 2013; 51:880–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Ávila-Ríos S, Garcia-Morales C, Matias-Florentino M, et al. . Pretreatment HIV-drug resistance in Mexico and its impact on the effectiveness of first-line antiretroviral therapy: a nationally representative 2015 WHO survey. Lancet HIV 2016; 3:e579–e91. [DOI] [PubMed] [Google Scholar]
- 24. Balduin M, Oette M, Däumer MP, Hoffmann D, Pfister HJ, Kaiser R. Prevalence of minor variants of HIV strains at reverse transcriptase position 103 in therapy-naive patients and their impact on the virological failure. J Clin Virol 2009; 45:34–8. [DOI] [PubMed] [Google Scholar]
- 25. Bansal V, Metzner KJ, Niederost B, et al. . Minority K65R variants and early failure of antiretroviral therapy in HIV-1-infected Eritrean immigrant. Emerg Infect Dis 2011; 17:1966–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Bansode V, McCormack GP, Crampin AC, et al. . Characterizing the emergence and persistence of drug resistant mutations in HIV-1 subtype C infections using 454 ultra deep pyrosequencing. BMC Infect Dis 2013; 13:52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Bellecave P, Recordon-Pinson P, Papuchon J, et al. . Detection of low-frequency HIV type 1 reverse transcriptase drug resistance mutations by ultradeep sequencing in naive HIV type 1-infected individuals. AIDS Res Hum Retroviruses 2014; 30:170–3. [DOI] [PubMed] [Google Scholar]
- 28. Bergroth T, Ekici H, Gisslén M, Hagberg L, Sönnerborg A. Difference in drug resistance patterns between minor HIV-1 populations in cerebrospinal fluid and plasma. HIV Med 2009; 10:111–5. [DOI] [PubMed] [Google Scholar]
- 29. Boltz VF, Bao Y, Lockman S, et al. ; OCTANE/A5208 Team Low-frequency nevirapine (NVP)-resistant HIV-1 variants are not associated with failure of antiretroviral therapy in women without prior exposure to single-dose NVP. J Infect Dis 2014; 209:703–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Buckton AJ, Prabhu D, Motamed C, et al. . Increased detection of the HIV-1 reverse transcriptase M184V mutation using mutation-specific minority assays in a UK surveillance study suggests evidence of unrecognized transmitted drug resistance. HIV Med 2011; 12:250–4. [DOI] [PubMed] [Google Scholar]
- 31. Casadellà M, Manzardo C, Noguera-Julian M, et al. ; ADVANZ and ADVANZ-3 Investigators Clinical value of ultradeep HIV-1 genotyping and tropism testing in late presenters with advanced disease. AIDS 2015; 29:1493–504. [DOI] [PubMed] [Google Scholar]
- 32. Casadellà M, van Ham PM, Noguera-Julian M, et al. ; SPREAD Programme Primary resistance to integrase strand-transfer inhibitors in Europe. J Antimicrob Chemother 2015; 70:2885–8. [DOI] [PubMed] [Google Scholar]
- 33. Chaillon A, Nakazawa M, Wertheim JO, et al. . No substantial evidence for sexual transmission of minority HIV drug resistance mutations in men who have sex with men. J Virol 2017; 91:pii: e00769-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Charpentier C, Laureillard D, Piketty C, et al. . High frequency of integrase Q148R minority variants in HIV-infected patients naive of integrase inhibitors. AIDS 2010; 24:867–73. [DOI] [PubMed] [Google Scholar]
- 35. Cheriro W, Kiptoo M, Kikuvi G, Mining S, Emonyi W, Songok E. High prevalence of HIV low abundance drug-resistant variants in a treatment-naive population in North Rift Kenya. AIDS Res Hum Retroviruses 2015; 31:1274–7. [DOI] [PubMed] [Google Scholar]
- 36. Clutter DS, Zhou S, Varghese V, et al. . Prevalence of drug-resistant minority variants in untreated HIV-1-infected individuals with and those without transmitted drug resistance detected by sanger sequencing. J Infect Dis 2017; 216:387–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Coovadia A, Hunt G, Abrams EJ, et al. . Persistent minority K103N mutations among women exposed to single-dose nevirapine and virologic response to nonnucleoside reverse-transcriptase inhibitor-based therapy. Clin Infect Dis 2009; 48:462–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Cozzi-Lepri A, Noguera-Julian M, Di Giallonardo F, et al. ; CHAIN Minority HIV-1 Variants Working Group Low-frequency drug-resistant HIV-1 and risk of virological failure to first-line NNRTI-based ART: a multicohort European case-control study using centralized ultrasensitive 454 pyrosequencing. J Antimicrob Chemother 2015; 70:930–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Cunningham E, Chan YT, Aghaizu A, et al. . Enhanced surveillance of HIV-1 drug resistance in recently infected MSM in the UK. J Antimicrob Chemother 2017; 72:227–34. [DOI] [PubMed] [Google Scholar]
- 40. Dalmat RR, Makhsous N, Pepper GG, et al. . Limited marginal utility of deep sequencing for HIV drug resistance testing in the age of integrase inhibitors. J Clin Microbiol 2018; 56:pii: e01443-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Derache A, Iwuji CC, Baisley K, et al. . Impact of next-generation sequencing defined human immunodeficiency virus pretreatment drug resistance on virological outcomes in the ANRS 12249 treatment-as-prevention trial. Clin Infect Dis 2019; 69:207–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Ekici H, Amogne W, Aderaye G, Lindquist L, Sönnerborg A, Abdurahman S. Minority drug-resistant HIV-1 variants in treatment naive East-African and Caucasian patients detected by allele-specific real-time PCR. PLoS One 2014; 9:e111042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Ekici H, Rao SD, Sönnerborg A, Ramprasad VL, Gupta R, Neogi U. Cost-efficient HIV-1 drug resistance surveillance using multiplexed high-throughput amplicon sequencing: implications for use in low- and middle-income countries. J Antimicrob Chemother 2014; 69:3349–55. [DOI] [PubMed] [Google Scholar]
- 44. Epaulard O, Signori-Schmuck A, Larrat S, et al. . Ultradeep sequencing of B and non-B HIV-1 subtypes: viral diversity and drug resistance mutations before and after one month of antiretroviral therapy in naive patients. J Clin Virol 2017; 95:13–9. [DOI] [PubMed] [Google Scholar]
- 45. Gega A, Kozal MJ, Chiarella J, et al. . Deep sequencing of HIV-1 variants from paired plasma and cerebrospinal fluid during primary HIV infection. J Virus Erad 2015; 1:264–8. [PMC free article] [PubMed] [Google Scholar]
- 46. Geretti AM, Fox ZV, Booth CL, et al. . Low-frequency K103N strengthens the impact of transmitted drug resistance on virologic responses to first-line efavirenz or nevirapine-based highly active antiretroviral therapy. J Acquir Immune Defic Syndr 2009; 52:569–73. [DOI] [PubMed] [Google Scholar]
- 47. Gonzalez S, Tully DC, Gondwe C, Wood C. Low-abundance resistant mutations in HIV-1 subtype C antiretroviral therapy-naive individuals as revealed by pyrosequencing. Curr HIV Res 2013; 11:43–9. [PMC free article] [PubMed] [Google Scholar]
- 48. Goodman DD, Zhou Y, Margot NA, et al. . Low level of the K103N HIV-1 above a threshold is associated with virological failure in treatment-naive individuals undergoing efavirenz-containing therapy. AIDS 2011; 25:325–33. [DOI] [PubMed] [Google Scholar]
- 49. Gupta S, Lataillade M, Kyriakides TC, et al. . Low-frequency NNRTI-resistant HIV-1 variants and relationship to mutational load in antiretroviral-naive subjects. Viruses 2014; 6:3428–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Hassan AS, Bibby DF, Mwaringa SM, et al. . Presence, persistence and effects of pre-treatment HIV-1 drug resistance variants detected using next generation sequencing: a retrospective longitudinal study from rural coastal Kenya. PLoS One 2019; 14:e0210559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Hofstra LM, Sánchez Rivas E, Nijhuis M, et al. . High rates of transmission of drug-resistant HIV in Aruba resulting in reduced susceptibility to the WHO recommended first-line regimen in nearly half of newly diagnosed HIV-infected patients. Clin Infect Dis 2017; 64:1092–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Inzaule SC, Hamers RL, Noguera-Julian M, et al. . Primary resistance to integrase strand transfer inhibitors in patients infected with diverse HIV-1 subtypes in sub-Saharan Africa. J Antimicrob Chemother 2018; 73:1167–72. [DOI] [PubMed] [Google Scholar]
- 53. Inzaule SC, Hamers RL, Noguera-Julian M, et al. ; PanAfrican Studies to Evaluate Resistance Clinically relevant thresholds for ultrasensitive HIV drug resistance testing: a multi-country nested case-control study. Lancet HIV 2018; 5:e638–46. [DOI] [PubMed] [Google Scholar]
- 54. Jakobsen MR, Tolstrup M, Søgaard OS, et al. . Transmission of HIV-1 drug-resistant variants: prevalence and effect on treatment outcome. Clin Infect Dis 2010; 50:566–73. [DOI] [PubMed] [Google Scholar]
- 55. Ji H, Liang B, Li Y, et al. . Low abundance drug resistance variants in transmitted HIV drug resistance surveillance specimens identified using tagged pooled pyrosequencing. J Virol Methods 2013; 187:314–20. [DOI] [PubMed] [Google Scholar]
- 56. Johnson JA, Li JF, Morris L, et al. . Emergence of drug-resistant HIV-1 after intrapartum administration of single-dose nevirapine is substantially underestimated. J Infect Dis 2005; 192:16–23. [DOI] [PubMed] [Google Scholar]
- 57. Johnson JA, Li JF, Wei X, et al. . Minority HIV-1 drug resistance mutations are present in antiretroviral treatment-naive populations and associate with reduced treatment efficacy. PLoS Med 2008; 5:e158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Keys JR, Zhou S, Anderson JA, et al. . Primer ID informs next-generation sequencing platforms and reveals preexisting drug resistance mutations in the HIV-1 reverse transcriptase coding domain. AIDS Res Hum Retroviruses 2015; 31:658–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Kozal MJ, Chiarella J, St John EP, et al. . Prevalence of low-level HIV-1 variants with reverse transcriptase mutation K65R and the effect of antiretroviral drug exposure on variant levels. Antivir Ther 2011; 16:925–9. [DOI] [PubMed] [Google Scholar]
- 60. Lataillade M, Chiarella J, Yang R, et al. . Prevalence and clinical significance of HIV drug resistance mutations by ultra-deep sequencing in antiretroviral-naive subjects in the CASTLE study. PLoS One 2010; 5:e10952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Le Nguyen H, Pitakpolrat P, Sirivichayakul S, Delaugerre C, Ruxrungtham K. Minority HIV-1 resistant variants in recent infection and in patients who failed first-line antiretroviral therapy with no detectable resistance-associated mutations in Thailand. J Med Virol 2012; 84:713–20. [DOI] [PubMed] [Google Scholar]
- 62. Leda AR, Hunter J, Oliveira UC, Azevedo IJ, Sucupira MCA, Diaz RS. Insights about minority HIV-1 strains in transmitted drug resistance mutation dynamics and disease progression. J Antimicrob Chemother 2018; 73:1930–4. [DOI] [PubMed] [Google Scholar]
- 63. Li JF, Linley L, Kline R, Ziebell R, Heneine W, Johnson JA. Sensitive sentinel mutation screening reveals differential underestimation of transmitted HIV drug resistance among demographic groups. AIDS 2016; 30:1439–45. [DOI] [PubMed] [Google Scholar]
- 64. Li JF, Lipscomb JT, Wei X, et al. . Detection of low-level K65R variants in nucleoside reverse transcriptase inhibitor-naive chronic and acute HIV-1 subtype C infections. J Infect Dis 2011; 203:798–802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Li JZ, Paredes R, Ribaudo HJ, et al. . Low-frequency HIV-1 drug resistance mutations and risk of NNRTI-based antiretroviral treatment failure: a systematic review and pooled analysis. JAMA 2011; 305:1327–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Mbisa JL, Kirwan P, Tostevin A, et al. ; UK HIV Drug Resistance Database Determining the origins of human immunodeficiency virus type 1 drug-resistant minority variants in people who are recently infected using phylogenetic reconstruction. Clin Infect Dis 2019; 69:1136–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Mbunkah HA, Marzel A, Schmutz S, et al. . Low prevalence of transmitted HIV-1 drug resistance detected by a dried blood spot (DBS)-based next-generation sequencing (NGS) method in newly diagnosed individuals in Cameroon in the years 2015-16. J Antimicrob Chemother 2018; 73:1917–29. [DOI] [PubMed] [Google Scholar]
- 68. Messiaen P, Verhofstede C, Vandenbroucke I, et al. . Ultra-deep sequencing of HIV-1 reverse transcriptase before start of an NNRTI-based regimen in treatment-naive patients. Virology 2012; 426:7–11. [DOI] [PubMed] [Google Scholar]
- 69. Metzner KJ, Allers K, Rauch P, Harrer T. Rapid selection of drug-resistant HIV-1 during the first months of suppressive ART in treatment-naive patients. AIDS 2007; 21:703–11. [DOI] [PubMed] [Google Scholar]
- 70. Metzner KJ, Giulieri SG, Knoepfel SA, et al. . Minority quasispecies of drug-resistant HIV-1 that lead to early therapy failure in treatment-naive and -adherent patients. Clin Infect Dis 2009; 48:239–47. [DOI] [PubMed] [Google Scholar]
- 71. Metzner KJ, Rauch P, Braun P, et al. . Prevalence of key resistance mutations K65R, K103N, and M184V as minority HIV-1 variants in chronically HIV-1 infected, treatment-naive patients. J Clin Virol 2011; 50:156–61. [DOI] [PubMed] [Google Scholar]
- 72. Metzner KJ, Scherrer AU, von Wyl V, et al. ; Swiss HIV Cohort Study Limited clinical benefit of minority K103N and Y181C-variant detection in addition to routine genotypic resistance testing in antiretroviral therapy-naive patients. AIDS 2014; 28:2231–9. [DOI] [PubMed] [Google Scholar]
- 73. Milne RS, Silverman RA, Beck IA, et al. . Minority and majority pretreatment HIV-1 drug resistance associated with failure of first-line nonnucleoside reverse-transcriptase inhibitor antiretroviral therapy in Kenyan women. AIDS 2019; 33:941–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Mitsuya Y, Varghese V, Wang C, et al. . Minority human immunodeficiency virus type 1 variants in antiretroviral-naive persons with reverse transcriptase codon 215 revertant mutations. J Virol 2008; 82:10747–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Mohamed S, Ravet S, Camus C, Khiri H, Olive D, Halfon P. Clinical and analytical relevance of NNRTIs minority mutations on viral failure in HIV-1 infected patients. J Med Virol 2014; 86:394–403. [DOI] [PubMed] [Google Scholar]
- 76. Moscona R, Ram D, Wax M, et al. . Comparison between next-generation and Sanger-based sequencing for the detection of transmitted drug-resistance mutations among recently infected HIV-1 patients in Israel, 2000–2014. J Int AIDS Soc 2017; 20:21846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Mzingwane ML, Tiemessen CT, Richter KL, Mayaphi SH, Hunt G, Bowyer SM. Pre-treatment minority HIV-1 drug resistance mutations and long term virological outcomes: is prediction possible? Virol J 2016; 13:170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Nanfack AJ, Redd AD, Bimela JS, et al. . Multimethod longitudinal HIV drug resistance analysis in antiretroviral-therapy-naive patients. J Clin Microbiol 2017; 55:2785–800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Neogi U, Sonnerborg A. Minor viral population with drug-resistant mutation and risk of persistent low-level viremia or ‘blips’ in HIV-1 subtype C. AIDS 2014; 28:2635–6. [DOI] [PubMed] [Google Scholar]
- 80. Nicot F, Sauné K, Raymond S, et al. . Minority resistant HIV-1 variants and the response to first-line NNRTI therapy. J Clin Virol 2015; 62:20–4. [DOI] [PubMed] [Google Scholar]
- 81. Nishizawa M, Hattori J, Shiino T, et al. . Highly-sensitive allele-specific PCR testing identifies a greater prevalence of transmitted HIV drug resistance in Japan. PLoS One 2013; 8:e83150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Ode H, Matsuda M, Matsuoka K, et al. . Quasispecies analyses of the HIV-1 near-full-length genome with Illumina MiSeq. Front Microbiol 2015; 6:1258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Paolucci S, Baldanti F, Campanini G, et al. . Analysis of HIV drug-resistant quasispecies in plasma, peripheral blood mononuclear cells and viral isolates from treatment-naive and HAART patients. J Med Virol 2001; 65:207–17. [DOI] [PubMed] [Google Scholar]
- 84. Paredes R, Lalama CM, Ribaudo HJ, et al. ; AIDS Clinical Trials Group (ACTG) A5095 Study Team Pre-existing minority drug-resistant HIV-1 variants, adherence, and risk of antiretroviral treatment failure. J Infect Dis 2010; 201:662–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Perrier M, Visseaux B, Landman R, et al. . No impact of HIV-1 protease minority resistant variants on the virological response to a first-line PI-based regimen containing darunavir or atazanavir. J Antimicrob Chemother 2018; 73:173–6. [DOI] [PubMed] [Google Scholar]
- 86. Pessôa R, Sanabani SS. High prevalence of HIV-1 transmitted drug-resistance mutations from proviral DNA massively parallel sequencing data of therapy-naive chronically infected Brazilian blood donors. PLoS One 2017; 12:e0185559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Pingen M, van der Ende ME, Wensing AM, et al. . Deep sequencing does not reveal additional transmitted mutations in patients diagnosed with HIV-1 variants with single nucleoside reverse transcriptase inhibitor resistance mutations. HIV Med 2013; 14:176–81. [DOI] [PubMed] [Google Scholar]
- 88. Porter DP, Daeumer M, Thielen A, et al. . Emergent HIV-1 drug resistance mutations were not present at low-frequency at baseline in non-nucleoside reverse transcriptase inhibitor-treated subjects in the STaR Study. Viruses 2015; 7:6360–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Raymond S, Nicot F, Pallier C, et al. ; French National Agency for Research on AIDS and Viral Hepatitis (ANRS) AC11 Resistance Study Group Impact of human immunodeficiency virus type 1 minority variants on the virus response to a rilpivirine-based first-line regimen. Clin Infect Dis 2018; 66:1588–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Ross LL, Weinberg WG, DeJesus E, et al. . Impact of low abundance HIV variants on response to ritonavir-boosted atazanavir or fosamprenavir given once daily with tenofovir/emtricitabine in antiretroviral-naive HIV-infected patients. AIDS Res Hum Retroviruses 2010; 26:407–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. SahBandar IN, Samonte G, Telan E, et al. . Ultra-deep sequencing analysis on HIV drug-resistance-associated mutations among HIV-infected individuals: first report from the Philippines. AIDS Res Hum Retroviruses 2017; 33:1099–106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Sili U, Aksu B, Tekin A, Hasdemir U, Soyletir G, Korten V. Assessment of transmitted HIV-1 drug resistance mutations using ultra-deep pyrosequencing in a Turkish Cohort. Curr HIV Res 2018; 16:216–21. [DOI] [PubMed] [Google Scholar]
- 93. Silver N, Paynter M, McAllister G, et al. . Characterization of minority HIV-1 drug resistant variants in the United Kingdom following the verification of a deep sequencing-based HIV-1 genotyping and tropism assay. AIDS Res Ther 2018; 15:18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Simen BB, Braverman MS, Abbate I, et al. ; 454 HIV Alphastudy Group An international multicenter study on HIV-1 drug resistance testing by 454 ultra-deep pyrosequencing. J Virol Methods 2014; 204:31–7. [DOI] [PubMed] [Google Scholar]
- 95. Stekler JD, Ellis GM, Carlsson J, et al. . Prevalence and impact of minority variant drug resistance mutations in primary HIV-1 infection. PLoS One 2011; 6:e28952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Su B, Zheng X, Liu Y, et al. . Detection of pretreatment minority HIV-1 reverse transcriptase inhibitor-resistant variants by ultra-deep sequencing has a limited impact on virological outcomes. J Antimicrob Chemother 2019; 74:1408–16. [DOI] [PubMed] [Google Scholar]
- 97. Telele NF, Kalu AW, Gebre-Selassie S, et al. . Pretreatment drug resistance in a large countrywide Ethiopian HIV-1C cohort: a comparison of Sanger and high-throughput sequencing. Sci Rep 2018; 8:7556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98. Todesco E, Charpentier C, Bertine M, et al. . Disparities in HIV-1 transmitted drug resistance detected by ultradeep sequencing between men who have sex with men and heterosexual populations. HIV Med 2017; 18:696–700. [DOI] [PubMed] [Google Scholar]
- 99. Van Eygen V, Thys K, Van Hove C, et al. . Deep sequencing analysis of HIV-1 reverse transcriptase at baseline and time of failure in patients receiving rilpivirine in the phase III studies ECHO and THRIVE. J Med Virol 2016; 88:798–806. [DOI] [PubMed] [Google Scholar]
- 100. Van Laethem K, De Munter P, Schrooten Y, et al. . No response to first-line tenofovir+lamivudine+efavirenz despite optimization according to baseline resistance testing: impact of resistant minority variants on efficacy of low genetic barrier drugs. J Clin Virol 2007; 39:43–7. [DOI] [PubMed] [Google Scholar]
- 101. Varghese V, Shahriar R, Rhee SY, et al. . Minority variants associated with transmitted and acquired HIV-1 nonnucleoside reverse transcriptase inhibitor resistance: implications for the use of second-generation nonnucleoside reverse transcriptase inhibitors. J Acquir Immune Defic Syndr 2009; 52:309–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102. Wang B, Dwyer DE, Chew CB, et al. . Sensitive detection of the K103N non-nucleoside reverse transcriptase inhibitor resistance mutation in treatment-naive HIV-1 infected individuals by rolling circle amplification. J Virol Methods 2009; 161:128–35. [DOI] [PubMed] [Google Scholar]
- 103. Xiaobai Z, Xi C, Tian H, et al. . Prevalence of WHO transmitted drug resistance mutations by deep sequencing in antiretroviral-naive subjects in Hunan Province, China. PLoS One 2014; 9:e98740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104. Zhou Z, Tang K, Zhang G, et al. . Detection of minority drug resistant mutations in Malawian HIV-1 subtype C-positive patients initiating and on first-line antiretroviral therapy. Afr J Lab Med 2018; 7:708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105. Zoufaly A, Jochum J, Hammerl R, et al. . Virological failure after 1 year of first-line ART is not associated with HIV minority drug resistance in rural Cameroon. J Antimicrob Chemother 2015; 70:922–5. [DOI] [PubMed] [Google Scholar]
- 106. Gianella S, Richman DD. Minority variants of drug-resistant HIV. J Infect Dis 2010; 202:657–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107. Beerenwinkel N, Günthard HF, Roth V, Metzner KJ. Challenges and opportunities in estimating viral genetic diversity from next-generation sequencing data. Front Microbiol 2012; 3:329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108. Di Giallonardo F, Zagordi O, Duport Y, et al. . Next-generation sequencing of HIV-1 RNA genomes: determination of error rates and minimizing artificial recombination. PLoS One 2013; 8:e74249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109. Jabara CB, Jones CD, Roach J, Anderson JA, Swanstrom R. Accurate sampling and deep sequencing of the HIV-1 protease gene using a primer ID. Proc Natl Acad Sci U S A 2011; 108:20166–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110. Seifert D, Di Giallonardo F, Töpfer A, et al. . A comprehensive analysis of primer IDs to study heterogeneous HIV-1 populations. J Mol Biol 2016; 428:238–50. [DOI] [PubMed] [Google Scholar]
- 111. Huber M, Metzner KJ, Geissberger FD, et al. . MinVar: A rapid and versatile tool for HIV-1 drug resistance genotyping by deep sequencing. J Virol Methods 2017; 240:7–13. [DOI] [PubMed] [Google Scholar]
- 112. St John EP, Simen BB, Turenchalk GS, et al. ; 454 HIV-1 Alpha Study Group A follow-up of the multicenter collaborative study on HIV-1 drug resistance and tropism testing using 454 ultra deep pyrosequencing. PLoS One 2016; 11:e0146687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113. World Bank. World Bank country and lending groups, June 2019 https://datahelpdesk.worldbank.org/knowledgebase/articles/906519-world-bank-country-and-lending-groups. Accessed 28 July 2019.
- 114. von Elm E, Altman DG, Egger M, Pocock SJ, Gøtzsche PC, Vandenbroucke JP; STROBE Initiative The Strengthening the Reporting of Observational Studies in Epidemiology (STROBE) statement: guidelines for reporting observational studies. Lancet 2007; 370:1453–7. [DOI] [PubMed] [Google Scholar]
- 115. Simen BB, Simons JF, Hullsiek KH, et al. ; Terry Beirn Community Programs for Clinical Research on AIDS Low-abundance drug-resistant viral variants in chronically HIV-infected, antiretroviral treatment-naive patients significantly impact treatment outcomes. J Infect Dis 2009; 199:693–701. [DOI] [PubMed] [Google Scholar]
- 116. Gunthard HF, Calvez V, Paredes R, et al. . Human immunodeficiency virus drug resistance: 2018 recommendations of the International Antiviral Society-USA Panel. Clin Infect Dis 2019;68:177–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117. Ji H, Enns E, Brumme CJ, et al. . Bioinformatic data processing pipelines in support of next-generation sequencing-based HIV drug resistance testing: the Winnipeg Consensus. J Int AIDS Soc 2018; 21:e25193. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.