SUMMARY
Reactive oxygen species (ROS) play critical roles in metabolism and disease, yet a comprehensive analysis of the cellular response to oxidative stress is lacking. To systematically identify regulators of oxidative stress, we conducted genome-wide Cas9/CRISPR and shRNA screens. This revealed a detailed picture of diverse pathways that control oxidative stress response, ranging from the TCA cycle and DNA repair machineries to iron transport, trafficking, and metabolism. Paradoxically, disrupting the pentose phosphate pathway (PPP) at the level of phosphogluconate dehydrogenase (PGD) protects cells against ROS. This dramatically alters metabolites in the PPP, consistent with rewiring of upper glycolysis to promote antioxidant production. In addition, disruption of peroxisomal import unexpectedly increases resistance to oxidative stress by altering the localization of catalase. Together, these studies provide insights into the roles of peroxisomal matrix import and the PPP in redox biology and represent a rich resource for understanding the cellular response to oxidative stress.
Graphical Abstract

In Brief
Despite its importance in metabolism and disease, a comprehensive analysis of the cellular response to oxidative stress is lacking. Here, Dubreuil et al. use genome-wide screens to identify cellular regulators of oxidative stress. They investigate paradoxical mechanisms by which disruption of the pentose phosphate and peroxisomal import pathways protect cells.
INTRODUCTION
Oxidative stress has diverse deleterious effects and can lead to tumorigenesis, cell death, neurological disease, and aging (Busciglio and Yankner, 1995; Conger and Fairchild, 1952; Cunningham et al., 1987; Finkel and Holbrook, 2000; Guo et al., 2011; Ishii et al., 2005; Liochev, 2013; Nagai et al., 2009; Sakurai et al., 2008; Totter, 1980; Wu et al., 2003). Conversely, reactive oxygen species (ROS) also have normal physiological roles and can promote autophagy (Chen et al., 2009; Scherz-Shouval et al., 2007) as well as signal proliferation and survival by activating various MAPK proteins (Ichijo et al., 1997; Matsuzawa et al., 2005; Meng et al., 2002; Ray et al., 2012). Diverse antioxidant systems help the cell maintain a redox environment permissive to normal metabolism and ROS signaling while preventing toxic ROS accumulation (Go and Jones, 2008). These systems include antioxidants such as vitamin C, reducing molecules such as NADPH and glutathione and antioxidant enzymes such as superoxide dismutase (SOD) and catalase. However, under conditions of metabolic or environmental stress, these mechanisms can be insufficient, and ROS levels can increase and cause DNA damage, protein dysfunction, and lipid oxidation (Kong and Chandel, 2018; Nathan and Cunningham-Bussel, 2013; Schieber and Chandel, 2014). Though a number of studies have begun to uncover the genetic effectors of ROS toxicity using model organisms and targeted screens in mammalian cells (Ayer et al., 2012; Kimura et al., 2008; Reczek et al., 2017; Ueno et al., 2012), much remains to be discovered, and a comprehensive screen in mammalian cells has not been performed.
Hydrogen peroxide (H2O2) is a ubiquitous ROS in biological systems. Endogenously, H2O2 is produced as a by-product of oxidative metabolism in peroxisomes and mitochondria and is converted from superoxide anion by SOD. Less reactive and longer lived than superoxide anion, H2O2 often acts as a membrane-permeable signaling molecule, promoting autophagy, growth, and survival in various contexts, including cancer (Moloney and Cotter, 2018). However, at higher concentrations, H2O2 can induce apoptosis and senescence as well as oxidative damage to proteins, lipids, and DNA (Kuehne et al., 2015; Nathan and Cunningham-Bussel, 2013; Nagai et al., 2009; de Oliveira et al., 2014; Pillai et al., 2005; Schuster and Feldstein, 2017; Sekine et al., 2012; Varani and Ward, 1994). H2O2 concentrations vary greatly in the human body. Though there is some disagreement regarding the level of H2O2 in blood and plasma, H2O2 levels have been found in the low micromolar range (Forman et al., 2016; Go and Jones, 2008; Roberts et al., 2005). H2O2 concentrations of 5–15 μM have been measured at sites of inflammation, which can induce oxidative stress in proximal cells (Buchmeier et al., 1995; Forman and Torres, 2002; Liu and Zweier, 2001; Test and Weiss, 1984; Varani and Ward, 1994; Weiss, 1980). Furthermore, UV radiation induces production of superoxide anion and H2O2 in melanocytes, creating localized H2O2 concentrations up to 1 mM in individuals with pigment deficiencies (Denat et al., 2014; Maresca et al., 1997; Schallreuter et al., 1999, 2012; Song et al., 2009). In addition, H2O2 levels have been shown to exceed 100 μM in human urine and are thought to fluctuate along the digestive tract (Go and Jones, 2008; Long and Halliwell, 2000; Long et al., 1999; Varma and Devamanoharan, 1990). Tumor cells are also known to produce high levels of ROS, although they typically upregulate antioxidant activity to counter increased ROS levels (Cairns et al., 2011; Szatrowski and Nathan, 1991). H2O2 thus represents an archetypical ROS that requires delicate control to maintain essential redox signaling without incurring cellular oxidative damage.
H2O2 toxicity is mediated by free (labile) iron or other transition metals, which decompose H2O2 into the highly reactive and damaging hydroxyl radical via the Fenton reaction (Halliwell and Gutteridge, 1990; Halliwell et al., 2000; Ueda et al., 1996). Iron is transported into cells via clathrin-mediated endocytosis of the transferrin receptor (TFRC), which binds to iron-bound transferrin. Labile iron is released in the early endosome (85%–95% of iron uptake) or when TFRC is degraded in the lysosome (5%–15% of iron uptake) (Tortorella and Karagiannis, 2014). The majority of labile iron is found in the lysosome, which is also responsible for the breakdown of the iron storage protein ferritin. Likely because of this high local free iron concentration, lysosomal membranes are sensitive to oxidative damage-induced permeabilization, which can initiate apoptosis (Turk and Turk, 2009). Thus, the transport and metabolism of iron are a key point of regulation for oxidative stress.
One of the first responses for a cell undergoing oxidative stress is to upregulate the pentose phosphate pathway (PPP) to increase production of the reducing agent NADPH (Kuehne et al., 2015). The PPP branches off of glycolysis, metabolizing glucose-6-phosphate to reduce NADP+ into NADPH and create purine nucleotide precursors (Patra and Hay, 2014). NADPH is a reducing agent that maintains the antioxidant capacities of the cell, including the glutathione antioxidant system and catalase (Kirkman et al., 1999; Lu, 2009, 2013). In some cells, the PPP acts as a first line of defense against ROS by increasing glucose flux through the oxidative branch of the PPP (Kuehne et al., 2015). Specifically, the metabolite 6-phosphogluconate (6PG), the substrate of phosphogluconate dehydrogenase (PGD), inhibits carbon flux through glycolysis, increasing carbon flux through the PPP (Parr, 1956). This inhibition of glycolysis quickly redirects the glucose flux of the cell into the oxidative PPP, producing NADPH, until the oxidative threat subsides. Thus, the PPP represents a first line of defense for the cell when undergoing oxidative stress.
Integral to the regulation of cellular redox balance, peroxisomes are single-membrane organelles that both detoxify and produce intracellular H2O2. Peroxisomes are centers for plasmalogen biosynthesis and the catabolism of diverse substrates, including fatty acids, select amino acids, xenobiotics, and glyoxylate (Schrader and Fahimi, 2006). These reactions, in particular the β-oxidation of fatty acids, produce H2O2. Peroxisomes also act as centers of antioxidant activity to detoxify these ROS. The majority of peroxisomal matrix proteins are imported into peroxisomes through the peroxisome targeting signal type-1 (PTS1) localization domain, which is recognized by PEX5, a cytosolic receptor that coordinates functions with eight other PEX proteins to import cargo and cycle back to the cytoplasm (Bottger et al., 2000; Fujiki et al., 2014; Harper et al., 2003; Miyazawa et al., 1989; Stanley et al., 2006). Catalase is key substrate of this pathway localized inside of peroxisomes, where it reduces H2O2 into water at high efficiency, potentially converting millions of H2O2 molecules per second under the proper conditions, thereby preventing ROS from escaping the peroxisome (Otera and Fujiki, 2012; Young and Woodside, 2001). Thus, while peroxisomes are a significant source of intracellular ROS, they also have substantial antioxidant roles in cells.
Despite the critical importance of oxidative stress, a comprehensive study of the genetic factors regulating the response in mammalian cells is lacking. Here, we use CRISPR/Cas9 knockout (KO) and short hairpin RNA (shRNA) knockdown screens (Barbie et al., 2009; Bassik et al., 2013; Koike-Yusa et al., 2014; Shalem et al., 2015; Silva et al., 2008; Wang et al., 2014; Zhou et al., 2014) to systematically identify the genetic factors that modulate cellular sensitivity to exogenous H2O2. We then validate these hits using targeted CRISPR-KO and CRISPRi libraries in HeLa and K562 cells, demonstrating conserved function across cell lines for a broad set of regulators of oxidative stress sensitivity. These screens identified several pathways with paradoxical ROS-sensitivity phenotypes that were inconsistent with their expected roles. In particular, we demonstrate that perturbation of the peroxisomal matrix protein import pathway protects cells from oxidative stress by causing the accumulation of catalase in the cytoplasm. We also investigate the function of PGD, a key member of the PPP; surprisingly, KO of this gene is highly protective against ROS and results in a dramatic increase of upstream PPP metabolites. Together, these results provide unexpected insights into the roles of peroxisomal matrix import pathway and the PPP in redox biology and a systematic and rich resource for studying the cellular response to oxidative stress.
RESULTS
Parallel Genome-wide CRISPR and shRNA Screens Identify Genetic Effectors of Cellular Sensitivity to Oxidative Stress
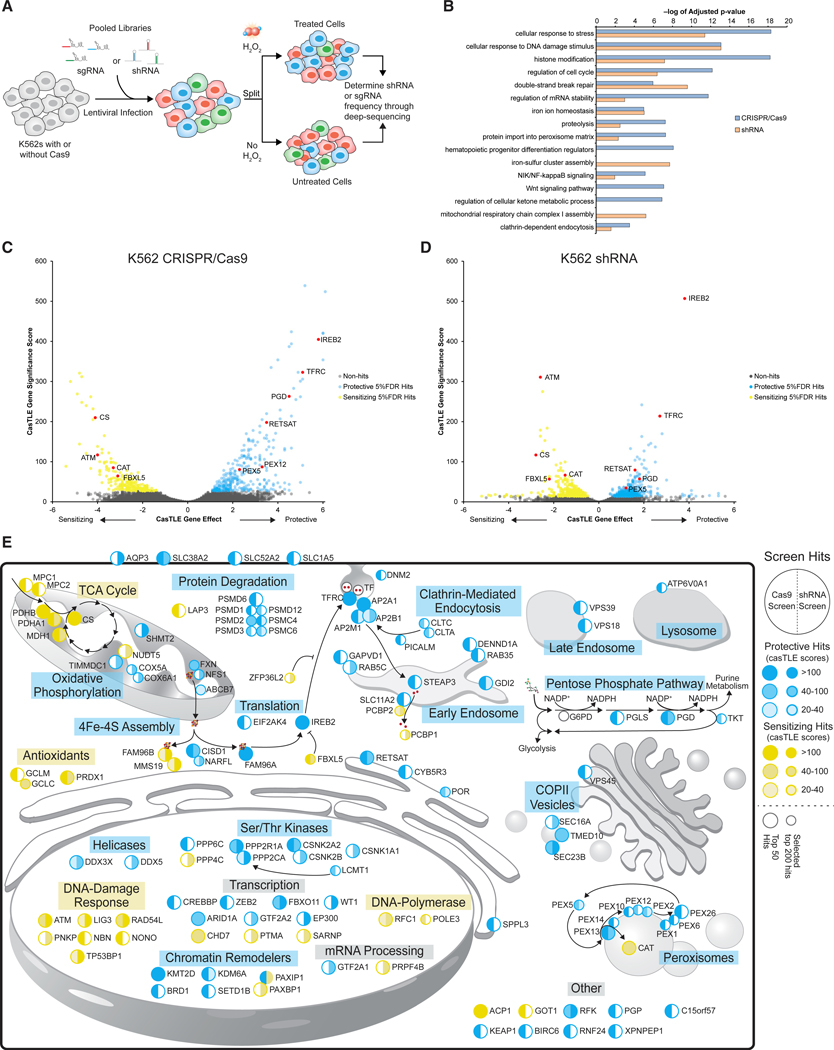
To systematically identify the genetic factors that modulate cellular sensitivity to oxidative stress, we performed genome-wide knockdown shRNA and KO single guide RNA (sgRNA)/Cas9 screens in K562 chronic myelogenous leukemia cells in the presence of H2O2. We infected K562 cells with genome-wide shRNA (Deans et al., 2016; Kampmann et al., 2015) or CRISPR/Cas9 (Morgens et al., 2017) libraries, then passaged the cells untreated or treated them with a 50% lethal concentration (LC50) dose of H2O2 repeatedly (allowing for recovery) (Figure 1A). In this manner, protective genes were enriched in the treated conditions relative to negative control cells, while sensitizing genes were depleted compared with negative control cells. The frequencies of the shRNA and sgRNA elements in each screening population were measured using high-throughput sequencing (Figure 1A) and analyzed using our previously published Cas9 high-throughput maximum likelihood estimator (casTLE) (Morgens et al., 2016). Using a 5% false discovery rate (FDR), we identified 511 significant gene hits with the CRISPR/Cas9 screen and 1,250 significant gene hits with the shRNA library (Figures 1C and 1D; Figures S1A and S1B; Table S1).
Figure 1. Parallel Genome-wide CRISPR and shRNA Screens Identify Genetic Effectors of Cellular Sensitivity to Oxidative Stress.
(A) Screening schematic for sgRNA and shRNA screens.
(B) GO term analysis for hits passing 5% FDR from genome-wide Cas9 knockout and genome-wide shRNA knockdown screens.
(D) Volcano plot showing casTLE analysis of independent K562 genome-wide Cas9 knockout screens (n = 2). Protective genes at 5% FDR cutoff are colored in blue, sensitizing genes at 5% FDR cutoff in yellow, and genes chosen from key pathways in red.
(E) Cell diagram showing the 50 most significant hits from shRNA and sgRNA genome-wide screens. Large circles represent hits in either screen’s top 50 most significant hits, and smaller circles represent relevant genes outside of the top 50 in either screen. Protective hits are shown in blue and sensitizing hits in yellow. Shades of colors are based on casTLE scores.
We and others have shown that KO and knockdown screen technologies often reveal distinct gene signatures because of differences in strength of on-target activity, off-target effects, efficacies of gene-targeting elements between libraries, compensatory mechanisms, and the ability to target essential genes (Barrangou et al., 2015; El-Brolosy et al., 2019; Frock et al., 2015; Grimm et al., 2006; Jackson and Linsley, 2010; Kaelin, 2012; Ma et al., 2019; Morgens et al., 2017; Pruett-Miller et al., 2009; Tsai et al., 2015; Weiss, 1980). By using both approaches, the genetic modifiers of a cellular phenomenon can be more comprehensively identified than if either approach was used alone (Morgens et al., 2016; Rosenbluh et al., 2017). Here we found that shRNA or CRISPR screens identified genes enriched in a range of pathways as measured by Gene Ontology (GO) term analysis, some of which were unique to a particular perturbation technology and some of which were shared by both (Figure 1B). Interestingly, the CRISPR/Cas9 screen had stronger signatures for cell cycle regulators, histone modifiers, members of the Wnt signaling pathway, and the peroxisomal matrix import pathway. In contrast, the shRNA screen identified regulators of iron-sulfur cluster assembly and mitochondrial respiratory chain complex I assembly genes.
These screens identified both known and unexpected genetic modifiers of cellular sensitivity to oxidative stress (Figure 1E). We found that perturbation of many of the known antioxidant systems sensitized cells to H2O2, including catalase (CAT), peroxiredoxin 1 (PRDX1), and the glutamate-cysteine ligase complex (GCLC and GCLM), which provides glutathione for the glutathione peroxidases. Disruption of genes involved in the cellular response to DNA damage, including ATM, TP53PB1, and MMS19, among others, was sensitizing to H2O2 (Figure 1B), consistent with previous observations that H2O2 can cause DNA damage and activate the ATM pathway (Driessens et al., 2009; Guo et al., 2010; Moiseeva et al., 2016). Furthermore, we also found that deletion of KEAP1 was protective, consistent with its role in negatively regulating NRF2, an oncogenic transcription factor that drives expression of antioxidant and prosurvival genes (Blake et al., 2010; DeNicola et al., 2011; Kobayashi et al., 2004).
Interestingly, both screening methods detected strong effects for genes involved in all aspects of cellular iron homeostasis (Figure 1E; Figure S1C). Iron can exacerbate the damaging effects of ROS by acting as a catalyst for the Fenton reaction, and thus, cells with less iron are more resistant to oxidative stress (Mantzaris et al., 2016; Mello Filho et al., 1984; Meneghini, 1997; Nishida, 2012). Indeed, one of the strongest protective hits was IREB2, a central iron homeostasis gene that regulates the translation of mRNAs that promote iron uptake and downregulates the translation of mRNAs regulating iron export and storage. Disrupting the expression of IREB2 would recapitulate high-iron conditions and decrease iron levels in the cell. In contrast, FBXL5 and ZFP36L2, which under high-iron conditions degrade IREB2 and TFRC mRNA, respectively, were both sensitizing hits in the screens (Patial et al., 2016; Vashisht et al., 2009). We also detected TFRC itself as a strongly protective hit, as well as the genes involved in clathrin-mediated endocytosis, the mechanism by which TFRC brings iron into the cell. These genes include members of the AP2 clathrin adapter complex, CLTC, CLTH, DNM2, PICALM, and RAB5C (Figure S1C; Gulbranson et al., 2019; Loerke et al., 2009; Tortorella and Karagiannis, 2014). Once TFRC is trafficked to the early endosome and releases transferrin and Fe3+, STEAP3 reduces Fe3+ to Fe2+, which is then transported out of the early endosomes by SLC11A2 (DMT2) as labile iron to iron chaperone PCBP2 (Bogdan et al., 2016). Disruption of both STEAP3 and SLC11A2 were protective against H2O2 toxicity.
Consistent with the toxic potential of intracellular labile iron, loss of the labile iron chaperones PCBP1 and PCBP2 had sensitizing effects. Both chaperones mediate direct metallation of non-heme-binding proteins and storage of labile iron in its non-toxic form within ferritin complexes (Bogdan et al., 2016; Leidgens et al., 2013; Shi et al., 2008; Yanatori et al., 2016). Finally, perturbation of genes involved in the assembly and formation of Fe/S clusters in the mitochondria (FXN, NFS1, ISCU, LYRM4, and ABCB7) were protective. Once Fe/S clusters are formed in the mitochondria, they are chaperoned by either FAM96A (CIA2A) or FAM96B (CIA2B) to distinct sets of proteins with differing effects on H2O2 toxicity. FAM96A, along with CISD1 and NARFL, had protective effects when perturbed and has been shown to directly bind to IREB2, potentially stabilizing its expression (Alvarez et al., 2017; Stehling et al., 2013). On the other hand, FAM96B and MMS19 negatively regulate IREB2 activity and chaperone Fe/S clusters to DNA damage repair proteins and enhanced H2O2 toxicity when perturbed in the genome-wide screens. Together, these results reveal a comprehensive picture of the diverse mechanisms of iron transport and metabolism that control cellular sensitivity to oxidative stress.
We also detected a number of seemingly paradoxical signatures. For example, although mitochondrial metabolism has been shown to be involved in the generation of ROS, it was somewhat surprising that our screens identify both sensitizing (tricarboxylic acid [TCA] cycle) and protective hits (mitochondrial complex I). Particularly striking among these pathways were protective effects observed upon deletion of genes involved in the peroxisomal import pathway and the PPP. Despite the strong antioxidant activity present in peroxisomes and their central role in redox regulation, our screen revealed that deletion of every member of the peroxisomal import pathway was highly protective against oxidative stress. Additionally, although the PPP is a significant source of reducing power, we found that perturbing the PPP at the level of PGD resulted in strong protection against oxidative stress. Given their prominent and puzzling effects, we further investigate the mechanisms of oxidative stress protection from perturbation of the peroxisomal import pathway and PGD below.
Hits from Genome-wide Screens Validate in Competitive Growth Assays and in Multiple Cell Lines
To further investigate the genome-wide screen hits in high throughput, we first performed batch retest screens. We and others have shown that smaller “batch” screens performed at higher coverage (cells/sgRNA) can reduce false positives and false negatives (Bassik et al., 2013; Han et al., 2017; Haney et al., 2018; Parnas et al., 2015). In this case, we sought to conduct batch retest screens not only with higher coverage but in additional cell types and using orthogonal perturbations in order to highlight robust hits for further investigation.
To do this, we designed a batch retest library designed to target the 511 hits passing 5% FDR from our genome-wide CRISPR screen, 282 manually identified genes from the literature that have been previously shown to be involved in redox biology or are related to pathways identified in the screens, and additional high-confidence genes from the genome-wide CRISPR screen, for a total of 1,000 genes (Figure S2A; Table S2). This set of 1,000 genes was used to generate both CRISPR KO libraries (Table S2) as well as knockdown libraries using CRISPRi (dCas9-KRAB; Table S2) both with ten sgRNAs per gene (Horlbeck et al., 2016; Morgens et al., 2017). CRISPR/Cas9 KO and CRISPRi/dCas9-KRAB knockdown K562 and HeLa cell lines were then lentivirally infected with the two batch retest libraries. After selection, the four libraries were pulsed four times with LC50 doses of H2O2, 30 μM and 300 μM for K562 and HeLa, respectively. After selection, libraries were prepared, sequenced, and analyzed as described above for the genome-wide screens.
The K562 batch KO screen correlated well with the genome-wide screen (r2 = 0.65), providing further validation of our initial genome-wide screen results (Figure 2B). Comparing results across cell lines, H2O2 doses, and CRISPR technologies, we found that genes representing major pathways showed consistent effects in both K562 and HeLa cells. Genes involved in iron homeostasis, vesicle trafficking, and the peroxisomal matrix import pathway all had protective effects across screens, while genes involved in the DNA damage response, the Krebs cycle, and antioxidants had sensitizing phenotypes across cell lines (Figure 2A; Figures S2B–S2E; Table S3). As expected for two different cell types, certain genetic pathways showed differences between HeLa and K562 cells. For example, although perturbation of genes involved in flavin metabolism and cytochrome P450 activity were protective in all cases, their magnitude of significance and effect was much greater in HeLa cells (Table S3). Nonetheless, our focused validation screens suggest that the genetic pathways implicated in oxidative stress from the genome-wide screens, including the seemingly paradoxical roles identified for the peroxisomal import and PPPs, are not specific to K562 cells and could be observed with multiple perturbation technologies.
Figure 2. Hits from Genome-wide Screens Validate in Competitive Growth Assays and in Multiple Cell Lines.
(A) CasTLE effect sizes for secondary Cas9 knockout and CRISPRi screens in K562 and HeLa cells ± 95% credible interval. Blue circle, K562 Cas9-KO; green circle, K562 CRISPRi; red square, HeLa Cas9-KO; yellow square, HeLa CRISPRi. Genes shown represent key hit pathways.
(B) Comparison of secondary screen and genome-wide Cas9 knockout screens in K562 (r2 = 0.65). Genes chosen from key pathways are in red.
(C) Results from sgRNA competitive growth assays. Bars represent mean across experiments (n = 3 per sgRNA) ± SD across sgRNAs. Dark blue dots show fold enrichment for individual experiments of one sgRNA, while light blue dots show fold enrichment for individual experiments for another sgRNA. **p < 0.01 and ***p < 0.001, by Student’s t test.
(D) Results from shRNA competitive growth assays. Bars represent mean across experiments (n = 3 per shRNA) ± SD across shRNAs. Dark orange dots show fold enrichment for individual experiments of one shRNA, while light orange dots show fold enrichment for individual experiments for another shRNA. *p < 0.05, **p <0.01, and ***p < 0.001, by Student’s t test.
To further confirm our screen results, we validated select hits from the genome-wide screen using competitive growth assays with individual sgRNAs. We mixed unlabeled control cells with fluorescently labeled cells expressing shRNAs or CRISPR sgRNAs targeting hits identified in the screen (Figure S2F). Using two sgRNAs or shRNAs per gene, we observed statistically significant protective and sensitizing effects for hits involved in iron homeostasis, iron trafficking, antioxidant activity, peroxisomal matrix import, and the PPP (Figures 2C and 2D), all of which corresponded to their phenotypes in the primary genome-wide and batch retest screens.
Disruption of the Peroxisomal Import Pathway Leads to Cytosolic Localization of Catalase and Protection against ROS
Strikingly, our results revealed that disruption of every peroxisomal matrix protein import pathway gene was protective against oxidative stress (PEX5, PEX13, PEX14, PEX2, PEX12, PEX10, PEX1, PEX6, and PEX26) (Figure 3A), despite the role of the peroxisome as a center for reducing activity. At the same time, we identified no peroxisomally localized proteins (other than the PEX genes themselves) whose loss of function could explain the observed effect, with the notable exception of catalase. Catalase, deletion of which was highly sensitizing, is imported into the peroxisome by the PEX5 pathway, where it detoxifies H2O2. Thus, we hypothesized that perturbation of the peroxisomal matrix protein import pathway causes localization of catalase to the cytoplasm, where it may more effectively detoxify cellular ROS.
Figure 3. Disruption of the Peroxisomal Import Pathway Leads to Cytosolic Localization of Catalase and Protection against ROS.
(A) Diagram showing peroxisomal matrix protein import pathway hits.
(B) Fold enrichment of knockout cells over control cells. Bars represent mean across experiments (n = 3) ± SE. **p < 0.01 and ***p < 0.001, by Student’s t test. Results from K562 cells are shown in blue and results from HeLa cells in red.
(C) Digitonin-mediated subcellular fractionation assay using negative control and PEX5-KO HeLa cells showing that catalase is enriched in the cytoplasm in PEX5-KO cells. Cells were fractionated into whole-cell extract (WCE), cytoplasm (C), and peroxisome-enriched pellet (P). LDH was immunoblotted as a cytoplasm control, and peroxisomal membrane proteins PEX13 and PEX14 were stained as peroxisomal controls. Peroxisomal matrix proteins with PTS1, such as ACOX1 and LONP2, were also immunoblotted. Only the B-chain of ACOX1 was shown in immunoblot with anti-ACOX1 antibody.
(D) Digitonin enzymatic release assay experimental diagram. Intact membranes are depicted with solid lines, and digitonin-permeabilized membranes are shown with dotted lines.
(E) Digitonin enzymatic release assay showing mean catalase enzymatic activity released (n = 3 for control and PEX5-KO, n = 2 for PEX12-KO) ± SE in HeLa cells. Curve represents hill equation regression modeling of data with r2 values for fit to data as well as the half maximal effective concentration (EC50) of digitonin for each cell line on the basis of fitted line.
(F) Representative immunofluorescence microscopy images for two negative control HeLa cell lines and HeLa cells expressing PEX5 and PEX12 sgRNAs. Cells were stained with DAPI to stain nuclei, PMP70 antibodies to stain peroxisomal membranes, and catalase antibodies. Scale bar represents 10 μm.
(G) Normalized area under the curve (A.U.C.) lethal fraction scores showing cell death of WT (gray) and CAT-KO (blue) cells expressing control and PEX sgRNAs after 48 h treated with 300 μM H2O2. Bars represent mean across experiments (n = 8) ± SE. NS, nonsignificant (p > 0.05); ***p < 0.001 (comparisons with negative control sgRNAs by Student’s t test).
To test whether perturbation of the peroxisomal matrix protein import pathway causes localization of catalase to the cytoplasm, we first created clonal KO lines of PEX5 and PEX12 and confirmed their protective effects in competitive growth assays (Figure 3B; Figure S3A). We then performed digitonin-permeabilization and subcellular fractionation to measure the localization of catalase in HeLa PEX5-KO or control cells (Figure 3C). We observed a complete shift of peroxisomal catalase to the cytoplasm in the PEX5-KO cells compared with the cells expressing a negative control sgRNA, while peroxisomal membrane proteins (PEX14 and PEX13) and a cytoplasmic marker (LDH) did not change localization. Impaired import of peroxisomal matrix proteins in PEX5-KO cells was verified by immunoblot indicating that B-chain of a PTS1 enzyme, ACOX1, derived from intraper-oxisomal processing of the full-length A-chain (Miyazawa et al., 1989) was not discernible and that another PTS1 protein, LONP2, was detectable only in the cytoplasm with lower stability (Figure 3C). Peroxisome function in PEX5-KO HeLa and K562 cells was assessed by analyzing plasmalogen levels by liquid chromatography-tandem mass spectrometry (LC-MS/MS). We found reduced levels of plasmalogens in both cell lines, suggesting that PEX5-KO results in non-functional peroxisomes (Figure S3B).
To further quantify catalase localization in the cytoplasm, we measured its enzymatic activity by selectively permeabilizing cellular membranes with digitonin and observing H2O2 degradation by the catalase released from the cells in negative control, PEX5-KO, and PEX12-KO HeLa cells (Tsukamoto et al., 1990). Because digitonin is a cholesterol-dependent membrane permeabilizing agent, lower concentrations of digitonin will selectively permeabilize the plasma membrane (which has higher cholesterol content), while only higher concentrations will permeabilize the peroxisomal membrane (Figure 3D). The assay is sensitive to catalase released into the assay buffer; thus the titration point of the catalase activity is dependent on the localization of catalase within the cell. We observed a much lower digitonin titration point for PEX5-KO and PEX12-KO HeLa cells than HeLa cells expressing a negative control sgRNA, strongly supporting that catalase is enriched in the cytoplasm in PEX5-KO and PEX12-KO cells (Figure 3E). Finally, we used immunofluorescence microscopy in PEX5 and PEX12 KO HeLa cells, which also demonstrated the localization of catalase to the cytoplasm, as well as the canonical “peroxisomal ghost” phenotype of these two KOs revealed by PMP70 staining (Figure 3F).
We next tested whether catalase is necessary for the protective effect of PEX5 and PEX12 sgRNAs. We created a clonal catalase-null (CAT-KO) HeLa cell line and expressed safe-targeting (control), PEX5, or PEX12 sgRNAs with Cas9 in either these cells or in HeLa-WT (wild-type) cells (Figure S3C). Cells were treated with 300 μM H2O2 and monitored using automated microscopy for 48 h post-H2O2 treatment to quantify cell death by uptake of SYTOX Green dye. We observed an overall greater sensitivity to the 300 μM H2O2 dose in the HeLa CAT-KO cells compared to the HeLa-WT cells, consistent with the screening results (Figure 3G). While sgRNAs targeting PEX5 and PEX12 were significantly protective in HeLa-WT cells treated with H2O2, they did not have a protective effect in HeLa-CAT-KO cells. Notably, we observed no major defects in peroxisomal number or morphology in CAT-KO cells using immunofluorescence staining for PMP70, which suggests that catalase KO cells do not have an aberrant peroxisomal phenotype (Figure S3D). Thus, only disruption of catalase-positive peroxisomes protects cells from ROS. Together, these data demonstrate that catalase is localized to the cytoplasm in peroxin gene KOs and is necessary for the protective effect observed in the context of oxidative stress.
Localization of Catalase to the CytoplasmIs Sufficient to Protect the Cell from ROS
To test whether non-peroxisomal catalase was sufficient to protect cells against oxidative stress, we examined if targeting CAT to different locations in the cell could protect against ROS (Figure 4A). We hypothesized that because PEX-KO cells have enriched cytoplasmic catalase and catalase is necessary for the PEX-KO protective effect, non-localized catalase should have an increased protective effect (Figure S4A). Native catalase has a non-canonical peroxisomal localization motif at its C terminus and localizes incompletely to the peroxisome (Purdue and Lazarow, 1996; Sheikh et al., 1998; Williams et al., 2012). We created expression constructs for catalase with its WT localization domain (K-A-N-L, WT-CAT), catalase with no localization domain (CAT-DKANL), and catalase with the canonical PTS1 peroxisomal localization domain (S-K-L, CAT-SKL), all in addition to an N-terminal FLAG epitope. Expression constructs were stably infected into HeLa cells using lentivirus (Figure S4B). Analysis of the expression of CAT variants by western blot showed levels similar to those of WT CAT (Figure 4B). Immunofluorescence microscopy confirmed the predicted localization of constructs in HeLa cells: WT-localized and PTS1 catalase localized to the peroxisomes, while the non-localized CAT-ΔDKANL localized diffusely across the whole cell (Figure 4C). We tested the sensitivity of HeLa cells expressing the differentially localized catalases to ROS by treating cells with 350 μM H2O2 (higher dose needed to achieve LD50 in catalase-expressing cells). Cells were monitored using automated microscopy for 48 h post-H2O2 treatment to quantify live cells by nuclear-localized mCherry and cell death by uptake of SYTOX Green dye (Figure 4D). Basal levels of cell death and growth were similar among untreated cell lines. However, HeLa cells expressing WT-CAT and CAT-ΔDKANL had significant reductions in cell death after H2O2 treatment compared with HeLa cells with no plasmid, by lethal fraction score (Forcina et al., 2017). Importantly, non-localized catalase had the lowest amount of cell death across any of the tested overexpression constructs. Interestingly, overexpressing CAT-SKL did not significantly decrease cell death compared with cells with no construct after H2O2 treatment. These results indicated that catalase localization determines its efficacy in protecting cells from oxidative damage and that cytoplasmic catalase is more protective against ROS than peroxisomal catalase.
Figure 4. Localization of Catalase to the Cytoplasm Is Sufficient to Protect the Cell from ROS.
(A) Diagram showing constructs used in differentially localized catalase panel.
(B) Western blot showing overexpression of catalase constructs in wild-type HeLa cells.
(C) Representative immunofluorescence microscopy images showing subcellular localization of catalase constructs. Cells were stained with DAPI to stain nuclei, PMP70 antibodies to stain peroxisomal membranes, and FLAG antibodies to stain FLAG-tagged catalase. Scale bar represents 10 μm.
(D) Normalized area under the curve (A.U.C.) lethal fraction scores showing cell death of HeLa cells expressing differentially localized catalase constructs or noplasmid. Bars represent mean across experiments (n = 9) ± SE. **p < 0.01 and ***p < 0.001, by Student’s t test.
PGD KO Decreases Glycolysis, Increases Upper PPP Glucose Flux, and Protects Cells against Oxidative Stress
Surprisingly, the genome-wide CRISPR/Cas9 and shRNA screens showed that deletion of specific members of the PPP protect cells against oxidative stress, including 6-phosphogluconolactonase (PGLS), PGD, and transketolase (TKT). This is counterintuitive given that the PPP metabolizes glucose-6-phosphate to reduce NADP+ into NADPH, which maintains the antioxidant capacity of the cell, and so removal of this pathway might be expected to be sensitizing (Kirkman et al., 1999; Lu, 2009; Patra and Hay, 2014). We generated KO lines for PGLS, PGD, and TKT using two sgRNAs per gene and validated their protective effects against H2O2 treatment by competitive growth assay (Figure 2C). PGD KO cells exhibited profound protection against H2O2 compared with other PPP KOs. Although PGD KO alone had a large negative growth effect, this was unlikely to be the cause of the protective effect, as there were many other genes with equivalently strong growth effects in the screen but no protection against ROS (Figure S5A). We also found no increase in the key antioxidant transcription factor, NRF2, in PGD-KO cells (Figures S5C and S5D). We hypothesized that this dramatic ROS-protective effect could be explained by the accumulation of the substrate of PGD, 6PG, which inhibits glycolysis and promotes glucose flux through the oxidative PPP, generating more NADPH during oxidative stress conditions compared with WT cells (Figure 5A; Kuehne et al., 2015; Parr, 1956). Indeed, we observed a very significant accumulation of 6PG in PGD KO cells using mass spectrometry: compared with negative control cells, 6-phosphogluconolactone (6PGL) and 6PG were present at 100and 20-fold higher levels, respectively (Figures 5B and 5C).
Figure 5. PGD Knockout Decreases Glycolysis, Increases Upper PPP Glucose Flux, and Protects Cells against Oxidative Stress.
(A) Diagram showing pentose phosphate pathway hits from genome-wide screens.
(B) Abundance of 6-phosphogluconolactone by mass spectrometry in K562 cells either untreated or treated with 30 μM H2O2 0, 5, 15, and 30 min before sample preparation. Bars represent mean across experiments (n = 3) ± SE.
(C) Abundance of 6-phosphogluconate by mass spectrometry in K562 cells either untreated or treated with 30 μM H2O2 0, 5, 15, and 30 min before sample preparation. Bars represent mean across experiments (n = 3) ± SE.
(D) Rate of anaerobic glycolysis and anaerobic glycolytic capacity measured by Seahorse glycolytic stress test assay of K562 cells expressing negative control orPGD sgRNAs. Bars represent mean extracellular acidification rate (ECAR) across experiments (n = 7) ± SE. **p < 0.01 and ***p < 0.001, by Student’s t test.
(E) CM-H2DCFDA fluorescence in PGD knockouts and negative control K562 cells. Background of non-dye cells was subtracted for each experiment. Bars represent mean across experiments (n = 5) ± SE. *p < 0.05 by Student’s t test.
(F) Western blot showing K562 polyclonal population knockouts of PGD and G6PD.
(G) Representative percentages for K562 competitive growth assay cultures showing the growth (left) and protective (right) effects of PGD knockout cells and the abrogation of the phenotype in the presence of a G6PD knockout. Bars represent mean across experiments (n = 3) ± SE. Experiment was performed twice with two sgRNAs per gene tested; full experimental results are shown in Figure S5B.
To test whether the accumulation of 6PG was responsible for the protective effect of PGD deletion against H2O2 treatment, we knocked out the upstream pathway member G6PD (which produces 6PGL, leading to 6PG production) in the background of PGD-KO cells (Figure 5F). Our hypothesis suggests that this should prevent both the accumulation of 6PG and the consequent upregulation of PPP flux, thus removing the protective phenotype of the PGD KO. Strikingly, whereas G6PD KO alone had little to no effect on growth rate or H2O2 sensitivity, the double G6PD/PGD KO almost completely reversed the phenotypes of the PGD KO, restoring both growth rate and sensitivity to H2O2 to WT levels (Figure 5G; Figure S5B). Interestingly, knocking out another upstream gene, PGLS, might also be expected to prevent accumulation of 6PG and cause protection against H2O2. However, we observed little effect on H2O2 sensitivity, possibly because of spontaneous hydrolysis of PGLS substrate, 6PGL, to 6PG, which had been previously observed (Miclet et al., 2001; Figure S5B). Overall, our data show that the growth phenotype and protective effect of PGD KO depend on G6PD activity.
To determine if the accumulation of 6PG in a PGD KO would decrease glycolysis (Figure 5A), we measured anaerobic glycolysis rates in K562 PGD-KO and negative control cells. We found decreased basal glycolysis rates and glycolytic capacity (Figure 5D; Figure S5E). We also found that G6PD KO decreased glycolysis rates, but to a lesser extent than PGD-KO cells. The double KO of PGD/G6PD had decreased glycolysis levels but was closer to the glycolytic rates of G6PD-KO compared with PGD-KO (Figure S5F). Thus, the decreased anaerobic glycolytic rate of PGD-KO cells and the partial rescue by KO of G6PD is consistent with inhibition of glycolysis rates by accumulation of 6PG in the oxidative branch of the PPP.
We next investigated the effect of PGD KO on cellular redox levels. We used a chloromethyl derivative of 2’,7’-dichlorodihydrofluorescein diacetate (CM-H2DCFDA), a ROS-sensitive fluorescent dye, to monitor the intracellular redox environment of K562 cells after H2O2 treatment. After H2O2 treatment, PGD-KO K562 cells had only minor changes to H2DCFDA fluorescence compared with untreated cells, while negative control cells had steadily increasing levels of ROS signal after H2O2 treatment (Figure 5E). This observation suggests that PGD-KO cells have stronger antioxidant responses to ROS compared with non-PGD-KO cells. Similarly, in HeLa cells basal levels of ROS were comparable between negative control and PGD-KO HeLa cells, but after 5 min of H2O2 treatment, PGD-KO cells had significantly lower ROS signal than negative control cells (Figure S5I). Interestingly, despite protection against H2O2 in PGD-KO HeLa cells (Figure 2A), time points at 15, 30, and 60 min showed no statistically significant difference in ROS levels between HeLa PGD-KOs and controls. The reduction in ROS was also reversed when G6PD and PGD were both deleted (Figures S5G–S5J), consistent with the reversal of PGD-KO phenotypes on growth, protection against H2O2, and glycolysis. Interestingly, there was no difference in steady-state NADPH/NADP+ ratio in PGD-KO cells in response to H2O2 treatment (Figures S5K and S5L).
Taken together, our data support a model in which PGD-KO protects cells against ROS toxicity through an accumulation of 6PG, which inhibits glucose-6-phosphate isomerase (GPI) and promotes glucose flux through the oxidative PPP at the expense of glucose flux through glycolysis. We observed increased PPP metabolites in PGD KO as well as decreased glycolysis, indicating an upregulation of PPP activity. Decreased ROS burden after H2O2 treatment, as shown by H2DCFDA fluorescence, suggests that PGD-KO cells have a stronger antioxidant response to acute increases in ROS compared with controls. Finally, we observed a complete buffering of the growth inhibition, ROS protection, and other phenotypes of PGD KO when G6PD was also deleted. These observations suggest that PGD is an important control point in the metabolic cellular response to acute oxidative stress.
DISCUSSION
In this study we conducted genome-wide shRNA and CRISPR screens to comprehensively identify diverse genetic factors that modulate cellular sensitivity to oxidative stress. We show through focused secondary sgRNA library screens that these major pathways are critical regulators of ROS response in both K562 and HeLa cells. Among these, we demonstrate roles for genes involved in DNA damage response, key antioxidants, iron homeostasis, peroxisomal protein import, and the PPP. We provide detailed insight into two pathways whose deletion showed unexpectedly protective phenotypes, contradictory to their annotated roles in redox biology: the peroxisomal matrix import pathway and the PPP. Although our results identify certain genes that were found in previous screens in model organisms (Ayer et al., 2012; Ueno et al., 2012), early RNAi screens (Kimura et al., 2008), and targeted screens in mammalian cells (Reczek et al., 2017), the majority of genes we found as significant hits were not identified in these screens.
One of the strongest protective signatures from our screens was one of the most puzzling: the peroxisomal matrix import pathway protects the cell from ROS when disrupted. Our screens identified all nine members of the PTS1 pathway involved in the import of catalase as protective hits. With three orthogonal methods, we show that perturbation of the peroxisomal matrix import pathway causes enrichment of non-peroxisomal catalase (Figures 3C–3F). Our data demonstrate that catalase-null cells have no protective phenotype against H2O2 when expressing sgRNAs targeting PEX genes, suggesting that catalase is necessary for the protective effect observed in PEX KOs (Figure 3G). Using a panel of differentially localized catalase constructs, we show that non-localized catalase is most protective in cells treated with H2O2 (Figure 4D). This suggests that the localization of catalase in the cell determines its efficacy in ameliorating oxidative stress.
Our results are consistent with an emerging model in the field of peroxisomal biology that catalase is actively transported outside of peroxisomes during periods of oxidative stress, where it confers a greater protective effect for the cell (Walton et al., 2017). A recent study identified redox-sensitive sites on PEX5 that inactivate peroxisomal protein import under oxidative conditions, which would prevent newly translated catalase from entering the peroxisome (Apanasets et al., 2014). Additional studies have suggested that BAK can permeabilize peroxisomal membranes during oxidative stress and may be involved in actively transporting catalase from the peroxisomes to the cytoplasm (Hosoi et al., 2017). Thus, although localization of catalase to the peroxisome is needed for detoxifying basal levels of ROS produced there, catalase export to the cytoplasm is likely a functional mechanism for ameliorating acute oxidative stress. Our results suggest that peroxisomal catalase export is a key control point in the cellular ROS response and that peroxisomes may act as antioxidant reservoirs for cells. Furthermore, delivery or manipulation of catalase localization could be a promising therapeutic strategy for diseases related to ROS (Barnham et al., 2004; Ebadi et al., 1996; Haney et al., 2015; Wu et al., 2003).
Another paradoxical finding was the protective effect of deleting the core PPP gene PGD. The role of the PPP in cellular survival and stress is complex. PGD has been shown to be upregulated in cancer metastases that are resistant to ROS (Bechard et al., 2018; McDonald et al., 2017; Shan et al., 2014), but inhibition of the PPP has also been shown to prevent erastin-induced ferroptosis, a form of cell death mediated by iron and ROS (Dixon et al., 2012). We show that deletion of PGD is strongly protective against H2O2 in HeLa and K562 cells in screens and individual competitive growth assays. Our data suggest that this is due to accumulation of the upstream metabolite 6PG (Figure 5C), which can inhibit GPI and thus glycolysis, increasing glucose flux through the PPP (Figure 5A; Kuehne et al., 2015). The increase of glucose flux through the oxidative PPP, which would result in increased NADPH production, could explain our observation that PGD-KO cells have significantly reduced ROS burden after H2O2 treatment (Figure 5E; Figures S5H and S5I). Finally, knocking out G6PD in PGD-KO cells buffered all of the phenotypes we observed (H2O2 protection, slow growth, reduced ROS), suggesting that the PGD-KO mechanism relies on the activity of G6PD and accumulation of PPP metabolites (Figure 5G). Although the precise mechanisms by which PGD-KO confers such dramatic cellular oxidative stress protection are unclear, it is interesting to note that we found no increase in the key antioxidant transcription factor, NRF2, in PGD-KO cells. This suggests that protection is not conferred by upregulation of this classic mechanism of antioxidant activity (Figures S5C and S5D). In addition, we find that the steady-state NADPH/NADP+ ratio in PGD-KO cells is unchanged, consistent with previous observations that perturbation of PGD protein levels does not affect the tightly controlled cytoplasmic NADPH/NADP+ ratio in cells (Figures S5K and S5L; Au et al., 2000; Christodoulou et al., 2019; Tao et al., 2017). Further investigation will be required to fully understand how PGD contributes to the control of glycolysis and cell survival, but our results suggest that it is a critical regulator of response to oxidative stress. Taken together, our results represent a rich resource for the study of oxidative stress and highlight a number of potential targets that might be exploited for therapeutic intervention.
STAR★METHODS
LEAD CONTACT AND MATERIALS AVAILABILITY
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Michael C. Bassik (bassik@stanford.edu). Plasmids generated during this study are deposited at Addgene, numbers available in key resources table.
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Rabbit FLAG antibody | Cell Signaling Technology | #14793; RRID:AB_2572291 |
| Mouse CAT antibody | Biomol International | YIF-LF-MA0003; RRID:AB_1611839 |
| Mouse GAPDH antibody | Invitrogen | AM4300; RRID:AB_437392 |
| Rabbit PGD antibody | Cell Signaling Technology | #13389; RRID:AB_2798202 |
| Rabbit NRF2 antibody | Cell Signaling Technology | #12721; RRID:AB_2715528 |
| Rabbit TKT antibody | Cell Signaling Technology | #8616; RRID:AB_10950823 |
| Rabbit CAT antibody | Abcam | ab16731; RRID:AB_302482 |
| Rabbit PMP70 antibody | Abcam | ab3421; RRID:AB_2219901 |
| Rabbit PEX12 antibody | Abcam | ab103456; RRID:AB_10711955 |
| Rabbit PGLS antibody | Abcam | ab127560; RRID:AB_11130290 |
| Rabbit b-actin antibody | Abcam | ab8227; RRID:AB_2305186 |
| Rabbit IREB2 antibody | Abcam | ab181153 |
| Mouse PMP70 antibodies | Sigma | SAB4200181; RRID:AB_10639362 |
| Rabbit PEX5 antibody | Otera et al., 2000 | N/A |
| Rabbit PEX13 antibody | Mukai and Fujiki, 2006 | N/A |
| Rabbit PEX14 antibody | Shimizu et al., 1999 | N/A |
| Rabbit ACOX1 antibody | Tsukamoto et al. 1990 | N/A |
| Rabbit LONP2 antibody | Okumoto et al., 2011 | N/A |
| Chemicals, Peptides, and Recombinant Proteins | ||
| Hydrogen Peroxide | Millipore-Sigma | #216763 |
| Herculase II Fusion DNA Polymerase | Agilent | Cat# 600675 |
| Digitonin | Wako Chemicals USA | #NC0141730 |
| Triton X-100 | Aqua Solutions | SKU# T9010 |
| SYTOX Green | Thermo Fisher Scientific | S7020 |
| CM-H2DCFDA | Thermo Fisher Scientific | C6827 |
| Critical Commercial Assays | ||
| Qiagen DNA Blood Maxi kit | Qiagen | Cat# 51194 |
| Promega NADP/NADPH-Glo Assay | Promega | G9081 |
| Seahorse glycolysis stress test | Agilent | #103017–100 |
| Deposited Data | ||
| Raw Screening Data | This paper | SRA: SRP235195 |
| List of CRISPR/Cas9 K562 genome-wide screen hits | This paper | N/A |
| List of shRNA genome-wide K562 hits | This paper | N/A |
| List of CRISPR/Cas9 K562 secondary batch screen hits | This paper | N/A |
| List of CRISPRi K562 secondary batch screen hits | This paper | N/A |
| List of CRISPR/Cas9 HeLa secondary batch screen hits | This paper | N/A |
| List of CRISPRi HeLa secondary batch screen hits | This paper | N/A |
| Experimental Models: Cell Lines | ||
| Human: K562 Cells | ATCC | CLL-243 |
| Human: HeLa Kyoto Cells | Hein et al., 2015 | CVCL_1922 |
| Oligonucleotides | ||
| Oligos for sgRNAs and shRNAs, see Table S4 | This paper | N/A |
| Primers for TIDE, see Table S4 | This paper | N/A |
| Primers for library sequencing, see Table S4 | This paper | N/A |
| Recombinant DNA | ||
| pMCB320 (pSico-pU6-sgRNA EF1A-Puro-T2A-mCherry) | Morgens et al., 2017 | Addgene Plasmid #89359 |
| pMCB306 (pSico-pU6-sgRNA EF1A-Puro-T2A-GFP) | Han et al., 2017 | Addgene: #89360 |
| pMCB280 (SFFV-Puro-T2A-mCherry-shRNA) | Deans et al., 2016 | N/A |
| pMCB309_pMK1221 (pSicoR-CMV-shRNA SFFV-puro-mCherry) | Kampmann et al., 2015 | Addgene: #84220 |
| pMMD650_UbC-G418-T2A-WT-CAT | This paper | Addgene: #137990 |
| pMMD651_UbC-G418-T2A-CAT-ΔKANL | This paper | Addgene: #137991 |
| pMMD652_UbC-G418-T2A-CAT-SKL | This paper | Addgene: #137992 |
| Software and Algorithms | ||
| casTLE screening analysis | Morgens et al., 2016 | https://bitbucket.org/dmorgens/castle |
| IncuCyt®e S3 Base Software | Essen Biosciences | V2016A/B |
| Excel | Microsoft | N/A |
| Progenesis QI 2.3 software | Progenesis | N/A |
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Cell Culture and antibiotics
K562 cells (ATCC) were grown in Roswell Park Memorial Institute (RPMI) 1640 Medium (Life Technologies, 11875093) supplemented with 10% Fetal Bovine Serum (Fisher, Cat# SH30910), 2 mM L-glutamine (Fisher, Cat# SH3003401) and 1% penicillin-streptomycin (Fisher, Cat#SV30010), and cultured at 37°C with 5% CO2. HeLa Kyoto cells (CVCL_1922) were grown in Dulbecco’s Modified Eagle’s Medium (Life Technologies, Cat# 11995073) supplemented with 10% FBS, 2 mM L-glutamine, and 1% penicillinstreptomycin, and cultured at 37°C with 5% CO2. Cell cultures were routinely tested and found negative for mycoplasma infection (MycoAlert, Lonza).
METHOD DETAILS
Genome Wide sgRNA Screen in K562 Cells
Lentiviral screening library preparation, lentiviral infection, library selection, genomic DNA extraction, PCR amplification, and casTLE analysis were performed as previously described (Deans et al., 2016; Jeng et al., 2019; Morgens et al., 2017). K562 cells, with a BFP-Cas9 lentiviral transgene, were lentivirally infected with a genome-wide sgRNA library as described (Morgens et al., 2017), containing ~200,000 sgRNAs targeting 20,549 protein-coding genes and 13,500 negative control sgRNAs at an MOI of 0.3–0.4 (as measured by the mCherry fluorescence from the lentiviral vector) with approximately 1,000x coverage per library element, and selected for lentiviral integration using puromycin (1 μg/ml) for 3 days as the cultures were expanded for the screens. In one flask, cells were grown with four pulses of 30 μM H2O2 (Millipore-Sigma #216763), the LC50 dose for K562-Cas9 cells, with a four day recovery in-between pulses or until cells began to double nearly daily again. The other flask was grown in the absence of H2O2. All conditions and replicates were maintained at a minimum of 1000x coverage per element for the duration of the screen. Untreated cells were diluted to a density of 500,000 cells/mL each day. H2O2-treated cells were diluted to a density of 500,000 cells/mL as needed. The cell culture screening lasted for 21 days total, including a final recovery period, with approximately 9.5 and 24 doublings for the treated and untreated populations, respectively. After the cell culture period, untreated and H2O2-treated cells were pelleted by centrifugation for genomic DNA extraction using the Qiagen DNA Blood Maxi kit (Cat# 51194), as described by manufacturer’s instructions. Approximately 2.5×108 cells were used for each genomic DNA extraction to maintain 1000x coverage per element. The sgRNA-encoding constructs were PCR-amplified using Agilent Herculase II Fusion DNA Polymerase (Cat# 600675) (See Table S4 for the primer sequences used). These libraries were then sequenced across an Illumina NextSeq flow cell (40 M reads per condition; ~200× coverage per library element). Computational analysis of genome-wide screen was performed as previously described using casTLE, which is a maximum likelihood estimator that uses a background of negative control sgRNAs as a null model to estimate gene effect sizes (Morgens et al., 2016). See Table S1 for the K562 genome-wide sgRNA screen results.
Genome Wide shRNA Screen for K562 Cells
Lentiviral screening library preparation, lentiviral infection, library selection, genomic DNA extraction, PCR amplification, and casTLE analysis were performed as previously described (Deans et al., 2016; Jeng et al., 2019; Morgens et al., 2017). K562 cells were lentirvirally infected with the genome-wide shRNA library as described previously (Deans et al., 2016), containing 506,995 sgRNAs targeting 19,128 protein-coding genes and 7,689 negative control shRNAs at an MOI of 0.3–0.4 (as measured by the mCherry fluorescence from the lentiviral vector) with approximately 1,000x coverage per library element, and selected for lentiviral integration using puromycin (1 mg/ml) for 3 days as the cultures were expanded for the screens. Infected cells were expanded and split into two flasks. Treated cells were grown with either 5 or 6 increasing doses of H2O2 (Millipore-Sigma #216763) (45 μM, 100 μM, 125 μM, 125 μM, and 125 μM or 30 μM, 35 μM, 40 μM, 50 μM, 100 μM, and 100 μM, respectively) until there was a roughly two-fold difference in doublings between the untreated and treated libraries (31.7 and 15.8 doublings or 29.9 and 17.7 doublings, respectively). The treated library was allowed to recover so cells were roughly doubling each day before the next treatment. All conditions and replicates were maintained at a minimum 1000x coverage per element for the duration of the screen. Untreated cells were diluted to a density of 500,000 cells/mL each day. H2O2-treated cells were diluted to a density of 500,000 cells/mL as needed. Cell culture screening lasted for 24 or 25 days total, including a final recovery period. After the cell culture period, untreated and H2O2-treated cells were pelleted by centrifugation for genomic DNA extraction using Qiagen DNA Blood Maxi kit (Cat# 51194), as described by manufacturer’s instructions. Approximately 5.0×108 cells were used for each genomic DNA extraction to maintain 1000x coverage per element. The shRNA-encoding constructs were PCR-amplified using Agilent Herculase II Fusion DNA Polymerase (Cat# 600675). These libraries were then sequenced across an Illumina NextSeq flow cell (~80 M reads per condition; ~200× coverage per library element). Computational analysis of genome-wide screen was performed as previously described using casTLE, which is a maximum likelihood estimator that uses a background of negative control shRNAs as a null model to estimate gene effect sizes (Morgens et al., 2016). See Table S1 for the K562 genome-wide shRNA screen results.
Batch Screen for K562 Cells
The batch screen library included the following, non-comprehensive sets of genes (1,000 genes in total, ~10 sgRNAs per gene, plus 5,000 negative control sgRNAs [2,000 non-targeting scrambled controls, 3,000 safe-targeting guides]): 511 hits passing 5% FDR from our genome-wide CRISPR screen, 282 manually identified genes from the literature that have been previously shown to be involved in redox biology or are related to pathways identified in the genome-wide screens, and finally 207 genes just outside the 5% FDR threshold in the genome-wide CRISPR screen for a total of 1,000 genes (see Table S2 for a complete list of genes and complete 14,917 element list). The same library of genes was used to generate a CRISPRi sgRNA library as well, with the same number of control sgRNAs (see Table S2 for complete 15,460 element list). The library oligos were synthesized by Agilent Technologies and cloned into pMCB320 using BstXI/BlpI overhangs after PCR amplification. Lentiviral screening library preparation, lentiviral infection, library selection, genomic DNA extraction, PCR amplification, and casTLE analysis were performed as previously described (Deans et al., 2016; Jeng et al., 2019; Morgens et al., 2017). The Cas9 and dCAS9-KRAB K562 cell lines were lentivirally infected with the secondary library at an MOI of 0.3–0.4 as described for the genome-wide screens. After puromycin selection (1 0μg/ml for 3 days) and expansion, 30 M (~2,000 coverage per library element) cells were treated with four pulses of 30 μM H2O2 (Millipore-Sigma, #216763), with a four day recovery in-between pulses or until cells began to double nearly daily again with a logarithmic growth (500,000 cells/ml) maintained as in the K562 genome-wide screen. Thirty million cells under each condition were used for genomic extractions, sequenced (~6–10 million reads per condition; 1000–20003 coverage per library element) and analyzed using casTLE as described for the genome-wide screens. See Table S3 for the K562 secondary screen results
Batch Screen for HeLa Cells
Lentiviral screening library preparation, lentiviral infection, library selection, genomic DNA extraction, PCR amplification, and casTLE analysis were performed as previously described (Deans et al., 2016; Jeng et al., 2019; Morgens et al., 2017). Cas9 and dCAS9-KRAB HeLa cell lines were lentivirally infected with the secondary library (14,917 and 15,460 elements, Table S2) at an MOI of 0.3–0.4 as described for the genome-wide screens. After puromycin selection (1 μg/mL for 3 days) and expansion, ×15 million Cas9 or dCas9-KRAB HeLa cells (~1,000× coverage of sgRNA library) were treated with three pulses of 300 μM H2O2 (Millipore-Sigma, #216763), with recovery in-between pulses until cells began to double nearly daily again with logarithmic growth. Cells were split every day to maintain logarithmic growth and prevent confluence. 15 million (~1,000× coverage) cells per condition were used for genomic extractions and sequencing as described for the genome-wide screens. See Table S2 for the HeLa secondary screen results.
Generation of Individual sgRNA-Expressing Cells/Stable Cell Lines
Lentivirus production and infection were performed as previously described (Morgens et al., 2016). Briefly HEK293T cells were transfected with third-generation packaging plasmids and the sgRNA-expressing vector. Lentivirus was harvested after 48 h and 72 h and filtered through a 0.45μm Polyvinylidene Fluoride (PVDF) filter (Millipore). K562 cells expressing lentiCas9-BFP were infected by centrifugation at 1000 x g for 2 h. HeLa cells expressing lentiCas9-BFP were infected by incubating the cells in lentivirus-containing media for 24 h. 3 days after infection, cells were selected with puromycin (1.2 μg/mL for 3 d for K562 and HeLa cells). Clonal knockout lines were generated for K562 PEX5-KO, K562 PEX12-KO, HeLa PEX5-KO, and HeLa PEX12-KO cell lines by single-cell sorting puromycin-selected cells into 96-well plates and expanding them for 2–3 weeks. Gene editing efficiency was determined by Sanger sequencing and analyzing the resulting chromatograms using TIDE software (Brinkman et al., 2014).
Western Blots
Live cells were lysed for 20 min at 4°C in protein extraction buffer (1% Triton X-100, 150 mM NaCl, 50 mM Tris pH 7.5, 1 mM EDTA) with protease inhibitors and centrifuged to at 21,000 x g to collect the supernatant lysate. For protein extraction buffer of nuclear lysates, cells were instead lysed with RIPA buffer (150 mM NaCl, 1% nonidet P-40, 0.5% sodium deoxycholate, 0.1% SDS, 25mM Tris pH 7.4 in ddH2O) and sheared with a needle to extract nuclear protein before centrifugation. The cell lysate was measured with Bradford reagent (Biorad). For each western blot, 4x LDS Sample Buffer (Invitrogen, NP0007) and DTT (to final added concentration of 125mM) were mixed with each cell lysate sample, which were then denatured at 70°C for 10 minutes. Protein lysates were then separated on 4–12% acrylamide SDS-PAGE gels (Invitrogen, NP0322BOX) and transferred to nitrocellulose membranes (BioRad, #162–0146). After blocking nitrocellulose membranes for 30 minutes with blocking buffer (LI-COR, #927–70001) and washing with TBST (137mM NaCL, 2.7mM KCl, and 19mM Tris base), membranes were stained with the primary antibody diluted in 5% milk in TBST, according to antibody manufacturer’s instructions, for 1 hour at room temperature. After washing and secondary antibody staining (45 minutes, room temperature), membranes were visualized on an Odyssey CLx and analyzed with Image Studio 5.x.
Immunofluorescence
Live HeLa cells were grown on glass coverslips overnight in normal cell culture conditions. Cells were fixed with 4% paraformaldehyde (EMS, #15710) for 15 minutes at room temperature. Cells were permeabilized with 0.1% Triton X-100 (Aqua Solutions, SKU# T9010–500ML) at room temperature for 30 minutes. Coverslips were then blocked with 1% BSA for 30 minutes at room temperature before adding primary antibody stain to coverslips for 1 hour at room temperature. After primary staining, cells were stained with secondary antibodies for 1 hour at room temperature. Coverslips were incubated with Hoechst stain (Thermo Fisher Scientific, H3570) at 10 mg/mL for 5 minutes. Coverslips were adhered to a microscopy slide (Corning #2947–75×25) using permafluor (Thermo Fisher Scientific, #TA-030-FM) according to manufacturer’s instructions. Coverslips were imaged on a Nikon Eclipse Ti.
Antibodies
Rabbit FLAG antibody (#14793, Cell Signaling Technology), mouse CAT antibody (YIF-LF-MA0003, Biomol International), mouse GAPDH antibody (AM4300, Invitrogen), rabbit PGD antibody (#13389, Cell Signaling Technology), rabbit TKT antibody (#8616, Cell Signaling Technology), rabbit NRF2 antibody (#12721, Cell Signaling Technology), rabbit CAT antibody (ab16731, Abcam), rabbit PMP70 antibody (ab3421, Abcam), rabbit PEX12 antibody (ab103456, Abcam), rabbit PGLS antibody (ab127560, Abcam), rabbit β-actin antibody (ab8227, Abcam), rabbit IREB2 antibody (ab181153, Abcam), mouse PMP70 antibodies (SAB4200181, Sigma). Rabbit antibodies to PEX5 (Otera et al., 2000), PEX13 (Mukai and Fujiki, 2006), PEX14 (Shimizu et al., 1999), ACOX1 (Tsukamoto et al., 1990), LONP2 (Okumoto et al., 2011) were used as described in corresponding references.
Gene editing measurements by Sanger sequencing
Both HeLa and K562 PEX5-KO and PEX12-KO clonal cells were harvested and total genomic DNA was isolated using QuickExtract DNA Extraction Solution (VWR, QE09050). PCR was prepared using 5x Phusion HF reaction buffer (Thermo Fisher Scientific, F-518) and Phusion DNA polymerase (Thermo Fisher Scientific, F-530L), 10cmM dNTPs and primers designed about 400–550cbp upstream (MMD371_TIDE_PEX5_fwd, 5′-CTAGGTATGGTCGGGCTGTT; MMD373_TIDE_PEX12_fwd, 5′-AGTCACTGTTGGACCCTGAA) and 400–550cbp downstream (MMD372_TIDE_PEX5_rev, 5′-CTCAGACCACCACCACCTG; MMD374_TIDE_PEX12_rev, 5′-TGACGTG CAGCATTTGACAA) of the predicted cut site. PCR reactions were run on a C1000 Touch Thermo Cycler (Bio-Rad). PCR products were then purified over an Econospin DNA column (Epoch, 1910–250) using buffers PB and PE (Qiagen, 19066 and 19065). Sanger sequencing was performed and applied biosystems sequence trace files (.ab1 files) were obtained from Quintara Biosciences. Editing efficiencies of knockout cell lines was analyzed using the Netherlands Cancer Institute’s online TIDE analysis software (Brinkman et al., 2014).
Digitonin Enzymatic Release Assay
Assay was conducted in similar fashion to protocol outlined in (Tsukamoto et al., 1990) and in Figure 3C. HeLa cells were plated at 2 × 106 cells in 6cm dish the day before conducting assay. HeLa cells were then trypsinized, resuspended, and count the cell number. Cells were kept on ice and washed cells in 1.5 mL tubes (1 × 106 cells per tube) twice with cold Hepes-sucrose buffer (HSB) (250 mM sucrose, 20 mM Hepes-KOH (pH 7.4)). Cells were then diluted to 500,000 cells/mL in HSB. A dilution series of digitonin (Wako Chemicals USA, #NC0141730) was made in HSB in 1.5 mL tubes and 50 μL of each digitonin concentration, including a 2% Triton X-100 control (Aqua Solutions, SKU# T9010–500ML), was pipetted into the bottom of glass test tube. 100 μL of HSB wad added to the blank/no cell control glass tubes. All tubes were placed in a crushed-ice water bath (at 0C). At the very start of the assay, 8.75 μL of 30% H2O2 to 25 mL of substrate solution (0.02 M imidazole, 0.1% BSA, and 250 mM sucrose) to create the active substrate solution for the assay. Next, 50 μL of the cell dilutions (500,000 cells/mL) was added to digitonin dilutions, mixed briefly with vortex (setting #1), and incubated in crushed-ice water (0°C) for 6 minutes. After 6 minutes, 1.0 mL of the active substrate solution was added to samples, mixed briefly by vortexing, and incubated in crushed-ice water (0°C) for 15 minutes. After 15 minutes, 1.0 mL of Ti(SO4)2 solution was added and solutions were briefly mixed by vortexing to stop catalase reaction. After vortexing, glass test tubes were incubated at RT for 10 minutes. Finally solutions were pipetted into a 96-well plate and the OD 410 was measured. EC50 curves were drawn and calculated by using the previously described Hill algorithm for each cell line (Gadagkar and Call, 2015).
Digitonin Subcellular Fractionation
HeLa cells, negative control and PEX5-KO, were plated the day before and grown to 80% confluence. Cells were washed twice with ice-cold PBS (4°C) and once with 4°C ice cold HSB, 1 mM EDTA, and protease inhibitors). Next, 400 μL of HSB containing 100 μg/mL digitonin (Wako Chemicals USA, #NC0141730) was added to cells. Cells and buffer were scraped into a 1.5 mL tube and incubated on ice for 10 minutes. Cells were divided into two different 1.5 mL tubes (180 μL/tube) for total lysate and cytosolic/membrane fraction lysates. The cytosolic/membrane fraction was centrifuged at 20,000 x g for 15 minutes at 4°C. After centrifugation, 150 μL of the supernatant was pipetted into a new 1.5 mL tube for the cytosolic fraction and 30 mL of 6x SDS-PAGE sample buffer (375 mM Tris-HCl, pH 6.8), 6% SDS, 6% b-mercaptoethanol, 30% glycerol) was added to sample. The supernatant left on the pellet was carefully removed and 50 μL of HSB was added to the pellet, or membrane fraction, which was further centrifuged at 20,000 x g for 15 minutes at 4°C to rinse the pellet. After carefully removing the HSB, 216 mL of 1x SDS-PAGE sample buffer was added to the membrane pellet, which was then sonicated in a sonic bath. Thirty-six μL of the 6x SDS-PAGE sample buffer was added to the 180 μL of total lysate fraction. Finally, all samples were boiled for 5 minutes before undergoing the standard western blot protocol as described above.
Incucyte Lethal Fraction Score Experiments
HeLa cell lines measured using the Incucyte were lentivirally infected with plasmid pMMD212-puro-3xNLS-mCherry. After puro selection (1 μg/ml for 3 days) and expansion, respective HeLa cell lines were trypsinized while in logarithmic growth and measured using a BD Accuri. After calculating live cells/mL, cell lines were diluted to 80,000 cells/mL and 0.5 mL was then plated in a 24 well plate (Corning #393047) and incubated under normal cell culture conditions for 2 hours. After two hours, 0.5 mL of DMEM medium with 40 ng/mL of SYTOX Green (Thermo Fisher Scientific, S7020) with or without 2x concentration of H2O2 (Millipore-Sigma, #216763) was added to treated or untreated wells. Cells were then placed in an Incucyte Zoom for 48 hours and were measured every 4 hours using exposure times of 400 ms for 488nm and 800 ms for 547nm.
Calculating Lethal Fraction Scores
To calculate lethal fraction score, we followed calculations outlined in (Forcina et al., 2017) combining live and dead cell counts every 4 hours across the experiment. Automated image analysis routines were optimized for HeLa cells using the Zoom software package (V2016A/B) and training data from untreated and H2O2 (Millipore-Sigma, #216763) treated samples.
HeLa CM-H2DCFDA
HeLa cells were plated at 40,000 cells/well 24 hours before initiating the experiment. Medium was replaced before beginning experiment and cells were treated with 300 μM H2O2 (Millipore-Sigma, #216763) for 5, 15, 30, and 60 minutes before washing 3 times with DMEM medium. After wash, cells were incubated in the dark at 37°C for 30 minutes with 5 μM CM-H2DCFDA (Thermo Fisher Scientific, C6827) DMEM medium. After incubation, cells were washed 3 times with warm DMEM medium and then imaged using an Incucyte S3 live-cell analysis system (Sartorius).
K562 CM-H2DCFDA
K562 cells were plated at 400,000 cells/well in a 24-well plate and adhered to plate using poly-L-lysine and a 5 minute 300 x g spin. Medium was replaced before beginning experiment and cells were treated with 30 μM H2O2 (Millipore-Sigma, #216763) for 5, 15, 30, and 60 minutes before washing 3 times with RPMI medium. After wash, cells were incubated in the dark at 37°C for 30 minutes with 5 μM CM-H2DCFDA (Thermo Fisher Scientific, C6827) RPMI medium. After incubation, cells were washed 3 times with warm RPMI medium and then imaged using an Incucyte S3 live-cell analysis system (Sartorius) or quantified using a BD Accuri C6 and BD Csampler software.
Seahorse Glycolysis Assays
K562 cells were plated at 30,000 cells/well in a Seahorse cell culture miniplate after washing twice with DPBS and adhered to plate using poly-L-lysine and a 5 minute 300 x g spin. Cells were incubated in Seahorse assay medium supplemented with 1% L-Glut at 37°C in a non-CO2 incubator for 45–60 minutes. Cells were then assayed using the Seahorse glycolysis stress test (Agilent, #103017100) according to kit instructions. Each assay was conducted in triplicate for each cell line.
Metabolite Extraction for Untargeted Metabolomics by LC-MS
K562 cells were grown in RPMI medium and split into untreated and 4 different lengths of incubation with 30 μM H2O2 (MilliporeSigma, #216763) per genetic background: 0 minutes (H2O2 added immediately prior to washing), 5 minutes, 15 minutes, and 30 minutes. After H2O2 incubation, cells were washed three times with 25 mL of ice-cold (4°C) PBS buffer to remove any remaining medium, and aliquoted into 5×106 cells and flash frozen in cell pellets. A previously described comprehensive untargeted metabolomics platform (Contrepois et al., 2015) was used to profile metabolites extracted from cell pellets. Cell pellets were re-suspended in 500 μl of 4:1 methanol:water including 17 internal standards to control for extraction efficiency and evaluate LC-MS performance. Re-suspended cells were vortexed for 30 s, sonicated using a bath sonicator for 30 s and incubated on ice for 30 s (repeated 3 times). Proteins were then precipitated by incubating samples at −20°C for two hours. Samples were centrifuged at 10,000 rpm for 10 min at 4°C, metabolite extracts were evaporated to dryness under nitrogen and reconstituted in 100 μl 1:1 methanol:water before analysis.
Data Acquisition for Untargeted Metabolomics by LC-MS
Each sample was analyzed four times using HILIC and RPLC separation in both positive and negative ionization modes. Data were acquired on a Thermo Q Exactive HF mass spectrometer for HILIC and a Thermo Q Exactive mass spectrometer (Thermo Fisher Scientific) for RPLC. Both mass spectrometers were equipped with a HESI-II probe and operated in full MS scan mode. For HILIC experiments, a ZIC-HILIC column (2.1 × 100 mm, 3.5 μm, 200 Å; EMD Millipore) was used with mobile phase solvents (A) 10 mM ammonium acetate in 50:50 acetonitrile:water and (B) 10 mM ammonium acetate in 95:5 acetonitrile:water. For RPLC experiments, a Zorbax-SB-aq column (2.1 × 50 mm, 1.7 μm, 100 Å; Agilent) used with mobile phase solvents (A) 0.06% acetic acid in water and (B) 0.06% acetic acid in methanol. Before running the sequences, LC-MS systems were equilibrated by injecting 12 and 6 pooled quality control samples (QCs) for HILIC and RPLC, respectively. MS/MS data were acquired on pooled QCs consisting of an equimolar mixture of all the samples in the study at normalized collision energies (NCE) of 25 and 35 for HILIC and 25 and 50 for RPLC. Multiple quality control measures were performed to ensure data quality. All samples were randomized prior to protein extraction and data acquisition. Further, mass accuracy, retention time and peak shape of internal standards were reviewed in each sample.
Data Processing for Untargeted Metabolomics by LC-MS
Raw data were imported into Progenesis QI 2.3 software (Water, Milford, MA, USA) to align and quantify chromatographic peaks. Data from each acquisition mode were processed independently and then merged. MS drift with time was corrected using the LOESS normalization method on pooled QCs injected every 10 injections in the sequence. A targeted list of metabolites containing 6-phosphogluconolactone and 6-phosphogluconate were formally identified by matching retention time and fragmentation spectra to authentic standards when possible.
Measurement of plasmalogens by LC-MS/MS
Total lipids were extracted by the Bligh and Dyer method (Bligh and Dyer, 1959). Briefly, 50 μg of total cellular proteins were suspended in methanol/chloroform/water at 2:1:0.8 (vol/vol/vol) and then 50 pmol of 1-heptadecanoyl-sn-glycero-3-phosphocholine (LPC; Avanti Polar Lipids #855676P), 1, 2-didodecanoyl-sn-glycero-3-phosphocholine (DDPC; Avanti Polar Lipids #850335P), and 1, 2-didodecanoyl-sn-glycero-3-phosphoethanolamine (DDPE; Avanti Polar Lipids #850702P) were added as internal standards, followed by incubating for 5 min at room temperature, adding 1 ml each of water and chloroform, centrifuging at 720 g for 5 min. The lower organic phase was collected. To re-extract lipids from the remaining water phase, 1 ml chloroform was added. The combined organic phase was evaporated under a nitrogen stream and the extracted lipids were dissolved in methanol. Total cellular plasmalogens were analyzed by LC-ESI-MS/MS (Abe et al., 2014) using a 4000 Q-TRAP quadrupole linear ion trap hybrid mass spectrometer (AB Sciex) with an ACQUITY UPLC system (Waters). The data were analyzed and quantified using Analyst software (AB Sciex).
Measuring NADPH/NADP+ Levels
NADP/NADPH-Glo™ Assay (G9081, Promega) was used to measure NADPH and NADP+ levels in HeLa cells according to instructions. HeLa cells were seeded into a 24-well plate at approximately 40,000 cells per well at the time the assay was performed. DMEM medium was replaced before beginning experiment and cells were treated with 300 μM H2O2 (Millipore-Sigma, #216763) for 5, 15, 30, and 60 minutes before quickly removing H2O2-treated media. 160 μL of PBS was added to each well before adding an additional 160 μL of PBS with 1% DTAB (Dodecyltrimethylammonium bromide) and 0.2N NaOH to lyse the cells for a final concentration of 0.5% DTAB and 0.1N NaOH in PBS. NADPH standards were diluted in the same concentration of DTAB and NaOH from freshly resuspended NADPH (#N8035, Sigma Aldrich). After lysis, each sample was divided into NADP+ and NADPH fractions. NADP+ samples were used directly, NADPH fractions were diluted by half using 0.5% DTAB, 0.1N NaOH PBS. 50 μL of each fraction for each sample was then added to a 96-well plate. 25 μL of 0.4 HCl was added to the NADP+ samples and all samples were then incubated in the covered plate for 15 minutes at 60°C, and then 10 minutes at room temperature. After incubation, 25 mL of 0.5M Trizma base was added to NADP+ samples and 50 μL of HCl/Trizma solution was added to NADPH samples. 24.5 μL of each sample was then transferred into two wells in a new white 96-well luminometer plate (#3917, Corning Costar). 24.5 μL of the kit NADP/NADPH-Glo detection reagent was then added to each well. After 30 minutes incubation at room temperature, luminescence from the plate was read on a TECAN Spark, using an integration time of 1,000 ms. Data was analyzed according to assay instructions.
QUANTIFICATION AND STATISTICAL ANALYSIS
Statistical analyses were performed using Microsoft Excel and python using the indicated tests and custom scripts where referenced. Lethal fraction scores calculated from live/dead cell counts using previously published formula (Forcina et al., 2017) and Incucyte Zoom and S3 software (Essen). Immunofluorescence image analysis was performed using NIS Elements. Western blots were imaged using an Odyssey CLx and analyzed with Image Studio 5.x. Statistical details of experiments can be found in the figure legends. N represents the number of replicates for each experiment. Randomization, blinding, and sample-size estimation were not applicable in this study.
DATA AND CODE AVAILABILITY
Sequencing data are available at Sequence Read Archive (SRA) accession number SRP235195 under BioProject accession number PRJNA594164.
Supplementary Material
Highlights.
Genome-wide shRNA and CRISPR screens identify modifiers of oxidative stress sensitivity
PEX gene perturbation protects cells via catalase redistribution
Disruption of the pentose phosphate pathway protects cells by metabolic rewiring
ACKNOWLEDGMENTS
We would like to thank Yuchi Abe for his analyses of plasmalogen levels of HeLa and K562 cells by LC-MS/MS. We also thank Gaelen Hess, Kim Tsui, Cameron Lee, Suzanne Pfeffer, Peter Jackson, the Dixon lab, and the Bassik lab for their technical expertise and helpful discussions. This work was funded by NIH Director’s New Innovator Award Program (1DP2HD084069-01 to M.C.B.), NIH grant 1R01GM122923 (to S.J.D.), and grant 2RM1HG00773506 (to M.P.S.). This work was supported in part by grants from the Ministry of Education, Culture, Sports, Science, and Technology of Japan; Grants-in-Aid for Scientific Research (JP26116007, JP15K14511, JP15K21743, and JP17H03675 to Y.F.); grants from the Takeda Science Foundation (to Y.F.), the Naito Foundation (to Y.F.), the Japan Foundation for Applied Enzymology (to Y.F.), and the Novartis Foundation (Japan) for the Promotion of Science (to Y.F.); and Kyushu University Progress 100 (to Y.F.).
Footnotes
DECLARATION OF INTERESTS
The authors declare no competing interests.
SUPPLEMENTAL INFORMATION
Supplemental Information can be found online at https://doi.org/10.1016/j.celrep.2020.01.013.
REFERENCES
- Abe Y, Honsho M, Nakanishi H, Taguchi R, and Fujiki Y. (2014). Very-long-chain polyunsaturated fatty acids accumulate in phosphatidylcholine of fibroblasts from patients with Zellweger syndrome and acyl-CoA oxidase1 deficiency. Biochim. Biophys. Acta - Mol. Cell Biol. Lipids 1841, 610–619. [DOI] [PubMed] [Google Scholar]
- Alvarez SW, Sviderskiy VO, Terzi EM, Papagiannakopoulos T, Moreira AL, Adams S, Sabatini DM, Birsoy K, and Possemato R. (2017). NFS1 undergoes positive selection in lung tumours and protects cells from ferroptosis. Nature 551, 639–643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Apanasets O, Grou CP, Van Veldhoven PP, Brees C, Wang B, Nordgren M, Dodt G, Azevedo JE, and Fransen M. (2014). PEX5, the shuttling import receptor for peroxisomal matrix proteins, is a redox-sensitive protein. Traffic 15, 94–103. [DOI] [PubMed] [Google Scholar]
- Au SWN, Gover S, Lam VMS, and Adams MJ (2000). Human glucose-6-phosphate dehydrogenase: the crystal structure reveals a structural NADP(+) molecule and provides insights into enzyme deficiency. Structure 8, 293–303. [DOI] [PubMed] [Google Scholar]
- Ayer A, Fellermeier S, Fife C, Li SS, Smits G, Meyer AJ, Dawes IW, and Perrone GG (2012). A genome-wide screen in yeast identifies specific oxidative stress genes required for the maintenance of sub-cellular redox homeostasis. PLoS ONE 7, e44278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barbie DA, Tamayo P, Boehm JS, Kim SY, Moody SE, Dunn IF, Schinzel AC, Sandy P, Meylan E, Scholl C, et al. (2009). Systematic RNA interference reveals that oncogenic KRAS-driven cancers require TBK1. Nature 462, 108–112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barnham KJ, Masters CL, and Bush AI (2004). Neurodegenerative diseases and oxidative stress. Biomed. Nat. Rev. Drug Discov 3, 205–214. [DOI] [PubMed] [Google Scholar]
- Barrangou R, Birmingham A, Wiemann S, Beijersbergen RL, Hornung V, and Smith A. van B. (2015). Advances in CRISPR-Cas9 genome engineering: lessons learned from RNA interference. Nucleic Acids Res. 43, 3407–3419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bassik MC, Kampmann M, Lebbink RJ, Wang S, Hein MY, Poser I, Weibezahn J, Horlbeck MA, Chen S, Mann M, et al. (2013). A systematic mammalian genetic interaction map reveals pathways underlying ricin susceptibility. Cell 152, 909–922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bechard ME, Word AE, Tran AV, Liu X, Locasale JW, and McDonald OG (2018). Pentose conversions support the tumorigenesis of pancreatic cancer distant metastases. Oncogene 37, 5248–5256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blake DJ, Singh A, Kombairaju P, Malhotra D, Mariani TJ, Tuder RM, Gabrielson E, and Biswal S. (2010). Deletion of Keap1 in the lung attenuates acute cigarette smoke-induced oxidative stress and inflammation. Am. J. Respir. Cell Mol. Biol 42, 524–536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bligh EG, and Dyer WJ (1959). A rapid method of total lipid extraction and purification. Can. J. Biochem. Physiol 37, 911–917. [DOI] [PubMed] [Google Scholar]
- Bogdan AR, Miyazawa M, Hashimoto K, and Tsuji Y. (2016). Regulators of iron homeostasis: new players in metabolism, cell death, and disease. Trends Biochem. Sci 41, 274–286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bottger G, Barnett P, Klein AT, Kragt A, Tabak HF, and Distel B. (2000). Saccharomyces cerevisiae PTS1 receptor Pex5p interacts with the SH3 domain of the peroxisomal membrane protein Pex13p in an unconventional, non-PXXP-related manner. Mol. Biol. Cell 11, 3963–3976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brinkman EK, Chen T, Amendola M, and van Steensel B. (2014). Easy quantitative assessment of genome editing by sequence trace decomposition. Nucleic Acids Res. 42, e168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buchmeier NA, Libby SJ, Xu Y, Loewen PC, Switala J, Guiney DG, and Fang FC (1995). DNA repair is more important than catalase for Salmonella virulence in mice. J. Clin. Invest 95, 1047–1053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Busciglio J, and Yankner BA (1995). Apoptosis and increased generation of reactive oxygen species in Down’s syndrome neurons in vitro. Nature 378, 776–779. [DOI] [PubMed] [Google Scholar]
- Cairns RA, Harris IS, and Mak TW (2011). Regulation of cancer cell metabolism. Nat. Rev. Cancer 11, 85–95. [DOI] [PubMed] [Google Scholar]
- Chen Y, Azad MB, and Gibson SB (2009). Superoxide is the major reactive oxygen species regulating autophagy. Cell Death Differ. 16, 1040–1052. [DOI] [PubMed] [Google Scholar]
- Christodoulou D, Kuehne A, Estermann A, Fuhrer T, Lang P, and Sauer U. (2019). Reserve flux capacity in the pentose phosphate pathway by NADPH binding is conserved across kingdoms. iScience 19, 1133–1144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conger AD, and Fairchild LM (1952). Breakage of chromosomes by oxygen. Proc. Natl. Acad. Sci. U S A 38, 289–299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Contrepois K, Jiang L, and Snyder M. (2015). Optimized analytical procedures for the untargeted metabolomic profiling of human urine and plasma by combining hydrophilic interaction (HILIC) and reverse-phase liquid chromatography (RPLC)–mass spectrometry. Mol. Cell. Proteomics 14, 1684–1695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cunningham ML, Peak JG, and Peak MJ (1987). Single-strand DNA breaks in rodent and human cells produced by superoxide anion or its reduction products. Mutat. Res 184, 217–222. [DOI] [PubMed] [Google Scholar]
- Deans RM, Morgens DW, Ökesli A, Pillay S, Horlbeck MA, Kampmann M, Gilbert LA, Li A, Mateo R, Smith M, et al. (2016). Parallel shRNA and CRISPR-Cas9 screens enable antiviral drug target identification. Nat. Chem. Biol 12, 361–366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denat L, Kadekaro AL, Marrot L, Leachman SA, and Abdel-Malek ZA (2014). Melanocytes as instigators and victims of oxidative stress. J. Invest. Dermatol 134, 1512–1518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeNicola GM, Karreth FA, Humpton TJ, Gopinathan A, Wei C, Frese K, Mangal D, Yu KH, Yeo CJ, Calhoun ES, et al. (2011). Oncogeneinduced Nrf2 transcription promotes ROS detoxification and tumorigenesis. Nature 475, 106–110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dixon SJ, Lemberg KM, Lamprecht MR, Skouta R, Zaitsev EM, Gleason CE, Patel DN, Bauer AJ, Cantley AM, Yang WS, et al. (2012). Ferroptosis: an iron-dependent form of nonapoptotic cell death. Cell 149, 1060–1072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Driessens N, Versteyhe S, Ghaddhab C, Burniat A, De Deken X, Van Sande J, Dumont JE, Miot F, and Corvilain B. (2009). Hydrogen peroxide induces DNA single- and double-strand breaks in thyroid cells and is therefore a potential mutagen for this organ. Endocr. Relat. Cancer 16, 845–856. [DOI] [PubMed] [Google Scholar]
- Ebadi M, Srinivasan SK, and Baxi MD (1996). Oxidative stress and antioxidant therapy in Parkinson’s disease. Prog. Neurobiol 48, 1–19. [DOI] [PubMed] [Google Scholar]
- El-Brolosy MA, Kontarakis Z, Rossi A, Kuenne C, Gu€nther S, Fukuda N, Kikhi K, Boezio GLM, Takacs CM, Lai SL, et al. (2019). Genetic compensation triggered by mutant mRNA degradation. Nature 568, 193–197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finkel T, and Holbrook NJ (2000). Oxidants, oxidative stress and the biology of ageing. Nature 408, 239–247. [DOI] [PubMed] [Google Scholar]
- Forcina GC, Conlon M, Wells A, Cao JY, and Dixon SJ (2017). Systematic quantification of population cell death kinetics in mammalian cells. Cell Syst. 4, 600–610.e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forman HJ, and Torres M. (2002). Reactive oxygen species and cell signaling: respiratory burst in macrophage signaling. Am. J. Respir. Crit. Care Med 166, S4–S8. [DOI] [PubMed] [Google Scholar]
- Forman HJ, Bernardo A, and Davies KJA (2016). What is the concentration of hydrogen peroxide in blood and plasma? Arch. Biochem. Biophys 603, 48–53. [DOI] [PubMed] [Google Scholar]
- Frock RL, Hu J, Meyers RM, Ho YJ, Kii E, and Alt FW (2015). Genome-wide detection of DNA double-stranded breaks induced by engineered nucleases. Nat. Biotechnol 33, 179–188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujiki Y, Okumoto K, Mukai S, Honsho M, and Tamura S. (2014). Peroxisome biogenesis in mammalian cells. Front. Physiol 5, 307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gadagkar SR, and Call GB (2015). Computational tools for fitting the Hill equation to dose-response curves. J. Pharmacol. Toxicol. Methods 71, 68–76. [DOI] [PubMed] [Google Scholar]
- Go YM, and Jones DP (2008). Redox compartmentalization in eukaryotic cells. Biochim. Biophys. Acta 1780, 1273–1290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grimm D, Streetz KL, Jopling CL, Storm TA, Pandey K, Davis CR, Marion P, Salazar F, and Kay MA (2006). Fatality in mice due to oversaturation of cellular microRNA/short hairpin RNA pathways. Nature 441, 537–541. [DOI] [PubMed] [Google Scholar]
- Gulbranson DR, Crisman L, Lee M, Ouyang Y, Menasche BL, Demmitt BA, Wan C, Nomura T, Ye Y, Yu H, et al. (2019). AAGAB controls AP2 adaptor assembly in clathrin-mediated endocytosis. Dev. Cell 50, 436–446.e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo JY, Chen HY, Mathew R, Fan J, Strohecker AM, Karsli-Uzunbas G, Kamphorst JJ, Chen G, Lemons JMS, Karantza V, et al. (2011). Activated Ras requires autophagy to maintain oxidative metabolism and tumorigenesis. Genes Dev. 25, 460–470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo Z, Kozlov S, Lavin MF, Person MD, and Paull TT (2010). ATM activation by oxidative stress. Science 330, 517–521. [DOI] [PubMed] [Google Scholar]
- Halliwell B, and Gutteridge JMC (1990). Role of free radicals and catalytic metal ions in human disease: an overview. Methods Enzymol. 186, 1–85. [DOI] [PubMed] [Google Scholar]
- Halliwell B, Clement MV, and Long LH (2000). Hydrogen peroxide in the human body. FEBS Lett. 486, 10–13. [DOI] [PubMed] [Google Scholar]
- Han K, Jeng EE, Hess GT, Morgens DW, Li A, and Bassik MC (2017). Synergistic drug combinations for cancer identified in a CRISPR screen for pairwise genetic interactions. Nat. Biotechnol 35, 463–474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haney MJ, Klyachko NL, Zhao Y, Gupta R, Plotnikova EG, He Z, Patel T, Piroyan A, Sokolsky M, Kabanov AV, et al. (2015). Exosomes as drug delivery vehicles for Parkinson’s disease therapy. J. Control. Release 207, 18–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haney MS, Bohlen CJ, Morgens DW, Ousey JA, Barkal AA, Tsui CK, Ego BK, Levin R, Kamber RA, Collins H, et al. (2018). Identification of phagocytosis regulators using magnetic genome-wide CRISPR screens. Nat. Genet 50, 1716–1727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harper CC, Berg JM, and Gould SJ (2003). PEX5 binds the PTS1 independently of Hsp70 and the peroxin PEX12. J. Biol. Chem 278, 7897–7901. [DOI] [PubMed] [Google Scholar]
- Hein MY, Hubner NC, Poser I, Cox J, Nagaraj N, Toyoda Y, Gak IA, Weisswange I, Mansfeld J, Buchholz F, et al. (2015). A human interactome in three quantitative dimensions organized by stoichiometries and abundances. Cell 163, 712–723. [DOI] [PubMed] [Google Scholar]
- Horlbeck MA, Gilbert LA, Villalta JE, Adamson B, Pak RA, Chen Y, Fields AP, Park CY, Corn JE, Kampmann M, et al. (2016). Compact and highly active next-generation libraries for CRISPR-mediated gene repression and activation. eLife 5, 1–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hosoi KI, Miyata N, Mukai S, Furuki S, Okumoto K, Cheng EH, and Fujiki Y. (2017). The VDAC2-BAK axis regulates peroxisomal membrane permeability. J. Cell Biol 216, 709–722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ichijo H, Nishida E, Irie K, ten Dijke P, Saitoh M, Moriguchi T, Takagi M, Matsumoto K, Miyazono K, and Gotoh Y. (1997). Induction of apoptosis by ASK1, a mammalian MAPKKK that activates SAPK/JNK and p38 signaling pathways. Science 275, 90–94. [DOI] [PubMed] [Google Scholar]
- Ishii T, Yasuda K, Akatsuka A, Hino O, Hartman PS, and Ishii N. (2005). A mutation in the SDHC gene of complex II increases oxidative stress, resulting in apoptosis and tumorigenesis. Cancer Res. 65, 203–209. [PubMed] [Google Scholar]
- Jackson AL, and Linsley PS (2010). Recognizing and avoiding siRNA off-target effects for target identification and therapeutic application. Nat. Rev. Drug Discov 9, 57–67. [DOI] [PubMed] [Google Scholar]
- Jeng EE, Bhadkamkar V, Ibe NU, Gause H, Jiang L, Chan J, Jian R, Jimenez-Morales D, Stevenson E, Krogan NJ, et al. (2019). Systematic identification of host cell regulators of Legionella pneumophila pathogenesis using a genome-wide CRISPR screen. Cell Host Microbe 26, 551–563.e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaelin WG (2012). Use and abuse of RNAi to study mammalian gene function. Science 337, 421–422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kampmann M, Horlbeck MA, Chen Y, Tsai JC, Bassik MC, Gilbert LA, Villalta JE, Kwon SC, Chang H, Kim VN, et al. (2015). Next-generation libraries for robust RNA interference-based genome-wide screens. Proc. Natl. Acad. Sci. U. S. A 112, E3384–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimura J, Nguyen ST, Liu H, Taira N, Miki Y, and Yoshida K. (2008). A functional genome-wide RNAi screen identifies TAF1 as a regulator for apoptosis in response to genotoxic stress. Nucleic Acids Res. 36, 5250–5259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirkman HN, Rolfo M, Ferraris AM, and Gaetani GF (1999). Mechanisms of protection of catalase by NADPH. J. Biol. Chem 274, 13908–13914. [DOI] [PubMed] [Google Scholar]
- Kobayashi A, Kang MI, Okawa H, Ohtsuji M, Zenke Y, Chiba T, Igarashi K, and Yamamoto M. (2004). Oxidative stress sensor Keap1 functions as an adaptor for Cul3-based E3 ligase to regulate proteasomal degradation of Nrf2. Mol. Cell. Biol 24, 7130–7139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koike-Yusa H, Li Y, Tan EP, Velasco-Herrera MDC, and Yusa K. (2014). Genome-wide recessive genetic screening in mammalian cells with a lentiviral CRISPR-guide RNA library. Nat. Biotechnol 32, 267–273. [DOI] [PubMed] [Google Scholar]
- Kong H, and Chandel NS (2018). Regulation of redox balance in cancer and T cells. J. Biol. Chem 293, 7499–7507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuehne A, Emmert H, Soehle J, Winnefeld M, Fischer F, Wenck H, Gallinat S, Terstegen L, Lucius R, Hildebrand J, et al. (2015). Acute activation of oxidative pentose phosphate pathway as first-line response to oxidative stress in human skin cells. Mol. Cell 59, 359–371. [DOI] [PubMed] [Google Scholar]
- Leidgens S, Bullough KZ, Shi H, Li F, Shakoury-Elizeh M, Yabe T, Subramanian P, Hsu E, Natarajan N, Nandal A, et al. (2013). Each member of the poly-r(C)-binding protein 1 (PCBP) family exhibits iron chaperone activity toward ferritin. J. Biol. Chem 288, 17791–17802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liochev SI (2013). Reactive oxygen species and the free radical theory of aging. Free Radic. Biol. Med 60, 1–4. [DOI] [PubMed] [Google Scholar]
- Liu X, and Zweier JL (2001). A real-time electrochemical technique for measurement of cellular hydrogen peroxide generation and consumption: evaluation in human polymorphonuclear leukocytes. Free Radic. Biol. Med 31, 894–901. [DOI] [PubMed] [Google Scholar]
- Loerke D, Mettlen M, Yarar D, Jaqaman K, Jaqaman H, Danuser G, and Schmid SL (2009). Cargo and dynamin regulate clathrin-coated pit maturation. PLoS Biol. 7, e1000057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long LH, and Halliwell B. (2000). Coffee drinking increases levels of urinary hydrogen peroxide detected in healthy human volunteers. Free Radic. Res 32, 463–467. [DOI] [PubMed] [Google Scholar]
- Long LH, Evans PJ, and Halliwell B. (1999). Hydrogen peroxide in human urine: implications for antioxidant defense and redox regulation. Biochem. Biophys. Res. Commun 262, 605–609. [DOI] [PubMed] [Google Scholar]
- Lu SC (2009). Regulation of glutathione synthesis. Mol. Aspects Med 30, 42–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu SC (2013). Glutathione synthesis. Biochim. Biophys. Acta, Gen. Subj 1830, 3143–3153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma Z, Zhu P, Shi H, Guo L, Zhang Q, Chen Y, Chen S, Zhang Z, Peng J, and Chen J. (2019). PTC-bearing mRNA elicits a genetic compensation response via Upf3a and COMPASS components. Nature 568, 259–263. [DOI] [PubMed] [Google Scholar]
- Mantzaris MD, Bellou S, Skiada V, Kitsati N, Fotsis T, and Galaris D. (2016). Intracellular labile iron determines H2O2-induced apoptotic signaling via sustained activation of ASK1/JNK-p38 axis. Free Radic. Biol. Med 97, 454–465. [DOI] [PubMed] [Google Scholar]
- Maresca V, Roccella M, Roccella F, Camera E, Del Porto G, Passi S, Grammatico P, and Picardo M. (1997). Increased sensitivity to peroxidative agents as a possible pathogenic factor of melanocyte damage in vitiligo. J. Invest. Dermatol 109, 310–313. [DOI] [PubMed] [Google Scholar]
- Matsuzawa A, Saegusa K, Noguchi T, Sadamitsu C, Nishitoh H, Nagai S, Koyasu S, Matsumoto K, Takeda K, and Ichijo H. (2005). ROS-dependent activation of the TRAF6-ASK1-p38 pathway is selectively required for TLR4-mediated innate immunity. Nat. Immunol 6, 587–592. [DOI] [PubMed] [Google Scholar]
- McDonald OG, Li X, Saunders T, Tryggvadottir R, Mentch SJ, Warmoes MO, Word AE, Carrer A, Salz TH, Natsume S, et al. (2017). Epigenomic reprogramming during pancreatic cancer progression links anabolic glucose metabolism to distant metastasis. Nat. Genet 49, 367–376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mello Filho AC, Hoffmann ME, and Meneghini R. (1984). Cell killing and DNA damage by hydrogen peroxide are mediated by intracellular iron. Biochem. J 218, 273–275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meneghini R. (1997). Iron homeostasis, oxidative stress, and DNA damage. Free Radic. Biol. Med 23, 783–792. [DOI] [PubMed] [Google Scholar]
- Meng T, Fukada T, and Tonks NK (2002). Reversible oxidation and inactivation of protein tyrosine phosphatases in vivo. Mol. Cell 9, 387–399. [DOI] [PubMed] [Google Scholar]
- Miclet E, Stoven V, Michels PAM, Opperdoes FR, Lallemand JY, and Duffieux F. (2001). NMR spectroscopic analysis of the first two steps of the pentose-phosphate pathway elucidates the role of 6-phosphogluconolactonase. J. Biol. Chem 276, 34840–34846. [DOI] [PubMed] [Google Scholar]
- Miyazawa S, Osumi T, Hashimoto T, Ohno K, Miura S, and Fujiki Y. (1989). Peroxisome targeting signal of rat liver acyl-coenzyme A oxidase resides at the carboxy terminus. Mol. Cell. Biol 9, 83–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moiseeva TN, Gamper AM, Hood BL, Conrads TP, and Bakkenist CJ (2016). Human DNA polymerase ε is phosphorylated at serine-1940 after DNA damage and interacts with the iron-sulfur complex chaperones CIAO1 and MMS19. DNA Repair (Amst.) 43, 9–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moloney JN, and Cotter TG (2018). ROS signalling in the biology of cancer. Semin. Cell Dev. Biol 80, 50–64. [DOI] [PubMed] [Google Scholar]
- Morgens DW, Deans RM, Li A, and Bassik MC (2016). Systematic comparison of CRISPR/Cas9 and RNAi screens for essential genes. Nat. Biotechnol 34, 634–636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morgens DW, Wainberg M, Boyle EA, Ursu O, Araya CL, Kimberly Tsui C, Haney MS, Hess GT, Han K, Jeng EE, et al. (2017). Genome-scale measurement of off-target activity using Cas9 toxicity in high-throughput screens. Nat. Commun 8, 1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukai S, and Fujiki Y. (2006). Molecular mechanisms of import of peroxisome-targeting signal type 2 (PTS2) proteins by PTS2 receptor Pex7p and PTS1 receptor Pex5pL. J. Biol. Chem 281, 37311–37320. [DOI] [PubMed] [Google Scholar]
- Nathan C, and Cunningham-Bussel A. (2013). Beyond oxidative stress: an immunologist’s guide to reactive oxygen species. Nat. Rev. Immunol 13, 349–361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishida Y. (2012). The chemical mechanism of oxidative stress due to the nontransferrin-bound iron (NTBI). Adv. Biosci. Biotechnol 3, 1076–1086. [Google Scholar]
- Nagai H, Noguchi T, Homma K, Katagiri K, Takeda K, Matsuzawa A, and Ichijo H. (2009). Ubiquitin-like sequence in ASK1 plays critical roles in the recognition and stabilization by USP9X and oxidative stress-induced cell death. Mol. Cell 36, 805–818. [DOI] [PubMed] [Google Scholar]
- Okumoto K, Kametani Y, and Fujiki Y. (2011). Two proteases, trypsin domain-containing 1 (Tysnd1) and peroxisomal lon protease (PsLon), cooperatively regulate fatty acid β-oxidation in peroxisomal matrix. J. Biol. Chem 286, 44367–44379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Oliveira S, López-Munoz A, Candel S, Pelegrĺn P, Calado Â, and Mulero V. (2014). ATP modulates acute inflammation in vivo through dual oxidase 1-derived H2O2 production and NF-kB activation. J. Immunol 192, 5710–5719. [DOI] [PubMed] [Google Scholar]
- Otera H, and Fujiki Y. (2012). Pex5p imports folded tetrameric catalase by interaction with Pex13p. Traffic 13, 1364–1377. [DOI] [PubMed] [Google Scholar]
- Otera H, Harano T, Honsho M, Ghaedi K, Mukai S, Tanaka A, Kawai A, Shimizu N, and Fujiki Y. (2000). The mammalian peroxin Pex5pL, the longer isoform of the mobile peroxisome targeting signal (PTS) type 1 transporter, translocates the Pex7p.PTS2 protein complex into peroxisomes via its initial docking site, Pex14p. J. Biol. Chem 275, 21703–21714. [DOI] [PubMed] [Google Scholar]
- Parnas O, Jovanovic M, Eisenhaure TM, Herbst RH, Dixit A, Ye CJ, Przybylski D, Platt RJ, Tirosh I, Sanjana NE, et al. (2015). A genomewide CRISPR screen in primary immune cells to dissect regulatory networks. Cell 162, 675–686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parr CW (1956). Inhibition of phosphoglucose isomerase. Nature 178, 1401. [DOI] [PubMed] [Google Scholar]
- Patial S, Stumpo DJ, Young WS, Ward JM, Flake GP, and Blackshear PJ (2016). Effects of combined tristetraprolin/tumor necrosis factor receptor deficiency on the splenic transcriptome. Mol. Cell. Biol 36, 1395–1411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patra KC, and Hay N. (2014). The pentose phosphate pathway and cancer. Trends Biochem. Sci 39, 347–354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pillai S, Oresajo C, and Hayward J. (2005). Ultraviolet radiation and skin aging: Roles of reactive oxygen species, inflammation and protease activation, and strategies for prevention of inflammation-induced matrix degradation A review. Int. J. Cosmet. Sci 27, 17–34. [DOI] [PubMed] [Google Scholar]
- Pruett-Miller SM, Reading DW, Porter SN, and Porteus MH (2009). Attenuation of zinc finger nuclease toxicity by small-molecule regulation of protein levels. PLoS Genet. 5, e1000376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Purdue PE, and Lazarow PB (1996). Targeting of human catalase to peroxisomes is dependent upon a novel COOH-terminal peroxisomal targeting sequence. J. Cell Biol 134, 849–862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ray PD, Huang BW, and Tsuji Y. (2012). Reactive oxygen species (ROS) homeostasis and redox regulation in cellular signaling. Cell. Signal 24, 981–990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reczek CR, Birsoy K, Kong H, Martínez-Reyes I, Wang T, Gao P, Sabatini DM, and Chandel NS (2017). A CRISPR screen identifies a pathway required for paraquat-induced cell death. Nat. Chem. Biol 13, 1274–1279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts CK, Barnard RJ, Sindhu RK, Jurczak M, Ehdaie A, and Vaziri ND (2005). A high-fat, refined-carbohydrate diet induces endothelial dysfunction and oxidant/antioxidant imbalance and depresses NOS protein expression. J. Appl. Physiol 98, 203–210. [DOI] [PubMed] [Google Scholar]
- Rosenbluh J, Xu H, Harrington W, Gill S, Wang X, Vazquez F, Root DE, Tsherniak A, and Hahn WC (2017). Complementary information derived from CRISPR Cas9 mediated gene deletion and suppression. Nat. Commun 8, 1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakurai T, He G, Matsuzawa A, Yu GY, Maeda S, Hardiman G, and Karin M. (2008). Hepatocyte necrosis induced by oxidative stress and IL-1 alpha release mediate carcinogen-induced compensatory proliferation and liver tumorigenesis. Cancer Cell 14, 156–165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schallreuter KU, Moore J, Wood JM, Beazley WD, Gaze DC, Tobin DJ, Marshall HS, Panske A, Panzig E, and Hibberts NA (1999). In vivo and in vitro evidence for hydrogen peroxide (H2O2) accumulation in the epidermis of patients with vitiligo and its successful removal by a UVB-activated pseudocatalase. J. Investig. Dermatol. Symp. Proc 4, 91–96. [DOI] [PubMed] [Google Scholar]
- Schallreuter KU, Salem MAEL, Gibbons NCJ, Martinez A, Slominski R, demann J, and Rokos H. (2012). Blunted epidermal L-tryptophan metabolism in vitiligo affects immune response and ROS scavenging by Fenton chemistry, part 1: epidermal H2O2/ONOO(−)-mediated stress abrogates tryptophan hydroxylase and dopa decarboxylase activities, leading to low seroton. FASEB J. 26, 2457–2470. [DOI] [PubMed] [Google Scholar]
- Scherz-Shouval R, Shvets E, Fass E, Shorer H, Gil L, and Elazar Z. (2007). Reactive oxygen species are essential for autophagy and specifically regulate the activity of Atg4. EMBO J. 26, 1749–1760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schieber M, and Chandel NS (2014). ROS function in redox signaling and oxidative stress. Curr. Biol 24, R453–R462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schrader M, and Fahimi HD (2006). Peroxisomes and oxidative stress. Biochim. Biophys. Acta 1763, 1755–1766. [DOI] [PubMed] [Google Scholar]
- Schuster S, and Feldstein AE (2017). NASH: novel therapeutic strategies targeting ASK1 in NASH. Nat. Rev. Gastroenterol. Hepatol. 14, 329–330. [DOI] [PubMed] [Google Scholar]
- Sekine Y, Hatanaka R, Watanabe T, Sono N, Iemura S, Natsume T, Kuranaga E, Miura M, Takeda K, and Ichijo H. (2012). The Kelch repeat protein KLHDC10 regulates oxidative stress-induced ASK1 activation by suppressing PP5. Mol. Cell 48, 692–704. [DOI] [PubMed] [Google Scholar]
- Shalem O, Sanjana NE, and Zhang F. (2015). High-throughput functional genomics using CRISPR-Cas9. Nat. Rev. Genet 16, 299–311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shan C, Elf S, Ji Q, Kang HB, Zhou L, Hitosugi T, Jin L, Lin R, Zhang L, Seo JH, et al. (2014). Lysine acetylation activates 6-phosphogluconate dehydrogenase to promote tumor growth. Mol. Cell 55, 552–565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheikh FG, Pahan K, Khan M, Barbosa E, and Singh I. (1998). Abnormality in catalase import into peroxisomes leads to severe neurological disorder. Proc. Natl. Acad. Sci. U S A 95, 2961–2966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi H, Bencze KZ, Stemmler TL, and Philpott CC (2008). A cytosolic iron chaperone that delivers iron to ferritin. Science 320, 1207–1210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimizu N, Itoh R, Hirono Y, Otera H, Ghaedi K, Tateishi K, Tamura S, Okumoto K, Harano T, Mukai S, et al. (1999). The peroxin Pex14p. cDNA cloning by functional complementation on a Chinese hamster ovary cell mutant, characterization, and functional analysis. J. Biol. Chem 274, 12593– 12604. [DOI] [PubMed] [Google Scholar]
- Silva JM, Marran K, Parker JS, Silva J, Golding M, Schlabach MR, Elledge SJ, Hannon GJ, and Chang K. (2008). Profiling essential genes in human mammary cells by multiplex RNAi screening. Science 319, 617–620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song X, Mosby N, Yang J, Xu A, Abdel-Malek Z, and Kadekaro AL (2009). alpha-MSH activates immediate defense responses to UV-induced oxidative stress in human melanocytes. Pigment Cell Melanoma Res. 22, 809–818. [DOI] [PubMed] [Google Scholar]
- Stanley WA, Filipp FV, Kursula P, Schu€ller N, Erdmann R, Schliebs W, Sattler M, and Wilmanns M. (2006). Recognition of a functional peroxisome type 1 target by the dynamic import receptor pex5p. Mol. Cell 24, 653–663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stehling O, Mascarenhas J, Vashisht AA, Sheftel AD, Niggemeyer B, Rösser R, Pierik AJ, Wohlschlegel JA, and Lill R. (2013). Human CIA2A-FAM96A and CIA2B-FAM96B integrate iron homeostasis and maturation of different subsets of cytosolic-nuclear iron-sulfur proteins. Cell Metab. 18, 187–198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szatrowski TP, and Nathan CF (1991). Production of large amounts of hydrogen peroxide by human tumor cells. Cancer Res. 51, 794–798. [PubMed] [Google Scholar]
- Tao R, Zhao Y, Chu H, Wang A, Zhu J, Chen X, Zou Y, Shi M, Liu R, Su N, et al. (2017). Genetically encoded fluorescent sensors reveal dynamic regulation of NADPH metabolism. Nat. Methods 14, 720–728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Test ST, and Weiss SJ (1984). Quantitative and temporal characterization of the extracellular H2O2 pool generated by human neutrophils. J. Biol. Chem 259, 399–405. [PubMed] [Google Scholar]
- Tortorella S, and Karagiannis TC (2014). Transferrin receptor-mediated endocytosis: a useful target for cancer therapy. J. Membr. Biol 247, 291–307. [DOI] [PubMed] [Google Scholar]
- Totter JR (1980). Spontaneous cancer and its possible relationship to oxygen metabolism. Proc. Natl. Acad. Sci. U S A 77, 1763–1767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai SQ, Zheng Z, Nguyen NT, Liebers M, Topkar VV, Thapar V, Wyvekens N, Khayter C, Iafrate AJ, Le LP, et al. (2015). GUIDE-seq enables genome-wide profiling of off-target cleavage by CRISPR-Cas nucleases. Nat. Biotechnol 33, 187–197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsukamoto T, Yokota S, and Fujiki Y. (1990). Isolation and characterization of Chinese hamster ovary cell mutants defective in assembly of peroxisomes. J. Cell Biol 110, 651–660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turk B, and Turk V. (2009). Lysosomes as “suicide bags” in cell death: myth or reality? J. Biol. Chem 284, 21783–21787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ueda J, Saito N, Shimazu Y, and Ozawa T. (1996). A comparison of scavenging abilities of antioxidants against hydroxyl radicals. Arch. Biochem. Biophys 333, 377–384. [DOI] [PubMed] [Google Scholar]
- Ueno S, Yasutake K, Tohyama D, Fujimori T, Ayusawa D, and Fujii M. (2012). Systematic screen for genes involved in the regulation of oxidative stress in the nematode Caenorhabditis elegans. Biochem. Biophys. Res. Commun 420, 552–557. [DOI] [PubMed] [Google Scholar]
- Varani J, and Ward PA (1994). Mechanisms of endothelial cell injury in acute inflammation. Shock 2, 311–319. [DOI] [PubMed] [Google Scholar]
- Varma SD, and Devamanoharan PS (1990). Excretion of hydrogen peroxide in human urine. Free Radic. Res. Commun 8, 73–78. [DOI] [PubMed] [Google Scholar]
- Vashisht AA, Zumbrennen KB, Huang X, Powers DN, Durazo A, Sun D, Bhaskaran N, Persson A, Uhlen M, Sangfelt O, et al. (2009). Control of iron homeostasis by an iron-regulated ubiquitin ligase. Science 326, 718–721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walton PA, Brees C, Lismont C, Apanasets O, and Fransen M. (2017). The peroxisomal import receptor PEX5 functions as a stress sensor, retaining catalase in the cytosol in times of oxidative stress. Biochim. Biophys. Acta 1864, 1833–1843. [DOI] [PubMed] [Google Scholar]
- Wang T, Wei JJ, Sabatini DM, and Lander ES (2014). Genetic screens in human cells using the CRISPR-Cas9 system. Science 343, 80–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiss SJ (1980). The role of superoxide in the destruction of erythrocyte targets by human neutrophils. J. Biol. Chem 255, 9912–9917. [PubMed] [Google Scholar]
- Williams C, Bener Aksam E, Gunkel K, Veenhuis M, and van der Klei IJ (2012). The relevance of the non-canonical PTS1 of peroxisomal catalase. Biochim. Biophys. Acta 1823, 1133–1141. [DOI] [PubMed] [Google Scholar]
- Wu DC, Teismann P, Tieu K, Vila M, Jackson-Lewis V, Ischiropoulos H, and Przedborski S. (2003). NADPH oxidase mediates oxidative stress in the 1-methyl-4-phenyl-1,2,3,6-tetrahydropyridine model of Parkinson’s disease. Proc. Natl. Acad. Sci. U S A 100, 6145–6150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yanatori I, Richardson DR, Imada K, and Kishi F. (2016). Iron export through the transporter ferroportin 1 is modulated by the iron chaperone PCBP2. J. Biol. Chem 291, 17303–17318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young IS, and Woodside JV (2001). Antioxidants in health and disease. J. Clin. Pathol 54, 176–186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Y, Zhu S, Cai C, Yuan P, Li C, Huang Y, and Wei W. (2014). High-throughput screening of a CRISPR/Cas9 library for functional genomics in human cells. Nature 509, 487–491. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Sequencing data are available at Sequence Read Archive (SRA) accession number SRP235195 under BioProject accession number PRJNA594164.