Abstract
The mechanisms by which truncating mutations in MYBPC3 (encoding cardiac myosin binding protein-C; cMyBPC) or myosin missense mutations cause hyper-contractility and poor relaxation in hypertrophic cardiomyopathy (HCM) are incompletely understood. Using genetic and biochemical approaches we explored how depletion of cMyBPC altered sarcomere function. We demonstrate that stepwise loss of cMyBPC resulted in reciprocal augmentation of myosin contractility. Direct attenuation of myosin function, via a damaging missense variant (F764L) that causes dilated cardiomyopathy (DCM) normalized the increased contractility from cMyBPC depletion. Depletion of cMyBPC also altered dynamic myosin conformations during relaxation - enhancing the myosin state that enables ATP hydrolysis and thin filament interactions while reducing the super relaxed conformation associated with energy conservation. MYK-461, a pharmacologic inhibitor of myosin ATPase, rescued relaxation deficits and restored normal contractility in mouse and human cardiomyocytes with MYBPC3 mutations. These data define dosage-dependent effects of cMyBPC on myosin that occur across the cardiac cycle as the pathophysiologic mechanisms by which MYBPC3 truncations cause HCM. Therapeutic strategies to attenuate cMyBPC activity may rescue depressed cardiac contractility in DCM patients, while inhibiting myosin by MYK-461 should benefit the substantial proportion of HCM patients with MYBPC3 mutations.
One Sentence Summary:
Analyses of cardiomyocytes with hypertrophic cardiomyopathy mutations in MYBPC3 reveal that these directly activate myosin contraction by disrupting myosin states of relaxation, and that genetic or pharmacological manipulation of myosin therapeutically abates the effects of MYBPC3 mutations.
Introduction
Hypertrophic cardiomyopathy (HCM) is a heritable disease of heart muscle affecting ~ 1 in 5001 individuals. Patient symptoms can be minimal or relentlessly progressive with resultant heart failure and/or sudden cardiac death2. Adverse clinical outcomes in HCM increase with disease duration, thereby underscoring the importance of therapeutic strategies to abate disease progression3.
Dominant pathogenic variants in eight sarcomere genes cause HCM, but predominate in MYBPC3 and MYH7 (encoding β cardiac myosin heavy chain)4. The overwhelming majority of HCM founder mutations5–11, including one affecting 4% of South Asians12 reside in MYBPC3. All HCM mutations in MYH7 encode missense substitutions4 and mutant myosins are incorporated into the sarcomere. By contrast, most MYBPC3 mutations are truncating and are predicted to cause haploinsufficiency of cMyBPC13, 14. The mechanisms by which distinctive mutations in these two sarcomere proteins uniformly produce hyperdynamic contraction and poor relaxation (diastolic dysfunction) in advance of the morphologic remodelling in HCM15–17 remain incompletely understood18.
Biophysical analyses demonstrate that HCM mutations in β-MHC, the molecular motor of the sarcomere can increase ATPase activity, actin-sliding velocity, and power. Structural analyses predict that these interfere with the myosin IHM (inter-head motif) shifting dynamic conformations of relaxed paired myosin molecules19, 20,21. These conformations are denoted as i) disordered relaxation (DRX), a state where only one myosin head could be active, able to hydrolyse ATP and potentiate force; and ii) super relaxation (SRX), a state of dual inactivation of myosins with both ATPases inhibited. The IHM is an evolutionarily conserved motif that is found in all muscle myosins and in primitive animals with non-muscle myosin II, indicating the importance of inhibiting myosin during relaxation22.
cMyBPC has structural and functional roles in sarcomere biology23. cMyBPC is generally thought to serve as a brake that limits cross bridge interactions23 through its biophysical interactions of its amino and carboxyl termini with both myosin22 and actin23. Phosphorylation of the amino terminus of cMyBPC reduces myosin interactions and increases ATPase activity and actin interactions to promote cross-bridge formation24, events that are reversed by calcium concentrations that maximally activate thin filaments25, 26. As such the phosphorylation state of cMyBPC is hypothesized to regulate the number of myosin heads available for force production24.
Interpreting these interactions in the context of human HCM mutations that reduce cMyBPC expression is complex for several reasons. Cardiac histopathology and in vivo function of heterozygous Mybpc3+/− mice, which genetically recapitulate human HCM mutations, are indistinguishable from wildtype. Homozygous Mybpc3−/− mice have a developmental defect in the normal pathways for cytokinesis that results increased numbers of cardiomyocytes that are mononuclear,27, 28 resulting in ventricular dilatation and decreased contractile force25, 29. Recent studies also demonstrate that loss of cMyBPC also alter proportions of relaxed myosin in DRX and SRX conformations30, 31, but whether or not this relates to contractility is unknown.
To better understand how MYBPC3 mutations cause HCM, we assessed sarcomere function in the setting of cMyBPC deficiency and genetically altered myosin or pharmacologic attenuation of myosin ATPase activity. In combination, these assays shed light on unifying mechanisms that drive HCM pathophysiology and demonstrate that a single pharmacologic manipulation of myosin corrects sarcomere dysfunction caused by MYBPC3 mutations, thereby providing a promising avenue for treating this prevalent cause of human HCM.
Results
We studied three mouse models with altered cMyBPC expression (Supplemental Figure 1). Mybpc3t/+ and Mybpc3t/t mice carry endogenous heterozygous or homozygous truncating mutations32, 33 and express graded reductions of cMyBPC protein. MyBPC-RNAi27 are wildtype (WT) mice transfected (P10) with a cardiotropic adeno-associated (serotype 9) virus carrying green fluorescent protein (GFP) and RNAi targeting Mybpc3 transcripts. Injection of 5 × 1011 viral genomes (vg)/kg reduced Mybpc3 transcripts to less than 10% of quantities in WT mice and abolished protein expression (Supplemental Figures 1– 4). This strategy bypassed the developmental defect in Mybpc3t/t mice and also excluded potential functional effects of a small amount of truncated residual cMyBPC protein in homozygous mice (Supplemental Figure 1 C– D). Sham-RNAi +/− denotes Mybpc3t /+ mice transfected with virus carry GFP alone. We also studied heterozygous (Myh6F764L/+) and homozygous (Myh6F764L/ F764L) mice, which model a human MYH7 mutation that causes dilated cardiomyopathy (DCM). This mutation significantly reduces both actin-activated myosin ATPase activity and actin filament velocity34, 35.
In vivo cardiac phenotypes in Mypbc3 mutant mice
Previous studies demonstrate that Mybpc3t/+ mice have normal cardiac contractility and minimally increased LV wall thickness in comparison to WT that is far less than occurs in other HCM mouse models3. cMyBPC phosphorylation in Mybpc3t/+ and WT mice was comparable (Supplemental Figure 1E–G). Mybpc3t/t mice have significantly increased LV volumes and mass but depressed contractile function due in part to increased numbers of cardiomyocytes from additional perinatal divisions prior to permanent exit from the cell cycle27. MyBPC-RNAi mice exhibit minimal LV hypertrophy (Supplemental Figure 2A–D)27 without changes in ventricular volumes or fractional shortening, an in vivo measure of contractility.
Contractility and Relaxation in Cardiomyocytes from Mypbc3 Mutant Mice.
As in vivo contractility and relaxation reflects sarcomere performance as well as myocardial geometry, histopathology, and hemodynamic load, we studied ex vivo cardiomyocytes to assess biophysical functions of sarcomeres with altered cMyBPC expression (Figure 1). Isolated cardiomyocytes from at least four mice of each genotype (cells numbers indicated in figure legends) were studied. Cardiomyocytes from Mybpc3t/t mice were more fragile and more heterogeneous than in other models.
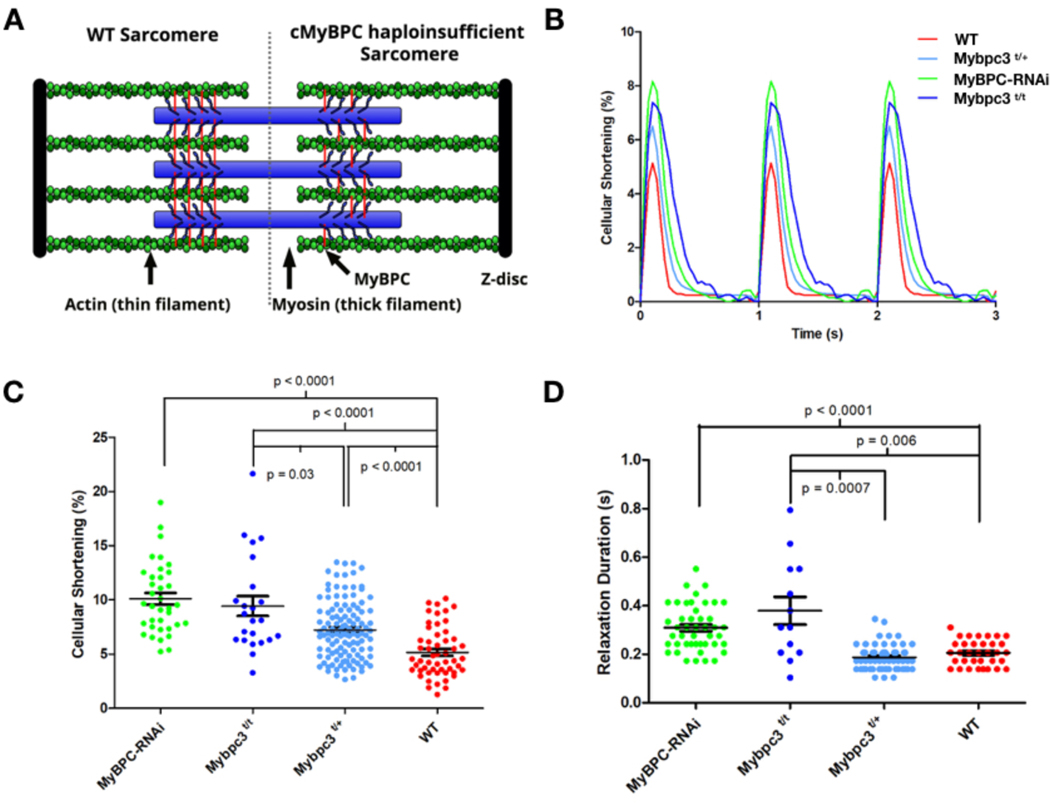
Figure 1: Contractile characterization of cMyBPC mouse models.
A) A schematic depiction of the WT sarcomere with normal cMyBPC integration (left half) and the consequences of mutations that deplete cMyBPC quantities in the sarcomere (right half). B) Representative contractile waveforms from isolated cardiomyocytes paced at 1Hz. Sarcomere lengths of isolated cardiomyocytes were tracked to define the percentage shortening per cell and duration of relaxation. Each trace is the averaged waveform across all cells analyzed for each treatment group. C) Comparisons of cellular shortening of isolated cardiomyocytes from four mice with different genotypes. (Cells analyzed: MyBPC-RNAi = 36; Mybpc3t/t = 23; Mybpc3t/+ = 118,WT= 53.) Data is plot as mean ± SEM. D) Measures of duration from peak contraction to relaxation in seconds plot as mean ± SEM (Cells analyzed: MyBPC-RNAi = 34; Mybpc3t/t = 13; Mybpc3t/+= 61,WT = 30).
Resting sarcomere lengths (Supplemental Figure 3) were comparable in cardiomyocytes from WT and Mybpc3t/+ mice, modestly decreased in cells from MyBPC-RNAi mice, and variable in cardiomyocytes from Mybpc3t/t mice. We assessed sarcomere lengths throughout the contractile cycle were determined to define percent shortening, a surrogate for systolic function (Figure 1B, C). Cardiomyocytes isolated from naïve or sham-RNAi +/− mice had comparable cell shortening thereby excluding an effect of AAV9 on contractility (Supplemental Figure 2D). Cardiomyocytes with altered cMyBPC expression had dosage-dependent increases in maximal cellular shortening. In comparison to WT, Mybpc3t/+ cardiomyocytes had 50% increased shortening (7.2 ± 0.25% p < 0.03), whilst cellular shortening was increased by 100% in cardiomyocytes lacking cMyBPC (MyBPC-RNAi: 10 ± 0.6%; Mybpc3t/t: 9.5 ± 0.9%; p<0.0001). Notably, augmentation of cellular shortening of isolated cardiomyocytes with cMyBPC deficiency did not result in increased contractility in vivo,27, 32 an observation that implies additional (biochemical, transcriptional, and morphologic) processes can modulate ensemble systolic performance of cardiomyocytes.
Human HCM is characterized by impaired diastolic performance, a parameter that is difficult to assess in mice. Instead, we tracked sarcomere lengths in isolated cardiomyocytes across the contractile cycle to assess the duration of relaxation as a quantitative proxy for diastolic function (Figure 1D). Relaxation was prolonged in MyBPC-RNAi and Mybpc3t/t cardiomyocytes (0.31 ± 0.02; p < 0.0001 and 0.38 ± 0.06; p < 0.0056) compared to WT cells (0.21 ± 0.02), while the duration of relaxation in Mybpc3t/+ cardiomyocytes was indistinguishable from WT (0.19 ± 0.02).
Genetic Repression of Myosin Function Corrects cMyBPC Deficiency
Rare myosin missense mutations cause DCM, a disorder characterized by ventricular enlargement and diminished cardiac contractility. Mice engineered to carry the human DCM mutation Myh6F764L/+ and Myh6F764L/ F764L mice recapitulate these phenotypes.34 Analyses of isolated cardiomyocytes from these models showed a genotype-dependent depression of cellular shortening (Figure 2A): contractility in Myh6F764L/+ and Myh6F764L/ F764L cardiomyocytes was 75% and 50% of normal (P < 0.0001 for each). DCM cardiomyocytes also had small, but significantly reduced durations of relaxation (Figure 2B) (Myh6F764L/+: 0.17 ± 0.01, p = 0.0002; Myh6F764L/ F764L: 0.14 ± 0.005, p < 0.0001) compared to WT cardiomyocytes (0.21 ± 0.02).
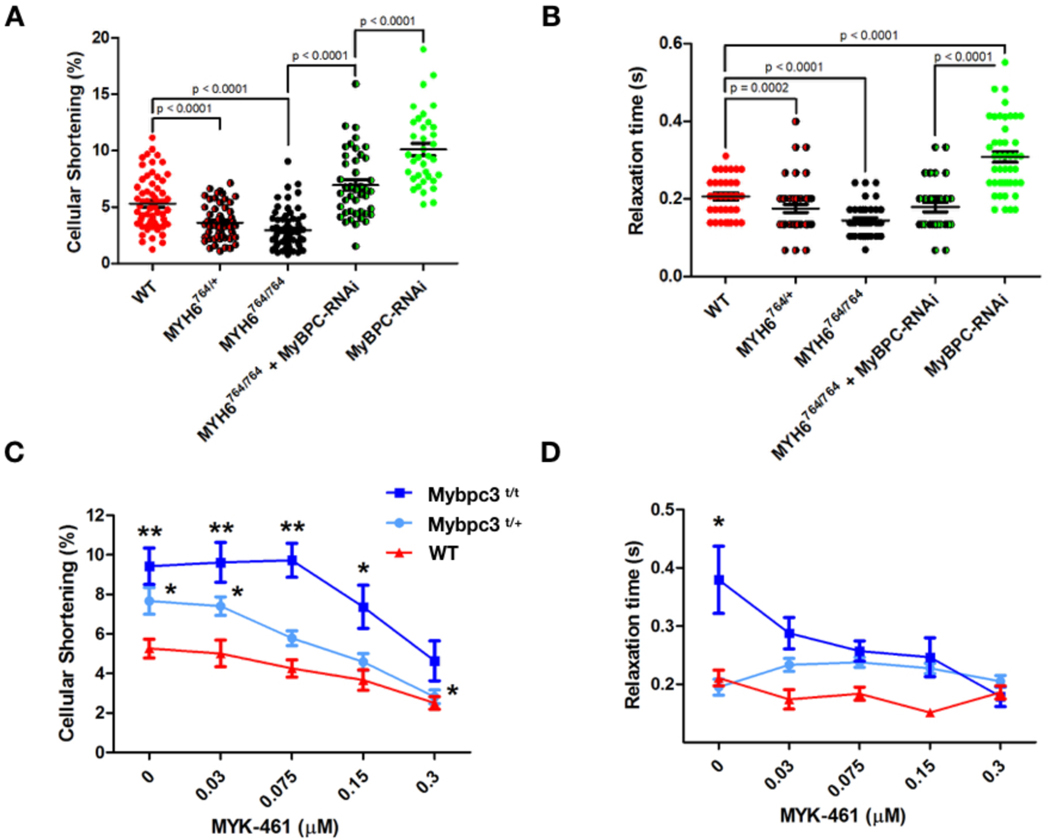
Figure 2: Genetic and pharmacological manipulation of cardiomyocytes depleted for cMyBPC.
A) Sarcomere contractility in isolated cardiomyocytes from WT, Myh6764/+, Myh6764/764, Myh6764/764 + MyBPC-RNAi, and MyBPC-RNAi mice. (Cells analyzed; WT = 63, Myh6764/+ = 55, Myh6764/764 = 71, Myh6764/764 + MyBPC-RNAi = 43, MyBPC-RNAi = 36) B) Sarcomere relaxation of isolated cardiomyocytes from WT, Myh6764/+, Myh6764/764, Myh6764/764 treated with MyBPC-RNAi, and MyBPC-RNAi mice. Individual data points are plot with mean ± SEM indicated. All significant p values are indicated on the graph. (Cells analyzed; WT = 30, Myh6764/+ = 45, Myh6764/764 = 29, Myh6764/764 + MyBPC-RNAi = 31, MyBPC-RNAi = 45) C) Sarcomere contractility of cardiomyocytes treated with 0.03 – 0.3 μM MYK-461. More than 20 cardiomyocytes were analyzed for each drug concentration and treatment group) D) Sarcomere relaxation of cardiomyocytes treated with 0.03 – 0.3 μM MYK-461. All data is displayed as mean ± SEM. *p < 0.01 and **p < 0.0001 denote comparisons with WT without MYK-461.
To determine if manipulation of myosin properties would alter contractile phenotypes in Mybpc3 mutant cardiomyocytes, we injected AAV9 carrying GFP and MyBPC-RNAi into P10 Myh6F764L/ F764L mice. Forty days post injection the cellular shortening and duration of relaxation (Figure 2A, B) were significantly improved in comparison to MyBPC-RNAi cardiomyocytes (p<0.0001); both contractile and relaxation parameters were indistinguishable from WT cardiomyocytes.
Inhibition of Myosin ATPase Corrects cMyBPC Defects in Cardiomyocytes and Cardiac Tissues
As genetic deficits in myosin normalized the performance of Mybc3-deficient cardiomyocytes, we hypothesized that MYK-461, a cardiac-selective, pharmacologic allosteric myosin ATPase inhibitor, would also be effective. To initially determined if MYK-461 (2.5 mg/kg per day via drinking water) elicited deleterious effects in Mybpc3t/+ and MyBPC-RNAi mice we used in vivo echocardiography to assess LV posterior wall dimensions (LVPW) and fractional shortening (FS) at five and 20 weeks after dosing. There were no significant changes in cardiac morphology when comparing treated and untreated mice within genotypes (Supplemental table 1).
Based on our findings that isolated cardiomyocytes from Mypbc3t/+ and Mypbc3t/t mice exhibited contractile differences that were not apparent from in vivo mouse imaging, we then acutely treated WT and mutant cardiomyocytes with MYK-461. The hyper-contractility in Mypbc3t/+ and Mypbc3t/t cardiomyocytes was reduced in a dose dependent manner (Figure 2C). Two-way ANOVA within treatment groups also showed significant reduction in cellular shortening after MYK-461 treatment. Contractile function was normalized in Mypbc3t/+ cardiomyocytes at 0.15 μM MYK-461 and at 0.3 μM in Mypbc3t/t cardiomyocytes. This higher dose of MYK-461 reduced sarcomere contractility by ~50% in both mutant genotypes and depressed contractility by ~30% in WT cardiomyocytes. Concurrently, 0.3 μM of MYK-461 normalized relaxation times in Mypbc3t/t cardiomyocytes (Figure 2D), but did not alter the duration of relaxation in Mypbc3t/+ or WT cardiomyocytes.
Increased Ratios of Myosin Heads in DRX:SRX caused by cMyBPC-deficiency are Normalized by Modulation of Myosin ATPase.
The proportions of myosin heads in DRX and SRX conformations correlate with the rate of ATP cycling in relaxed muscle, which can be measured by the decay of a fluorescent, non-hydrolyzable ATP (Mant-ATP) from skinned muscle36 or cell fibers (Figure 3 and Supplemental Figure 5). Myosin heads in the DRX configuration have ~5x more ATPase activity than myosin heads in the SRX configuration36. Hence the fraction of myosin heads in the SRX configuration and DRX configuration can be estimated from the fraction of Mant-ATP that is released rapidly (DRX) or slowly (SRX).
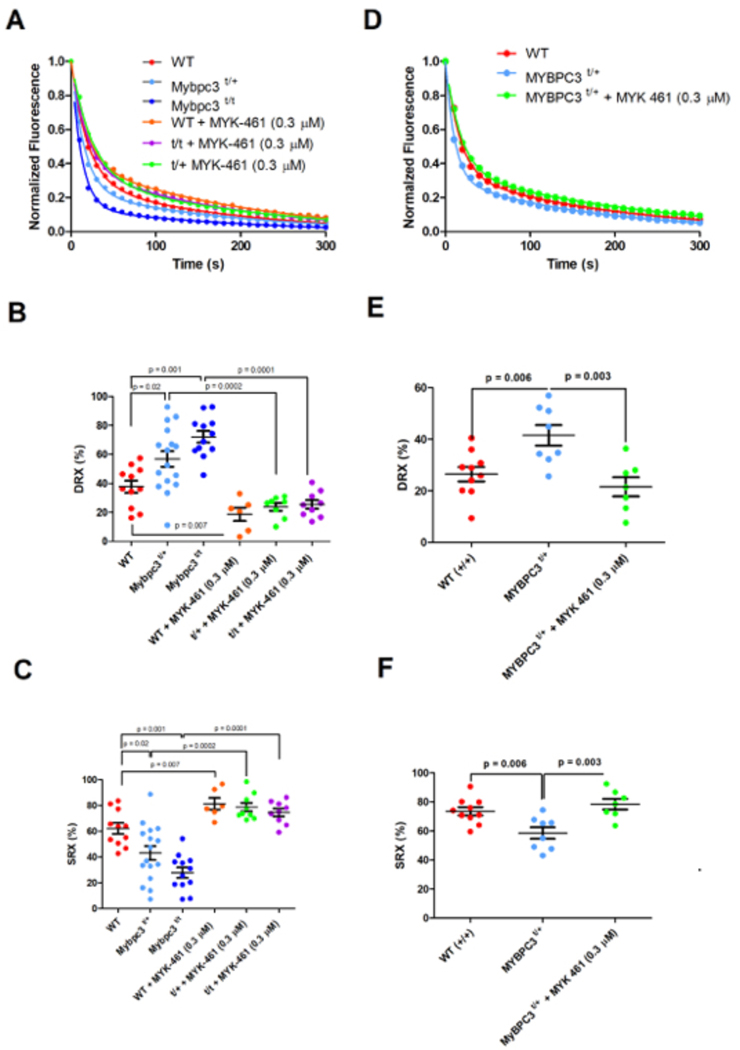
Figure 3: Mant-ATP assessment and correction of SRX and DRX ratios in mouse and human myocardium.
A) Raw average Mant-ATP fluorescence decay curves plot from fluorescence decay due to dark ATP wash, acquisition duration 5 minutes. Data points are the mean of ~12 separate experiments from 3 separate individuals in each genotype/treatment group. Data is fit by a double exponential decay to assess ratios of DRX and SRX heads (Methods). B) Plot of the initial rapid decay amplitude corresponding to DRX heads. C) Plot of the second exponents slow decay amplitude corresponding to SRX heads. D) Average Mant-ATP fluorescence decay curves of unrelated human hearts: three without HCM (WT) and three HCM heart with MYBPC3 t/+ mutations. Each curve is the average of 12 experiments from three separate samples in each treatment group. Data is fit by double exponential decay to assess ratios of DRX and SRX heads in the myocardium. E) Plot of the initial rapid decay amplitude corresponding to DRX heads. F) Plot of the second exponents slow decay amplitude corresponding to SRX heads. All data is presented in each panel, plotted as mean ± SEM with significances indicated with p values.
Assays of skinned cardiac fibers from WT, Mypbc3t/+ and Mypbc3t/t mice showed significantly different proportions of myosins in DRX and SRX confirmations (Figure 3 A–C). Compared to WT cardiac tissues, Mypbc3t/+ and Mypbc3t/t had a 50% increase (p = 0.02) and 94% increase (p = 0.001), respectively, of myosins in DRX (Figure 3B), changes that paralleled the dose-dependent increase in cardiomyocyte contractility (Figure 1C).
The proportions of SRX and DRX found in Myh6F764L/ F764L (Supplemental Figure 6) were comparable to those of wildtype mice (p =0.053). This finding, combined with prior biophysical analyses of Myh6F764L/ F764L molecules34, 35, indicated that the depressed contractility associated with this DCM genotype largely reflects intrinsic deficits in the mutant myosin, rather than a major shift in ratio of SRX and DRX conformations. Consistent with this model, fibers from MyBPC-RNAi treated Myh6F764L/ F764L mice had improved proportions of myosins in SRX and DRX than fibers from MyBPCt/t mice, but were not corrected to the physiologic proportions found in WT fibers p =0.004 (Supplemental Figure 6).
We then asked if an allosteric inhibitor of myosin ATPase, MYK-461, influenced the dynamic ratios of myosin heads in the SRX and DRX conformations. Skinned cardiac fibers from WT and cMyBPC-deficient mice treated with MYK-461 (0.3 μM) increased the proportion of myosins in SRX and reduced myosins in DRX by 60% in WT (p = 0.006), 65% in Mypbc3t/+ (p = 0.0001) and by 70% in Mypbc3t/t fibers (p = 0.0001) in comparison to untreated corresponding genotypes (Figure 3 A–C). Notably this MYK-461 dose normalized cardiomyocyte contractility (Figure 2C, D), indicating a direct relationship between the proportion of myosins in DRX and cellular hyper-contractility and relaxation rates.
Analyses of Mant-ATP release from skinned human HCM heart fibers with heterozygous MYBPC3 truncations showed abnormalities comparable to those in mutant mouse hearts. The proportion of myosin in DRX was increased (~50%, p = 0.006) compared to normal human heart fibers (Figure 3 D–F). Moreover, treatment of MYBPC3-mutant fibers with 0.3 μM MYK-461 normalized the ratio of DRX/SRX, by reciprocally reducing the proportion in DRX and increasing the proportion in SRX by 40% (p = 0.002, vs. untreated).
Taken together, these observations indicate that MYBPC3 mutations in humans and mice disrupted normal myosin conformations, resulting in the increased contractility, diminished relaxation, and excessive ATP consumption – prototypic findings in HCM. These abnormalities can be pharmacologically corrected with the myosin ATPase inhibitor, MYK-461.
Discussion
Truncating germline mutations in one or both Mybpc3 alleles and RNAi silencing of Mybpc3 transcripts caused comparable abnormalities in cardiomyocyte contraction and relaxation, thereby supporting the conclusion that MYBPC3 mutations cause HCM by haploinsufficiency. We show that the severity of cardiomyocyte phenotypes is dependent on cMyBPC quantities; truncation of one allele that reduced protein levels without altering MyBPC phosphorylation (Supplemental figure 1) produced milder abnormalities in systolic and diastolic performance than biallelic mutations. The phenotypes of MyBPC-RNAi cardiomyocytes confirmed this dose-dependent relationship; extinguishing post-natal protein expression substantially amplified hypercontractility and impaired relaxation, evidencing a direct role of cMyBPC across the cardiac cycle.
Our studies uncovered a dichotomous reduction of systolic contraction in Mypbc3t/t mice despite prominent hypercontractility in isolated cardiomyocytes from Mypbc3t/t or MyBPC-RNAi mice. Several factors may account for these observations. The hypercontractility due to profoundly depressed cMyBPC quantities in Mypbc3t/t mice may evoke life-long compensatory mechanisms such as reduced phosphorylation of regulatory light chains that could normalize sarcomere performance21, 37–40. Compensatory effects in other model systems with germline mutations have similarly mitigated the expected phenotype41. Second, the Mypbc3t/t cardiomyocytes with impaired post-natal cytokinesis that increases numbers of cardiomyocytes, particularly mononuclear cardiomyocytes27 may be dysfunctional. Increased numbers of fragile mononuclear cardiomyocytes, as well as hypercontractility of binuclear cells, and altered myocardial geometry may each increase energy demands, which when unmet, could compromise contraction, promote cardiomyocyte death and fibrosis, and thereby diminish in Mypbc3t/t hearts. By eliminating these factors, our analyses of isolated Mypbc3t/t and post-natal MyBPC-RNAi cardiomyocytes provided a more proximal readout, revealing that reduced cMyBPC increased sarcomere contractility.
Through genetic and pharmacological approaches we demonstrate that myosin dysregulation provides a unifying mechanism by which thick filament gene mutations in MYH7 and MYPBC3 cause HCM (Figure 4). Genetic repression of myosin’s motor function, as occurs in Myh6764/764 cardiomyocytes34, improved the hypercontractile phenotype of cMyBPC deficiency, a finding that centrally positions myosin dysregulation in the pathogenicity of cMyBPC mutations. Conversely, depletion of cMyBPC protein levels activated sarcomere performance similar to the effects associated with phosphorylation or the regulatory light chain21, 37–40. These observations indicate that strategies to reduce cMyBPC can, at least transiently, increase myosin contractility.
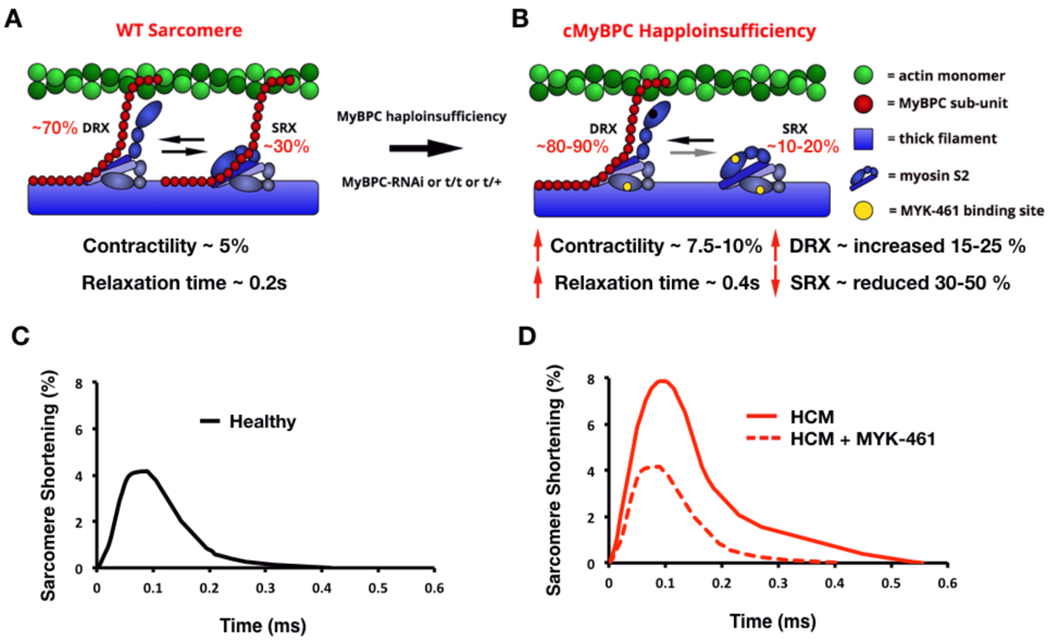
Figure 4: Schematic of the mechanism by which haploinsufficiency of cMyBPC causes HCM.
A) Schematic of a WT sarcomere with normal cMyBPC quantities, and physiologic contractility and relaxation due to appropriate proportions myosins in state of super relaxation (SRX) with low energy consumption or disordered relaxation (DRX) with high energy consumption. B) Schematic of an HCM sarcomere with reduced cMyBPC quantities that dysregulates the proportions of myosins in DRX (increased) and SRX (reduced). The increased proportion of DRX myosins causes inappropriate sarcomere hypercontractility. Yellow denotes the approximate interaction site of MYK-461 on myosin, which abates the hypercontractile phenotype and shifts the myosin DRX:SRX equilibrium back toward normal. C) Contractile waveform of an individual cardiomyocyte isolated from a healthy individual, showing normal sarcomere shortening and normal relaxation duration. D) Contractile waveform from a cardiomyocyte isolated from a HCM patient with cMyBPC haploinsufficiency, showing hypercontractility with increased sarcomere shortening and slowed relaxation. MYK-461 normalizes the HCM phenotypes of hypercontractility by restoring physiologic balance of myosin DRX:SRX.
Depletion of MyBPC also slowed cardiac relaxation, as has been previously observed42–45, an abnormality that precedes the development of hypertrophy in HCM patients with heterozygous MYBPC3 mutations46–49. Although we could not demonstrate significant relaxation deficits in Mypbc3t/+ cardiomyocytes, diastolic abnormalities were prominent in Mypbc3t/t and MyBPC-RNAi cardiomyocytes. While Myh6764/764 reduced the increased duration of relaxation associated with cMyBPC deficiency (Figure 2B, we demonstrated that pharmacologic treatment with MYK-461 more effectively improved relaxation.
Mant-ATP experiments provided a mechanism for these observations. Our experiments and those by others30, 31 show altered proportions of myosins in DRX and SRX in mouse and human myocardium with truncating cMyBPC mutations. Loss of cMyBPC increased the proportions of myosin in the more active DRX conformation. Importantly, MYK-461 shifted DRX/SRX proportions in mice and human tissues, at concentrations that alleviated enhanced cardiomyocyte contractility, data that strongly suggests that increases in the proportion of myosins in DRX contributes to the hyper-contractile phenotype of HCM. As MYK-461 was acutely administered to permeabilized myocardium in our study, its binding to myosin (and not secondary signalling events) is most likely the driver of these beneficial biophysical changes. That MYK-461 represses filamentous function both by directly decreasing myosin contractile function and increasing relaxation properties provides evidence that MYK-461 modulates muscle function via dual mechanisms. We propose that shifting the DRX/SRX ratio to favor the normal SRX abundance reduces the pool of myosin heads available for strong cross-bridge formation, and diminishes the abundance of active heads that must detach from actin to allow relaxation, thereby shortening the time for cardiomyocytes to restore resting sarcomere length.
Importantly we demonstrate the therapeutic potential of targeting myosin in patients with MYBPC3 truncating mutations. As previously shown in HCM mice with myosin mutations3, MYK-461-treatment of mouse or human heart tissues with cMyBPC mutations resulted in dose-dependent attenuation of hypercontractility. As MyBPCt/+ mice do not exhibit in vivo morphologic or hemodynamic parameters of human HCM, these models can only confirm that MYK-461 is well tolerated (Supplemental table 1). However, in combination with correction of cellular abnormalities in isolated mouse MyBPCt/+ cardiomyocytes and normalized rates of ATP cycling in human HCM hearts with MYBPC3 mutations, we expect that MYK-461 will also be effective in patients.
We recognize several limitations in this study. Mypbc3t/+ mice do not recapitulate the extent of hypertrophy nor the degree of relaxation deficits found in patients with heterozgyous MYBPC3 mutations, factors that diminish the value of longitudinal treatment trials in mice. Contractility measurements and relaxation assays were performed in un-loaded cardiomyocytes. Despite these limitations, we suggest that these preclinical data support the need for detailed studies in human HCM patients.
In summation, cMyBPC truncation causes HCM by a mechanism of haploinsufficiency, wherein myosin SRX conformations are destabilized, leading to deleterious ratios of DRX/SRX conformations that drive hyper-contractility, impair relaxation, and increase energy consumption. This triad of abnormalities accurately explains the clinical phenotypes of hyper-dynamic contraction, diastolic dysfunction, and energy deprivation in HCM hearts. In addition, these observations endorse the conclusion that myosin dysregulation is a central mechanism of HCM pathophysiology in MyBPC haploinsufficiency, and substantially contributes to diastolic dysfunction. By targeting myosin functions, genetically or pharmacologically, these phenotypes are normalized in cardiomyocytes and will likely reduce disease pathogenesis in vivo. Demonstrating that myosin is a central player in the pathophysiology of cMyBPC truncation may extend the therapeutic utility of MYK-461 to the proportionally largest subset of HCM patients - those with MYBPC3 mutations.
Materials and Methods
Study Design
We hypothesize that cardiac contractile abnormalities observed in patients with MYBPC3 mutations that truncate the MyBPC protein could be modeled at the cellular level. To test this we studied mouse cardiomyocytes (from n > 3 randomized animals per genotype and treatment group), interrogating cells with heterozygous mutations and cells depleted of MyBPC. No data exclusion criteria were set and outliers were not removed, all analysis was performed under blinded conditions. Cardiomyocyte contractility was sampled until 80% of initial contractility was observed, at which point analysis was halted. Cellular replicates were performed (>10 per experiment) and experiments were performed at least thrice for all samples.
After establishing a contractile phenotype in cardiomyocytes with decreased levels of MyBPC protein we hypothesized that myosin activities were involved. We tested this model by studying cardiomyocytes with a damaging mutation in myosin and with a myosin allosteric inhibitor MYK-461. Pharmacologic perturbations were performed to test dose-dependence of MYK-461 on contractile function.
Based on experimental observations we hypothesized that depletion of MyBPC caused contractile phenotypes by disturbing the balance of relaxed conformational states of myosin DRX/SRX and that MYK-461 corrected this imbalance. We extended this model by comparing the proportions of each confirmation in human heart tissue from patients with heterozygous mutations that truncate MyBPC (n=3) and donor tissues (n=2) without MYBPC3 mutations at baseline and after treatment with MYK-461.
MyBPC truncation
All animal protocols were compliant with the approved protocols of the Association for the Assessment and Accreditation of Laboratory Animal Care and Harvard Medical School. Mypbc3t/+, Mypbc3t/t and WT mice were studied (129SvEv background) with histopathology being previously described in detail25, 32. The truncated Mybpc3 alleles were created by PGK-neomycin resistance gene insertion into exon 30 creating a predicted truncation at amino acid 1,064 of the 1,270 residues of cMyBPC. Homozygous mice express ~10% of the amount of MyBP-C protein in wildtype myofibrillar extracts32. It should be noted that a greater proportion of cells in the Mypbc3t/t cohort exhibited fibrillation and were excluded from contractile measures as they could not be reliably paced, making measures in Mypbc3t/t cardiomyocytes more challenging. Mypbc3t/+ and WT mice were administered MYK-461 (2.5 mg/kg per day via drinking water) as described.3 Echocardiograms of mice were obtained at baseline (age, five weeks) and every 5 weeks through 20 weeks of age.
RNAi
RNAi was delivered at P10 by AAV vector using AAV9 capsid packaging by triple transfection as described27. The AAV9 vector contains an shRNA construct that specifically targets specific 21-base-pair sequence targeted to Mybpc3 exon19 and an enhanced GFP plasmid (Addgene). Vector was injected into the thoracic cavity of P1 t/+ neonates [5 × 1013 viral genomes (vg)/kg], GFP fluorescence was used to identify isolated cardiomyocytes that had taken up the vector. GFP fluorescence was evident from 48 hours post-injection for at least 5 months, RNAi reduced cMyBPC expression to ~10% of normal 27.
Human myectomy samples
Human myectomy samples were obtained after written informed consent from three HCM patients with distinct heterozygous frame shift truncating variants (Gln981fs, Leu1014fs, and Lys1209fs) in MYBPC3. Myectomy samples of the septum were flash frozen and stored in liquid nitrogen and tissue preparation for Mant-ATP experiments were performed as described below.
Cardiomyocyte isolation
Cardiomyocytes were isolated from 8–20 week old mice by rapid explantation and aortic cannulation on a Langendorff apparatus for perfusion with Enzyme Buffer (EB composition: 135 mM NaCl, 4 mM KCl, 0.33 mM NaH2PO4, 1 mM MgCl2, and 10 mM HEPES, pH 7.40, which incorporated Collagenase D, Collagenase B and Protease XIV) for 10 minutes. After perfusion the atria and right ventricle were removed and the left ventricle was minced in TA buffer (Composition: 135 mM NaCl, 4 mM KCl, 0.33 mM NaH2PO4, 1 mM MgCl2, and 10 mM HEPES, pH 7.40, which incorporated bovine serum albumin) and passed through a 100 μm filter into a 50ml conical tube. Tissue settled for 15 minutes to allow myocytes to pellet by gravity. The pellet was then sequentially resuspended every 10 minutes through an increasing calcium gradient (5%, 20%, 50%, 100% calcium tyrode) to provide a cell fraction enriched in myocytes with a final experimental buffer (Composition: NaCl 137 mM, KCl 5.4 mM, CaCl2 1.2 mM, MgCl2 0.5 mM, HEPES 10 mM at pH 7.4, which incorporated glucose).
Contractile measures of myocyte function
Isolated cardiomyocytes were placed in wells of a 6-well plate that had been precoated with laminin. Laminin coating was performed for two hours before cardiomyocyte introduction at a concentration of 10 ug/mL in PBS (Composition: KH2PO4 1mM, NaCl 155 mM, Na2HPO4 3mM, at pH 7.4). Laminin coating solution was washed once with PBS before cells were introduced into the wells. Once cells were introduced they were left to incubate for 10 minutes to equilibrate to experimental temperature (30°C). Cells were imaged using a Keyence BZ-X710 microscope using a Nikon 40X/0.65 NA objective. Cells were kept at 30°C using microscope specific incubation chamber that was also used to deliver 20% O2 and 5% CO2 to the experimental chamber. Cells were paced at 1Hz using custom-built electrodes hooked up to a pacing unit (Pulsar 6i, FHC Brunswick, ME, USA) delivering 20V. Movies were acquired at 29 frames per second for 5 seconds (5 contractile cycles).
An ImageJ plugin SarCoptiM was used to track sarcomere lengths during contractile cycles 50. Sarcomere tracking was then used to calculate cellular shortening (%), relaxed and contracted sarcomere lengths (μm), contractile cycle and relaxation durations (seconds).
For experiments incorporating MYK-461, drug was applied in concentrations ranging 0.05 – 0.3 μM in the experimental buffer. MYK-461 was incubated with cells for a minimum of 10 minutes before data acquisition.
Mant-ATP experiments
Mice were sacrificed by rapid cervical dislocation, atria and right ventricle were removed and samples were flash frozen in liquid nitrogen. Mant-ATP protocols were adapted from publications30,36: 20 mg left ventricular human or mouse myocardial samples were thawed in permeabilization buffer (Composition: NaCl 100 mM, MgCl2 8 mM, EGTA 5 mM, K2HPO4 5 mM, KH2PO4 5 mM, NaN3 3 mM, ATP 5 mM, DTT 1 mM, BDM 20 mM, Triton-X 100 0.1%, at pH 7.0). Samples were permeabilized for 6 hours on ice on a rocker solution changes occurring every two hours. At the completion of this step samples were stored overnight at −20°C in glycerinating solution (Composition: K acetate, 120 mM; Mg acetate, 5 mM; K2HPO4, 2.5 mM; KH2PO4, 2.5 mM; MOPS, 50 mM; ATP, 5 mM; BDM, 20 mM; DTT, 2 mM; glycerol, 50% (v/v), pH 6.8.) for dissection within 2 days.
Once glycerinated ventricular myocardium was dissected into ~ 90 × 400 μm pieces that were held under two pins in a chamber constructed from a slide and coverslip. These samples were permeabilized using the permeabilization buffer for a further 30 minutes on ice prior to experimentation. After secondary permeabilization chambers were flushed with glycerinating buffer.
For fluorescence acquisition a Nikon TE2000-E was used with a Nikon 20X/0.45 objective, using a Hammamatsu C9100 EM-CCD. Frames were acquired every 10 seconds with a 20 ms acquisition and exposure time using a DAPI filter set, images were collected for 15 minutes. Prior to acquisition each chamber was flushed with ATP buffer (Composition: K acetate 120 mM, Mg acetate 5 mM, K2HPO4 2.5 mM, KH2PO4 2.5 mM, ATP 4 mM, MOPS 50 mM, DTT 2 mM at pH 6.8) to remove glycerol. This buffer was replaced with two chamber volumes of rigor buffer (Composition: K acetate 120 mM, Mg acetate 5 mM, K2HPO4 2.5 mM, KH2PO4 2.5 mM, MOPS 50 mM, DTT 2 mM at pH 6.8). Rigor buffer was incubated for 5 minutes to allow rigor to set in. Initial fluorescence acquisition was simultaneous with the addition of one chamber volume of rigor buffer with 250 μM Mant-ATP to visualize fluorescent Mant-ATP wash in. At the end of a 15-minute acquisition, a chamber volume of ATP buffer (Rigor buffer + 4 mM ATP) was added to the chamber with simultaneous acquisition of the Mant-ATP chase. For experiments with MYK-461 all experimental solutions contained 0.3 μM MYK-461.
Mant-ATP analyses
Similar to protocols previously described for analysis30 three regions of each myocardial tissue strip were sampled for fluorescence decay using the ROI manager in ImageJ. The final data point of fluorescence wash in defined the y-intercept. Subtraction of non-myosin bound Mant-ATP fluorescence signal was made using a correction factor of 52% as indicated previously36. All data was plot as a normalized intensity of initial fluorescent intensity from the three sampled regions. These data are fit to an unconstrained double exponential decay using Sigmaplot:
Where A1 is the amplitude of the initial rapid decay approximating the ‘disordered relaxed state’ (DRX) with T1 as the time constant for this decay. A2 is the slower second decay approximating the proportion of myosin heads in the ‘super relaxed state’ (SRX) with its associated time constant T2.
Each individual experiment was fit using this double exponential decay with all values determined and plot. Statistical analysis was performed using 2-way ANOVA with multiple comparisons tests.
Statistics
Where appropriate students t-tests were employed. In instances where experimental hypothesis were tested amongst multiple treatment groups one-way analysis of variance (ANOVA) was used. For multiple comparisons post-hoc Bonferroni corrections were used with a significance cut off of p < 0.05.
Supplementary Material
Table S1. Echocardiographic parameters with MYK-461 administration
Fig. S1. Representation of Mybpc3 mouse models
Fig. S2. In vivo cardiac function and proteomic characterization in Mybpc3 mouse models
Fig. S3. The protein and function effects of increasing MyBPC-RNAi titers
Fig. S4. Analysis of Mant-ATP video files
Fig. S5. Mant-ATP assays in cardiac tissues for Myh6764/764 mice with DCM
Supplementary Movie S1. Mant-ATP fluorescence decay
Acknowledgments:
We thank Dr. Roger Cooke for assistance with the Mant-ATP assays. This work was supported in part by grants from a Sir Henry Wellcome postdoctoral fellowship from the Wellcome Trust (206466/Z/17/Z C.N.T.) the Sarnoff Foundation (A.G.), National Medical Research Council (NMRC) and Ministry of Education, Singapore for D.L. and J.J., American Heart Association Midwest Predoctoral Fellowship (15PRE22430028 to TLL), National Institutes of Health grants (R01HL130356, R01HL105826, and K02HL114749 to SS),Research council of Norway (221707 to IGL). British Heart Foundation (Programme grant RG/12/16/29939) and the British Heart Foundation Centre of Research Excellence (Oxford) to H.W. and C.S.R. The National Heart Blood and Lung Institutes (HL084553 and HL080494 to J.G.S. and C.E.S.), and the Howard Hughes Medical Institute (C.E.S.)
Footnotes
Competing Financial Interest
C.E.S. and J.G.S. are founders and own shares in Myokardia Inc., a startup company that is developing therapeutics that target the sarcomere.
References and Notes:
- 1.Maron BJ, Gardin JM, Flack JM, Gidding SS, Kurosaki TT and Bild DE. Prevalence of hypertrophic cardiomyopathy in a general population of young adults. Echocardiographic analysis of 4111 subjects in the CARDIA Study. Coronary Artery Risk Development in (Young) Adults. Circulation. 1995;92:785–9. [DOI] [PubMed] [Google Scholar]
- 2.Maron BJ, Casey SA, Poliac LC, Gohman TE, Almquist AK and Aeppli DM. Clinical course of hypertrophic cardiomyopathy in a regional United States cohort. JAMA. 1999;281:650–5. [DOI] [PubMed] [Google Scholar]
- 3.Green EM, Wakimoto H, Anderson RL, Evanchik MJ, Gorham JM, Harrison BC, Henze M, Kawas R, Oslob JD, Rodriguez HM, Song Y, Wan W, Leinwand LA, Spudich JA, McDowell RS, Seidman JG and Seidman CE. A small-molecule inhibitor of sarcomere contractility suppresses hypertrophic cardiomyopathy in mice. Science. 2016;351:617–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Walsh R, Thomson KL, Ware JS, Funke BH, Woodley J, McGuire KJ, Mazzarotto F, Blair E, Seller A, Taylor JC, Minikel EV, Exome Aggregation C, MacArthur DG, Farrall M, Cook SA and Watkins H. Reassessment of Mendelian gene pathogenicity using 7,855 cardiomyopathy cases and 60,706 reference samples. Genet Med. 2017;19:192–203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Moolman-Smook JC, De Lange WJ, Bruwer EC, Brink PA and Corfield VA. The origins of hypertrophic cardiomyopathy-causing mutations in two South African subpopulations: a unique profile of both independent and founder events. Am J Hum Genet. 1999;65:1308–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kubo T, Kitaoka H, Okawa M, Matsumura Y, Hitomi N, Yamasaki N, Furuno T, Takata J, Nishinaga M, Kimura A and Doi YL. Lifelong left ventricular remodeling of hypertrophic cardiomyopathy caused by a founder frameshift deletion mutation in the cardiac Myosin-binding protein C gene among Japanese. J Am Coll Cardiol. 2005;46:1737–43. [DOI] [PubMed] [Google Scholar]
- 7.Teirlinck CH, Senni F, Malti RE, Majoor-Krakauer D, Fellmann F, Millat G, Andre-Fouet X, Pernot F, Stumpf M, Boutarin J and Bouvagnet P. A human MYBPC3 mutation appearing about 10 centuries ago results in a hypertrophic cardiomyopathy with delayed onset, moderate evolution but with a risk of sudden death. BMC Med Genet. 2012;13:105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Saltzman AJ, Mancini-DiNardo D, Li C, Chung WK, Ho CY, Hurst S, Wynn J, Care M, Hamilton RM, Seidman GW, Gorham J, McDonough B, Sparks E, Seidman JG, Seidman CE and Rehm HL. Short communication: the cardiac myosin binding protein C Arg502Trp mutation: a common cause of hypertrophic cardiomyopathy. Circ Res. 2010;106:1549–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sabater-Molina M, Saura D, Garcia-Molina Saez E, Gonzalez-Carrillo J, Polo L, Perez-Sanchez I, Olmo MD, Oliva-Sandoval MJ, Barriales-Villa R, Carbonell P, Pascual-Figal D and Gimeno JR. A Novel Founder Mutation in MYBPC3: Phenotypic Comparison With the Most Prevalent MYBPC3 Mutation in Spain. Rev Esp Cardiol (Engl Ed). 2017;70:105–114. [DOI] [PubMed] [Google Scholar]
- 10.Adalsteinsdottir B, Teekakirikul P, Maron BJ, Burke MA, Gudbjartsson DF, Holm H, Stefansson K, DePalma SR, Mazaika E, McDonough B, Danielsen R, Seidman JG, Seidman CE and Gunnarsson GT. Nationwide study on hypertrophic cardiomyopathy in Iceland: evidence of a MYBPC3 founder mutation. Circulation. 2014;130:1158–67. [DOI] [PubMed] [Google Scholar]
- 11.van Velzen HG, Schinkel AFL, Oldenburg RA, van Slegtenhorst MA, Frohn-Mulder IME, van der Velden J and Michels M. Clinical Characteristics and Long-Term Outcome of Hypertrophic Cardiomyopathy in Individuals With a MYBPC3 (Myosin-Binding Protein C) Founder Mutation. Circ Cardiovasc Genet. 2017;10. [DOI] [PubMed] [Google Scholar]
- 12.Dhandapany PS, Sadayappan S, Xue Y, Powell GT, Rani DS, Nallari P, Rai TS, Khullar M, Soares P, Bahl A, Tharkan JM, Vaideeswar P, Rathinavel A, Narasimhan C, Ayapati DR, Ayub Q, Mehdi SQ, Oppenheimer S, Richards MB, Price AL, Patterson N, Reich D, Singh L, Tyler-Smith C and Thangaraj K. A common MYBPC3 (cardiac myosin binding protein C) variant associated with cardiomyopathies in South Asia. Nat Genet. 2009;41:187–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Marston S, Copeland O, Jacques A, Livesey K, Tsang V, McKenna WJ, Jalilzadeh S, Carballo S, Redwood C and Watkins H. Evidence from human myectomy samples that MYBPC3 mutations cause hypertrophic cardiomyopathy through haploinsufficiency. Circ Res. 2009;105:219–22. [DOI] [PubMed] [Google Scholar]
- 14.van Dijk SJ, Dooijes D, dos Remedios C, Michels M, Lamers JM, Winegrad S, Schlossarek S, Carrier L, ten Cate FJ, Stienen GJ and van der Velden J. Cardiac myosin-binding protein C mutations and hypertrophic cardiomyopathy: haploinsufficiency, deranged phosphorylation, and cardiomyocyte dysfunction. Circulation. 2009;119:1473–83. [DOI] [PubMed] [Google Scholar]
- 15.Ho CY, Sweitzer NK, McDonough B, Maron BJ, Casey SA, Seidman JG, Seidman CE and Solomon SD. Assessment of diastolic function with Doppler tissue imaging to predict genotype in preclinical hypertrophic cardiomyopathy. Circulation. 2002;105:2992–7. [DOI] [PubMed] [Google Scholar]
- 16.Forsey J, Benson L, Rozenblyum E, Friedberg MK and Mertens L. Early changes in apical rotation in genotype positive children with hypertrophic cardiomyopathy mutations without hypertrophic changes on two-dimensional imaging. J Am Soc Echocardiogr. 2014;27:215–21. [DOI] [PubMed] [Google Scholar]
- 17.Russel IK, Brouwer WP, Germans T, Knaapen P, Marcus JT, van der Velden J, Gotte MJ and van Rossum AC. Increased left ventricular torsion in hypertrophic cardiomyopathy mutation carriers with normal wall thickness. J Cardiovasc Magn Reson. 2011;13:3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Viswanathan SK, Sanders HK, McNamara JW, Jagadeesan A, Jahangir A, Tajik AJ and Sadayappan S. Hypertrophic cardiomyopathy clinical phenotype is independent of gene mutation and mutation dosage. PLoS One. 2017;12:e0187948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Alamo L, Ware JS, Pinto A, Gillilan RE, Seidman JG, Seidman CE and Padron R. Effects of myosin variants on interacting-heads motif explain distinct hypertrophic and dilated cardiomyopathy phenotypes. Elife. 2017;6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Moore JR, Leinwand L and Warshaw DM. Understanding cardiomyopathy phenotypes based on the functional impact of mutations in the myosin motor. Circ Res. 2012;111:375–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Trivedi DV, Adhikari AS, Sarkar SS, Ruppel KM and Spudich JA. Hypertrophic cardiomyopathy and the myosin mesa: viewing an old disease in a new light. Biophys Rev. 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lee KH, Sulbaran G, Yang S, Mun JY, Alamo L, Pinto A, Sato O, Ikebe M, Liu X, Korn ED, Sarsoza F, Bernstein SI, Padron R and Craig R. Interacting-heads motif has been conserved as a mechanism of myosin II inhibition since before the origin of animals. Proc Natl Acad Sci U S A. 2018;115:E1991–E2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pfuhl M and Gautel M. Structure, interactions and function of the N-terminus of cardiac myosin binding protein C (MyBP-C): who does what, with what, and to whom? J Muscle Res Cell Motil. 2012;33:83–94. [DOI] [PubMed] [Google Scholar]
- 24.Kensler RW, Shaffer JF and Harris SP. Binding of the N-terminal fragment C0-C2 of cardiac MyBP-C to cardiac F-actin. J Struct Biol. 2011;174:44–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.McConnell BK, Fatkin D, Semsarian C, Jones KA, Georgakopoulos D, Maguire CT, Healey MJ, Mudd JO, Moskowitz IP, Conner DA, Giewat M, Wakimoto H, Berul CI, Schoen FJ, Kass DA, Seidman CE and Seidman JG. Comparison of two murine models of familial hypertrophic cardiomyopathy. Circ Res. 2001;88:383–9. [DOI] [PubMed] [Google Scholar]
- 26.Previs MJ, Mun JY, Michalek AJ, Previs SB, Gulick J, Robbins J, Warshaw DM and Craig R. Phosphorylation and calcium antagonistically tune myosin-binding protein C’s structure and function. Proc Natl Acad Sci U S A. 2016;113:3239–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jiang J, Burgon PG, Wakimoto H, Onoue K, Gorham JM, O’Meara CC, Fomovsky G, McConnell BK, Lee RT, Seidman JG and Seidman CE. Cardiac myosin binding protein C regulates postnatal myocyte cytokinesis. Proc Natl Acad Sci U S A. 2015;112:9046–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Farrell ET, Grimes AC, de Lange WJ, Armstrong AE and Ralphe JC. Increased Postnatal Cardiac Hyperplasia Precedes Cardiomyocyte Hypertrophy in a Model of Hypertrophic Cardiomyopathy. Front Physiol. 2017;8:414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Korte FS, McDonald KS, Harris SP and Moss RL. Loaded shortening, power output, and rate of force redevelopment are increased with knockout of cardiac myosin binding protein-C. Circ Res. 2003;93:752–8. [DOI] [PubMed] [Google Scholar]
- 30.McNamara JW, Li A, Smith NJ, Lal S, Graham RM, Kooiker KB, van Dijk SJ, Remedios CGD, Harris SP and Cooke R. Ablation of cardiac myosin binding protein-C disrupts the super-relaxed state of myosin in murine cardiomyocytes. J Mol Cell Cardiol. 2016;94:65–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.McNamara JW, Li A, Lal S, Bos JM, Harris SP, van der Velden J, Ackerman MJ, Cooke R and Dos Remedios CG. MYBPC3 mutations are associated with a reduced super-relaxed state in patients with hypertrophic cardiomyopathy. PLoS One. 2017;12:e0180064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.McConnell BK, Jones KA, Fatkin D, Arroyo LH, Lee RT, Aristizabal O, Turnbull DH, Georgakopoulos D, Kass D, Bond M, Niimura H, Schoen FJ, Conner D, Fischman DA, Seidman CE and Seidman JG. Dilated cardiomyopathy in homozygous myosin-binding protein-C mutant mice. J Clin Invest. 1999;104:1235–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Watkins H, Conner D, Thierfelder L, Jarcho JA, MacRae C, McKenna WJ, Maron BJ, Seidman JG and Seidman CE. Mutations in the cardiac myosin binding protein-C gene on chromosome 11 cause familial hypertrophic cardiomyopathy. Nat Genet. 1995;11:434–7. [DOI] [PubMed] [Google Scholar]
- 34.Schmitt JP, Debold EP, Ahmad F, Armstrong A, Frederico A, Conner DA, Mende U, Lohse MJ, Warshaw D, Seidman CE and Seidman JG. Cardiac myosin missense mutations cause dilated cardiomyopathy in mouse models and depress molecular motor function. Proc Natl Acad Sci U S A. 2006;103:14525–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kamisago M, Sharma SD, DePalma SR, Solomon S, Sharma P, McDonough B, Smoot L, Mullen MP, Woolf PK, Wigle ED, Seidman JG and Seidman CE. Mutations in sarcomere protein genes as a cause of dilated cardiomyopathy. N Engl J Med. 2000;343:1688–96. [DOI] [PubMed] [Google Scholar]
- 36.Hooijman P, Stewart MA and Cooke R. A new state of cardiac myosin with very slow ATP turnover: a potential cardioprotective mechanism in the heart. Biophys J. 2011;100:1969–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Toepfer C, Caorsi V, Kampourakis T, Sikkel MB, West TG, Leung MC, Al-Saud SA, MacLeod KT, Lyon AR, Marston SB, Sellers JR and Ferenczi MA. Myosin regulatory light chain (RLC) phosphorylation change as a modulator of cardiac muscle contraction in disease. J Biol Chem. 2013;288:13446–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Toepfer CN, West TG and Ferenczi MA. Revisiting Frank-Starling: regulatory light chain phosphorylation alters the rate of force redevelopment (ktr ) in a length-dependent fashion. J Physiol. 2016;594:5237–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Nag S, Trivedi DV, Sarkar SS, Adhikari AS, Sunitha MS, Sutton S, Ruppel KM and Spudich JA. The myosin mesa and the basis of hypercontractility caused by hypertrophic cardiomyopathy mutations. Nat Struct Mol Biol. 2017;24:525–533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Spudich JA, Aksel T, Bartholomew SR, Nag S, Kawana M, Yu EC, Sarkar SS, Sung J, Sommese RF, Sutton S, Cho C, Adhikari AS, Taylor R, Liu C, Trivedi D and Ruppel KM. Effects of hypertrophic and dilated cardiomyopathy mutations on power output by human beta-cardiac myosin. J Exp Biol. 2016;219:161–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.El-Brolosy MA and Stainier DYR. Genetic compensation: A phenomenon in search of mechanisms. PLoS Genet. 2017;13:e1006780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Pohlmann L, Kroger I, Vignier N, Schlossarek S, Kramer E, Coirault C, Sultan KR, El-Armouche A, Winegrad S, Eschenhagen T and Carrier L. Cardiac myosin-binding protein C is required for complete relaxation in intact myocytes. Circ Res. 2007;101:928–38. [DOI] [PubMed] [Google Scholar]
- 43.Harris SP, Bartley CR, Hacker TA, McDonald KS, Douglas PS, Greaser ML, Powers PA and Moss RL. Hypertrophic cardiomyopathy in cardiac myosin binding protein-C knockout mice. Circ Res. 2002;90:594–601. [DOI] [PubMed] [Google Scholar]
- 44.Fraysse B, Weinberger F, Bardswell SC, Cuello F, Vignier N, Geertz B, Starbatty J, Kramer E, Coirault C, Eschenhagen T, Kentish JC, Avkiran M and Carrier L. Increased myofilament Ca2+ sensitivity and diastolic dysfunction as early consequences of Mybpc3 mutation in heterozygous knock-in mice. J Mol Cell Cardiol. 2012;52:1299–307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.van Dijk SJ, Kooiker KB, Napierski NC, Touma KD, Mazzalupo S and Harris SP. Point mutations in the tri-helix bundle of the M-domain of cardiac myosin binding protein-C influence systolic duration and delay cardiac relaxation. J Mol Cell Cardiol. 2018;119:116–124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ho CY, Carlsen C, Thune JJ, Havndrup O, Bundgaard H, Farrohi F, Rivero J, Cirino AL, Andersen PS, Christiansen M, Maron BJ, Orav EJ and Kober L. Echocardiographic strain imaging to assess early and late consequences of sarcomere mutations in hypertrophic cardiomyopathy. Circ Cardiovasc Genet. 2009;2:314–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Nagueh SF, Bachinski LL, Meyer D, Hill R, Zoghbi WA, Tam JW, Quinones MA, Roberts R and Marian AJ. Tissue Doppler imaging consistently detects myocardial abnormalities in patients with hypertrophic cardiomyopathy and provides a novel means for an early diagnosis before and independently of hypertrophy. Circulation. 2001;104:128–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Nagueh SF, McFalls J, Meyer D, Hill R, Zoghbi WA, Tam JW, Quinones MA, Roberts R and Marian AJ. Tissue Doppler imaging predicts the development of hypertrophic cardiomyopathy in subjects with subclinical disease. Circulation. 2003;108:395–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Poutanen T, Tikanoja T, Jaaskelainen P, Jokinen E, Silvast A, Laakso M and Kuusisto J. Diastolic dysfunction without left ventricular hypertrophy is an early finding in children with hypertrophic cardiomyopathy-causing mutations in the beta-myosin heavy chain, alpha-tropomyosin, and myosin-binding protein C genes. Am Heart J. 2006;151:725 e1–725 e9. [DOI] [PubMed] [Google Scholar]
- 50.Pasqualin C, Gannier F, Yu A, Malecot CO, Bredeloux P and Maupoil V. SarcOptiM for ImageJ: high-frequency online sarcomere length computing on stimulated cardiomyocytes. Am J Physiol Cell Physiol. 2016;311:C277–83. [DOI] [PubMed] [Google Scholar]
- 51.Toepfer CN, Wakimoto H, Garfinkel AC, McDonough B, Liao D, Jiang J, Tai A, Joshua G, Lunde I, Lun M, Lynch T, Sadayappan S, Redwood C, Watkins H, Seidman J and Seidman C. MYBPC3 Mutations cause Hypertrophic Cardiomyopathy by Dysregulating Myosin: Implications for Therapy. bioRxiv. 2018. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1. Echocardiographic parameters with MYK-461 administration
Fig. S1. Representation of Mybpc3 mouse models
Fig. S2. In vivo cardiac function and proteomic characterization in Mybpc3 mouse models
Fig. S3. The protein and function effects of increasing MyBPC-RNAi titers
Fig. S4. Analysis of Mant-ATP video files
Fig. S5. Mant-ATP assays in cardiac tissues for Myh6764/764 mice with DCM
Supplementary Movie S1. Mant-ATP fluorescence decay