Abstract
Offspring born preterm (i.e., before 37 weeks gestation) are more likely to die or experience longstanding illness compared to full term offspring. Maternal genetic variants (i.e., heritable, stable variations in the genetic code) and epigenetic modifications (i.e., chemical modifications to the genetic code that can affect which genes are turned on or off) in response to stress have been implicated in preterm birth. Fetal genetic variants have been linked to preterm birth; though, the role of offspring epigenetics in preterm birth remains understudied. This systematic review synthesizes the literature examining associations among stress during pregnancy and epigenetic modifications to offspring DNA, with 25 reports identified. Ten reports examined DNA methylation (i.e., addition/removal of methyl groups to/from DNA) across the epigenome. The remainder examined DNA methylation near genes of interest, primarily genes linked to hypothalamic-pituitary-adrenal axis function (NR3C1, FKBP51), growth/immune function (IGF2), and socioemotional regulation (SLC6A4, OXTR). The majority of reports noted associations among stress and offspring DNA methylation, primarily when perceived stress, anxiety, or depression served as the predictor. Findings suggest that differences in offspring epigenetic patterns may play a role in stress-associated preterm birth and serve as targets for novel interventions.
Introduction
Preterm birth, or birth before 37 weeks of an expected 40 week gestation, is unique in the sense that it is defined solely on the basis of the timing of birth. The definition of preterm birth highlights the recognized importance of adequate time in utero, as hastened birth clearly introduces risk to the offspring. For example, an infant born before 37 weeks gestation is nearly 20 times more likely to die in the first year of life than an infant carried to at least 39 weeks gestation.1 Among surviving preterm offspring, the odds of repeated hospital admission during infancy are nearly 18-fold greater than infants born at 39 weeks gestation or later.2 By five years of age, offspring who were born preterm are at 3.4-fold greater odds of experiencing a longstanding illness than offspring who were born at or beyond 39 weeks.2 This being said, the definition of preterm birth also introduces significant challenge to its study and prevention, as women arrive at this outcome under a number of different circumstances.3,4 In fact, preterm birth is best described as a syndrome, or set of co-occurring signs or symptoms, that can take multiple forms and arise for multiple reasons. As such, the study of preterm birth requires careful classification and consideration of diverse biological mechanisms.5
As the research and clinical communities continue their efforts in addressing a high and rising rate of preterm birth, which now exceeds one in ten United States births,6 factors that appear to contribute to but do not fully explain preterm birth risk are being considered in terms of additive and potentially interactive effects on perinatal biology. A prime example is the examination of genetic and environmental exposures that may work together to shape preterm birth risk. For example, a recent study of over 1,000 spontaneous births before 34 weeks gestation noted that 32.3% of women show some evidence of familial risk for preterm birth.4 In the same study, 56.6% of women showed evidence of maternal stress.4 Maternal exposures to stressful events, perceptions of stress, and emotional responses to stress during pregnancy have been consistently implicated as risk factors for preterm birth, with some studies suggesting that such exposures increase the odds of preterm birth by up to roughly 3.5-fold.7 Examination of the extant literature shows significant potential for genetic and environmental exposures such as stress to affect similar biological systems (e.g., sympathetic nervous, neuroendocrine, and immune systems) and the development of preterm birth within and across generations.
Specifically, maternal genetic variants (i.e., heritable, stable variations in the genetic code) within genes important to, for example, the sympathetic and neuroendocrine response to stress and function of the immune system have been weakly but consistently implicated in preterm birth.8,9 Stress has also been shown to affect similar biological systems, with a growing body of literature suggesting that this may occur in part through effects on epigenetic patterns (i.e., chemical modifications to the genetic code that can affect which genes are turned on or off).10,11 Maternal differences in epigenetic patterns have, in turn, been linked to risk for preterm birth.12,13 Moreover, while there is certainly a degree of overlap between maternal and fetal genetic and epigenetic variation,14,15 evidence suggests that the fetus contributes uniquely, shaping risk for premature delivery among the offspring and the offspring’s children.8,15,16 As such, the purpose of this systematic review is to synthesize the literature examining associations among maternal stress during pregnancy and epigenetic modifications to offspring DNA, providing direction for future work examining effects of maternal and offspring genetic variation and environmental exposures on maternal and offspring health, including preterm birth risk within and across generations.
Methods
A systematic review and critical appraisal of the literature was conducted according to the guidelines of the Preferred Reporting Items for Systematic Reviews and Meta-Analysis (PRISMA) Statement.17 The protocol was registered with PROSPERO (CRD pending). Reports were identified and assessed for eligibility independently by two authors (AN, SG). Reports were also reviewed for data extraction and outcomes (AN, SG) and assessed for risk of bias (AM, SG) by two authors. All discrepancies were addressed through active discussion with at least one additional author.
Information Sources and Search Strategy
A systematic search of the literature was conducted using Scopus, PubMed, CINAHL, and PsychINFO on September 19, 2019 and all identified records were exported and subsequently screened. Included reports were published or posted online ahead of print by the date of the initial search. Search terms were clustered and combined to identify reports examining associations among stress of the mother during pregnancy and epigenetic patterns in offspring cord blood collected at birth (i.e., (pregnancy OR pregnant OR prenatal OR maternal OR gestation OR gestational) AND (stress OR distress OR anxiety OR anxious OR depression OR depressive OR depressed OR trauma OR traumatic OR maltreatment OR mood) AND (methylation OR histone OR chromatin OR epigenetic OR epigenetics OR ncrna OR lncrna OR “noncoding RNA” OR mirna OR microrna OR sirna OR “short interfering RNA”) AND (umbilical OR cord OR fetal OR newborn OR infant) AND (blood OR leukocyte OR leukocytes)). Four systematic reviews identified through the search process and addressing the area of interest were also reviewed in detail to identify additional records for inclusion. None were identified. Only original research reports were included in the current systematic review.
Eligibility Criteria and Study Selection
Decided a priori to improve the quality of identified reports, prospective cohort studies or case-control studies nested within a prospective cohort design were included in this systematic review. Case studies, reviews, and commentaries were excluded. Eligible reports assessed stress occurring and measured during pregnancy and prior to birth, with the conceptualization of stress defined according to the Transactional Model of Stress and Coping.18,19 Therefore, reports quantifying stress stimuli (e.g., life events, chronic difficulties), participant appraisal of stress stimuli (e.g., perceived stress), and/or emotional responses to stress stimuli (e.g., prenatal distress, depressive or anxiety symptoms) were included. Eligible reports were also required to assess offspring epigenetic patterns across the epigenome or via targeted methods, with examples including DNA methylation (DNAm), histone modification, or expression of non-coding RNA. The epigenome also differs according to which cells and/or tissues are evaluated20 and may be susceptible to environmental exposures during early life.21,22 As such, included studies were limited to those assessing epigenetic modifications in cord blood collected at birth, a source for offspring DNA. The search parameters were left open in terms of years of publication, as the study team was not aware of any prior systematic reviews on this specific topic. Studies employing animal models of stress during pregnancy, using cell lines, or experimentally manipulating cells cultured ex vivo were excluded. Full reports not available in English were excluded.
Data Extraction and Synthesis
Study design, sample size, type of stress assessed, method of stress assessment, epigenetic marker(s) assessed, method of epigenetic marker assessment and key findings were extracted for each report meeting eligibility criteria. To achieve consistency in study design classifications, a priori were created definitions for potential study designs.23 Prospective cohort studies were defined as those in which stress during pregnancy was measured prior to birth, with ≥80% of participants analyzed. Prospective cohort studies with limited data were defined as those for which the variables of interest were only available among less than 80% of maternal-offspring dyads or use of a subset was noted with no parent cohort size reported. Case-control studies nested within a prospective cohort design were defined as those in which stress during pregnancy was measured prior to birth but only a subset of participants was analyzed according to a stress-related case-control definition. For data synthesis, included reports were divided first according to the stress parameter and then according to the epigenetic marker(s) under study.
Assessment of Risk of Bias
Reports were assessed using the Newcastle-Ottawa Scale (NOS) for quality assessment of cohort studies,24 adapted for use for the topic of interest. Quality is rated on a scale of 0-9, with a greater number of stars signifying higher quality ratings. Specifically, the NOS supports the rating of cohort studies across three domains—selection, comparability, and outcome. Reports can earn up to four stars on the basis of selection under the categories of representativeness of the exposed (e.g., high stress) cohort, selection of the non-exposed (e.g., low stress) cohort, ascertainment of the exposure (i.e., stress during pregnancy), and demonstration that the outcome of interest (i.e., cord blood epigenetics at birth) was not present at the start of the study. Reports can earn up to two stars based on comparability of cohorts per study design or analysis. Here, focus was placed on matching or statistical adjustment for maternal smoking and body mass index. A number of studies have shown epigenetic patterns to be particularly susceptible to smoking and nutrition, which may confound results in examining associations among stress during pregnancy and epigenetic modifications to offspring DNA.25,26 Lastly, reports can earn up to three stars on the basis of outcome under the categories of assessment of outcome (i.e., quantification of epigenetic marker(s)), length of follow-up (i.e., pregnancy through birth), and adequacy of follow-up (i.e., how loss-to-follow-up or limited availability of data was addressed and reported).
Results
Study Selection
Study selection is summarized in Figure 1. Searching Scopus, PubMed, CINAHL, and PsychINFO, a total of 702 records were identified. After removing duplicates, 516 unique records remained. After exclusion on the basis of title and abstract, 36 records remained for full-text review. After exclusion on the basis of full text review, 25 articles were included in the final review. One additional record (a correction to an original report) was not included as one of the final 25 articles but was reviewed to ensure correct reporting of the final results of this systematic review.
Figure 1.
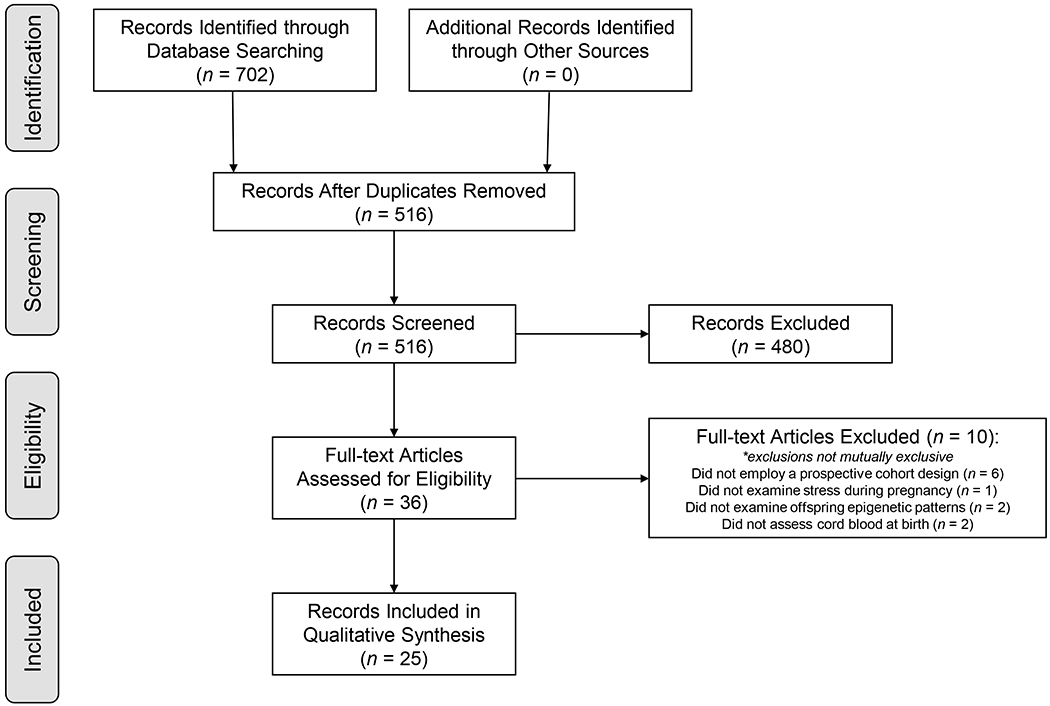
Record Selection
Study Characteristics
The characteristics of the included reports are shown in Table 1. All reports were published between the years of 2008 and 2019. Per a detailed review of methods, the 25 included reports appeared to analyze data from 17 cohorts, including the larger Project Viva (n = 1),27 Generation R (n = 2),27,28 Prediction and Prevention of Preeclampsia Intrauterine Growth Restriction (PREDO; n = 2),29,30 Boston Birth (n = 1),31 and Avon Longitudinal Study of Parents and Children (ALSPAC; n = 2)28,32 cohorts. The majority of reports (18/25) analyzed data from prospective cohort studies. Of the prospective cohort analyses, most (14/18) were missing data on stress during pregnancy and/or cord blood from more than 20% of participants. The remaining studies employed a nested case-control design in which a subset of samples were assessed using data that was prospectively collected over time. Sample sizes varied widely, with the largest study analyzing data among 2,244 maternal-offspring dyads.30 Reports most commonly assessed symptoms of depression and/or anxiety during pregnancy as the predictor of interest and all studies assessed DNAm of offspring cord blood (i.e., the addition or removal of methyl groups to or from DNA) as the outcome of interest. Results from epigenome-wide analyses were included for ten reports. The remainder focused on the methylation status of specific sites or regions that were in proximity to genes of interest.
Table 1.
Characteristics of studies examining stress during pregnancy and epigenetic modifications to offspring DNA (n = 25)
| First Author, Year [Ref] | Design | Sample Sizea | Stress Assessed | Method of Stress Assessment | Epigenetic Marker(s) Assessed | Method of Epigenetic Marker Assessment | NOS Rating (0 – 9)b |
|---|---|---|---|---|---|---|---|
| Cardenas et al., 2019 [27] | Prospective cohort with limited data | N = 2128; Analyzed = 479 | Anxiety; Depression | PRAS; EDS | Epigenome-wide DNA methylation | Infinium 450K BeadChip | 8 |
| Chen et al., 2014 [37] | Prospective cohort with limited data | N = not stated; Analyzed = 50 | Life events; Perceived stress; Anxiety; Depression | LEI; PSS; STAI; Structured interviews for DSM-IV diagnoses | DNA methylation of 5 CpG sites co-locating with IGF2 and H19 | Pyrosequencing | 6 |
| Czamara et al., 2019 [30] | Prospective cohort | N = 2365; Analyzed = 2244 | Anxiety; Depression | STAI; CES-D | Epigenome-wide DNA methylation | Infinium 450K BeadChip; Infinium 850K BeadChip | 9 |
| Devlin et al., 2010 [48] | Prospective cohort | N = 98; Analyzed = 82 | Depression | HAM-D; EDS | DNA methylation of 22 CpG sites co-locating with SLC6A4 and BDNF | Pyrosequencing | 5 |
| Duis et al., 2018 [31] | Nested Case-control | N = ~6,000; Cases analyzed = 30; Controls analyzed = 30 | Anxiety; Depression | Abstracted from medical record ICD-9 codes | DNA methylation of 65 CpG sites co-locating with FKBP5 | Pyrosequencing | 8 |
| Dukal et al., 2015 [34] | Nested Case-control | N = 410; Cases analyzed = 45; Controls analyzed = 45 | Life events; Perceived stress; Distress; Anxiety; Depression; Psychiatric disorders | LES; PSS; PDQ; STAI; EDS; M.I.N.I. | DNA methylation of 5 CpG sites co-locating with SLC6A4 | Pyrosequencing | 6 |
| Gurnot et al., 2015 [53] | Nested Case-control | N = 91; Cases analyzed = 12; Controls analyzed = 12 | Depression | HAM-D | Epigenome-wide DNA methylation | Infinium 27K BeadChip | 6 |
| Hompes et al., 2013 [42] | Prospective cohort with limited data | N = 170; Analyzed = 83 | Anxiety; Depression | PRAQ; STAI; EDS | DNA methylation of 31 CpG sites co-locating with NR3C1 | MassARRAY | 8 |
| Liu et al., 2012 [47] | Prospective cohort with limited data | N = 1700; Analyzed = 508 | Depression | CES-D | IGF2, H19, MEG3-IG, MEG3, PEG3, MEST, PEG10, NNAT, PLAGL1 DMRs | Pyrosequencing | 8 |
| Mansell et al., 2016a [40] | Prospective cohort with limited data | N = 1074; Analyzed = 576 | Perceived stress; Anxiety; Depression | PSS; EDS | IGF2 and H19 DMRs | MassARRAY | 7 |
| Mansell et al., 2016b [38] | Prospective cohort with limited data | N = 1074; Analyzed = 481 | Perceived stress; Anxiety; Depression | PSS; EDS | DNA methylation of 47 CpG sites co-locating with NR3C1 | MassARRAY | 7 |
| Nemoda et al., 2015 [46] | Nested Case-control | N = 127; Cases analyzed = 29; Controls analyzed = 15 | Depression | EDS; MADRS; Self-report | Epigenome-wide DNA methylation | Infinium 450K BeadChip | 7 |
| Nieratschker et al., 2014 [33] | Nested Case-control | N = 180; Cases analyzed = 9; Controls analyzed = 8 | Life events; Perceived stress; Distress; Anxiety; Depression; Psychiatric disorders | LES; PSS; PDQ; STAI; EDS; M.I.N.I. | Epigenome-wide promoter DNA methylation; DNA methylation of MORC1 promoter region | Methylated DNA Immunoprecipitation; Pyrosequencing | 6 |
| Non et al., 2014 [49] | Nested Case-control | N = 1941; Cases analyzed = 35; Controls analyzed = 23 | Anxiety; Depression | Abstracted from medical records | Epigenome-wide DNA methylation | Infinium 450K BeadChip | 8 |
| Oberlander et al., 2008 [44] | Prospective cohort | N = 98; Analyzed = 82 | Anxiety; Depression | HAM-A; HAM-D; EDS | LINE-1 global DNA methylation; DNA methylation of 13 CpG sites co-locating with NR3C1 | Pyrosequencing | 6 |
| Rijlaarsdam et al., 2016 [28] | Prospective cohort with limited data | N = 24,319; Analyzed = 1,740 | Life events; Chronic difficulties; Distress | LES; Long lasting difficulties questionnaire; Family assessment device; Pregnancy outcome questionnaire | Epigenome-wide DNA methylation | Infinium 450K BeadChip | 7 |
| Schroeder et al., 2012 [50] | Prospective cohort | N = 201; Analyzed = 178 | Depression | Structured interviews for DSM-IV diagnoses; HRSD17; BDI | Epigenome-wide DNA methylation | Infinium 27K BeadChip | 7 |
| Soubry et al., 2011 [52] | Prospective cohort with limited data | N = 690; Analyzed = 436 | Depression | Self-report with medical record verification | IGF2 and H19 DMRs | Pyrosequencing | 8 |
| Suarez et al., 2018 [29] | Prospective cohort with limited data | N = 1083; Analyzed = 814 | Depression | CES-D; self-report | DNA methylation gestational age | Infinium 450K BeadChip | 6 |
| Unternaehrer et al., 2016 [36] | Prospective cohort with limited data | N = 100; Analyzed = 39 | Life events; Depression | ILE; EDS | DNA methylation of 22 CpG sites co-locating with OXTR | MassARRAY | 6 |
| Vangeel et al., 2015 [45] | Prospective cohort with limited data | N = 170; Analyzed = 80 | Anxiety; Depression | PRAQ; STAI; EDS | DNA methylation of amplicons co-locating with IGF2 DMR0, IGF2AS, and GNASXL | MassARRAY | 8 |
| Vangeel et al., 2017 [43] | Nested case-control with full cohort validation | N = 170; Cases analyzed = 23; Controls analyzed = 33; Validation = 80 | Anxiety | PRAQ | Epigenome-wide DNA methylation; GABBR1 DMRs | Infinium 450K BeadChip; MassARRAY | 8 |
| Vidal et al, 2014 [41] | Prospective cohort with limited data | N = 1700; Analyzed = 537 | Perceived stress | PSS | IGF2, H19, MEG3-IG, MEG3, PEG3, MEST, SGCE/PEG10, NNAT, PLAGL1 DMRs | Pyrosequencing | 9 |
| Viuff et al., 2018 [32] | Prospective cohort with limited data | N = 14,541; Analyzed = 844 | Depression | EDS | Epigenome-wide DNA methylation | Infinium 450K BeadChip | 8 |
| Wu et al., 2018 [35] | Prospective cohort with limited data | N = 1054; Analyzed = 247 | Life events; Anxiety; Depression | Exposure to violence questionnaire; Crisis in Family Systems-Revised Survey; STAI; EDS | DNA methylation of 7 CpG sites co-locating with TNFA, IL6, and IL8 | Pyrosequencing | 7 |
maternal-offspring dyads;
NOS Quality Rating, based on scores produced across the domains of selection, comparability, and outcome. Higher scores indicate higher quality (high quality ≥ 8; fair quality 5 – 7; low quality ≤ 4).
PRAS = Pregnancy-related anxiety scale; EDS = Edinburgh depression scale; LEI = Life Experiences Interview; PSS = Perceived stress scale; STAI = State-trait anxiety interview; CES-D = Center for Epidemiologic Studies Depression Scale; HAM-D = Hamilton rating scale for depression; LES = Life experiences survey; PDQ = Prenatal distress questionnaire; M.I.N.I. = Mini International Neuropsychiatric Interview; PRAQ = Pregnancy-related anxiety questionnaire; MADRS = Montogomery-Asberg Depression Rating Scale; HAM-A = Hamilton rating scale for anxiety; HRSD17 = Hamilton Rating Scale for Depression; BDI = Beck Depression Inventory; ILE = Inventory of life events; CpG = phosphate-linked cytosine-guanine dinucleotide; DMR = differentially methylated region
Study Findings
Composite measures of stress.
Five reports examined associations among composite measures that included one or more aspect of stress, including stimuli, appraisal, and emotional responses, and offspring cord blood DNAm at birth. Using this method, one report found stress during pregnancy to be associated with differences in the methylation of offspring DNA at many sites across the epigenome (i.e., epigenome-wide), including 3,405 sites that were in close proximity to known genes.33 This report also noted an association between stress during pregnancy and DNAm at a site examined due to its’ proximity with a gene of interest, MORC1.33 Yet, reports also note the lack of an association when examining many sites across the epigenome (i.e., epigenome-wide),28 regions across the epigenome (i.e., differentially methylated regions),28 and specific sites within close proximity to genes of interest (i.e., candidate genes; SLC6A4;34 TNFα, IL6, IL835). Czamara et al.30 also assembled models combining gene and environmental factors (including anxiety and depression), predicting offspring cord blood DNA methylation at regions that showed differences in methylation.
Stress stimuli.
Two reports examined associations among stress stimuli (i.e., life events) and offspring cord blood DNA methylation at birth, both of which examined sites within close proximity to genes of interest. A greater number of stressful life events in the two years preceding assessment at 21-32 weeks of pregnancy was associated with methylation of offspring DNA in an area of the OXTR gene that is responsible for coding oxytocin (i.e., protein-coding region).36 Alternatively, stressful life events related to relationship or health difficulties during pregnancy failed to predict offspring DNAm at five sites examined due to their proximity with regions that control whether the genes IGF2/H19 are turned on or off (i.e., imprinting control regions).37
Participant appraisal of stress stimuli.
Three reports examined associations among participant appraisal of stress stimuli (i.e., perceived stress) and offspring cord blood DNAm at birth. All studies examining perceived stress focused on specific sites or regions due to their proximity with genes of interest. Perceived stress during pregnancy was associated with methylation of a site that is involved in the initial step of turning on the NR3C1 gene (i.e., promoter region),38 region that control whether the gene IGF2 is turned on or off (i.e., imprinting control region),39,40 and methylation levels in a region of the MEST gene41 in offspring samples. Vidal et al.41 examined an additional eight regions involved in turning genes on or off (i.e., imprinted differentially methylated regions), failing to note an association between perceived stress and cord blood DNAm.
Emotional responses to stress stimuli.
Nineteen reports examined associations among emotional responses to stress stimuli (e.g., anxiety symptoms, depressive symptoms) and offspring cord blood DNAm at birth. Symptoms of anxiety during pregnancy, including anxiety specific to pregnancy,42 predicted differences in patterns of DNAm when many sites and regions were examined across the epigenome (i.e., 13 sites,27 901 regions43) and DNAm of sites examined due to proximity to genes of interest (i.e., NR3C1,38,42,44 IGF2,39,40,45 GNAS45). In examining depressive symptoms during pregnancy, four reports reported associations when examining many sites and regions across the epigenome (i.e., 3 sites,27 163 sites,46 2 sites, 39 regions,32 DNAm age which is calculated by examining the methylation of multiple sites29). Depressive symptoms during pregnancy were also associated with offspring DNAm at several sites examined due to proximity with genes of interest (i.e., NR3C1,38,44 IGF2,45 MEG3,47 SLC6A4,48 OXTR36 DNA methylation). In examining the DNAm of many sites across the epigenome, 42 sites showed differences among offspring born to women diagnosed with anxiety and/or depression compared to offspring born to women lacking these diagnoses.49 Nine reports examined but did not find associations among emotional responses to stress stimuli during pregnancy and offspring cord blood DNAm (i.e., anxiety or depression and FKBP5 DNAm;31 depression and epigenome-wide DNAm;50 depression and NR3C1 DNAm in multivariable models,42 BDNF,48 IGF2,39,40,51,52 or CYP2E153 DNAm).
Assessment of Risk of Bias
The twenty-five reports included in this systematic review were assessed for risk of bias by two authors using the Newcastle-Ottawa Scale for the assessment of cohort studies. The average total rating was 7.16 out of a possible 9 stars. Of note, recruitment of representative samples was rare (e.g., Generation R27 set in Rotterdam of the Netherlands). Alternatively, most studies recruited convenience samples of pregnant women through outpatient obstetric settings. Moreover, a number of studies did not appear to account for maternal smoking and/or body mass index. The number of total covariates considered and assessed varied widely. Also of note, the potential for risk of bias introduced through inadequate follow-up was difficult to determine for several studies reporting that data were missing for a significant proportion of the parent sample. For most of these studies, demographic and clinical characteristics among mothers for whom data were available versus unavailable were not compared. The Vidal et al. report was an exception to this pattern, analyzing 537 of the initial 1700 enrollees but providing a description and comparison of those included versus excluded. Results of the reviewed reports must be considered with this in mind, particularly in terms of generalizability.
Discussion
The majority but not all of the reports identified through this systematic review provide evidence of associations among maternal stress during pregnancy and epigenetic modifications to offspring DNA at birth, specifically the methylation of DNA. Associations among stress during pregnancy and DNAm were most commonly noted when perceived stress, anxiety, or depression served as a predictor, suggesting that maternal perceptions of and emotional responses to stress may be particularly important determinants of maternal and offspring health.18 Among studies examining the methylation of sites and regions due to their proximity with genes of interest, the majority focused on genes pertinent to offspring hypothalamic-pituitary-adrenal (HPA) axis function (NR3C1, FKBP51), growth and immune function (IGF2), and socioemotional regulation (SLC6A4, OXTR), the expression of which may have implications for preterm birth risk as well as the general health of the offspring.
Specifically, four of the reviewed studies examined the epigenetic modification of DNA within HPA axis-related genes, which coordinate the neuroendocrine response to stress.31,38,42,44 Under basal conditions, the HPA axis maintains a circadian rhythm, secreting cortisol under the control of a negative feedback loop in which high circulating levels of the hormone inhibits further secretion. Under stressful conditions, cortisol release can be heightened and inappropriately maintained.54 Cortisol exerts its biological actions through the glucocorticoid receptor, which is encoded by the NR3C1 gene.54 Upon binding, the glucocorticoid receptor moves to the nucleus, affecting which genes are turned on and off as well as a myriad of physiological processes in nearly all cell types.55 NR3C1 activity is also regulated by a number of intracellular molecules. For example, the FKBP51 gene encodes a protein involved in maintaining the glucocorticoid receptor in an inactive but high-affinity state, leaving the receptor poised to bind to cortisol.56
This systematic review identified three studies reporting associations among greater perceived stress, symptoms of anxiety, or symptoms of depression, and higher levels of NR3C1 cord blood methylation.38,42,44 Of note, associations did not withstand adjustment for multiple comparison or alternative indices of stress in several instances, including in studies that examined many sites across the epigenome.38,42 While a direct effect of stress was not supported, Duis et al.31 noted a stronger positive association between genotype at a specific position (denoted rs1360780 as its accession number for identification purposes) of the FKBP51 gene and cord blood methylation of three sites within the FKBP51 gene in the context of a maternal anxiety or depression diagnosis. If replicated, these findings would suggest that stress during pregnancy may hinder one’s ability to produce the glucocorticoid receptor and regulate glucocorticoid signaling in the offspring, potentially impairing a number of physiologic processes, including the ability of cortisol to inhibit further release of cortisol when levels are already high. Indeed, Oberlander et al.44 went on to report that when high levels of methylation were noted in association with the NR3C1 gene among offspring at birth, offspring responded to a stressful task at three months of age by producing greater amounts of salivary cortisol than infants with lower levels of methylation, suggesting an impaired ability to dampen cortisol release.
While the implications of such findings are only beginning to be considered, there is biological plausibility that hyperactivity of both the maternal and fetal HPA axis during gestation increases the risk for preterm birth. Specifically, it has been proposed that stress during pregnancy hastens the expected pregnancy-associated rise in maternal cortisol levels, prematurely enhancing placental corticotropin-releasing hormone release and the production of pro-labor peptides.57 Work in this area has focused largely on maternal contributions to stress-induced dysregulation of the hypothalamic-pituitary-adrenal-placental circuit. However, the fetus is capable of synthesizing cortisol de novo by mid-pregnancy and the cortisol-secreting zona fasciculata of the adrenal gland is fully formed by the third trimester, providing the basis for a fetal stress response that is independent from that of the mother.58–60 The potential for fetal contributions to birth timing has certainly been considered, with studies of fetal genetic variation identifying differences in the genetic code within genes pertinent to neuroendocrine function as potential antecedents to preterm birth.61 Replication of association studies has been problematic, highlighting the likelihood of multifactorial contributions to complications of pregnancy.62 With numerous studies implicating heightened maternal HPA axis activity and the studies within this systematic review implicating heightened fetal HPA axis activity in preterm birth, novel intervention studies targeting the HPA axis, potentially by correcting methylation profiles, are warranted.
Also of interest to studies identified in this review was epigenetic modification of genes studied primarily due to their pertinence to growth in utero but also known to affect immune function, particularly IGF2 which encodes insulin-like growth factor 2. IGF2 is an imprinted gene, unique in the sense that the paternal form of the gene (i.e., paternal allele) is expressed while the maternal form of the gene (i.e., maternal allele) is silenced by adding methyl groups to the DNA (i.e., via methylation).63 Due to this pattern of expression, even slight changes in epigenetic patterns may result in serious health consequences.64 Imprinted genes are dependent on epigenetic control when eggs and sperm are formed, and as a result appear to be particularly susceptible to environmental insults.65
Most but not all studies included in this systematic review suggest that stress during pregnancy, particularly perceived stress, anxiety, and depression, is associated with DNAm of the IGF2 gene in offspring cord blood.37,40,45,47,52,66 Specifically, investigators focused on the region of the epigenome that controls whether the gene IGF2 is turned on or off (i.e., the imprinting control region). Also noted were associations among stress during pregnancy and cord blood methylation of several additional imprinted genes, including GNAS, MEST, and MEG3.41,45,51 Several studies have reported associations among offspring methylation of IGF2 and birth weight,47,67,68 highlighting the potential implications for fetal growth, though lack of support for an association was also noted in one study.45 Genetic variation within the IGF1 gene of the mother has also been consistently implicated in preterm birth, with its effects on immune function hypothesized to contribute to the development of early birth.8
There is some evidence to suggest that levels of SLC6A4 and OXTR methylation are susceptible to stress and affect mood,10,69 leading to examination of these regions in the identified reports of cord blood methylation. The SLC6A4 gene encodes the serotonin transporter protein, which regulates the uptake of serotonin, a well-studied regulator of mental health.70 OXTR encodes the oxytocin receptor, which promotes labor and contributes to lactation as well as maternal-offspring bonding. Two of three reviewed studies provided evidence supporting the presence of an association between greater stress during pregnancy, particularly stressful life events and depressive symptoms, and lower SLC6A4- and OXTR-associated cord blood methylation.48,71 It has been posited that demethylation of such genes may serve as an adaptive response to adversity that ultimately holds negative long-term implications for socioemotional wellbeing among the offspring.72 There is some evidence to support the notion that methylation associated with SLC6A4 and OXTR may affect future mental health, such as risk for depression, though much work remains to replicate such findings and uncover the biologic pathways conveying risk.10,73 Such findings are important to the general health of the offspring. Though, DNAm of OXTR, in particular, has also been implicated in preterm birth.13,16 In fact, oxytocin antagonists such as atosiban have been examined as maintenance tocolytics after an episode of preterm labor but question remains as to their ability to prevent preterm birth.74
Strengths and Limitations
Reports included in this systematic review were required to use a prospective cohort or nested case-control design, ensuring that stress data were collected prior to epigenetic data for all studies. Included studies were also limited to those assessing offspring epigenetic markers in cord blood samples collected at birth, standardizing the source tissue and timing of sampling. As such, included reports were primarily assessed to be of high quality. The findings of this systematic review must also be considered with several limitations in mind. First, there was a notable lack of consistency in findings among included reports, highlighting the potential for differences in findings dependent upon how stress was assessed and the portion of the epigenome that was examined. There was also limited overlap between the sites and regions identified in studies looking across the epigenome versus at specific sites or regions examined due to their proximity with known genes. Many studies also had significant missing data with minimal explanations for why the data were missing, introducing potential for bias. Results of the reviewed reports must be considered with this in mind, particularly in terms of generalizability. Additionally, some of the reports are limited in their description of methods, also making the risk of bias difficult to assess.
Conclusion
Taken together, the extant literature and reports identified through this systematic review suggest that maternal environmental exposures, including stress during pregnancy, are capable of affecting maternal and offspring epigenetic patterns, namely the addition or removal of methyl groups to or from DNA (i.e., DNAm). Therefore, maternal and offspring genetic variants (i.e., heritable, stable variations in the genetic code) and epigenetic patterns (i.e., chemical marks to the genetic code that can affect which genes are turned on or off) are likely to both contribute to the health of the mother and offspring, including by shaping risk for preterm birth. While much work remains, these findings suggest that differences in epigenetic patterns among the mother and the offspring may play important roles in preterm birth following maternal stress and therefore serve as targets for novel interventions aiming to reduce risk.
Acknowledgments
Sources of Funding: The writing of this report was supported by the National Institute of Nursing Research of the National Institutes of Health under award numbers F31NR018363 (PI Nowak), T32NR014225 (MPI Pickler & Melnyk), and K23NR017902 (PI Gillespie).
Footnotes
Conflicts of Interest: None declared.
Contributor Information
Alexandra L. Nowak, Martha S. Pitzer Center for Women, Children, & Youth, College of Nursing, The Ohio State University, Columbus, Ohio, USA.
Cindy M. Anderson, Martha S. Pitzer Center for Women, Children, & Youth, College of Nursing, The Ohio State University, Columbus, Ohio, USA.
Amy R. Mackos, College of Nursing, The Ohio State University, Columbus, Ohio, USA.
Emily Neiman, Wexner Medical Center, The Ohio State University, Columbus, Ohio, USA.
Shannon L. Gillespie, Martha S. Pitzer Center for Women, Children, & Youth, College of Nursing, The Ohio State University, Columbus, Ohio, USA.
References
- 1.Matthews TJ, MacDorman MF, Thoma ME. Infant Mortality Statistics From the 2013 Period Linked Birth/Infant Death Data Set. Natl Vital Stat Rep. 2015;64(9):1–30. [PubMed] [Google Scholar]
- 2.Boyle EM, Poulsen G, Field DJ, et al. Effects of gestational age at birth on health outcomes at 3 and 5 years of age: population based cohort study. BMJ. 2012;344:e896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Meertens LJE, van Montfort P, Scheepers HCJ, et al. Prediction models for the risk of spontaneous preterm birth based on maternal characteristics: a systematic review and independent external validation. Acta Obstet Gynecol Scand. 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Manuck TA, Esplin MS, Biggio J, et al. The phenotype of spontaneous preterm birth: application of a clinical phenotyping tool. Am J Obstet Gynecol. 2015;212(4):487.e481–487.e411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Villar J, Papageorghiou AT, Knight HE, et al. The preterm birth syndrome: A prototype phenotypic classification. Am J Obstet Gynecol. 2012;206(2):119–123. [DOI] [PubMed] [Google Scholar]
- 6.Martin JA, Hamilton BE, Osterman MJK. Births in the United States, 2018. NCHS Data Brief. 2019(346):1–8. [PubMed] [Google Scholar]
- 7.Staneva A, Bogossian F, Pritchard M, Wittkowski A. The effects of maternal depression, anxiety, and perceived stress during pregnancy on preterm birth: A systematic review. Women and Birth : Journal of the Australian College of Midwives. 2015;28(3):179–193. [DOI] [PubMed] [Google Scholar]
- 8.Dolan SM, Hollegaard MV, Merialdi M, et al. Synopsis of preterm birth genetic association studies: the preterm birth genetics knowledge base (PTBGene). Public Health Genomics. 2010;13(7-8):514–523. [DOI] [PubMed] [Google Scholar]
- 9.Uzun A, Dewan AT, Istrail S, Padbury JF. Pathway-based genetic analysis of preterm birth. Genomics. 2013;101(3):163–170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Park C, Rosenblat JD, Brietzke E, et al. Stress, epigenetics and depression: A systematic review. Neurosci Biobehav Rev. 2019;102:139–152. [DOI] [PubMed] [Google Scholar]
- 11.Li M, D’Arcy C, Li X, Zhang T, Joober R, Meng X. What do DNA methylation studies tell us about depression? A systematic review. Transl Psychiatry. 2019;9(1):68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Knijnenburg TA, Vockley JG, Chambwe N, et al. Genomic and molecular characterization of preterm birth. Proceedings of the National Academy of Sciences - PNAS. 2019;116(12):5819–5827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Toure DM, ElRayes W, Barnes-Josiah D, Hartman T, Klinkebiel D, Baccaglini L. Epigenetic modifications of human placenta associated with preterm birth: a systematic review. J Matern Fetal Neonatal Med. 2018;31(4):530–541. [DOI] [PubMed] [Google Scholar]
- 14.Parets SE, Conneely KN, Kilaru V, Menon R, Smith AK. DNA methylation provides insight into intergenerational risk for preterm birth in African Americans. Epigenetics. 2015;10(9):784–792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wang X-M, Tian F-Y, Fan L-J, Xie C-B, Nui Z-Z, Chen W-Q. Comparison of DNA methylation profiles associated with spontaneous preterm birth in placenta and cord blood. BMC Med Genomics. 2019;12(1). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Barcelona de Mendoza V, Wright ML, Agaba C, et al. A systematic review of DNA methylation and preterm birth in African American women. Biol Res Nurs. 2017;19(3):308–317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Moher D, Shamseer L, Clarke M, et al. Preferred reporting items for systematic review and meta-analysis protocols (PRISMA-P) 2015 statement. Syst Rev. 2015;4(1). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lazarus RS, Folkman S. Transactional theory and research on emotions and coping. EJP. 1987;1(3):141–169. [Google Scholar]
- 19.Ibrahim SM, Lobel M. Conceptualization, measurement, and effects of pregnancy-specific stress: review of research using the original and revised Prenatal Distress Questionnaire. J Behav Med. 2019. [DOI] [PubMed] [Google Scholar]
- 20.Zhang B, Zhou Y, Lin N, et al. Functional DNA methylation differences between tissues, cell types, and across individuals discovered using the M&M algorithm. Genome Res. 2013;23(9):1522–1540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Alvarado-Cruz I, Alegría-Torres JA, Montes-Castro N, Jimenez-Garza O, Qunitanilla-Vega B. Environmental epigenetic changes, as risk factors for the development of diseases in children: A systematic review. Ann Glob Health. 2018;84(2):212–224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chan JC, Nugent BM, Bale TL. Parental advisory: Maternal and paternal stress can impact offspring neurodevelopment. Biological Psychiatry (1969). 2018;83(10):886–894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Salkind NJ. Encyclopedia of Research Design. Thousand Oaks, CA, 1: SAGE Publications, Inc.; 2010. [Google Scholar]
- 24.Wells GA, Shea B, O’Connell D, Peterson J, Welch V, Losos M. The Newcastle-Ottawa Scale (NOS) for assessing the quality of nonrandomised studies in meta-analyses. 2012. [Google Scholar]
- 25.Kaur G, Begum R, Thota S, Batra S. A systematic review of smoking-related epigenetic alterations. Arch Toxicol. 2019;93(10):2715–2740. [DOI] [PubMed] [Google Scholar]
- 26.Dunford AR, Sangster JM. Maternal and paternal periconceptional nutrition as an indicator of offspring metabolic syndrome risk in later life through epigenetic imprinting: A systematic review. Diabetes Metab Syndr. 2017;11 Suppl 2:S655–s662. [DOI] [PubMed] [Google Scholar]
- 27.Cardenas A, Faleschini S, Cortes Hidalgo A, et al. Prenatal maternal antidepressants, anxiety, and depression and offspring DNA methylation: Epigenome-wide associations at birth and persistence into early childhood. Clin Epigenetics. 2019;11(1):56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rijlaarsdam J, Pappa I, Walton E, et al. An epigenome-wide association meta-analysis of prenatal maternal stress in neonates: A model approach for replication. Epigenetics. 2016;11(2):140–149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Suarez A, Lahti J, Czamara D, et al. The epigenetic clock at birth: Associations with maternal antenatal depression and child psychiatric problems. J Am Acad Child Adolesc Psychiatry. 2018;57(5):321–328. e322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Czamara D, Eraslan G, Page CM, et al. Integrated analysis of environmental and genetic influences on cord blood DNA methylation in new-borns. Nat Commun. 2019;10(1):2548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Duis J, Cox OH, Ji Y, Seifuddin F, Lee RS, Wang X. Effect of genotype and maternal affective disorder on intronic methylation of FK506 binding protein 5 in cord blood DNA. Front Genet. 2018;9:648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Viuff AC, Sharp GC, Dai D, et al. Maternal depression during pregnancy and cord blood DNA methylation: Findings from the Avon Longitudinal Study of Parents and Children. Transl Psychiatry. 2018;8(1). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nieratschker V, Massart R, Gilles M, et al. MORC1 exhibits cross-species differential methylation in association with early life stress as well as genome-wide association with MDD. Transl Psychiatry. 2014;4:e429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Dukal H, Frank J, Lang M, et al. New-born females show higher stress- and genotype-independent methylation of SLC6A4 than males. Borderline Personal Disord Emot Dysregul. 2015;2:8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wu S, Gennings C, Wright RJ, et al. Prenatal stress, methylation in inflammation-related genes, and adiposity measures in early childhood: The programming research in obesity, growth environment and social stress cohort study. Psychosom Med. 2018;80(1):34–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Unternaehrer E, Bolten M, Nast I, et al. Maternal adversities during pregnancy and cord blood oxytocin receptor (OXTR) DNA methylation. Soc Cogn Affect Neurosci. 2016;11(9):1460–1470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chen J, Li Q, Rialdi A, et al. Influences of maternal stress during pregnancy on the epigenome: Comparison of placenta and umbilical cord blood. J Depress Anx. 2014;3(2). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mansell T, Vuillermin P, Ponsonby AL, Collier F, Saffery R, Ryan J. Maternal mental well-being during pregnancy and glucocorticoid receptor gene promoter methylation in the neonate. Dev Psychopathol. 2016;28(4pt2):1421–1430. [DOI] [PubMed] [Google Scholar]
- 39.Chen BH, Marioni RE, Colicino E, et al. DNA methylation-based measures of biological age: Meta-analysis predicting time to death. Aging (Albany N Y). 2016;8(9):1844–1865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mansell T, Novakovic B, Meyer B, et al. The effects of maternal anxiety during pregnancy on IGF2/H19 methylation in cord blood. Transl Psychiatry. 2016;6:e765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Vidal AC, Benjamin Neelon SE, Liu Y, et al. Maternal stress, preterm birth, and DNA methylation at imprint regulatory sequences in humans. Genetics & Epigenetics. 2014;6(6):37–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hompes T, Izzi B, Gellens E, et al. Investigating the influence of maternal cortisol and emotional state during pregnancy on the DNA methylation status of the glucocorticoid receptor gene (NR3C1) promoter region in cord blood. J Psychiatr Res. 2013;47(7):880–891. [DOI] [PubMed] [Google Scholar]
- 43.Vangeel EB, Pishva E, Hompes T, et al. Newborn genome-wide DNA methylation in association with pregnancy anxiety reveals a potential role for GABBR1. Clin Epigenetics. 2017;9:107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Oberlander TF, Weinberg J, Papsdorf M, Grunau R, Misri S, Devlin AM. Prenatal exposure to maternal depression, neonatal methylation of human glucocorticoid receptor gene (NR3C1) and infant cortisol stress responses. Epigenetics. 2008;3(2):97–106. [DOI] [PubMed] [Google Scholar]
- 45.Vangeel EB, Izzi B, Hompes T, et al. DNA methylation in imprinted genes and is associated with prenatal maternal stress Prenatal stress and imprinted genes. Genes, Brain and Behav. 2015;14(8):573–582. [DOI] [PubMed] [Google Scholar]
- 46.Nemoda Z, Massart R, Suderman M, et al. Maternal depression is associated with DNA methylation changes in cord blood T lymphocytes and adult hippocampi. Transl Psychiatry. 2015;5:e545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Liu Y, Murphy SK, Murtha AP, et al. Depression in pregnancy, infant birth weight and DNA methylation of imprint regulatory elements. Epigenetics. 2012;7(7):735–746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Devlin AM, Brain U, Austin J, Oberlander TF. Prenatal exposure to maternal depressed mood and the MTHFR C677T variant affect SLC6A4 methylation in infants at birth. PLoS One. 2010;5(8):e12201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Non AL, Binder AM, Kubzansky LD, Michels KB. Genome-wide DNA methylation in neonates exposed to maternal depression, anxiety, or SSRI medication during pregnancy. Epigenetics. 2014;9(7):964–972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Schroeder JW, Smith AK, Brennan PA, et al. DNA methylation in neonates born to women receiving psychiatric care. Epigenetics. 2012;7(4):409–414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Liu Y, Murphy SK, Murtha AP, et al. Depression in pregnancy, infant birth weight and DNA methylation of imprint regulatory elements Epigenetics. 2012;7(7):735–746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Soubry A, Murphy S, Huang Z, et al. The effects of depression and use of antidepressive medicines during pregnancy on the methylation status of the IGF2 imprinted control regions in the offspring. Clin Epigenetics. 2011;3:2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Gurnot C, Martin-Subero I, Mah SM, et al. Prenatal antidepressant exposure associated with CYP2E1 DNA methylation change in neonates. Epigenetics. 2015;10(5):361–372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.McEwen BS, Wingfield JC. The concept of allostasis in biology and biomedicine. Horm Behav. 2003;43(1):2–15. [DOI] [PubMed] [Google Scholar]
- 55.Binder EB. The role of FKBP5, a co-chaperone of the glucocorticoid receptor in the pathogenesis and therapy of affective and anxiety disorders. Psychoneuroendocrinology. 2009;34(suppl. 1):S186–S195. [DOI] [PubMed] [Google Scholar]
- 56.Oakley RH, Cidlowsk JA. The biology of the glucocorticoid receptor: New signaling mechanisms in health and disease. J Allergy Clin Immunol. 2013;132(5):1033–1044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Sandman CA. Prenatal CRH: An integrating signal of fetal distress. Dev Psychopathol. 2018;30(3):941–952. [DOI] [PubMed] [Google Scholar]
- 58.Sheikh IA, Ahmad E, Jamal MS, et al. Spontaneous preterm birth and single nucleotide gene polymorphisms: A recent update. BMC Genomics. 2016;17(Suppl 9):759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Chen Y, He Z, Chen G, Liu M, Wang H. Prenatal glucocorticoids exposure and fetal adrenal developmental programming. Toxicology. 2019;428:152308. [DOI] [PubMed] [Google Scholar]
- 60.Gitau R, Fisk NM, Teixeira JM, Cameron A, Glover V. Fetal hypothalamic-pituitary-adrenal stress responses to invasive procedures are independent of maternal responses. J Clin Endocrinol Metab. 2001;86(1):104–109. [DOI] [PubMed] [Google Scholar]
- 61.Wadhwa PD, Entringer S, Buss C, Lu MC. The contribution of maternal stress to preterm birth: Issues and considerations. Clin Perinatol. 2011;38(3):351–384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Rappoport N, Toung J, Hadley D, et al. A genome-wide association study identifies only two ancestry specific variants associated with spontaneous preterm birth. Sci Rep. 2018;8(1):226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Mulligan C, D’Errico NC, Stees J, Hughes DA. Methylation changes at NR3C1 in newborns associate with maternal prenatal stress exposure and newborn birth weight. Epigenetics. 2012;7(8):853–857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Charalambous M, Ferron SR, da Rocha ST, et al. Imprinted gene dosage is critical for the transition to independent life. Cell Metab. 2012;15(2):209–221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Kappil M, Lambertini L, Chen J. Environmental influences on genomic imprinting. Curr Environ Health Rep. 2015;2(2):155–162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Vidal AC, Murphy SK, Murtha AP, et al. Associations between antibiotic exposure during pregnancy, birth weight and aberrant methylation at imprinted genes among offspring. Int J Obes (Lond). 2013;37(7):907–913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Montoya-Williams D, Quinlan J, Clukay C, Rodney NC, Kertes DA, Mulligan CJ. Associations between maternal prenatal stress, methylation changes in IGF1 and IGF2, and birth weight. J Dev Orig Health Dis. 2018;9(2):215–222. [DOI] [PubMed] [Google Scholar]
- 68.Su R, Wang C, Feng H, et al. Alteration in expression and methylation of IGF2/H19 in placenta and umbilical cord blood are associated with macrosomia exposed to intrauterine hyperglycemia. PLoS One. 2016;11(2):e0148399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Provenzi L, Giorda R, Beri S, Montirosso R. SLC6A4 methylation as an epigenetic marker of life adversity exposures in humans: A systematic review of literature. Neurosci Biobehav Rev. 2016;71:7–20. [DOI] [PubMed] [Google Scholar]
- 70.Baudry A, Pietri M, Launay JM, Kellermann O, Schneider B. Multifaceted regulations of the serotonin transporter: Impact on antidepressant response. Front Neurosci. 2019;13:91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Unternaehrer E, Luers P, Mill J, et al. Dynamic changes in DNA methylation of stress-associated genes (OXTR, BDNF ) after acute psychosocial stress. Transl Psychiatry. 2012;2:e150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Gluckman PD, Hanson MA, Spencer HG. Predictive adaptive responses and human evolution. Trends Ecol Evol. 2005;20(10):527–533. [DOI] [PubMed] [Google Scholar]
- 73.Kader M, Naim-Shuchana. Physical activity and exercise during pregnancy. Eur J Physiother. 2014;16(1):2–9. [Google Scholar]
- 74.Papatsonis DN, Flenady V, Liley HG. Maintenance therapy with oxytocin antagonists for inhibiting preterm birth after threatened preterm labour. Cochrane Database Syst Rev. 2013(10):Cd005938. [DOI] [PMC free article] [PubMed] [Google Scholar]


