Abstract
Protective effects of liraglutide on H9C2 cells cultured using CoCl2 and its mechanism of action were investigated. Hypoxia model was established using CoCl2-treated H9C2 cells. With liraglutide as the treatment factor, apoptosis, changes in nitric oxide (NO) and reactive oxygen species (ROS) activity, mitochondrial membrane potential and change in cell autophagy level were detected via Hoechst staining, enzyme-linked immunosorbent assay (ELISA), polymerase chain reaction (PCR) and western blotting (WB), respectively. Liraglutide ameliorated the CoCl2-induced decrease in H9C2 cell viability, the increases in cytotoxicity and percentage of apoptotic cells as well as oxidative stress in cells. Moreover, it stimulated the elevation of cell autophagy level. However, the protective effects of liraglutide on H9C2 cells were attenuated remarkably after adding the cell autophagy inhibitor. Liraglutide can ameliorate the CoCl2-induced oxidative stress and apoptosis in H9C2 cells via regulating cell autophagy.
Keywords: liraglutide, oxidative stress, apoptosis, autophagy
Introduction
After ischemia and hypoxia in myocardial cells, the metabolism is inhibited significantly, the biological processes such as DNA transcription and translation as well as protein assembly and transport are restricted, and the cell viability is reduced. Furthermore, the raised production of reactive oxygen species (ROS), enhanced permeability of inner and outer mitochondrial membranes, release of cytochrome c, calcium overload and opening of mitochondrial permeability transition pore can lead to reduced generation of adenosine triphosphate (ATP) and energy metabolism disturbance in mitochondria and activate cell apoptosis and necrosis pathways (1,2). Mitochondrion is a major organelle for energy metabolism through cell respiration, and NAD+ and NADP+ are involved in the electron transfer and ATP synthesis in respiratory chain complex. Besides, the level of mitochondrial membrane potential can reflect the integrity of mitochondrial membrane structure and function, and mitochondrial damage is the key link of cellular hypoxic-ischemic damage (3).
The role of glucagon-like peptide-1 (GLP-1), a natural hypoglycemic hormone, in regulating blood glucose has been relatively well investigated, and theoretical bases for the cardiovascular protective effects of GLP-1 have been provided through studies (4,5). Liraglutide is an analogue of GLP produced via gene recombination and belongs to GLP-1 receptor agonist. It shows homology of 97% with natural physiological GLP-1 and favorable safety. A study manifested that liraglutide can promote the survival of cardiomyocytes (6), but there are few studies on the role of liraglutide in inhibiting cell apoptosis.
Autophagy is an intracellular route of ‘self-digestion’ that can maintain cellular homeostasis. It can not only degrade the misfolded or denatured proteins and polymers in cells but also break down the impaired mitochondria and other organelles (7). Moreover, autophagy, also known as ‘autophagic flux’, is regarded as a dynamic process. Studies have manifested that autophagy exerts certain effects in the occurrence and development of ischemic cardiomyopathy. Liraglutide can regulate the autophagy levels of liver cells and pancreatic β-cells (8), but whether liraglutide can exert mycardial protective effect by enhancing the autophagic flux in cardiomyocytes has not been confirmed yet.
This investigation observed the protective effects of different concentrations of liraglutide on cardiomyocytes under anoxic conditions by culturing primary cardiomyocytes under CoCl2-induced chemical hypoxia.
Materials and methods
Experimental materials
H9C2 cells were purchased from Shanghai Institute of Biochemistry and Cell Biology (Shanghai, China). The cell suspension was transferred into a 10 ml centrifuge tube and added with 5 ml of Dulbecco's modified Eagle's medium (DMEM) containing 10% fetal bovine serum (FBS) (both from Gibco; Thermo Fisher Scientific, Inc.). After centrifugation at 4˚C, 950 x g for 10 min, the supernatant was discarded, and an appropriate amount of DMEM containing 10% FBS was added, pipetted and mixed evenly. Then the cell density was adjusted to 1x105/ml, the cells were seeded into a 25 ml culture flask for culture in an incubator with 5% CO2 at 37˚C, and the medium was replaced after 24 h.
Model and grouping
The hypoxia model was established using CoCl2 (500 µmol/l for 24 h). The H9C2 cells were divided into blank control group (Cont group), liraglutide groups (Lira group) (1, 10, 100 and 1,000 nmol/l). CoCl2 group (CoCl2 group) and CoCl2 + liraglutide groups (CoCl2 + Lira group) (1, 10, 100 and 1,000 nmol/l).
Cell counting kit-8 (CCK-8)
Cells in the logarithmic growth phase were digested and collected, prepared into cell suspension with a concentration of 1x105/ml, and inoculated into a 96-well plate (100 µl/well). In the experiment, 3 duplicated wells and blank controls were set. After inoculation overnight, the good cell adherence was confirmed under a microscope. Next, 20 µl of 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) (Sigma-Aldrich; Merck KGaA) was added into each well after the cells were treated by groups, followed by culture at 37˚C for 4 h. Later, the supernatant was absorbed carefully, and 150 µl of dimethyl sulfoxide (DMSO) was added into each well and mixed by shaking. Finally, the optical density (OD) value at the wavelength of 570 nm in each well was measured using a microplate reader.
Lactate dehydrogenase (LDH)
Cells in the logarithmic growth phase were digested and collected, prepared into cell suspension with a concentration of 1x105/ml, and inoculated into the 96-well plate (100 µl/well). Three duplicated wells and control group were set for the experiment. After cell adherence, the culture plate was placed in an incubator with 5% CO2 for culture at 37˚C for 24 h. Subsequently, the supernatant was taken for each group (20 µl/well), and corresponding reagents were added according to the kit instructions, followed by mixing, placing at room temperature for 3 min and zeroing using double distilled water and cuvette (optical path, 1 cm) at the wavelength of 440 nm. Finally, the OD value in each tube was determined using the microplate reader. Pyruvic acid (1 gmol) produced in the reaction system after reaction with matrix in 1,000 ml of culture solution at 37˚C for 15 min was defined as 1 unit. The content of LDH in the medium was calculated according to the formula.
Caspase-3 activity
Cells in each group were collected and washed with phosphate-buffered saline (PBS). Then the cells were lysed with trypsin via ice bath, and the cell lysate was extracted to aspirate the cell culture solution for later use. After that, the adherent cells were digested using trypsin and collected into the spare cell culture solution. Finally, Ac-DEVD-pNA (2 mM) was added, mixed and incubated at 37˚C for 60-120 min. The absorbance could be detected when there was relatively apparent color change.
ROS concentration
Cells cultured to a density of 6-8x104/ml in each group were taken and added with culture solution. After passage for 24 h, different concentrations of stimuli were added for reaction for 8 h, then the culture solution was discarded, and the probe CM-H2DCFDA (final concentration: 5 pmol/l) for total ROS in cells was added for incubation in the dark at 37˚C for 30 min. Next, the probe was washed with PBS, and a laser scanning confocal microscope (excitation wavelength: 488 nm and emission wavelength: 515 nm) was employed for detection, under which green fluorescence was visible. Screenshots were taken with 8-10 cells/high-power field (x600), and the fluorescence intensity was analyzed via software.
Nitric oxide (NO) concentration
The concentration was detected using the NO kit provided by Applygen Technologies Inc. DMEM solution was changed before pharmacological preconditioning, and the supernatant of the cells cultured was taken and centrifuged at 4˚C, 950 x g for 10 min for the measurement of NO content. All the operations were conducted in strict accordance of the methods in the kit instructions. Finally, the OD value at the wavelength of 540 nm was measured by adding Griess Reagent I and Griess Reagent II in sequence.
Mitochondrial membrane potential
After treatment, the cells were collected and resuspended in 0.5 ml of cell culture solution with serum available. Subsequently, 0.5 ml of JC-1 staining solution was added and mixed by inverting several times, followed by incubation in the incubator away from light at 37˚C for 30 min and centrifugation at 4˚C, 1,050 x g for 10 min. Then the supernatant was discarded, and the cells were washed with 1X JC-1 staining buffer twice. Later, the cells were resuspended in 1X JC-1 staining buffer and centrifuged at 4˚C, 10,500 x g for 10 min, and the aforementioned washing was repeated once after the supernatant was discarded. Finally, the fluorescence intensity of the cells was detected using a flow cytometer (FACSCalibur; BD Biosciences).
Polymerase chain reaction (PCR)
The treated cells in each group were collected to extract the total ribonucleic acid (RNA) using TRIzol (Invitrogen; Thermo Fisher Scientific, Inc.). After the concentration of the samples was measured, the reverse transcription system was added according to the concentration to perform the reverse transcription. The former 40 cycles were utilized to synthesize the cDNA, and the reverse transcription reaction conditions were set for PCR amplification. Real-time fluorescence signal was collected after each cycle, and the amplification and melting curves were recorded.
Western blotting (WB)
Cells in each group were fetched and washed twice with D-Hank's solution, which was then absorbed using absorbent paper. Next, 150 µl of ice-cold lysis buffer was added into each group and then placed on ice for lysis for 30 min. The proteins in each group were collected into an Eppendorf (EP) tube using a cell scraper, followed by centrifugation at 4˚C, 10,500 x g for 10 min. Then, the supernatant was sucked and transferred into a new EP tube, and 5X loading buffer was added and mixed after the protein concentration was determined via bicinchoninic acid (BCA) method (Pierce; Thermo Fisher Scientific, Inc.), followed by heating at 100˚C for 6 min. Then 30 µl of proteins was added into prepared separation gel and spacer gel loading wells, which were subjected to electrophoresis in the electrophoresis buffer under a proper voltage. Then, the gel was stuck closely to a polyvinylidene fluoride (PVDF) membranes (Millipore), followed by transfer in transmembrane solution at 0˚C under a constant voltage (100 V) for 60 min. After the PVDF membranes were blocked in 5% skim milk powder at room temperature for 1 h, it was clipped according to the molecular weight and then sealed in primary antibodies in a refrigerator at 4˚C overnight. The PVDF membrane was taken out the next day and rinsed with Tris-buffered saline-Tween-20 (TBST), followed by addition of secondary antibody IgG (1:5,000) for incubation at room temperature for 1 h. After that, the membrane was rinsed with TBST again, and Tannon 5200 immunofluorescence development system was applied for image development as well as measurement and calculation of gray scale.
Statistical analysis
The data are presented as mean ± SD (standard deviation) and percentage to those in control group, and analyzed using Statistical Product and Service Solutions (SPSS) 13.0 software (SPSS Inc.). The t-test was used for analyzing measurement data. Differences between two groups were analyzed by using the Student's t-test. Comparison between multiple groups was done using One-way ANOVA test followed by Post Hoc Test (Least Significant Difference). P<0.05 indicates that the difference in data is statistically significant.
Results
Impact of different concentrations of liraglutide on H9C2 cell viability and cytotoxicity
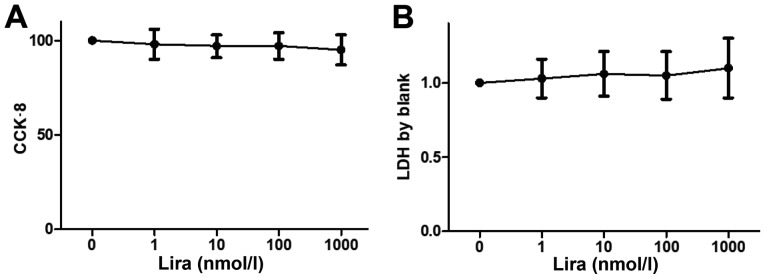
Cell viability was detected via CCK-8, and the LDH concentration was measured to determine the impact on cytotoxicity. The results showed that there was no obvious decline in cell viability and cytotoxicity between Lira group and Cont groups (Fig. 1).
Figure 1.
Impacts of liraglutide on cardiomyocytes. (A) Cell viability in Cont group and Lira group (1, 10, 100 and 1,000 nmol/l) detected via CCK-8. (B) Cytotoxicity in Cont group and Lira group (1, 10, 100 and 1,000 nmol/l) detected via LDH. *P<0.05 vs. Cont groups. CCK-8, cell counting kit-8; LDH, lactate dehydrogenase.
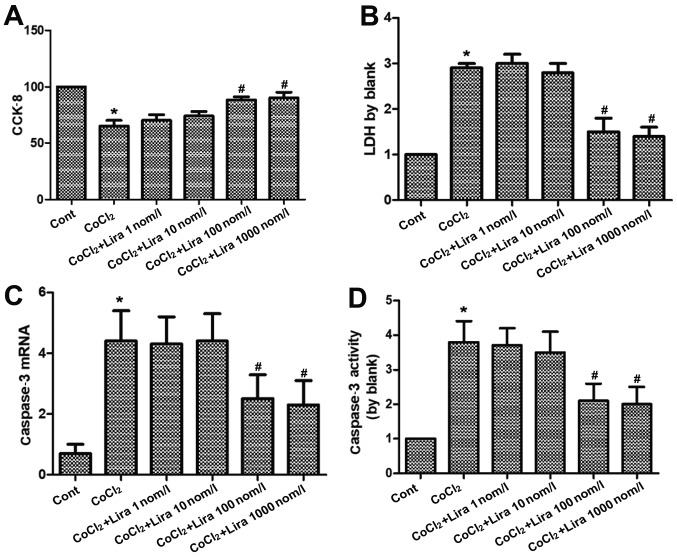
Liraglutide relieves apoptosis of hypoxic H9C2 cells
The cell viability was detected via CCK-8, the LDH concentration was measured to determine the impact on cytotoxicity, the RNA expression of caspase-3 was determined via PCR, and the activation level of caspase-3 was detected using the kit. According to the results, the cell viability was weakened markedly in CoCl2 group compared to that in Cont group (Fig. 2A; P<0.05), while it was strengthened in Lira groups in comparison to that in CoCl2 group. Moreover, 100 nmol/l group had higher cell viability than 10 nmol/l group, displaying a statistically significant difference (P<0.05). Cell viability in 1,000 nmol/l group was higher than that in 100 nmol/l group, but the difference was not statistically significant. Cytotoxicity was increased evidently in CoCl2 group compared with that in Cont group (Fig. 2B; P<0.05), while it was decreased after treatment with liraglutide in comparison to that in CoCl2 group. Moreover, 100 nmol/l group manifested a more apparent change than 10 nmol/l group, with a statistically significant difference (P<0.05), but no statistical difference was detected between 1,000 nmol/l group and 100 nmol/l group. Both RNA expression and activation level of caspase-3 were raised remarkably in CoCl2 group, and the changes were more prominent than those in Cont group (Fig. 2C and D; P<0.05). Liraglutide reduced the RNA expression and activation level of caspase-3 in a dose-dependent manner. 100 nmol/l group had more obvious changes than 10 nmol/l group (P<0.05), while there were no statistically significant differences between 1,000 nmol/l group and 100 nmol/l group. Therefore, 100 nmol/l was selected as the therapeutic concentration for subsequent research. It was proven that liraglutide is able to protect the hypoxic cells.
Figure 2.
Impact of liraglutide on apoptosis of hypoxic cardiomyocytes. (A) Cell viability in Cont group, CoCl2 group and CoCl2 + Lira group (1, 10, 100 and 1,000 nmol/l) detected via CCK-8. (B) Cytotoxicity in Cont group, CoCl2 group and CoCl2 + Lira group (1, 10, 100 and 1,000 nmol/l) detected via LDH. (C) Caspase-3 mRNA detected in each group via PCR. (D) Caspase-3 activity detected in each group. *P<0.05 vs. Cont groups; #P<0.05 vs. CoCl2 groups. CCK-8, cell counting kit-8; LDH, lactate dehydrogenase.
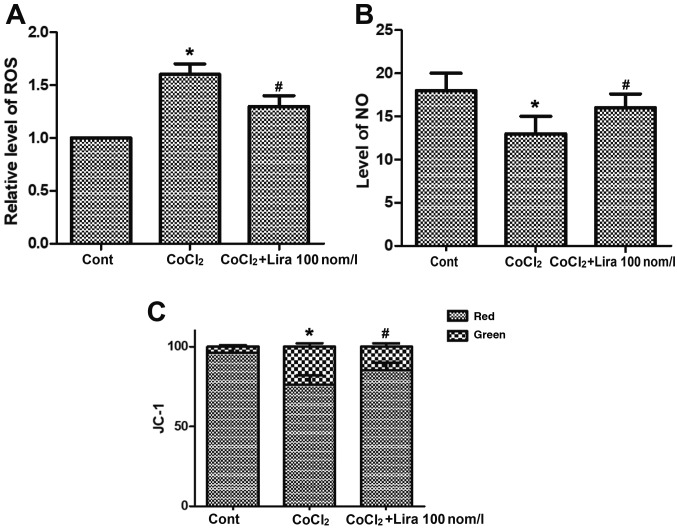
Liraglutide ameliorates oxidative stress in hypoxic H9C2 cells
The concentrations of ROS and NO and change in mitochondrial membrane potential were detected. CoCl2 increased the concentration of ROS notably (Fig. 3A; P<0.05), while the CoCl2-induced ROS increase could be relieved by liraglutide. CoCl2 group exhibited obviously declined NO concentration in cells (Fig. 3B; P<0.05) and destroyed mitochondrial membrane potential, and the mitochondrial membrane potential was decreased markedly compared with that in Cont group (Fig. 3C; P<0.05). In Lira group, however, the lowered NO concentration triggered by CoCl2 was improved notably, and the mitochondrial membrane potential was stabilized, displaying apparent changes in comparison with CoCl2 group (P<0.05). These results verified that liraglutide is capable of ameliorating the oxidative stress in hypoxic cells.
Figure 3.
Impact of liraglutide on oxidative stress in hypoxic cells. (A) ROS concentration detected in Cont group, CoCl2 group and CoCl2 + Lira group. (B) NO concentration detected in Cont group, CoCl2 group and CoCl2 + Lira group. (C) Change in mitochondrial membrane potential detected in Cont group, CoCl2 group and CoCl2 + Lira group. *P<0.05 vs. Cont groups; #P<0.05 vs. CoCl2 groups. ROS, reactive oxygen species; NO, nitric oxide.
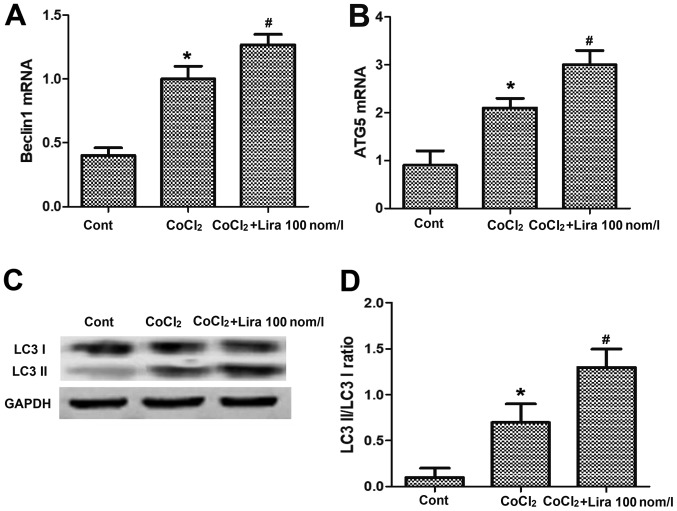
Liraglutide induces autophagy of hypoxic H9C2 cells
PCR was performed to detect the messenger RNA (mRNA) expression of Beclin1 and autophagy-related-5 (Atg-5), and WB assay was conducted to determine the expression of autophagy-related proteins. It was indicated that after the cell hypoxia was simulated by adding CoCl2, the mRNA expression of autophagy-related genes Beclin1 and Atg-5 were raised evidently in comparison with those in Cont group (Fig. 4A and B; P<0.05). After the addition of liraglutide, the mRNA expression of Beclin1 and Atg-5 were further increased, obviously higher than those in CoCl2 group (Fig. 4A and B; P<0.05). The results of WB assay revealed that the light chain 3 (LC3) II/LC3 I ratio was increased. After treatment with liraglutide, the expression of Beclin1 and Atg-5 were further elevated, and the LC3 II/LC3 I ratio was raised compared with those in CoCl2 group (Fig. 4C and D; P<0.05), demonstrating that liraglutide can induce the increased autophagy of hypoxic cells.
Figure 4.
Impact of liraglutide on autophagy of hypoxic cells. (A) mRNA expression of Beclin1 in Cont group, CoCl2 group and CoCl2 + Lira group detected via PCR. (B) mRNA expression in each group detected via PCR. (C) Protein expression of LC3 I, LC3 II and GAPDH in each group detected via WB. (D) Statistical analysis of LC3 II/LC3 I ratio. *P<0.05 vs. Cont groups; #P<0.05 vs. CoCl2 groups. WB, western blotting; Atg-5, autophagy-related-5.
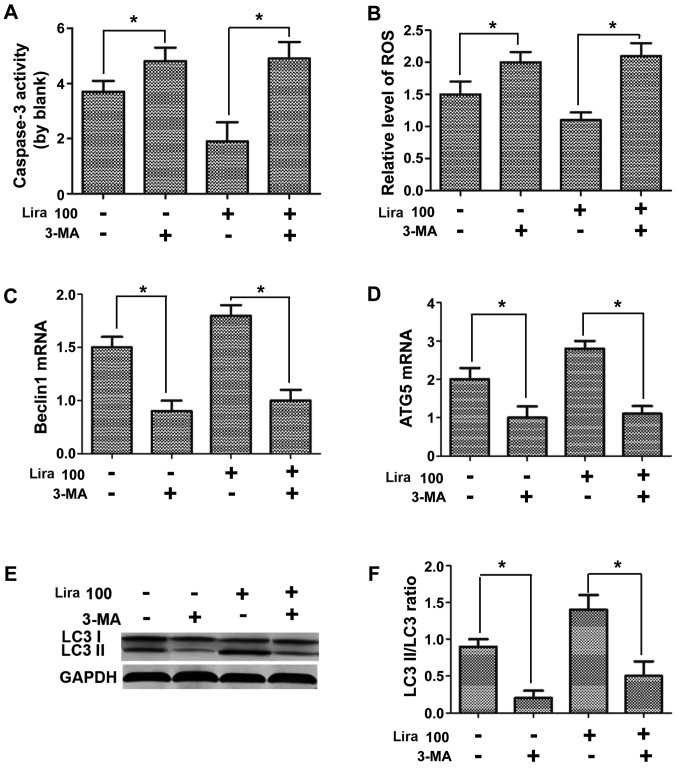
Autophagy inhibitor represses the protective effects of liraglutide on hypoxic H9C2 cells
Caspase-3 activity and expression of ROS and NO were detected, the mRNA expression of Beclin1 and Atg-5 were measured via PCR, and the expression of autophagy-related proteins were determined through WB assay. According to the results, compared with those in Lira group, caspase-3 activity and ROS expression were increased (Fig. 5A and B; P<0.05), while the NO expression was reduced (Fig. 5C; P<0.05) in hypoxic cardiomyocytes after adding 3-MA inhibitor. The detection of expression of autophagy-related genes indicated that 3-MA could prominently repress the increased mRNA expression of Beclin1 and Atg-5 induced by liraglutide (Fig. 5D and E; P<0.05), and the LC3 II/LC3 I ratio was decreased notably (Fig. 5F; P<0.05), confirming that the inhibition on cell autophagy can suppress the protective effects of liraglutide on hypoxic cells.
Figure 5.
Impacts of autophagy inhibitor on protective effects of liraglutide on hypoxic cells. (A) Caspase-3 activity detected in each group. (B) ROS activity detected in each group. (C) mRNA expression of Beclin1 detected in each group. (D) mRNA expression of Atg-5 detected in each group. (E) Protein expression of LC3 I, LC3 II and GAPDH in each group detected via WB. (F) Statistical analysis of LC3 II/LC3 I ratio. *P<0.05 between two groups. ROS, reactive oxygen species; WB, western blotting.
Discussion
CoCl2 is a very important hypoxia inducible factor. It was shown in this study that liraglutide was able to alleviate the CoCl2-induced cardiomyocyte injury, whose major mechanism was to relieve intracellular oxidative stress, mitochondrial damage and cardiomyocyte apoptosis by promoting cell autophagy.
As an analogue of human GLP-1, liraglutide can exert potential cardiovascular protective effects through both GLP-1 receptor-dependent and -independent pathways (9), but its mechanism remains unclear. Currently, most studies have argued that the GLP-1 receptor is expressed in myocardial tissues (10). It has been pointed out in various studies that in in vitro experiments, the GLP-1 receptor agonist exerts the protective effects by directly activating the GLP-1 receptor on the myocardium (11,12).
Ischemic injury can reduce the content of ATP in myocardial cells, resulting in energy stress and excessive production of ROS (2). Ischemia causes hypoxia of myocardial cells, leading to serious or irreversible cardiac injury (3). Excessive ROS from mitochondria is closely related to the pathogenesis of cardiovascular diseases, such as atherosclerosis, myocardial infarction and heart failure (1,2). In this study, we found that liraglutide relieved the ROS increase, improved the lowered NO concentration and stabilized the mitochondrial membrane potential, which indicated that liraglutide is capable of ameliorating the oxidative stress in hypoxic cells.
Autophagy, an intracellular protective mechanism, can degrade the misfolded proteins and damaged organelles such as mitochondria in the cells, thus enabling the mitochondria to synthesize ATP and maintain energy homeostasis in cells (13). Whether the autophagy has protective effects on hypoxic myocardium still remains controversial. Some research teams argued that it is beneficial in increasing the autophagy level in myocardium (14), but it is also reported that the excess increase in autophagy will damage the myocardium (15).
According to the experimental results in this research, liraglutide elevated the autophagy level in myocardium and had certain mycardial protective effects at the same time. Therefore, it was considered in this study that properly increased autophagy in CoCl2-induced hypoxic myocardium is conducive to eliminating the damaged substances in cells, reducing oxidative stress, ameliorating mitochondrial damage and producing cardioprotective effects. Interestingly, the expression of Beclin and Atg-5 also increased in CoCl2 treated cells. However, no protective effect was found in CoCl2 treated cells. This phenomenon further demonstrates the complexity of autophagy.
The autophagy inhibitor 3-MA was applied to further prove that the protective effects of liraglutide on the CoCl2-induced hypoxic myocardium is associated with cell autophagy (16). It was found that after the introduction of 3-MA, the autophagy level and the effects of liraglutide on cardiomyocyte autophagy induced were repressed. Furthermore, the analyses on oxidative stress, mitochondrial damage and apoptosis change in cells revealed that the effects of liraglutide in alleviating oxidative stress in cardiomyocytes, protecting the mitochondria and resisting cell apoptosis were inhibited significantly after the addition of 3-MA. The results confirmed that the protective effects of liraglutide can be inhibited by 3-MA, that is, the protective effects of liraglutide on hypoxic cardiomyocytes are triggered by inducing cardiomyocyte autophagy.
In conclusion, liraglutide ameliorates the CoCl2-induced oxidative stress in hypoxic cardiomyocytes, relieve mitochondrial damage and reduce apoptosis via inducing cell autophagy.
Acknowledgements
Not applicable.
Funding
Not funding was received.
Availability of data and materials
All data generated or analyzed during this study are included in this published article.
Authors' contributions
ZP and SL designed the study and performed the experiments, TW, YL and LW collected the data, JY and HD analyzed the data, ZP and SL prepared the manuscript. All authors read and approved the inal manuscript.
Ethics approval and consent to participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Lai YF, Wang L, Liu WY. Nicotinamide pretreatment alleviates mitochondrial stress and protects hypoxic myocardial cells via AMPK pathway. Eur Rev Med Pharmacol Sci. 2019;23:1797–1806. doi: 10.26355/eurrev_201902_17143. [DOI] [PubMed] [Google Scholar]
- 2.Cheng Y, Liu DZ, Zhang CX, Cui H, Liu M, Zhang BL, Mei QB, Lu ZF, Zhou SY. Mitochondria-targeted antioxidant delivery for precise treatment of myocardial ischemia-reperfusion injury through a multistage continuous targeted strategy. Nanomedicine (Lond) 2019;16:236–249. doi: 10.1016/j.nano.2018.12.014. [DOI] [PubMed] [Google Scholar]
- 3.Li Y, Qiu L, Liu X, Hou Z, Yu B. PINK1 alleviates myocardial hypoxia-reoxygenation injury by ameliorating mitochondrial dysfunction. Biochem Biophys Res Commun. 2017;484:118–124. doi: 10.1016/j.bbrc.2017.01.061. [DOI] [PubMed] [Google Scholar]
- 4.Arturi F, Succurro E, Miceli S, Cloro C, Ruffo M, Maio R, Perticone M, Sesti G, Perticone F. Liraglutide improves cardiac function in patients with type 2 diabetes and chronic heart failure. Endocrine. 2017;57:464–473. doi: 10.1007/s12020-016-1166-4. [DOI] [PubMed] [Google Scholar]
- 5.Inoue T, Inoguchi T, Sonoda N, Hendarto H, Makimura H, Sasaki S, Yokomizo H, Fujimura Y, Miura D, Takayanagi R. GLP-1 analog liraglutide protects against cardiac steatosis, oxidative stress and apoptosis in streptozotocin-induced diabetic rats. Atherosclerosis. 2015;240:250–259. doi: 10.1016/j.atherosclerosis.2015.03.026. [DOI] [PubMed] [Google Scholar]
- 6.Chang G, Liu J, Qin S, Jiang Y, Zhang P, Yu H, Lu K, Zhang N, Cao L, Wang Y, et al. Cardioprotection by exenatide: A novel mechanism via improving mitochondrial function involving the GLP-1 receptor/cAMP/PKA pathway. Int J Mol Med. 2018;41:1693–1703. doi: 10.3892/ijmm.2017.3318. [DOI] [PubMed] [Google Scholar]
- 7.Wang Y, Liu J, Tao Z, Wu P, Cheng W, Du Y, Zhou N, Ge Y, Yang Z. Exogenous HGF prevents cardiomyocytes from apoptosis after hypoxia via up-regulating cell autophagy. Cell Physiol Biochem. 2016;38:2401–2413. doi: 10.1159/000445592. [DOI] [PubMed] [Google Scholar]
- 8.Wang J, Wu J, Wu H, Liu X, Chen Y, Wu J, Hu C, Zou D. Liraglutide protects pancreatic β-cells against free fatty acids in vitro and affects glucolipid metabolism in apolipoprotein E-/- mice by activating autophagy. Mol Med Rep. 2015;12:4210–4218. doi: 10.3892/mmr.2015.3944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ban K, Noyan-Ashraf MH, Hoefer J, Bolz SS, Drucker DJ, Husain M. Cardioprotective and vasodilatory actions of glucagon-like peptide 1 receptor are mediated through both glucagon-like peptide 1 receptor-dependent and -independent pathways. Circulation. 2008;117:2340–2350. doi: 10.1161/CIRCULATIONAHA.107.739938. [DOI] [PubMed] [Google Scholar]
- 10.Wei Y, Mojsov S. Distribution of GLP-1 and PACAP receptors in human tissues. Acta Physiol Scand. 1996;157:355–357. doi: 10.1046/j.1365-201X.1996.42256000.x. [DOI] [PubMed] [Google Scholar]
- 11.Hu SY, Zhang Y, Zhu PJ, Zhou H, Chen YD. Liraglutide directly protects cardiomyocytes against reperfusion injury possibly via modulation of intracellular calcium homeostasis. J Geriatr Cardiol. 2017;14:57–66. doi: 10.11909/j.issn.1671-5411.2017.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bose AK, Mocanu MM, Carr RD, Brand CL, Yellon DM. Glucagon-like peptide 1 can directly protect the heart against ischemia/reperfusion injury. Diabetes. 2005;54:146–151. doi: 10.2337/diabetes.54.1.146. [DOI] [PubMed] [Google Scholar]
- 13.Mizushima N, Levine B, Cuervo AM, Klionsky DJ. Autophagy fights disease through cellular self-digestion. Nature. 2008;451:1069–1075. doi: 10.1038/nature06639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.He C, Zhu H, Li H, Zou MH, Xie Z. Dissociation of Bcl-2-Beclin1 complex by activated AMPK enhances cardiac autophagy and protects against cardiomyocyte apoptosis in diabetes. Diabetes. 2013;62:1270–1281. doi: 10.2337/db12-0533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kobayashi S, Xu X, Chen K, Liang Q. Suppression of autophagy is protective in high glucose-induced cardiomyocyte injury. Autophagy. 2012;8:577–592. doi: 10.4161/auto.18980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yang J, Pi C, Wang G. Inhibition of PI3K/Akt/mTOR pathway by apigenin induces apoptosis and autophagy in hepatocellular carcinoma cells. Biomed Pharmacother. 2018;103:699–707. doi: 10.1016/j.biopha.2018.04.072. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All data generated or analyzed during this study are included in this published article.