Abstract
IL-1 family cytokines play a dual role in the gut, with different family members contributing either protective or pathogenic effects. IL-36γ is an IL-1 family cytokine involved in polarizing type-1 immune responses. However, its function in the gut, including in colorectal cancer pathogenesis, is not well appreciated. In a murine model of colon carcinoma, IL-36γ controls tertiary lymphoid structure formation and promotes a type-1 immune response concurrently with a decrease in expression of immune checkpoint molecules in the tumor microenvironment. Here, we demonstrate that IL-36γ plays a similar role in driving a pro-inflammatory phenotype in human colorectal cancer. We analyzed a cohort of 33 primary colorectal carcinoma tumors using imaging, flow cytometry, and transcriptomics to determine the pattern and role of IL-36γ expression in this disease. In the colorectal tumor microenvironment, we observed IL-36γ to be predominantly expressed by M1 macrophages and cells of the vasculature, including smooth muscle cells and high endothelial venules. This pattern of IL-36γ expression is associated with a CD4+ central memory T cell infiltrate and an increased density of B cells in tertiary lymphoid structures, as well as with markers of fibrosis. Conversely, expression of the antagonist to IL-36 signaling, IL-1F5, was associated with intratumoral expression of checkpoint molecules, including PD-1, PD-L1, and CTLA4, which can suppress the immune response. These data support a role for IL-36γ in the physiologic immune response to colorectal cancer by sustaining inflammation within the tumor microenvironment.
Electronic supplementary material
The online version of this article (10.1007/s00262-018-2259-0) contains supplementary material, which is available to authorized users.
Keywords: Tertiary lymphoid structure, Interleukin (IL)-36 g, Colorectal cancer, Memory T cells, M1 classically activated macrophages
Introduction
Colorectal cancer is the third most common form of cancer worldwide [1]. In 2017, it is estimated that over 50,000 Americans will die from the disease [2], and in France, approximately 17,500 colorectal cancer-associated deaths are reported each year [3]. While the rate of mortality from colorectal cancer is currently decreasing in both countries, only a subset of patients is likely to respond to therapeutic intervention, with most patients having limited or invasive treatment options available [4].
We recently reported the efficacy of an IL-36γ-based therapy in delaying tumor progression in the MC38 murine model of colon adenocarcinoma [5]. The IL-36 cytokines are an IL-1 subfamily [6] consisting of three agonists that signal through a common heterodimeric receptor, IL-36R [7, 8]. Signaling through the IL-36R can be inhibited by the full receptor antagonist, IL-1F5 (aka IL-36RA), which blocks the recruitment of IL-1RAcP, the IL-1 family receptor accessory protein, required for signaling through the IL-36R [7]. IL-36R is expressed on endothelial cells as well as cells of the immune system, including T cells and DC [9–12]. Through its effects on immune cells, IL-36γ is involved in polarizing towards type-1 immune responses [13, 14]. In particular, it is a downstream target of the type-1 transactivator Tbet [13] and can induce Tbet expression in target cells [5]. The therapeutic introduction of IL-36γ into the tumor microenvironment using a DC-based vector delayed tumor progression in conjunction with a rapid (within 4–10 h) recruitment of T cells into the tumor microenvironment and the formation of tumor-associated tertiary lymphoid structures (TLS). IL-36γ-overexpressing DC expressed elevated levels of TLS-promoting chemokines, including LTβR agonists LTA and LIGHT, and CCR7 agonist CCL21. When introduced intratumorally, IL-36γ-based therapy also decreased the level of PD-1, CTLA4, and TIM-3 on CD3+ TIL [5].
It has recently been described that in humans, some colorectal cancers also present with a high immune infiltrate [15], which is sometimes organized into TLS. The presence of TLS within the tumor microenvironment is a positive prognostic marker in colorectal carcinoma and many other solid tumors [16–18]. TLS form at sites of persistent inflammation, such those found in tissues impacted by chronic viral infections, autoimmune diseases, or cancer. These structures are often marked by a germinal center with dense B cell infiltrate and follicular DCs [19, 20]. The principal histologic marker used to identify TLS in colorectal cancer is, therefore, CD20, which identifies these B cells [21, 22]. In addition, TLS contain DC-LAMP+ mature DCs in a T cell zone, follicular DCs, and are surrounded by blood and lymphatic vessels [21, 22] including high endothelial venules (HEV) [17] that allow naïve and central memory lymphocytes to be recruited into TLS.
In this study, we attempt to translate our findings from the mouse model of colon carcinoma into a human by investigating the pattern of expression of IL-36γ in colorectal cancer, and whether IL-36γ expression is associated with TLS components and the infiltration of immune cells into the tumor microenvironment.
Materials and methods
Public transcriptomic datasets
Transcriptomic data from colorectal cancer tumors [23] was downloaded from Gene Expression Omnibus (accession code GSE39582). The data from Affymetrix Human Genome U133 Plus 2.0 Array was normalized using the frozen RMA method with the R package frma [24]. Normalized sorted cells transcriptomic data was obtained from Becht et al. [25] (Gene Expression Omnibus accession code GSE86362). The expression fold-change of a gene was computed as the difference between the median log2 gene expression for the positive samples (defined as all samples from the considered cell population) and all negative samples (defined as all other samples).
Patient cohort
A cohort of 33 primary colorectal tumors was collected between October 2, 2014 and March 3, 2016 from patients operated at the Ambroise Paré hospital (Boulogne Billancourt, France). Patient characteristics are summarized in Table 1.
Table 1.
Patient characteristics
| Median age (range), years | 80 (45–94) |
| Gender, no. (%) | |
| Female | 18 (54.5) |
| Male | 15 (45.5) |
| T stage primary tumor, no. (%) | |
| 0 | 0 (0) |
| 1 | 0 (0) |
| 2 | 2 (6.1) |
| 3 | 21 (63.6) |
| 4 | 10 (30.3) |
| N stage primary tumor, no. (%) | |
| 0 | 20 (60.6) |
| 1 | 11 (30.3) |
| 2 | 2 (6.1) |
| M stage primary tumor, no. (%) | |
| n.a. | 33 (100) |
| Localization primary tumor, no. (%) | |
| Caecum | 5 (15.2) |
| Left colon | 3 (9.1) |
| Rectum | 1 (3.0) |
| Right colon | 13 (39.4) |
| Sigmoid | 8 (24.2) |
| Transverse | 2 (6.1) |
| Upper rectum | 1 (3.0) |
| Microsatellite status, no. (%) | |
| MSS | 25 (75.8) |
| MSI | 8 (24.2) |
IHC, immunofluorescence and image quantification
Immunohistochemical (IHC) staining was performed as previously described [26]. Serial 5-µm formalin-fixed paraffin-embedded tissue sections generated from colorectal cancer were stained using the Dako Autostainer Plus (Agilent). Antigen retrieval and deparaffinization were carried out on a PT-Link (Dako) using the EnVision FLEX Target Retrieval Solutions (Dako). The antibodies used are listed in Supplementary Table 1 and negative control staining for the IL-36γ antibody is demonstrated in Supplementary Fig. 1. Signal intensity was amplified using Envision + System HRP labelled polymers (Dako) or ImmPRESS HRP Polymer Detection Kit (Vector). For IHC staining, peroxidase activity was detected using diaminobenzidine (DAB) substrate (Dako), counterstained with hematoxylin (Dako), and mounted with Glycergel Mounting Medium (Dako). The degree of smooth muscle actin (SMA) expression in the tumor stroma was quantified according to the following grading system: (1) scarce fibroblasts; (2) continuous layer of fibroblast between tumor nests with overall thickness inferior to three cells; (3) continuous layer of fibroblast between tumor nests with overall thickness superior to three cells and fibroblast area < 50% of tumor area; and (4) continuous layer of fibroblast between tumor nests with overall thickness superior to three cells and fibroblast area > 50% of tumor area.
For immunofluorescent staining, signals were detected by Tyramide SuperBoost Signal Amplification (Life Technologies) and slides were counterstained and mounted using ProLong Gold Antifade reagent with DAPI (Molecular Probes). Slides visualized by IHC were digitalized with a NanoZoomer scanner (Hamamatsu) and digitally quantified with Calopix software (Tribvn). Slides visualized by immunofluorescence were digitalized with an Axioscan scanner (Zeiss) and digitally quantified with Visiopharm Integration System (VIS) software (Visiopharm). Individual cells were identified by DAPI+ nuclei. Cell densities are presented as cell number per mm2 of tissue.
Tumor processing, surface staining and cell sorting
Flow cytometry staining was performed as previously described [26]. Briefly, tumors were dilacerated and incubated for 1 h at 4 °C with Cell Recovery Solution (Fisher Scientific); mixtures were filtrated and TILs separated with Ficoll-Paque PLUS (GE Healthcare Life Science). Cells were then stained with the monoclonal antibodies as listed in Supplementary Table 2. Samples were acquired in a FACS Fortessa cytometer with FACSDiva software (BD Bioscience) and data analyzed with FlowJo 7.9.4 software (Tree Star, Inc.). The fraction of cells co-expressing multiple markers was calculated in SPICE 5.3033 (Exon), a data mining software application that normalizes and analyzes large FlowJo datasets [27].
Gene array analyses
Total RNA was isolated from fresh tumor tissue using Quiashredder columns and the RNeasy Mini Kit (both from Qiagen). RNA quality was validated using the RNA 6000 Nano Chip (Agilent) and read on a 2100 Bioanalyzer (Agilent). Transcript analysis was assayed with nCounter (Nanostring). The list of gene targets evaluated in this study is listed in Supplementary Table 3.
Statistical analyses
All data were analyzed using R (R Foundation for Statistical Computing). For categorical variables, data were analyzed using the Mann–Whitney test in the case of two groups; for more than two groups, analyses were carried out using the Kruskal–Wallis test for overall comparison and Dunn test for pairwise comparison (with the use of the R package dunn.test), with Benjamini–Hochberg (False Discovery Rate) correction method for multiple testing. For comparisons of two continuous variables, data were analyzed by Pearson correlation. Test results were considered significant when p < 0.05. For correlations, data were considered biologically meaningful if the absolute value of the correlation value was > 0.5.
Results
IL-36γ is detected in the immune and vascular compartments in the tumor microenvironment
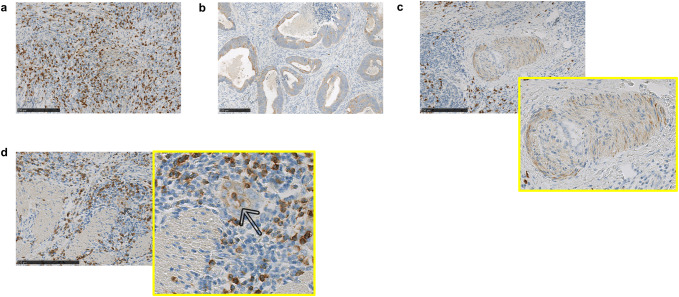
We first used IHC staining to identify and localize IL-36γ+ cells within patient colon carcinoma primary tumors. IL-36γ was detected on a variety of cell types in the tumor microenvironment, including immune cells (Fig. 1a), tumor cells (Fig. 1b) and vascular/perivascular cells (Fig. 1c, d). We noted that within the vascular compartment, both smooth muscle cells (SMC; Fig. 1c) and vascular endothelial cells (VEC; Fig. 1d, indicated by an arrow and inset) were IL-36γ positive, though coordinate detection in the same vessel was uncommon: SMC surrounding large vessels, and VEC of smaller vessels, were found to be IL-36γ+. We did not observe significant differences in IL-36γ expression between tumors with or without lymph node metastases (Supplementary Fig. 2a) or between tumors with MSI versus MSS microsatellite status (Supplementary Fig. 2b). Because the prognosis and treatment modalities for tumors localized to the colon versus the rectum differs clinically, we also compared the levels of IL-36γ expression in tumors at these two sites, and observed that they fell within the same range (Supplementary Fig. 2c).
Fig. 1.
IL-36γ is expressed by a variety of cell populations in the tumor microenvironment. FFPE tumor sections were stained for IL-36γ by IHC as described in “Materials and methods” for expression of IL-36γ. Expression of this protein was observed in immune cells (a), tumor cells (b), and cells of the vasculature (c, d). Both smooth muscle (c) and endothelial cells (d) of the vasculature were observed to express IL-36γ. Bars 250 μm
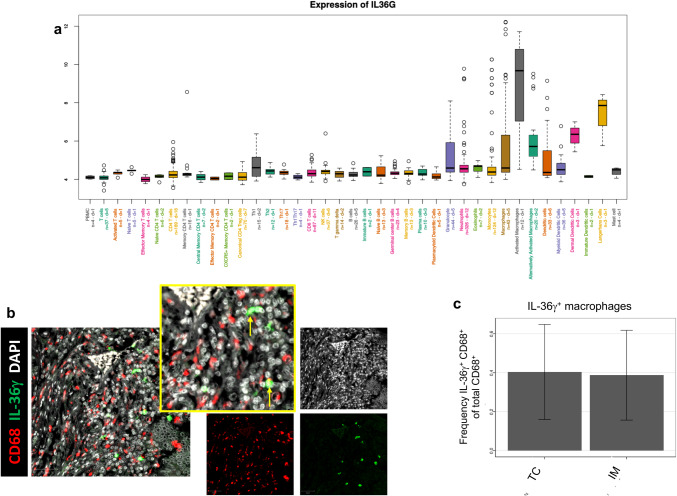
To further investigate the distinct subtypes of immune cells expressing IL-36γ in the tumor microenvironment, we analyzed transcriptomic data of purified immune cell populations from MCP-transcriptomes of 81 public datasets (Fig. 2a). IL36G was found to have a higher transcription level in activated macrophages, also referred to as classically activated or M1 macrophages, than in all other immune cell types, including “alternatively activated” M2 macrophages (log2-fold change versus other cells 5.35, see “Materials and methods”). The two other IL-36γ transcript positive cell types—dermal DCs and Langerhans cells—are found in other organs than the colon. Immunofluorescence imaging showed that CD68+ macrophages are capable of expressing IL-36γ protein within the colorectal cancer tumor microenvironment (Fig. 2b). After quantitation, it was determined that 40.4% of CD68+ cells in the TC and 38.7% in the invasive margin (IM) of tumors expressed IL-36γ (Fig. 2c).
Fig. 2.
M1 macrophages express high levels of IL-36γ. In a, Affymetrix microarray data from the Gene Expression Omnibus (GSE39582) was analyzed for expression of IL36G in various human immune cell subsets. Data are presented on a log2 scale. Results indicate an elevated level of IL-36γ expression by activated M1 macrophages compared to the median expression by all other cell types. In b, IL-36γ was visualized by immunofluorescence imaging in conjunction with CD68+ macrophages. Bars 50 μm. Data were analyzed as described in “Materials and methods”, and the average frequency of IL-36γ+ macrophages, identified as DAPI+ nuclei surrounded by colocalized CD68 and IL-36γ signal, in both the TC and IM across the cohort is presented in c
IL-36γ expression by macrophages is associated with markers of inflammation
We next investigated whether a correlation could be made between IL-36γ expression by macrophages and other previously established prognostic markers for patients with colon cancer, using a prospective cohort of 33 primary tumors. In particular, we were interested in determining whether IL-36γ+ macrophages were linked to a fibroblastic signature within the tumor microenvironment and/or to an intratumoral T cell response.
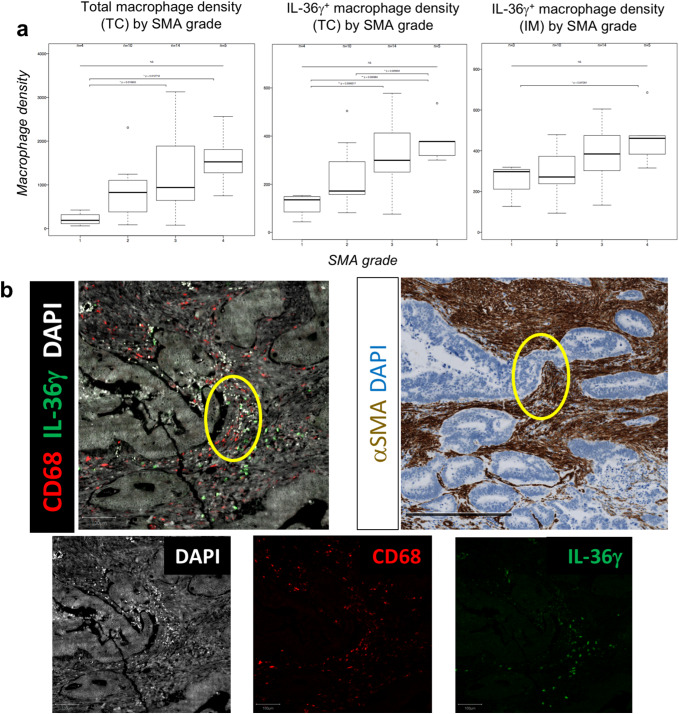
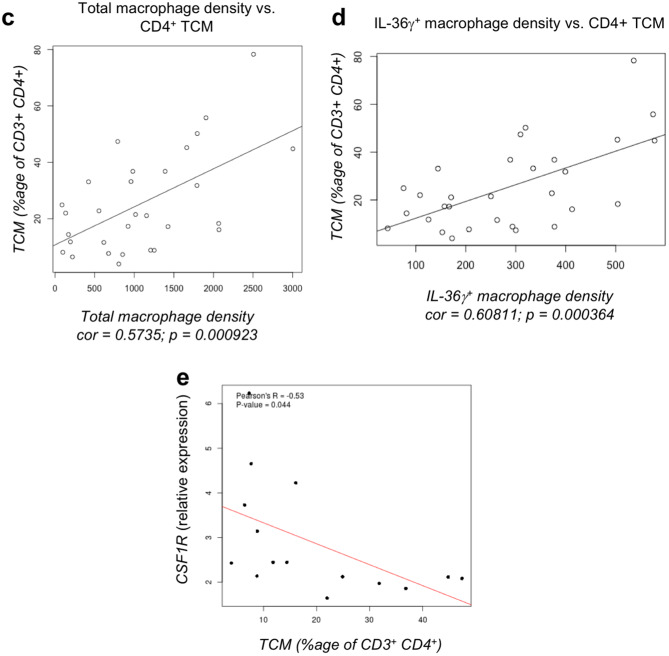
Alpha-smooth muscle actin (SMA) was used to detect cancer-associated fibroblasts by IHC grading, as described in “Materials and methods” [28]. In this cohort, 15% (5/33) of tumors were classified as SMA grade 1, 27% (9/33) were grade 2, 42% (14/33) were grade 3, and 15% (5/33) were grade 4. As shown in Fig. 3a, tumor-associated macrophages located in the tumor core (TC) were positively correlated with an increase in SMA grade. When just the IL-36γ+ macrophage subset was analyzed, this correlation became stronger in the TC, with a similar correlation noted in the IM. Immunofluorescence imaging suggested a close contact between (IL-36γ+) macrophages and SMA+ cells in the tumor microenvironment, indicating that macrophages can be predominantly found within the stromal compartment in colorectal tumors (Fig. 3b). TIL subsets from the same tumors were phenotyped by flow cytometric analysis of T cells isolated from fresh tissues. We identified four subsets each of CD4+ and CD8+ T cells: naïve (CCR7+ CD45RA+), effector memory (TEM; CCR7− CD45RA−), central memory (TCM; CCR7+ CD45RA−), and effector memory RA (TEMRA; CCR7− CD45RA+) (Table 2) [26]. We found a positive correlation between CD4+ central memory T cells (identified by flow cytometry; Supplementary Fig. 3) and macrophages in the TC (identified by immunofluorescent imaging; r = 0.574 and p = 0.000923; Fig. 3c), but not the IM (data not shown). When just the IL-36γ+ macrophage subset in the TC was studied, the correlation with CD4+ TCM was improved (r = 0.608 and p = 0.000364; Fig. 3d). We also analyzed the expression of immune genes (Supplementary Table 3) in whole tumor samples, and the gene expression studies showing negative correlation between CD4+ TCM and the CSF1R transcript, a marker of immunosuppressive M2 macrophages [29] (p = 0.044; Fig. 3e) confirmed these data. No significant correlations were found between IL-36γ+ macrophages in the TC and any other naïve or memory CD4+ or CD8+ TIL subsets (Supplementary Fig. 4). Thus, it appears that IL-36γ+ macrophages are associated with both an increased fibroblastic signature—a negative prognostic marker—and a memory immune response—a positive prognostic marker in the setting of colorectal cancer.
Fig. 3.
IL-36γ+ macrophages are associated with a proinflammatory tumor microenvironment. FFPE tumor sections were visualized by IHC for expression of alpha-SMA or by immunofluorescence for IL-36γ+ CD68+ macrophages. A positive correlation between increased alpha-SMA grade and intratumoral density of IL-36γ+ macrophages is shown in a. In b, tumors were visualized either by immunofluorescence for CD68 and IL-36γ (top left) or by IHC for alpha-SMA (top right). Single-color stains corresponding to the composite immunofluorescence image are shown below. CD68+ macrophages and alpha-SMA+ cells were observed in close contact with each other within the tumor microenvironment, indicating a localization of macrophages to the tumor stroma. Bar 250 μm for immunofluorescent image and 500 μm for immunohistochemical image. The overall density of macrophages is positively-correlated with the presence of CD4+ central memory T cells in tumors (c), but that the strength of this correlation is increased when just the IL-36γ+ macrophage subset is analyzed (d). Furthermore, the presence of CD4+ central memory T cell infiltrate is inversely correlated with the expression of CSF1R, a biomarker of M2 macrophages (e)
Table 2.
TIL subset frequencies
| Mean (%) | SD (%) | |
|---|---|---|
| CD4 | ||
| Naïve | 2.422 | 5.380 |
| TCM | 25.448 | 17.756 |
| TEM | 69.357 | 18.542 |
| TEMRA | 2.780 | 5.397 |
| CD8 | ||
| Naïve | 1.697 | 3.217 |
| TCM | 8.805 | 9.760 |
| TEM | 81.143 | 14.020 |
| TEMRA | 6.530 | 7.367 |
The frequency of TIL populations is denoted as the percentage of naïve, TCM, TEM, or TEMRA cells out of total C3+ CD4+ or CD3+ CD8+ T cells
The predominant IL-36γ-expressing cells in TLS are HEV-associated VEC
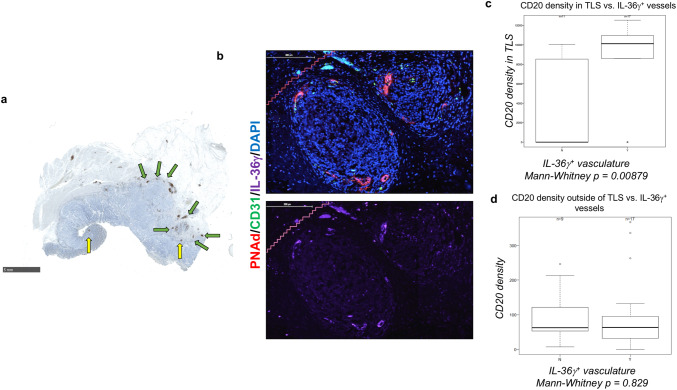
CD4+ TCM have been reportedly found primarily in TLS within the confines of human tumors [30]. Since IL-36γ expression was correlated with CD4+ TCM, we next investigated TLS in the colorectal cancer tumor microenvironment, using the B cell marker CD20. Indeed, we found a series of dense CD20+ aggregates in tumors from this patient cohort, most being located in the IM (Fig. 4a). TLS can also be marked by the presence of peripheral node addressin (PNAd)+ HEV (Fig. 4b), i.e. specialized CD31+ VEC involved in the recruitment of CD62L/L-selectin+ naïve or central memory lymphocytes from the peripheral blood circulation. We next investigated the expression of IL-36γ within these structures. We found that IL-36γ was principally expressed on HEV themselves, with minimal expression by the constituent immune cells or by “normal” CD31+ PNAd− VEC (n = 7; Fig. 4b). Since IL-36γ has not been reported by other groups to be expressed by the vasculature, we next sought to further investigate this pattern of expression.
Fig. 4.
Expression of CD20 suggests the presence of TLS in colorectal tumors. FFPE tumors were visualized by IHC for the presence of CD20 (a), a B cell marker used to identify TLS. TLS in the IM are annotated with green arrows; TLS in the TC are marked by yellow arrows. Tumor sections were probed with antibodies against CD31 and PNAd to identify HEV, together with an antibody reactive against IL-36γ (b). Bars 5 mm in a and 200 μm in b. IL-36γ expression on the vasculature correlated with an increased density of B cells within TLS (c), but not outside of these structures (d) within the tumor microenvironment
IL-36γ expression on the vasculature is associated with maintenance of TLS structures
To investigate the vascular expression of IL-36γ, we divided our cohort into two groups: patients with IL-36γ expression on any vessels in the tumor (i.e. including HEV or arteries, n = 21), and those devoid of IL-36γ expression in vascular structures (n = 12). Within these cohorts, we then analyzed tissues for correlations between vascular expression of IL-36γ and immune cell infiltrate into the tumors. We observed that in the IM, the density of CD20+ B cells in the TLS of patients with IL-36γ+ blood vessels was significantly higher than in patients without any IL-36γ+ vessels (p = 0.00879; Fig. 4c). We did not discern a significant correlation between IL-36γ expression on blood vessels and the density of intratumoral B cells outside of TLS (p = 0.829; Fig. 4d) or absolute numbers of TLS within tumors (data not shown). These data suggest that IL-36γ expression on the vasculature may be involved in the maintenance of TLS in the tumor microenvironment. A larger cohort of patients presenting with TLS will be required to parse the effects of HEV versus vasculature outside of TLS.
IL-1F5 expression in the tumor microenvironment is associated with immunosuppressive markers
Tissue expression of IL-1F5 (aka IL-36RA), the natural antagonist to the IL-36 receptor, was probed using IHC to investigate whether this negative regulatory member of the IL-36R signaling pathway might be associated with suppression of either TIL function or TLS organization in our cohort. IL-1F5 expression was found on the tumor vasculature (Supplementary Fig. 5a) as well as vasculature in the tonsil, a secondary lymphoid organ (Supplementary Fig. 4b). The density of B cells in IM-localized TLS was not significantly different in tumors whether blood vessels expressed IL-1F5 or not (Supplementary Fig. 5c). We next investigated whether the presence of intratumoral IL-1F5 correlated with other markers of the tumor microenvironment as detected by transcriptome analysis. IL-1F5 (but not IL-36γ) expression in the IM of tumors was found to be positively correlated with local expression of the PDCD1 (i.e. PD-1), CTLA4, and CD274 (PD-L1) immune checkpoint markers, but not with expression of LAG3, ICOS, or ICOSL (Supplementary Fig. 6).
Discussion
In this study, we report a link between intratumoral expression of IL-36γ and markers of an ongoing anti-tumor immune response in the tumor microenvironment. IL-36γ+ macrophage density was found to correlate with CD4+ TCM frequency in TILs. CD4+ TCM infiltrate was also associated with a decreased M2 macrophage marker in the tumor microenvironment. In human lung [30] and breast cancers [31] cancer patients, CD4+ TCM have been reported to be found predominantly within tumor-associated TLS, though further studies in colorectal cancer should be done to verify in this disease. The presence of TLS within the tumor microenvironment has been associated with an ongoing local anti-tumor immune response [18]. It has previously been reported as a positive prognostic marker in colorectal cancer [16, 22, 32], with TLS most commonly identified by dense aggregates of CD20+ B cells resembling the germinal centers found in lymph nodes [33].
Our studies also suggest a link between IL-36γ+ macrophages and SMA grade in our tumors. In colorectal cancer, the grade of SMA is associated with poor progression-free and overall survival rates [15, 28]. In the setting of esophageal squamous cell carcinoma, expression of fibroblast-associated genes, including SMA, are associated with poor overall and progression-free survival [34]. In this latter study, the authors observed that the increased prevalence of tumor-infiltrating macrophages was associated with an increased fibroblast signature and poor prognosis. Thus, we would predict that SMA grade 4 patients are most likely to also exhibit robust IL-36γ+ macrophage infiltrates and type-1 pro-inflammatory/anti-tumor immune responses. Indeed, it has previously been reported in a murine model of atherosclerosis that M1 macrophages can act indirectly as lymphoid tissue inducer cells that lead to the formation of TLS by the secretion of chemokines and cytokines that act on SMA+ vascular smooth muscle cells and convey a lymphoid tissue organizer phenotype onto these cells [35]. Further studies should investigate a role for IL-36γ+ macrophages in TLS organization in the tumor microenvironment, and whether cancer patients presenting with a high stromal cell signature along with a high IL-36γ+ macrophage or TLS signature have a better prognostic outcome than their counterparts with a high stromal cell signature but lacking a type-1 immune infiltrate.
Release of IL-36γ in IBD and colitis leads to IL-36R-mediated signaling in colonic fibroblasts and to secondary production of chemokines/cytokines (i.e. GM-CSF, CCL1, CCL2) known to recruit and differentiate monocytes/macrophages [36], and IL-36γ treatment has been shown in in vitro models to lead to the secretion of chemokines including CXCL1, CXCL2, and CXCL8 by myofibroblasts [37]. Furthermore, IL-36R signaling promotes healing of the mucosa following damage. These results are consistent with our current findings suggesting a positive correlation between IL-36γ+ macrophages and SMA in the tumor microenvironment. Notably, fibrosis is a mechanism involved with the healing of damaged tissues [38], and tumors have long been referred to as “wounds that do not heal” [39]. One might, therefore, anticipate that signaling through the IL-36R on colonic fibroblasts could coordinately promote macrophage recruitment and mucosal healing mechanisms that drive local fibrosis. However, in the context of cancer, fibrosis is classically viewed as a promoter of disease progression [40]. In addition to fibroblasts, a common cell type involved with the progression of fibrosis is the myofibroblast [41]. Interestingly, IL-36γ can be intrinsically expressed by healthy colonic myofibroblasts as a consequence of IL-1β-induced signaling [42]. Furthermore, IL-1β is a key cytokine in the transition of stromal cells including fibroblasts, smooth muscle cells, and pericytes to become myofibroblasts [43]. Together, these results suggest that in addition to the IL-36γ+ SMA+ cells within the tumor that were determined by pathologic characterization to be muscle fibers (Fig. 1c), a portion of the IL-36γ+ SMA+ cells may represent mature myofibroblasts. Myofibroblasts represent an “activated” fibroblast phenotype that has been reported to be present in colorectal cancer that has metastasized to the lymph node [44]. Future studies should investigate whether the frequency of IL-36γ-expressing fibroblasts in the primary tumor is differentially associated with disease progression/metastasis or the induction of an adaptive immune response. Such studies should also include analyses of IL-36α, ανοτηερ αγονιστ οφ ΙΛ−36Ρ, that has been reported to independently predict increased overall survival amongst patients with colorectal cancer [45].
To the best of our knowledge, we are the first to characterize the expression of IL-36γ by cells of the tumor-associated vasculature, with this cytokine found located on VEC in HEV and on SMC surrounding large blood vessels. We observed vascular expression of IL-36γ associated with a higher density of B cells in TLS in the IM (but not with TLS number), suggesting a role for vascular expression of IL-36γ in maintaining the cellular composition of existing TLS. Signaling through the IL-36R on VEC can result in upregulated expression of VCAM-1 and ICAM-1 and the production of chemokines, such as IL-8, CCL2, and CCL20 [11]. Following stimulation of with IL-36γ, T cells exhibit increased migratory capability towards VEC [11]. These data suggest that the VEC of HEV may be able to both produce and respond to IL-36γ in an autocrine manner, a phenomenon which has previously been shown to occur in myeloid cells [46–48]. Thus, a positive feedback signaling mechanism may increase the “recruiting” capacity of IL-36γ+ HEV for protective/therapeutic immune cell populations into existing TLS, either from the periphery or from elsewhere within colorectal cancer lesions.
In conclusion, our findings support a role for macrophage- and VEC-produced IL-36γ in recruiting and maintaining intratumoral immune responses, independent of other factors known to promote anti-tumor immunity. The promotion of a memory T cell response and the maintenance of TLS are both predictors of a positive prognosis in colorectal cancer and are both associated with increased IL-36γ production within the tumor microenvironment. In line with these findings, tumors presenting with elevated levels of the IL-36 receptor antagonist, IL-1F5, generally express less IL-36γ (data not shown) and were observed to have lower levels of central memory T cell infiltrates and lower densities of intratumoral B cells. IL-1F5 expression also was associated with elevated levels of the immune checkpoint molecules PD-1, PD-L1, and CTLA4 in the tumor microenvironment. In other studies, expression of IL-1F5 has been associated with a poor prognosis in colorectal cancer [49]. Because IL-36γ has a 100- to 1000-fold higher binding affinity for the IL-36R than does the receptor antagonist IL-1F5 [10], administration of an IL-36γ-based therapy would be expected to at least partially reverse the inhibition mediated by endogenous IL-1F5, leading to enhanced tumor infiltration by beneficial immune cell populations. Because of the correlation between IL-1F5 expression and an upregulation of immune checkpoint molecules, the co-application of an IL-36γ-based therapy with checkpoint blockade would be expected to also enhance the anti-tumor efficacy of these treatments. Future studies should determine whether the presence of IL-36γ in the tumor microenvironment at baseline is predictive of superior response to immunotherapeutic intervention.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Abbreviations
- EMRA
Effector memory T cells that express CD45RA
- HEV
High endothelial venule
- IHC
Immunohistochemistry
- IM
Invasive margin
- MSI
Microsatellite instable
- PNAd
Peripheral node addressin
- SMA
Alpha-smooth muscle actin
- SMC
Smooth muscle cells
- TC
Tumor core
- TCM
Central memory T cells
- TLS
Tertiary lymphoid structures
- VEC
Vascular endothelial cells
Author contributions
AMW, NAG, WHF, and CS-F designed experiments. AMW, NAG, LL, and LM performed experiments. AMW, NAG, FP, WHF, and CS-F analyzed data. AMW, NAG, WHF, WJS, and CS-F wrote the manuscript. FP, CJ and J-FE designed clinical protocol and acquired clinical samples. All authors approved the final version of this manuscript.
Funding
This work was supported by the Institut National de la Santé et de la Recherche Médicale, the University Paris-Descartes, the University Pierre et Marie Curie, Labex Immuno-Oncology (LAXE62_9UMRS972 FRIDMAN) and Cancer Research for Personalized Medecine programmes (CARPEM T8), and grants from Institut du Cancer (INCa) HTE Plan Cancer (C1608DS) and NIH RO1s CA169118 and CA204419. Aliyah M. Weinstein was supported by a Chateaubriand Fellowship of the Office for Science and Technology of the Embassy of France in the United States and Florent Petitprez by CARPEM doctorate fellowship.
Compliance with ethical standards
Conflict of interest
The authors declare that they have no conflict of interest.
Ethical approval and ethical standards
The research project “Reincolon” was approved by the Institutional Review Board of Hôpitaux Universitaires Paris-Ile de France Ouest-Ambroise Paré (IO-ACA-CRB-FM002). The study was conducted according to the recommendations in the Helsinki Declaration.
Informed consent
All the included patients signed an informed consent form prior to inclusion in the study.
References
- 1.Jorgensen ML, Young JM, Solomon MJ. Optimal delivery of colorectal cancer follow-up care: improving patient outcomes. Patient Relat Outcome Meas. 2015;6:127–138. doi: 10.2147/PROM.S49589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Siegel RL, Miller KD, Fedewa SA, et al. Colorectal cancer statistics, 2017. CA Cancer J Clin. 2017;67:177–193. doi: 10.3322/caac.21395. [DOI] [PubMed] [Google Scholar]
- 3.Ribassin-Majed L, Le Teuff G, Hill C. La fréquence des cancers en 2016 et leur évolution. Bull Cancer. 2017;104:20–29. doi: 10.1016/j.bulcan.2016.11.003. [DOI] [PubMed] [Google Scholar]
- 4.Iqbal A, George TJ. Randomized clinical trials in colon and rectal cancer. Surg Oncol Clin N Am. 2017;26:689–704. doi: 10.1016/j.soc.2017.05.008. [DOI] [PubMed] [Google Scholar]
- 5.Weinstein AM, Chen L, Brzana EA, et al. Tbet and IL-36γ cooperate in therapeutic DC-mediated promotion of ectopic lymphoid organogenesis in the tumor microenvironment. Oncoimmunology. 2017 doi: 10.1080/2162402X.2017.1322238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Busfield SJ, Comrack CA, Yu G, et al. Identification and gene organization of three novel members of the IL-1 family on human chromosome 2. Genomics. 2000;66:213–216. doi: 10.1006/geno.2000.6184. [DOI] [PubMed] [Google Scholar]
- 7.Debets R, Timans JC, Homey B, et al. Two novel IL-1 family members, IL-1 and IL-1, function as an antagonist and agonist of NF-B activation through the orphan IL-1 receptor-related protein 2. J Immunol. 2001;167:1440–1446. doi: 10.4049/jimmunol.167.3.1440. [DOI] [PubMed] [Google Scholar]
- 8.Towne JE, Garka KE, Renshaw BR, et al. Interleukin (IL)-1F6, IL-1F8, and IL-1F9 signal through IL-1Rrp2 and IL-1RAcP to activate the pathway leading to NF-kappaB and MAPKs. J Biol Chem. 2004;279:13677–13688. doi: 10.1074/jbc.M400117200. [DOI] [PubMed] [Google Scholar]
- 9.Mutamba S, Allison A, Mahida Y, et al. Expression of IL-1Rrp2 by human myelomonocytic cells is unique to DCs and facilitates DC maturation by IL-1F8 and IL-1F9. Eur J Immunol. 2012;42:607–617. doi: 10.1002/eji.201142035. [DOI] [PubMed] [Google Scholar]
- 10.Vigne S, Palmer G, Lamacchia C, et al. IL-36R ligands are potent regulators of dendritic and T cells. Blood. 2011;118:5813–5823. doi: 10.1182/blood-2011-05-356873. [DOI] [PubMed] [Google Scholar]
- 11.Bridgewood C, Stacey M, Alase A, et al. IL-36γ has proinflammatory effects on human endothelial cells. Exp Dermatol. 2017;26:402–408. doi: 10.1111/exd.13228. [DOI] [PubMed] [Google Scholar]
- 12.Penha R, Higgins J, Mutamba S, et al. IL-36 receptor is expressed by human blood and intestinal T lymphocytes and is dose-dependently activated via IL-36β and induces CD4+ lymphocyte proliferation. Cytokine. 2016;85:18–25. doi: 10.1016/j.cyto.2016.05.023. [DOI] [PubMed] [Google Scholar]
- 13.Bachmann M, Scheiermann P, Härdle L, et al. IL-36γ/IL-1F9, an innate T-bet target in myeloid cells. J Biol Chem. 2012;287:41684–41696. doi: 10.1074/jbc.M112.385443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Vigne S, Palmer G, Martin P, et al. IL-36 signaling amplifies Th1 responses by enhancing proliferation and Th1 polarization of naive CD4+ T cells. Blood. 2012;120:3478–3487. doi: 10.1182/blood-2012-06-439026. [DOI] [PubMed] [Google Scholar]
- 15.Guinney J, Dienstmann R, Wang X, et al. The consensus molecular subtypes of colorectal cancer. Nat Med. 2015;21:1350–1356. doi: 10.1038/nm.3967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Remark R, Alifano M, Cremer I, et al. Characteristics and clinical impacts of the immune environments in colorectal and renal cell carcinoma lung metastases: influence of tumor origin. Clin Cancer Res. 2013;19:4079–4091. doi: 10.1158/1078-0432.CCR-12-3847. [DOI] [PubMed] [Google Scholar]
- 17.Di Caro G, Bergomas F, Grizzi F, et al. Occurrence of tertiary lymphoid tissue is associated with T-cell infiltration and predicts better prognosis in early-stage colorectal cancers. Clin Cancer Res. 2014;20:2147–2158. doi: 10.1158/1078-0432.CCR-13-2590. [DOI] [PubMed] [Google Scholar]
- 18.Dieu-Nosjean M-C, Giraldo NA, Kaplon H, et al. Tertiary lymphoid structures, drivers of the anti-tumor responses in human cancers. Immunol Rev. 2016;271:260–275. doi: 10.1111/imr.12405. [DOI] [PubMed] [Google Scholar]
- 19.Gatto D, Brink R. The germinal center reaction. J Allergy Clin Immunol. 2010;126:898–907. doi: 10.1016/j.jaci.2010.09.007. [DOI] [PubMed] [Google Scholar]
- 20.Dieu-Nosjean M-C, Antoine M, Danel C, et al. Long-term survival for patients with non-small-cell lung cancer with intratumoral lymphoid structures. J Clin Oncol. 2008;26:4410–4417. doi: 10.1200/JCO.2007.15.0284. [DOI] [PubMed] [Google Scholar]
- 21.Bergomas F, Grizzi F, Doni A, et al. Tertiary intratumor lymphoid tissue in colo-rectal cancer. Cancers (Basel) 2011;4:1–10. doi: 10.3390/cancers4010001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Coppola D, Nebozhyn M, Khalil F, et al. Unique ectopic lymph node-like structures present in human primary colorectal carcinoma are identified by immune gene array profiling. Am J Pathol. 2011;179:37–45. doi: 10.1016/j.ajpath.2011.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Marisa L, de Reyniès A, Duval A, et al. Gene expression classification of colon cancer into molecular subtypes: characterization, validation, and prognostic value. PLoS Med. 2013;10:e1001453. doi: 10.1371/journal.pmed.1001453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.McCall MN, Bolstad BM, Irizarry RA. Frozen robust multiarray analysis (fRMA) Biostatistics. 2010;11:242–253. doi: 10.1093/biostatistics/kxp059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Becht E, Giraldo NA, Lacroix L, et al. Estimating the population abundance of tissue-infiltrating immune and stromal cell populations using gene expression. Genome Biol. 2016;17:218. doi: 10.1186/s13059-016-1070-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Giraldo NA, Becht E, Vano Y, et al. Tumor-infiltrating and peripheral blood T-cell immunophenotypes predict early relapse in localized clear cell renal cell carcinoma. Clin Cancer Res. 2017;23:4416–4428. doi: 10.1158/1078-0432.CCR-16-2848. [DOI] [PubMed] [Google Scholar]
- 27.Roederer M, Nozzi JL, Nason MC. SPICE: exploration and analysis of post-cytometric complex multivariate datasets. Cytometry A. 2011;79:167–174. doi: 10.1002/cyto.a.21015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Becht E, de Reyniès A, Giraldo NA, et al. Immune and stromal classification of colorectal cancer is associated with molecular subtypes and relevant for precision immunotherapy. Clin Cancer Res. 2016;22:4057–4066. doi: 10.1158/1078-0432.CCR-15-2879. [DOI] [PubMed] [Google Scholar]
- 29.Ohashi T, Aoki M, Tomita H, et al. M2-like macrophage polarization in high lactic acid-producing head and neck cancer. Cancer Sci. 2017;108:1128–1134. doi: 10.1111/cas.13244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.de Chaisemartin L, Goc J, Damotte D, et al. Characterization of chemokines and adhesion molecules associated with T cell presence in tertiary lymphoid structures in human lung cancer. Cancer Res. 2011;71:6391–6399. doi: 10.1158/0008-5472.CAN-11-0952. [DOI] [PubMed] [Google Scholar]
- 31.Martinet L, Garrido I, Filleron T, et al. Human solid tumors contain high endothelial venules: association with T- and B-lymphocyte infiltration and favorable prognosis in breast cancer. Cancer Res. 2011;71:5678–5687. doi: 10.1158/0008-5472.CAN-11-0431. [DOI] [PubMed] [Google Scholar]
- 32.Meshcheryakova A, Tamandl D, Bajna E, et al. B cells and ectopic follicular structures: novel players in anti-tumor programming with prognostic power for patients with metastatic colorectal cancer. PLoS One. 2014;9:e99008. doi: 10.1371/journal.pone.0099008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Germain C, Gnjatic S, Tamzalit F, et al. Presence of B cells in tertiary lymphoid structures is associated with a protective immunity in patients with lung cancer. Am J Respir Crit Care Med. 2014;189:832–844. doi: 10.1164/rccm.201309-1611OC. [DOI] [PubMed] [Google Scholar]
- 34.Ha SY, Yeo S-Y, Xuan Y, Kim S-H. The prognostic significance of cancer-associated fibroblasts in esophageal squamous cell carcinoma. PLoS One. 2014;9:e99955. doi: 10.1371/journal.pone.0099955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Guedj K, Khallou-Laschet J, Clement M, et al. M1 macrophages act as LTβR-independent lymphoid tissue inducer cells during atherosclerosis-related lymphoid neogenesis. Cardiovasc Res. 2014;101:434–443. doi: 10.1093/cvr/cvt263. [DOI] [PubMed] [Google Scholar]
- 36.Scheibe K, Backert I, Wirtz S, et al. IL-36R signalling activates intestinal epithelial cells and fibroblasts and promotes mucosal healing in vivo. Gut. 2016;66:823–838. doi: 10.1136/gutjnl-2015-310374. [DOI] [PubMed] [Google Scholar]
- 37.Kanda T, Nishida A, Takahashi K, et al. Interleukin(IL)-36α and IL-36γ induce proinflammatory mediators from human colonic subepithelial myofibroblasts. Front Med. 2015;2:69. doi: 10.3389/fmed.2015.00069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wynn TA, Vannella KM. Macrophages in tissue repair, regeneration, and fibrosis. Immunity. 2016;44:450–462. doi: 10.1016/j.immuni.2016.02.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Dvorak HF. Tumors: wounds that do not heal-redux. Cancer Immunol Res. 2015;3:1–11. doi: 10.1158/2326-6066.CIR-14-0209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rybinski B, Franco-Barraza J, Cukierman E. The wound healing, chronic fibrosis, and cancer progression triad. Physiol Genom. 2014;46:223–244. doi: 10.1152/physiolgenomics.00158.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Cirri P, Chiarugi P. Cancer associated fibroblasts: the dark side of the coin. Am J Cancer Res. 2011;1:482–497. [PMC free article] [PubMed] [Google Scholar]
- 42.Takahashi K, Nishida A, Shioya M et al (2015) Interleukin (IL)-1β Is a strong inducer of IL-36γ expression in human colonic myofibroblasts. 10:e0138423. 10.1371/journal.pone.0138423 [DOI] [PMC free article] [PubMed]
- 43.Kendall RT, Feghali-Bostwick CA. Fibroblasts in fibrosis: novel roles and mediators. Front Pharmacol. 2014;5:123. doi: 10.3389/fphar.2014.00123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yeung TM, Buskens C, Wang LM, et al. Myofibroblast activation in colorectal cancer lymph node metastases. Br J Cancer. 2013;108:2106–2115. doi: 10.1038/bjc.2013.209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wang Z-S, Cong Z-J, Luo Y, et al. Decreased expression of interleukin-36α predicts poor prognosis in colorectal cancer patients. Int J Clin Exp Pathol. 2014;7:8077–8081. [PMC free article] [PubMed] [Google Scholar]
- 46.Barksby HE, Nile CJ, Jaedicke KM, et al. Differential expression of immunoregulatory genes in monocytes in response to Porphyromonas gingivalis and Escherichia coli lipopolysaccharide. Clin Exp Immunol. 2009;156:479–487. doi: 10.1111/j.1365-2249.2009.03920.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Dietrich D, Martin P, Flacher V, et al. Interleukin-36 potently stimulates human M2 macrophages, Langerhans cells and keratinocytes to produce pro-inflammatory cytokines. Cytokine. 2016;84:88–98. doi: 10.1016/j.cyto.2016.05.012. [DOI] [PubMed] [Google Scholar]
- 48.Foster AM, Baliwag J, Chen CS, et al. IL-36 promotes myeloid cell infiltration, activation, and inflammatory activity in skin. J Immunol. 2014;192:6053–6061. doi: 10.4049/jimmunol.1301481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Smith JJ, Deane NG, Wu F, et al. Experimentally derived metastasis gene expression profile predicts recurrence and death in patients with colon cancer. Gastroenterology. 2010;138:958–968. doi: 10.1053/j.gastro.2009.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.